Corrigendum: Long Non-coding RNA Structure and Function: Is There a Link?
- 1King’s British Heart Foundation Centre, King’s College London, London, United Kingdom
- 2Faculty of Science and Technology, Middlesex University, London, United Kingdom
- 3Department of Informatics, King’s College London, London, United Kingdom
RNA has emerged as the prime target for diagnostics, therapeutics and the development of personalized medicine. In particular, the non-coding RNAs (ncRNAs) that do not encode proteins, display remarkable biochemical versatility. They can fold into complex structures and interact with proteins, DNA and other RNAs, modulating the activity, DNA targets or partners of multiprotein complexes. Thus, ncRNAs confer regulatory plasticity and represent a new layer of epigenetic control that is dysregulated in disease. Intriguingly, for long non-coding RNAs (lncRNAs, >200 nucleotides length) structural conservation rather than nucleotide sequence conservation seems to be crucial for maintaining their function. LncRNAs tend to acquire complex secondary and tertiary structures and their functions only impose very subtle sequence constraints. In the present review we will discuss the biochemical assays that can be employed to determine the lncRNA structural configurations. The implications and challenges of linking function and lncRNA structure to design novel RNA therapeutic approaches will also be analyzed.
Introduction
The HUMAN GENOME project has transformed our understanding of the basic unit of genetic information with RNA emerging as a versatile regulator of central cellular processes (Thum and Condorelli, 2015). The non-coding RNAs (ncRNAs), transcripts that do not encode proteins comprise the biggest class and are arbitrarily divided into small (<200 nucleotides) and long non-coding RNAs (lncRNA (>200 nucleotides). MicroRNAs (miRNAs) are the best studied small ncRNAs, representing an additional layer of posttranscriptional regulators that absorb perturbations and ensure the robustness of biological systems (Liu and Olson, 2010; Ebert and Sharp, 2012; Rotllan et al., 2016).
Substantial effort has now been directed toward dissecting the function of lncRNAs. In the cardiovascular system, lncRNAs were reported to play key roles in physiology and disease and targeting lncRNAs for novel therapeutic interventions has been explored (Uchida and Dimmeler, 2015; Boon et al., 2016; Buhrke et al., 2018). Here we will discuss the experimental tools to determine the RNA structure that can offer unique insights into the lncRNA function in the cardiovascular system.
Challenges in Assessing lncRNA Functionality
The unique features of lncRNA have been extensive investigated (Guttman and Rinn, 2012; Ulitsky and Bartel, 2013; Bar et al., 2016; Ulitsky, 2016). Several characteristics of lncRNAs make functional evaluation challenging. Typically, lncRNAs display poor conservation across species showing only “patches” of conserved bases surrounded by large seemingly unconstrained sequences (Ponjavic et al., 2007; Guttman et al., 2009; Necsulea et al., 2014; Washietl et al., 2014). Additionally, lncRNAs exhibit low abundance that restricts their mode and sites of action (Mercer et al., 2008; Cabili et al., 2011, 2015; Washietl et al., 2014; Ulitsky, 2016; Wilk et al., 2016; Jandura and Krause, 2017). In terms of the modes of function, both cis-and trans-regulatory activity have been described (Mercer and Mattick, 2013). As cis-regulators, lncRNAs exert their function on neighboring genes on the same allele from which they are transcribed, displaying expression correlation and perturbation in an allele-specific manner. CARMEN, an enhancer associated lncRNA and a crucial regulator of cardiac specification in human cardiac progenitor cells was shown to act in cis to control the expression of miR-143/145 (Ounzain et al., 2015). On the other hand, acting in trans-lncRNAs can control gene expression at a distance from their transcription site, by altering the chromatin state, influencing the nuclear structure or regulating protein function (Vance and Ponting, 2014; Kopp and Mendell, 2018).
Intriguingly, for some low abundance lncRNAs the act of transcription seems to be more important than the transcript itself. In a seminal study, Engreitz et al. (2016) genetically manipulated 12 genomic loci that produce lncRNAs to find that 5 loci influenced the expression of a neighboring gene in cis. The expression of the lncRNAs transcripts themselves was not required but instead processes associated with their transcription were critical (Engreitz et al., 2016).
The RNA Interactome
The above functional versatilities of lncRNAs stem from their ability to conform to different structures and molecular interactions with proteins, RNA and DNA (Guttman and Rinn, 2012; Marchese et al., 2017). In ribonucleoprotein complexes (RNPs), lncRNAs may act as scaffolds to stabilize the complexes, directing them to specific subcellular loci or the DNA. In endothelial cells, interaction of the lncRNA MANTIS with the ATPase catalytic subunits confers specificity to the switch/sucrose non-ferentable (SWI/SNF) chromatin remodeling complex directing it to a subset of angiogenic genes and facilitating nucleosome remodeling and transcription initiation (Leisegang et al., 2017; Zampetaki and Mayr, 2017). In fact binding of lncRNA to specific ATPase subunits of the SWI/SNF complex is a common regulatory mechanism (Cajigas et al., 2015; Zhu et al., 2016).
Interaction of lncRNAs with chromatin complexes is particularly important as these lncRNA-RNPs can trigger chromatin modifications through interference with the chromatin-modifying machinery (Tsai et al., 2010; Brockdorff, 2013; Simon et al., 2013). In the heart, Chaer a cardiac enriched lncRNA acts as an epigenetic switch by interfering with the polycomb repressive complex 2 (PRC2) and inhibiting H3K27m3 at genes involved in cardiac hypertrophy (Wang et al., 2016), while mesoderm faith determining lncRNA Fendrr can bind to both PRC2 and Trithorax group/MLL (TrxG/MLL) complexes acting as a fine tuner (Grote et al., 2013).
Apart from proteins, interaction of lncRNAs with DNA has also been described. This can lead to the formation of RNA-DNA triplex, a structure that is widespread in vivo and facilitates target gene recognition by lncRNAs (Mondal et al., 2015). This interaction was elegantly demonstrated in MEG3, a cardiac fibroblast enriched lncRNA that promotes fibrosis (Piccoli et al., 2017). MEG3 interacts with the PRC2 complex and forms RNA-DNA triplex structures through GA-rich sequence binding sites. Chromatin RNA immunoprecipitation revealed that MEG3 modulates the activity of TGF-b pathway genes and target recognition occurs via the triplex structures (Mondal et al., 2015).
Long non-coding RNA regulatory functions also rely on RNA–RNA interactions. Crosstalk with miRNAs creates an intricate network that exerts post-transcriptional regulation of gene expression. LncRNAs can harbor miRNA binding sites and act as molecular decoys or sponges that sequester miRNAs away from other transcripts. Noteworthy, competition between lncRNAs and miRNAs for binding to target mRNAs has been reported and leads to de-repression of gene expression (Yoon et al., 2014; Ballantyne et al., 2016). Finally, lncRNAs may contain embedded miRNA sequences and serve as a source of miRNAs (Piccoli et al., 2017).
Linking RNA Structure to Function
RNA molecules adopt higher order tertiary interactions (Staple and Butcher, 2005; Wan et al., 2011). Although links between structure and function are emerging, the structural domains that dominate the RNA interactome are still not well defined. The functional implications of transcript structure are better understood in the processing the primary miRNAs (primiRNAs) to mature miRNAs. Using multiple mutagenesis assays, the secondary structures such as stem length, hairpin pairing, bulge size and position, and apical loop size that contribute to effective miRNA biogenesis were defined (Auyeung et al., 2013; Fang and Bartel, 2015; Nguyen et al., 2015; Roden et al., 2017). In clustered miRNAs that consist of multiple miRNA genes, the tertiary structure was also proposed to contribute to the processing to individual mature miRNAs. An autoregulatory role for the tertiary structure of miR-17∼92 cluster in its maturation and binding of auxiliary factors to conserved terminal loops was shown (Chakraborty et al., 2012). Recently, in the miR-497∼195 cluster, mutations in miR-195a hairpin were reported to affect the processing of miR-497a that resides in the same cluster. Computational analysis highlighted differences in the tertiary structure of the primiRNA in mutants that may affect the maturation process (Lataniotis et al., 2017). On a different note, in primiR-30c-1 the tertiary structure promotes the interaction with SRSF3, an SR protein family member that facilitates primiRNA recognition and processing. A single G/A sequence variation leads to a structural rearrangement of the apical region of the primiRNA affecting the conserved residues placed at the basal part of the stem and mature miRNA generation (Fernandez et al., 2017).
In lncRNAs, selection acting on structure rather than primary sequence may explain the rapid rate of evolution, that led to the “RNA modular code” hypothesis based on the view that selection acts on structural domains (Wutz et al., 2002; Tsai et al., 2010; Guttman and Rinn, 2012). Some experimental evidence supports this concept. The MEG3 lncRNA gene contains three distinct structure modules M1, M2, and M3. Deletion analysis showed that motifs M2 and M3 are important for p53 activation. Intriguingly, a hybrid MEG3 transcript in which half of the primary sequence in the M2 motif was replaced by an entirely unrelated artificial sequence that displayed a similar secondary structure was fully functional in stimulating p53-mediated transcription (Zhang et al., 2010).
RNA Structure Determination Methods
Chemical and enzymatic probing methods can provide an understanding of the secondary structure of RNA (Ehresmann et al., 1987). Enzymatic probing relies on nucleases that bind to paired and unpaired RNA and digest it to generate RNA fragments that can be analyzed. On the other hand, in chemical probing small size chemicals that react and covalently modify solvent accessible nucleotides are used. Following modification or cleavage, positions are typically mapped by reverse transcription, which either stops or introduces a mutation into the cDNA (Wilkinson et al., 2006). An analysis of the resulting cDNA is then used to determine the nucleotide position and modification frequency. Next generation sequencing (NGS) can be applied to directly sequence the cDNA products. This allows RNA structural characterization at a transcriptome-wide level in a single experiment (Lucks et al., 2011; Incarnato et al., 2014; Loughrey et al., 2014; Rouskin et al., 2014). Although initially the technologies were established to analyse RNA structure in vitro, structural characterization in vivo mainly through the use of probes that can diffuse quickly across membranes has also been reported (Spitale et al., 2013; Ding et al., 2014; Spitale et al., 2015; Flynn et al., 2016).
Enzymatic Probing
PARS
PARS (parallel analysis of RNA structure) is a high-throughput enzymatic probing method that measures the structural properties of isolated polyadenylated transcript pools that are renatured in vitro and treated with RNase V1 or S1. RNase V1 and RNase S1 cleave the 3′ phosphodiester bonds of double-stranded and single-stranded RNA, respectively, allowing evaluation of the double- or single-stranded conformation (Kertesz et al., 2010).
Frag-Seq
Frag-Seq (fragmentation sequencing) is an enzymatic method that uses a nuclease P1 to specifically cleave single-stranded RNA. High-throughput sequencing then analyses the fragments generated. This workflow provides an “RNA accessibility profile” that is likened to the DNase hypersensitivity assays on chromatin (Underwood et al., 2010). Noteworthy, Frag-seq isolates fragments <200 bases after RNase P1 cleavage, hence large RNAs maybe underrepresented. As Frag-seq and PARS can provide complementary data a combined approach could improve the accuracy of genome-wide RNA structure measurements (Wan et al., 2011).
Chemical Probing
DMS Probing
The dimethyl sulfate (DMS) is a base specific reagent that can bind and alter the methylation state of unpaired adenosine and cytosine nucleotides (Tijerina et al., 2007; Rouskin et al., 2014). DMS footprinting is optimized for structural analysis of RNA. Protein binding to RNA generates a “footprint” that can be traced due to alterations in the RNA structure. The transcript size that can be evaluated is rather small (<500 nt) but this method can be performed both in vitro and in vivo as DMS can easily penetrate the cell membrane. DMS-seq that combines DMS methylation with NGS was recently performed in vivo (Ding et al., 2014; Rouskin et al., 2014).
Targeted Structure-Seq
Targeted Structure-Seq relies on RNA methylation by DMS being performed in vivo. Subsequently, RNA is isolated from cells and the methylation sites are determined by employing gene specific primers for the reverse transcription reaction. Sequencing of the DMS derived fragments can be used to assess the cellular conformation of the RNA. Based on this method, structural models of elements within Xist were developed (Fang et al., 2015). Although initially reported using DMS this workflow can be adapted for other probing reagents.
SHAPE
SHAPE (selective 2′-hydroxyl acylation by primer extension) can interrogate the RNA structure both in vitro and in vivo using the chemical NMIA and its derivatives to detect flexible regions in RNA secondary structure (Wilkinson et al., 2006; Weeks and Mauger, 2011). Several SHAPE reagents have been tested in order to improve the signal to background ratio (Lee et al., 2017). In SHAPE, the 2′-hydroxyl groups of all four nucleotides are selectively acylated when flexible and unpaired. This results in the formation of covalent SHAPE adducts that block the reverse transcription leading to truncated cDNA fragments. SHAPE reactivities can then be used to model secondary structures and quantify any process that modulates RNA dynamics.
SHAPE-MaP
SHAPE-MaP (SHAPE and mutational profiling) was the first to combine the SHAPE protocol with NGS. Initially performed and reported to define the HIV-1 RNA genome, SHAPE-MaP is a highly sensitive technique that allowed rapid, de novo discovery and direct validation of new functional motifs (Siegfried et al., 2014; Mustoe et al., 2018).
In-cell SHAPE-Seq
In-cell SHAPE-Seq is a modification of the SHAPE-Seq technique that combines the SHAPE-seq with gene expression measurements to elucidate the association of RNA structure and function in vivo. It revealed translational regulatory mechanisms in E. coli in vivo (Watters et al., 2016).
icSHAPE-seq
icSHAPE-seq (in vivo click SHAPE sequencing) uses the in-cell SHAPE chemical NAI-N3 followed by selective chemical enrichment of NAI-N3-modified RNA that provides an improved signal-to-noise ratio (Flynn et al., 2016). Follow-up NGS allows accurate identification at single-nucleotide resolution. In mouse embryonic stem cells it was shown that in vitro RNA folding is programmed entirely by the sequence, whereas in vivo, the RNA structure depends on the context of intracellular environment and interaction with RNA binding proteins that may lead to focal structural rearrangements (Spitale et al., 2015). Hence, this assay offers the exciting possibility of viewing the RNA structurome in vivo in the presence or absence of stimulation.
RNA Structurome and Interactome Determination
PARIS
PARIS (psoralen analysis of RNA interactions and structures) was recently developed to determine both RNA structure and interactions in vivo. It uses the highly specific and reversible nucleic acid crosslinker psoralen-derivative 4′-aminomethyltrioxsalen to fix base pairs in living cells. Subsequently, partial RNase and complete proteinase digestion lead to purification of a set of small crosslinked and directly base-paired RNA fragments. Purification of the crosslinked fragments using 2D electrophoresis, followed by proximity ligation of duplex RNA fragments, reversal of crosslinks, and high throughput sequencing reveals the direct base pairing between fragments. Based on these reads, models of RNA structures and interactions can be generated with high specificity and sensitivity (Lu et al., 2016). Using this approach a model for the higher order structure of Xist was interrogated (Lu et al., 2016). Encouragingly, these findings are in agreement with crystallographic studies of the defined domains in vitro (Arieti et al., 2014).
lncRNA Structure Determination
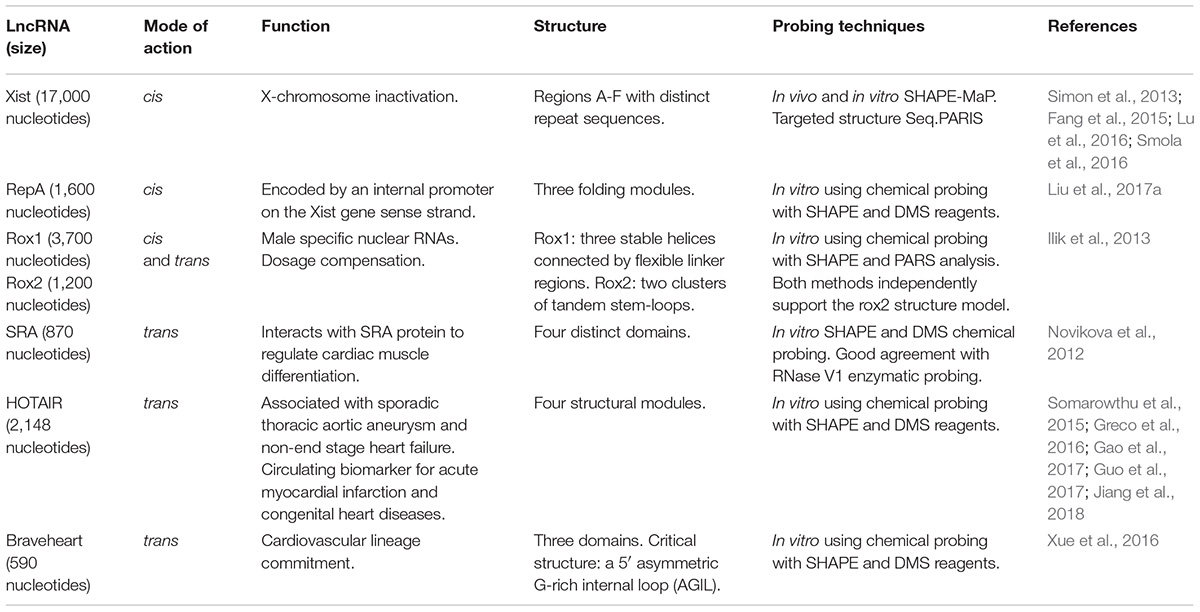
Structure determination of lncRNA in vivo is extremely challenging as they are highly heterogenic with regions with well-defined base-pairing, others without base-pairing and regions with multiple structures. Additionally, lncRNAs may stretch across thousands of nucleotides, they are expressed in low abundance and tend to be part of multicomponent complexes (Busan and Weeks, 2017). Nevertheless, the structure of several lncRNAs has been experimentally determined (Table 1).
Xist
This is a very long lncRNA (17,000 nucleotides) controlling X chromosome inactivation. It spreads across the entire chromosome while triggering stable epigenetic modifications through recruitment of the PRC2 complex and enrichment for the H3K27me3 repressive chromatin modification (Simon et al., 2013; Fang et al., 2015; Smola et al., 2016). In vivo SHAPE data identified 33 regions in Xist that form well-defined secondary structures linked by structurally variable and dynamic regions.
RepA
This is a 1,600 nucleotides mouse lncRNA encoded by an internal promoter on the Xist-gene sense strand. Applying SHAPE and DMS chemical probing in vitro, an intricate structure of three independently folding modules was revealed. Phylogenetic analysis and computational 3D modeling demonstrated a defined tertiary architecture that can form autonomously in the absence of protein partners (Liu et al., 2017a).
Rox1/Rox2
In Drosophila dosage compensation is achieved using two lncRNAs that are transcribed from the X chromosome. RNA on the X 1 and 2 (roX1 and roX2) are 3,700 and 1,200 nucleotides in length, respectively. In vitro SHAPE probing and PARS analysis revealed common, conserved and distinct structural motifs that may function as targeting sites and assembly platforms for the male specific lethal complex (Ilik et al., 2013).
SRA
The human steroid receptor RNA activator (SRA) is an 870 nucleotide lncRNA that is derived from a gene encoding both lncRNA and protein coding transcripts. The structure of SRA was experimentally interrogated using SHAPE and DMS chemical probing in vitro. In parallel, RNase V1 enzymatic probing was performed. It was shown that SRA consists of four distinct domains with a variety of secondary structures (Novikova et al., 2012). More importantly, comparative structural analysis between mouse and human strongly suggested that a large number of evolutionary changes had minimal mutational effect on the protein derived from the locus while stabilizing the RNA structural core (Novikova et al., 2012).
HOTAIR
Associated with Sporadic Thoracic Aortic Aneurysm through regulation of extracellular matrix deposition and human aortic smooth muscle cells apoptosis, this lncRNA plays a key role in the cardiovascular system (Guo et al., 2017). In non-end stage heart failure patients HOTAIR was among a panel of lncRNAs that were significantly modulated (Greco et al., 2016). A protective role of HOTAIR in cardiomyocytes (Gao et al., 2017) and as a circulating biomarker for acute myocardial infarction and congenital heart diseases were also proposed (Jiang et al., 2018). Hotair is 2,148 nucleotides long making the structural determination extremely challenging. To address this issue a non-denaturing purification protocol to obtain a homogeneous and monodisperse form was established. Structural modules and distinct evolutionary conserved elements were determined in vitro using chemical probing with SHAPE and DMS reagents (Somarowthu et al., 2015).
Braveheart
Braveheart is a 590 nucleotide lncRNA that acts in trans to regulate cardiovascular lineage commitment. Its secondary structure was experimentally assessed using SHAPE and DMS probing in vitro. It emerged that Braveheart is organized into a highly intricate modular structure comprising of three domains, consisting of 12 helices, 8 terminal loops, 5 sizeable internal loops, and a five-way junction. Intriguingly, it includes a 5′ asymmetric G-rich internal loop (AGIL) and a 55 nucleotide stretch at the 3′ end exhibiting high reactivity suggesting low probability of structure. Genetic deletion of this specific 11 nucleotide fragment demonstrated that the AGIL motif is essential for mouse embryonic stem cell differentiation to cardiomyocytes through binding of the zing-finger protein CNBP/ZNF9 (Xue et al., 2016).
Future Directions
RNA structure determination combined with genetic manipulations can elucidate the important functional domains of lncRNAs. To this end, advanced experimental tools, bioinformatics and genome engineering should be integrated. The CRISPR/Cas9 gene editing system emerged as a robust technology that can be used to generate targeted modifications at precise genomic loci. Cas9, a nuclease that can induce double-stranded breaks (DSBs) to the DNA, can be guided in the immediate vicinity of the proto-adjacent motif NGG by an RNA molecule (sgRNA) consisting of a small 20 nucleotide long variable sequence and an adaptor transactivating RNA. Precise insertions, deletions, or base substitutions can be introduced at a DSB site (Lin et al., 2014) in primary cells and in vivo in mouse models of disease (Platt et al., 2014; Abrahimi et al., 2015). A modified version of the CRISPR/Cas9 system has recently been employed for genome scale screenings of functional lncRNAs. This CRISPR interference approach uses a nuclease dead Cas9 (dCas9) that is not capable of inducing DSB to the DNA. Fused to a repressor domain (e.g., KRAB) (Liu et al., 2017b) or an activation domain (e.g., VP64) (Konermann et al., 2015; Bester et al., 2018) dCas9 and can be guided by sgRNAs to specific loci in the upstream regulatory region to trigger repression or activation of lncRNA transcription, respectively. Such approaches are extremely useful to test the functionality of lncRNAs in a high throughput manner.
Once specific lncRNAs are identified, technologies that can define the lncRNA structure in vivo are critical to determine lncRNA modules and structural domains. RNA structure determination can be coupled with comparative genomics analysis that will take into consideration the positional conservation and the fact that lncRNAs may rely on short elements rather than long stretches of conserved sequences. Genetic studies that can target precisely these structural domains while maintaining the expression of the lncRNA (Matsumoto et al., 2017) will delineate the functional impact of these motifs. The use of CRISPR/Cas9 gene editing in induced pluripotent stem cells that can be clonally expanded, engineered to harbor defined deletions of the structural motifs and differentiated to other cell types (Cochrane et al., 2017; Granata et al., 2017) can provide conclusive evidence for the functional impact of these domains in the cardiovascular system. The potential of these elements as novel targets could be explored further for precise interventions suitable for therapeutic applications.
Author Contributions
AZ initiated the study, designed its structure, and wrote the manuscript. AA provided conceptual advice and revised the manuscript. KS designed the review structure, provided conceptual advice, and revised the manuscript. All authors read and approved the submitted version.
Funding
This work was funded by the British Heart Foundation. AZ is an Intermediate Fellow of the British Heart Foundation (FS/13/18/30207).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abrahimi, P., Chang, W. G., Kluger, M. S., Qyang, Y., Tellides, G., Saltzman, W. M., et al. (2015). Efficient gene disruption in cultured primary human endothelial cells by CRISPR/Cas9. Circ. Res. 117, 121–128. doi: 10.1161/CIRCRESAHA.117.306290
Arieti, F., Gabus, C., Tambalo, M., Huet, T., Round, A., and Thore, S. (2014). The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic Acids Res. 42, 6742–6752. doi: 10.1093/nar/gku277
Auyeung, V. C., Ulitsky, I., McGeary, S. E., and Bartel, D. P. (2013). Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152, 844–858. doi: 10.1016/j.cell.2013.01.031
Ballantyne, M. D., McDonald, R. A., and Baker, A. H. (2016). lncRNA/MicroRNA interactions in the vasculature. Clin. Pharmacol. Ther. 99, 494–501. doi: 10.1002/cpt.355
Bar, C., Chatterjee, S., and Thum, T. (2016). Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation 134, 1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686
Bester, A. C., Lee, J. D., Chavez, A., Lee, Y. R., Nachmani, D., Vora, S., et al. (2018). An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 173, 649e.20–664.e20. doi: 10.1016/j.cell.2018.03.052
Boon, R. A., Jae, N., Holdt, L., and Dimmeler, S. (2016). Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 67, 1214–1226. doi: 10.1016/j.jacc.2015.12.051
Brockdorff, N. (2013). Noncoding RNA and polycomb recruitment. RNA 19, 429–442. doi: 10.1261/rna.037598.112
Buhrke, A., Bar, C., and Thum, T. (2018). [Non-coding RNA: innovative regulators with therapeutic perspective]. Herz 43, 115–122. doi: 10.1007/s00059-017-4660-4
Busan, S., and Weeks, K. M. (2017). Visualization of RNA structure models within the integrative genomics viewer. RNA 23, 1012–1018. doi: 10.1261/rna.060194.116
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16:20. doi: 10.1186/s13059-015-0586-4
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Cajigas, I., Leib, D. E., Cochrane, J., Luo, H., Swyter, K. R., Chen, S., et al. (2015). Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 142, 2641–2652. doi: 10.1242/dev.126318
Chakraborty, S., Mehtab, S., Patwardhan, A., and Krishnan, Y. (2012). Pri-miR-17-92a transcript folds into a tertiary structure and autoregulates its processing. RNA 18, 1014–1028. doi: 10.1261/rna.031039.111
Cochrane, A., Kelaini, S., Tsifaki, M., Bojdo, J., Vila-Gonzalez, M., Drehmer, D., et al. (2017). Quaking is a key regulator of endothelial cell differentiation, neovascularization, and angiogenesis. Stem Cells 35, 952–966. doi: 10.1002/stem.2594
Ding, Y., Tang, Y., Kwok, C. K., Zhang, Y., Bevilacqua, P. C., and Assmann, S. M. (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. doi: 10.1038/nature12756
Ebert, M. S., and Sharp, P. A. (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524. doi: 10.1016/j.cell.2012.04.005
Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J. P., and Ehresmann, B. (1987). Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128.
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Fang, R., Moss, W. N., Rutenberg-Schoenberg, M., and Simon, M. D. (2015). Probing Xist RNA structure in cells using targeted structure-Seq. PLoS Genet. 11:e1005668. doi: 10.1371/journal.pgen.1005668
Fang, W., and Bartel, D. P. (2015). The menu of features that define primary micrornas and enable de novo design of MicroRNA genes. Mol. Cell 60, 131–145. doi: 10.1016/j.molcel.2015.08.015
Fernandez, N., Cordiner, R. A., Young, R. S., Hug, N., Macias, S., and Caceres, J. F. (2017). Genetic variation and RNA structure regulate microRNA biogenesis. Nat. Commun. 8:15114. doi: 10.1038/ncomms15114
Flynn, R. A., Zhang, Q. C., Spitale, R. C., Lee, B., Mumbach, M. R., and Chang, H. Y. (2016). Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat. Protoc. 11, 273–290. doi: 10.1038/nprot.2016.011
Gao, L., Liu, Y., Guo, S., Yao, R., Wu, L., Xiao, L., et al. (2017). Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol. Biochem. 44, 1497–1508. doi: 10.1159/000485588
Granata, A., Serrano, F., Bernard, W. G., McNamara, M., Low, L., Sastry, P., et al. (2017). An iPSC-derived vascular model of Marfan syndrome identifies key mediators of smooth muscle cell death. Nat. Genet. 49, 97–109. doi: 10.1038/ng.3723
Greco, S., Zaccagnini, G., Perfetti, A., Fuschi, P., Valaperta, R., Voellenkle, C., et al. (2016). Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 14:183. doi: 10.1186/s12967-016-0926-5
Grote, P., Wittler, L., Hendrix, D., Koch, F., Wahrisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi: 10.1016/j.devcel.2012.12.012
Guo, X., Chang, Q., Pei, H., Sun, X., Qian, X., Tian, C., et al. (2017). Long non-coding RNA-mRNA correlation analysis reveals the potential role of HOTAIR in pathogenesis of sporadic thoracic aortic aneurysm. Eur. J. Vasc. Endovasc. Surg. 54, 303–314. doi: 10.1016/j.ejvs.2017.06.010
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. doi: 10.1038/nature10887
Ilik, I. A., Quinn, J. J., Georgiev, P., Tavares-Cadete, F., Maticzka, D., Toscano, S., et al. (2013). Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 51, 156–173. doi: 10.1016/j.molcel.2013.07.001
Incarnato, D., Neri, F., Anselmi, F., and Oliviero, S. (2014). Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 15:491. doi: 10.1186/s13059-014-0491-2
Jandura, A., and Krause, H. M. (2017). The new RNA World: growing evidence for long noncoding RNA functionality. Trends Genet. 33, 665–676. doi: 10.1016/j.tig.2017.08.002
Jiang, Y., Mo, H., Luo, J., Zhao, S., Liang, S., Zhang, M., et al. (2018). HOTAIR is a potential novel biomarker in patients with congenital heart diseases. Biomed. Res. Int. 2018:2850657. doi: 10.1155/2018/2850657
Kertesz, M., Wan, Y., Mazor, E., Rinn, J. L., Nutter, R. C., Chang, H. Y., et al. (2010). Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107. doi: 10.1038/nature09322
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. doi: 10.1038/nature14136
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Lataniotis, L., Albrecht, A., Kok, F. O., Monfries, C. A. L., Benedetti, L., Lawson, N. D., et al. (2017). CRISPR/Cas9 editing reveals novel mechanisms of clustered microRNA regulation and function. Sci. Rep. 7:8585. doi: 10.1038/s41598-017-09268-0
Lee, B., Flynn, R. A., Kadina, A., Guo, J. K., Kool, E. T., and Chang, H. Y. (2017). Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23, 169–174. doi: 10.1261/rna.058784.116
Leisegang, M. S., Fork, C., Josipovic, I., Richter, F. M., Preussner, J., Hu, J., et al. (2017). Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 136, 65–79. doi: 10.1161/CIRCULATIONAHA.116.026991
Lin, S., Staahl, B. T., Alla, R. K., and Doudna, J. A. (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766. doi: 10.7554/eLife.04766
Liu, F., Somarowthu, S., and Pyle, A. M. (2017a). Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 13, 282–289. doi: 10.1038/nchembio.2272
Liu, S. J., Horlbeck, M. A., Cho, S. W., Birk, H. S., Malatesta, M., He, D., et al. (2017b). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355:aah7111. doi: 10.1126/science.aah7111
Liu, N., and Olson, E. N. (2010). MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510–525.
Loughrey, D., Watters, K. E., Settle, A. H., and Lucks, J. B. (2014). SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 42:e165. doi: 10.1093/nar/gku909
Lu, Z., Zhang, Q. C., Lee, B., Flynn, R. A., Smith, M. A., Robinson, J. T., et al. (2016). RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. doi: 10.1016/j.cell.2016.04.028
Lucks, J. B., Mortimer, S. A., Trapnell, C., Luo, S., Aviran, S., Schroth, G. P., et al. (2011). Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. U.S.A. 108, 11063–11068. doi: 10.1073/pnas.1106501108
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18:206. doi: 10.1186/s13059-017-1348-2
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721. doi: 10.1073/pnas.0706729105
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6:7743. doi: 10.1038/ncomms8743
Mustoe, A. M., Busan, S., Rice, G. M., Hajdin, C. E., Peterson, B. K., Ruda, V. M., et al. (2018). Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173, 181.e18–195.e18. doi: 10.1016/j.cell.2018.02.034
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U., et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. doi: 10.1038/nature12943
Nguyen, T. A., Jo, M. H., Choi, Y. G., Park, J., Kwon, S. C., Hohng, S., et al. (2015). Functional anatomy of the human microprocessor. Cell 161, 1374–1387. doi: 10.1016/j.cell.2015.05.010
Novikova, I. V., Hennelly, S. P., and Sanbonmatsu, K. Y. (2012). Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 40, 5034–5051. doi: 10.1093/nar/gks071
Ounzain, S., Micheletti, R., Arnan, C., Plaisance, I., Cecchi, D., Schroen, B., et al. (2015). CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell Cardiol. 89, 98–112. doi: 10.1016/j.yjmcc.2015.09.016
Piccoli, M. T., Gupta, S. K., Viereck, J., Foinquinos, A., Samolovac, S., Kramer, F. L., et al. (2017). Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 121, 575–583. doi: 10.1161/CIRCRESAHA.117.310624
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. doi: 10.1016/j.cell.2014.09.014
Ponjavic, J., Ponting, C. P., and Lunter, G. (2007). Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17, 556–565. doi: 10.1101/gr.6036807
Roden, C., Gaillard, J., Kanoria, S., Rennie, W., Barish, S., Cheng, J., et al. (2017). Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 27, 374–384. doi: 10.1101/gr.208900.116
Rotllan, N., Price, N., Pati, P., Goedeke, L., and Fernandez-Hernando, C. (2016). microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis 246, 352–360. doi: 10.1016/j.atherosclerosis.2016.01.025
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M., and Weissman, J. S. (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. doi: 10.1038/nature12894
Siegfried, N. A., Busan, S., Rice, G. M., Nelson, J. A., and Weeks, K. M. (2014). RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. doi: 10.1038/nmeth.3029
Simon, M. D., Pinter, S. F., Fang, R., Sarma, K., Rutenberg-Schoenberg, M., Bowman, S. K., et al. (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. doi: 10.1038/nature12719
Smola, M. J., Christy, T. W., Inoue, K., Nicholson, C. O., Friedersdorf, M., Keene, J. D., et al. (2016). SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 113, 10322–10327. doi: 10.1073/pnas.1600008113
Somarowthu, S., Legiewicz, M., Chillon, I., Marcia, M., Liu, F., and Pyle, A. M. (2015). HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361. doi: 10.1016/j.molcel.2015.03.006
Spitale, R. C., Crisalli, P., Flynn, R. A., Torre, E. A., Kool, E. T., and Chang, H. Y. (2013). RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20. doi: 10.1038/nchembio.1131
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. doi: 10.1038/nature14263
Staple, D. W., and Butcher, S. E. (2005). Pseudoknots: RNA structures with diverse functions. PLoS Biol. 3:e213. doi: 10.1371/journal.pbio.0030213
Thum, T., and Condorelli, G. (2015). Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 116, 751–762. doi: 10.1161/CIRCRESAHA.116.303549
Tijerina, P., Mohr, S., and Russell, R. (2007). DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2, 2608–2623. doi: 10.1038/nprot.2007.380
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
Uchida, S., and Dimmeler, S. (2015). Long noncoding RNAs in cardiovascular diseases. Circ. Res. 116, 737–750. doi: 10.1161/CIRCRESAHA.116.302521
Ulitsky, I. (2016). Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614. doi: 10.1038/nrg.2016.85
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
Underwood, J. G., Uzilov, A. V., Katzman, S., Onodera, C. S., Mainzer, J. E., Mathews, D. H., et al. (2010). FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7, 995–1001. doi: 10.1038/nmeth.1529
Vance, K. W., and Ponting, C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30, 348–355. doi: 10.1016/j.tig.2014.06.001
Wan, Y., Kertesz, M., Spitale, R. C., Segal, E., and Chang, H. Y. (2011). Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 12, 641–655. doi: 10.1038/nrg3049
Wang, Z., Zhang, X. J., Ji, Y. X., Zhang, P., Deng, K. Q., Gong, J., et al. (2016). The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139. doi: 10.1038/nm.4179
Washietl, S., Kellis, M., and Garber, M. (2014). Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 24, 616–628. doi: 10.1101/gr.165035.113
Watters, K. E., Abbott, T. R., and Lucks, J. B. (2016). Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 44:e12. doi: 10.1093/nar/gkv879
Weeks, K. M., and Mauger, D. M. (2011). Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 44, 1280–1291. doi: 10.1021/ar200051h
Wilk, R., Hu, J., Blotsky, D., and Krause, H. M. (2016). Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 30, 594–609. doi: 10.1101/gad.276931.115
Wilkinson, K. A., Merino, E. J., and Weeks, K. M. (2006). Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616. doi: 10.1038/nprot.2006.249
Wutz, A., Rasmussen, T. P., and Jaenisch, R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. doi: 10.1038/ng820
Xue, Z., Hennelly, S., Doyle, B., Gulati, A. A., Novikova, I. V., Sanbonmatsu, K. Y., et al. (2016). A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell 64, 37–50. doi: 10.1016/j.molcel.2016.08.010
Yoon, J. H., Abdelmohsen, K., and Gorospe, M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 34, 9–14. doi: 10.1016/j.semcdb.2014.05.015
Zampetaki, A., and Mayr, M. (2017). Long noncoding RNAs and angiogenesis: regulatory information for chromatin remodeling. Circulation 136, 80–82. doi: 10.1161/CIRCULATIONAHA.117.028398
Zhang, X., Rice, K., Wang, Y., Chen, W., Zhong, Y., Nakayama, Y., et al. (2010). Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151, 939–947. doi: 10.1210/en.2009-0657
Keywords: non-coding RNA, lncRNA, RNA structure, gene editing, cardiovascular diseases
Citation: Zampetaki A, Albrecht A and Steinhofel K (2018) Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 9:1201. doi: 10.3389/fphys.2018.01201
Received: 05 June 2018; Accepted: 10 August 2018;
Published: 24 August 2018.
Edited by:
Vincenzo Lionetti, Scuola Sant’Anna di Studi Avanzati, ItalyReviewed by:
Elena Guzzolino, Consorzio Pisa Ricerche, ItalyAmalia Forte, Università degli Studi della Campania Luigi Vanvitelli, Italy
Elisabetta Cervio, Cardiocentro Ticino, Switzerland
Copyright © 2018 Zampetaki, Albrecht and Steinhofel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Zampetaki, YW5uYS56YW1wZXRha2lAa2NsLmFjLnVr
 Anna Zampetaki
Anna Zampetaki Andreas Albrecht
Andreas Albrecht Kathleen Steinhofel
Kathleen Steinhofel