- 1Division of Infectious Diseases, Department of Medicine, Center for Genome Sciences and Systems Biology, Washington University School of Medicine, St. Louis, MO, United States
- 2Section of Infectious Diseases, Department of Pediatrics, Baylor College of Medicine, Houston, TX, United States
As the intestinal microbiota has become better appreciated as necessary for maintenance of physiologic homeostasis and also as a modulator of disease processes, there has been a corresponding increase in manipulation of the microbiota in mouse models. While germ-free mouse models are generally considered to be the gold standard for studies of the microbiota, many investigators turn to antibiotics treatment models as a rapid, inexpensive, and accessible alternative. Here we describe and compare these two approaches, detailing advantages and disadvantages to both. Further, we detail what is known about the effects of antibiotics treatment on cell populations, cytokines, and organs, and clarify how this compares to germ-free models. Finally, we briefly describe recent findings regarding microbiota regulation of infectious diseases and other immunologic challenges by the microbiota, and highlight important future directions and considerations for the use of antibiotics treatment in manipulation of the microbiota.
Introduction
Over the past several decades, there has been a dramatic increase in both scientific and popular interest in the effects of the intestinal microbiota on human health. The microbiota, consisting of the bacteria, viruses, fungi, and archaea that inhabit different niches in the human body, has been implicated in regulation of inflammatory, infectious and metabolic diseases, and appears to play a critical role in potentially causing, propagating, or preventing human illnesses (Lai et al., 2014; Norman et al., 2014; Palm et al., 2015). With the surge of enthusiasm to understand this new and massively complex factor in human health has come the need to effectively model it. Specifically, the development of small animal models of the microbiota permits testing of subsets of the microbiota as causative vs. correlative factors in disease states, as well as offering a system to uncover putative therapeutics.
Two main methods have emerged to explore the effects of the microbiota on physiology and disease in mice: germ-free models and antibiotics treatment regimens. Both approaches have strengths and weaknesses. Here we will discuss commonly used regimens and methods to deplete the microbiota, the effects of these approaches on host physiology including cellular composition, signaling pathways, and organ function, and briefly describe what has been found using these two different methods to model the effects of the microbiota on human disease.
Germ-Free and Antibiotics Treatment Models
Germ-free mice are bred in isolators which fully block exposure to microorganisms, with the intent of keeping them free of detectable bacteria, viruses, and eukaryotic microbes. Initially conceptualized by Louis Pasteur in 1885, colonies of germ-free rodents were first established in the 1940s (Yi and Li, 2012; Al-Asmakh and Zadjali, 2015). Germ-free mice allow for study of the complete absence of microbes or for the generation of gnotobiotic animals exclusively colonized by known microbes. However, generating and maintaining these mice requires specialized facilities, and the cost, labor, and skills required to maintain them can make these models inaccessible to many researchers. Germ-free mice must be monitored regularly for contamination using a combination of culturing, microscopy, serology, gross morphology, and sequencing-based detection techniques (Fontaine et al., 2015; Nicklas et al., 2015). For example, Charles River, one common germ-free vendor, routinely uses a serologic assay to test for viral contamination including murine norovirus, mouse rotavirus, and mouse cytomegalovirus; PCR (both 16S and pathogen-specific), microscopy, and culturing to test for bacteria; and gross examination of animals to test for parasites (Charles River Germ-Free Mouse Report). Additionally, any unique mouse strain to be studied under germ-free conditions must be rederived in these facilities, and this limits the number of different genotypes that are feasible to study. Further, maintenance of mice in isolators may make it impractical or challenging to conduct some studies (for example, behavioral testing or pathogen infections).
An alternate method that has emerged to avoid some of these complications has been the use of antibiotics treatment (Figure 1). Treatment with broad-spectrum antibiotics is commonly used to deplete the gut microbiota of mice, and can be readily applied to any genotype or condition of mouse. Unlike germ-free conditions, under which complete sterility is maintained throughout life, antibiotics can deplete bacterial populations in mice which were normally colonized since birth. Germ-free animals are broadly impaired in many aspects of development and early immune education, whereas antibiotics treatment in adult mice specifically allows for study of the role of bacteria in maintaining cell functionality and signaling pathways after development. Alternatively, some studies deliver antibiotics in drinking water to pregnant dams to limit maternal transfer of microbes and then maintain the cage on the regimen during weaning to study the effects of bacterial depletion early in development (Lamousé-Smith et al., 2011; Deshmukh et al., 2014; Gonzalez-Perez et al., 2016; Li et al., 2017).
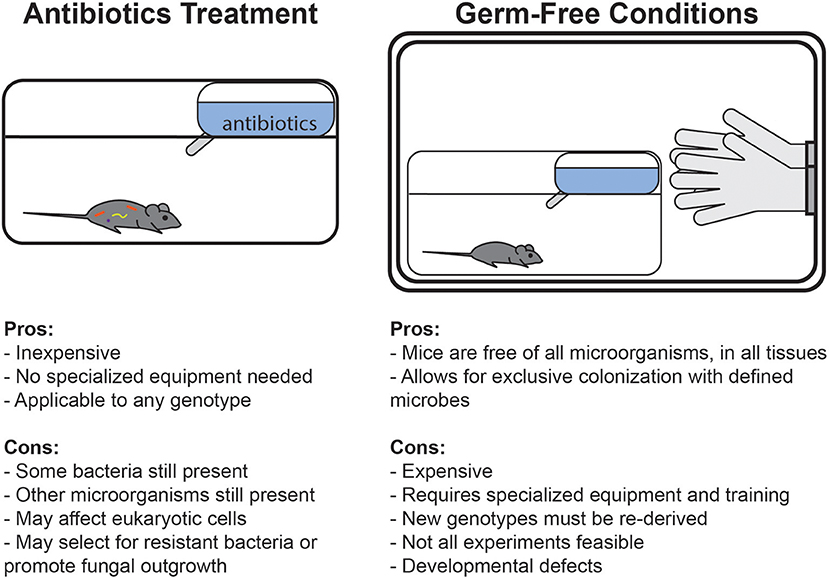
Figure 1. Comparison of the advantages and disadvantages of germ-free and antibiotics-treated mouse model systems.
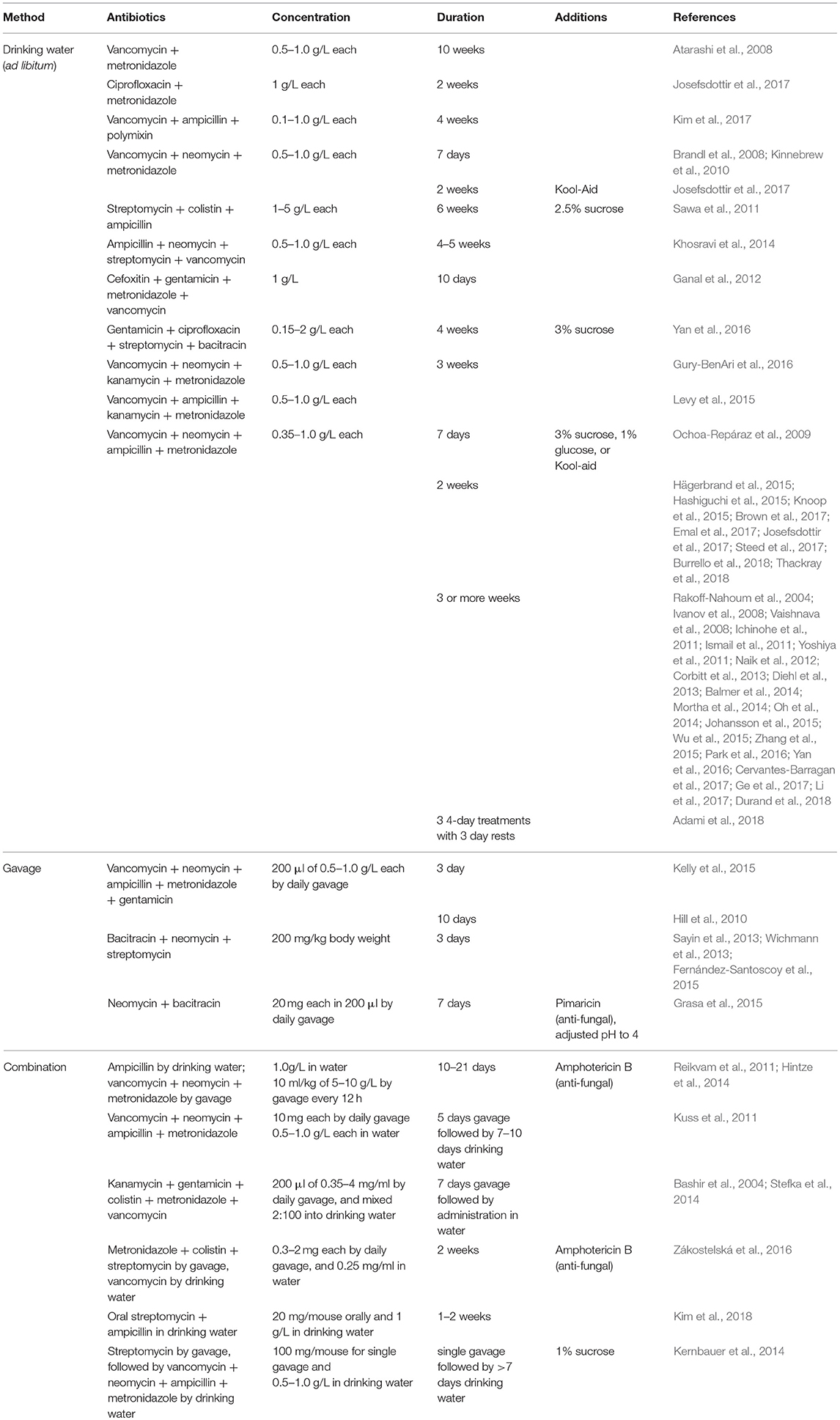
Due to differences in mechanism of action, antibiotics can selectively deplete different members of the microbiota. For example, metronidazole and clindamycin both target anaerobes, vancomycin is only effective against gram-positive bacteria, and polymyxin B specifically targets gram-negative bacteria (Atarashi et al., 2011; Schubert et al., 2015). Individual antibiotics can be used to shift the composition of the gut microbiota in order to identify classes of bacteria relevant to different phenotypes (Schubert et al., 2015; Zackular et al., 2016). In contrast, a cocktail of different classes of antibiotics can be used to broadly deplete the gut microbiota. Researchers have used various regimens which differ in antibiotic combination, dose, and length of treatment (Table 1). All of these combinations broadly target Gram-positive, Gram-negative, and anaerobic bacteria. Often, antibiotics are diluted in drinking water and mice are allowed to drink ad libitum throughout the course of treatment; therefore, actual delivered doses can vary. Some protocols additionally include antifungals in the cocktail to avoid fungal overgrowth during treatment (Reikvam et al., 2011; Grasa et al., 2015; Zákostelská et al., 2016). Many also add sweeteners such as sugar, Splenda, or Kool-aid to mask any bitterness and ensure mice drink the antibiotics-containing water (Abt et al., 2012; Baldridge et al., 2015; Emal et al., 2017). However, there are reports of mice avoiding water and becoming dehydrated when provided antibiotics in this manner (Hill et al., 2010; Reikvam et al., 2011; Zákostelská et al., 2016). Daily oral gavage can prevent dehydration and allow delivery of a precise dose of antibiotics, so this method is sometimes used alone or in combination with delivery in drinking water, though it is more labor-intensive (Kuss et al., 2011; Reikvam et al., 2011).
Validation of bacterial depletion can be performed with culture-based methods by assessing the colony-forming units (CFUs) from fecal samples plated in aerobic and/or anaerobic conditions on non-selective media. However, this method only accounts for cultivatable microbes. Quantitative PCR of the gene encoding 16S rRNA allows for culture-independent assessment of gastrointestinal bacterial load. Broad-spectrum antibiotics treatment can decrease bacterial load by multiple orders of magnitude in 2 weeks of treatment or less (Baldridge et al., 2015; Gonzalez-Perez et al., 2016; Brown et al., 2017). Both germ-free and antibiotics-treated mice allow for the introduction of microbes in which the contributions of defined bacterial species or consortia can be studied (Tan et al., 2016; Staley et al., 2017). Although some phenotypes seen with antibiotics treatment are attributed to removal of a single bacterial species, many differences that occur are due to broad decreases in bacterial load. Consistent with this, treating bacterially-depleted mice with conserved pattern recognition receptor ligands such as flagellin (Ichinohe et al., 2011; Oh et al., 2014) or CpG dinucleotides (Ichinohe et al., 2011; Hill et al., 2012) can restore some defects, even in the absence of microbiota restoration.
Although most studies attribute phenotypic differences after antibiotics treatment to the depletion of gut microbes, some studies have assessed how regimens affect commensal populations at other sites. Oral antibiotic regimens can decrease culturable bacteria in the respiratory tract (Ichinohe et al., 2011; Abt et al., 2012; Brown et al., 2017) and the vagina of mice (Oh et al., 2016), but do not affect skin bacterial communities (Naik et al., 2012). Importantly, though rarely quantified, antibiotics treatment also likely drastically affects bacteriophage populations, though there is debate in the literature about whether phage play important roles in transfer of antibiotic resistance genes (Modi et al., 2013; Enault et al., 2017; Górska et al., 2018). Antibiotics treatment can allow for the outgrowth of commensal fungal species, potentially confounding results as these organisms can alter immune function, hence the inclusion of antifungals in some antibiotics treatment regimens (Noverr et al., 2004; Kim et al., 2014). An increasing appreciation for important roles for the virome and mycobiome may lead to enhanced interrogation of these effects in the future, as well as the potential impact of antibiotics on eukaryotic viruses and archaea (Norman et al., 2014). An important final potential disadvantage of antibiotics treatment can be the evolution or development of antibiotic-resistant bacteria and their subsequent selection and outgrowth in mouse intestines (Zhang et al., 2013; Morgun et al., 2015). Depending upon the starting bacterial composition, antibiotic cocktail, duration of treatment, and phenotypic read-out, antibiotic-resistance may confound findings in experiments, especially if resistant bacteria are present in only a subset of tested mice. Longitudinal analysis of bacterial populations in all experimental groups can aid in detection of resistance and analysis of whether resistant bacteria may affect experimental results.
Mice on antibiotics are not completely cleared of bacteria, but significant reductions in bacterial load are associated with shifts in cell populations, signaling pathways, and organ morphology, with results often paralleling what is seen in germ-free mice.
Effects of the Microbiota on Cell Populations and Cytokines
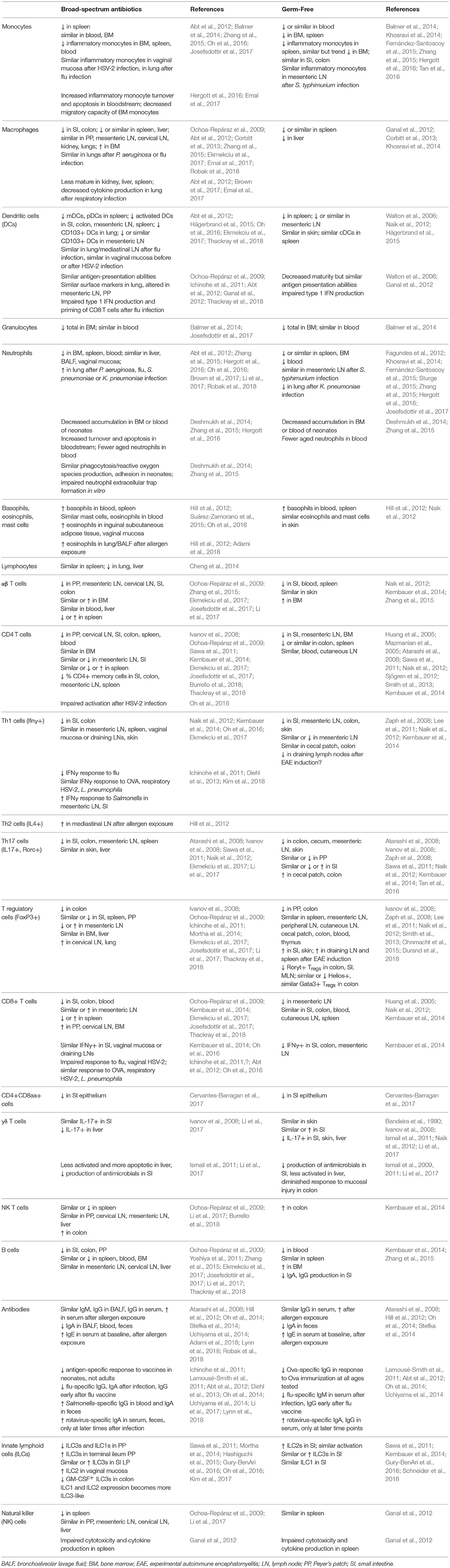
Although many aspects of murine physiology are affected by microbial populations, the effects of antibiotics treatment on immune cell populations are some of the most well-studied (Figure 2). The immune system constantly responds to both pathogenic and commensal microbial populations, and shifts after antibiotics treatment reflect the dependence of cell populations and function on bacterial signals. While results of cell composition analysis are not uniform across studies, we will describe the most prominent and consistent observations (see Table 2 for exact findings by different groups).
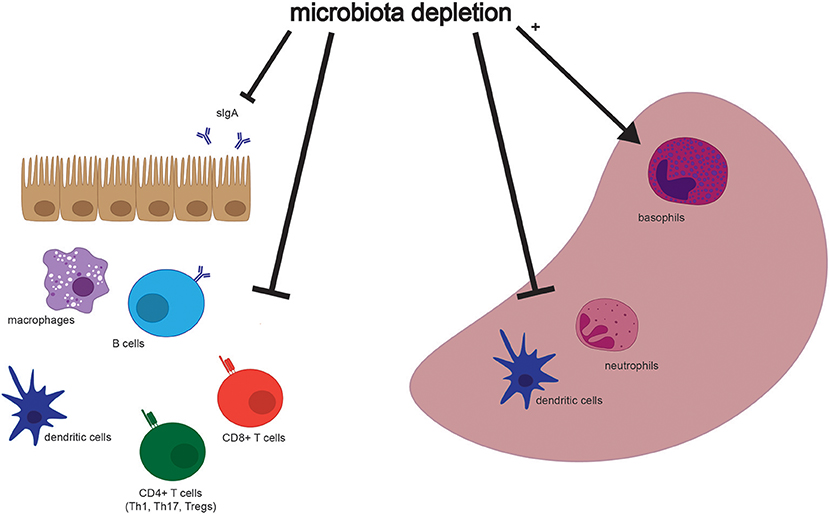
Figure 2. Selected effects of microbiota depletion on cells in the gastrointestinal tract and spleen. Populations of different cell types are altered in association with depletion of the microbiota in both the gastrointestinal tract (Left) and the spleen (Right). Both secretory IgA (sIgA) and immune cell types are depleted in the intestine, while dendritic cells and neutrophils are depleted, but basophils are enriched, in the spleen. Please see Table 2 for more detailed findings by different groups and in other tissues.
Myeloid Cells
Innate immune cells lack antigen-specific receptors, responding instead to broadly conserved microbial patterns. As innate cells continuously interface with the microbial populations constituting the microbiota, sensing of these microbes via pattern recognition receptors is essential for maintenance of normal host physiology (Chu and Mazmanian, 2013; Fawkner-Corbett et al., 2017).
Myeloid cell populations, which include macrophages, monocytes, and granulocytes, are broadly decreased in systemic sites after antibiotics treatment, similar to what is seen in germ-free mice (Khosravi et al., 2014). Although monocytes are generally not diminished in the bone marrow or peripheral blood of mice receiving antibiotics (Zhang et al., 2015; Josefsdottir et al., 2017), these cells have a reduced migratory capacity consistent with their decreased presence in peripheral tissues (Zhang et al., 2015; Emal et al., 2017). In contrast, the effects of antibiotics treatment on inflammatory monocytes and macrophages are more variable, with some groups reporting decreases in blood, bone marrow, and peripheral tissues (Zhang et al., 2015; Hergott et al., 2016; Ekmekciu et al., 2017) and others reporting no significant differences at baseline or after infection (Abt et al., 2012; Oh et al., 2016; Brown et al., 2017; Robak et al., 2018). Even when cell numbers are similar, macrophages are often less mature after antibiotics treatment, impairing their responses to pathogens (Abt et al., 2012; Brown et al., 2017; Emal et al., 2017).
Bulk granulocytes decrease in the bone marrow of antibiotics-treated mice, though their numbers in peripheral blood are similar (Balmer et al., 2014; Josefsdottir et al., 2017). Neutrophils decrease in bone marrow and in peripheral sites, with an increased rate of apoptosis and decrease in aged neutrophils in the bloodstream after microbiota depletion (Deshmukh et al., 2014; Zhang et al., 2015; Hergott et al., 2016). However, neutrophil populations are not diminished at the site of infection after pathogen exposure in antibiotics-treated mice (Abt et al., 2012; Oh et al., 2016; Brown et al., 2017; Robak et al., 2018). In contrast to neutrophils, the proliferation of basophil precursors in the bone marrow is increased after antibiotics treatment, associated with increased basophils in the periphery and an enhanced response to allergen exposure (Hill et al., 2012). Similarly, eosinophils in various tissues are enhanced at baseline (Suárez-Zamorano et al., 2015; Oh et al., 2016), and in the lung after inhaled allergen exposure (Hill et al., 2012). These alterations in granulocyte populations are consistent with a shift from type 1 to type 2 immune responses after depletion of the commensal microbiota.
Various dendritic cell subsets are also reduced after antibiotics treatment at both mucosal and systemic sites (Ichinohe et al., 2011; Ekmekciu et al., 2017; Thackray et al., 2018), although these differences may not be apparent after infection (Abt et al., 2012; Oh et al., 2016). Differences in dendritic cell numbers have not been reported in germ-free mice, though impairment in priming has been seen (Walton et al., 2006; Ganal et al., 2012).
Reductions in innate immune cell number and function, characteristic of both germ-free mice and antibiotics-treated mice, may be explained by diminished cytokine and chemokine levels, which are necessary for normal cell recruitment, differentiation, and functionality (Mortha et al., 2014; Brown et al., 2017). Reductions in myeloid populations are likely not explained by decreases in progenitor populations: although antibiotic exposure beginning in utero can reduce postnatal granulocytosis (Deshmukh et al., 2014), treatment in adult mice does not reduce myeloid progenitor populations in the bone marrow (Josefsdottir et al., 2017; Thackray et al., 2018).
Lymphoid Cells
In contrast to what is seen with myeloid progenitors, common lymphoid progenitors are reduced in the bone marrow after microbiota depletion (Josefsdottir et al., 2017; Thackray et al., 2018), consistent with what is seen in some, but not all, germ-free models (Balmer et al., 2014; Iwamura et al., 2017). Total lymphocytes are similarly reduced in the peripheral blood after antibiotics treatment (Josefsdottir et al., 2017).
The effects of the microbiota in regulating differentiated T cell populations has been widely explored, but results found are somewhat variable. αβ T cells generally decrease in peripheral organs (Ochoa-Repáraz et al., 2009; Zhang et al., 2015; Ekmekciu et al., 2017), although not in the bone marrow or blood (Zhang et al., 2015; Josefsdottir et al., 2017). Similarly, many reports suggest that CD4+ T helper cells decrease in tissues (Ochoa-Repáraz et al., 2009; Kernbauer et al., 2014; Ekmekciu et al., 2017; Josefsdottir et al., 2017; Thackray et al., 2018), as do CD4+ T memory cells (Ekmekciu et al., 2017), although others see either no difference or increases in specific tissues (Ivanov et al., 2008; Sawa et al., 2011; Ekmekciu et al., 2017; Josefsdottir et al., 2017; Burrello et al., 2018). Th1 cells tend to decrease in the gastrointestinal tract (Naik et al., 2012; Kernbauer et al., 2014; Ekmekciu et al., 2017), but not in extra-intestinal tissues (Naik et al., 2012; Oh et al., 2016; Ekmekciu et al., 2017), whereas Th17 cells decrease in most tissues studied (Atarashi et al., 2008; Ivanov et al., 2008; Sawa et al., 2011; Naik et al., 2012; Ekmekciu et al., 2017). The effects of microbial depletion on Th2 cells are less well-studied, although they have been seen to increase in lymph nodes after allergen exposure (Hill et al., 2012). Results with regulatory T cells are inconsistent across studies, with some citing decreases in different tissues (Ochoa-Repáraz et al., 2009; Mortha et al., 2014; Ekmekciu et al., 2017; Thackray et al., 2018), others seeing similar numbers regardless of antibiotics treatment (Ivanov et al., 2008; Ichinohe et al., 2011; Josefsdottir et al., 2017; Li et al., 2017), and still others seeing increases in some sites (Ochoa-Repáraz et al., 2009; Ichinohe et al., 2011).
Similar to CD4+ T cells, cytotoxic CD8+ T cells generally decrease in the intestine after antibiotics, though results at other sites are more varied (Ochoa-Repáraz et al., 2009; Kernbauer et al., 2014; Ekmekciu et al., 2017; Josefsdottir et al., 2017; Thackray et al., 2018). Proinflammatory cytokine production from cytotoxic T cells is not diminished at baseline after antibiotics treatment, but has been reported to decrease in response to infection with some pathogens (Ichinohe et al., 2011; Abt et al., 2012; Kernbauer et al., 2014; Oh et al., 2016).
Reports of B cell population shifts are varied, with some groups seeing declines in the blood, bone marrow, and tissues after antibiotics treatment, and others noting similar numbers regardless of microbiota depletion (Ochoa-Repáraz et al., 2009; Yoshiya et al., 2011; Zhang et al., 2015; Ekmekciu et al., 2017; Josefsdottir et al., 2017; Li et al., 2017; Thackray et al., 2018). Likewise, shifts in antibody responses are inconsistent—in general, total IgG and IgM levels remain similar in different sites analyzed, but secretory and serum IgA levels tend to decrease and serum IgE levels increase after microbiota depletion (Hill et al., 2010; Oh et al., 2014; Uchiyama et al., 2014; Adami et al., 2018; Lynn et al., 2018; Robak et al., 2018). Antigen-specific response to infection or vaccination vary by pathogen, mouse age, and time point after exposure analyzed, but neonates in particular generally produce a less robust response to vaccination after exposure to antibiotics (Ichinohe et al., 2011; Lamousé-Smith et al., 2011; Abt et al., 2012; Diehl et al., 2013; Oh et al., 2014; Uchiyama et al., 2014; Li et al., 2017; Lynn et al., 2018).
Innate-like lymphocytes (including CD8αα+ T cells, γδ T cells, NK T cells) and innate lymphoid cells (ILCs, including natural killer cells) localize to barrier sites and are influenced by the presence of commensal microbes (Constantinides, 2018). Double-positive CD4+CD8αα+ T cells serve regulatory functions in the small intestinal epithelium and are diminished in antibiotics-treated mice, associated with a reduction in the bacterium Lactobacillus reuteri which induces this cell type (Cervantes-Barragan et al., 2017). γδ T cells are present in epithelial tissues, mediating tissue repair and monitoring microbial populations. Although studies in germ-free mice suggest that the microbiota is dispensable for these cells to home to the intestine or skin (Bandeira et al., 1990; Ismail et al., 2011; Naik et al., 2012), both germ-free and antibiotics-treated models indicate that microbial colonization is necessary for normal activation and production of antimicrobial compounds by these cells (Ivanov et al., 2008; Ismail et al., 2009, 2011; Naik et al., 2012; Li et al., 2017). Levels of NK T cells are generally similarly maintained in tissues after antibiotics treatment, though their activation has not been well-studied (Ochoa-Repáraz et al., 2009; Li et al., 2017; Burrello et al., 2018). Likewise, ILCs have not been extensively evaluated after antibiotics treatment, although multiple studies report shifts in the representation or function of ILC subsets at mucosal surfaces (Ochoa-Repáraz et al., 2009; Sawa et al., 2011; Ganal et al., 2012; Mortha et al., 2014; Hashiguchi et al., 2015; Gury-BenAri et al., 2016; Oh et al., 2016; Kim et al., 2017; Li et al., 2017).
Cytokines
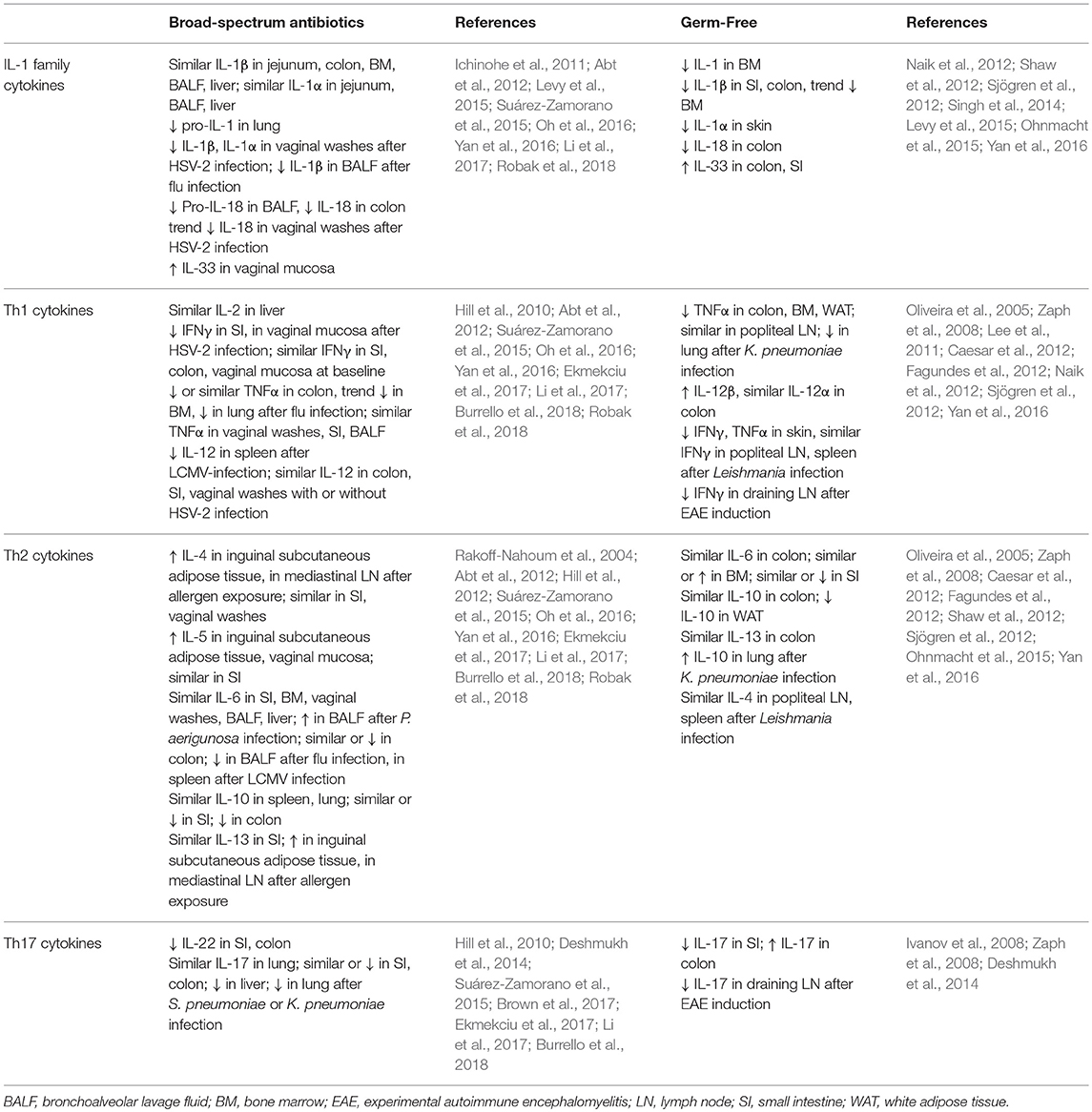
Although shifts in cytokine levels after antibiotics treatment are variable (Table 3), most studies that report differences describe a shift away from proinflammatory cytokines. Many associate microbiota depletion with decreases in the production of IL-1 family cytokines, Th1 cytokines such as IFNγ and TNFα, and IL-17 family cytokines. The production of these cytokines is often similar or reduced specifically in the gastrointestinal tract in naïve animals, but the diminished response becomes apparent after challenge with a pathogen, often at the site of infection. For example, although IL-1 family cytokine levels are similar, pro-IL-1 and pro-IL-18 are reduced in the vaginal mucosa and lung in antibiotics-treated mice at baseline, associated with a reduced production of IL-1 family cytokines in response to infection at each site (Ichinohe et al., 2011; Abt et al., 2012; Oh et al., 2016; Robak et al., 2018). Similarly, type 1 cytokines such as IFNγ and TNFα are generally present at similar levels in tissues after antibiotics treatment in uninfected mice but reduced at the site of infection in microbiota-depleted mice (Abt et al., 2012; Oh et al., 2016; Robak et al., 2018). Inflammatory Th17 cytokines such as IL-17 and IL-22 are generally reduced in the intestines even at baseline in antibiotics-treated mice (Hill et al., 2010; Deshmukh et al., 2014; Ekmekciu et al., 2017), whereas differences in tissues such as the lung become apparent after infection at that site (Deshmukh et al., 2014; Suárez-Zamorano et al., 2015). In parallel with this decrease in inflammatory cytokines, some reports suggest that there is an increase in the expression of Th2 family cytokines such as IL-4, IL-5, and IL-13, especially after allergen exposure, consistent with a shift from Th1 to Th2-type immunity after microbiota depletion (Hill et al., 2010; Suárez-Zamorano et al., 2015; Oh et al., 2016).
Broadly, there is reasonable concordance between germ-free and antibiotics treatment mouse models in alterations of cellular compartments and cytokines. However, different groups have reported disparate findings with different antibiotics treatment regimens, making it challenging to definitively categorize microbiota-mediated modulatory effects. We propose that distinct starting microbiota composition and distinct regimens likely underlie this variability, and highlight this as an area of much-needed standardization.
Microbiota Effects at the Organ Level
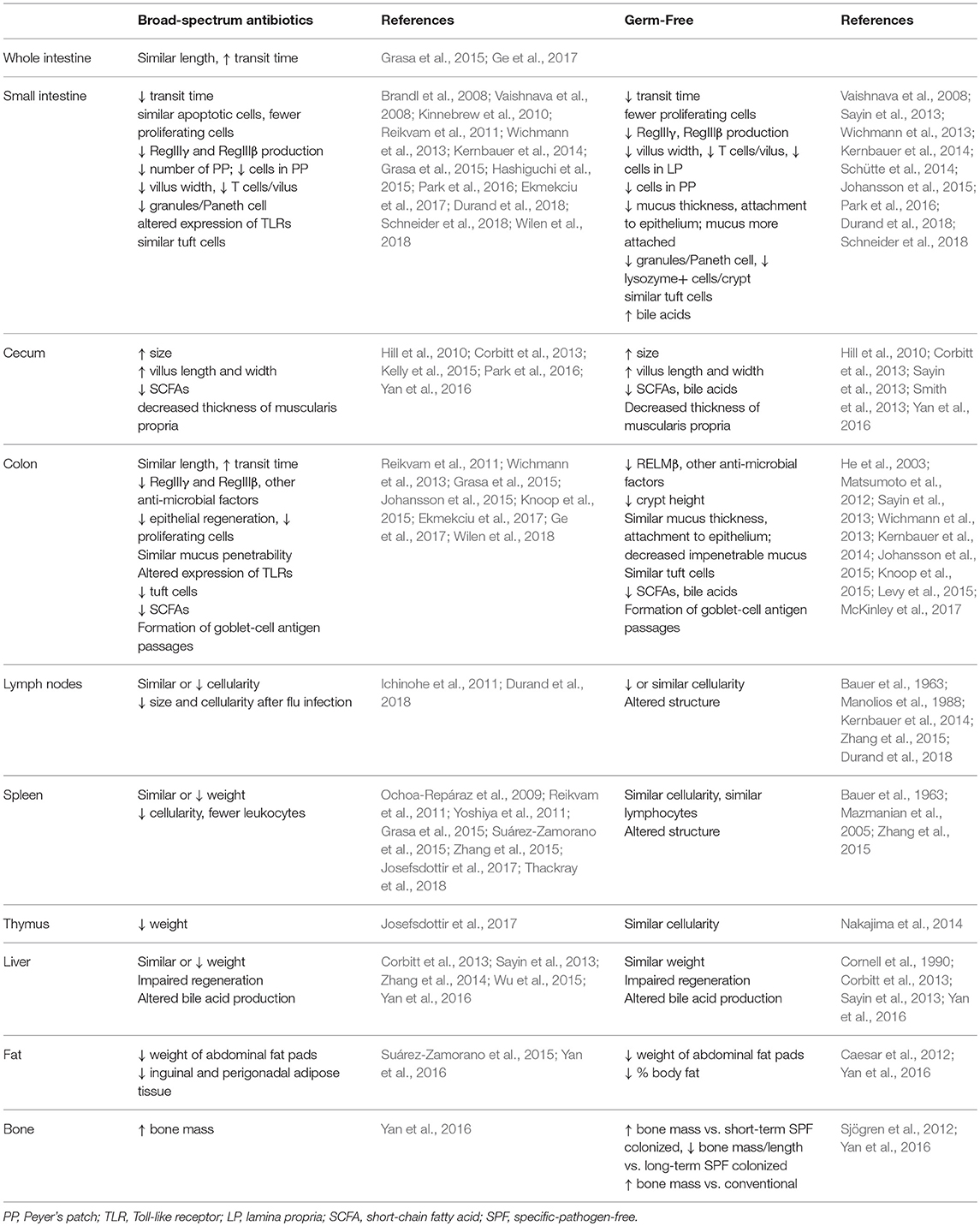
In addition to shifts in cell populations and signaling pathways, antibiotics treatment has been seen to affect organ morphology more broadly, both in the gastrointestinal tract as well as in extra-intestinal organs (Table 4). As the bulk of commensals reside in the gastrointestinal tract where they assist with digestion and interact closely with epithelial cells, it is not surprising that many changes are seen in intestinal physiology after microbial depletion. The length of the whole intestine or the colon is not affected, but the cecum becomes dramatically larger, transit time increases, and fecal pellet frequency and consistency can be altered (Grasa et al., 2015; Suárez-Zamorano et al., 2015; Ge et al., 2017). Moreover, villi become narrower (Kernbauer et al., 2014), cellular proliferation decreases (Reikvam et al., 2011; Ekmekciu et al., 2017), and features such as tuft cells (Wilen et al., 2018) or goblet-cell antigen passages (Knoop et al., 2015) are affected in specific regions of the gastrointestinal tract. Immune function in the intestines is also affected, as the production of antimicrobial peptides is reduced (Brandl et al., 2008; Vaishnava et al., 2008; Kinnebrew et al., 2010; Reikvam et al., 2011), Paneth cells granules are diminished (Kernbauer et al., 2014), Peyer's patches become less abundant and decrease in cellularity (Reikvam et al., 2011; Grasa et al., 2015; Hashiguchi et al., 2015), expression of Toll-like receptors is altered (Grasa et al., 2015), and tolerance to the commensal intestinal microbiota is impaired (Kim et al., 2018).
Non-gastrointestinal organs also depend on ongoing bacterial signals to maintain normal morphology. The spleen (Ochoa-Repáraz et al., 2009; Reikvam et al., 2011; Yoshiya et al., 2011; Zhang et al., 2015; Josefsdottir et al., 2017; Thackray et al., 2018), thymus (Josefsdottir et al., 2017), and lymph nodes (Ichinohe et al., 2011; Durand et al., 2018) may decrease in size and/or cellularity after antibiotics treatment. Liver regeneration is impaired in antibiotics-treated mice (Wu et al., 2015) and bile acid synthesis is altered (Sayin et al., 2013; Zhang et al., 2014). Additionally, fat pads diminish and bone mass increases, consistent with a role for the microbiota in maintaining normal body composition (Suárez-Zamorano et al., 2015; Yan et al., 2016). There have been a number of intriguing studies recently exploring the role of the microbiota in regulating brain function and behavior via the gut-brain axis; this complex topic has been recently reviewed elsewhere (Cryan and Dinan, 2012; Liu and Zhu, 2018).
Microbiota Regulation of Immune Challenges
As might be expected, given the systemic and tissue-specific differences in immune function, antibiotics-treated mice are more susceptible to a variety of pathogens. For example, microbiota-depleted mice are more susceptible to bacterial pathogens such as vancomycin-resistant enterococcus, Salmonella, and Clostridium difficile in the gastrointestinal tract (Kinnebrew et al., 2010; Fernández-Santoscoy et al., 2015; Theriot et al., 2016), a variety of pneumonia-causing bacteria in the respiratory tract (Brown et al., 2017; Robak et al., 2018), and systemic Escherichia coli (Deshmukh et al., 2014). Additionally, after antibiotics treatment, mice are impaired in their response to vaginal HSV-2 (Oh et al., 2016), flaviviruses (Thackray et al., 2018), influenza (Ichinohe et al., 2011; Abt et al., 2012), and cutaneous Leishmania (Naik et al., 2012). However, microbiota-depleted mice actually become less susceptible to enteric viral pathogens including murine norovirus and poliovirus (Kuss et al., 2011; Uchiyama et al., 2014; Baldridge et al., 2015), possibly due to direct interactions between viral pathogens and enteric bacteria or due to loss of specific cell types required for viral infection. Antibiotics-treated mice are additionally impaired in their development of tolerance to food antigens (Bashir et al., 2004; Kim et al., 2018) and are more prone to allergic diseases (Hill et al., 2012; Adami et al., 2018).
Many of the effects after antibiotics treatment in mice are consistent with what is seen in germ-free models, suggesting that these are dependent on regular signals from the microbiota. However, it is important to note that antibiotics can have effects on eukaryotes independently of the microbiota, as treatment of germ-free mice with antibiotics can replicate some findings seen when treating normally colonized mice (Han et al., 2015; Gopinath et al., 2018). Replicating key findings in germ-free mice can help confirm that the differences seen after antibiotics treatment are indeed caused by microbial depletion.
Considerations for the Future
While antibiotics treatment offers an inexpensive and accessible alternative to germ-free models, results obtained using these regimens come with the caveats of potential off-target drug effects and incomplete or inconsistent ablation of microbes. Additionally, because so many groups use distinct treatment regimens and mouse microbial populations may be institution-specific, antibiotics studies are much more challenging to compare than germ-free mouse studies.
We suggest that some standardization of antibiotics treatment regimens would be helpful; for example, if a standard cocktail were employed to demonstrate an initial finding, this could be compared to other studies, and subsequent follow-up experiments could be done with modified cocktails as necessary. Additionally, we suggest that at least a limited assessment of the replicability of findings in antibiotics-treated mice and germ-free mice would be of high value for most studies, to rule in or out potential off-target drug effects or developmental differences between germ-free and standard pathogen-free mice that may be important for a phenotype. Finally, it will be critical for investigators to ensure that microbial loads are consistently monitored in both antibiotics treatment and germ-free models to identify any effects of contaminants or antibiotic-resistant microbes.
Ensuring that we are able to interpret the contribution of an individual study to the field of microbiota research will require careful planning and execution of these experiments on the part of investigators. As we continue to uncover additional health and disease states in which the microbiota plays a role, the use of these models will become increasingly common.
Author Contributions
EK and MB wrote and edited the manuscript. KK edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MB was supported by NIH grant K22 AI127846, Digestive Diseases Research Core Centers P30 DK052574, and the Global Probiotics Council's Young Investigator Grant for Probiotics Research. KK was supported by NIH grants R01 HL136333 and R01 HL134880. EK was supported by NSF Graduate Research Fellowship DGE-1745038.
References
Abt, M. C., Osborne, L. C., Monticelli, L. A., Doering, T. A., Alenghat, T., Sonnenberg, G. F., et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. doi: 10.1016/j.immuni.2012.04.011
Adami, A. J., Bracken, S. J., Guernsey, L. A., Rafti, E., Maas, K. R., Graf, J., et al. (2018). Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr. Res. doi: 10.1038/s41390-018-0031-y [Epub ahead of print].
Al-Asmakh, M., and Zadjali, F. (2015). Use of germ-free animal models in microbiota-related research. J. Microbiol. Biotechnol. 25, 1583–1588. doi: 10.4014/jmb.1501.01039
Atarashi, K., Nishimura, J., Shima, T., Umesaki, Y., Yamamoto, M., Onoue, M., et al. (2008). ATP drives lamina propria TH17 cell differentiation. Nature 455, 808–812. doi: 10.1038/nature07240
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells. Science 331, 337–342. doi: 10.1126/science.1198469
Baldridge, M. T., Nice, T. J., McCune, B. T., Yokoyama, C. C., Kambal, A., Wheadon, M., et al. (2015). Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347, 266–269. doi: 10.1126/science.1258025
Balmer, M. L., Schürch, C. M., Saito, Y., Geuking, M. B., Li, H., Cuenca, M., et al. (2014). Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193, 5273–5283. doi: 10.4049/jimmunol.1400762
Bandeira, A., Mota-Santos, T., Itohara, S., Degermann, S., Heusser, C., Tonegawa, S., et al. (1990). Localization of gamma/delta T cells to the intestinal epithelium is independent of normal microbial colonization. J. Exp. Med. 172, 239–244. doi: 10.1084/jem.172.1.239
Bashir, M. E. H., Louie, S., Shi, H. N., and Nagler-Anderson, C. (2004). Toll-Like Receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J. Immunol. 172, 6978–6987. doi: 10.4049/jimmunol.172.11.6978
Bauer, H., Horowitz, R., Levenson, S., and Popper, H. (1963). The response of the lymphatic tissue to the microbial flora. Am. J. Pathol. 42, 471–483.
Brandl, K., Plitas, G., Mihu, C. N., Ubeda, C., Jia, T., Fleisher, M., et al. (2008). Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807. doi: 10.1038/nature07250
Brown, R. L., Sequeira, R. P., and Clarke, T. B. (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 8:1512. doi: 10.1038/s41467-017-01803-x
Burrello, C., Garavaglia, F., Cribiù, F. M., Ercoli, G., Bosari, S., Caprioli, F., et al. (2018). Short-term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural killer T and conventional CD4+T cells. Front. Med. 5:21. doi: 10.3389/fmed.2018.00021
Caesar, R., Reigstad, C. S., Backhed, H. K., Reinhardt, C., Ketonen, M., Lunden, G., et al. (2012). Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 61, 1701–1707. doi: 10.1136/gutjnl-2011-301689
Cervantes-Barragan, L., Chai, J. N., Tianero, M. D., Di Luccia, B., Ahern, P. P., Merriman, J., et al. (2017). Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357, 806–810. doi: 10.1126/science.aah5825
Charles River Germ-Free Mouse Report (2018). Available online at: https://www.criver.com/HealthData/na/H01W8G0M.pdf
Cheng, M., Qian, L., Shen, G., Bian, G., Xu, T., Xu, W., et al. (2014). Microbiota modulate tumoral immune surveillance in lung through a γδT17 immune cell-dependent mechanism. Cancer Res. 74, 4030–4041. doi: 10.1158/0008-5472.CAN-13-2462
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Constantinides, M. G. (2018). Interactions between the microbiota and innate and innate-like lymphocytes. J. Leukoc. Biol. 103, 409–419. doi: 10.1002/JLB.3RI0917-378R
Corbitt, N., Kimura, S., Isse, K., Specht, S., Chedwick, L., Rosborough, B. R., et al. (2013). Gut bacteria drive kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. Am. J. Pathol. 182, 180–191. doi: 10.1016/j.ajpath.2012.09.010
Cornell, R. P., Liljequist, B. L., and Bartizal, K. F. (1990). Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology 11, 916–922. doi: 10.1002/hep.1840110603
Cryan, J. F., and Dinan, T. G. (2012). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. doi: 10.1038/nrn3346
Deshmukh, H. S., Liu, Y., Menkiti, O. R., Mei, J., Dai, N., O'Leary, C. E., et al. (2014). The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530. doi: 10.1038/nm.3542
Diehl, G. E., Longman, R. S., Zhang, J. X., Breart, B., Galan, C., Cuesta, A., et al. (2013). Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX 3 CR1 hi cells. Nature 494, 116–120. doi: 10.1038/nature11809
Durand, A., Audemard-Verger, A., Guichard, V., Mattiuz, R., Delpoux, A., Hamon, P., et al. (2018). Profiling the lymphoid-resident T cell pool reveals modulation by age and microbiota. Nat. Commun. 9:68. doi: 10.1038/s41467-017-02458-4
Ekmekciu, I., von Klitzing, E., Fiebiger, U., Escher, U., Neumann, C., Bacher, P., et al. (2017). Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front. Immunol. 8:397. doi: 10.3389/fimmu.2017.00397
Emal, D., Rampanelli, E., Stroo, I., Butter, L. M., Teske, G. J., Claessen, N., et al. (2017). Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 28, 1450–1461. doi: 10.1681/ASN.2016030255
Enault, F., Briet, A., Bouteille, L., Roux, S., Sullivan, M. B., and Petit, M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247. doi: 10.1038/ismej.2016.90
Fagundes, C. T., Amaral, F. A., Vieira, A. T., Soares, A. C., Pinho, V., Nicoli, J. R., et al. (2012). Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 188, 1411–1420. doi: 10.4049/jimmunol.1101682
Fawkner-Corbett, D., Simmons, A., and Parikh, K. (2017). Microbiome, pattern recognition receptor function in health and inflammation. Best Pract. Res. Clin. Gastroenterol. 31, 683–691. doi: 10.1016/j.bpg.2017.11.001
Fernández-Santoscoy, M., Wenzel, U. A., Yrlid, U., Cardell, S., Bäckhed, F., and Wick, M. J. (2015). The gut microbiota reduces colonization of the mesenteric lymph nodes and IL-12-independent IFN-γ production during salmonella infection. Front. Cell. Infect. Microbiol. 5:93. doi: 10.3389/fcimb.2015.00093
Fontaine, C. A., Skorupski, A. M., Vowles, C. J., Anderson, N. E., Poe, S. A., and Eaton, K. A. (2015). How free of germs is germ-free? Detection of bacterial contamination in a germ free mouse unit. Gut Microbes 6, 225–233. doi: 10.1080/19490976.2015.1054596
Ganal, S. C., Sanos, S. L., Kallfass, C., Oberle, K., Johner, C., Kirschning, C., et al. (2012). Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 37, 171–186. doi: 10.1016/j.immuni.2012.05.020
Ge, X., Ding, C., Zhao, W., Xu, L., Tian, H., Gong, J., et al. (2017). Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J. Transl. Med. 15:13. doi: 10.1186/s12967-016-1105-4
Gonzalez-Perez, G., Hicks, A. L., Tekieli, T. M., Radens, C. M., Williams, B. L., and Lamousé-Smith, E. S. N. (2016). Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J. Immunol. 196, 3768–3779. doi: 10.4049/jimmunol.1502322
Gopinath, S., Kim, M. V., Rakib, T., Wong, P. W., van Zandt, M., Barry, N. A., et al. (2018). Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat. Microbiol. 3, 611–621. doi: 10.1038/s41564-018-0138-2
Górska, A., Peter, S., Willmann, M., Autenrieth, I., Schlaberg, R., and Huson, D. H. (2018). Dynamics of the human gut phageome during antibiotic treatment. Comput. Biol. Chem. 74, 420–427. doi: 10.1016/j.compbiolchem.2018.03.011
Grasa, L., Abecia, L., Forcén, R., Castro, M., de Jalón, J. A. G., Latorre, E., et al. (2015). Antibiotic-induced depletion of murine microbiota induces mild inflammation and changes in toll-like receptor patterns and intestinal motility. Microb. Ecol. 70, 835–848. doi: 10.1007/s00248-015-0613-8
Gury-BenAri, M., Thaiss, C. A., Serafini, N., Winter, D. R., Giladi, A., Lara-Astiaso, D., et al. (2016). The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13. doi: 10.1016/j.cell.2016.07.043
Hägerbrand, K., Westlund, J., Yrlid, U., Agace, W., and Johansson-Lindbom, B. (2015). MyD88 signaling regulates steady-state migration of intestinal CD103 + dendritic cells independently of TNF-α and the gut microbiota. J. Immunol. 195, 2888–2899. doi: 10.4049/jimmunol.1500210
Han, D., Walsh, M. C., Kim, K. S., Hong, S. W., Lee, J., Yi, J., et al. (2015). Microbiota-independent ameliorative effects of antibiotics on spontaneous Th2-associated pathology of the small intestine. PLoS ONE 10:e0133787. doi: 10.1371/journal.pone.0118795
Hashiguchi, M., Kashiwakura, Y., Kojima, H., Kobayashi, A., Kanno, Y., and Kobata, T. (2015). Peyer's patch innate lymphoid cells regulate commensal bacteria expansion. Immunol. Lett. 165, 1–9. doi: 10.1016/j.imlet.2015.03.002
He, W., Wang, M. -L., Jiang, H. -Q., Steppan, C. M., Shin, M. E., Thurnheer, M. C., et al. (2003). Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology 125, 1388–1397. doi: 10.1016/j.gastro.2003.07.009
Hergott, C. B., Roche, A. M., Tamashiro, E., Clarke, T. B., Bailey, A. G., Laughlin, A., et al. (2016). Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471. doi: 10.1182/blood-2015-10-675173
Hill, D. A., Hoffmann, C., Abt, M. C., Du, Y., Kobuley, D., Kirn, T. J., et al. (2010). Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158. doi: 10.1038/mi.2009.132
Hill, D. A., Siracusa, M. C., Abt, M. C., Kim, B. S., Kobuley, D., Kubo, M., et al. (2012). Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18, 538–546. doi: 10.1038/nm.2657
Hintze, K. J., Cox, J. E., Rrompato, G., Benninghoff, A. D., Ward, R., Broadbent, J., et al. (2014). Broad scope method for creating humanized animal models for animal health and disease research through antibiotic treatment and human fecal transfer. Gut Microbes 5, 183–191. doi: 10.4161/gmic.28403
Huang, T., Wei, B., Velazquez, P., Borneman, J., and Braun, J. (2005). Commensal microbiota alter the abundance and TCR responsiveness of splenic naïve CD4+T lymphocytes. Clin. Immunol. 117, 221–230. doi: 10.1016/j.clim.2005.09.012
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract in fluenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Ismail, A. S., Behrendt, C. L., and Hooper, L. V. (2009). Reciprocal Interactions between commensal bacteria and intraepithelial lymphocytes during mucosal injury. J. Immunol. 182, 3047–3054. doi: 10.4049/jimmunol.0802705
Ismail, A. S., Severson, K. M., Vaishnava, S., Behrendt, C. L., Yu, X., Benjamin, J. L., et al. (2011). Intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc. Natl. Acad. Sci. U.S.A. 108, 8743–8748. doi: 10.1073/pnas.1019574108
Ivanov, I. I., de Frutos, R. L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. doi: 10.1016/j.chom.2008.09.009
Iwamura, C., Bouladoux, N., Belkaid, Y., Sher, A., and Jankovic, D. (2017). Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood 129, 171–176. doi: 10.1182/blood-2016-06-723742
Johansson, M. E. V., Jakobsson, H. E., Holmén-Larsson, J., Schütte, A., Ermund, A., Rodríguez-Piñeiro, A. M., et al. (2015). Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18, 582–592. doi: 10.1016/j.chom.2015.10.007
Josefsdottir, K. S., Baldridge, M. T., Kadmon, C. S., and King, K. Y. (2017). Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129, 729–739. doi: 10.1182/blood-2016-03-708594
Kelly, C. J., Zheng, L., Campbell, E. L., Saeedi, B., Scholz, C. C., Bayless, A. J., et al. (2015). Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671. doi: 10.1016/j.chom.2015.03.005
Kernbauer, E., Ding, Y., and Cadwell, K. (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. doi: 10.1038/nature13960
Khosravi, A., Yáñez, A., Price, J. G., Chow, A., Merad, M., Goodridge, H. S., et al. (2014). Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15, 374–381. doi: 10.1016/j.chom.2014.02.006
Kim, M., Galan, C., Hill, A. A., Wu, W. J., Fehlner-Peach, H., Song, H. W., et al. (2018). Critical role for the microbiota in CX3CR1+intestinal mononuclear phagocyte regulation of intestinal t cell responses. Immunity 49, 151.e5–163.e5. doi: 10.1016/j.immuni.2018.05.009
Kim, S. H., Cho, B. H., Kiyono, H., and Jang, Y. S. (2017). Microbiota-derived butyrate suppresses group 3 innate lymphoid cells in terminal ileal Peyer's patches. Sci. Rep. 7:3980. doi: 10.1038/s41598-017-02729-6
Kim, Y. G., Udayanga, K. G. S., Totsuka, N., Weinberg, J. B., Núñez, G., and Shibuya, A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 15, 95–102. doi: 10.1016/j.chom.2013.12.010
Kinnebrew, M. A., Ubeda, C., Zenewicz, L. A., Smith, N., Flavell, R. A., and Pamer, E. G. (2010). Bacterial flagellin stimulates toll-like receptor 5–dependent defense against vancomycin-resistant enterococcus infection. J. Infect. Dis. 201, 534–543. doi: 10.1086/650203
Knoop, K. A., McDonald, K. G., McCrate, S., McDole, J. R., and Newberry, R. D. (2015). Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8, 198–210. doi: 10.1038/mi.2014.58
Kuss, S. K., Best, G. T., Etheredge, C. A., Pruijssers, A. J., Frierson, J. M., Hooper, L. V., et al. (2011). Intestinal microbiota promote enteric virus replication and systemic Pathogenesis. Science 334, 249–252. doi: 10.1126/science.1211057
Lai, H. -C., Young, J., Lin, C. -S., Chang, C. -J., Lu, C. -C., Martel, J., et al. (2014). Impact of the gut microbiota, prebiotics, and probiotics on human health and disease. Biomed. J. 37, 259–268. doi: 10.4103/2319-4170.138314
Lamousé-Smith, E. S., Tzeng, A., and Starnbach, M. N. (2011). The intestinal flora is required to support antibody responses to systemic immunization in infant and germ free mice. PLoS ONE 6:e27662. doi: 10.1371/journal.pone.0027662
Lee, Y. K., Menezes, J. S., Umesaki, Y., and Mazmanian, S. K. (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108, 4615–4622. doi: 10.1073/pnas.1000082107
Levy, M., Thaiss, C. A., Zeevi, D., Dohnalová, L., Zilberman-Schapira, G., Mahdi, J. A., et al. (2015). Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443. doi: 10.1016/j.cell.2015.10.048
Li, F., Hao, X., Chen, Y., Bai, L., Gao, X., Lian, Z., et al. (2017). The microbiota maintain homeostasis of liver-resident γδ T-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 7:13839. doi: 10.1038/ncomms13839
Liu, L., and Zhu, G. (2018). Gut-brain axis and mood disorder. Front. Psychiatry 9:1. doi: 10.3389/fpsyt.2018.00001
Lynn, M. A., Tumes, D. J., Choo, J. M., Sribnaia, A., Blake, S. J., Leong, L. E. X., et al. (2018). Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe 23, 653–660.e5. doi: 10.1016/j.chom.2018.04.009
Manolios, N., Geczy, C. L., and Schrieber, L. (1988). High endothelial venule morphology and function are inducible in germ-free mice: a possible role for interferon-gamma. Cell. Immunol. 117, 136–151. doi: 10.1016/0008-8749(88)90083-4
Matsumoto, M., Kibe, R., Ooga, T., Aiba, Y., Kurihara, S., Sawaki, E., et al. (2012). Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2:233. doi: 10.1038/srep00233
Mazmanian, S. K., Cui, H. L., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
McKinley, E. T., Sui, Y., Al-Kofahi, Y., Millis, B. A., Tyska, M. J., Roland, J. T., et al. (2017). Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight 2:93487. doi: 10.1172/jci.insight.93487
Modi, S. R., Lee, H. H., Spina, C. S., and Collins, J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222. doi: 10.1038/nature12212
Morgun, A., Dzutsev, A., Dong, X., Greer, R. L., Sexton, D. J., Ravel, J., et al. (2015). Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 64, 1732–1743. doi: 10.1136/gutjnl-2014-308820
Mortha, A., Chudnovskiy, A., Hashimoto, D., Bogunovic, M., Spencer, S. P., Belkaid, Y., et al. (2014). Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343:1249288. doi: 10.1126/science.1249288
Naik, S., Bouladoux, N., Wilhelm, C., Molloy, M. J., Salcedo, R., Kastenmuller, W., et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119. doi: 10.1126/science.1225152
Nakajima, A., Negishi, N., Tsurui, H., Kadowaki-Ohtsuji, N., Maeda, K., Nanno, M., et al. (2014). Commensal bacteria regulate thymic Aire expression. PLoS ONE 9:e105904. doi: 10.1371/journal.pone.0105904
Nicklas, W., Keubler, L., and Bleich, A. (2015). Maintaining and monitoring the defined microbiota status of gnotobiotic rodents. ILAR J. 56, 241–249. doi: 10.1093/ilar/ilv029
Norman, J. M., Handley, S. A., and Virgin, H. W. (2014). Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 146, 1459–1469. doi: 10.1053/j.gastro.2014.02.001
Noverr, M. C., Noggle, R. M., Toews, G. B., and Huffnagle, G. B. (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72, 4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004
Ochoa-Repáraz, J., Mielcarz, D. W., Ditrio, L. E., Burroughs, A. R., Foureau, D. M., Haque-Begum, S., et al. (2009). Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 183, 6041–6050. doi: 10.4049/jimmunol.0900747
Oh, J. E., Kim, B. -C., Chang, D. -H., Kwon, M., Lee, S. Y., Kang, D., et al. (2016). Dysbiosis-induced IL-33 contributes to impaired antiviral immunity in the genital mucosa. Proc. Natl. Acad. Sci. U.S.A. 113, E762–E771. doi: 10.1073/pnas.1518589113
Oh, J. Z., Ravindran, R., Chassaing, B., Carvalho, F. A., Maddur, M. S., Bower, M., et al. (2014). TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492. doi: 10.1016/j.immuni.2014.08.009
Ohnmacht, C., Park, J. -H., Cording, S., Wing, J. B., Atarashi, K., Obata, Y., et al. (2015). The microbiota regulates type 2 immunity through ROR t+ T cells. Science 349, 989–993. doi: 10.1126/science.aac4263
Oliveira, M. R., Tafuri, W. L., Afonso, L. C. C., Oliveira, M. A. P., Nicoli, J. R., Vieira, E. C., et al. (2005). Germ-free mice produce high levels of interferon-gamma in response to infection with Leishmania major but fail to heal lesions. Parasitology 131, 477–488. doi: 10.1017/S0031182005008073
Palm, N. W., de Zoete, M. R., and Flavell, R. A. (2015). Immune-microbiota interactions in health and disease. Clin. Immunol. 159, 122–127. doi: 10.1016/j.clim.2015.05.014
Park, J., Kotani, T., Konno, T., Setiawan, J., Kitamura, Y., Imada, S., et al. (2016). Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS ONE 11:e0156334. doi: 10.1371/journal.pone.0156334
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S., and Medzhitov, R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. doi: 10.1016/j.cell.2004.07.002
Reikvam, D. H., Erofeev, A., Sandvik, A., Grcic, V., Jahnsen, F. L., Gaustad, P., et al. (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS ONE 6:e17996. doi: 10.1371/journal.pone.0017996
Robak, O. H., Heimesaat, M. M., Kruglov, A. A., Prepens, S., Ninnemann, J., Gutbier, B., et al. (2018). Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 128, 3535–3545. doi: 10.1172/JCI97065
Sawa, S., Lochner, M., Satoh-Takayama, N., Dulauroy, S., Bérard, M., Kleinschek, M., et al. (2011). RORγt+innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 12, 320–328. doi: 10.1038/ni.2002
Sayin, S. I., Wahlström, A., Felin, J., Jäntti, S., Marschall, H. U., Bamberg, K., et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235. doi: 10.1016/j.cmet.2013.01.003
Schneider, C., O'Leary, C. E., von Moltke, J., Liang, H. E., Ang, Q. Y., Turnbaugh, P. J., et al. (2018). A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271.e14–284.e14. doi: 10.1016/j.cell.2018.05.014
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against clostridium difficile. mBio 6:e00974–15. doi: 10.1128/mBio.00974-15
Schütte, A., Ermund, A., Becker-Pauly, C., Johansson, M. E. V., Rodriguez-Pineiro, A. M., Backhed, F., et al. (2014). Microbial-induced meprin cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc. Natl. Acad. Sci. U.S.A. 111, 12396–12401. doi: 10.1073/pnas.1407597111
Shaw, M. H., Kamada, N., Kim, Y. -G., and Núñez, G. (2012). Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state T H 17 cells in the intestine. J. Exp. Med. 209, 251–258. doi: 10.1084/jem.20111703
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Sjögren, K., Engdahl, C., Henning, P., Lerner, U. H., Tremaroli, V., Lagerquist, M. K., et al. (2012). The gut microbiota regulates bone mass in mice. J. Bone Miner. Res. 27, 1357–1367. doi: 10.1002/jbmr.1588
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Staley, C., Kaiser, T., Beura, L. K., Hamilton, M. J., Weingarden, A. R., Bobr, A., et al. (2017). Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5:87. doi: 10.1186/s40168-017-0306-2
Steed, A. L., Christophi, G. P., Kaiko, G. E., Sun, L., Goodwin, V. M., Jain, U., et al. (2017). The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502. doi: 10.1126/science.aam5336
Stefka, A. T., Feehley, T., Tripathi, P., Qiu, J., McCoy, K., Mazmanian, S. K., et al. (2014). Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. U.S.A. 111, 13145–13150. doi: 10.1073/pnas.1412008111
Sturge, C. R., Burger, E., Raetz, M., Hooper, L. V., and Yarovinsky, F. (2015). Cutting edge: developmental regulation of IFN-γ production by mouse neutrophil precursor cells. J. Immunol. 195, 36–40. doi: 10.4049/jimmunol.1500366
Suárez-Zamorano, N., Fabbiano, S., Chevalier, C., Stojanović, O., Colin, D. J., Stevanović, A., et al. (2015). Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 21, 1497–1501. doi: 10.1038/nm.3994
Tan, T. G., Sefik, E., Geva-Zatorsky, N., Kua, L., Naskar, D., Teng, F., et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E8141–E8150. doi: 10.1073/pnas.1617460113
Thackray, L. B., Handley, S. A., Gorman, M. J., Poddar, S., Bagadia, P., Briseño, C. G., et al. (2018). Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 22, 3440–3453.e6. doi: 10.1016/j.celrep.2018.03.001
Theriot, C. M., Bowman, A. A., and Young, V. B. (2016). Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045–15. doi: 10.1128/mSphere.00045-15
Uchiyama, R., Chassaing, B., Zhang, B., and Gewirtz, A. T. (2014). Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 210, 171–182. doi: 10.1093/infdis/jiu037
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L., and Hooper, L. V. (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105, 20858–20863. doi: 10.1073/pnas.0808723105
Walton, K. L. W., He, J., Kelsall, B. L., Sartor, R. B., and Fisher, N. C. (2006). Dendritic cells in germ-free and specific pathogen-free mice have similar phenotypes and in vitro antigen presenting function. Immunol. Lett. 102, 16–24. doi: 10.1016/j.imlet.2005.07.001
Wichmann, A., Allahyar, A., Greiner, T. U., Plovier, H., Lundén, G. Ö., Larsson, T., et al. (2013). Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14, 582–590. doi: 10.1016/j.chom.2013.09.012
Wilen, C. B., Lee, S., Hsieh, L. L., Orchard, R. C., Desai, C., Hykes, B. L., et al. (2018). Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208. doi: 10.1126/science.aar3799
Wu, X., Sun, R., Chen, Y., Zheng, X., Bai, L., Lian, Z., et al. (2015). Oral Ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology 62, 253–264. doi: 10.1002/hep.27791
Yan, J., Herzog, J. W., Tsang, K., Brennan, C. A., Bower, M. A., Garrett, W. S., et al. (2016). Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. U.S.A. 113, E7554–E7563. doi: 10.1073/pnas.1607235113
Yi, P., and Li, L. J. (2012). The germfree murine animal: an important animal model for research on the relationship between gut microbiota and the host. Vet. Microbiol. 157, 1–7. doi: 10.1016/j.vetmic.2011.10.024
Yoshiya, K., Lapchak, P. H., Thai, T. -H., Kannan, L., Rani, P., Lucca, J. J. D., et al. (2011). Depletion of gut commensal bacteria attenuates intestinal ischemia/reperfusion injury. Am. J. Physiol. Liver Physiol. 301, G1020–G1030. doi: 10.1152/ajpgi.00239.2011
Zackular, J. P., Baxter, N. T., Chen, G. Y., and Schloss, P. D. (2016). Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere 1, e00001–15. doi: 10.1128/mSphere.00001-15
Zákostelská, Z., Málková, J., Klimešová, K., Rossmann, P., Hornová, M., Novosádová, I., et al. (2016). Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing Th17 response. PLoS ONE 11:e0159539. doi: 10.1371/journal.pone.0159539
Zaph, C., Du, Y., Saenz, S. A., Nair, M. G., Perrigoue, J. G., Taylor, B. C., et al. (2008). Commensal-dependent expression of IL-25 regulates the IL-23–IL-17 axis in the intestine. J. Exp. Med. 205, 2191–2198. doi: 10.1084/jem.20080720
Zhang, D., Chen, G., Manwani, D., Mortha, A., Xu, C., Faith, J. J., et al. (2015). Neutrophil ageing is regulated by the microbiome. Nature 525, 528–532. doi: 10.1038/nature15367
Zhang, L., Huang, Y., Zhou, Y., Buckley, T., and Wang, H. H. (2013). Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 57, 3659–3666. doi: 10.1128/AAC.00670-13
Keywords: antibiotics, microbiota, gnotobiotic, microbiome, immunity
Citation: Kennedy EA, King KY and Baldridge MT (2018) Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 9:1534. doi: 10.3389/fphys.2018.01534
Received: 03 August 2018; Accepted: 11 October 2018;
Published: 31 October 2018.
Edited by:
Stephen J. Pandol, Cedars-Sinai Medical Center, United StatesReviewed by:
Aline Silva Miranda, Universidade Federal de Minas Gerais, BrazilBreck A. Duerkop, University of Colorado, United States
Copyright © 2018 Kennedy, King and Baldridge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Megan T. Baldridge, bWJhbGRyaWRnZUB3dXN0bC5lZHU=
 Elizabeth A. Kennedy1
Elizabeth A. Kennedy1 Katherine Y. King
Katherine Y. King Megan T. Baldridge
Megan T. Baldridge