- 1School of Biology, Scottish Oceans Institute, University of St. Andrews, St Andrews, United Kingdom
- 2Serra Húnter Fellow, Cell Biology Physiology and Immunology Department, School of Biology, Universitat de Barcelona, Barcelona, Spain
Seasonal temperature changes markedly effect the swimming performance of some cyprinid fish acutely tested at different temperatures, involving a restructuring of skeletal muscle phenotype including changes in contractile properties and myosin heavy chain expression. We analyzed the transcriptome of fast myotomal muscle from goldfish (Carassius auratus L.) acclimated to either 8 or 25∘C for 4 weeks (12 h light: 12 h dark) and identified 10 myosin heavy chains (myh) and 13 myosin light chain (myl) transcripts. Goldfish orthologs were classified based on zebrafish nomenclature as myhz1.1α, myhz1.1β, myhz1.1γ, myha, myhb, embryo_myh1, myh9b, smyh2, symh3, and myh11 (myosin heavy chains) and myl1a, myl1b, myl2, myl9a, myl9b, myl3, myl13, myl6, myl12.1a, myl12.1b, myl12.2a, myl12.2b, and myl10 (myosin light chains). The most abundantly expressed transcripts myhz1.1α, myhz1.1β, myhz1.1γ, myha, myl1a, myl1b, myl2, and myl3) were further investigated in fast skeletal muscle of goldfish acclimated to either 4, 8, 15, or 30∘C for 12 weeks (12 h light:12 h dark). Total copy number for the myosin heavy chains showed a distinct optimum at 15∘C (P < 0.01). Together myhz1.1α and myhz1.1β comprised 90 to 97% of myhc transcripts below 15∘C, but only 62% at 30∘C. Whereas myhz1.1α and myhz1.1β were equally abundant at 4 and 8∘C, myhz1.1β transcripts were 17 and 12 times higher than myhz1.1α at 15 and 30∘C, respectively, (P < 0.01). Myhz1.1γ expression was at least nine-fold higher at 30∘C than at cooler temperatures (P < 0.01). In contrast, the expression of myha and myosin light chains showed no consistent pattern with acclimation temperature. A phylogenetic analysis indicated that the previously reported ability of goldfish and common carp to alter contractile properties and myofibrillar ATPase activity with temperature acclimation was related to the duplication of a single myhz1.1 fast muscle myosin heavy chain found in basal cyprinids such as the zebrafish (Danio rerio).
Introduction
The temperature of freshwater lakes and all but the smallest ponds changes more slowly seasonally than the air temperature providing a stable cue that allows phenotypic plasticity to act as a compensatory mechanism to adjust physiology and behavior to the prevailing environmental conditions (Johnston and Temple, 2002; Seebacher et al., 2012; Jeffries et al., 2016). These phenomena have been particularly studied in skeletal muscle because of the relative ease of interpreting changes at the molecular and sarcomere levels in terms of altered contractile performance and swimming behavior (reviewed in Johnston, 2006). Such phenotypic plasticity is commonly observed in cyprinid fish including goldfish, crucian carp, common carp, and grass carp that experience relatively large difference between summer and winter temperatures of 25°C or more (Fry and Hart, 1948; Johnston et al., 1975; Johnston and Maitland, 1980; Imai et al., 1996; Tao et al., 2005). In contrast, species of the same family inhabiting habitats with a seasonal range of 15°C or less including the model zebrafish (Danio rerio) have temperature-performance profiles that are unaltered by several months acclimation to the average maximum or minimum seasonal temperature (McClelland et al., 2006).
One of the earliest papers documenting temperature acclimation responses in fish measured the maximum speed that goldfish (Carassius auratus) could maintain in a rotating swimming chamber at various test temperatures for a fixed duration of 2 min (Fry and Hart, 1948). They found that temperature-maximum performance curves were markedly different in fish acclimated for several weeks to different test temperatures. Cold-acclimation improved performance at low temperatures but resulted in reduced swimming performance at high test temperatures and vice versa (Fry and Hart, 1948). Cold-acclimation was subsequently shown to result in a more aerobic phenotype in both fast and slow muscle fibers (Johnston and Maitland, 1980; Sidell, 1980) and higher myofibrillar ATPase activity at low test temperatures (Johnston et al., 1975). Contractile phenotype was also profoundly modified by temperature acclimation in the common carp (Cyprinus carpio) including the amount of sarcoplasmic reticulum, twitch contraction times (Fleming et al., 1990) and the force-velocity relationship (Johnston et al., 1985). At the molecular level temperature acclimation was found to modify the predominant isoform of myosin heavy chain expressed in fast muscle in both common carp (Imai et al., 1996) and grass carp (Ctenopharyngodon idella) acclimated to either 10 or 30°C for 5–8 weeks (Tao et al., 2009). In both species, three myosin heavy chains were expressed in an acclimation-temperature dependent manner and named according to their predominant expression temperature as 10°C-type, intermediate-type and 30°C-type (Imai et al., 1996; Tao et al., 2005, 2009). In contrast, temperature acclimation did not alter the sub-unit composition of myosin light chains (Hwang et al., 1990).
In the present study, the transcriptome of fast myotomal muscle was characterized in goldfish acclimated to either 8 or 30°C for 4 weeks to identify the most abundant myosin heavy chain (myh) and myosin light chains (myl) expressed. Phylogenetic analysis was used to provide an insight into the evolution of myosin heavy chains in the Cypriniformes lineage that have expression patterns which are seasonally regulated. Myosin genes in the goldfish were named according to the nomenclature used for the zebrafish (Danio rerio), a more basal cyprinid (Wang et al., 2007) with a more restricted temperature range (15–30°C) that is unable to modify swimming performance following temperature acclimation (McClelland et al., 2006). To further investigate the transcriptional regulation of myosin, the copy number of the predominantly expressed myh and myl chain transcripts was then quantified in a second experiment for goldfish acclimated to either 4, 8, 15, or 30°C for a period of 12 weeks.
Materials and Methods
Fish and Experimental Conditions
All experimental procedures were approved by the Animal Welfare and Ethics Committee (AWEC) of the University of St Andrews. Goldfish (Carassius auratus L.) were obtained from accredited suppliers and maintained in 200 L tanks of temperature-controlled freshwater with 96% recirculation (12 h light: 12 h dark) in the Scottish Oceans Institute aquarium facilities. Fish were hand fed with a commercial diet at maintenance level. In a preliminary study of the fast muscle transcriptome goldfish of 20–30 g body mass were acclimated to either 8 or 25°C for 4 weeks. The main experiment comprised four groups of 30 fish of similar body size that were maintained in duplicated tanks and acclimated to 15°C for 2 weeks. The temperature of each replicate tank was adjusted at the rate of 1°C day-1 to final acclimation temperatures of 4, 8, 15, or 30°C (12 h light: 12 h dark). Fish were maintained at these temperatures for 12 weeks before being humanely sacrificed using a schedule-1 United Kingdom Home Office approved protocol involving a sharp blow in the head followed by sectioning of the spinal cord. The body size of the different temperature acclimation groups was not significantly different at the end of the experiment (P > 0.5; Supplementary File S1).
Tissue Sampling
All tissue samples were taken from deep layers of the dorsal myotomal muscle at the level of the dorsal fin which consists of pure fast muscle fibers. Around 1 g of fast muscle was dissected per fish for RNA extraction, snap frozen in liquid nitrogen and stored at -80°C until further analysis.
RNA Extraction
Between 20 and 40 mg of pure fast skeletal muscle was used for total RNA extraction using TriSure reagent (Bioline, London, United Kingdom) following the manufacturer’s recommendations. Briefly, tissue was homogenized in 1 ml TriSure using MatrixD (MP Biomedicals), vigorously mixed with 200 μl of chloroform and centrifuged. The upper liquid phase was recovered, mixed with 500 μl of 2-propanol and centrifuged again to precipitate the RNA. The resulting pellet was washed three times with 1 ml of cold 70% ethanol, dried and diluted in 50 μl of RNAse free water. Total RNA concentration, 260/280 and 260/230 ratios were estimated using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, United Kingdom) and integrity was evaluated in a 1.5% (m/v) ethidium bromide agarose gel. All samples used had non-degraded RNA with 260/280 and 260/230 ratios over 2.
Fast Muscle Transcriptome
Equal amounts of total RNA from 5 goldfish per acclimation temperature (8 and 25°C for 4 weeks) were pooled. The two pools were pair-end sequenced on one lane of an Illumina Miseq using v3 chemistry by Eurofins Genomics (ENA Accession Number: PRJEB17982). Low quality reads and adaptors were removed and remaining reads assembled using Eurofins in-house scripts combining Velvet (Zerbino and Birney, 2008) and Oases (Schulz et al., 2012) assembler software. In order to reduce redundancy from the initial assembly, contigs were re-clustered based on sequence identity using cd-hit-EST (Li and Godzik, 2006) with an identity threshold of 99%. Contigs obtained after the clustering were blasted (BLASTx) against the NCBI non-redundant database. Gene Ontology (GO) annotation was performed using BLAST2GO software (Götz et al., 2008) with an e-value threshold of 10-3.
Sarcomere protein components were identified in the de novo transcriptome using two complementary strategies: (1) amino acid sequences for myosin heavy chain and myosin light chain orthologs for zebrafish were retrieved from the available genome1 and BLAST (tBLASTn) against the de novo goldfish transcriptome (2) in parallel all nucleotide sequences of myosin sub-units available for common carp (Cyprinus carpio) were also retrieved from NCBI public database2 and genome assembly3 and BLAST (BLASTn) against the goldfish transcriptome. Positive hits from both approaches were retrieved and re-BLAST against the NCBI non-redundant (nr) database to confirm their identity.
Orthologs for myosin heavy chain and myosin light chain genes were retrieved from the Ensembl database1 for Oreochromis niloticus, Gasterosteus aculeatus, Danio rerio, Astyanax mexicanus, Oryzias latipes, Tetraodon nigroviridis, Homo sapiens, and Mus musculus (all accession number are provided in Supplementary File S2). Nucleotide sequences were aligned using the GUIDANCE2 online server (Sela et al., 2015) with PRANK as the multi-sequence alignment algorithm. Columns below the 0.93 GUIDANCE2 quality alignment score were removed and remaining aligned sequences used for phylogenetic reconstruction. The best evolutionary model for each alignment was estimated using MEGA7 (Kumar et al., 2016). Bayesian MCMC phylogenetic trees, following a Yule speciation process model and UPGMA starting tree, were generated using BEAST v1.7.5 software with 10,000,000 random seeds (Drummond et al., 2012). Final Bayesian trees were generated using TreeAnnotator v1.7.5 with a burning value of 1,000. All trees were visualized using FigTree v1.3.1.
Validation of Paralogous Sequences
Potential gene paralogs identified in the de novo transcriptome were experimentally validated. In brief, primers targeting amplicons between 500–1000 bp were design in the most divergent region of the potential paralogs in order to favor the amplification of an individual gene (Supplementary File S3). Paralogs were amplified using standard PCR procedures using the following protocol: 95°C 5 min, 25 × (95°C 30 s, 60°C 30 s, and 72°C 1.5 min) followed by a final extension step of 7 min at 72°C. PCR reactions were resolved in a 2% (m/v) ethidium bromide gel and bands of expected size were cut-out and stored at -20°C prior to extraction. PCR amplicons were extracted, from agarose gels using the QIAquick extraction kit (QIAGEN) and ligated to a TOPO® vector (Thermofisher, Paisley, United Kingdom) containing a resistance to ampicillin, transformed into chemically competent E. coli by thermal shock and incubated 1 h at 37°C in 200 μl of SOC medium following the manufacturer’s recommendations (Thermofisher). A total of 100 μl of the transformed E. coli were plated in LB-agarose plates containing 75 ng/ml ampicillin (Thermofisher) and incubated for 16 h at 37°C. A total of 8 clones per product were growth in LB-broth medium at 37°C for 16 h and 1 μl of bacteria was amplified by PCR using T7/T3 primers to confirm the presence of the right-sized insert in all selected clones. Miniprep extractions were performed for those clones with the correct insert size and plasmid concentration estimated by Nanodrop spectrometry. All clones were Sanger sequenced by the University of Dundee Sequencing Service.
cDNA Synthesis and Quantitative PCR
1 μg of total RNA per individual was reverse transcribed using Quantitect cDNA synthesis kit (Qiagen) including a gDNA wipe-out step to remove any remnants of genomic DNA. Six microliters per sample were mixed with 7.5 μl of SensiFast SYBR Lo-ROX 2x master mix (Bioline, London, United Kingdom) containing 400 nmol-1 sense/antisense primers. Reactions were performed in duplicate using a Mx3005P Thermocycler (Agilent, Berkshire, United Kingdom), with one cycle of 2 min at 95°C and 40 cycles of 5 s at 95°C and 20 s at 65°C, followed by a dissociation curve analysis, which resulted in a single peak for all qPCR reactions analyzed.
Primers were designed using Primer 3 (Untergasser et al., 2012) to anneal at 60°C and amplify products between 100–250 bp (Supplementary File S3). NetPrimer (PremierBiosoft) was used to test for primer hairpins, self-dimmers, and cross-dimmers. In the case of confirmed paralogs pairs, primers were designed in the most divergent fragments targeting differences in the 3′ region of the sequence as previously described (Garcia de la serrana et al., 2012; Garcia de la serrana and Johnston, 2013). Gene efficiency was estimated by a dilution series of a pooled sample. Housekeeping genes beta actin (ß-actin), elongation factor 1 alpha (ef1a), and ribosomal protein 27 (rpl27) were tested for stability using BestKeeper (Pfaffl et al., 2004) and rpl27 was selected for gene expression normalization. All qPCR reactions were resolved in a 2% (m/v) ethidium-bromide agarose gel and specific bands were purified from the gel using a QIAgel extraction kit (QIAGEN) and sequenced at the University of Dundee Sequencing Service. For absolute quantification of transcript abundance, qPCR fragments were cloned in plasmids and purified as described above. A calibration curve with 10, 1, 0.1, 0.01, and 0.001 nanograms of plasmid for each insert were constructed and the qPCR protocol described above followed. The calibrations curves were used to translate Ct from each of the genes analyzed to number of copies per μg of cDNA. Transcript abundance of myosin heavy chains and myosin light chains was normalized against by dividing the gene total number of transcripts by the housekeeping (rpl27) total number of transcripts.
Statistical Analysis
All statistical analyses were conducted using R-Studio v.1.1.419. Shapiro-Wilk was used to test normality of the gene expression data. Differences in gene expression were analyzed using normalized expression values by ANOVA test with temperature as factor followed by a Tukey post hoc test. P-values were corrected by multiple comparison applying a false discovery rate (FDR) correction. Differences were considered significant when FDR < 0.05. All graphs were produced using the ggplot2 R-build package (Wickham, 2016).
Results
Fast Skeletal Muscle Transcriptome
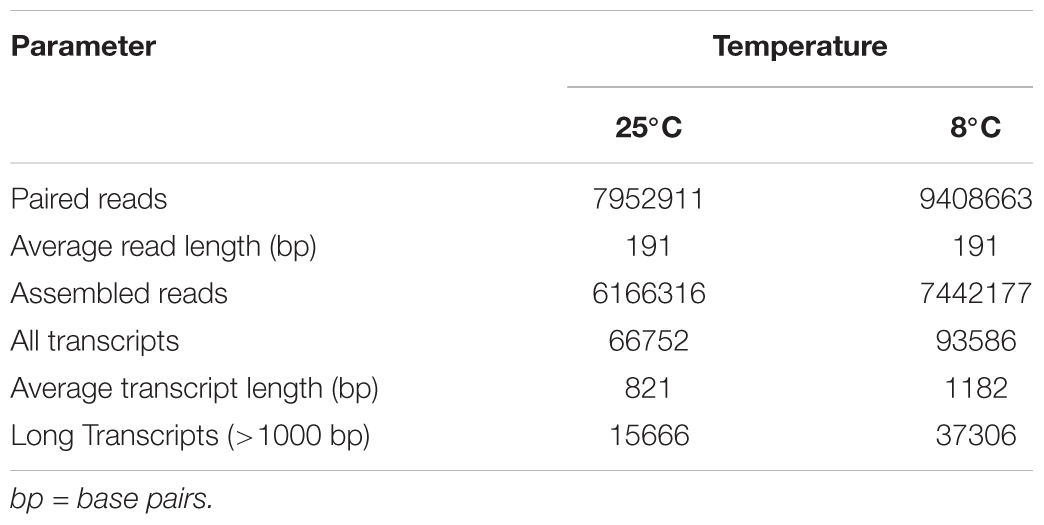
A total of 7.5 and 10.0 million paired-end reads (190 bp) were obtained by Illumina sequencing of cDNA libraries prepared from the fast myotomal muscle of goldfish acclimated to either 25 or 8°C, respectively. Following assembly, the number or transcripts obtained was 66,752 (25°C library) and 93,586 (8°C library) (Table 1). Transcripts longer than 1,000 bp comprised 23% of the 25°C library and 40% of the 8°C library (Table 1). The transcripts coding for subunits of myosin were exhaustively characterized using the combined transcriptome.
Myosin Heavy Chains (myh)
To identify myh transcripts the transcriptome was successively BLAST against the zebrafish genome (vz9) and myhc transcripts from the common carp (Cyprinus carpio) another cyprinid fish which differentially regulates myh expression with temperature acclimation (Imai et al., 1996). A total of 10 different myosin heavy chain transcripts were identified of which 8 were of sufficient length for further validation by phylogenetic analysis (Supplementary File S4). The goldfish myosin heavy chain orthologs were named according to the zebrafish nomenclature as myhz1.1α, myhz1.1β, myhz1.1γ, myha, myhb, smyh2, smyh3, embryo_myh1, myh10, and myh11 (Figure 1). In the case of embryo_myh1 gene, blast results indicated a equal similarity to zebrafish myhz2, myhz1.3, and myhc4 (data not show) but since the phylogenetic analysis did not revolve its identity (Figure 1) the previous cyprinid nomenclature was maintained. The myhz1.1α, myhz1.1β, and myhz1.1γ transcripts were orthologous to the major fast muscle myosin heavy chain isoforms expressed in the common carp at acclimation temperatures of 10, 15, and 30°C, respectively, (Imai et al., 1996) and to a single ortholog of myhz1.1 in zebrafish. Myha and myhb are also characterized as fast muscle genes in the zebrafish genome assembly (Nord et al., 2014).
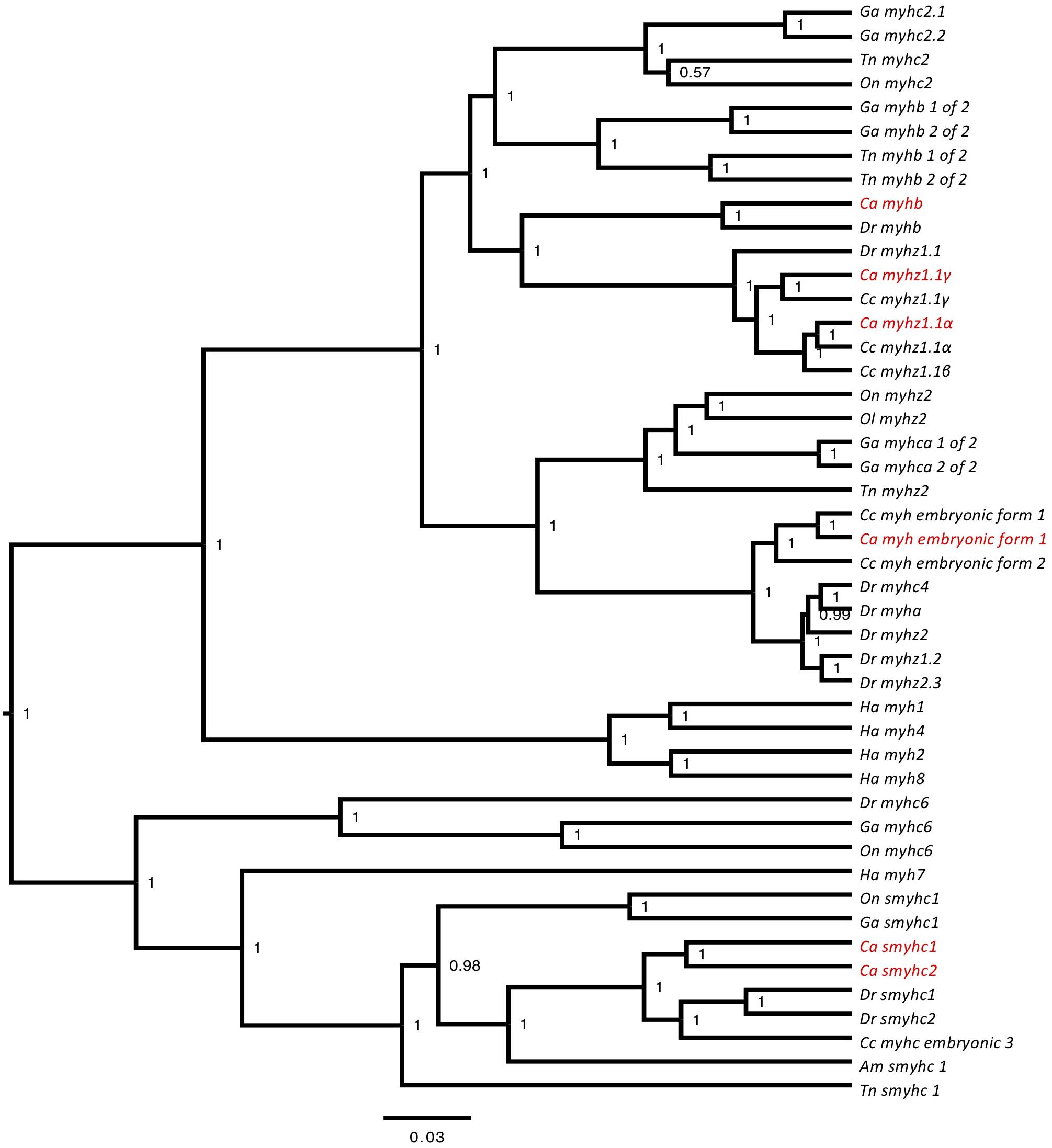
FIGURE 1. Phylogenetic tree constructed for myosin heavy chain genes. Bayesian phylogenetic relationships for the myosin heavy chain orthologs. Phylogenetic tree node numbers represent posterior values. To facilitate interpretation, goldfish orthologs (Carassius auratus, Ca) are highlighted in red. Myosin heavy chain orthologs for zebrafish (Danio rerio, Dr), stickleback (Gasterosteus aculeatus, Ga), common carp (Cyprinus carpio, Cc), green spotted pufferfish (Tetraodon nigroviridis, Tn), tilapia (Oreochromis niloticus, On), medaka (Oryzias latipes, Ol), and human (Homo sapiens, Ha) were also included.
Myosin Light Chains (myl)
13 myosin light chain (myl) genes were identified: myosin light chain 1a (myl1a), myosin light chain 1b (myl1b), myosin light chain 2 (myl2), myosin light chain 9a (myl9a), myosin light chain 9b (myl9b), myosin light chain 3 (myl3), myosin light chain 13 (myl13), myosin light chain 6 (myl6), myosin light chain 12.1a (myl12.1a), myosin light chain 12.1b (myl12.1b), myosin light chain 12.2a (myl12.2a), myosin light chain 12.2b (myl12.2b), and myosin light chain 10 (myl10) (Supplementary File S4). The abundantly expressed sequences myl2, myl3, myl1a, and myl1b clustered in monophyletic branches with their zebrafish orthologs following phylogenetic analysis (Figure 2).
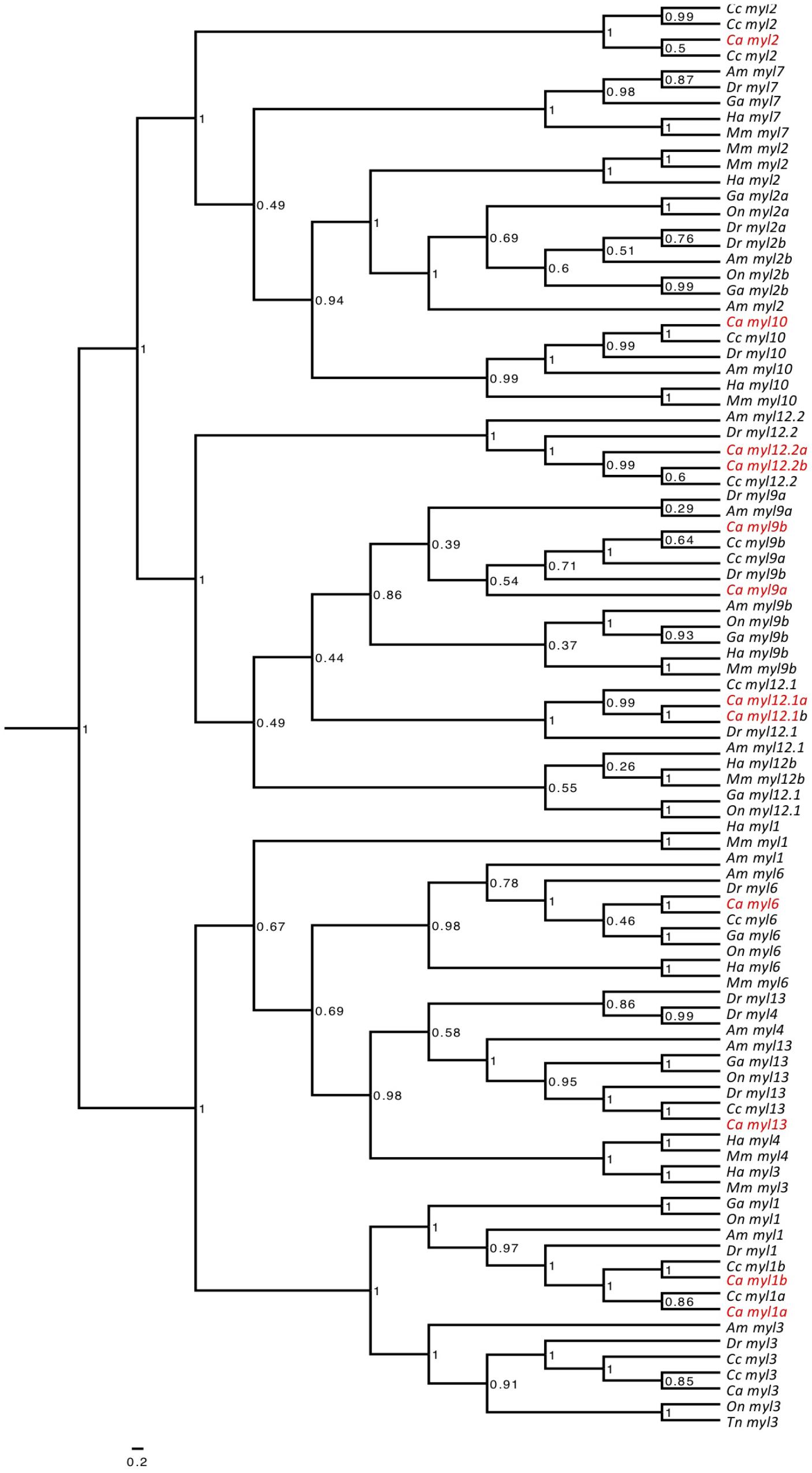
FIGURE 2. Phylogenetic tree constructed for myosin light chain genes. Bayesian phylogenetic relationships for the myosin light chain orthologs. Phylogenetic tree node numbers represent posterior values. To ease interpretation, goldfish orthologs (Carassius auratus, Ca) are highlighted in red. Myosin heavy chain orthologs for zebrafish (Danio rerio, Dr), stickleback (Gasterosteus aculeatus, Ga), common carp (Cyprinus carpio, Cc), green spotted pufferfish (Tetraodon nigroviridis, Tn), tilapia (Oreochromis niloticus, On), cavefish (Astyanax mexicanus, Am), medaka (Oryzias latipes, Ol) and human (Homo sapiens, Ha) were also included.
Myosin Heavy but Not Light Chain Transcripts Are Differentially Regulated With Temperature Acclimation
Four myh (myhz1.1α, myhz1.1β, myhz1.1γ, and myha) and four myl (myl2, myl3, myl1a, and myl1b) constituted > 96% of the transcripts expressed for these proteins in fast myotomal muscle. Total transcript copy number for the four abundantly expressed myh transcripts showed a distinct optimum in fish acclimated to 15°C for 12 weeks and were 63% lower in the 30°C group and 75% lower at 4 and 8°C (P < 0.01) (Figure 3). Myhz1.1α and myhz1.1β comprised 90, 96, and 97% of myh transcripts at 4, 8, and 15°C, falling to 62% at an acclimation temperature of 30°C. The ratio of myhz1.1α and myhz1.1β transcripts was similar in the 4 and 8°C acclimation groups whereas myhz1.1β was 17 and 12 times more highly expressed in 15 and 30°C acclimation groups relative to myhz1.1α (P < 0.01) (Figures 3B,C). The myhz1.1γ transcript copy number was a minimum of nine-fold higher in the 30°C-group than at any other acclimation temperature (P < 0.01) (Figure 3D). The copy number for myha was much lower than the other more abundant myh transcripts and showed no consistent pattern with acclimation temperature (Figure 3). Each myosin heavy chain peptide is associated with two myosin light chain peptides, but this was not reflected in relative transcript abundances. For example, total transcript abundance for myosin light chains was lower at acclimation temperatures of 4 and 8°C than at 15 and 30°C (Figure 4). In contrast to myosin heavy chains the relative proportions of transcripts for the different orthologs of myosin light chains was largely independent of acclimation temperature (Figure 4).
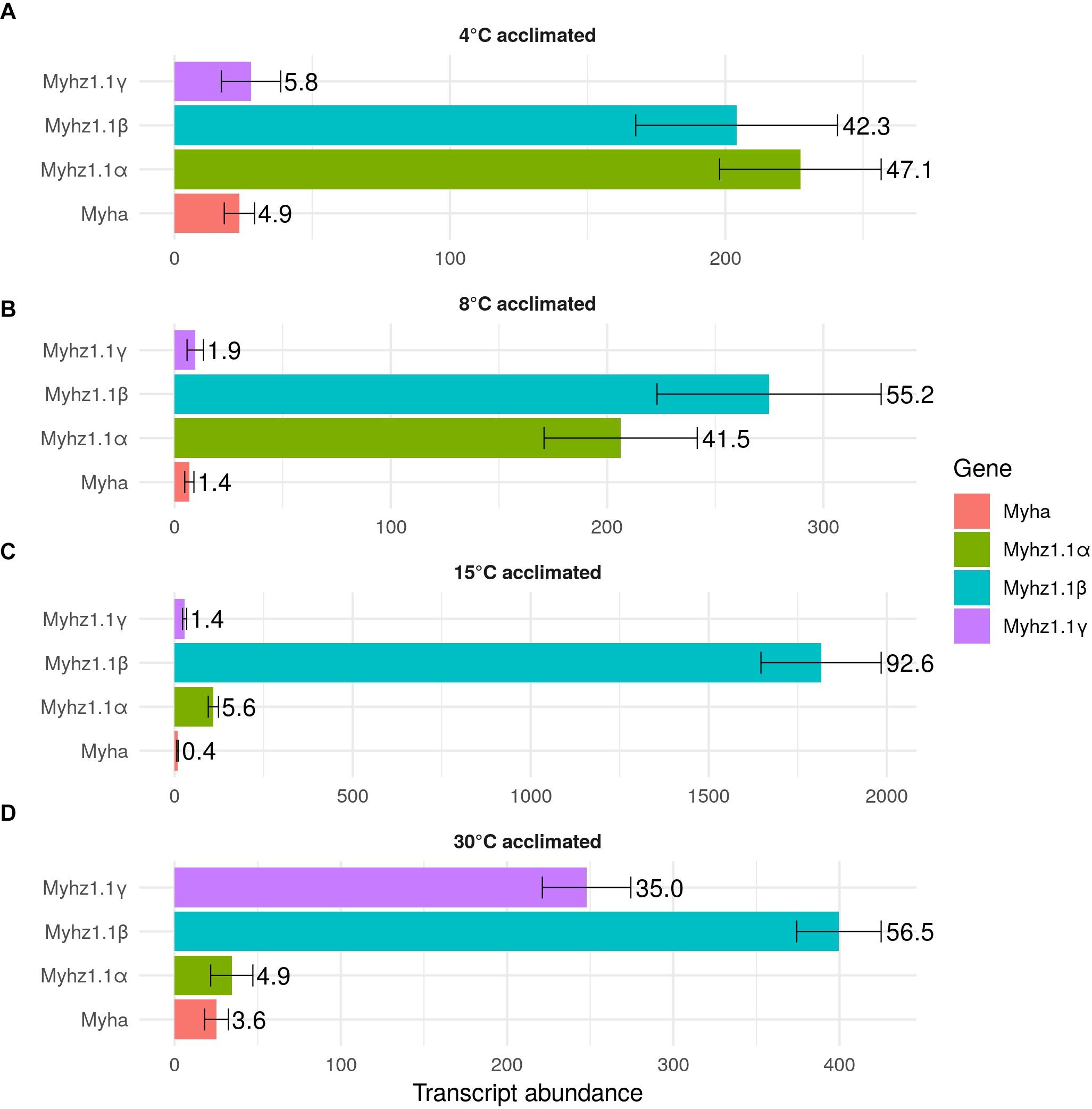
FIGURE 3. Transcript abundance of the main myosin heavy chain transcripts in goldfish fast skeletal muscle. Values represent Mean ± SD transcript copy number normalized to the reference gene. The transcripts of myosin heavy chain orthologs myhz1.1α (green), myhz1.1β (blue), myhz1.1γ (purple), and myha (red) in response to long-term acclimation to 4, 8, 15, and 30°C are illustrated. To ease interpretation, the percentage of each transcript is shown to the right of each bar. (A) 4°C acclimated, (B) 8°C acclimated, (C) 15°C acclimated and (D) 30°C acclimated.
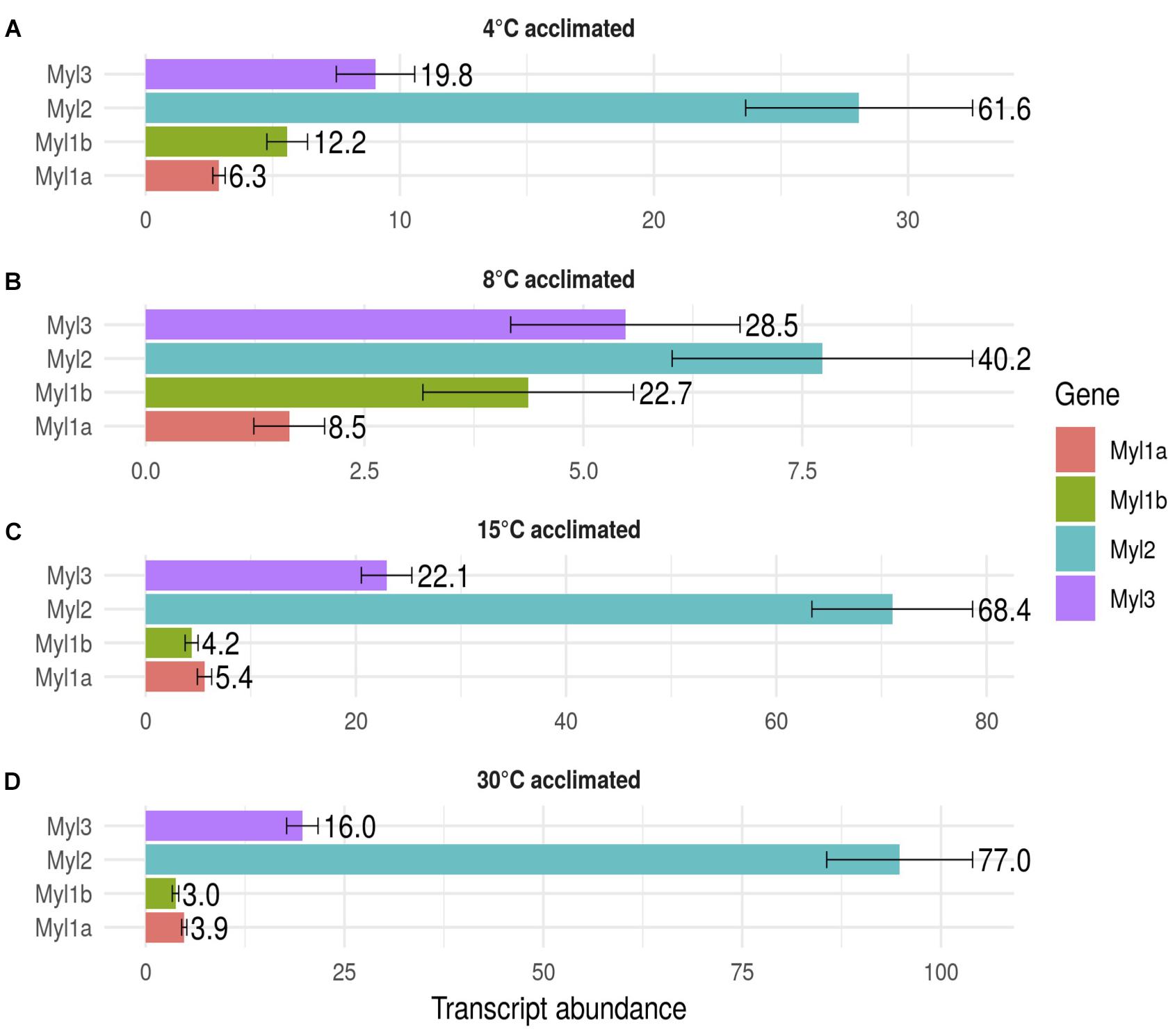
FIGURE 4. Transcript abundance of the main myosin light chain isoforms in goldfish fast skeletal muscle. Values represent Mean ±SD transcript copy number normalized to the reference gene. The transcripts of myosin light chain orthologs myl1a (red), myl1b (green), myl2 (blue), and myl3 (purple) in response to long-term acclimation to 4, 8, 15, and 30°C are illustrated. To ease interpretation, the percentage of each transcript is shown to the right of each bar. (A) 4°C acclimated, (B) 8°C acclimated, (C) 15°C acclimated and (D) 30°C acclimated.
Discussion
The advent of genome sequencing and RNA-seq has illustrated the extraordinary diversity of myosin heavy and light chain genes that are expressed even in single muscle fiber types of vertebrates (Lynch et al., 2015). In the present study, 11 myosin heavy chains (myh) and 13 myosin light chain (myl) genes were identified in myotomal samples that only contained fast twitch muscle fibers. Just four transcripts (myhz1.1α, myhz1.1β, myhz1.1γ, and myha) comprised more than 96% of all the expressed myosin heavy chains. Myotubes are continuously produced in the fast myotomal muscle of teleost fish until 44% of the maximum adult body length (Weatherley et al., 1988). In situ hybridization studies in zebrafish have shown that myotubes/immature muscle fibers express five developmental-stage specific myosin heavy chain genes which become downregulated as fiber diameter and body size increase (Johnston et al., 2009). Since juvenile goldfish were used in our study some of the minor myosin heavy chain components e.g., emb1_myh probably corresponded to transcripts expressed in differentiating fast muscle myotubes. Other minor components were transcripts highly expressed in slow twitch or cardiac muscle (Supplementary File S4). For example, smyh2 and smyh3 are paralogs of human MYH7 which is highly expressed in slow-twitch muscle (Elworthy et al., 2008; Naganawa and Hirata, 2011) and myha is an trunk, tail, and cranial muscles (Nord et al., 2014). Although myofibers constitute the dominant component of fast muscle it also contains capillaries and motor neurones as well as other cell types such as macrophages, all of which express myosin heavy chain genes. For example, we detected myh9 and myh10, non-muscle myosin heavy chains, which are thought to play a role in cytokinesis (Yang et al., 2012) and myh11 a myosin heavy chain highly expressed in smooth muscle (Wallace et al., 2005).
The Cyprinidae family including goldfish, common carp, and the killifish Fundulus heteroclitus all show large increases in maximum swimming performance at low temperatures following a period of several weeks cold-acclimation at the expense of reduced performance at warm-acclimation temperatures (Fry and Hart, 1948; Johnson and Bennett, 1995). Seasonal plasticity of swimming performance in these species reflects a profound and complex remodeling of muscle phenotype at the level of proteins and organelles which serve to modify twitch contraction times, force production, shortening speed, and metabolic characteristics according to the prevailing environmental conditions (reviewed in Johnston and Temple, 2002). Differential expression of myosin heavy chains, but not myosin light chains, at the transcript and protein levels has been shown to be an important component of the mechanism underlying adjustments in myofibrillar ATPase activity and contractile properties in goldfish (Johnson and Bennett, 1995) and common carp (Imai et al., 1996). In Fundulus heteroclitus which is unable to modify myosin heavy chain composition with temperature acclimation much more modest adjustments in twitch contraction times and myofibrillar ATPase activity were observed (Johnson and Bennett, 1995). The short-horned sculpin (Myoxocephalus scorpius) of the family Cottidae showed a modest increase in the maximum shortening speed of fast muscle fibers at 15°C following acclimation from 5 to 15°C for 1–2 months, but unlike Cyprinidae, contractile performance (Ball and Johnston, 1996) and swimming behavior (Temple and Johnston, 1998) were unaffected by cold acclimation. Increased maximum muscle shortening speed of fast fibers with warm acclimation in short-horned sculpin was associated with altered expression of myosin light chains independently of myosin heavy chain composition (Ball and Johnston, 1996). These results indicate considerable species diversity in the response of the musculoskeletal system to temperature acclimation at the whole animal, tissue and molecular levels. However, a transition to a more aerobic phenotype is generally observed in all species following cold acclimation regardless of whether or not this is accompanied by altered locomotory performance (Johnston and Maitland, 1980; Egginton and Sidell, 1989; McClelland et al., 2006).
In our study on goldfish the total copy number of the most abundantly expressed myosin heavy chain transcripts showed a distinct optimum at 15°C with the relative proportions of myhz1.1α, myhz1.1β, and myhz1.1γ transcripts differing markedly in proportion above or below this temperature. Acclimation to temperatures at or above 15°C resulted in a marked increase in the relative expression of myhz1.1β and myhz1.1γ at the expense of myhz1.1α. The sequences of myhz1.1α, myhz1.1β, and myhz1.1γ in goldfish are orthologous to the 10, 15, and 30°C type myosin heavy chain genes described in the common carp according to the temperature at which they were expressed (Imai et al., 1996). Goldfish and common carp cluster on a common lineage of the Cyprinidae phylogenetic tree which split after the lineage leading to zebrafish (Huang et al., 2017). Our analysis shows that the three goldfish myhz1.1 paralogs arose through the duplication of a single myhz1.1 found in zebrafish which is a more basal cyprinid which has a more modest temperature range of 15 to 30°C and shows a shift to a more aerobic phenotype with cold acclimation, but no ability to modify maximum swimming speed (McClelland et al., 2006).
Local duplications seem to be common in cyprinids, for example zebrafish have three copies of the myhz1 (myhz1.1, myhz1.2, and myhz1.3), three copies of the slow myosin heavy chain (smyh1, smyh2, and smyh3) and several copies of cryptochrome genes all of them originated by local duplication (McGuigan et al., 2004; Liu et al., 2015). There is also evidence of a whole genome duplication event (WGD) event occurring on the cyprinids lineage 8 Million years ago (Li et al., 2015). Because we cannot distinguish between local duplication and whole genome duplication from the current analysis we named the duplicated genes as α, β, and γ following the annotation method suggested by Garcia de la serrana and Macqueen (2018). The medaka Oryzias latipes, can also modify myosin heavy chain expression following acclimation to either 10 or 30°C. In this species, luciferase reporter constructs involving deletions and site mutations of the 5′ flanking region of the gene established that a MEF2 transcription factor binding site was essential for expression in an acclimation-temperature dependent manner (Liang et al., 2008). Together these results indicate that a single duplication event followed by sub-functionalization may have been responsible for the extreme ability of these Cyprinidae species to modify maximum swimming with seasonal temperature acclimation.
Author Contributions
DG and KW were responsible for animal maintenance, sampling, and data collection. DG was responsible for gene expression analysis. IJ and DG planned the experimental procedures and wrote the manuscript.
Funding
This work received funding from the MASTS pooling initiative (The Marine Alliance for Science and Technology for Scotland) and their support was gratefully acknowledged. MASTS was funded by the Scottish Funding Council (Grant Reference HR09011) and contributing institutions. DG is a Serra Húnter Fellow of the University of Barcelona.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Christopher Hollenbeck for his help with the construction of the graphs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01724/full#supplementary-material
FILE S1 | Length and weight of experimental animals.
FILE S2 | Sequences used for phylogenetic reconstruction of myosin heavy chains.
FILE S3 | Primers used for Sequencing and qPCR analysis.
FILE S4 | Goldfish myosin heavy chains and myosin light chains sequences reported.
Footnotes
References
Ball, D., and Johnston, I. A. (1996). Molecular mechanisms underlying the plasticity of muscle contractile properties with temperature acclimation in the marine fish Myoxocephalus scorpius. J. Exp. Biol. 199, 1363–1373.
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Egginton, S., and Sidell, B. D. (1989). Thermal acclimation induces adaptive changes in subcellular structure of fish skeletal muscle. Am. J. Physiol. Integr. Comp. Physiol. 256, R1–R9. doi: 10.1152/ajpregu.1989.256.1.R1
Elworthy, S., Hargrave, M., Knight, R., Mebus, K., and Ingham, P. W. (2008). Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for hedgehog and Prdm1 activity. Development 135, 2115–2126. doi: 10.1242/dev.015719
Fleming, J., Crockford, T., Altringham, J., and Johnston, I. (1990). Effects of temperature acclimation on muscle relaxation in the carp: a mechanical, biochemical, and ultrastructural study. J. Exp. Zool. 255, 286–295. doi: 10.1002/jez.1402550306
Fry, F. E., and Hart, J. (1948). The relation of temperature to oxygen consumption in the goldfish. Biol. Bull. 94, 66–77. doi: 10.2307/1538211
Garcia de la serrana, D., and Johnston, I. A. (2013). Expression of heat shock protein (Hsp90) paralogues is regulated by amino acids in skeletal muscle of atlantic salmon. PLoS One 8:e74295. doi: 10.1371/journal.pone.0074295
Garcia de la serrana, D., and Macqueen, D. J. (2018). Insulin-like growth factor-binding proteins of teleost fishes. Front. Endocinol. 9:80. doi: 10.3389/fendo.2018.00080
Garcia de la serrana, D., Vieira, V. L. A., Andree, K. B., Darias, M., Estévez, A., Gisbert, E., et al. (2012). Development temperature has persistent effects on muscle growth responses in gilthead sea bream. PLoS One 7:e51884. doi: 10.1371/journal.pone.0051884
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Huang, S.-P., Wang, F., and Wang, T.-Y. (2017). Molecular phylogeny of the opsariichthys group (Teleostei:Cypriniformes) based on complete mitochondrial genomes. Zool. Stud. 56:40. doi: 10.6620/ZS.2017.56-40
Hwang, G. C., Watabe, S., and Hashimoto, K. (1990). Changes in carp myosin ATPase induced by temperature acclimation. J. Comp. Physiol. B 160, 233–239. doi: 10.1007/BF00302588
Imai, J.-I., Hirayama, Y., Kikuchi, K., Katinuma, M., and Watabe, S. (1996). cDNA cloning of myosin heavy chain isoforms from carp fast skeletal muscle and their gene expression associated with temperature acclimation. J. Exp. Biol. 200, 27–37.
Jeffries, K. M., Connon, R. E., Davis, B. E., Komoroske, L. M., Britton, M. T., Sommer, T., et al. (2016). Effects of high temperatures on threatened estuarine fishes during periods of extreme drought. J. Exp. Biol. 219, 1705–1716. doi: 10.1242/jeb.134528
Johnson, T., and Bennett, A. (1995). The thermal acclimation of burst escape performance in fish: an integrated study of molecular and cellular physiology and organismal performance. J. Exp. Biol. 198, 2165–2175.
Johnston, I., and Maitland, B. (1980). Temperature acclimation in crucian carp, Carassius carassius L., morphometric analyses of muscle fibre ultrastructure. J. Fish Biol. 17, 113–125. doi: 10.1111/j.1095-8649.1980.tb02746.x
Johnston, I., and Temple, G. (2002). Thermal plasticity of skeletal muscle phenotype in ectothermic vertebrates and its significance for locomotory behaviour. J. Exp. Biol. 205, 2305–2322.
Johnston, I. A. (2006). Environment and plasticity of myogenesis in teleost fish. J. Exp. Biol. 209, 2249–2264. doi: 10.1242/jeb.02153
Johnston, I. A., Davison, W., and Goldspink, G. (1975). Adaptations in Mg2+-activated myofibrillar ATPase activity induced by temperature acclimation. FEBS Lett. 50, 293–295. doi: 10.1016/0014-5793(75)80512-6
Johnston, I. A., Lee, H.-T., Macqueen, D. J., Paranthaman, K., Kawashima, C., Anwar, A., et al. (2009). Embryonic temperature affects muscle fibre recruitment in adult zebrafish: genome-wide changes in gene and microRNA expression associated with the transition from hyperplastic to hypertrophic growth phenotypes. J. Exp. Biol. 212, 1781–1793. doi: 10.1242/jeb.029918
Johnston, I. A., Sidell, B. D., and Driedzic, W. R. (1985). Force-velocity characteristics and metabolism of carp muscle fibres following temperature acclimation. J. Exp. Biol. 119, 239–249.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, J. T., Hou, G. Y., Kong, X. F., Li, C. Y., Zeng, J. M., Li, H., et al. (2015). The fate of recent duplicated genes following a fourth-round whole genome duplication in a tetraploid fish, common carp (Cyprinus carpio). Sci. Rep. 5, 1–9. doi: 10.1038/srep08199
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Liang, C. S., Ikeda, D., Kinoshita, S., Shimizu, A., Sasaki, T., Asakawa, S., et al. (2008). Myocyte enhancer factor 2 regulates expression of medaka Oryzias latipes fast skeletal myosin heavy chain genes in a temperature-dependent manner. Gene 15, 42–53. doi: 10.1016/j.gene.2007.09.016
Liu, C., Hu, J., Qu, C., Wang, L., Huang, G., Niu, P., et al. (2015). Molecular evolution and functional divergence of zebrafish (Danio rerio) cryptochrome genes. Sci. Rep. 5:8113. doi: 10.1038/srep08113
Lynch, C. J., Xu, Y., Hajnal, A., Salzberg, A. C., and Kawasawa, Y. I. (2015). RNA sequencing reveals a slow to fast muscle fiber type transition after olanzapine infusion in rats. PLoS One 10:e0123966. doi: 10.1371/journal.pone.0123966
McClelland, G. B., Craig, P. M., Dhekney, K., and Dipardo, S. (2006). Temperature- and exercise-induced gene expression and metabolic enzyme changes in skeletal muscle of adult zebrafish (Danio rerio). J. Physiol. 577, 739–751. doi: 10.1113/jphysiol.2006.119032
McGuigan, K., Phillips, P. C., and Postlethwait, J. H. (2004). Evolution of sarcomeric myosin heavy chain genes: evidence from fish. Mol. Biol. Evol. 21, 1042–1056. doi: 10.1093/molbev/msh103
Naganawa, Y., and Hirata, H. (2011). Developmental transition of touch response from slow muscle-mediated coilings to fast muscle-mediated burst swimming in zebrafish. Dev. Biol. 355, 194–204. doi: 10.1016/j.ydbio.2011.04.027
Nord, H., Burguiere, A.-C., Muck, J., Nord, C., Ahlgren, U., von Hofsten, J., et al. (2014). Differential regulation of myosin heavy chains defines new muscle domains in zebrafish. Mol. Biol. Cell 25, 1384–1395. doi: 10.1091/mbc.E13-08-0486
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Schulz, M. H., Zerbino, D. R., Vingron, M., and Birney, E. (2012). Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092. doi: 10.1093/bioinformatics/bts094
Seebacher, F., Holmes, S., Roosen, N. J., Nouvian, M., Wilson, R. S., and Ward, A. J. W. (2012). Capacity for thermal acclimation differs between populations and phylogenetic lineages within a species. Funct. Ecol. 26, 1418–1428. doi: 10.1111/j.1365-2435.2012.02052.x
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Sidell, B. D. (1980). Responses of goldfish (Carassius auratus, L.) muscle to acclimation temperature: alterations in biochemistry and proportions of different fiber types. Physiol. Zool. 53, 98–107. doi: 10.1086/physzool.53.1.30155778
Tao, Y., Kobayashi, M., Fukushima, H., and Watabe, S. (2005). Changes in enzymatic and structural properties of grass carp fast skeletal myosin induced by the laboratory-conditioned thermal acclimation and seasonal acclimatization. Fish Sci. 71, 195–204. doi: 10.1111/j.1444-2906.2005.00948.x
Tao, Y., Wang, S.-Y., Liang, C.-S., Fukushima, H., and Watabe, S. (2009). Structural differences in the motor domain of temperature-associated myosin heavy chain isoforms from grass carp fast skeletal muscle. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 248–254. doi: 10.1016/j.cbpb.2009.06.013
Temple, G. K., and Johnston, I. A. (1998). Testing hypotheses concerning the phenotypic plasticity of escape performance in fish of the family cottidae. J. Exp. Biol. 201, 317–331.
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Wallace, K. N., Dolan, A. C., Seiler, C., Smith, E. M., Yusuff, S., Chaille-Arnold, L., et al. (2005). Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev. Cell 8, 717–726. doi: 10.1016/j.devcel.2005.02.015
Wang, X., Li, J., and He, S. (2007). Molecular evidence for the monophyly of east asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 42, 157–170. doi: 10.1016/j.ympev.2006.06.014
Weatherley, A. H., Gill, H. S., and Lobo, A. F. (1988). Recruitment and maximal diameter of axial muscle fibres in teleosts and their relationship to somatic growth and ultimate size. J. Fish Biol. 33, 851–859. doi: 10.1111/j.1095-8649.1988.tb05532.x
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. doi: 10.1007/978-3-319-24277-4
Yang, F., Wei, Q., Adelstein, R. S., and Wang, P. J. (2012). Non-muscle myosin IIB is essential for cytokinesis during male meiotic cell divisions. Dev. Biol. 369, 356–361. doi: 10.1016/j.ydbio.2012.07.011
Keywords: skeletal muscle, teleost, myosin heavy chain, temperature, gene duplication
Citation: Garcia de la serrana D, Wreggelsworth K and Johnston IA (2018) Duplication of a Single myhz1.1 Gene Facilitated the Ability of Goldfish (Carassius auratus) to Alter Fast Muscle Contractile Properties With Seasonal Temperature Change. Front. Physiol. 9:1724. doi: 10.3389/fphys.2018.01724
Received: 29 September 2018; Accepted: 15 November 2018;
Published: 04 December 2018.
Edited by:
Arjan P. Palstra, Wageningen University & Research, NetherlandsReviewed by:
Bozidar Raskovic, University of Belgrade, SerbiaMichael L. Fine, Virginia Commonwealth University, United States
Copyright © 2018 Garcia de la serrana, Wreggelsworth and Johnston. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Garcia de la serrana, Z2FyY2lhZGVsYXNlcnJhbmFAdWIuZWR1
 Daniel Garcia de la serrana
Daniel Garcia de la serrana Kristin Wreggelsworth1
Kristin Wreggelsworth1 Ian A. Johnston
Ian A. Johnston