- 1State Key Laboratory of Wheat and Maize Crop Science, College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Helicoverpa armigera can develop resistance to Bacillus thuringiensis (Bt), which threaten the long-term success of Bt crops. In the present study, RNAseq was employed to investigate the midgut genes response to strains with different levels of resistance (LF5, LF10, LF20, LF30, LF60, and LF120) in H. armigera. Results revealed that a series of differentially expressed unigenes (DEGs) were expressed significantly in resistant strains compared with the LF-susceptible strain. Nine trypsin genes, ALP2, were downregulated significantly in all the six resistant strains and further verified by qRT-PCR, indicating that these genes may be used as markers to monitor and manage pest resistance in transgenic crops. Most importantly, the differences in DEG functions in the different resistant strains revealed that different resistance mechanisms may develop during the evolution of resistance. The immune and detoxification processes appear to be associated with the low-level resistance (LF5 strain). Metabolic process-related macromolecules possibly lead to resistance to Cry1Ac in the LF10 and LF20 strains. The DEGs involved in the “proton-transporting V-type ATPase complex” and the “proton-transporting two-sector ATPase complex” were significantly expressed in the LF30 strain, probably causing resistance to Cry1Ac in the LF30 strain. The DEGs involved in binding and iron ion homeostasis appear to lead to high-level resistance in the LF60 and LF120 strains, respectively. The multiple genes and different pathways seem to be involved in Cry1Ac resistance depending on the levels of resistance. Although the mechanisms of resistance are very complex in H. armigera, a main pathway seemingly exists, which contributes to resistance in each level of resistant strain. Altogether, the findings in the current study provide a transcriptome-based foundation for identifying the functional genes involved in Cry1Ac resistance in H. armigera.
Introduction
The Cry1Ac toxin from Bacillus thuringiensis (Bt) is harmless to most organisms and considered as an environmentally friendly pesticide. Hence, Cry1Ac toxin is used commercially as a bio-insecticide and expressed in transgenic plants for controlling insect pests (Wu et al., 2008; Hutchison et al., 2010; Edgerton et al., 2012; Lu et al., 2012; Klümper and Qaim, 2014). The area of Bt transgenic crops planted worldwide has rapidly increased to 98.5 million hectares in 2016 and has accumulated more than 830 million hectares since 1996 (James, 2016). Although Bt crops have brought great economic and environmental benefits (Wu et al., 2008; Hutchison et al., 2010; Edgerton et al., 2012; Lu et al., 2012; Klümper and Qaim, 2014), the development of resistance to Bt toxins can reduce or even eliminate these benefits (Tabashnik et al., 2013; Van den Berg et al., 2013; Farias et al., 2014; Gassmann et al., 2014; Tabashnik and Carrière, 2017). Unfortunately, the cumulative number of cases of practical resistance to the Bt toxins in transgenic crops surged from 3 in 2005 to 16 in 2016 according to a report from Tabashnik and Carrière (2017) attracting researcher's attention.
Understanding the mode of Bt action and the mechanisms that confer resistance to Bt toxins can help to sustain and even enhance their efficacy to control pests. Models of Bt action agree that Bt protoxins are first converted to activated toxins by insect midgut proteases and then the activated toxins bind to the insect midgut receptors, finally leading to insect death (Gill et al., 1992; Pardo-López et al., 2013; Adang et al., 2014). Four main functional receptors of the Cry1Ac toxin have been identified and verified from the brush border epithelium, including alkaline phosphatase (ALP) (Flores-Escobar et al., 2013), cadherin (Chen et al., 2014), aminopeptidase (APN) (Zhang et al., 2009; Tiewsiri and Wang, 2011; Valaitis, 2011; Flores-Escobar et al., 2013; Wei et al., 2016b), and ATP-binding cassette transporter proteins (ABCs) (Tanaka et al., 2017). The identification of Cry1A receptors has broadened our understanding of Cry1Ac action. However, the mode of action of Cry1Ac is very complex and a change in any step of the toxicology process will inevitably lead to insect resistance. The most common Bt-resistant mechanisms had been reported in Lepidoptera, including a reduced binding capacity of Bt toxins to midgut receptors by a decrease in the activity and transcription of ALP or APN as well as mutations of APN, cadherin, and ABCC2 (Xu et al., 2005; Zhang et al., 2009, 2012; Gahan et al., 2010; Baxter et al., 2011; Jurat-Fuentes et al., 2011; Atsumi et al., 2012; Xiao et al., 2014; Chen et al., 2015) and a reduced conversion of the protoxin to the toxin by downregulation of trypsin (Rajagopal et al., 2009; Cao et al., 2013; Liu et al., 2014; Wei et al., 2016a). These studies indicate that many genes are involved in resistance of insects to Bt toxins. In addition, it sounds like it is hard to identify the common marker to monitor and manage pest resistance in transgenic crops.
Most importantly, different genes may show different contributions to resistance at different development levels. The decreasing protoxin activation in Ostrinia nubilalis (HD-1 Bt kurstaki-resistant strain) caused a 47-fold resistance to Dipel, which contained Cry1Ab, Cry1Aa, Cry1Ac, and Cry2Aa (Li et al., 2004, 2005). In a 100-fold more Cry1Ab-resistant Diatraea saccharalis strain, the resistance to the Cry1Ab toxin is due to the lower gene expression level of cadherin (Yang et al., 2011). In the H. armigera GYBT-resistant strain, a deletion between exons 8 and 25 of the cadherin gene resulted in a 564-fold resistance to Cry1Ac-activated toxin (Xu et al., 2005). For the Cry1Ac-resistant BtR strain, it was identified that a deletion mutation of APN3 and the downregulation of cadherin lead to Cry1Ac resistance. A subsequent study confirmed that a deletion mutant in the APN1 gene caused a more than 2,971-fold resistance to Cry1Ac in the BtR strain (Wang et al., 2005; Zhang et al., 2009). Recently, the mutations in the ABCC2 transporter and the cadherin genes were reported to have a different affect on the binding of Cry toxins to the proteins on the brush border membrane vesicle (BBMV) from H. virescens (Gahan et al., 2010). The mutations of the ABCC2 transporter showed a higher level of resistance to Cry toxins. These studies demonstrate that different genes have different impacts on resistance levels.
Moreover, the resistant pathways were studied also to find the key factors that regulated the expression of resistant genes. Guo et al. (2015b) reported a novel transregulatory signaling mechanism in which the mitogen-activated protein kinase (MAPK) signaling pathway was confirmed to be responsible for regulating the expressions of ALP and ABCC genes in a field-evolved resistant strain of P. xylostella. Also, the constitutively transcriptionally activated upstream gene, MAP4K4, in the MAPK signaling pathway is responsible for this transregulatory signaling mechanism (Guo et al., 2015b). Importantly, the key resistant factor, MAP4K4, may be used for molecular control of the Cry1Ac resistance.
However, the genes and pathways affected the Cry1Ac resistance depending on the Cry1Ac selection stresses in H. armigera have not been comprehensively assessed. The next-generation DNA sequencing provides a research technique to study the changes in gene expression in the midgut transcriptomes of Bt-resistant and -susceptible strains; this technology can detect differentially expressed genes in biochemical pathways involved in Bt resistance and provide new insights into resistance mechanisms (Lei et al., 2014; Nanoth Vellichirammal et al., 2015; Zhang et al., 2017). In this study, a susceptible LF (a laboratory strain collected from Langfang) strain and six resistant LF strains were selected for an analysis of related resistance genes, in particular, six substrains of LF came from the LF strain by selecting a series of gradually increasing resistant strains (Cao et al., 2013, 2014; Liu et al., 2014; Xiao et al., 2014; Chen et al., 2015; Wei et al., 2016a). First, RNA sequencing was employed to construct a complete and comprehensive reference transcriptome database from midgut samples of these seven strains. The differentially expressed genes were detected further among these seven strains by digital gene expression analysis (DGE). These data provide a foundation for understanding the systemic differences between Cry1Ac-resistant strains and Cry1Ac-susceptible strain and might aid in finding candidate resistance. Most importantly, the analysis of differently expressed genes among the seven strains will uncover the role of different genes in the different resistance phases and might explain how selection can cause fixed changes of the expression levels of numerous genes.
Materials and Methods
Insect Strains
The LF-susceptible H. armigera strain was established from a field population by collecting from the Langfang County, Hebei Province of China in 1998. The LF strain was reared in a lab environment without exposure to any insecticides (Wu and Guo, 2004). The six LF substrains came from the LF-susceptible strain via a series of selections: LF5, LF10, LF20, LF30, LF60, and LF120 (Cao et al., 2013, 2014; Liu et al., 2014; Xiao et al., 2014; Chen et al., 2015; Wei et al., 2016a; Table 1). The strain name is in accordance to the selection concentration for each strain, where the number from 5 to 120 follows LF: such as LF5 was selected with a 5 μg/ml Cry1Ac protoxin artificial diet (Liang et al., 2008). In this study, LF5, LF10, LF20, LF30, LF60, and LF120 had been selected for 60, 52, 42, 38, 21, and 17 generations with corresponding Cry1Ac diets, respectively (Table 1).
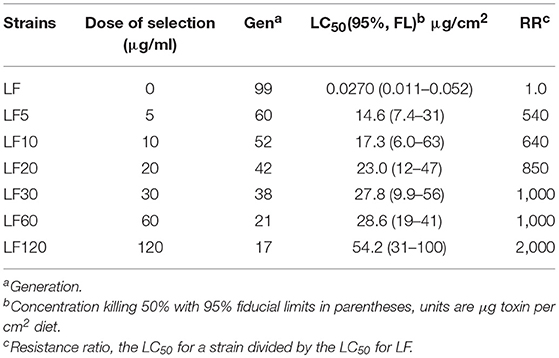
Table 1. Responses to Cry1Ac of the susceptible strain (LF) and six resistant strains (LF5, LF10, LF20, LF30, LF60, and LF120) of H. armigera.
Bioassay of Resistance to Cry1Ac Toxin
Larval responses to Cry1Ac toxin were evaluated using the methods reported by Wei et al. (2015). The Cry1Ac protoxin crystals were obtained from the HD-73 strain of B. thuringiensis (kindly supplied by Biotechnology Research Group, Institute of Plant Protection, Chinese Academy of Agricultural Sciences). Totally, 72 neonates per concentration for each treatment were tested. About 7 days later, dead insects and those that were still first instars were scored as dead. Five or more toxin concentrations were used to calculate the LC50 values of each strain (Table 1).
Dissection of Midgut and Extraction of RNA
Larvae from different colonies (LF, LF5, LF10, LF20, LF30, LF60, and LF120) were reared with a non-Bt toxin diet under standard rearing conditions. The midgut tissues of larvae in fifth instars (n = 30 per pool) were dissected from different strains. The lumen was then rapidly washed with a solution of 0.7% NaCl (w/v) to remove debris. Two biological replicates were employed. Total RNA from every replicate was extracted separately from each pool (LF, LF5, LF10, LF20, LF30, LF60, and LF120) using the Trizol reagent according to manufacturer's suggestions (Invitrogen, CA). In order to remove genomic DNA contamination, resulting RNA was treated with DNase I (Promega, Madison, WI, USA) following manufacturer's instructions. Quantity and quality of total RNA were assessed by denaturing gel electrophoresis and spectrophotometry on a Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA).
Preparation of Library for Analysis of Transcriptome
Sequencing libraries were generated from 3 μg total RNA per sample using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's recommendations. Briefly, mRNA was separated from total RNA using poly-T oligo-attached magnetic beads. First-strand cDNA was generated using random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH-) followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. The 3′ ends of DNA fragments were firstly adenylated and then NEBNext Adaptors with hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). About 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion high-fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. The PCR products were purified finally (AMPure XP system) and the quality of library was assessed on the Agilent Bioanalyzer 2100 system.
Analysis of Results of Illumina Sequencing
The clustering of the index-coded samples was carried out on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2000 platform and paired-end reads were generated. Transcriptome assembly was accomplished by using Trinity r20121005 (Grabherr et al., 2011) with min_kmer_cov set to 2 by default and all other parameters set default. Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. Clean data (clean reads) were then obtained by removing noise signals (reads containing adapter, reads containing ploy-N, and low-quality reads) from raw data. The following data analyses were performed based on the clean data. After eliminating redundancy using cd-hit and cap3 software, these data were mixed with our unpublic data of cotton bollworm transcriptome database. The resulting unigene database was used as a reference transcriptome database for subsequent analysis of DGE. The homology searches of all unigenes were performed based on BLASTx and BLASTn programs against the GenBank non-redundant protein (nr) and nucleotide sequence (nt) database at NCBI (v2.2.28). Matches of an E-value < 1.0E-5 were considered to be significant (Altschul et al., 1997). Gene ontology term (GO, http://www.geneontology.org/) annotations were assigned by Blast2GO software (b2g4pipe_v2.5) (Götz et al., 2008). The KOG (euKaryotic Ortholog Groups) and KEGG (Kyoto Encyclopedia of Genes and Genomes) annotations were performed using Blastall software against the KOG database (http://www.ncbi.nlm.nih.gov/COG/) and KEGG database (http://www.genome.jp/kegg/), respectively.
DGE Library Preparation and Sequencing
Library for DGE sequencing was prepared according to earlier-mentioned method (see “Library preparation for transcriptome analysis”). After cluster generation, the library sequencing was performed on an Illumina Hiseq 2000 platform and 100 bp single-end reads were generated.
Analysis and Mapping of DGE
After removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data, the clean data were then obtained. All analyses were performed according to the clean data. For unigene DEG, single-end clean reads were aligned to the unigene sequences by Bowtie v0.12.9. The HTSeq v0.5.4p3 was used to count unigene DEG numbers mapped to each unigene. Reads per kilobase of exon model per million mapped reads (RPKM) of each gene were calculated based on the length of the gene and reads count mapped to this gene (Mortazavi et al., 2008). The differentially expressed unigenes were used for mapping and annotation.
Evaluation of DGE Libraries
The frequency of each unigene in the different cDNA libraries was analyzed to compare gene expression in different strains. The DEGSeq R package (1.12.0) was used to analyze the differential expression of two conditions. The P-values were adjusted using the Benjamini & Hochberg method. Significant differential expression genes were obtained using set threshold values [corrected P-value of 0.005 and log2 (Fold-change) of 1]. For pathway-enrichment analysis, we mapped all the differentially expressed genes to terms in the GO data database and KEGG database. The GO-enrichment analysis of differentially expressed genes was implemented by the GOseq R package, in which gene length bias was corrected. The GO terms with corrected P-value < 0.05 were considered significantly enriched by differentially expressed genes. We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways.
Validation of qRT-PCR
The first-strand cDNA of each strain was used as the template for real-time PCR analysis. Each strain of H. armigera included 90 larvae (30 larvae per biological replicate). The mRNA expression levels of ALP-like, ALP2, APN5, and APN1 in different strains were analyzed by a quantitative real-time PCR (qRT-PCR). The β-actin and GAPDH of H. armigera were used as internal reference genes (Liu et al., 2014). The primers of the said genes used for qRT-PCR analysis are listed in Table S1. Each qRT-PCR (TaqMan) (TIANGEN, FP206, China) reaction was performed individually in a 20-μL system containing 1 μL of the template cDNA, 10 μL of the 2 × SuperReal PreMix (Probe), 0.6 μL of the 10uM of each primer, 0.4 μL of the 10 uM of the probe, 0.2 μL of the 50 × ROX Reference Dye*3, and 7.2 μL of the RNase-Free ddH2O. The thermal cycler conditions used for real-time PCR were: 40 cycles of 3 s at 95°C and 30 s at 60°C. The mRNA expression levels of trypsin genes were tested by SYBR Green Supermix (TaKaRa). The primers of trypsin genes used for qRT-PCR analysis can be found in Table S2. The H. armigera 18S (Du et al., 2017) and EF1-α (Yuan et al., 2006) were used as internal reference genes. The qRT-PCR was performed at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 20 s. Real-time PCR of trypsin and reference genes was done in a 20-μL reaction system containing 10 μL of 2 × SYBR Mix and 10 μM forward primer and reverse primer (1.0 μL each), 1 μL template cDNA, and 7.0 μL nuclease-free water. All qRT-PCR reactions were performed in 96-well optical plates in an ABI 7500 Real-time PCR System (Applied Biosystems).
The expression levels of all the earlier-mentioned genes were calculated with their amplification efficiency (E) and mean Ct, and the expression levels of the candidate genes were normalized with the geometric mean of the expression of each of the two reference genes (GAPDH and EF-1α/18S and EF1-α; Livak and Schmittgen, 2001; Vandesompele et al., 2002; Liu et al., 2015). The results of each gene among different strains were determined with one-way analysis of variance (ANOVA), followed by Tukey's honestly significance difference (HSD) test for mean comparison. All statistical analysis was performed with SPSS v.18.0 (SPSS Inc., Chicago, IL, USA) at P < 0.05 level of significance.
Results
Insect Resistance Levels
After 60 generations of Cry1Ac selection, the LF5 laboratory colony had an estimated resistance ratio of 540 compared with the susceptible LF strain (Table 1). The LF10 was divided from LF5; then, after selection for 52 generations using diets containing 10-μg/mL Cry1Ac toxin, the resistance ratio of the LF10 strain cotton bollworm reached 640 for Cry1Ac toxin (Table 1). The LF20 was divided from LF10 and after selection for 42 generations on diets containing 20 μg/mL Cry1Ac toxin, the resistance ratio of LF20 strain cotton bollworm increased to 850. Similarly, LF30 was divided from LF20 and selected for 38 generations on diets containing 30-μg/mL Cry1Ac toxin; in addition, LF60 was divided from LF30 and selected for 21 generations on diets containing 60-μg/mL Cry1Ac toxin. The resistance ratio of these two strains was 1,000 when compared with the susceptible-LF strain (Table 1). The LF120 was divided from LF60 and selected for 17 generations on diets containing 120-μg/mL Cry1Ac toxin; the LF120 strain showed the highest resistance levels (2,000-fold) (Table 1). Generally, with the increase of the selection concentration of Cry1Ac toxin, the resistance levels were improved correspondingly (Table 1).
Illumina Sequencing and Transcriptome Assembly
In total, 77,422,352 clean reads were obtained from transcriptomics analysis of samples obtained from the midguts of the seven strains and were assembled into 66,502 transcripts. The mean length of the transcripts was 1,324 bp with lengths ranging from 201 to 49,954 bp. After mixing with our private cotton bollworm transcriptome database, a total of 139,012 unigenes were obtained. The size distribution of these unigenes is shown in Figure S1.
Annotation of Predicted Proteins
Annotation of gene function was performed by running Blast on the following databases: Nr (NCBI non-redundant protein sequences), Pfam (Protein family), Nt (NCBI non-redundant nucleotide sequences), Swiss-Prot (A manually annotated and reviewed protein sequence database), KOG (euKaryotic Ortholog Groups), GO (Gene Ontology), and KO (KEGG Ortholog database). The results demonstrated that 64.96% unigenes were annotated in NR, 35.14% were annotated in NT, 15.83% were annotated in KO, 43.77% were annotated in SwissPort, 41.29% were annotated in PFAM, 48.11% were annotated in GO, and 32.96% were annotated in KOG. In total, 6.12% unigenes were annotated in all Databases. Finally, most of the 139,012 unigenes (72.89%) were matched to known genes. This transcriptome database will be used as a reference database to analyze differences in gene expression among different strains of cotton bollworm.
Classification of Gene Ontology (GO)
The GO classification demonstrated that 66,886 sequences could be assigned into 48 functional groups (Figure 1A). In the three main categories of the GO classification, “metabolic process,” “binding,” and “cell and cell part” terms were dominant, respectively.
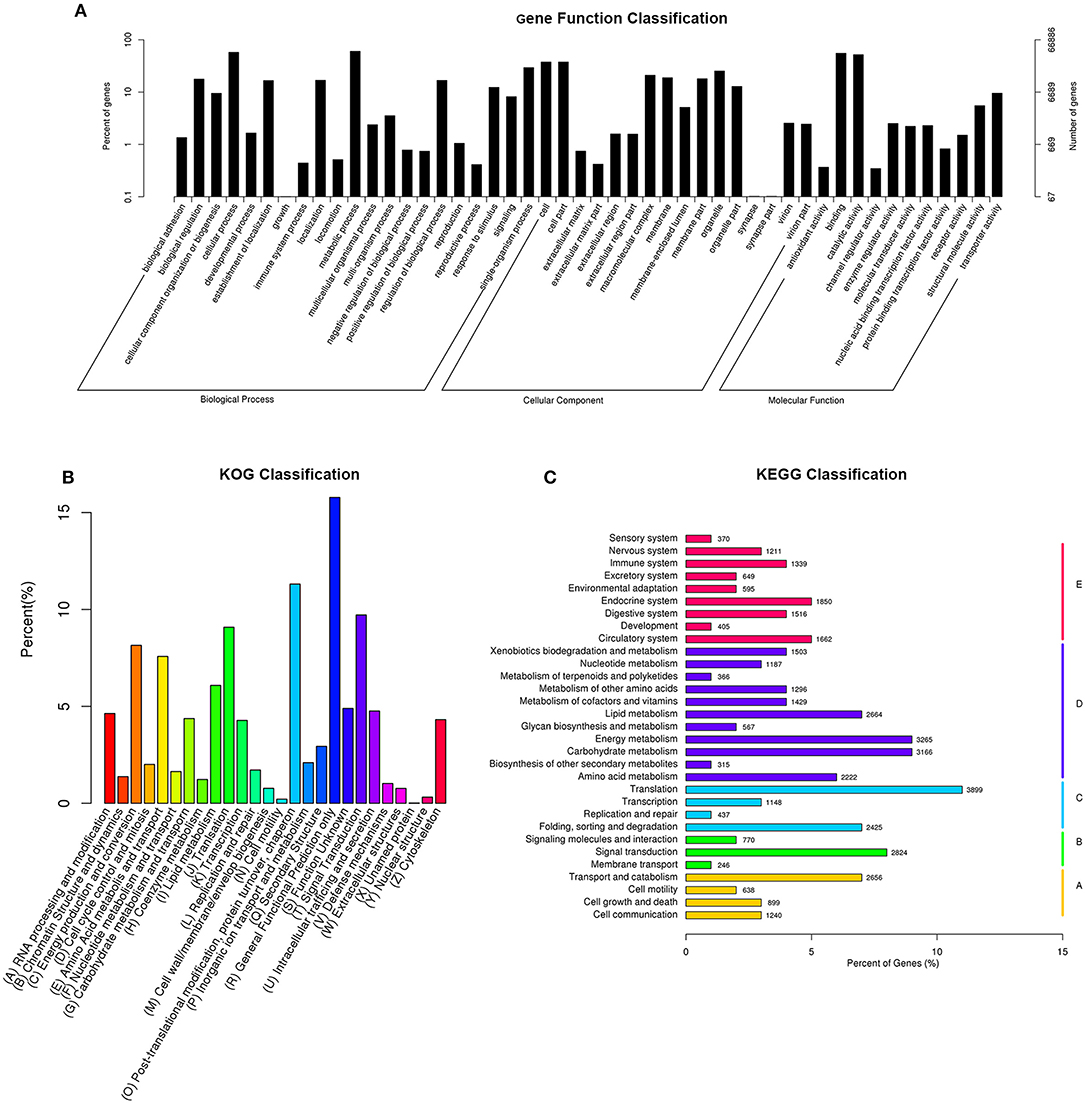
Figure 1. Histogram of gene ontology classification (A), Clusters of Orthologous Groups of proteins (B), and KEGG Ortholog database (C). (A) The results are summarized in three main categories: biological process, cellular Component, and molecular function. The right y-axis indicates the number of genes in a category. The left y-axis indicates the percentage of a specific category of genes in that main category. (B) The X-axis indicates 26 group categories; the Y-axis indicates the percentage of a specific category of genes in that main category. (C) The Y-axis is the enrichment of the KEGG term, the X-axis indicates the percentage of a specific category of genes in that main category. According to participation in KEGG metabolic pathways, genes can be divided into five branches: A, cellular processes; B, environmental information processing; C, genetic-based information processing; D, metabolism; E, organismal systems.
The GO analysis showed that the functions of the identified genes involved various biological processes. Totally, 40,256 unigenes were annotated in the “metabolic process” category, 38,541 unigenes were annotated in the “cellular process” category, and 36,898 unigenes were annotated in the “binding” category (Figure 1A).
KOG Classification
In total, 45,832 unigenes were categorized into 26 functional groups (Figure 1B). The main groups were “post-translational modification, protein turnover, chaperone” (5,184 unigenes), “general functional prediction only” (7,232 unigenes), and “signal transduction” (4,453 unigenes). These results demonstrated that based on high-throughput sequencing, novel genes that might play roles in CryAc resistance can be identified (Figure 1B).
Functional Classification by KEGG
The KEGG classification revealed that 35,802 annotated unigenes were mapped to the reference canonical pathways in KEGG and categorized into 5 KEGG pathways. The unigenes were clustered into various classifications, including metabolism (25,537 members), organismal systems (13,554 members), genetic information processing (8,519 members), cellular processes (6,849 members), and environmental information processing (5,583 members). These annotations of unigenes provide a valuable resource for investigating functions, specific processes, and pathways in cotton bollworm Cry1Ac-resistance research (Figure 1C).
Estimates of Differential Expression Among the Midgut Transcripts
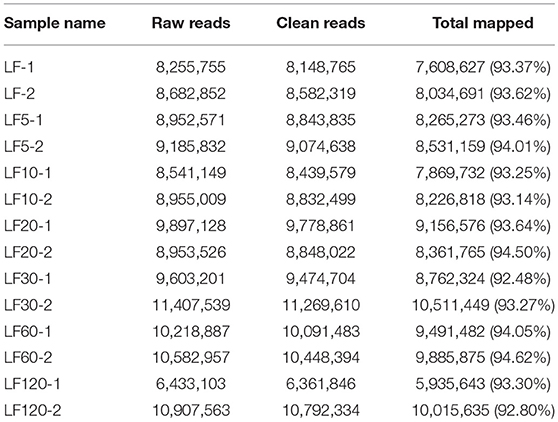
The DGE was used to analyze gene expression among the seven strains, including one susceptible and six Cry1Ac-resistant strains. Fourteen DGE libraries (containing two biological replicates): LF-1, LF-2, LF5-1, LF5-2, LF10-1, LF10-2, LF20-1, LF20-2, LF30-1, LF30-2, LF60-1, LF60-2, LF120-1, and LF120-2 were sequenced and between 6.4 and 11.3 million clean reads were generated. The number of clean read entities with unique nucleotide sequences ranged from 5,935,643 to 10,511,449 (Table 2). Moreover, 94.62% (9,885,875) of the sequences in the transcriptome database were unequivocally identified by unique genes (Table 2).
Differentially Expressed Genes in Different Resistant Developmental Strains
The DEG numbers detected to confer resistance level in six resistant LF strains did not increase along with the increase of resistance level to Cry1Ac (Table 1; Figure 2); the changes in the trends in the numbers of DEGs in six resistant LF strains are similar to the letter “N” in Figure 2A. Commonly, more upregulated genes than downregulated genes were detected in each of the six resistant LF strains (Figure 2A). Compared with the susceptible LF strain, 3,688 unigenes were expressed differentially in the LF5 strain, which presents the lowest resistance level. The highest numbers of DGEs occurred in the LF10 strain (9,712) followed by the LF20 stain (8,558) (Figure 2B). The lowest numbers of DGEs occurred in the LF30 strain (2,085) (Figure 2). Although LF60 showed the same resistance level as LF30, more DEGs were found in the LF60 strain (4,990), possibly due to the greater exposure to Cry1Ac toxin for the LF60 strain (Table 1; Figure 2). In total, 6,859 unigenes were expressed differentially in the LF120 strain, although the larvae of this strain showed the highest resistance. Comparing two neighboring strains, more genes showed significant differences in expression levels between LF10 vs. LF5 and between LF20 vs. LF10, and fewer genes showed significant differences in expression levels between LF30 vs. LF20 and between LF60 vs. LF30 (Figure 2A). However, the declining trend did not continue between LF120 vs. LF60, possibly due at least in part to the greater number of mutations in more genes or alleles involved in conferring a higher resistance level in the LF120 strain (Figure 2A). To analyze the function of DEGs between the LF and LF-resistant strains, these genes were classified in GO terms. The 30 significantly enriched (according to the corrected pValue) GO terms are shown in Figure S2. The differentially expressed genes showed significant enrichment in “catalytic activity,” “endopeptidase activity,” “aminopeptidase activity,” “serine-type endopeptidase activity,” “proteolysis,” “biological process,” “metabolic process,” “peptidase activity,” “serine-type peptidase activity,” “metallopeptidase activity,” “exopeptidase activity,” “hydrolase activity,” “serine hydrolase activity,” “protein metabolic process,” “peptidase activity,” “acting on L-amino acid peptides,” and “organic substance metabolic process” terms in all resistant strains (Table 3). These DEGs may help the cotton bollworm to enhance their physiology to adapt to the Cry1Ac toxin. The LF10, LF20, LF30, and LF60 have moderate resistance level and some of the differentially expressed genes in these four resistant strains were significantly enriched in “ribosome,” “translation,” “non-membrane-bounded organelle,” “ribonucleoprotein complex,” “structural constituent of ribosome,” and “intracellular non-membrane-bounded organelle” (Table 3). The LF5 has the least resistance and some of the differentially expressed genes had the functions related to xenobiotics because they showed significant enrichment in “xenobiotic metabolic process,” “response to xenobiotic stimulus,” “antioxidant activity,” “cis-stilbene-oxide hydrolase activity,” “coenzyme binding,” “cellular response to chemical stimulus,” and “cellular response to xenobiotic stimulus” (Table 3; Figure S2). As the resistance level increased, the genes of the cotton bollworms showed some significant differences in macromolecule metabolic processes in the LF10 and LF20 strains (Table 3; Figure S3). Different from other strains, some genes involved in “proton-transporting V-type ATPase complex” and “proton-transporting two-sector ATPase complex” showed significant changes in the LF30 strain (Table 3; Figure S2). For the LF60 strain, some genes involved in “chitin binding,” “sterol binding,” and “alcohol binding” showed more significant expression differences than other Go terms, and this was specifically true in this strain (Table 3; Figure S2). As the highest resistance-level strain LF120, the differentially expressed genes were enriched more significantly in “cellular iron ion homeostasis,” “ferric iron binding,” “hexachlorocyclohexane metabolic process,” “chlorinated hydrocarbon metabolic process,” “halogenated hydrocarbon metabolic process,” “cellular transition metal ion homeostasis,” “transition metal ion homeostasis,” and “iron ion homeostasis” (Table 3; Figure S2). The enrichment of DEGs in the same or in different pathways provides information that can aid in understanding the development of resistance and the resistance mechanisms in different strains.
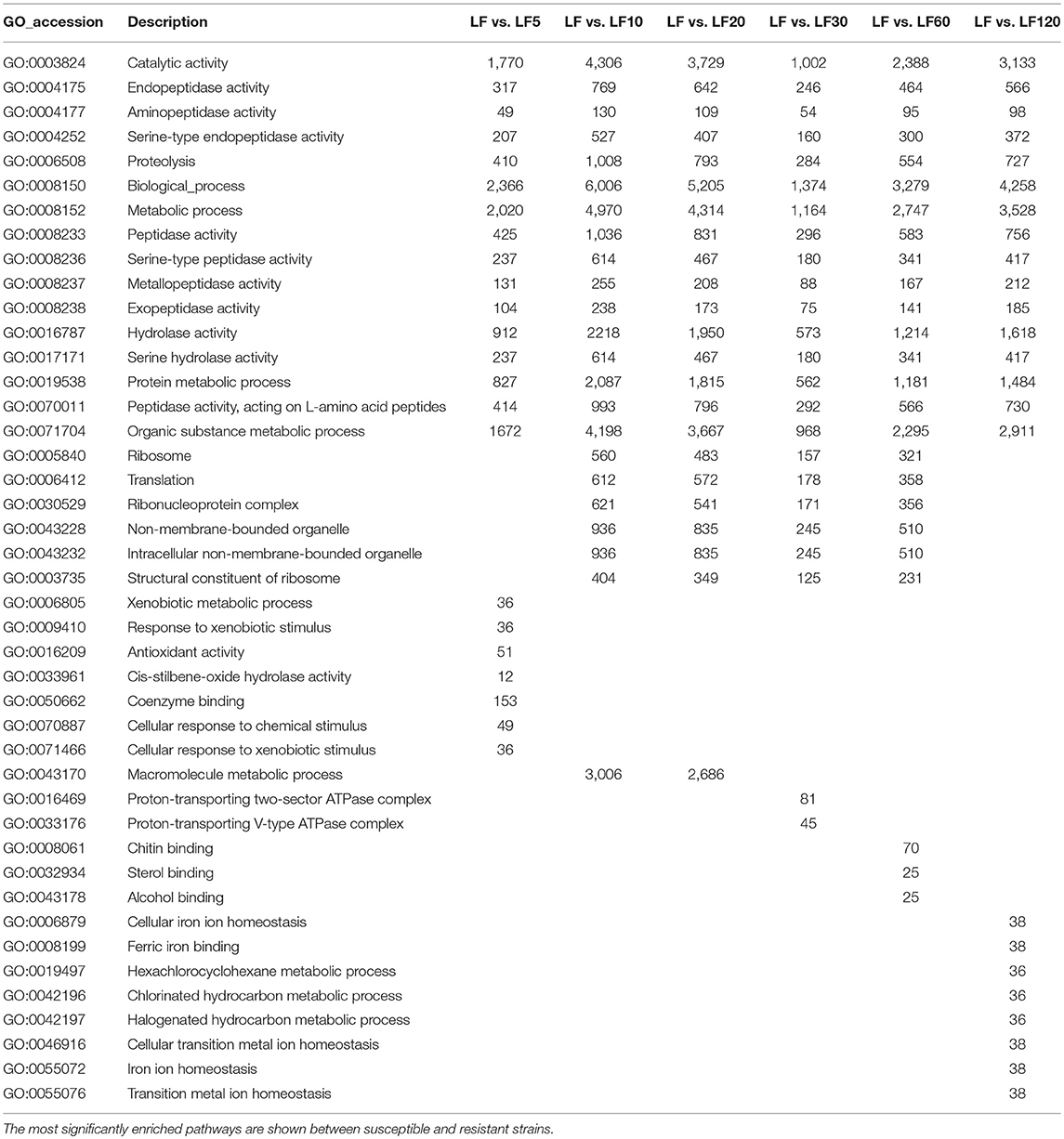
Table 3. Gene ontology (GO) classification of differentially expressed genes in susceptible and resistant strains.
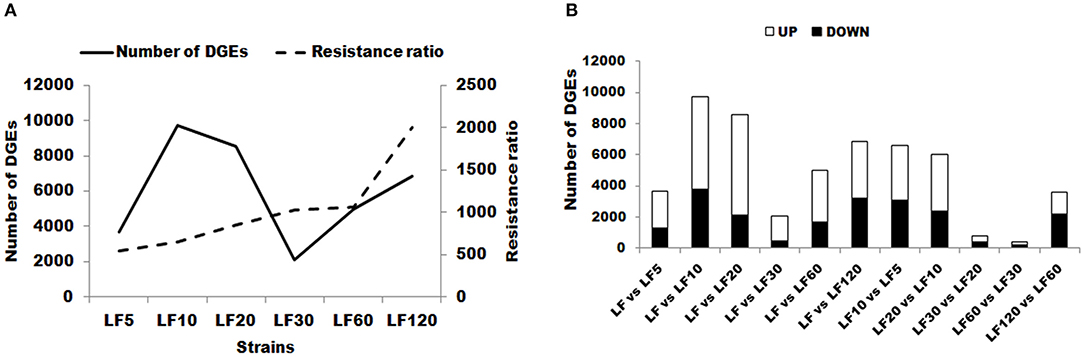
Figure 2. Changes in gene expression profiles among the different resistant strains. (A) The number of upregulated and downregulated genes between LF and LF5; LF and LF10; LF and LF20; LF and LF30; LF and LF60; LF and LF120; LF10 and LF5; LF20 and LF10; LF30 and LF20; LF60 and LF30; and LF120 and LF60 are summarized. Up: upregulated in LF-resistant strains; Down: downregulated in LF-resistant strains. (B) The number of total changes in gene expression in each resistant strain and resistance ratio of each resistant strain are summarized. DEGs, differentially expressed genes.
In organisms including cotton bollworm, different genes possess special biological functions and coordinate with each other. In another way, through KEGG pathway analysis, significant enrichment can identify DGEs that are involved in the main biochemical pathways and signal transduction pathways. The most significantly enriched (according to the corrected p-value) 20 KEGG pathways are shown in Figure S3. The results indicated that the DEGs were enriched more significantly in “propanoate metabolism,” “two-component system,” and “protein processing” in the endoplasmic reticulum in all resistant strains (Table 4). In the five least-resistant strains, the differentially expressed genes were enriched more significantly in “citrate cycle (TCA cycle)” and “carbon fixation pathways in prokaryotes.” For the LF5 strain, some DEGs were enriched significantly in “starch and sucrose metabolism” and in “plant-pathogen interaction” pathways. For the LF10 strain, some differentially expressed genes were enriched significantly in “ABC transporters” pathways (Table 4) and other genes that differentially expressed between from LF5 and LF 10 were involved significantly in “glutathione metabolism,” a pathway that may help to detoxify Cry1Ac toxins. Some DEGs from LF30 were found to be involved significantly in the “mTOR signaling pathway” (Table 4), which is a crucial signaling pathway that mediated cell growth and proliferation (Kazuyoshi Yonezawaa, 2004). These results indicated that these genes in LF30 may affect larval growth. Further details of differentially expressed genes that were enriched significantly in the KEGG pathway are shown in Table 4 and Figure S3.
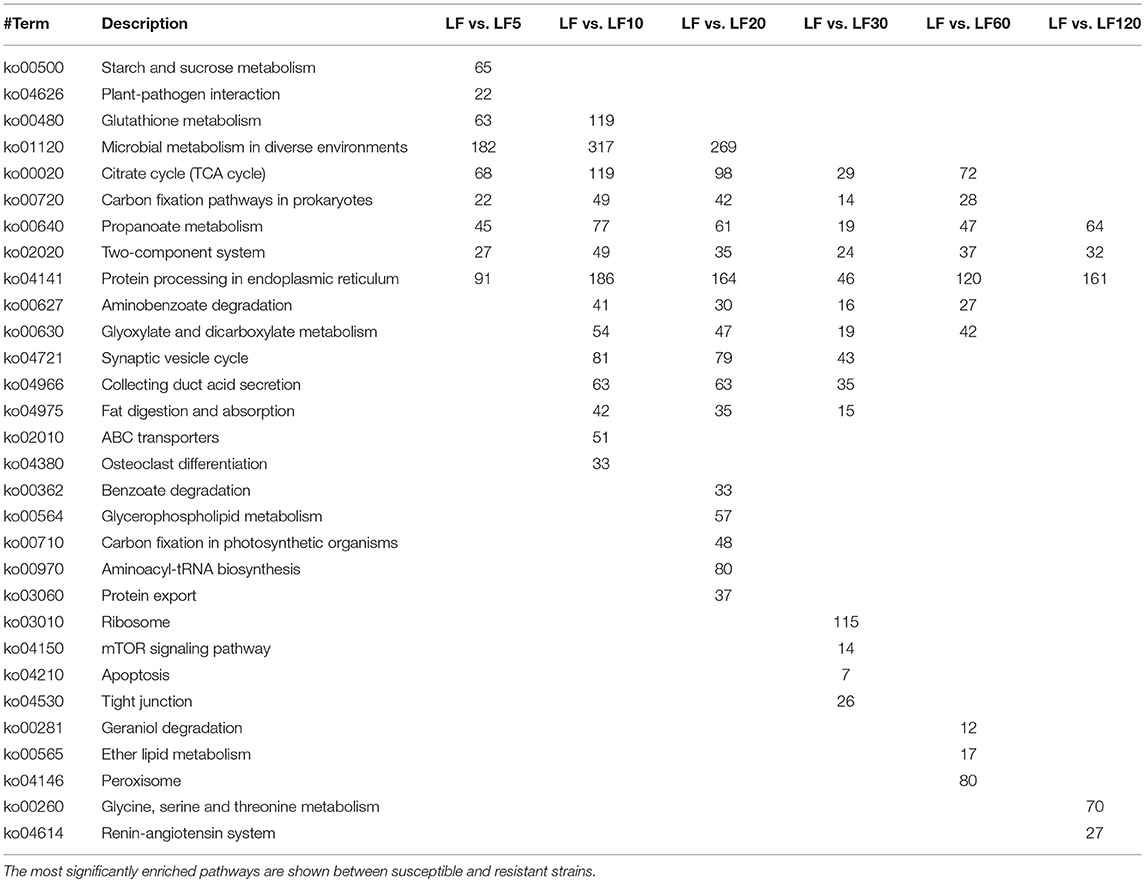
Table 4. The KEGG ortholog classification of differentially expressed genes between the susceptible and the resistant strains.
Expression Level of Trypsin Genes Involved in Development of Resistance
The trypsin family is present widespread in animals and plays a variety of roles, especially in the digestive system. Lower expression and activity of trypsin proteases result in decreased activation of the Cry1Ac protoxin and is a mechanism of resistance to Cry1Ac in H. armigera (Liu et al., 2014; Wei et al., 2016a). Fourteen unigenes (comp35781_c0_seq1, my_s32485, comp41058_c1_seq5, Unigene12105, Unigene47996, Unigene20462, Unigene36451, Unigene15742, Unigene43312, Unigene8736, comp41058_c4_seq1, Unigene31995, my_rep_c15473, and my_rep_c25150) were matched to nine corresponding candidate trypsin genes (Gene bank: XM_021340600, XM_021340599, XM_021340597, XM_021333008, XM_021329499, XM_021338512, XM_021344340.1, XM_021344341.1, and XM_021344468.1), and the mRNA levels of these genes were found to be decreased significantly in resistant strains in comparison with the susceptible LF strain (Table 5), consistent with the qRT-PCR results obtained in all six resistant strains (Figure 3).
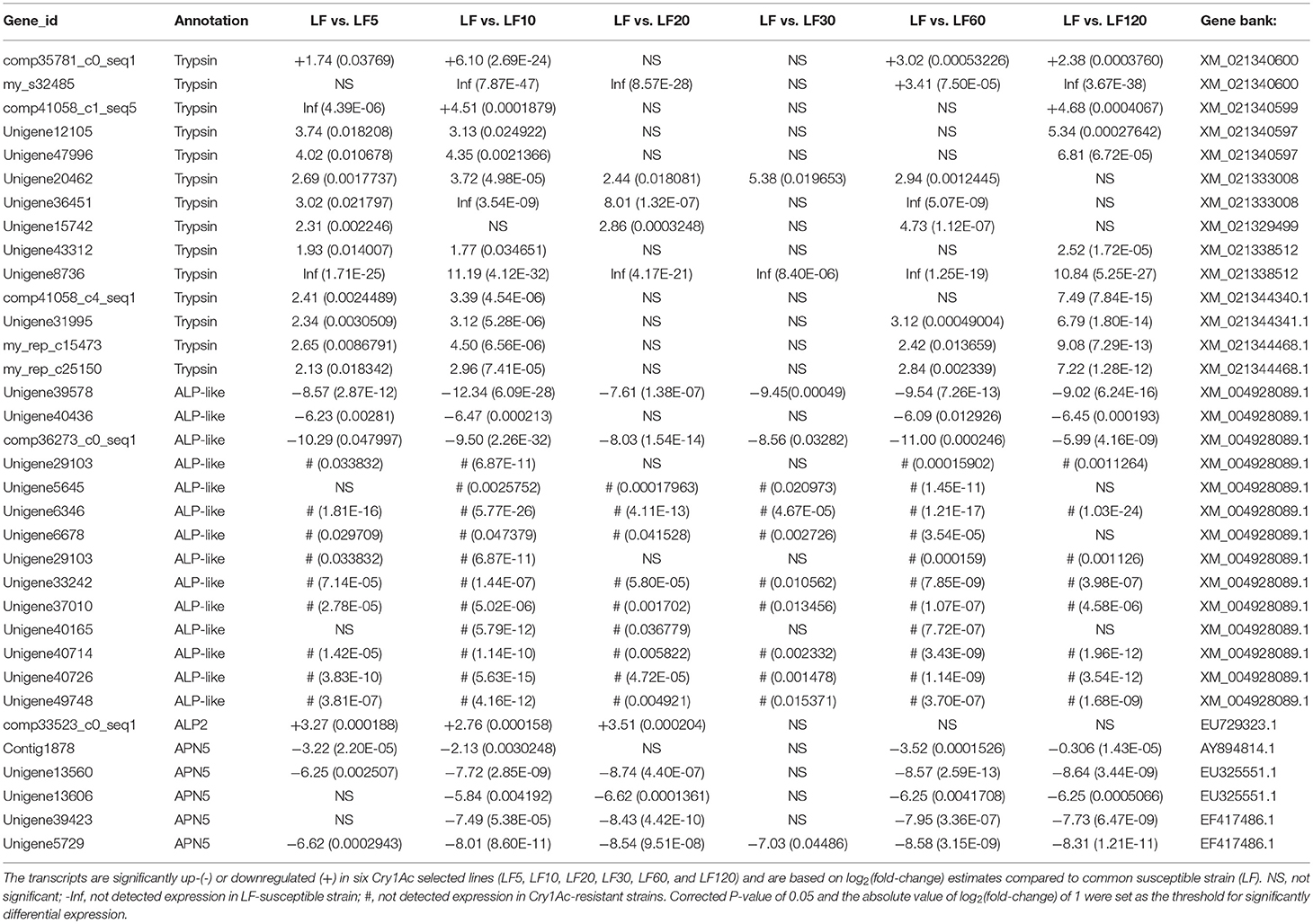
Table 5. Estimates of transcripts levels (expression) among candidate Cry1Ac toxin-resistance genes in H. armigera midgut based on DGE sequence mapping to each unigene.
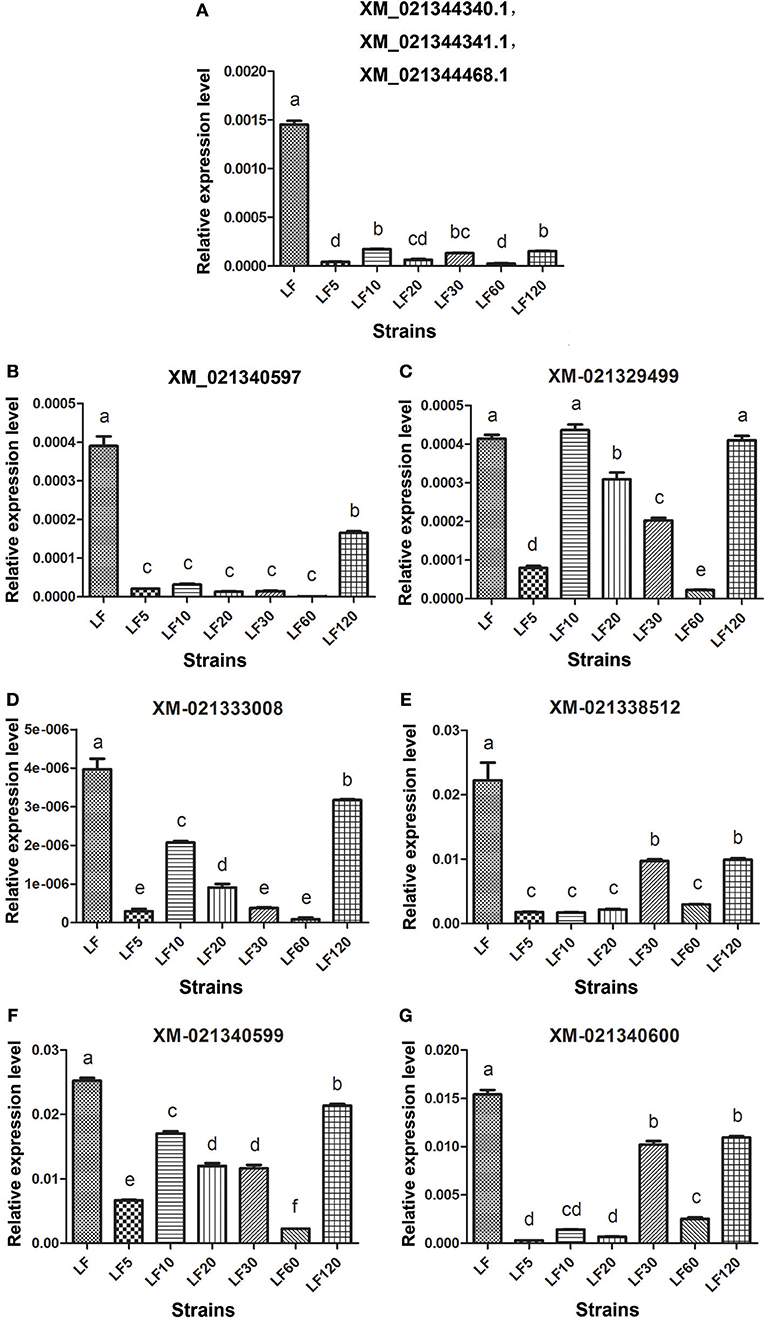
Figure 3. The qRT-PCR analysis of transcript abundances among candidate Bt resistance trypsin genes within H. armigera-susceptible (LF) and -resistant (LF5, LF10, LF20, LF30, LF60, and LF120) strains. (A) The three genes ID numbers represent the three trypsin genes in NCBI; a pair of primers in the conserved region of these three trypsin genes were used to analyze transcript abundances. (B-G) Represent the relative expression levels of trypsin genes of XM_021340597, XM_021329499, XM_021333008, XM_021338512, XM_021340599 and XM_021340600 in resistance strains, respectively. Values shown are means and standard errors. Different letters indicate significant differences between treatments (P < 0.05; HSD test).
Expression Level of Cry1Ac-Receptors Genes Involved in Development of Resistance
Several known Bt receptors and Bt-resistance genes including ALP-like (XM_004928089.1), ALP2 (EU729323.1), and APN5 (AY894814.1, EU325551.1, and EF417486.1) showed the same changed trend in all resistance strains (Table 5). The ALP-like genes encoded by fourteen unigenes (Unigene39578, Unigene40436, comp36273_c0_seq1, Unigene29103, Unigene5645, Unigene6346, Unigene6678, Unigene29103, Unigene33242, Unigene37010, Unigene40165, Unigene40714, Unigene40726, and Unigene49748) were found to be upregulated significantly based on DGE results (Table 5) and these results were further verified by qRT-PCR in all six resistant strains (Figure 4A). Another ALP gene, ALP2 (comp33523_c0_seq1), was significantly upregulated in the LF5, LF10, and LF20 strains, but there was no significant change in LF30, LF60, and LF120. However, qRT-PCR analysis demonstrated that this ALP2 (comp33523_c0_seq1) of H. armigera was ubiquitous and significantly reduced in all six resistant strains (Figure 4B). The APN5 (Contig1878, Unigene13560, Unigene13606, Unigene39423, and Unigene5729) was upregulated significantly in the LF-resistant strains according to DGE results (Table 3), However, qRT-PCR analysis indicated that this gene was upregulated significantly in the LF5, LF20, and LF30 strains, unchanged in the LF120 strain, and significantly downregulated in the LF10 and LF60 strains (Figure 4C). In contrast, APN1 (AF441377) was downregulated significantly in all these six resistance strains according to the qRT-PCR analysis, but not according to the DGE results (Figure 4D).
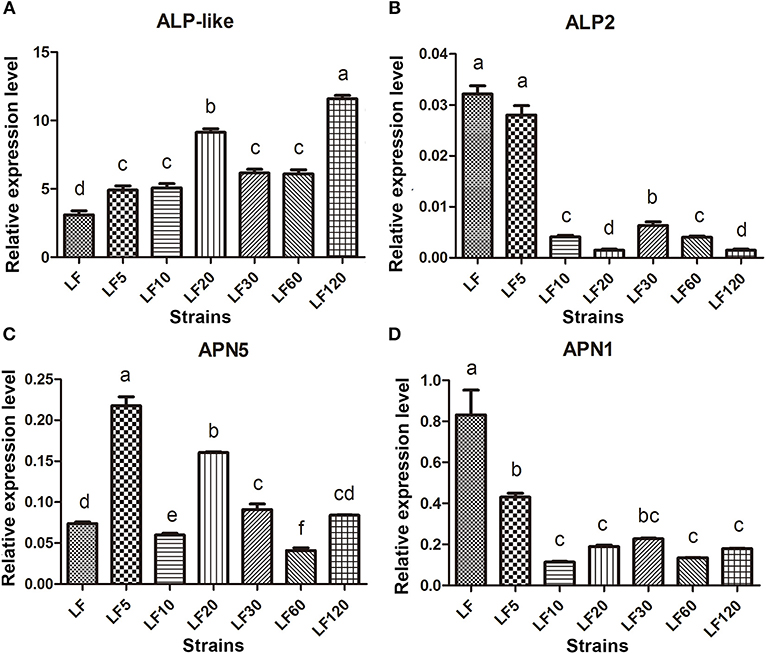
Figure 4. The qRT-PCR estimates of transcript abundances among candidate Bt-resistance receptor genes within H. armigera-susceptible (LF) and -resistant (LF5, LF10, LF20, LF30, LF60, and LF120) strains. (A-D) Represent the relative expression levels of ALP-like, ALP2, APN5 and APN1 in resistance strains, respectively. Values shown are means and standard errors. Different letters indicate significant differences between treatments (P < 0.05; HSD test).
Discussion
Resistance to Cry1Ac is controlled by multiple genes involved in fitness costs and in the selection of recessive or dominant receptors (and even alleles with different types of mutations on the same locus) and their interactions (Tabashnik et al., 2005; Xu et al., 2005; Zhang et al., 2009, 2012; Gahan et al., 2010; Baxter et al., 2011; Jurat-Fuentes et al., 2011; Atsumi et al., 2012; Xiao et al., 2014; Chen et al., 2015). Fitness costs contain longer period of development and reduction in survival, pupal weight, and fecundity (Sayyed et al., 2008). Fitness costs are expected to increase steadily with the development of increased resistance (Cao et al., 2014). According to Cao et al. (2014), who established multiple regressions to predict overall fitness cost and resistance level with fitness costs, these LF-resistant strains may use a second phase of resistance. In this stage, resistance gene-encoding enzymes, such as digestive enzymes, hydrolase, detoxification enzymes, and catalytic enzymes are considered the most important factor to produce fitness cost (Rajagopal et al., 2009; Zhu et al., 2011; Guo et al., 2012; Cao et al., 2013; Lei et al., 2014; Liu et al., 2014; Wei et al., 2016a; Zhang et al., 2017). As a significant and universal phenomenon, we found in this study that a significant portion of DEGs were enriched predominantly in “catalytic activity,” “endopeptidase activity,” “aminopeptidase activity,” “serine-type endopeptidase activity,” “proteolysis,” “biological process,” “metabolic process,” “peptidase activity,” “metallopeptidase activity,” “serine-type peptidase activity,” “exopeptidase activity,” “serine hydrolase activity,” “hydrolase activity,” “protein metabolic process,” “acting on L-amino acid peptides,” “peptidase activity,” and “organic substance metabolic process” for all the resistant strains (Table 3; Figure S2). High-level expression of the genes involved in the earlier-mentioned pathways will help resistant insects to avoid Cry1Ac damage. However, in compensation, these resistant insects develop a lower hatching rate, a lower copulation rate, a lower emergence rate, and even a lower survival rate (Cao et al., 2014). These findings suggest that the resistance to Cry1Ac in H. armigera might also be associated with increased catalytic activity, digestive activity, hydrolase activity, and detoxification activity.
The current understanding of Bt-toxin resistance in insects is associated generally with either conversion of Bt protoxins to activated toxins by insect midgut proteases (Rajagopal et al., 2009; Cao et al., 2013; Liu et al., 2014; Wei et al., 2016a) or by altered binding capacity of toxins to midgut proteins (Xu et al., 2005; Zhang et al., 2009, 2012; Gahan et al., 2010; Baxter et al., 2011; Jurat-Fuentes et al., 2011; Atsumi et al., 2012; Xiao et al., 2014; Chen et al., 2015). Decreased transcript levels of trypsins have been associated with reduced Cry1Ac protoxin activation (Rajagopal et al., 2009; Liu et al., 2014). This result is consistent with our present results, in which nine transcripts encoding trypsin serine protease were found to be downregulated in six LF-resistant strains (Table 5; Figure 3). Previous studies also found that reduced trypsinlike activity is correlated with reduced expression levels of trypsin gene transcripts in the LF120 strain (Wei et al., 2016a). Similar results were also reported in Ostrinia nubilalis, in which defense against Bt toxins was considered as main mechanism of resistance (Yao et al., 2012; Nanoth Vellichirammal et al., 2015). These results revealed that reduced protoxin activation is considered generally as a resistance mechanism against Bt proteins. More importantly, for the first time, we revealed that these nine trypsin serine proteases downregulated in Cry1Ac-resistant H. armigera was probably a common phenomenon. This result indicated that some trypsin activators may be used to improve the toxicity of Bt to a certain degree.
High levels of resistance are most commonly associated with mutations that disrupt the binding of Cry proteins to midgut receptors, decreased expression of these specific receptors in the midgut, and decreases of the toxin binding to its midgut proteins in the resistant strain (Ferré and Van Rie, 2002; Wu, 2014). While investigating the universal mechanism of resistance to Bt proteins, downregulation of ALP2 and APN1 and upregulation of ALP-like were found in these LF-resistant strains (Table 5; Figure 4). Reduced activity and transcription of ALP, which binds Cry1Ac, caused the resistance to Cry1Ac in the LF10, LF30, LF60, and LF120 strains when compared with a 96S-susceptible strain (Chen et al., 2015). The ALP1 and ALP2 all had been reported as the receptors of H. armigera (Ning et al., 2010; Chen et al., 2015); however, the total ALP activity was decreased and the transcription of the conserved region of HaALP2 (accession no. EU729323) and HaALP1 (accession no. EU729322) isoforms was also reduced in the resistance strains. It was difficult to know whether ALP1 or ALP2 caused the resistance to Cry1Ac. Here, we pinpoint accurately that the downregulation of ALP2 transcription leads to Cry1Ac resistance in LF resistance. Moreover, in LF Cry1Ac-resistant H. armigera larvae, a decrease in ALP activity may be correlated with reduced levels of ALP2 transcripts. However, ALP1 seems not to be a gene that importantly associated with resistance to Bt Cry1Ac since ALP1 was verified to be upregulated in all the LF-resistant strains (Zhang, 2013, PhD thesis).
In our study, ALP1 and ALP-like (Table 5; Figure 4) were found to be upregulated in all these LF-resistant strains. The increased expression of both genes in the LF-resistant strains was associated probably with the gut defensive response to Cry1Ac intoxication. In fact, ALP expression is considered as a marker for stem cell proliferation, which is crucial to gut defensive responses to Cry toxins (Singh et al., 2012). Moreover, midgut regeneration has been proposed as a mechanism of Cry1Ac resistance in H. virescens (Forcada et al., 1999). Further studies are needed to determine the molecular mechanisms responsible for the upregulation of ALP1 and ALP-like in these LF-resistant larvae.
Various studies with glycosylphophatidylinositol (GPI)-anchored APN1 from lepidopteran insects consistently demonstrated that APN1 is one of the midgut receptors for Cry1Ac and related to the resistance to Bt toxins (Zhang et al., 2009; Tiewsiri and Wang, 2011; Valaitis, 2011; Flores-Escobar et al., 2013; Wei et al., 2016b). However, our DGE data analysis demonstrated that the expression level of APN1 transcript was not significantly different between the LF-susceptible and -resistant strains. This result contradicted our qRT-PCR analysis (Figure 4), which demonstrated that APN1 transcript was decreased significantly in the LF-resistant strains (Figure 4). The discrepancy between the DGE analysis and the qRT-PCR analysis may be due to the sensitivity of qRT-PCR, which is higher than that for DEG. The APN5 can also bind to Cry1Ac in H. armigera (Wang et al., 2005), but its involvement in Cry1Ac resistance has not been documented. In this study, we found that APN5 is widely upregulated in the LF-resistant strains using DGE analysis, and this result was confirmed further by qRT-PCR in the LF5, LF20, LF30, and LF120 strains. The upregulation of APN6 has been reported to act as a compensation of APN1 loss in order to minimize the fitness costs of resistance in Trichoplusia ni (Tiewsiri and Wang, 2011). Whether a similar function of APN5 exists in LF5, LF20, LF30, and LF120 H. armigera warrants further study. Other Cry1Ac receptors (cadherin and ABC transports) and related genes showed different expression levels in individual LF-resistant strains, but they did not show a universal mechanism of resistance to Cry1Ac in all LF-resistant strains. This finding suggests that changes in expression of one or more of the Cry1Ac receptors and related genes can influence Cry toxin resistance traits to different degrees. Indeed, additional study is required to decipher the individual roles of the interactions between Bt receptors within the framework of toxin modes of actions. Nevertheless, our results provide evidence that the downregulation of ALP2 and APN1 affected the resistance in both lower and higher levels of resistance. Therefore, these genes may be used as markers to monitor and manage pest resistance in transgenic crops.
Lei et al. (2014) identified unigenes that are differentially expressed between Cry1Ac-susceptible and two resistant Plutella xylostella strains by RNA-seq analysis, and further analysis found that the higher resistance strain showed the greater number of EDUs. However, the higher resistance to Cry1Ac in insects does not always involve the use of more DEGs to adapt to more toxins. Our results showed the numbers of DEGs increased from LF5 to LF10 and LF30, LF60 to LF120, consistent with the resistance ratios of these strains (Figure 2B). However, the DEG numbers in the LF30, LF60, and LF120 strains were all lower than those in the LF10 and LF20 and a negative correlation was found between DEG numbers and resistance ratios in the LF10, LF20, and LF30 strains (Figure 2). Obviously, our data indicate that this is in response to different selection pressures. Different genes are used to adapt to the new environment, including the evolution of resistance to Cry1Ac, finally developing different resistant mechanisms. This conclusion was confirmed further by previous studies in these LF-resistant strains. For example, the cis-mediated downregulation of HaTryR expression is considered as the main resistance mechanism in the LF5 strain (Liu et al., 2014). In the LF60 strain, an ABCC2 mutant (in which a 6bp deletion in genomic DNA introduces a premature stop codon) leads to the resistance to Cry1Ac (Xiao et al., 2014). Decreased ALP activity and transcription are considered to cause the Cry1Ac resistance in LF10, LF30, LF60, and LF120 (Chen et al., 2015). Although trypsin activity was decreased significantly in LF120, the high level of resistance to protoxin and activated toxin indicated that reduced activation of the protoxin was not a major Cry1Ac-resistance mechanism in LF120 (Wei et al., 2016a). Interestingly, the finding of the lowest number of DGEs indicated that a new domain mechanism of resistance has evolved in the LF30 strain (Figure 2). This result indicated that different genes and pathways were involved in Cry1Ac resistance in H. armigera. Also, these pathways seem to be differently affected depending on the level of resistance.
The GO and KEGG category analyses provided an important cue to uncover the different mechanisms involved in the development of resistance. First, the initial resistance appeared to increase immune and detoxification processes as shown by the series of DEGs in LF5 strains, predominantly DEGs involved in “xenobiotic metabolic process,” “response to xenobiotic stimulus,” “antioxidant activity,” “cis-stilbene-oxide hydrolase activity,” “coenzyme binding,” “cellular response to xenobiotic stimulus,” and “cellular response to chemical stimulus” (Table 3). These DEGs in the LF5 strain lead to increased immune and detoxification functions in the body, thus initiating defense to Bt invasion. Similar results were found in the KEGG pathway analysis, in which a series of DEGs were also enriched significantly in “plant-pathogen interaction,” “glutathione metabolism,” and “microbial metabolism in diverse environments” (Table 4). The changes of these DEGs in the LF5 stain indicated that low-level resistance is probably associated with insect immune and detoxification processes. With the increase of selection pressure involving Cry1Ac, the LF10 and LF20 strains showed different gene changes to that in the LF5 strain. These genes were found to participate in macromolecule metabolic process (Table 3) involved in the degradation, metabolism, transport, secretion, and absorption of macromolecular substances (Table 4; Figure S2). In particular, the 51 unigenes that encode ABC transporters were found to be expressed significantly in the LF10 strain. The ABC transporters, such as ABCG1, ABCC2, and ABCC3 have been confirmed to be Bt receptors and are related to the resistance to Bt (Xiao et al., 2014; Guo et al., 2015a,b; Tanaka et al., 2017). These results indicate that these genes are associated probably with insect resistance to Bt in the LF10 and LF20 strains. Convincing evidence shows that ABC transporters have been verified to be involved in the mode of Cry1Ac action and the mechanism of resistance to Cry1Ac.
The DEGs involved in “proton-transporting V-type ATPase complex” and “proton-transporting two-sector ATPase complex” were found to be expressed significantly in the LF30 strain (Table 3). It is not surprising that V-ATPase takes part in the resistance to Bt because previous reports have demonstrated that a number of V-ATPase subunits can bind to different Bt proteins, including Cry1Ab, Cry1Ac, and Cry4Ba (Bayyareddy et al., 2009; Chen et al., 2010; Xu et al., 2013). Similar results were reported in a study of a Cry1F-resistant strain, in which seven transcripts encoding V-ATPase subunits were identified significantly in downregulation (Nanoth Vellichirammal et al., 2015). It has been reported that V-ATPase subunits are involved in maintaining the alkaline conditions of the midgut (Onken et al., 2008). Further work should be carried out to verify the effect of V-ATPase subunits on midgut pH in the LF30 strain, as the results may provide further insight into resistance mechanisms used by this resistant strain.
At high levels of resistance to Cry1Ac, receptor mutations may be the main reasons underlying the resistance. However, the resistance mechanisms in LF60 have been identified (Xiao et al., 2014). In addition, other factors must be involved in the resistance to Cry1Ac because our results identified 4,990 DEGs that were expressed significantly in the LF60 strain, compared with the LF-susceptible stain. Interestingly, some DGEs were found to act in “chitin binding,” “sterol binding,” and “alcohol binding” (Table 3). Other DGEs were found to function in “geraniol degradation” and “ether lipid metabolism” based on the KEGG ortholog classification. Future studies are needed to determine the roles of these DGEs in the development of resistance in the LF60 strain.
Our previous study confirmed that trypsin activity was decreased significantly in LF120 (Wei et al., 2016a). Correspondingly, in the present study, nine trypsins were found to be significantly downregulated in the LF120 strain (Table 3). However, LF120 has the highest levels of mRNA for different trypsins among all resistant strains. These results indicate that the reduced activation of protoxin seems not to be a main mechanism of resistance to Bt proteins in this strain. Meanwhile, DGEs in LF120 were enriched significantly in “cellular iron ion homeostasis,” “ferric iron binding,” “cellular transition metal ion homeostasis,” “transition metal ion homeostasis,” and “iron ion homeostasis” (Table 3). This result suggested to us that ion homeostasis in the insect's body may play a important role in affecting the resistance to Bt, because unbalanced ion homeostasis can impair the normal functions of proteins within cells, hinder pore formation, and lead to cell death. As reported, Cry toxins can induce the formation of non-selective channels and then lead to imbalance of cations, anions, neutral solutes, and water, finally causing cell swelling and lysis (Knowles and Ellar, 1987). The role of ions, especially iron, in the mechanisms of resistance in the LF120 strain will be explored in future studies.
From LF5 strain to LF120 strain, many pathways seemingly exist. However, a domain pathway contributes hugely to Cry1Ac resistance. The upregulated or downregulated genes may not be fully illustrated in the resistance mechanisms that occur in the LF-resistant strains. Importantly, key genes in different pathways regulating the expressions of resistance-associated genes should be identified further. Also, these key genes may be used to modify via gene edition (CRISPR) for molecular control of the resistance. For example, the interplay between ALP and ABCC is controlled by MAP4K4 in the MAPK signaling pathway in P. xylostella (Guo et al., 2015b). However, as the firstly discovered pathways, V-ATPase, ABC transporters, or ion homeostasis, which are involved in Cry1Ac resistance, are lesser known, and more functional experiments need to be carried out in the future.
Conclusion
Based on our observations, several factors are associated with Cry1Ac resistance in the LF-resistant H. armigera strains. Changes in catalytic activity, digestive activity, hydrolase activity, and detoxification activity and in the downregulations of receptors and related genes, including ALP2, APN1, and trypsin, unavoidably result in resistance to Cry1Ac. The ALP2 and APN1 can, therefore, be considered as probes to monitor the resistance of H. armigera to first-generational Cry1Ac crops in the field. Most importantly, the results here revealed multiple genes and pathways that are probably involved in resistance. Also, these pathways seem to be differently affected depending on the level of resistance. For controlling the lower level Cry1Ac-resistance, some enzyme inhibitors or activators can be explored to improve the toxicity of Bt. As the resistance increases, the catalytic activity, digestive activity, hydrolase activity, and detoxification activity may not be the main role of resistance. Special pathways and genes may be involved in the resistance, such as V-ATPase, ABC transporters, or ion homeostasis, and they seem to differently contribute to resistance depending on the level of resistance. The identification of the key genes that regulate the main pathway contributing to resistance are underway. Also, these genes could be used via gene edition (CRISPR) for molecular control of resistance.
Data Accessibility Statement
Original sequencing reads are available from the GenBank Under accession numbers: PRJNA451313.
Author Contributions
JW and GL conceived and designed the experiments. JW, LC, and SY performed the experiments. GL, SA, and JW analyzed the data. JW, XL, MD, SA, and GL wrote the manuscript. JW, SA, and GL shared the microscopic observations and writing responsibilities. All authors have read and approved the manuscript for publication.
Funding
This research was supported by the Key Project for National Key R&D Program of China (2017YFD0201900), Breeding Genetically Modified Organisms (grant number 2016ZX08011–002), the National Natural Science Funds of China (grant number 31621064), and the State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201708).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01653/full#supplementary-material
References
Adang, M. J., Crickmore, N., and Jurat-Fuentes, J. L. (2014). Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv. Insect Physiol. 47, 39–87. doi: 10.1016/B978-0-12-800197-4.00002-6
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Atsumi, S., Miyamoto, K., Yamamoto, K., Narukawa, J., Kawai, S., Sezutsu, H., et al. (2012). Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 109, E1591–E1598. doi: 10.1073/pnas.1120698109
Baxter, S. W., Badenes-Pérez, F. R., Morrison, A., Vogel, H., Crickmore, N., Kain, W., et al. (2011). Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics 189, 675–679. doi: 10.1534/genetics.111.130971
Bayyareddy, K., Andacht, T. M., Abdullah, M. A., and Adang, M. J. (2009). Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem. Mol. Biol. 39, 279–286. doi: 10.1016/j.ibmb.2009.01.002
Cao, G., Feng, H., Guo, F., Wu, K., Li, X., Liang, G., et al. (2014). Quantitative analysis of fitness costs associated with the development of resistance to the bt toxin Cry1Ac in Helicoverpa armigera. Sci. Rep. 4:5629. doi: 10.1038/srep05629
Cao, G., Zhang, L., Liang, G., Li, X., and Wu, K. (2013). Involvement of nonbinding site proteinases in the development of resistance of Helicoverpa armigera (Lepidoptera: Noctuidae) to Cry1Ac. J. Econ. Entomol. 106, 2514–2521. doi: 10.1603/EC13301
Chen, L., Liang, G., Zhang, J., Wu, K., Guo, Y., and Rector, B. (2010). Proteomic analysis of novel Cry1Ac binding proteins in Helicoverpa armigera (Hübner). Arch. Insect Biochem. 73, 61–73. doi: 10.1002/arch.20340
Chen, R., Ren, X., Han, Z., Mu, L., Guo, Q., Ma, Y., et al. (2014). A cadherin-like protein from the beet armyworm Spodoptera exigua (Lepidoptera: noctuidae) is a putative Cry1Ac receptor. Arch. Insect Biochem. 1, 58–71. doi: 10.1002/arch.21163
Chen, W., Liu, C., Xiao, Y., Zhang, D., Zhang, Y., Li, X., et al. (2015). A toxin-binding alkaline phosphatase fragment synergizes Bt toxin Cry1Ac against susceptible and resistant Helicoverpa armigera. PLoS ONE 10:e0126288. doi: 10.1371/journal.pone.0126288
Du, M., Liu, X., Ma, N., Liu, X., Wei, J., Yin, X., et al. (2017). Calcineurin-mediated dephosphorylation of acetyl-coA carboxylase is required for PBAN-induced sex pheromone biosynthesis in Helicoverpa armigera. Mol. Cell. Proteomics 16:2138. doi: 10.1074/mcp.RA117.000065
Edgerton, M. D., Fridgen, J., Anderson, J. R. Jr., Ahlgrim, J., Criswell, M., Dhungana, P., et al. (2012). Transgenic insect resistance traits increase corn yield and yield stability. Nat. Biotechnol. 30, 493–496. doi: 10.1038/nbt.2259
Farias, J. R., Andow, D. A., Horikoshi, R. J., Sorgatto, R. J., Fresia, P., Santos, A. C. D., et al. (2014). Field-evolved resistance of Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 64, 150–158. doi: 10.1016/j.cropro.2014.06.019
Ferré, J., and Van Rie, J. (2002). Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47, 501–533. doi: 10.1146/annurev.ento.47.091201.145234
Flores-Escobar, B., Rodríguez-Magadan, H., Bravo, A., Soberón, M., and Gómez, I. (2013). Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl. Environ. Microbiol. 79:4543. doi: 10.1128/AEM.01062-13
Forcada, C., Alcacer, E., Garcera, M. D., Tato, A., and Martinez, R. (1999). Resistance to Bacillus thuringiensis Cry1Ac toxin in three strains of Heliothis virescens: proteolytic and SEM study of the larval midgut. Arch. Insect Biochem. 42, 51–63. doi: 10.1002/(SICI)1520-6327(199909)42:1<51::AID-ARCH6>3.0.CO;2-6
Gahan, L. J., Pauchet, Y., Vogel, H., and Heckel, D. G. (2010). An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248. doi: 10.1371/journal.pgen.1001248
Gassmann, A. J., Petzold-Maxwell, J. L., Clifton, E. H., Dunbar, M. W., Hoffmann, A. M., Ingber, D. A., et al. (2014). Field evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 111, 5141–5146. doi: 10.1073/pnas.1317179111
Gill, S. S., Cowles, E. A., and Pietrantonio, P. V. (1992). The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 37, 615–636. doi: 10.1146/annurev.en.37.010192.003151
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, S. H., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, Z., Cheng Zhu, Y., Huang, F., Luttrell, R., and Leonard, R. (2012). Microarray analysis of global gene regulation in the Cry1Ab-resistant and Cry1Ab-susceptible strains of Diatraea Fsaccharalis. Pest Manag. Sci. 68, 718–730. doi: 10.1002/ps.2318
Guo, Z., Kang, S., Chen, D., Wu, Q., Wang, S., Xie, W., et al. (2015b). MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 11:e1005124. doi: 10.1371/journal.pgen.1005124
Guo, Z., Kang, S., Zhu, X., Xia, J., Wu, Q., Wang, S., et al. (2015a). Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Molec. 59, 30–40. doi: 10.1016/j.ibmb.2015.01.009
Hutchison, W. D., Burkness, E. C., Mitchell, P. D., Moon, R. D., Leslie, T. W., Fleischer, S. J., et al. (2010). Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 330, 222–225. doi: 10.1126/science.1190242
James, C. (2016). Global Status of Commercialized Biotech/GM Crops: International Service for the Acquisition of Agri-Biotech Applications Brief No 52. Uthaca, NY: ISAAA.
Jurat-Fuentes, J. L., Karumbaiah, L., Jakka, S. R. K., Ning, C., Liu, C., Wu, K., et al. (2011). Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS ONE 6:e17606. doi: 10.1371/journal.pone.0017606
Kazuyoshi Yonezawaa, B. (2004). mTOR signaling pathway. Hepatol. Res. 30, 9–13. doi: 10.1016/j.hepres.2004.08.011
Klümper, W., and Qaim, M. (2014). A meta-analysis of the impacts of genetically modified crops. PLoS ONE 9:e111629. doi: 10.1371/journal.pone.0111629
Knowles, B. H., and Ellar, D. J. (1987). Colloid-osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis, δ-endotoxins with different insect specificity. BBA Gen. Subjects 924, 509–518.
Lei, Y., Zhu, X., Xie, W., Wu, Q., Wang, S., Guo, Z., et al. (2014). Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 533, 180–187. doi: 10.1016/j.gene.2013.09.091
Li, H., Oppert, B. R., Higgins, R. A., Buschman, L., and Zhu, K. (2005). Susceptibility of Dipel-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae) to individual Bacillus thuringiensis protoxins. J. Econ. Entomol. 98, 1333–1340. doi: 10.1603/0022-0493-98.4.1333
Li, H., Oppert, B. R., Higgins, R. A., Huang, F., Zhu, K. Y., and Bushman, L. L. (2004). Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 34, 753–762. doi: 10.1016/j.ibmb.2004.03.010
Liang, G., Wu, K., Yu, H., Li, K., Feng, X., and Guo, Y. (2008). Changes of inheritance mode and fitness in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) along with its resistance evolution to Cry1Ac toxin. J. Invertebr. Pathol. 97, 142–149. doi: 10.1016/j.jip.2007.09.007
Liu, C., Xiao, Y., Li, X., Oppert, B., Tabashnik, B. E., and Wu, K. (2014). Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci. Rep. 4:7219. doi: 10.1038/srep07219
Liu, S., Wang, M., and Li, X. (2015). Overexpression of tyrosine hydroxylase and dopa decarboxylase associated with pupal melanization in Spodoptera exigua. Sci. Rep. 5:11273. doi: 10.1038/srep11273
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Y., Wu, K., Jiang, Y., Guo, Y., and Desneux, N. (2012). Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 487, 362–365. doi: 10.1038/nature11153
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nanoth Vellichirammal, N., Wang, H., Eyun, S., Moriyama, E., Coates, B., Miller, N., et al. (2015). Transcriptional analysis of susceptible and resistant European corn borer strains and their response to Cry1F protoxin. BMC Genomics 16:558. doi: 10.1186/s12864-015-1751-6
Ning, C., Wu, K., Liu, C., Gao, Y., Jurat-Fuentes, J. L., and Gao, X. (2010). Characterization of a Cry1Ac toxin-binding alkaline phosphatase in the midgut from Helicoverpa armigera (HÜbner) larvae. J. Insect Physiol. 56, 666–672. doi: 10.1016/j.jinsphys.2010.02.003
Onken, H., Moffett, S. B., and Moffett, D. F. (2008). Alkalinization in the isolated and perfused anterior midgut of the larval mosquito, Aedes aegypti. J. Insect Sci. 8, 1–20. doi: 10.1673/031.008.4601
Pardo-López, L., Soberón, M., and Bravo, A. (2013). Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37, 3–22. doi: 10.1111/j.1574-6976.2012.00341.x
Rajagopal, R., Arora, N., Sivakumar, S., Rao, N. G. V., Nimbalkar, S. A., and Bhatnagar, R. K. (2009). Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem. J. 419, 309–316. doi: 10.1042/BJ20081152
Sayyed, A. H., Ahmad, M., and Crickmore, N. (2008). Fitness costs limit the development of resistance to indoxacarb and deltamethrin in Heliothis virescens (Lepidoptera: Noctuidae). J. Econ. Entomol. 101, 1927–1933. doi: 10.1603/0022-0493-101.6.1927
Singh, U., Quintanilla, R. H., Grecian, S., Gee, K. R., Rao, M. S., and Lakshmipathy, U. (2012). Novel live alkaline phosphatase substrate for identification of pluripotent stem cells. Stem Cell Rev. 8, 1021–1029. doi: 10.1007/s12015-012-9359-6
Tabashnik, B. E., Brévault, T., and Carrière, Y. (2013). Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31:510–521. doi: 10.1038/nbt.2597
Tabashnik, B. E., and Carrière, Y. (2017). Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 35, 926–935. doi: 10.1038/nbt.3974
Tabashnik, B. E., Dennehy, T. J., and Carrière, Y. (2005). Delayed resistance to transgenic cotton in pink bollworm. Proc. Natl. Acad. Sci. USA. 102, 15389–15393. doi: 10.1073/pnas.0507857102
Tanaka, S., Endo, H., Adegawa, S., Iizuka, A., Imamura, K., Kikuta, S., et al. (2017). Bombyx mori abc transporter c2 structures responsible for the receptor function of Bacillus thuringiensis Cry1Aa toxin. Insect Biochem. Mol. 91:44–54. doi: 10.1016/j.ibmb.2017.11.002
Tiewsiri, K., and Wang, P. (2011). Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. U.S.A. 34, 14037–14042. doi: 10.1073/pnas.1102555108
Valaitis, A. P. (2011). Localization of Bacillus thuringiensis Cry1A toxin-binding molecules in gypsy moth larval gut sections using fluorescence microscopy. J Invertebr. Pathol. 108, 69–75. doi: 10.1016/j.jip.2011.07.001
Van den Berg, J., Hilbeck, A., and Bøhn, T. (2013). Pest resistance to Cry1Ab Bt maize: field resistance, contributing factors and lessons from South Africa. Crop Prot. 54, 154–160. doi: 10.1016/j.cropro.2013.08.010
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by genometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Wang, G., Liang, G., Wu, K., and Guo, Y. (2005). Gene cloning and sequencing of aminopeptidase N3, a putative receptor for Bacillus thuringiensis insecticidal Cry1Ac toxin in Helicoverpa armigera (Lepidoptera: Noctuidae). Eur. J. Entomol. 102, 13–19. doi: 10.14411/eje.2005.002
Wei, J., Guo, Y., Liang, G., Wu, K., Zhang, J., and Tabashnik, B. E. (2015). Cross-resistance and interactions between Bt toxins Cry1Ac and Cry2Ab against the cotton bollworm. Sci. Rep. 5:7714. doi: 10.1038/srep07714
Wei, J., Liang, G., Wang, B., Zhong, F., Chen, L., Khaing, M. M., et al. (2016a). Activation of Bt protoxin Cry1Ac in resistant and susceptible cotton bollworm. PLoS ONE 11:e0156560. doi: 10.1371/journal.pone.0156560
Wei, J., Zhang, M., Liang, G., Wu, K., Guo, Y., Ni, X., et al. (2016b). APN1 is a functional receptor of Cry1Ac but not Cry2Ab in Helicoverpa zea. Sci. Rep. 6:19179. doi: 10.1038/srep19179
Wu, K., and Guo, Y. (2004). Changes in susceptibility to conventional insecticides of a Cry1Ac-selected population of Helicoverpa armigera (Hubner) (Lepidoptera:Noctuidae). Pest Manage. Sci. 60, 680–684. doi: 10.1002/ps.848
Wu, K. M., Lu, Y. H., Feng, H. Q., Jiang, Y. Y., and Zhao, J. Z. (2008). Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321, 1676–1678. doi: 10.1126/science.1160550
Wu, Y. (2014). Detection and mechanisms of resistance evolved in insects to Cry toxins from Bacillus thuringiensis. Adv. Insect Physiol. 47, 297–342. doi: 10.1016/B978-0-12-800197-4.00006-3
Xiao, Y., Zhang, T., Liu, C., Heckel, D. G., Li, X., Tabashnik, B. E., et al. (2014). Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci. Rep. 4:6184. doi: 10.1038/srep06184
Xu, L., Ferry, N., Wang, Z., Zhang, J., Edwards, M. G., Gatehouse, A. M., et al. (2013). A proteomic approach to study the mechanism of tolerance to Bt toxins in Ostrinia furnacalis larvae selected for resistance to Cry1Ab. Transgenic Res. 22, 1155–1166. doi: 10.1007/s11248-013-9718-3
Xu, X., Yu, L., and Wu, Y. (2005). Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 71, 948–954. doi: 10.1128/AEM.71.2.948-954.2005
Yang, Y., Zhu, Y. C., Ottea, J., Husseneder, C., Leonard, B. R., Abel, C., et al. (2011). Down regulation of a gene for cadherin, but not alkaline phosphatase, associated with Cry1Ab resistance in the sugarcane borer Diatraea saccharalis. PLoS ONE 6:e25783. doi: 10.1371/journal.pone.0025783
Yao, J., Buschman, L. L., Oppert, B., Khajuria, C., and Zhu, K. (2012). Characterization of cDNAs encoding serine proteases and their transcriptional responses to Cry1Ab protoxin in the gut of Ostrinia nubilalis larvae. PLoS ONE 7:e44090. doi: 10.1371/journal.pone.0044090
Yuan, J. S., Reed, A., Chen, F., and Stewart, C. N. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85
Zhang, H., Wu, S., Yang, Y., Tabashnik, B. E., and Wu, Y. (2012). Non-recessive Bt toxin resistance conferred by an intracellular cadherin mutation in field-selected populations of cotton bollworm. PLoS ONE 7:e53418. doi: 10.1371/journal.pone.0053418
Zhang, S., Cheng, H., Gao, Y., Wang, G., Liang, G., and Wu, K. (2009). Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 39, 421–429. doi: 10.1016/j.ibmb.2009.04.003
Zhang, T. (2013). Resistance Mechanism of Helicoverpa armigera to Bt and the Changes of Cotton Transcriptomic After Helicoverpa armigera Feeding. Dissertation's thesis. Guangxi University, Guangxi, China.
Zhang, T., Coates, B. S., Wang, Y., Wang, Y., Bai, S., Wang, Z., et al. (2017). Down-regulation of aminopeptidase N and ABC transporter subfamily G transcripts in Cry1Ab and Cry1Ac resistant Asian corn borer, Ostrinia furnacalis (Lepidoptera: Crambidae). Int. J. Biol. Sci. 13, 835–851. doi: 10.7150/ijbs.18868
Zhu, Y. C., Guo, Z., Chen, M., Zhu, K., Liu, X., and Scheffler, B. (2011). Major putative pesticide receptors, detoxification enzymes, and transcriptional profile of the midgut of the tobacco budworm, Heliothis virescens (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 106, 296–307. doi: 10.1016/j.jip.2010.10.007
Keywords: Helicoverpa armigera, DGE, Cry1Ac, trypsin, receptors, mechanisms of resistance
Citation: Wei J, Yang S, Chen L, Liu X, Du M, An S and Liang G (2018) Transcriptomic Responses to Different Cry1Ac Selection Stresses in Helicoverpa armigera. Front. Physiol. 9:1653. doi: 10.3389/fphys.2018.01653
Received: 11 May 2018; Accepted: 02 November 2018;
Published: 22 November 2018.
Edited by:
Su Wang, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Gustavo Bueno Rivas, University of Florida, United StatesDandan Wei, Southwest University, China
Copyright © 2018 Wei, Yang, Chen, Liu, Du, An and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiheng An, YW5zaGloZW5nQGFsaXl1bi5jb20=
Gemei Liang, Z21saWFuZ0BpcHBjYWFzLmNu
 Jizhen Wei
Jizhen Wei Shuo Yang1
Shuo Yang1 Lin Chen
Lin Chen Xiaoguang Liu
Xiaoguang Liu Shiheng An
Shiheng An