
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 14 November 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01614
This article is part of the Research Topic Physiological Adaptations of Insects Exposed to Different Stress Conditions, Volume I View all 38 articles
A correction has been applied to this article in:
Corrigendum: Selection and Validation of Reference Genes for RT-qPCR Analysis of the Ladybird Beetle Henosepilachna vigintioctopunctata
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a momentous technique for quantifying expression levels of the targeted genes across various biological processes. Selection and validation of appropriate reference genes for RT-qPCR analysis are a pivotal precondition for reliable expression measurement. Henosepilachna vigintioctopunctata is one of the most serious insect pests that attack Solanaceae plants in Asian countries. Recently, the transcriptomes of H. vigintioctopunctata were sequenced, promoting gene functional studies of this insect pest. Unfortunately, the reference genes for H. vigintioctopunctata have not been selected and validated. Here, a total of 7 commonly used reference genes, namely, Actin, GAPDH, RPL13, RPL6, RPL32, RPS18, and ATPB, were selected and assessed for suitability under four experimental conditions, namely, developmental stage, tissue, temperature, and host plant, using RefFinder, which integrates four different analytical tools (Normfinder, geNorm, the ΔCt method, and BestKeeper). The results displayed that RPL13 and RPS18 were the best suitable reference genes for each experimental condition. The relative transcript levels of 2 target genes, lov and TBX1, varied greatly according to normalization with the two most- and least-suited reference genes. Our results will be helpful for improving the accuracy of the RT-qPCR analysis for future functional investigations of target gene expression in H. vigintioctopunctata.
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a frequently used technique for gene expression studies on account of its high specificity, high sensitivity, high throughput, and low cost (Hellemans and Vandesompele, 2014). However, many factors relevant to biological and technical variations, for instances, RNA isolation, integrity, purity; reverse transcription; PCR efficiency, can affect the precision of RT-qPCR analysis (Bustin et al., 2005, 2009). Generally, RT-qPCR involves standardization to the expression of a battery of appropriately stable reference genes concurrent. In spite of reference gene transcript levels should be stably expressed in a serious of different biological or experimental conditions, previous studies have shown that many frequently used reference genes differ observably in different treatments (Li et al., 2013; Yang et al., 2014, 2015a,b,c, 2016, 2018; Pan et al., 2015a,b). Therefore, a systematic and customized study for each tested species is recommended for identifying appropriate reference genes.
Henosepilachna vigintioctopunctata (Fabricius) (Coleoptera: Coccinellidae) is one of the most serious insect pests in Asian countries (Ghosh and Senapati, 2001; Shinogi et al., 2005; Venkatesha, 2006; Zhou et al., 2015). H. vigintioctopunctata colonizes many different species of plants, for example, solanaceous plants such as eggplant, tomato, potato, and pepper; cucurbitaceous plants such as cucumber, white gourd, and loofah. In addition, it attacks many weeds, such as the black nightshade, winter cherry, thorn apple, and tobacco (Pang and Mao, 1979). The destructive potential of H. vigintioctopunctata is high at both the adult and larval stages, leading to up to 60% loss of fruit production (Sharma et al., 2012; Kawazu, 2014).
In China, H. vigintioctopunctata is widely distributed from Hainan Province in the south to Heilongjiang Province in the north and from Gansu Province in the west to Shanghai City in the east (Pang and Mao, 1979). Recently, because of climate warming, development of trade, and expansion of the cultivated area for protected vegetables, the food for H. vigintioctopunctata is constantly increasing throughout the year, and the occurrence and damage of H. vigintioctopunctata have become significant (Li et al., 2006; Zhou et al., 2015; Wang et al., 2017). Recently, we sequenced the transcriptomes of H. vigintioctopunctata at different developmental stages (unpublished data) and obtained a huge amount of genes involved in the development, energy metabolism, and reproduction. Recently, RNA interference (RNAi) has been used widely in the study of functional genomics, resulting in the development of new modes of action insecticides for insect pest management (Baum et al., 2007; Bellés, 2010; Burand and Hunter, 2013; Scott et al., 2013; Zhang et al., 2017). To identify novel target genes for controlling H. vigintioctopunctata, accurate gene expression of this pest under different biotic and abiotic conditions is necessary.
In this study, to discern the solidly expressed reference genes of H. vigintioctopunctata for RT-qPCR investigation under four experimental conditions (developmental stage, tissue, temperature, and host plant), 7 most commonly used reference genes, namely, actin (Actin), ribosomal protein L13 (RPL13), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), ribosomal protein L6 (RPL6), ribosomal protein L32 (RPL32), ribosomal protein S18 (RPS18), and vacuolar-type H+-ATPase subunit B (ATPB), were selected from the transcriptomes of H. vigintioctopunctata. The expression stability of each candidate reference gene was assessed under 2 biotic (developmental stage and tissue) and 2 abiotic (temperature and host plant) conditions by using RefFinder, which integrates four different analytical tools (Normfinder, geNorm, the ΔCt method, and BestKeeper). Finally, jim lovell (lov) and T-box transcription factor (TBX1) were used as the target genes to verify our findings. The results will be useful in improving the accuracy of RT-qPCR analysis for future functional investigations of target gene expression in H. vigintioctopunctata.
In April 2018, the adults of H. vigintioctopunctata were collected from Solanum nigrum (L.) in Guangzhou City, Guangdong Province, China, and reared in the incubator at 25 ± 0.5°C temperature, 14L:10D photoperiod, and 80% relative humidity in petri dishes by using S. nigrum and S. melongena leaves.
All stages of H. vigintioctopunctata were sampled: eggs, four larval instars, pupae, and female and male adults (collected on the first day of each stage). The number of sampled individuals for each replicate across the different developmental stage was as follows: 20 eggs for the egg stage; 10 individuals for the first instar; 5 individuals for the second instar; 3 individuals for the third instar; 1 individual for the fourth instar; 1 pupa for the pupal stage; and 1 male or female individual for the adult male or female stage. Different body tissues, namely, the Malpighian tubule, fat body, midgut, and cuticle, were dissected from the fourth instar larvae, about 40 individuals were dissected for each replicate. The tissue samples were placed in RNAlater® (Thermo Fisher Scientific Inc., United Stats) and stored at 4°C before total RNA isolation.
For the temperature treatment, 5 s instars were exposed to 8, 25, and 35°C for 3 h. For the host plant treatment, 2 third instars larvae were collected as one sample that reared on S. nigrum and eggplant.
Each experiment was replicated 3 times. All the samples, except the ones used for the host plant treatment, were collected from the colony reared on S. nigrum. All the samples, except the tissue ones, were placed in 1.5 ml RNA free centrifuge tubes, rapidly frozen in liquid nitrogen, and stored at -80°C before the total RNA extraction.
The total RNAs of egg, Malpighian tubule, and fat body were isolated using TRIzol reagent (Invitrogen, United States), in line with our previously described methods (Pan et al., 2015a). Total RNAs from the other samples were isolated using a HiPure Total RNA Micro Kit (Magen, China), according to the manufacturer’s instructions. A NanoDrop OneC spectrophotometer (Thermo Fisher Scientific, Waltham, MA United States) was used to ascertain the RNA concentration. The total RNA was dissolved in 20–100 μl of ddH2O, and the concentrations were as follows: 494.3 ± 16.7 ng/μl [mean ± standard error of the mean (SEM)] for the eggs, 427.6 ± 28.6 ng/μl for the first instars, 683.9 ± 79.8 ng/μl for the second instars, 567.1 ± 38.7 ng/μl for the third instars, 619.8 ± 61.9 ng/μl for the fourth instars, 818.6 ± 59.1 ng/μl for the pupae, 760.2 ± 36.6 ng/μl for the male adults, 763.4 ± 78.9 ng/μl for the female adults, 677.5 ± 68.0 ng/μl for the cuticles, 565.3 ± 38.1 ng/μl for the fat body, 870.3 ± 22.6 ng/μl for the midguts, 483.8 ± 74.9 ng/μl for the Malpighian tubules, 468.4 ± 6.8 ng/μl for the second-instar under 15°C, 683.9 ± 79.8 ng/μl for the second-instar under 25°C, 356.1 ± 7.9 ng/μl for the second-instar under 35°C, and 422.1 ± 68.4 ng/μl for the third-instar under eggplant. The OD260/280 value of all samples was 1.9–2.1. The PrimeScript RT kit (containing gDNA Eraser, Perfect Real Time, TaKaRa, China) was used for preparing the first-strand cDNA for gene expression investigation. The cDNA was diluted 10-fold for the pursuant RT-qPCR experiments.
In our study, 7 candidate reference genes that are most frequently used in RT-qPCR investigations were assessed (Table 1). The primers were designed based on the PrimerQuest Tool1, according to the sequences obtained from our recently sequenced transcriptomes for H. vigintioctopunctata (unpublished data).
The PCR reaction system and parameters were used according to our previous study (Yang et al., 2014). Amplicons of the expected lengths were purified using the TIANgel Midi Purification Kit (Tiangen, China), and subcloned into the pClone007 Blunt vector before transformation into Escherichia coli DH5α competent cells (Tsingke, China) for sequencing by Tsingke company. The reference genes were confirmed using sequence analysis.
The RT-qPCR reactions and program were conducted, the melting curve and standard curve for each reference gene were generated according to our previous study (Yang et al., 2014, 2015a,b,c, 2016, 2018). The homologous RT-qPCR efficiencies (E) were calculated according to the equation: E = (10[-1/slope] -1) × 100.
The reference gene expression stability was assessed using RefFinder2, which integrates four different analytical tools, NormFinder (Andersen et al., 2004), geNorm (Vandesompele et al., 2002), the ΔCt method (Silver et al., 2006), and BestKeeper (Pfaffl et al., 2004). The optimal number of reference genes for target gene expression normalization was decided by pairwise variation (Vn/Vn+1). A Vn/Vn+1 cutoff value of 0.15 indicates the additional 1 more reference gene is not necessary, i.e., the starting n reference genes are enough for the target gene normalization; the V-values were calculated using geNorm (Vandesompele et al., 2002).
Lov is a nuclear protein has roles in various larval and adult behaviors (Bjorum et al., 2013). TBX1 plays a vital role in the upgrowth of Drosophila heart (Griffin et al., 2000). The stability of these reference genes was investigated using lov and TBX1 as the target genes. The primer sequences of these two target genes were as follows: lov, forward (5′-CTCCCGCCCAACACTTTAT-3′) and reverse (5′-TCGCTTTGCGGTAGTAGATG-3′); TBX1, forward (5′-GAAACACCTCTGGGACGAAT-3′) and reverse (5′-TCGGAGTGCAAGTCTAAACC-3′). Lov and TBX1 expression levels in different tissues were computed on the basis of normalization to the 2 most- and 2 least-stable candidates. The relative gene expression of lov and TBX1was computed using the 2-ΔΔCt method (Livak and Schmittgen, 2001). One-way analysis of variance was used to detect significances in lov and TBX1 expression levels among different tissues (SPSS 17.0).
All of these candidate reference genes were expressed in H. vigintioctopunctata and intuitional with a single amplicon of the expected size for each gene (Supplementary Figure S1). Gene-specific amplification of all reference genes was verified by a single peak in the melting curve analysis (Figure 1). The PCR efficiency ranged between 92 and 100% (Table 1). The standard curve for each reference gene is also provided (Supplementary Figure S2).
The quantification cycle (Cq) values for all the reference genes under the 4 experimental conditions ranged from 20 to 29. RPS18 and RPL13 were the most abundant reference genes, whereas RPL32 and RPL6 were the least expressed ones (Figure 2).
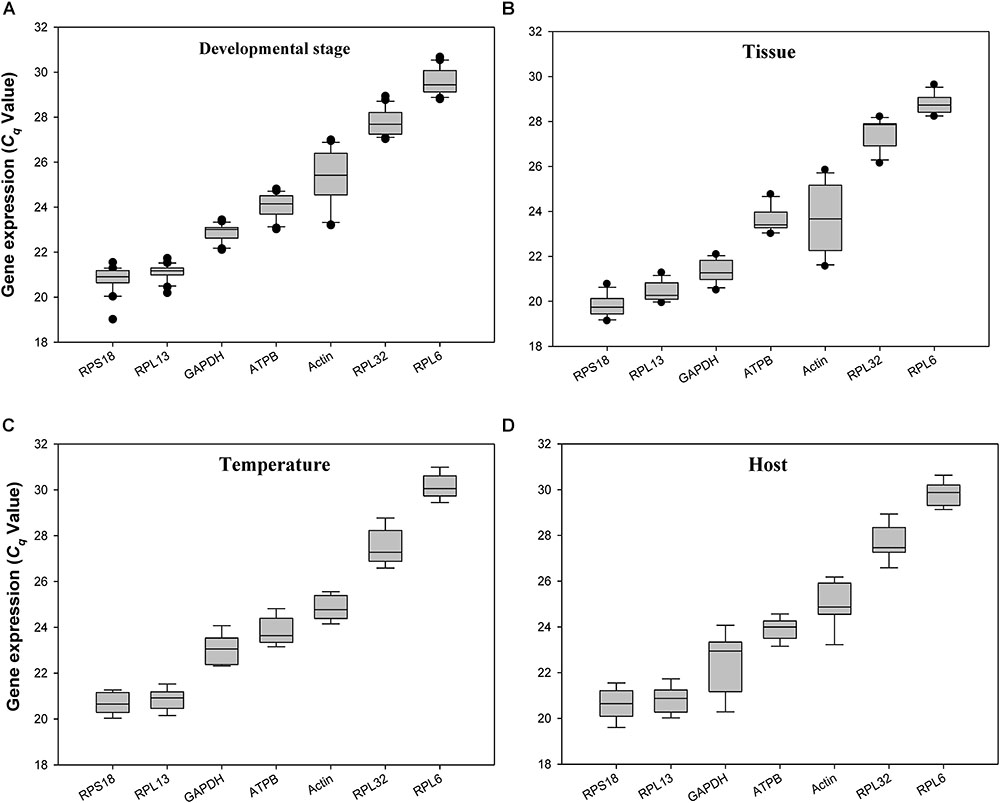
FIGURE 2. Expression profiles of the 7 candidate reference genes in all 4 experiments for H. vigintioctomaculata. The expression levels of the reference genes are shown in terms of the Cq-value for each experimental condition. (A) Development stage, (B) Tissue, (C) Temperature, (D) Host plant.
According to RefFinder, across different development stages, the comprehensive reference gene rankings from the best to the least stable were as follows: RPL13, RPS18, ATPB, GAPDH, RPL6, RPL32, and Actin (Figure 3A). For the tissue treatment, the comprehensive reference gene rankings were as follows: RPL13, RPS18, RPL6, RPL32, ATPB, GAPDH, and Actin (Figure 3B). Under different temperature conditions, the integrated reference gene rankings were as follows: RPL13, RPS18, RPL6, ATPB, Actin, RPL32, and GAPDH (Figure 3C). For the host plant treatment, the comprehensive reference gene rankings were as follows: RPL13, RPS18, ATPB, RPL32, RPL6, GAPDH, and Actin (Figure 3D). The expression stability value for each gene was also computed using Normfinder, geNorm, the ΔCt method, and BestKeeper under each experimental condition (Table 2).
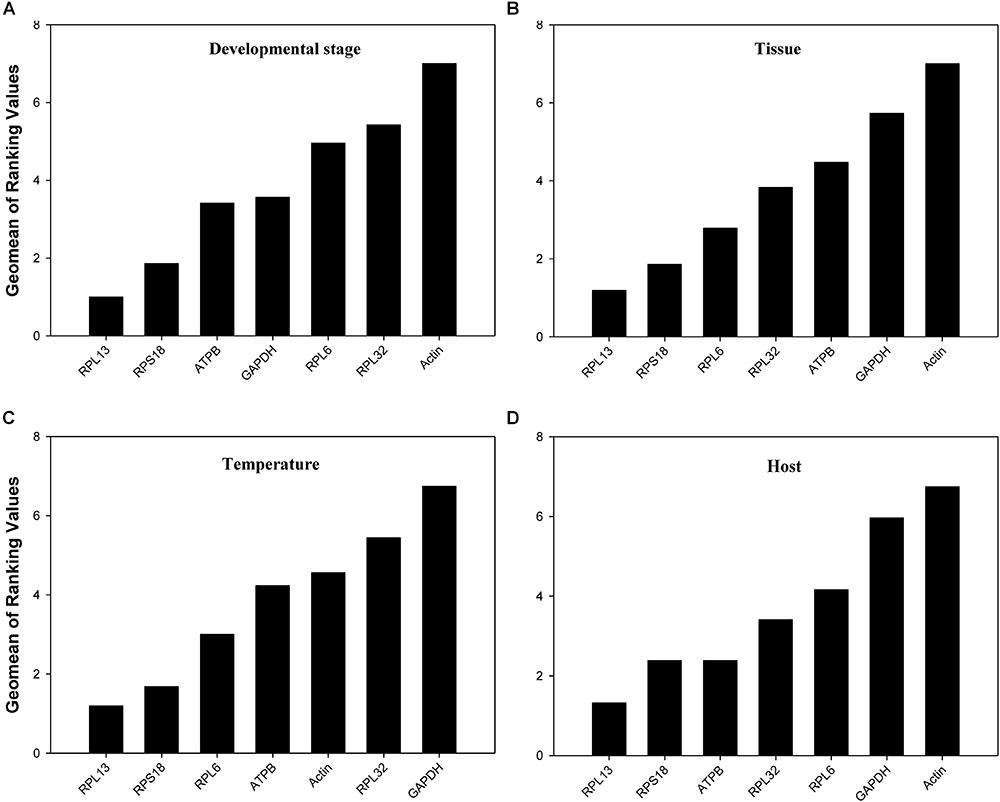
FIGURE 3. Stability of the 7 candidate reference gene expressions in H. vigintioctomaculata under different treatment conditions analyzed using RefFinder. (A) Development stage, (B) Tissue, (C) Temperature, (D) Host plant.
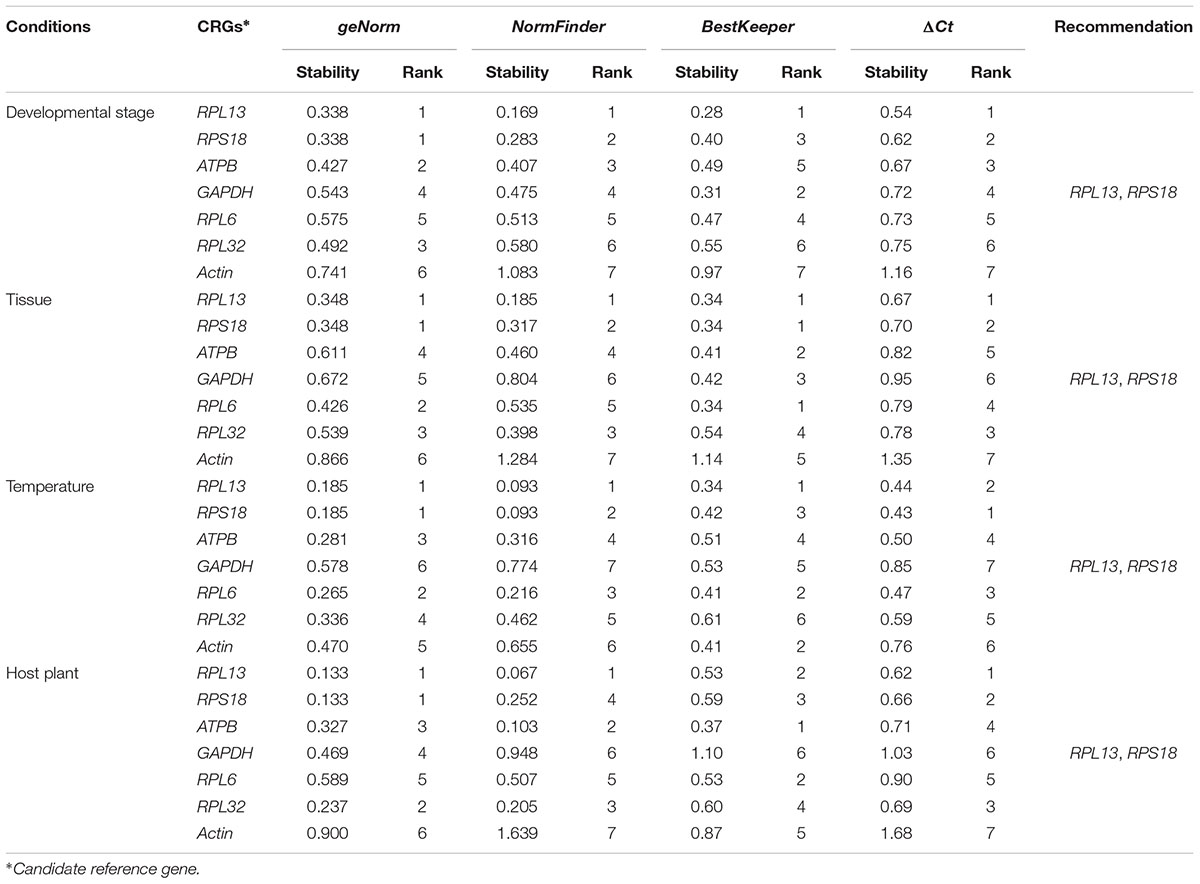
TABLE 2. Stability of 7 candidate reference gene expression in H. vigintioctomaculata under different experimental conditions calculated by the 4 different analytical tools geNorm, Normfinder, BestKeeper, and the ΔCt method, respectively.
For each of four different experimental conditions, the initial V-value < 0.15 were all emerged at V2/3, respectively, suggesting 2 reference genes were enough for the target gene normalization. By coincidence, RPL13 and RPS18 were the best stable reference genes for each experimental condition, respectively (Figure 4).
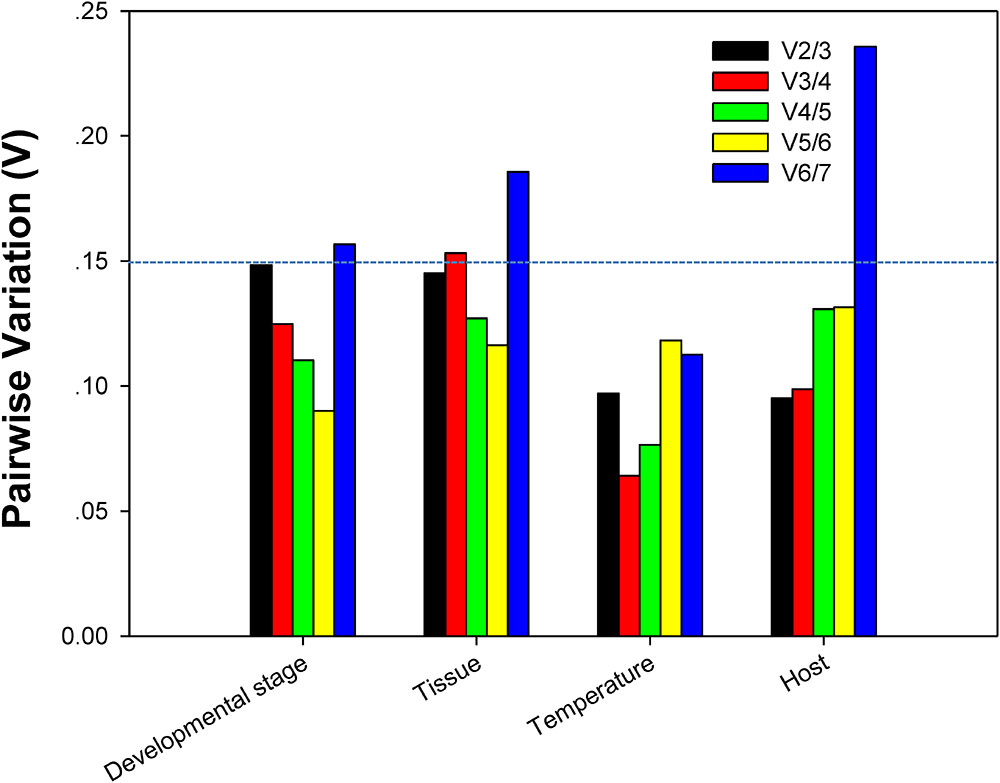
FIGURE 4. Optimum number of reference genes required for accurate normalization of gene expression. Pairwise variation (V) values in the 4 groups using geNorm.
The relative expression levels of lov and TBX1 were used to validate the reference genes among the different tissues. The Lov expression patterns were similar, however, lov expression was about 15- and 99-fold higher in the fat body than in the Malpighian tubule when normalized to the 2 most- and least-stable reference genes, respectively (Figure 5). In contrast, TBX1 expression patterns were inconsistent among the different tissues when normalized to the 2 most- and least-stable reference genes. Different degrees of gene expression were obtained in each tissue under the 2 normalization conditions (Figure 6).
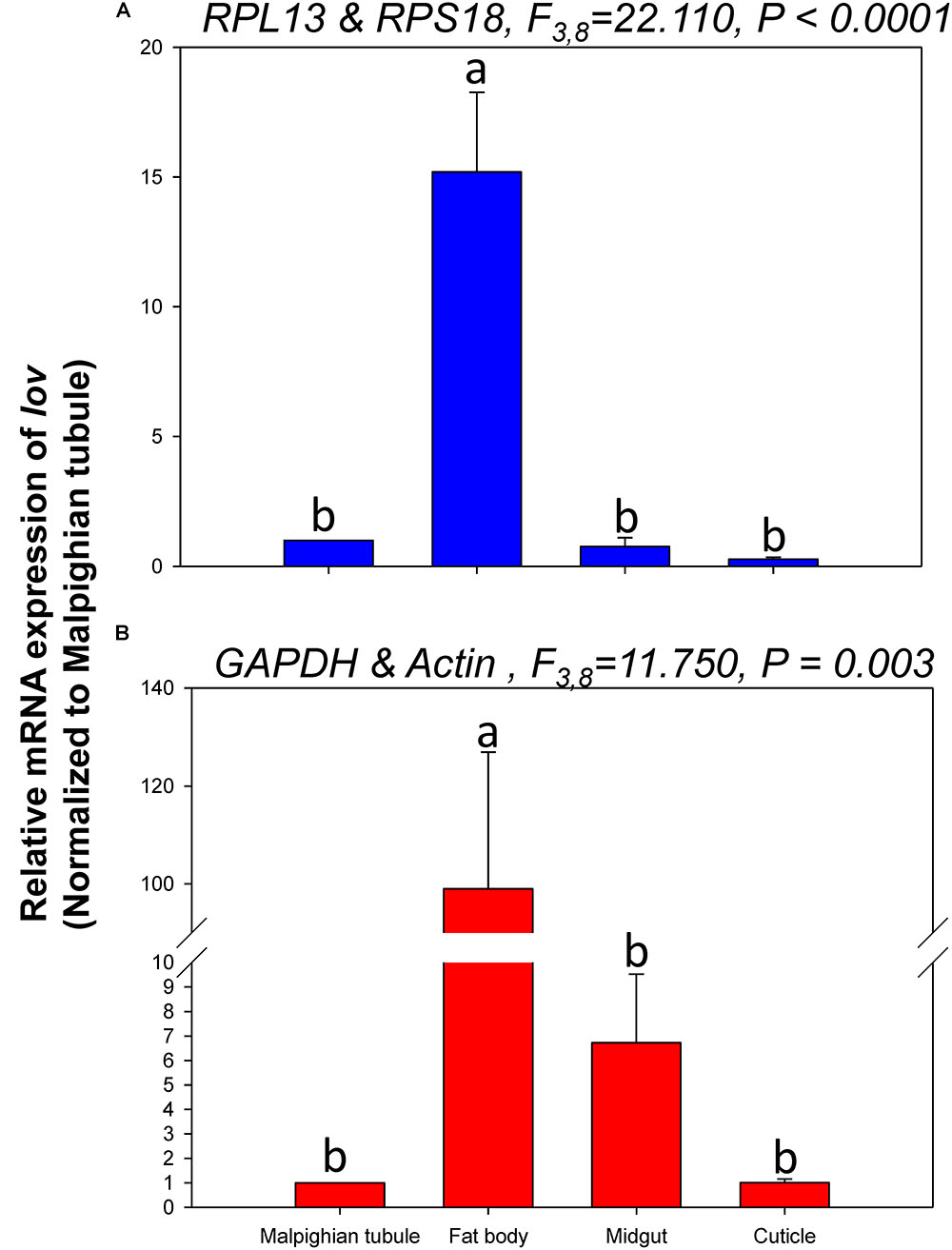
FIGURE 5. Relative gene expression of lov in different tissues of H. vigintioctomaculata. The relative abundance of lov in the Malpighian tubule, fat body, midgut, and cuticle were normalized to the best stable (A, RPL13 and RPS18) and least stable (B, GAPDH and Actin) reference genes, respectively. The values are means + SE. Different letters indicate significant differences in gene expression among different tissues of H. vigintioctomaculata (P < 0.05).
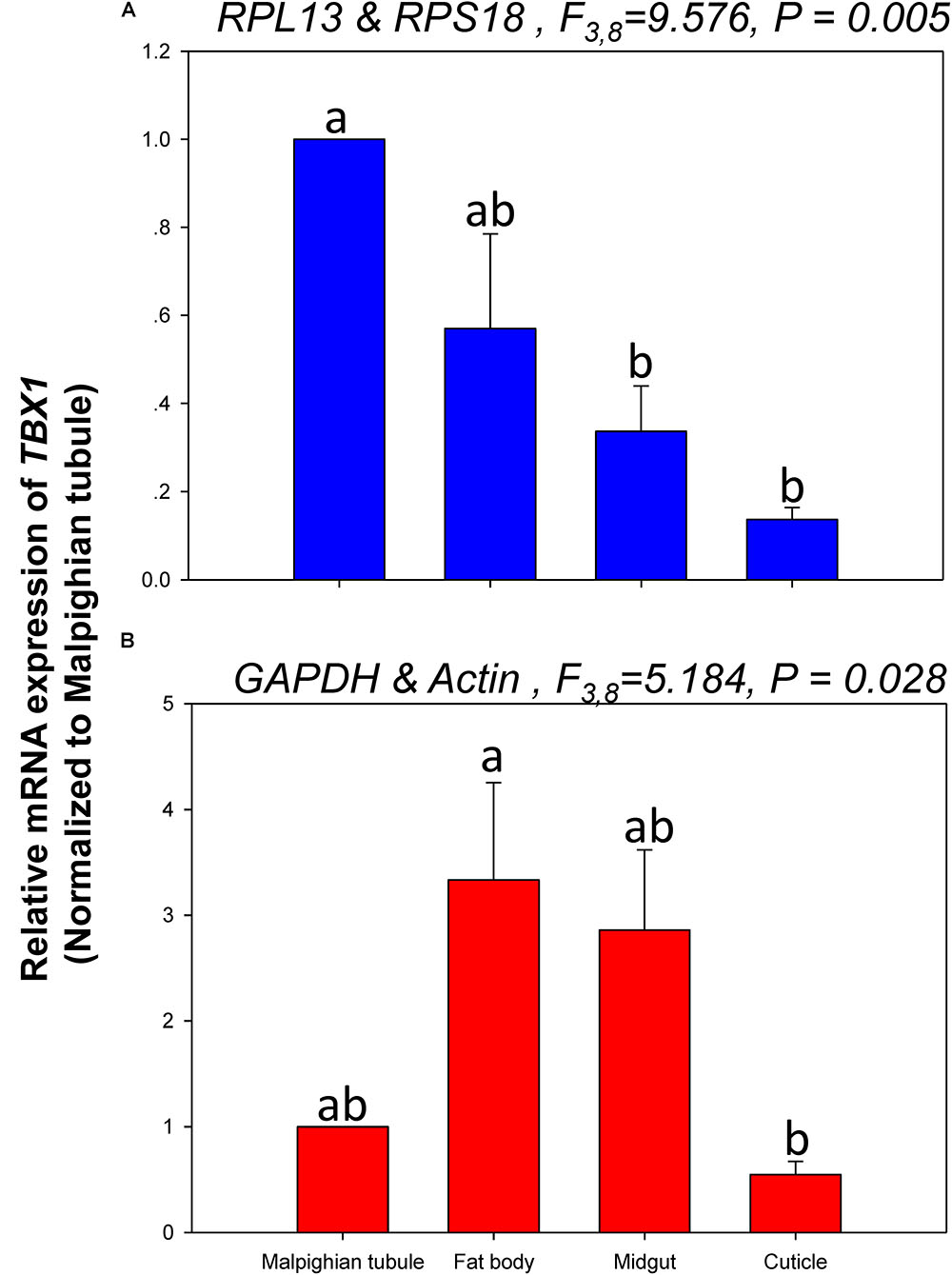
FIGURE 6. Relative gene expression of TBX1 in different tissues of H. vigintioctomaculata. The relative gene expression levels of TBX1 in the Malpighian tubule, fat body, midgut, and cuticle were normalized to the best stable (A, RPL13 and RPS18) and least stable (B, GAPDH and Actin) reference genes, respectively. The values are means + SE. Different letters indicate significant differences in gene expression among different tissues of H. vigintioctomaculata (P < 0.05).
Previous studies have demonstrated there is no “universal” reference gene applicable for various test conditions, even for the same insect species. For example, five papers have been published for the reference gene selection of the whitefly, Bemisia tabaci, a notorious and invasive insect species, in the past 5 years (Li et al., 2013; Su et al., 2013; Collins et al., 2014; Liang et al., 2014; Dai et al., 2017). It is pivotal to select and validate the reference genes expression stability under various experimental conditions precedence using them for normalizing gene expression.
We sequenced the transcriptomes for the developmental stages of H. vigintioctopunctata (unpublished data). In the future, we will use RNA interference to investigate the gene functions, and RT-qPCR will be widely applied for evaluating the gene expression changes. However, the reference genes have not been previously selected and validated in this insect pest. Therefore, in this study, seven frequently used reference genes were picked out and their stability was investigated using five software programs under four experimental conditions. The results displayed that the reference gene transcript levels vary with the experimental conditions. Thus, it is no doubt that the expression profiles of the target genes will show substantive variations relying on the reference gene and experimental treatments (Yang et al., 2016).
Our results certified that 5 computational methods yielded disparate stability rankings for the seven reference genes (Table 2 and Figure 3). Coincidentally, the two most-suited references genes were the same under each experimental condition (Figure 3). Recently, researchers have been more receptive to the use of multiple reference genes to replace a single normalizer in the RT-qPCR analysis (Vandesompele et al., 2002; Li et al., 2013; Yang et al., 2014, 2015a,b,c, 2016, 2018; Pan et al., 2015a,b), and the pairwise variations suggest that 2 reference genes were adequate for normalization under each experimental condition. Thus, RPL13 and RPS18 are suitable for use as the reference genes under each of the four experimental conditions.
In order to further validate the reference genes in H. vigintioctopunctata, the relative gene expression levels of lov and TBX1 were evaluated in different tissues. Our results showed that lov expression was sevenfold higher in the Malpighian tubule than in the fat body when normalized to the 2 least stable reference genes GAPDH and Actin than when normalized to the 2 most stable reference ones RPL13 and RPS18 (Figure 5). In addition, TBX1 expression patterns were inconsistent in the different tissues when normalized to the two best- and least-stable reference genes. The expression level was different in each tissue under the two normalization conditions (Figure 6). These results indicated that the unreasonable use of reference genes may give rise to inaccurate results for target genes. Therefore, selection and validation of the best reference genes are crucial for determining the veracity of the expression results. Thus, our study provides a more strict way to normalize RT-qPCR data in H. vigintioctopunctata, which will boost our comprehension of the target gene functions in this serious pest.
We identified stable reference genes for RT-qPCR analysis of H. vigintioctopunctata. RPL13 and RPS18 combinations are proposed as the reference genes for each experimental condition. This study on behalf of the initial step to establish the standardized RT-qPCR analyses of H. vigintioctopunctata and could contribute to the in-depth functional genomic dissection of H. vigintioctopunctata.
HP and CY conceived and designed research. JL, SC, MG, and CY conducted experiments. BQ and JW contributed reagents. HP and JL analyzed data. HP and CY wrote the manuscript.
This research was supported by the National Key R&D Program of China (2017YFD0200900), project supported by GDUPS (2017), and a start-up fund from the South China Agricultural University. The granting agencies have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01614/full#supplementary-material
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Bellés, X. (2010). Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 55, 111–128. doi: 10.1146/annurev-ento-112408-085301
Bjorum, S. M., Simonette, R. A., Alanis, R. Jr., Wang, J. E., Lewis, B. M., Trejo, M. H., et al. (2013). The Drosophila BTB domain protein Jim Lovell has roles in multiple larval and adult behaviors. PLoS One 8:e61270. doi: 10.1371/journal.pone.0061270
Burand, J. P., and Hunter, W. B. (2013). RNAi: future in insect management. J. Invertebr. Pathol. 112, S68–S74. doi: 10.1016/j.jip.2012.07.012
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR-a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755
Collins, C., Patel, M. V., Colvin, J., Bailey, D., Seal, S., and Wolfner, M. (2014). Identification and evaluation of suitable reference genes for gene expression studies in the whitefly Bemisia tabaci (Asia I) by reverse transcription quantitative real time PCR. J. Insect Sci. 14:63. doi: 10.1093/jis/14.1.63
Dai, T. M., Lü, Z. C., Liu, W. X., and Wan, F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS One 12:e0173821. doi: 10.1371/journal.pone.0173821
Ghosh, S. K., and Senapati, S. K. (2001). Biology and seasonal fluctuation of Henosepilachna vigintioctopunctata Fabr. on brinjal under Terai region of West Bengal. Indian J. Agric. Res. 35, 149–154.
Griffin, K. J. P., Stoller, J., Gibson, M., Chen, S., Yelon, D., Stainier, D. Y. R., et al. (2000). A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev. Biol. 218, 235–247. doi: 10.1006/dbio.1999.9571
Hellemans, J., and Vandesompele, J. (2014). Selection of reliable reference genes for RT-qPCR analysis. Methods Mol. Biol. 1160, 19–26. doi: 10.1007/978-1-4939-0733-5_3
Kawazu, K. (2014). Rearing the 28-spotted ladybird beetle, Henosepilachna vigintioctopunctata (Coleoptera: Coccinelidae), with a switchover from host plant leaves to artificial diet. Appl. Entomol. Zool. 49, 359–362. doi: 10.1007/s13355-014-0250-6
Li, R., Xie, W., Wang, S., Wu, Q., Yang, N., Yang, X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8:e53006. doi: 10.1371/journal.pone.0053006
Li, Y. X., Meng, Z. Q., Liu, D. M., Su, Z. F., Song, J. H., and Li, A. L. (2006). Produce harm characteristics and control technology of Henosepilachna vigintioctopunctata. China’s Rural Well-off Technol. 3:45.
Liang, P., Guo, Y., Zhou, X., and Gao, X. (2014). Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLoS One 9:e87514. doi: 10.1371/journal.pone.0087514
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Pan, H., Yang, X., Bidne, K., Hellmich, R. L., Siegfried, B. D., and Zhou, X. (2015a). Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio-indicator. PLoS One 10:e0129482. doi: 10.1371/journal.pone.0129482
Pan, H., Yang, X., Siegfried, B. D., and Zhou, X. (2015b). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10:e0125868. doi: 10.1371/journal.pone.0125868
Pang, X. F., and Mao, J. L. (1979). Economic Insects of China, 14, Coleoptera-Coccinellidae, II. 170. 16 Pls. Beijing: Science Press.
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Sharma, A., Thakur, A., Kaur, S., and Pati, P. K. (2012). Effect of Alternaria alternata on the coccinellid pest Henosepilachna vigintioctopunctata and its implications for biological pest management. J. Pest Sci. 85, 513–518. doi: 10.1007/s10340-012-0432-3
Shinogi, T., Hamanishi, Y., Otsu, Y., Wang, Y. Q., Nonomura, T., Matsuda, Y., et al. (2005). Role of induced resistance in interactions of Epilachna vigintioctopunctata with host and non-host plant species. Plant Sci. 168, 1477–1485. doi: 10.1016/j.plantsci.2005.01.022
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Su, Y. L., He, W. B., Wang, J., Li, J. M., Liu, S. S., and Wang, X. W. (2013). Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 106, 1446–1455. doi: 10.1603/EC12459
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–research0034.11. doi: 10.1186/gb-2002-3-7-research0034
Venkatesha, M. G. (2006). Seasonal occurrence of Henosepilachna vigintioctopunctata (F.) (Coleoptera: Coccinellidae) and its parasitoid on Ashwagandha in India. J. Asia Pac. Entomol. 9, 265–268. doi: 10.1016/S1226-8615(08)60301-5
Wang, Z. L., Li, C. R., Yuan, J. J., Li, S. X., Wang, X. P., and Chi, H. (2017). Demographic comparison of Henosepilachna vigintioctopunctata (F.) (Coleoptera: Coccinellidae) reared on three cultivars of Solanum melongena L. and a wild host plant Solanum nigrum L. J. Ecol. Entomol. 110, 2084–2091. doi: 10.1093/jee/tox207
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2014). Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphididae). PLoS One 9:e110454. doi: 10.1371/journal.pone.0110454
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2015a). Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS One 10:e0130593. doi: 10.1371/journal.pone.0130593
Yang, C., Pan, H., Noland, J. E., Zhang, D., Zhang, Z., Liu, Y., et al. (2015b). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5:18201. doi: 10.1038/srep18201
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2015c). Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS One 10:e0130593. doi: 10.1371/journal.pone.0130593
Yang, C., Preisser, E. L., Zhang, H., Liu, Y., Dai, L., Pan, H., et al. (2016). Corrigendum: Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 7:1672. doi: 10.3389/fpls.2016.01672
Yang, X., Pan, H., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. doi: 10.1038/s41598-018-20612-w
Zhang, J., Khan, S. A., Heckel, D. G., and Bock, R. (2017). Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 35, 871–882. doi: 10.1016/j.tibtech.2017.04.009
Keywords: Henosepilachna vigintioctopunctata, RT-qPCR analysis, reference gene, RefFinder, geNorm
Citation: Lü J, Chen S, Guo M, Ye C, Qiu B, Wu J, Yang C and Pan H (2018) Selection and Validation of Reference Genes for RT-qPCR Analysis of the Ladybird Beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9:1614. doi: 10.3389/fphys.2018.01614
Received: 06 September 2018; Accepted: 25 October 2018;
Published: 14 November 2018.
Edited by:
Su Wang, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Yingjun Cui, University of Missouri, United StatesCopyright © 2018 Lü, Chen, Guo, Ye, Qiu, Wu, Yang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunxiao Yang, eWFuZ2NodW54aWFvQHNjYXUuZWR1LmNu Huipeng Pan, cGFuaHVpcGVuZ0BzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.