- Department of Physiological Sciences, College of Veterinary Medicine, University of Florida, Gainesville, FL, United States
Emerging evidence suggests an associative link between gut dysbiosis, the autonomic nervous system (ANS) and the immune system in pathophysiology of neurogenic hypertension (HTN). However, the close interplay between these three systems presents us with difficulties in deciphering the cause-effect relationship in disease. The present study utilized beta 1 and 2 adrenergic receptor knock out (AdrB1tm1BkkAdrB2tm1Bkk/J KO) mice to isolate the effects of reduced overall sympathetic drive on gut microbiota and systemic immune system. We observed the following: (i) Diminished beta adrenergic signaling mainly reflects in shifts in the Firmicutes phyla, with a significant increase in abundance of largely beneficial Bacilli Lactobacillales in the KO mice; (ii) This was associated with increased colonic production of beneficial short chain fatty acids (SCFAs) butyrate, acetate and propionate, confirming functional microbiota shifts in the KO mice; (iii) Dampened systemic immune responses in the KO mice reflected in reduction on circulating CD4+.IL17+ T cells and increase in young neutrophils, both previously associated with shifts in the gut microbiota. Taken together, these observations demonstrate that reduced expression of beta adrenergic receptors may lead to beneficial shifts in the gut microbiota and dampened systemic immune responses. Considering the role of both in hypertension, this suggests that dietary intervention may be a viable option for manipulation of blood pressure via correcting gut dysbiosis.
Introduction
Hypertension (HTN), or high blood pressure (BP), is one of the most modifiable risk factors of cardiovascular disease (CVD), which is among the leading causes of death in the United States. HTN is regarded as the “silent killer” as there are no obvious symptoms until development of overt clinical signs. Untreated or uncontrolled HTN typically results in end-organ damage such as hemorrhagic stroke, heart disease, peripheral arterial disease, retinopathy, and renal failure (Chobanian et al., 2003). A significant proportion of all HTN cases are either resistant or refractory to currently available antihypertensive treatments, and many of them have no clear etiology, thus characterized by primary or essential HTN (Calhoun et al., 2008). One of the characteristics of essential HTN is its neurogenic component, reflecting in overactivation of the sympathetic nervous system (SNS) that is involved in initiation and maintenance of elevated BP via activation of adrenergic receptors (Esler et al., 1981; Anderson et al., 1989; Rumantir et al., 2000; Schlaich et al., 2004). In addition, evidence links rodent models of neurogenic HTN with exaggerated SNS activity to chronic low-grade inflammation and neuronflammation (Harrison, 2014; Singh et al., 2014; Zubcevic et al., 2014), and most recently gut dysbiosis (Yang et al., 2015, 2017; Adnan et al., 2017; Raizada et al., 2017; Santisteban et al., 2017).
Human gastrointestinal (GI) tract provides one of the best examples of symbiosis in nature. The gut represents a complex ecosystem of host-bacteria interaction comprised of approximately 100 trillion bacteria (Chow et al., 2010). Despite the dynamic range of bacteria present on earth, the gut microbial community is mainly dominated by four phyla: (1) Firmicutes, (2) Bacteroidetes, (3) Actinobacteria and (4) Proteobacteria (Chow et al., 2010). Existing studies have illustrated that gut microbiota has a profound impact on host biology ranging from systemic immunity and morphological integrity of intestinal epithelial cells, to endocrine and absorptive functions (Bry et al., 1996; Hooper et al., 2001; Pamer, 2007; Smith et al., 2007; Round and Mazmanian, 2009; Chow et al., 2010). In order to establish a healthy steady state, host-microbiota interactions require a delicate balance (Sansonetti, 2004; Pamer, 2007; Smith et al., 2007), with regulatory mechanisms in place that serve to regulate bacterial colonization of the gut, while simultaneously enabling immune responses against harmful antigens (Chow et al., 2010).
There is no absolute definition of a “healthy” gut microbiota as yet, however, evidence suggests that the ratio of Firmicutes (F) to Bacteroidetes (B), the F/B ratio as a biomarker for pathological conditions (Mariat et al., 2009; Sanz and Moya-Pérez, 2014; Yang et al., 2015, 2017; Adnan et al., 2017; Santisteban et al., 2017). Recent data suggests that changes in gut microbiota might also impact disorders such as metabolic syndrome, diabetes, and obesity (Tilg and Kaser, 2011; Everard and Cani, 2013), as well as neurogenic diseases and conditions including but not limited to anxiety, sleep apnea, and HTN, all marked by significantly altered sympathetic activity (Cryan and Dinan, 2012; Zubcevic et al., 2014; Mell et al., 2015; Yang et al., 2015, 2017; Adnan et al., 2017; Santisteban et al., 2017). To defend the host against pathologic threats and maintain physiological homeostasis, the SNS and immune system are reciprocally regulated (Bellinger and Lorton, 2014; Jänig, 2014). Moreover, the host-microbiota interaction also reflects in direct communication between the gut bacteria and the host immune system, and dysregulation of the immune system has been associated with intestinal diseases such as colorectal cancer and inflammatory bowel disease (Baumgart and Carding, 2007; Gao et al., 2015). Given the regulatory role of the SNS on immune system and BP (Zubcevic et al., 2014; Yang et al., 2017) and the presence of gut dysbiosis and exaggerated immune system in conditions such as HTN (Santisteban et al., 2015, 2017; Yang et al., 2015; Adnan et al., 2017), we hypothesized that blockade of the effects of sympathetic drive will result in beneficial changes in the immune system responses and shifts in the gut microbiota richness and diversity, leading to functional changes such as production of gut microbiota metabolites. To test our current hypothesis, we utilized beta 1 and beta 2 adrenergic receptor (AdrB1tm1BkkAdrB2tm1Bkk/J) knockout (b1.b2 Adr KO) mice, characterized by reduced BP and heart rate responses (Rohrer et al., 1999) resulting from reduction of the effects of the SNS on the whole body system including the gut and the immune system. Our findings support a role for SNS in modulation of gut microbiota and mainly that of the Firmicutes phylum. Moreover, we note a beneficial reduction in circulating CD4+.IL17+ T cells and increase in circulating young neutrophils in the b1.b2 Adr KO mice, both previously linked with shifts in the gut microbiota (Zhang et al., 2015; Wilck et al., 2017). Thus, the current may be useful in explaining some of the HTN and similar inflammatory phenotypes.
Methods
Animal Models
All experimental procedures were approved by the University of Florida Institutional Animal Care and Use Committee and complied with the standards stated in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mice were housed in a temperature-controlled room (22°C to 23°C) with a 12:12-h light-dark cycle, in specific-pathogen free cages, and had access to standard mice chow and water ad libitum. Twenty-week old male heterozygous beta 1 and beta 2 adrenergic receptor (AdrB1tm1BkkAdrB2tm1Bkk/J) knockout (KO) mice (#003810, Jax Labs) and their wild type (WT) C57BL/6J controls (#000664, Jax Labs) were used in all experiments. Genoytyping was performed prior to all experiments to confirm the strain differences, as per the Jacksonville Laboratories protocols. Briefly, mouse tails were snipped and lysed in 200 μl of Direct PCR Lysis Reagent (Viagen Biotech cat#101-T) supplemented with 4 μl of 200 mg/ml Proteinase K solution (Fisher cat#BP1700-100). Following 6 h of incubation at 55°C, and additional 45 min incubation at 85°C, the samples were briefly centrifuged and the DNA lysate was used for PCR. The following primers were used in regular PCR reaction, as per The Jackson Laboratory protocol for Adrb1tm1Bkk-Probe (see https://www2.jax.org/protocolsdb/f?p=116:1:0::NO:RP::) (i) to confirm the KO strain: F: TTG GGA AGA CAA TAG CAG GC, and R: CGC TGT CCA CAG TGG TTG T; (ii) to confirm the wild type strain: F: ATC TGG TCA TGG GAT TGC TG, and R: ACA CAG CAC ATC TAC CGA AG. The PCR reaction was performed using Amplitaq Gold 360 Master Mix (Invitrogen cat#4398876), and products were run on a 1% agarose gels. Using this protocol, the KO strain showed one band at 113 bps, while the wild type strain showed two bands, one at 113 bps, and one and 225 bps, as per the Jacksonville Laboratories instructions.
RNA Extraction, cDNA Preparation, and Real Time PCR (qPCR) in Colon of WT and KO Mice
Strains were confirmed by genotyping as per The Jackson Laboratory protocols prior to commencement of experiments. RNA extraction was performed as previously described (Yang et al., 2017). Briefly, extraction of RNA from proximal/distal colon (n = 7 per strain) was performed using 1mL TRIzol® Reagent (#15596018, Life Technologies, Carlsbad, CA), as per manufacturer's protocol. Final RNA pellets were resuspended in 30μL of RNAse-DNAse free water to avoid potential genomic contamination. RNA concentration was determined using the NanoDrop-2000 spectrophotometer (Thermo Scientific), then subsequently diluted to 100 ng/μL. After dilution, 1 μg of RNA was used for cDNA synthesis using the High Capacity cDNA Reverse Transcriptase Kit (#4368814, Thermo Scientific). PCR was performed to evaluate the expression of beta 1 and 2 adrenergic receptors (B1AdrR: forward GAA GGC GCT CAA GAC ACT GG; reverse: CCA GGT CGC GGT GGA A; B2AdrR: forward ATG TCG GTT ATC GTC CTG GC; reverse: AAG TCC AGA ACT CGC ACC AG) using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, California). Two additional transcripts were analyzed in order to evaluate potential presence of overall inflammatory markers in the colon: TNFalpha (TNF-α; forward: AGT GGT GCC AGC CGA TGG GTT GT; reverse: GCT GAG TTG GTC CCC CTT CTC CAG) and IL-6 (forward: AAA GAG TTG TGC AAT GGC AAT TCT; reverse: AAG TGC ATC ATC GTT GTT CAT ACA). The conditions of the PCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, primer annealing at 60°C for 30 s, and 72°C for 30 s. Dissociation curves were generated, starting at 75°C and ending at 95°C with increments of 0.5°C every 5 s, to confirm the specificity of reaction. Results were analyzed using the 2-ΔCt method as previously described (Zubcevic et al., 2017) by normalizing to the housekeeping gene GAPDH (forward: GGT GAA GGT CGG TGT GAA CG; reverse: CTC GCT CCT GGA AGA TGG TG), as an endogenous control. Additional no reverse transcriptase control was used.
Fecal DNA Extraction From Colon of WT and KO Mice
All mice were euthanized with 4% isoflurane in a mixture of 95%/5% of O2/CO2. Fecal samples were collected from the proximal and distal colon region from all WT and KO mice separately at end point (n = 5–6 per strain). Bacterial DNA was extracted using ZR Fecal DNA MiniPrep (Zymo Research, Irvine, CA) as per manufacturer's protocols. DNA concentrations were determined using the NanoDrop-2000 spectrophotometer (Thermo Scientific).
16 rDNA Sequencing and Analyses
The analyses of microbial composition were performed, as previously described (Yang et al., 2015, 2017; Santisteban et al., 2017). Briefly, bacterial 16S rDNA V4-V5 regions were obtained using Illumina Miseq compatible primers (Table 1). PCR amplicons were purified (Qiagen, Madison, WI) following separation on an agarose gel, and quantified by the Qubit Fluorometer (Invitrogen, Grand Island, NY). Equal amounts of amplicon for each sample were qualified with Kapa SYBR fast qPCR kit (Kapa Biosystems, Inc., Woburn, MA). Individual samples were run as a library on the Illumina Miseq platform using Miseq v2 reagent kit (Illumina, Inc., San Diego, CA). Primers used for amplification are listed in Table 1. The bioinformatics analyses were performed using QIIME 1.9.2 as described previously (Yang et al., 2015, 2017; Santisteban et al., 2017). Briefly, sequenced reads were demultiplexed according to a combination of forward and reverse indices. Subsequently, obtained reads were filtered based on additional requirements: exact match to sequencing primers and quality score of 30 or more. Filtered reads were applied against Greengene database, assigned taxonomic classifications at a 97% clustering threshold, and summarized in a refOTU table. Alpha, beta diversity, and principal coordinate analyses (PCoA) measures were created using QIIME. Linear discriminant analysis (LDA) along with effect size measurements (LEfSe) in Galaxy was used to distinguish differentially significant features at each taxonomic level. Bacterial genera were identified based on 16S sequencing data (Yang et al., 2015, 2017; Santisteban et al., 2017).
Short Chain Fatty Acid (SCFA) Extraction and High Performance Liquid Chromatography (HPLC) For Determination of Fecal SCFA Levels
SCFAs were extracted from colonic fecal pellets from all mice (n = 4/strain) as previously described (Yang et al., 2017). Briefly, 500 μL of 12M HCl was added to fecal homogenates to preserve the volatile SCFAs. Samples were then vortexed vigorously to suspend the fecal mass. SCFAs were extracted using 5 mL of methylene chloride with gentle rotation at room temperature for 20 min. The supernatant was discarded after centrifugation at 3,500 rpm for 5 min. 500 μL of 1M NaOH were added to the organic pellets, samples were rotated for an additional 20-min at room temperature. Following centrifugation at 3,500 rpm for 5 min, the top aqueous layer was collected and mixed with 100 μL of 12M HCl before being filtered for injection. Standard curves were generated using serial dilutions prepared for each SCFA separately in a similar manner. For example, a 1M stock of sodium butyrate was prepared by dissolving 0.55g of sodium butyrate (#303410-100g, Sigma-Aldrich) into 5 mL of molecular grade water. The solution was then serially diluted to 1 mM. A 1 μM sodium butyrate standard was prepared by mixing 1.5 μL of the 1 mM sodium butyrate stock with 500 μL of molecular grade water and 1000 μL of methanol. A blank standard was also prepared by mixing 500 μL of molecular grade water with 1000 μL of methanol. Each injection used a 50 μL aliquot. The chromatographic separation was performed at room temperature using Perkin Elmer Series 200 HPLC System (Perkin Elmer Instruments, Norwalk, CT), equipped with an autosampler, quaternary pump, and 200 series UV/VIS detector.
Flow Cytometry in Blood of WT and KO Mice
Flow cytometry analysis was performed in all mice (n = 6/strain) to quantify the levels of circulating inflammatory cells (ICs) as previously described (Ahmari et al., 2016; Yang et al., 2017). Briefly, total mononuclear cells (MNCs) were isolated from blood and prepared in a concentration of 0.5-1 × 10E6 cells/100 ul in PBS+2% FBS+1mM EDTA mixture media. Cells were incubated with antibodies in a 1:250 ratio, for 30 min at 4°C. The following markers were used to identify specific inflammatory cells: (i) CD4+IL17+ (T cells; #100406, BioLegend®; #559502, BD Biosciences), (ii) CD8a (T cells; #553033, BD Biosciences), (iii) CD3e (T cells; #553067, BD Biosciences), (iv) CD11b (monocyte/macrophages; #101205, BioLegend®), (v) Ly6+CD184+CD62L+ (neutrophils; #551460, BD Biosciences; #551966, BD Biosciences; #553152, BD Biosciences). In the case of IL17, which is an intracellular marker, following incubation with anti-CD4, cells were washed in normal PBS and fixed in 1% PFA. Following this, cells were incubated with 0.5% Tween20 in normal PBS for 1 h at RT, in order to permeabilize the cell membrane and allow penetration of anti-IL17 inside the cell. Following this, cells were washed in normal PBS, and incubated with anti-IL17 at 1:250 ratio for 30 min at 4°C. SYTOX blue cell stain (ThermoFisher Scientific, #S34857) was added to all samples per manufacturer protocol for exclusion of dead cells during flow cell analysis. Following incubation with antibodies, the cells were washed in normal PBS and fixed with 1% PFA, apart from IL17-stained samples, which were processed as described above. All samples were read on an LSR-II (BD Biosystems) in University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) and the data were analyzed with FACS Diva software, version 6.1.2. Fractions of young (Ly6+CD184+CD62Lhi) and old (Ly6+CD184+CD62Llo) neutrophils was determined as previously described (Zhang et al., 2015).
Statistical Methods
Statistical analysis was performed using GraphPad Prizm and two-tailed unpaired T-test was used for comparison between the two experimental groups. P < 0.05 was considered significant. In addition, linear discriminant analysis (LDA) along with effect size measurements (LEfSe) in Galaxy and ANOSIM in R were used to distinguish differentially significant features at each taxonomic level.
Results
Confirmation of Beta Adrenergic Gene Downregulation in the Colon of KO Mice
To confirm the relative expression levels of beta 1 adrenergic receptors in colon of WT and KO mice, we performed quantitative RT PCR. We observed a significant downregulation in beta 1 adrenergic receptor relative expression levels (Mean ± SEM: 1 ± 0.237 (WT) and 0.289 ± 0.018 (KO); p < 0.05; Table 2) and beta 2 adrenergic receptor relative expression levels (Mean ± SEM: 0.0045 ± 0.0001 (WT) and 0.00085 ± 0.00026 (KO); p < 0.05; Table 2) in the colon of KO compared to WT mice.
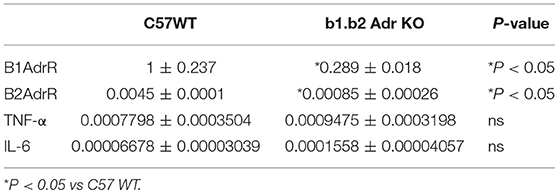
Table 2. Relative expression levels of genes in proximal colon of WT C57 and b1.b2 Adr KO mice as obtained by real time qPCR.
Relative Expression Levels of TNF-α and IL-6 in the Colon of WT vs. KO Mice
We performed quantitative RT PCR to assess the relative gene expression levels of select inflammatory genes in the colon of WT and KO mice (Table 2). Both cytokines have previously linked to proinflammatory responses in the gut and subsequent shifts in the microbiota diversity (Rosser et al., 2014; Jones-Hall et al., 2015; Burton et al., 2017). We observed no significant differences in relative expression of either of the genes investigated: (i) TNF-α: 0.0007798 ± 0.0003504, n = 5 (WT) vs. 0.0009475 ± 0.0003198 (KO) (Table 1); (ii) IL-6: 0.00006678 ± 0.00003039, n = 7 (WT) and 0.0001558 ± 0.00004057 (KO) (Table 2).
Alterations in the Gut Microbiota Reflect in Increased Abundance of Bacilli Class of Firmicutes in the KO Mice
The rodent gut microbiota is generally dominated by Firmicutes and Bacteroidetes phyla, with Proteobacteria, Actinobacteria, and Tenericutes constituting the remaining phyla (Yang et al., 2015, 2017; Santisteban et al., 2017). In our sample, we also observed a small population of Cyanobacteria (Figure 1C). Shifts in the bacterial proportions and phylotypes are widely used to evaluate gut homeostasis. Our 16S rRNA amplicon sequencing data suggested that Chao richness and Shannon diversity indices did not differ between C57 and b1/2-ARs KO mice (Figure 1A), indicating that deficiency in b1/2-AdrRs signaling did not induce significant changes in the microbiota species richness or biodiversity. In addition, we did not observe any difference in the Firmicutes to Bacteroidetes (F/B) ratios (Figure 1A, far right panel) or total species observed (Figure 1B) between WT and KO mice, indicating absence of dysbiosis in the KO strain. Moreover, there were no significant differences at phylum level between the two strains (Figure 1C). However, analyses of gut microbial community structure by unweighted UniFrac principal coordinate analysis (PCoA) revealed two separate clusters, indicating dissimilarities in microbial community structure between the KO and WT mice (Figure 1D, P < 0.05). Analysis of taxonomy indicated a significant increase in the abundance of Bacilli class of Firmicutes, and more specifically in the order of Lactobacillales, with significantly increased abundance of families of Lactobacillaeae and Carnobacteriaceae in the KO compared to WT mice (Figure 2, in green). Moreover, a depletion of Anaerofustis genera of Eubacteriaceae family, the Peptococcaceae family, and Anaerovorax genera, all of Firmicutes phyla, were also observed in the KO vs. WT mice (Figure 2, in red). Thus, overall deficiency in beta adrenergic signaling led to distinct alterations in specific gut Firmicutes with no significant dysbiosis.
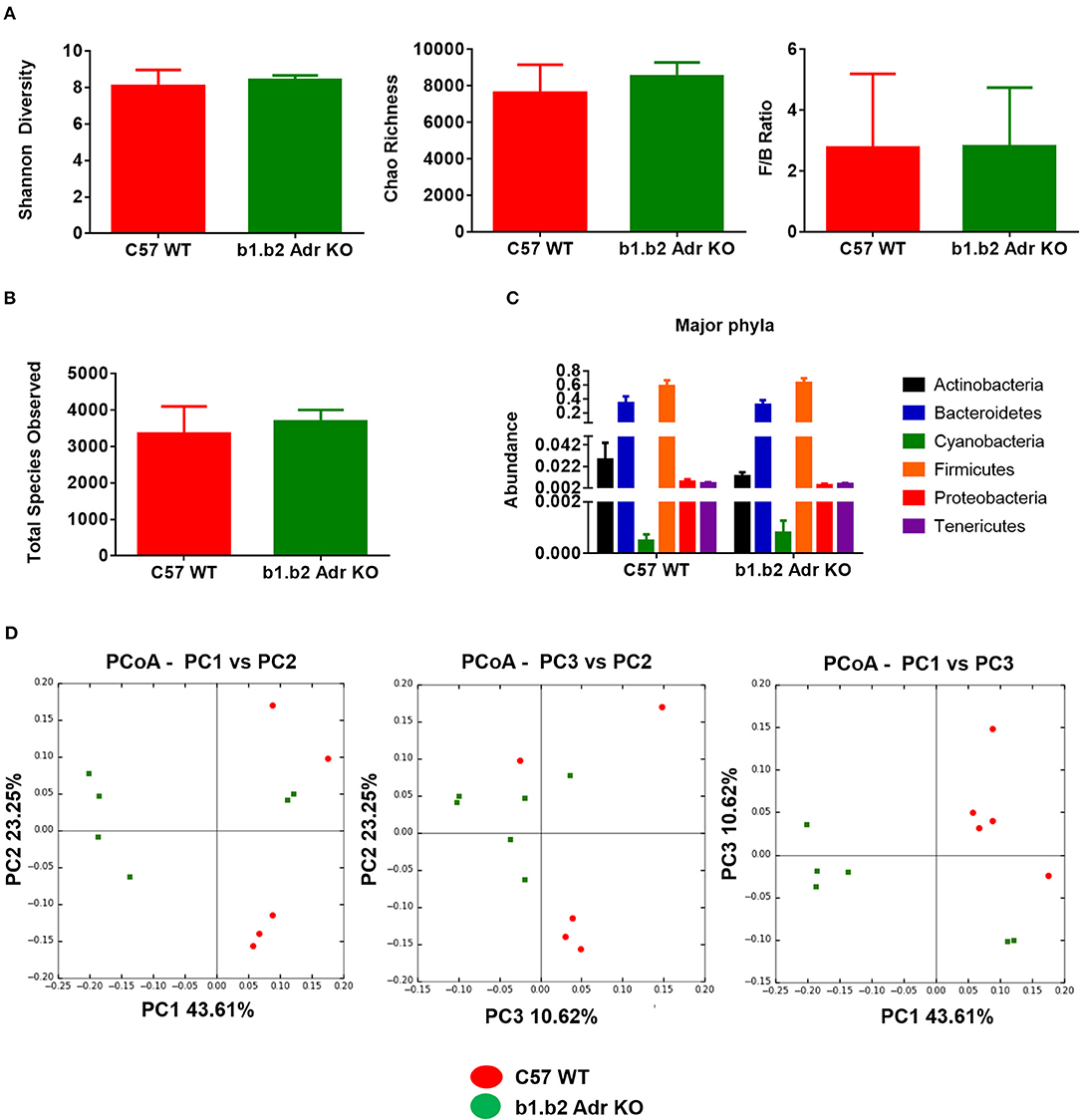
Figure 1. Analysis of gut microbiota in b1.b2 Adr KO and C57 WT mice. (A) No significant difference in gut microbiota richness, diversity, or F/B ratios between the KO and WT mice. (B) No significant differences in the total bacterial species observed in the KO and WT mice. (C) No significant difference in abundance of major bacterial phyla. (D) Unweighted Principal coordinate analysis (PCoA) shows a significant separation of red and green clusters representing the WT and KO mice groups, respectively (ANOSIM in R, P < 0.05 vs. C57 WT). N = 5–6 per strain.
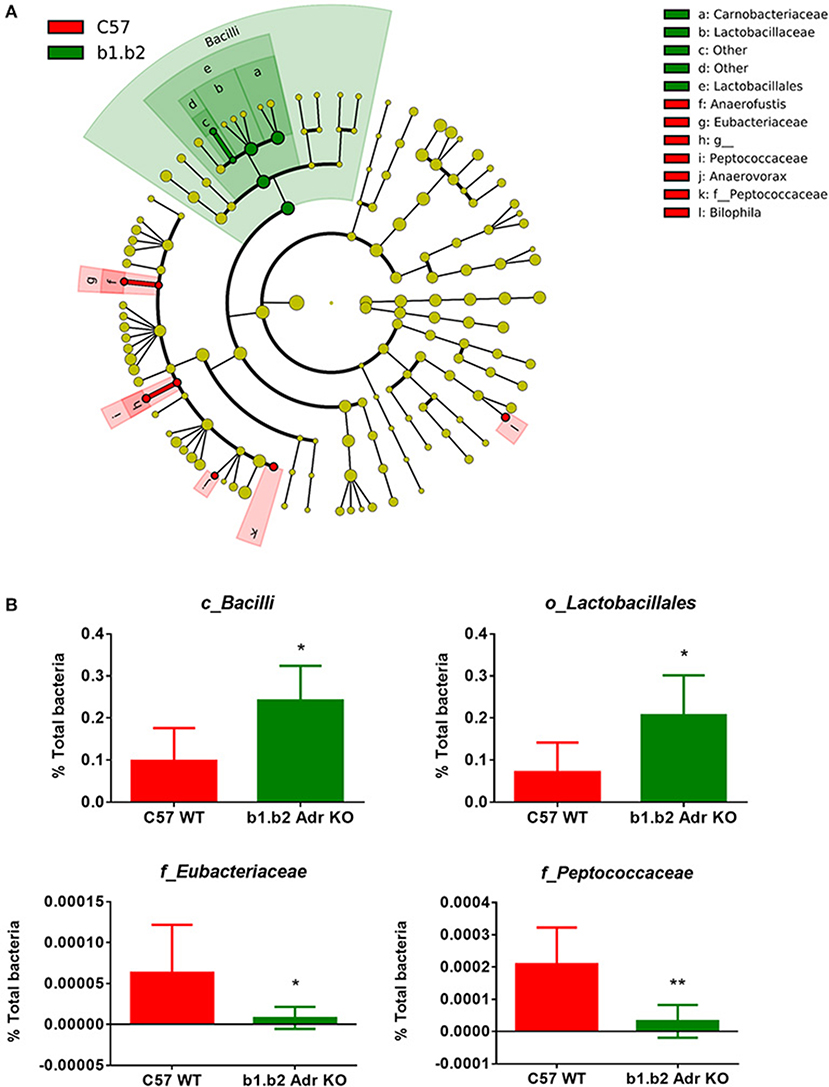
Figure 2. Expansion of Bacilli class of the Firmicutes phylum in the b1.b2 Adr KO mice. (A) Taxonomic cladogram was generated by performing linear discriminant analysis (LDA) of effect size (LEfSe) in Galaxy. Each circle dot represents a bacterial taxon with its diameter proportional to the taxon's relative abundance. Differences are represented in colors where red indicates significantly higher abundance in the WT mice, and green indicates significantly higher abundance in the KO mice (N = 5–6 per strain). Yellow indicates no significant differences between the two strains. (B) Unpaired two-tailed T test analyses of specific class (c_), order (o_) and family (f_) from the Firmicutes phyla, with significant differences between the KO and WT mice. N = 5–6 per strain, *P < 0.05 and **P < 0.01 vs. C57 WT.
Increased Levels of Bacterial Short Chain Fatty Acid (SCFA) Metabolites in Feces of KO Mice
To functionally confirm observed differences in the gut microbiota between the two strains, we performed HPLC analysis in fecal samples to determine levels of different SCFA bacterial metabolites. We observed a significant increase in the levels of fecal sodium butyrate in KO mice compared to WT (mean ± SEM: 2.7 ± 0.4 μmol/g (WT) vs. 5.5 ± 1 μmol/g (KO), p < 0.05; Figure 3A). Furthermore, our analysis revealed a significant increase in fecal acetate levels in KO compared to WT mice (mean ± SEM: 29.9 ± 1.4 μmol/g (WT) and 34.5 ± 1.4 μmol/g (KO), p < 0.05; Figure 3A). Lastly, we observed a significant increase in fecal propionate levels in KO compared to WT mice (mean ± SEM: 0.8 ± 0.08 μmol/g (WT) vs. 2.2 ± 0.2 μmol/g (KO), p < 0.0001; Figure 3A). Figure 3B shows examples of raw data from HPLC analysis. This suggests a potential beneficial functional shift in gut microbiota in the KO mice.
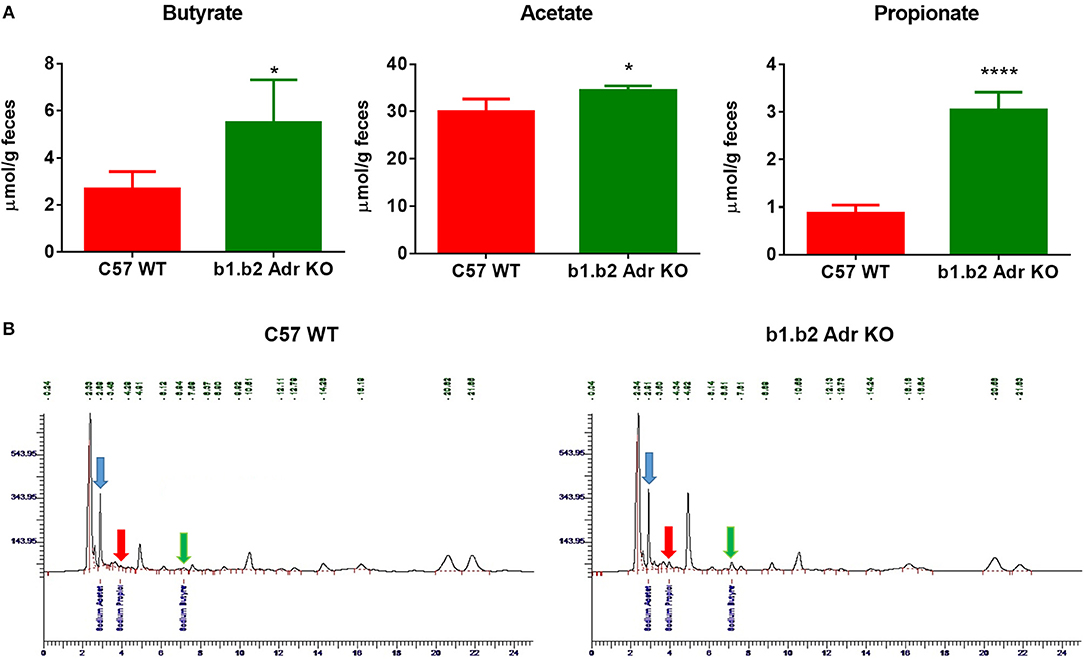
Figure 3. Relative amounts of SCFAs in fecal samples from C57 WT and b1.b2 Adr KO mice. In (A): significantly higher levels of sodium butyrate (left panel), acetate (middle panel) and propionate (right panel) were observed in the KO vs. WT mice. In (B): representative images of butyrate peaks (marked by green arrows), acetate peaks (marked by blue arrows), and propionate peaks (marked by red arrows) in the C57 WT (left panel) and b1.b2 Adr KO mice (right panel), as measured by HPLC. N = 4 per strain; *P < 0.05 and ****P < 0.0001 vs. C57 WT.
KO Mice Exhibit Shifts in Levels of Circulating CD4+.IL17+ T Cells and “Young” Neutrophils
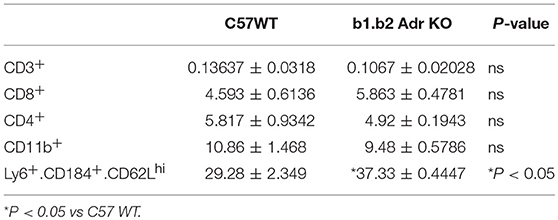
Flow cytometry analysis was performed to assess the levels of circulating ICs (summarized in Table 2 and Figure 4). Our analysis did not reveal any significant differences in circulating levels of several T cell subtypes: (i) CD3+: Mean ± SEM: 0.13637% ± 0.0318% (WT) and 0.1067% ± 0.02028% (KO); (ii) CD8+: Mean ± SEM: 4.593% ± 0.6136% (WT) and 5.863% ± 0.4781% (KO); and (iii) CD4+: Mean ± SEM: 5.817% ± 0.9342% (WT) and 4.92% ± 0.1943% (KO) (Table 3). Furthermore, we observed no significant differences in levels of circulating CD11b+ monocytes/macrophages between the two strains (Mean ± SEM: 10.86% ± 1.468% (WT) and 9.48% ± 0.5786% (KO) (Table 3). However, we observed a significant reduction in circulating levels of CD4+.IL17+ T cells in the KO mice (Mean ± SEM: 0.34% ± 0.1% (WT) and 0.11% ± 0.06% (KO); p < 0.05; Figure 4). Moreover, there was a significant increase in circulating levels of “young” neutrophils (Ly6+.CD184+.CD62Lhi) in the KO mice (Mean ± SEM: 29.28% ± 2.349% (WT) and 37.33% ± 0.4447% (KO); p < 0.05, Table 3).
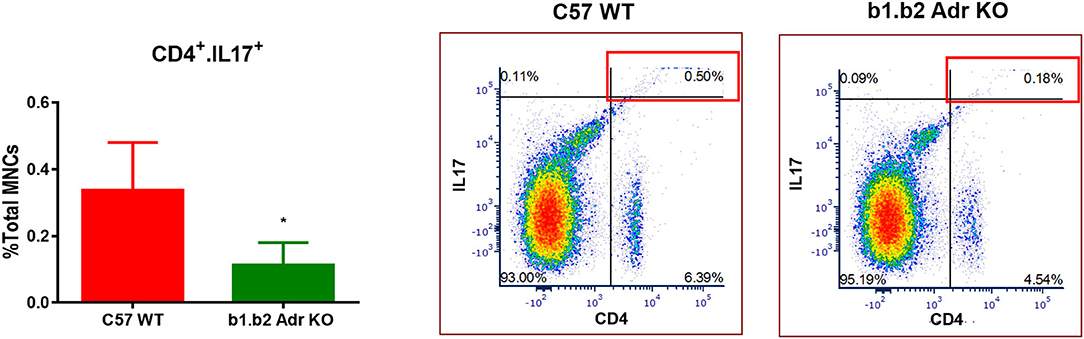
Figure 4. Quantification of circulating inflammatory cells in C57 WT and b1.b2 Adr KO mice. Significantly decreased levels of CD4+.IL17+ T cells in b1.b2 Adr KO, with representative gating shown in the right panel. N = 6 per strain; *p < 0.05 vs. C57 WT.
Discussion
Our study highlights important host-microbiota interactions in a mouse model characterized by reduced overall effects of the sympathetic drive. Our major findings are as follows: (i) we observed no major differences in the diversity, richness or F/B ratio in b1.b2 Adr KO mice compared to C57 WT controls, suggesting no presence of gut dysbiosis in these mice. Moreover, there were no differences in the abundance of the major phyla between the two strains. (ii) However, unifrac unweighted PCoA revealed a significant separation between microbiota of C57 WT and b1.b2 Adr KO mice. This was reflected in significantly increased abundance of the Bacilli class of Firmicutes in the b1.b2 Adr KO mice, and specifically the family of Lactobacillaceae. Conversely, we observed an increased abundance of family of Peptococcaceae and Eubacteriaceae of the Firmicutes phyla, among several others, in the b1.b2 Adr KO mice. (iii) The increase in levels of butyrate, acetate and propionate in the feces of b1.b2 Adr KO mice, linked to anti-inflammatory and anti-hypertensive effects (Natarajan and Pluznick, 2014; Natarajan et al., 2016; Ohira et al., 2017; Pluznick, 2017), suggested beneficial functional shifts in the gut microbiota in these mice. (iv) While we observed no significant differences in inflammatory cytokines between the two strains, suggesting no major difference in local gut inflammatory profile, our data does show a reduction in circulating CD4+.IL17+ T cells in the b1.b2 Adr KO mice, previously linked to rodent and human HTN (Wang et al., 2016; Wilck et al., 2017; Zubcevic et al., 2017; Kim et al., 2018), while an observed increase in the circulating levels of young neutrophils in these mice has also previously been directly linked to beneficial changes in the gut microbiota (Zhang et al., 2015).
Previous studies have characterized HTN with alterations in the gut microbiota and sympathetic dysregulation (Zubcevic et al., 2014; Yang et al., 2015; Adnan et al., 2017; Santisteban et al., 2017). In the present study, we investigated the close association between the SNS, immune system and gut microbiota in a b1.b2 Adr KO mouse model characterized by reduced BP and heart rate responses upon stimulation (Rohrer et al., 1999). This allowed isolation of the effects of one variable, the SNS, in the complex host-microbiota communication in regulation of BP. Our previous data showed that reduced SNS effects specifically on the bone marrow produced a marked immunosuppression and resulted in significant shifts in the microbiota and reduction in BP (Ahmari et al., 2016; Yang et al., 2017). However, these effects were mainly reflected in decreased abundance of specific members of Bacteriodetes (Yang et al., 2017). While an increase in abundance of some members of the Firmicutes was also observed following blockade of bone marrow SNS only (Yang et al., 2017), these changes do not overlap with the microbiota alterations in the full b1.b2 Adr KO mice examined in the present study. Interestingly, a significant decrease in the order of Lactobacillales of the Bacilli family was observed in the spontaneously hypertensive rat (SHR), a rodent model of HTN characterized by both exaggerated SNS and immune system (Zubcevic et al., 2014; Yang et al., 2015; Santisteban et al., 2017). Since this is in exact opposition to our present finding, we propose that the SNS may contribute to regulation of abundance of Lactobacillales order of the Bacilli class. Others have suggested that norepinephrine (NE), one of the neurotransmitters released following stimulation of SNS, can directly modulate bacterial growth (Freestone et al., 2000; Doherty et al., 2009). However, our model is characterized by lack of beta-adrenergic receptors, thus resulting in reduced effects of SNS rather than the inherent differences in SNS innervation or NE release. Thus, the changes in the gut microbiota observed in the b1.b2 Adr KO mice in the present study may be due to modified gut homeostasis resulted from loss of beta-adrenergic receptors. This may reflect in reduced blood supply to the gut, or suppression of the renin-angiotensin-aldosterone system, which could also be a cause of reduced adrenergic receptor activation, and could results in changes in gut microbiota composition. Further studies are required to investigate the role of SNS in gut homeostasis, but previous data from SHR show association between overactive SNS and breakdown in gut epithelium with reduced levels of Lactobacillaceae (Yang et al., 2015; Santisteban et al., 2017). Thus, supplementation with specific Lactobacillales, and especially family of Lactobacillaceae in the SHR may counteract the effects of increased SNS to the gut in this strain. Indeed, previously published research has noted positive cardiovascular effects following supplementation with members of the Lactobacillaceae family or their metabolic products (Sonestedt et al., 2011; Wilck et al., 2017; Hidalgo et al., 2018).
Lactobacillaceae fermentation has been shown to produce powerful bioactive peptides with the capability to inhibit ACE (Fuglsang et al., 2003; Barla et al., 2016), a key enzyme of the renin-angiotensin system (RAS) positively correlated with HTN (Wang et al., 2011; He et al., 2013). Indeed, there has been several clinical studies which have noted reduced BP in hypertensive patients supplemented with specific Lactobacillaceae (Nakajima et al., 1995; Seppo et al., 2003; Jauhiainen et al., 2005; Tanida et al., 2005; Wilck et al., 2017). The increase in fecal butyrate, acetate and propionate confirms a general beneficial shift in the gut microbiota in the b1.b2 Adr KO mice. These short chain fatty acids are metabolite products of gut bacteria with suggested anti-inflammatory and anti-hypertensive properties (Natarajan and Pluznick, 2014; Natarajan et al., 2016; Ohira et al., 2017; Pluznick, 2017). These findings are again in direct opposition to those observed in the SHR, which is characterized by reduced levels of butyrate-, acetate- and propionate-producing gut bacteria (Yang et al., 2015).
Moreover, dietary supplementation with specific members of Lactobacillales has been shown to reduce the effects of increased dietary salt by dampening the IL17-dependent immune responses in rodent models of HTN and human HTN (Wilck et al., 2017). In studies of inflammatory bowel disease, elevated levels of Th17 cells and an upregulation of IL17 have been shown (Chen et al., 2015), and emerging evidence illustrates that certain Lactobacillales strains have the potential to reduce inflammation by modulating IL17 expression (Wells, 2011; Jan et al., 2012; Noto Llana et al., 2013; Wilck et al., 2017). Our present data confirms the positive correlation between elevated abundance of Lactobacillales and reduced CD4+.IL17+ T cells in circulation of b1.b2 Ard KO mice. Thus, reduced effects of SNS may cause changes in the gut microbiota that may have a direct effect on these immune cells. Evidence arising from experimental models of inflammatory bowel disease suggests that CD4+ T cells are key players in the initiation and regulation of the immunopathologic processes, partly through production of proinflammatory cytokines such as tumor necrosis factor-α (TNF-α) (Wells, 2011). Strains of Lactobacillales have also been shown to elicit adaptive and innate immune responses in the host (Noto Llana et al., 2013). However, we did not observe significant differences in expression levels of TNF-α or IL-6 cytokines in the gut of our mice. We also observed a depletion of Eubacteriaceae and Peptococcaceae in the order of Clostridiales and in the phyla of Firmicutes, in our b1.b2 Adr KO mice. These strains are largely considered to be beneficial, with Peptococcaceae a sulfate-reducing bacteria (Petzsch et al., 2015), a property suggested to be important in regulation of BP (Ahmad et al., 2012; Yu et al., 2017). Similarly, we saw a reduction in Bilophila in the b1.b2 Adr KO mice, which are, however considered to be largely pathogenic, which may be related to reduced presence of hydrogen sulfide (da Silva et al., 2008). Further studies are needed to confirm the relevance of these bacterial shifts in cardiovascular and immune homeostasis.
A significant increase in circulating young neutrophils in our b1.b2 Adr KO mice may also be driven by changes in the microbiota in these mice. Neutrophils represent a large population of leucocytes in the mouse, and they have a unique property in that they follow daily cycles of release from the bone marrow into circulation as “young” neutrophils, where their “aging” is positively correlated with pro-inflammatory properties (Casanova-Acebes et al., 2013). Recent work by Zhang et al (Zhang et al., 2015) suggested that neutrophil aging in circulation is regulated by the microbiome, with shifts in abundance of different Bacilli, Clostridiales and Proteobacteria species associated with reduction in circulating aged neutrophils thus dramatically reducing inflammatory organ damage in certain immune disorders (Zhang et al., 2015). Based on this, the observed increase in circulating young neutrophils in our b1.b2 Adr KO mice may be considered anti-inflammatory and microbiota-driven, and may be related to the reduced effects of the SNS. Future data should compare the effects of fecal matter transplant from b1.b2 Adr KO mice on the immune system and SNS, to further characterize role of SNS in the host-microbiota interactions.
In summary, our data illustrates an association between reduced beta-adrenergic receptor signaling, associated with dampened effects of the SNS on the whole body including the gut and the immune system, and a significant shift in Lactobacillaceae family in the gut microbiota and reduced immune responses in mice with reduced BP and heart rate responses (Rohrer et al., 1999). Given the current research interest in the use of probiotics in treating inflammation and gut dysbiosis, our data offers further evidence to why administration of various strains of Bacilli has a positive effect on BP in preclinical and clinical setting.
Ethics Statement
This study was carried out in accordance with the recommendations and approval by the IACUC protocol number 201708217.
Author Contributions
AB and TY designed and performed experiments and analyzed the data. They also contributed to writing of the manuscript. RA, WM, RL, and KM contributed to collection and analysis of data. TV contributed to writing and editing of the manuscript. JZ designed the experiments, oversaw analysis of the data and interpretation of results, and contributed to writing and final editing of the manuscript. Authors would like to thank Niousha Ahmari for technical assistance with the manuscript.
Funding
This work was supported by AHA14SDG18300010 and UF CVM Start up funds to JZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adnan, S., Nelson, J. W., Ajami, N. J., Venna, V. R., Petrosino, J. F., Bryan, R. M., et al. (2017). Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genomics 49, 96–104. doi: 10.1152/physiolgenomics.00081.2016
Ahmad, F. U., Sattar, M. A., Rathore, H. A., Abdullah, M. H., Tan, S., Abdullah, N. A., et al. (2012). Exogenous hydrogen sulfide (H2S) reduces blood pressure and prevents the progression of diabetic nephropathy in spontaneously hypertensive rats. Ren. Fail. 34, 203–210. doi: 10.3109/0886022X.2011.643365
Ahmari, N., Schmidt, J. T., Krane, G. A., Malphurs, W., Cunningham, B. E., Owen, J. L., et al. (2016). Loss of bone marrow adrenergic beta 1 and 2 receptors modifies transcriptional networks, reduces circulating inflammatory factors, and regulates blood pressure. Physiol. Genomics 48, 526–36. doi: 10.1152/physiolgenomics.00039.2016
Anderson, E. A., Sinkey, C. A., Lawton, W. J., and Mark, A. L. (1989). Elevated sympathetic nerve activity in borderline hypertensive humans. evidence from direct intraneural recordings. Hypertension 14, 177–83. doi: 10.1161/01.HYP.14.2.177
Barla, F., Koyanagi, T., Tokuda, N., et al. (2016). The γ-aminobutyric acid-producing ability under low pH conditions of lactic acid bacteria isolated from traditional fermented foods of Ishikawa Prefecture, Japan, with a strong ability to produce ACE-inhibitory peptides. Biotechnol. Rep. 10, 105–110. doi: 10.1016/j.btre.2016.04.002
Baumgart, D. C., and Carding, S. R. (2007). Inflammatory bowel disease: cause and immunobiology. Lancet 369, 1627–1640. doi: 10.1016/S0140-6736(07)60750-8
Bellinger, D. L., and Lorton, D. (2014). Autonomic regulation of cellular immune function. Auton. Neurosci. 182, 15–41. doi: 10.1016/j.autneu.2014.01.006
Bry, L., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1996). A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383. doi: 10.1126/science.273.5280.1380
Burton, K. J., Rosikiewicz, M., Pimentel, G., Bütikofer, U., von Ah, U., Voirol, M. J., et al. (2017). Probiotic yogurt and acidified milk similarly reduce postprandial inflammation and both alter the gut microbiota of healthy, young men. Br. J. Nutr. 117, 1312–1322. doi: 10.1017/S0007114517000885
Calhoun, D. A., Jones, D., Textor, S., Goff, D. C., Murphy, T. P., Toto, R. D., et al. (2008). Resistant hypertension: diagnosis, evaluation, and treatment. Hypertension 51, 1403–19. doi: 10.1161/HYPERTENSIONAHA.108.189141
Casanova-Acebes, M., Pitaval, C., Weiss, L. A., Nombela-Arrieta, C., Chèvre, R. A.-, et al. (2013). Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 doi: 10.1016/j.cell.2013.04.040
Chen, L., Zou, Y., Peng, J., Lu, F., Yin, Y., Li, F., et al. (2015). Lactobacillus acidophilus suppresses colitis-associated activation of the IL-23/Th17 axis. J. Immunol. Res. 2015:909514. doi: 10.1155/2015/909514
Chobanian, A. V., Bakris, G. L., Black, H. R., Cushman, W. C., Green, L. A., Izzo, J. L., et al. (2003). The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure the JNC 7 report. J. Am. Med. Assoc. 289, 2560. doi: 10.1001/jama.289.19.2560
Chow, J., Lee, S. M., Shen, Y., Khosravi, A., and Mazmanian, S. K. (2010). Host-bacterial symbiosis in health and disease. Adv. Immunol. 107, 243–274. doi: 10.1016/B978-0-12-381300-8.00008-3
Cryan, J. F., and Dinan, T. G. (2012). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. doi: 10.1038/nrn3346
da Silva, S. M., Venceslau, S. S., Fernandes, C. L., Valente, F. M., and Pereira, I. A. (2008). Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Van Leeuwenhoek. 93, 381–90. doi: 10.1007/s10482-007-9215-x
Doherty, N. C., Tobias, A., Watson, S., and Atherton, J. C. (2009). The effect of the human gut-signalling hormone, norepinephrine, on the growth of the gastric pathogen Helicobacter pylori. Helicobacter 14, 223–230. doi: 10.1111/j.1523-5378.2009.00682.x
Esler, M., Jackman, G., Bobik, A., Leonard, P., Kelleher, D., Skews, H., et al. (1981). Norepinephrine kinetics in essential hypertension. Defective neuronal uptake of norepinephrine in some patients. Hypertension 3, 149–156. doi: 10.1161/01.HYP.3.2.149
Everard, A., and Cani, P. D. (2013). Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 27, 73–83. doi: 10.1016/j.bpg.2013.03.007
Freestone, P. P., Lyte, M., Neal, C. P., Maggs, A. F., Haigh, R. D., and Williams, P. H. (2000). The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growthin the presence of transferrin or lactoferrin. J. Bacteriol. 182, 6091–6098. doi: 10.1128/JB.182.21.6091-6098.2000
Fuglsang, A., Rattray, F. P., Nilsson, D., and Nyborg, N. C. (2003). Lactic acid bacteria: inhibition of angiotensin converting enzyme in vitro and in vivo. Antonie Van Leeuwenhoek. 83, 27–34 doi: 10.1023/A:1022993905778
Gao, Z., Guo, B., Gao, R., Zhu, Q., and Qin, H. (2015). Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 6:20. doi: 10.3389/fmicb.2015.00020
Harrison, D. G. (2014). The immune system in hypertension. Trans. Am. Clin. Climatol. Assoc. 125, 130–140.
He, Q., Fan, C., Yu, M., Wallar, G., Zhang, Z. F., Wang, L., et al. (2013). Associations of ACE gene insertion/deletion polymorphism, ACE activity, and ACE mRNA expression with hypertension in a Chinese population. PLoS ONE 8:e75870. doi: 10.1371/journal.pone.0075870.
Hidalgo, M., Prieto, I., Abriouel, H., Villarejo, A. B., Ramírez-Sánchez, M., Cobo, A., et al. (2018). Changes in gut microbiota linked to a reduction in systolic blood pressure in spontaneously hypertensive rats fed an extra virgin olive oil-enriched diet. Plant Foods Hum. Nutr. 73, 1–6. doi: 10.1007/s11130-017-0650-1
Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G., and Gordon, J. I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. doi: 10.1126/science.291.5505.881.
Jan, R. L., Yeh, K. C., Hsieh, M. H., Lin, Y. L., Kao, H. F., Li, P. H., et al. (2012). Lactobacillus gasseri suppresses Th17 pro-inflammatory response and attenuates allergen-induced airway inflammation in a mouse model of allergic asthma. Br. J. Nutr. 108, 130–139. doi: 10.1017/S0007114511005265
Jänig, W. (2014). Sympathetic nervous system and inflammation: a conceptual view. Auton. Neurosci. 182, 4–14. doi: 10.1016/j.autneu.2014.01.004
Jauhiainen, T., Vapaatalo, H., Poussa, T., Kyrönpalo, S., Rasmussen, M., and Korpela, R (2005). Lactobacillus helveticus fermented milk lowers blood pressure in hypertensive subjects in 24-h ambulatory blood pressure measurement. Am. J. Hypertens. 8, 1600–1605. doi: 10.1016/j.amjhyper.2005.06.006
Jones-Hall, Y. L., Kozik, A., and Nakatsu, C. (2015). Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS ONE 10:e0119441. doi: 10.1371/journal.pone.0119441
Kim, S., Goel, R., Kumar, A., Qi, Y., Lobaton, G., Hosaka, K., et al. (2018). Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin. Sci. 132, 701–718. doi: 10.1042/CS20180087
Mariat, D., Firmesse, O., Levenez, F., Guimarăes, V., Sokol, H., Doré, J., et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9:123. doi: 10.1186/1471-2180-9-123
Mell, B., Jala, V. R., Mathew, A. V., Byun, J., Waghulde, H., Zhang, Y., et al. (2015). Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genomics. 47, 187–197. doi: 10.1152/physiolgenomics.00136.2014
Nakajima, K, Hata, Y., Osono, Y., Hamura, M., Kobayashi, S., and Watanuki, M (1995). Antihypertensive effect of extracts of Lactobacillus casei in patients with hypertension. J. Clin. Biochem. Nutr. 18, 181–187. doi: 10.3164/jcbn.18.181
Natarajan, N., Hori, D., Flavahan, S., Steppan, J., Flavahan, N. A., Berkowitz, D. E., et al. (2016). Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol. Genomics 48, 826–834. doi: 10.1152/physiolgenomics.00089.2016
Natarajan, N., and Pluznick, J. L. (2014). From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am. J. Physiol. Cell Physiol. 307, C979–C985. doi: 10.1152/ajpcell.00228.2014
Noto Llana, M., Sarnacki, S. H., Aya Castañeda M del, R., Bernal, M. I., Giacomodonato, M. N., and Cerquetti, M. C. (2013). Consumption of Lactobacillus casei fermented milk prevents salmonella reactive arthritis by modulating IL-23/IL-17 Expression. PLoS ONE 8:e82588. doi: 10.1371/journal.pone.0082588
Ohira, H., Tsutsui, W., and Fujioka, Y. (2017). Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis?. J. Atheroscler. Thromb. 24, 660–672. doi: 10.5551/jat.RV17006
Pamer, E. G. (2007). Immune responses to commensal and environmental microbes. Nat. Immunol. 8, 1173–1178. doi: 10.1038/ni1526
Petzsch, P., Poehlein, A., Johnson, D. B., Daniel, R., Schlömann, M., and Mühling, M. (2015). Genome Sequence of the Acidophilic Sulfate-Reducing Peptococcaceae Strain CEB3. Genome Announc. 3:e00886-15. doi: 10.1128/genomeA.00886-15
Pluznick, J. L (2017) Microbial short-chain fatty acids and blood pressure regulation. Curr. Hypertens. Rep. 19:25. doi: 10.1007/s11906-017-0722-5.
Raizada, M. K., Joe, B., Bryan, N. S., Chang, E. B., Dewhirst, F. E., Borisy, G. G., et al. (2017). Report of the national heart, lung, and blood institute working group on the role of microbiota in blood pressure regulation: current status and future directions. Hypertension. doi: 10.1161/HYPERTENSIONAHA.117.09699. [Epub ahead of print].
Rohrer, D. K., Chruscinski, A., Schauble, E. H., Bernstein, D., and Kobilka, B. K. (1999). Cardiovascular and metabolic alterations in mice lacking both beta1- and beta2-adrenergic receptors. J. Biol. Chem. 274, 16701–16708. doi: 10.1074/jbc.274.24.16701
Rosser, E. C., Oleinika, K., Tonon, S., Doyle, R., Bosma, A., Carter, N. A., et al. (2014). Regulatory B cells are induced by gut microbiota–driven interleukin-1β and interleukin-6 production. Nat. Med. 20, 1334–1339. doi: 10.1038/nm.3680
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi: 10.1038/nri2515
Rumantir, M. S., Kaye, D. M., Jennings, G. L., Vaz, M., Hastings, J. A., and Esler, M. D. (2000). Phenotypic evidence of faulty neuronal norepinephrine reuptake in essential hypertension. Hypertension 36, 824–829. doi: 10.1161/01.HYP.36.5.824
Sansonetti, P. J. (2004). War and peace at mucosal surfaces. Nat. Rev. Immunol. 4, 953–964. doi: 10.1038/nri1499
Santisteban, M. M., Ahmari, N., Carvajal, J. M., Zingler, M. B., Qi, Y., Kim, S., et al. (2015). Involvement of bone marrow cells and neuroinflammation in hypertension. Circ. Res. 117, 178–91. doi: 10.1161/CIRCRESAHA.117.305853
Santisteban, M. M., Qi, Y., Zubcevic, J., Kim, S., Yang, T., Shenoy, V., et al. (2017). Hypertension-linked pathophysiological alterations in the gut. Circ. Res. 120, 312–323. doi: 10.1161/CIRCRESAHA.116.309006
Sanz, Y., and Moya-Pérez, A. (2014). Microbiota, inflammation and obesity. Adv. Exp. Med. Biol. 817, 291–317. doi: 10.1007/978-1-4939-0897-4_14
Schlaich, M. P., Lambert, E., Kaye, D. M., Krozowski, Z., Campbell, D. J., Lambert, G., et al. (2004). Sympathetic augmentation in hypertension. Hypertension 43, 169–175 doi: 10.1161/01.HYP.0000103160.35395.9E
Seppo, L., Jauhiainen, T., Poussa, T., and Korpela, R. (2003). A fermented milk high in bioactive peptides has a blood pressure-lowering effect in hypertensive subjects. Am. J. Clin. Nutr. 77, 326–330. doi: 10.1093/ajcn/77.2.326
Singh, M. V, Chapleau, M. W., Harwani, S. C., and Abboud, F. M. (2014). The immune system and hypertension. Immunol. Res. 59, 243–253. doi: 10.1007/s12026-014-8548-6
Smith, K., McCoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi: 10.1016/j.smim.2006.10.002
Sonestedt, E., Wirfält, E., Wallström, P., Gullberg, B., Orho-Melander, M., and Hedblad, B. (2011). Dairy products and its association with incidence of cardiovascular disease: the Malmö diet and cancer cohort. Eur. J. Epidemiol. 26, 609–618. doi: 10.1007/s10654-011-9589-y
Tanida, M., Yamano, T., Maeda, K., Okumura, N., Fukushima, Y., and Nagai, K. (2005). Effects of intraduodenal injection of Lactobacillus johnsonii La1 on renal sympathetic nerve activity and blood pressure in urethane-anesthetized rats. Neurosci. Lett. 389, 109–114. doi: 10.1016/j.neulet.2005.07.036
Tilg, H., and Kaser, A. (2011). Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 121, 2126–2132. doi: 10.1172/JCI58109
Wang, G., Lai, F. M., Kwan, B. C., Lai, K. B., Chow, K. M., Li, P. K., et al. (2011). Expression of ACE and ACE2 in patients with hypertensive nephrosclerosis. Kidney Blood Press. Res. 34, 141–149. doi: 10.1159/000324521
Wang, Z., Shi, W., Liang, X., Wang, W., and Liang, J. (2016). Association of interleukin 17 / angiotensin II with refractory hypertension risk in hemodialysis patients. Afr. Health Sci. 16, 766–771. doi: 10.4314/ahs.v16i3.17
Wells, J. M. (2011). Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 10 (Suppl. 1):S17. doi: 10.1186/1475-2859-10-S1-S17
Wilck, N., Matus, M. G., Kearney, S. M., Olesen, S. W., Forslund, K., Bartolomaeus, H., et al. (2017). Salt responsive gut commensal modulates TH 17 axis and disease. Nature 551, 585–589. doi: 10.1038/nature24628
Yang, T., Ahmari, N., Schmidt, J. T., Redler, T., Arocha, R., Pacholec, K., et al. (2017). Shifts in the gut microbiota composition due to depleted bone marrow beta adrenergic signaling are associated with suppressed inflammatory transcriptional networks in the mouse colon. Front. Physiol. 8:220. doi: 10.3389/fphys.2017.00220
Yang, T., Santisteban, M. M., Rodriguez, V., Li, E., Ahmari, N., Carvajal, J. M., et al. (2015). Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–40. doi: 10.1161/HYPERTENSIONAHA.115.05315
Yu, W., Liao, Y., Huang, Y., Chen, S. Y., Sun, Y., Sun, C., et al. (2017). Endogenous hydrogen sulfide enhances carotid sinus baroreceptor sensitivity by activating the transient receptor potential cation channel subfam ily V member 1 (TRPV1). Channel. J. Am. Heart Assoc. 6:e004971. doi: 10.1161/JAHA.116.004971
Zhang, D., Chen, G., Manwani, D., Mortha, A., Xu, C., Faith, J. J., et al. (2015). Neutrophil ageing is regulated by the microbiome. Nature 525, 528–32. doi: 10.1038/nature15367
Zubcevic, J., Jun, J. Y., Kim, S., Perez, P. D., Afzal, A., Shan, Z., et al. (2014). Altered inflammatory response is associated with an impaired autonomic input to the bone marrow in the spontaneously hypertensive rat. Hypertens 63, 542–550. doi: 10.1161/HYPERTENSIONAHA.113.02722
Keywords: microbiota, IL17, colon, sympathetic nervous system, adrenergic receptors
Citation: Bartley A, Yang T, Arocha R, Malphurs WL, Larkin R, Magee KL, Vickroy TW and Zubcevic J (2018) Increased Abundance of Lactobacillales in the Colon of Beta-Adrenergic Receptor Knock Out Mouse Is Associated With Increased Gut Bacterial Production of Short Chain Fatty Acids and Reduced IL17 Expression in Circulating CD4+ Immune Cells. Front. Physiol. 9:1593. doi: 10.3389/fphys.2018.01593
Received: 04 June 2018; Accepted: 24 October 2018;
Published: 13 November 2018.
Edited by:
Maurizio Acampa, Azienda Ospedaliera Universitaria Senese, ItalyReviewed by:
Lu Liu, University of New South Wales, AustraliaMarta Toral, Universidad de Granada, Spain
Marcello Pastorelli, Università degli Studi di Siena, Italy
Copyright © 2018 Bartley, Yang, Arocha, Malphurs, Larkin, Magee, Vickroy and Zubcevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jasenka Zubcevic, amFzZW5rYXpAdWZsLmVkdQ==
 Akeem Bartley
Akeem Bartley Tao Yang
Tao Yang Rebeca Arocha
Rebeca Arocha Riley Larkin
Riley Larkin Jasenka Zubcevic
Jasenka Zubcevic