
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 10 July 2018
Sec. Gastrointestinal Sciences
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.00888
This article is part of the Research Topic Xenobiotics and the gut microbiome in health and disease View all 15 articles
Humans and their associated microbiomes are exposed to numerous xenobiotics through drugs, dietary components, personal care products as well as environmental chemicals. Most of the reciprocal interactions between the microbiota and xenobiotics, such as halogenated compounds, occur within the human gut harboring diverse and dense microbial communities. Here, we provide an overview of the flux of halogenated compounds in the environment, and diverse exposure routes of human microbiota to these compounds. Subsequently, we review the impact of halogenated compounds in perturbing the structure and function of gut microbiota and host cells. In turn, cultivation-dependent and metagenomic surveys of dehalogenating genes revealed the potential of the gut microbiota to chemically alter halogenated xenobiotics and impact their fate. Finally, we provide an outlook for future research to draw attention and attract interest to study the bidirectional impact of halogenated and other xenobiotic compounds and the gut microbiota.
The term xenobiotic is usually used in the context of environmental pollutants to refer to synthetic compounds produced in large volumes for industrial, agricultural and domestic use (Atashgahi et al., 2018c). Xenobiotics can enter the environment at high (μg/L to mg/L range) or at “micropollutant” concentrations (ng/L to μg/L range) (Schwarzenbach et al., 2006; Meckenstock et al., 2015). One important group of xenobiotics comprise halogenated compounds with diverse sources and sinks. Halogenated organic compounds, organohalogens, are usually synthesized for industrial, agricultural and pharmaceutical applications (Häggblom and Bossert, 2003). It has also been shown that over 5000 organohalogens are naturally produced from biogenic and geogenic sources (Gribble, 2010). Inorganic halogenated compounds such as chlorine dioxide, hypochlorite, and chlorite are commonly applied as bleaching agents and disinfectants (Liebensteiner et al., 2016). In turn, halogenated compounds can be used as carbon sources, electron donors and acceptors by a diverse array of aerobic and anaerobic microorganisms in growth-dependent and co-metabolic modes (Janssen et al., 2001; Van Pée and Unversucht, 2003; Schneidewind et al., 2014; Peng et al., 2017). As such, microbial degradation represents an important sink of halogenated compounds.
Xenobiotics are also considered as chemical substances from natural or synthetic sources found within an organism that are not naturally produced by the organism or expected to be present. As such, the human body is exposed to variety of (halogenated) xenobiotic compounds, such as persistent organic compounds (POPs), pesticides, pharmaceuticals and personal care products (PPCPs), and food additives. Site-specific microbiomes associated with the gut, skin, or respiratory tract are the first to encounter xenobiotics and mediate “first pass” metabolism prior to compound absorption to internal organ systems (Dietert and Silbergeld, 2015). Among these portals of entry, most interactions between xenobiotics and the human microbiota occur within the human gut (Sousa et al., 2008; Dietert and Silbergeld, 2015). The gut microbiota is a diverse and dense microbial community composed of bacteria, fungi, archaea, and viruses (Li et al., 2014; Nielsen et al., 2014). Its immense metabolic diversity is encoded by the intestinal metagenome, that contains genetic information for multiple xenobiotic detoxification and sequestration functions (Haiser and Turnbaugh, 2013; Spanogiannopoulos et al., 2016). The anoxic environment of the gut is well-suited for a reductive and hydrolytic metabolism. This will generate non-polar low-molecular weight by-products that can be absorbed by the host cells. In contrast, the readily absorbed non-polar xenobiotics are transported and metabolized in the liver by a rich collection of oxidative and conjugative enzymes. Such hepatic metabolism will generate hydrophilic, polar and high-molecular weight metabolites. The latter are secreted via the bile and reach the gut where they can be re-metabolized by reductive and hydrolytic enzymes (Sousa et al., 2008; Claus et al., 2016; Koppel et al., 2017). Thus, xenobiotic metabolism by gut microbiota can exert a profound influence on the toxicity and bioavailability of xenobiotics entering the gut via different routes. The outcome of xenobiotic metabolism may be beneficial (Shin et al., 2013), detrimental or even lethal (Okuda et al., 1998) to the host. In turn, exposure to xenobiotics can alter gut microbiota composition and change metabolic activity (Maurice et al., 2013). This may increase predisposition to various diseases (Wang et al., 2011; Lee et al., 2014; Lu et al., 2015).
Extensive research of the last decades has provided insight into the metabolism of halogenated xenobiotics and opened avenues to harness the metabolic machinery of microbes for bioremediation (Smidt and de Vos, 2004; Sutton et al., 2015; Atashgahi et al., 2017, 2018c; Weatherill et al., 2018). In contrast, much less is known about the flux, impact, and fate of halogenated xenobiotics in host-associated ecosystems like the human gut. As such, there is only very limited information about specific microorganisms, genes, and enzymes responsible for halogenated xenobiotic metabolism in the human gut. This contrasts with the enormous and expanding interest in understanding the role of the gut microbiome in health and disease.
Here, we address the present state of the art on the flux, impact and fate of halogenated compounds in the gut. We first provide an overview of the flux of halogenated compounds to highlight their environmental sources and diverse exposure routes of human microbiota to these compounds. Subsequently, we provide an overview of the impact of halogenated compounds on the structure and function of the gut microbiota and host cells. Lastly, we review the fate and metabolism of halogenated compounds in the gut based on published experimental data and a metagenomic survey of the dehalogenation genes in gut microbiome. In a larger context, we provide a rationale for studying the bidirectional impact of (halogenated) xenobiotic compounds and the gut microbiota, i.e., toxicant-microbiota interactions.
The main sources of halogenated (micro)pollutants are from industry (e.g., POPs), agriculture (e.g., pesticides), domestic use (e.g., PPCPs) and disinfection by-products (DBPs). Some examples of the halogenated (micro)pollutants that are discussed in this review are shown in Figure 1. Humans come into contact with halogenated xenobiotics through: (i) oral exposure (eventually ending up in the gut), (ii) inhalation (via nose and lungs), (iii) dermal exposure (through the skin), and (iv) ocular exposure (through the eyes) (Figure 2). Oral ingestion is the main exposure route of the general population to halogenated xenobiotics. This is especially the case for the chronic exposure to micropollutant concentrations of residues in food, vegetables, fruits, and drinking water (Boxall et al., 2006, 2012; Damalas and Eleftherohorinos, 2011). In contrast, dermal/inhalation exposure due to showering, bathing, and swimming through daily-life and/or recreational activities is a more important route of exposure to DBPs than oral exposure (Villanueva et al., 2006a,b). Inhalation, dermal and ocular routes are more relevant to occupational exposure of workers in- or nearby residents of- industrial production plants, farms or greenhouses that produce and use halogenated xenobiotics (Damalas and Eleftherohorinos, 2011; Besis and Samara, 2012).
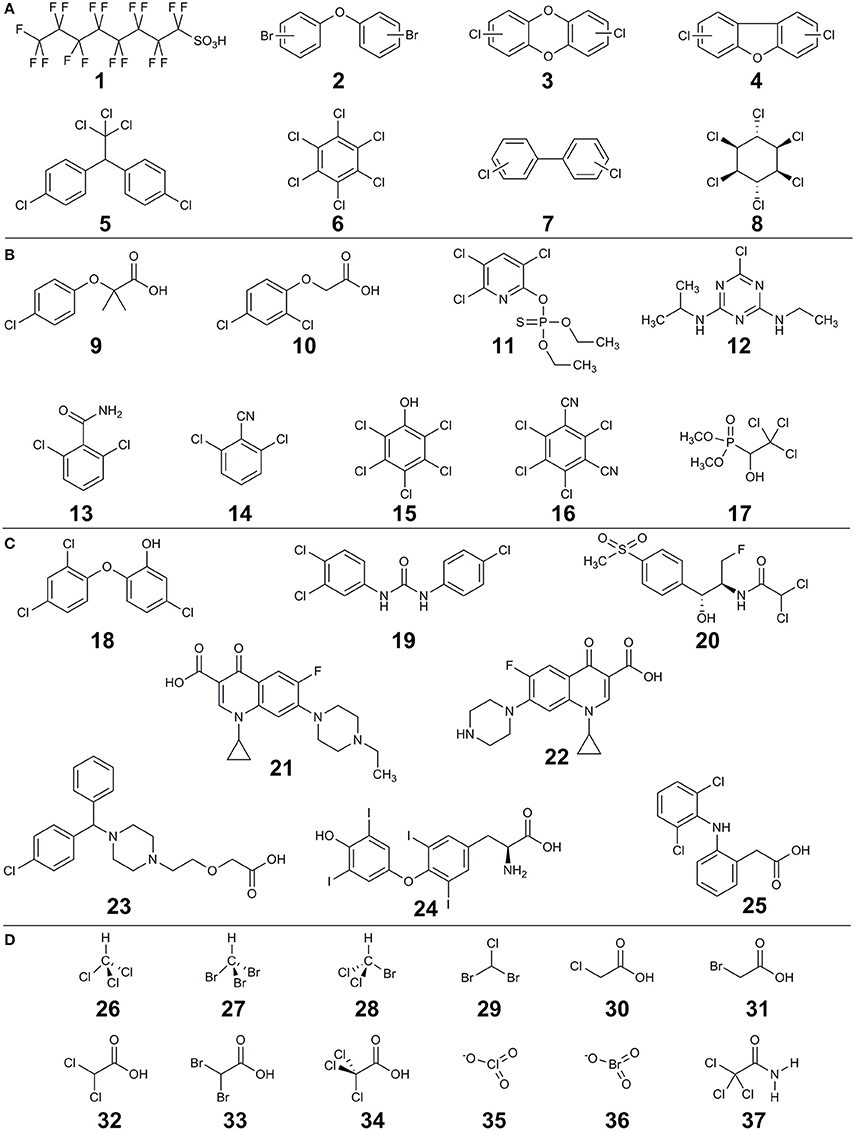
Figure 1. Examples of halogenated compounds discussed in this review. (A) persistent organic compounds (POPs), (B) pesticides, (C) pharmaceuticals and personal care products (PPCP) and (D) disinfection by-products (DBPs). Compounds are: perfluorooctanesulfonic acid (1), polybrominated diphenyl ethers (2), polychlorinated dibenzo-p-dioxins (3), polychlorinated dibenzofurans (4), dichlorodiphenyltrichloroethane (5), hexachlorobenzene (6), polychlorinated biphenyls (7), γ-hexachlorocyclohexane (lindane) (8), clofibric acid (9), 2,4-dichlorophenoxyacetic acid (10), chlorpyrifos (11), atrazine (12), 2,6-dichlorobenzamide (13), 2,6-dichlorobenzonitrile (14), pentachlorophenol (15), chlorothalonil (16), trichlorfon (17), triclosan (18), triclocarbon (19), florfenicol (20), enrofloxacin (21), ciprofloxacin (22), cetirizine (23), thyroxine (L) (24), diclofenac (25), chloroform (26), bromoform (27), bromodichloromethane (28), chlorodibromomethane (29), chloroacetic acid (30), bromoacetic acid (31), dichloroacetic acid (32), dibromoacetic acid (33), trichloroacetic acid (34), chlorate (35), bromate (36), trichloroacetamide (37).
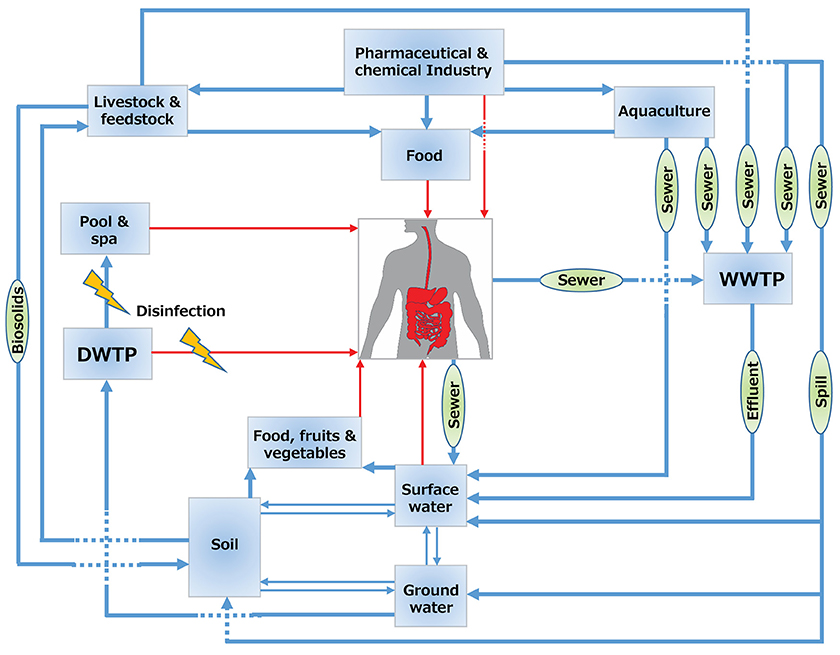
Figure 2. Flux of halogenated compounds. Blue arrows show the flux in the environment and red arrows show the flux to humans. WWTP, wastewater treatment plant; DWTP, drinking water treatment plant.
Raw sewer and treated effluents from wastewater treatment plants play a key role in the spread of halogenated compounds in the water cycle (Figure 2). Most current waste- and drinking water treatment plants are not (optimally) designed for the removal of halogenated compounds or their transformation products (Heidler and Halden, 2009; Noguera-Oviedo and Aga, 2016). The latter is particularly relevant as sometimes the transformation products can be more toxic than the parent compounds. For example, perfluorooctanesulfonic acid (compound number 1 in Figure 1) is a POP that can also be produced through biotransformation of other synthetic chemicals such as perfluoroalkyl acids in wastewater treatment plants (Guerra et al., 2014). Similarly, biodegradation of clofibric acid (9) leads to production of the more toxic 4-chlorophenol (Salgado et al., 2012). Trace concentrations of such contaminants of emerging concern may eventually end up in finished drinking water due to their toxicity and persistence (Benotti et al., 2008).
Biosolids from the treated sewage sludge represent concentrated sources of hydrophobic organohalogens such as the flame retardant polybrominated diphenyl ethers (2), and the biocidal compounds triclosan (18) and triclocarbon (19). These compounds can desorb and contaminate soil and water, once biosolids are applied as fertilizer (Andrade et al., 2015) (Figure 2). Similarly, organohalogen antibiotics such as fluoroquinolones added to animal feed for disease prevention or for growth promotion may end up in biosolids from livestock production (Martínez-Carballo et al., 2007). Once applied as manure for fertilization of arable land, these compounds can reach humans via different routes. For example, veterinary organohalogen drugs such as florfenicol (20) and enrofloxacin (21) can be taken up by plants such as lettuce (Boxall et al., 2006).
Recent non-targeted analysis has further highlighted the environmental footprint of organohalogens. For example, monitoring of bottlenose dolphins has identified 327 organohalogens of synthetic as well as natural origins (Shaul et al., 2015). This indicates severe bioaccumulation of organohalogens in marine food webs. A similar non-targeted screening of the sediments of Lake Michigan has identified 1,593 organobromine compounds, many of which were not known previously (Peng et al., 2015). The deposited organohalogens in sediments can be chronically released (Yamashita et al., 2000) and bioaccumulate in organisms in food webs and thereby reach humans.
POPs are long lived organic compounds that resist biological, chemical, and photolytic degradation. They exhibit high lipid solubility and hence bioaccumulate in fatty tissues and become concentrated as they move up the food chain. POPs are dominated by organohalogens such as polychlorinated dibenzo-p-dioxins and furans (3, 4), dichlorodiphenyltrichloroethane (5), hexachlorobenzene (6), polychlorinated biphenyls (7), γ-hexachlorocyclohexane (lindane, 8), perfluorooctanesulfonic acid, polybrominated diphenyl ethers, etc.
Although application of organochlorine pesticides and polychlorinated biphenyls has been restricted since the late 1970s, they are still among the most ubiquitous and concerning environmental pollutants due to their persistence, toxicity and bioaccumulation (Xu et al., 2017). Moreover, electronic waste (e-waste) continues to produce POPs such as polychlorinated dibenzo-p-dioxins and furans, polybrominated diphenyl ethers, and polychlorinated biphenyls. The e-waste recycling industry in developing countries has drawn the world's attention as a new source of environmental contamination by POPs (Leung et al., 2007; Zhang et al., 2010). POPs can also be produced naturally. For example, polychlorinated dibenzo-p-dioxins and furans are formed as by-products of high-temperature processes, such as volcano outbursts, forest fires and waste incineration. The endocrine-disrupting flame retardant polybrominated diphenyl ethers have natural counterparts that are produced by marine sponges (Agarwal et al., 2017) at levels that can exceed 10% of the sponge tissue dry weight (Unson et al., 1994). Naturally produced organohalogens were reported to bioaccumulate in marine mammals (Vetter et al., 2002; Teuten and Reddy, 2007) and humans (Wan et al., 2010; Wang et al., 2012) indicating human contact through the marine food web. Accordingly, POPs can come into contact with humans primarily through dietary intake including fatty fish, red meat and poultry (Kiviranta et al., 2004; Schecter et al., 2010), but also through inhalation and dermal absorption (Besis and Samara, 2012).
Pesticides such as herbicides, insecticides, and fungicides have been used since the 1940s for agricultural and non-agricultural purposes (Schwarzenbach et al., 2010). Chlorinated phenoxy acid herbicides such as 2,4-dichlorophenoxyacetic acid (10), the broad-spectrum chlorinated organophosphate chlorpyrifos (11) and the herbicide atrazine (12) are among the most intensively used pesticides worldwide (Bradberry et al., 2000; Arias-Estévez et al., 2008; John and Shaike, 2015). Pesticide contamination from specific point sources (μg/L to mg/L range) can be due to accidental releases at manufacturing plants, spills on farm yards and from wastewater treatment plant effluent (Vandermaesen et al., 2016). In contrast, diffuse contamination (ng/L to μg/L range) originates from actual pesticide application that results in large-scale contamination of groundwater through leaching, and contamination of surface water through runoff, erosion, drainage, and drifting (Holvoet et al., 2007). Moreover, contamination can be due to transformation products. For example, 2,6-dichlorobenzamide (13) is a highly mobile and persistent groundwater pollutant that originates from transformation of the widely used herbicide dichlobenil (2,6-dichlorobenzonitrile) (14) (Horemans et al., 2017). Besides oral ingestion as the main route of exposure, human contact with pesticides could be through: (i) inhalation by breathing the mobile pesticides e.g., during on farm pesticide spraying, (ii) dermal, and (iii) ocular routs e.g., during accidental splashing or spraying pesticides on unprotected skin/eyes of agricultural workers and workers in the pesticide industry (Damalas and Eleftherohorinos, 2011).
Pharmaceuticals are used to treat or prevent disease or as feed additives in animal farming, whereas personal care products are used in personal hygiene and for beautification, and include products such as shampoos, toothpastes, moisturizers, deodorants, lipsticks, perfumes, etc (Boxall et al., 2012). Whereas the main route of PPCPs release into environment is generally excretion to the sewage system following use, manufacturing facilities can be important local point sources (Ebele et al., 2017) (Figure 2). For example, effluent from a wastewater treatment plant of a major drug manufacturer contained ciprofloxacin (22) at a concentration of up to 31 mg/L, that exceeds levels toxic to some bacteria by over 1000-fold (Larsson et al., 2007). A subsequent study found ciprofloxacin (up to 6.5 mg/L) and cetirizine (23) (up to 1.2 mg/L) in two lakes in the same region impacted by the wastewater treatment plant effluent (Fick et al., 2009). These mg/L concentrations are 100,000 to 1 million times higher than reported levels of fluoroquinolones in surface waters contaminated by effluents from wastewater treatment plants (Kolpin et al., 2002; Xiao et al., 2008).
Triclosan and triclocarban are broad-spectrum phenolic organochlorine biocides with activity against both bacteria and fungi. These chemicals are found in a wide variety of consumer products, including soaps, detergents, toothpaste, medical devices, plastics, and textiles (Pycke et al., 2014). As a result of common and widespread use, humans are exposed to these chemicals via different routes including absorption (e.g., soaps, toothpaste), ingestion (e.g., drinking water, food), inhalation (e.g., aerosols, dust), and injection/implantation (e.g., medical sutures and devices) (Halden, 2016). For example, according to a survey in 2003-2004, triclosan was found in about three-quarters of urine samples analyzed in the USA at concentrations of 2.4–3790 μg/L (Calafat et al., 2008). A later analysis of triclosan, triclocarban, thier metabolites and by-products in maternal urine and cord plasma in an urban population in the USA has shown widespread fetal exposure to these compounds (Pycke et al., 2014). Triclosan and triclocarban have been detected in aquatic environments such as groundwater, drinking water, wastewater, sewage sludge, and in some food sources representing environmental sources, besides direct consumer-product use (Lindström et al., 2002; Singer et al., 2002; Halden and Paull, 2005).
Water disinfection during the production of drinking water has been widely implemented to protect human health against waterborne diseases like cholera, typhoid, dysentery, etc (Richardson and Ternes, 2018). To this end, strong oxidants such as free chlorine, chlorine dioxide, chloramines, and ozone are used, which efficiently kill pathogens. However, disinfectants can unintentionally form DBPs by further reacting with other constituents found in waters i.e., natural organic matter, anthropogenic organic contaminants, and halide ions (chloride, bromide, iodide) (Richardson et al., 2007; Gonsior et al., 2014; Postigo and Richardson, 2014). Moreover, DBPs in pool and spa waters are formed by the reaction of disinfectants with organic matter including natural organic matter from source water and human inputs such as urine, sweat and PPCPs (Daiber et al., 2016; Jmaiff Blackstock et al., 2017). For instance, more than 100 DBPs were recently found in swimming pools and hot tubs, and organic extracts from those samples were more mutagenic than the corresponding tap water extracts (Daiber et al., 2016). Although ~700 DBP have been identified, only three classes are regularly monitored: trihalomethanes (e.g., chloroform, bromoform, bromodichloromethane, chlorodibromomethane) (26–29), haloacetic acids (e.g., chloro-, bromo-, dichloro-, dibromo-, and trichloro-acetic acid) (30–34), and oxyhalides (e.g., chlorate and bromate) (35–36) (Richardson and Ternes, 2018). People are exposed to a diverse range of DBPs by drinking, uptake through the skin upon contact, and inhalation of volatile DBPs e.g., in indoor swimming facilities (Gonsior et al., 2014; Daiber et al., 2016). Epidemiological studies have reported a relation between human ingestion of drinking water containing DBPs and increased spontaneous abortions, stillbirth, birth defects and bladder cancer in particular (Richardson et al., 2007).
The negative impact of halogenated compounds has been known since long from a toxicological point of view. However, the role of the (gut) microbiota has not been well-incorporated into the study of interactions between environmental exposures and health outcomes (Dietert and Silbergeld, 2015).
The impact of halogenated compounds on gut microbiota has mostly been studied using rodent models (Table 1). For example, dietary exposure to 2,3,7,8-tetrachlorodibenzofuran has been shown to induce inflammation and decrease the Firmicutes to Bacteroidetes ratio in mice (Zhang L. et al., 2015). A similar decreased Firmicutes/Bacteroidetes ratio in mice was reported due to exposure to trichloroacetamide (37) (Zhang L. et al., 2015) or chlorpyrifos (11) (Zhao et al., 2016), and in juvenile goldfish due to exposure to pentachlorophenol (15) (Kan et al., 2015) (Table 1). In contrast, the Firmicutes/Bacteroidetes ratio increased in mice after 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure (Lefever et al., 2016). Changes in the Firmicutes/Bacteroidetes ratio was first observed in obesity studies and was subsequently addressed by various studies, confirming or challenging its impact, or pointing to technical artifacts (Ley et al., 2005; Schwiertz et al., 2010; Bahl et al., 2012). Nevertheless, these ratios may in any case indicate different levels of short chain fatty acids production, pH and activity in the gut, and hence could be meaningful when methodological bias is excluded (Duncan et al., 2009; Kolmeder et al., 2015). Accordingly, treatment of mice with 2,3,7,8-tetrachlorodibenzofuran enriched Butyrivibrio spp., common butyrate-producing gut microbes, coupled with elevation of butyrate and propionate in feces and cecal contents (Zhang L. et al., 2015). In another study, oral exposure to polychlorinated biphenyls decreased the overall abundance of bacterial species in mice gut microbiota primarily by decreasing the levels of Proteobacteria (Choi et al., 2013). Interestingly, exercise attenuated alterations of mice gut microbiota composition (Choi et al., 2013). Exercise has been shown to increase beneficial metabolites, such as butyrate in the rat cecum (Matsumoto et al., 2008).
Organohalogen POPs accumulate in adipose tissue in humans because of their lipophilicity. These POPs have been shown to bind to aryl hydrocarbon receptor (AHR), that is a transcription factor involved in the regulation of biological responses to planar aromatic (aryl) hydrocarbons (Arsenescu et al., 2008). This xenobiotic sensor modulates the activity of immune and nonimmune cells in the gut, and may represent an important link between the environment and immune system perturbations (Monteleone et al., 2012). In line with this, ahr-knockout (Ahr−/−) mice did not show large shifts in gut microbial composition in response to 2,3,7,8-tetrachlorodibenzofuran exposure (Zhang L. et al., 2015). Modulation of gut microbiota by AHR was proposed to play an important role in the induction of obesity by chronic POP exposure (Myre and Imbeault, 2014). For instance, exposure to polychlorinated biphenyls has been shown to impair glucose homeostasis in mice (Baker et al., 2013). Furthermore, higher body burden of dioxins, and polychlorinated biphenyls was reported in obese people as opposed to lean people (Kim et al., 2011). A comprehensive literature review on the impact of chlorinated POPs in humans showed that rather than a few individual POPs, background exposure to low-dose POP mixtures may promote type 2 diabetes and obesity (Lee et al., 2014). In another survey, evaluation of 72 epidemiological studies revealed the strongest positive correlation of diabetes with organochlorine than non-organochlorine POPs (Taylor et al., 2013).
In a recent in vitro study, the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) model was inoculated with feces from healthy humans, and subsequently exposed to chronic and low-doses of the insecticide chlorpyrifos (11). This induced major changes in the microbial community, in particular, increased numbers of Enterococcus and Bacteroides spp., and decreased numbers of lactobacilli and bifidobacteria, the latter including probiotics commonly associated with health benefits (Joly et al., 2013). Compositional shifts in intestinal bacterial community structure and distortion of their metabolic functions were similarly reported due to exposure of rats to chlorpyrifos (Zhao et al., 2016; Fang et al., 2018) (Table 1). In another study, oral exposure of bees to the organochlorine fungicide chlorothalonil (16) induced microbial changes, increased putative genes for oxidative phosphorylation and declined sugar metabolism and peptidase potential (Kakumanu et al., 2016). In contrast to these reports, early-life exposure to the estimated environmental concentration of atrazine (12) (200 μg/L) did not affect gut bacterial diversity or community composition of tadpoles (in vivo or in vitro) or adult frogs (Knutie et al., 2018). Discrepancies may arise due to dose, timing, route of exposure, host type and metabolism, and the applied chemical differences.
Besides the known impact of (halogenated) antibiotics in inducing major but partly reversible changes in gut microbiota composition (Dethlefsen and Relman, 2011; Vrieze et al., 2014), an increasing number of studies have shown microbiota perturbations by non-antibiotic biocides. For example, a recent comprehensive screening of more than 1,000 non-antibiotic drugs against 40 representative gut bacterial strains showed that 24% of the drugs with human targets inhibited the growth of at least one strain in vitro (Maier et al., 2018). Some intensively used organohalogen drugs such as thyroxine (L) (24), a medication used to treat thyroid hormone deficiency, and the anti-inflammatory diclofenac (25) were among the tested compounds (Maier et al., 2018). In another study, adolescent rats receiving triclosan orally at levels comparable to human exposures showed lower gut microbiota diversity and more noticeable compositional changes, whereas these differences were diminished in adult rats (Hu et al., 2016). Moreover, triclosan exposure was reported to reduce alpha diversity in the gut microbiota of rats (Kennedy et al., 2016; Gao et al., 2017) (Table 1). In contrast, triclosan exposure experiments in humans have not shown major perturbations in the gut and oral microbiota (Poole et al., 2016; Ribado et al., 2017) (Table 1). Since humans experience much lower triclosan exposures in products such as soap and toothpaste that are rinsed off immediately, the impacts observed in high-dose and acute animal exposures might not be observed in humans. However, a positive correlation was reported between the exposure to triclosan and the occurrence of Staphylococcus aureus as an opportunistic pathogen in the human nasal microbiota (Syed et al., 2014). There are also concerns regarding contribution of non-antibiotic antimicrobials to antibiotic resistance due to cross-resistance (Hartmann et al., 2016; Maier et al., 2018). For example, prolonged exposure to triclosan was associated with developing resistance and cross-resistance to ampicillin and/or ciprofloxacin in S. aureus and Escherichia coli (Wesgate et al., 2016). Exposure of a susceptible Pseudomonas aeruginosa strain to triclosan has been shown to select multidrug-resistance mediated by multidrug efflux pumps (Chuanchuen et al., 2001). Similarly, abundance of several multidrug-resistance efflux pump genes was reported to significantly increase after triclosan exposure (Gao et al., 2017). Recent studies on the mouse gut indicated the selective pressure of 2,3,7,8-tetrachlorodibenzo-p-dioxin in promoting blooms of Enterobacteriaceae, that harbor antimicrobial resistance genes (Stedtfeld et al., 2017) (Table 1). Similar increased levels of Enterobacteriaceae were reported in: (i) the gut of honey bees exposed to the organochlorine fungicide chlorothalonil (16) (Kakumanu et al., 2016), and (ii) the human gut due to gastrointestinal infection (Lupp et al., 2007) and antibiotic therapies (Sekirov et al., 2008). Finally, DBPs have the potential to select for antibiotic resistance (Li et al., 2016; Zhang et al., 2016). These reports indicate similar impacts despite different types of stressors and hosts.
Extensive studies have been performed to understand the fate of xenobiotics by oxidative and conjugative enzymes in the liver (Zanger et al., 2008; Zanger and Schwab, 2013). Among the hepatic enzymes, cytochrome P-450s are the major oxidative enzymes for transformation of (halogenated) lipophilic xenobiotics and drugs e.g., PCDDs (Hu and Bunce, 1999) and diclofenac (Leemann et al., 1993). In contrast, much less is known about the dehalogenation and/or degradation of halogenated xenobiotics by intestinal microbiota that employ hydrolytic and reductive mechanisms (Sousa et al., 2008).
Understanding of the fate of halogenated xenobiotics using cultivation-based studies has been derived from the exposure of the intestinal contents or specific microbial isolates of intestinal origin. For example, incubation of chloramphenicol, that contains a nitrobenzene group and an amide of dichloroacetic acid, with human fecal bacteria led to the hydrolysis of the amide linkage and reduction of the nitro group to an amine on the aromatic ring (Figure 3A) (Holt, 1967). Nitroreductases reducing nitro (–NO2) functional groups to the corresponding amines are an important group of enzymes identified for the gut microbial xenobiotic metabolism (Rickert et al., 1981; Claus et al., 2016). Another example of nitroreductive metabolism of organohalogens was shown for clonazepam, a medication used to prevent and treat seizures and panic disorder, that was converted to 7-aminoclonazepam by rat intestinal lumen microbiota (Figure 3B) (Elmer and Remmel, 1984). In contrast to such amine group formation, amine group removal by human intestinal microbiota was reported for the anti-fungal 5-fluorocytosine (Figure 3C) (Harris et al., 1986; Vermes et al., 2003). Susceptible fungi contain a cytosine deaminase which converts 5-fluorocytosine to 5-fluorouracil (Figure 3C). The latter is further metabolized to 5-fluorodeoxyuridylic acid, an inhibitor of thymidylate synthetase and subsequently DNA synthesis (Vermes et al., 2000). Although human host cells lack the deaminase enzyme, 5-fluorocytosine conversion to 5-fluorouracil by the human intestinal microbiota plays an important role in the development of hematologic and gastrointestinal toxicity (Harris et al., 1986). Co-administration of 5-fluorocytosine with the antiviral drug sorivudine led to 18 acute deaths due to an unknown lethal gut microbial metabolism (Okuda et al., 1998). Further research has revealed that intestinal Bacteroides species, namely Bacteroides vulgatus, B. thetaiotaomicron, B. fragilis, B. uniformis, and B. eggerthii can convert sorivudine to (E)-5-(2-bromovinyl)uracil (Nakayama et al., 1997) (Figure 3D), whereas the latter was barely detected in the plasma of germ-free rats (Ashida et al., 1993). A key liver enzyme that regulates the systemic 5-fluorocytosine level is subsequently inactivated by (E)-5-(2-bromovinyl)uracil, leading to toxic levels of 5-fluorocytosine and death in rats and humans (Okuda et al., 1998). This is an important example of the role of gut microbiota in toxification processes.
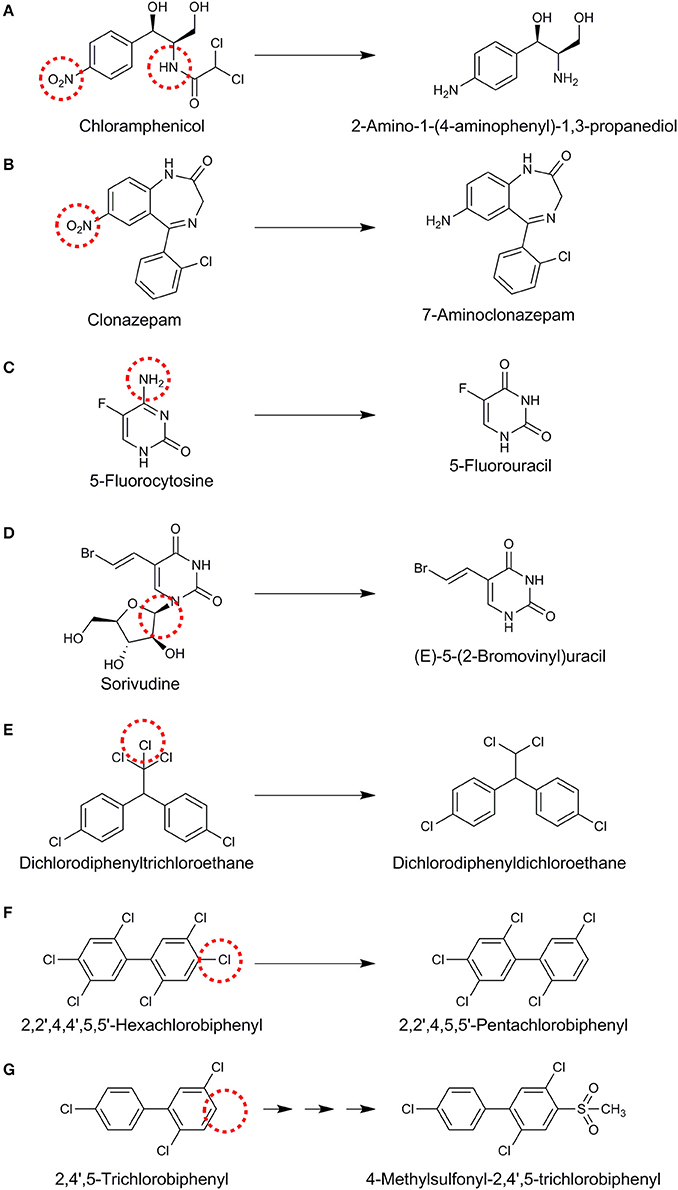
Figure 3. Proposed reactions for different organohalogens (A–G) mediated by gut microbiota. Reaction sites are indicated with red dashed circles.
Reductive dehalogenation of dichlorodiphenyltrichloroethane to dichlorodiphenyldichloroethane (Figure 3E) has been shown in anoxic incubations of the strictly anaerobic human intestinal bacterium Eubacterium limosum (Yim et al., 2008) and rat intestinal microbiota (Mendel and Walton, 1966). However, it is not known if this is a bioactivation or detoxification mechanism as dichlorodiphenyldichloroethane is still an endocrine disruptor (Claus et al., 2016). Reductive dehalogenation can be mediated co-metabolically by vitamin B12 (cobalamin) that is synthesized by some human gut microbes (Degnan et al., 2014). In contrast, metabolic reductive dehalogenation is mediated by specific bacterial groups that can use organohalogens as their terminal electron acceptors (Smidt and de Vos, 2004; Atashgahi et al., 2016). A co-culture of Clostridium perfringens and C. beijerinckii was also shown to reductively dehalogenate hexachlorobiphenyl to pentachlorobiphenyl (Figure 3F) and tetrachlorobiphenyl to trichlorobiphenyl (De et al., 2006). Gut microbiota was also shown to be involved in generation of methylsulfone (MeSO2) metabolites from polychlorinated biphenyls (PCBs) through a series of reactions in combination with the host cells (Figure 3G) (Bakke et al., 1982; Brandt et al., 1982). MeSO2–PCBs can bind to specific proteins and accumulate in the lipophylic tissues with adverse effects (Shigematsu et al., 1978). Exposure to 2,3,7,8-tetrachlorodibenzofuran has been shown to enhance the level of Flavobacteria in the gut of mice (Zhang L. et al., 2015). These bacteria are reported to possess glutathione-dependent reductive dehalogenase activity (Xun et al., 1992), although 2,3,7,8-tetrachlorodibenzofuran dehalogenation by the gut microbiota has not been shown yet.
Intestinal lactobacilli have been reported to degrade organohalogens in vitro. For instance, Lactobacillus lactis, L. fermentum, L. plantarum, E. coli, and Enterococcus faecalis were tested for chlorpyrifos degradation potential. The results indicated that besides E. coli, L. lactis and L. fermentum could grow in the presence of over 1.5 mg/mL chlorpyrifos (Harishankar et al., 2013). Similarly, four lactic acid bacteria isolated from kimchi fermentation in the presence of 200 mg/L chlorpyrifos were reported to use this pesticide as the sole source of carbon and phosphorus (Cho et al., 2009). Moreover, lactic acid bacteria seeded to skimmed milk were shown to degrade the insecticide trichlorfon (17) (Zhao and Wang, 2012). However, the actual impact of lactobacilli on organohalogen fate in the gut is not known, especially since this bacterial group seems particularly sensitive to organohalogen exposure (Table 1).
As a complementary approach to cultivation, metagenomic approaches can also be used to infer xenobiotic metabolism potential by the gut microbiota (Haiser and Turnbaugh, 2013; Spanogiannopoulos et al., 2016). Due to environmental persistence and toxicity, great attention has been given to understand microbial transformation of halogenated xenobiotics in environmental studies with the end goal of bioremediation. Indeed a variety of microbial dehalogenation mechanisms have been described that can remove halogens from organic compounds by oxidation, reduction and substitution mechanisms that are employed in co-metabolic and/or energy-yielding modes (van Pée, 1996; Fetzner, 1998; Janssen et al., 2001; Smidt and de Vos, 2004).
Although a metagenomic view of xenobiotic metabolism in the gut has been provided (Haiser and Turnbaugh, 2013; Spanogiannopoulos et al., 2016), specific information on the prevalence and diversity of known dehalogenase-encoding genes in the gut metagenome is lacking. Therefore, we surveyed the Joint Genome Institute Integrated Microbial Genomes & Microbiome System (JGI-IMG/MER) database for the occurrence of different dehalogenating gene classes (Table S1) in 670 bacterial and archaeal genomes from fecal origin (Table S2), 254 metagenomes obtained from human fecal samples (Table S3) and 86 metagenomes obtained from gut/rumen fluid/fecal samples of animals (Table S4). The metadata and criteria used for the selection of genomes and metagenomes are listed in Tables S2–4. The codes used for the analysis and visualization are provided in supplementary information. The results showed that at least one dehalogenating gene was present in 32.2% of the bacterial and archaeal genomes, and in 61 and 75.6% of the metagenomes derived from human and non-human origins, respectively (Tables S5–S7). Among these, five types of genes were found in the bacterial genomes (Figure 4, Table S5) and human metagenomes (Table S6), and six types of genes in non-human metagenomes (Table S7). Four types of genes were shared among the three datasets and were predicted to code for (S)-2-haloacid dehalogenase (EC:3.8.1.2), haloacetate dehalogenase (EC:3.8.1.3), haloalkane dehalogenase (EC:3.8.1.5), and reductive dehalogenase (EC:1.21.99.5). In contrast, a canonical gene for chlorate dismutase (EC: 1.13.11.49) was not found in the human metagenomes whereas that for atrazine chlorohydrolase (EC:3.8.1.8) was absent from the bacterial genomes. Atrazine chlorohydrolase catalyzes the conversion of the herbicide atrazine to hydroxyatrazine, the first step in the atrazine degradation pathway (Mandelbaum et al., 1995).
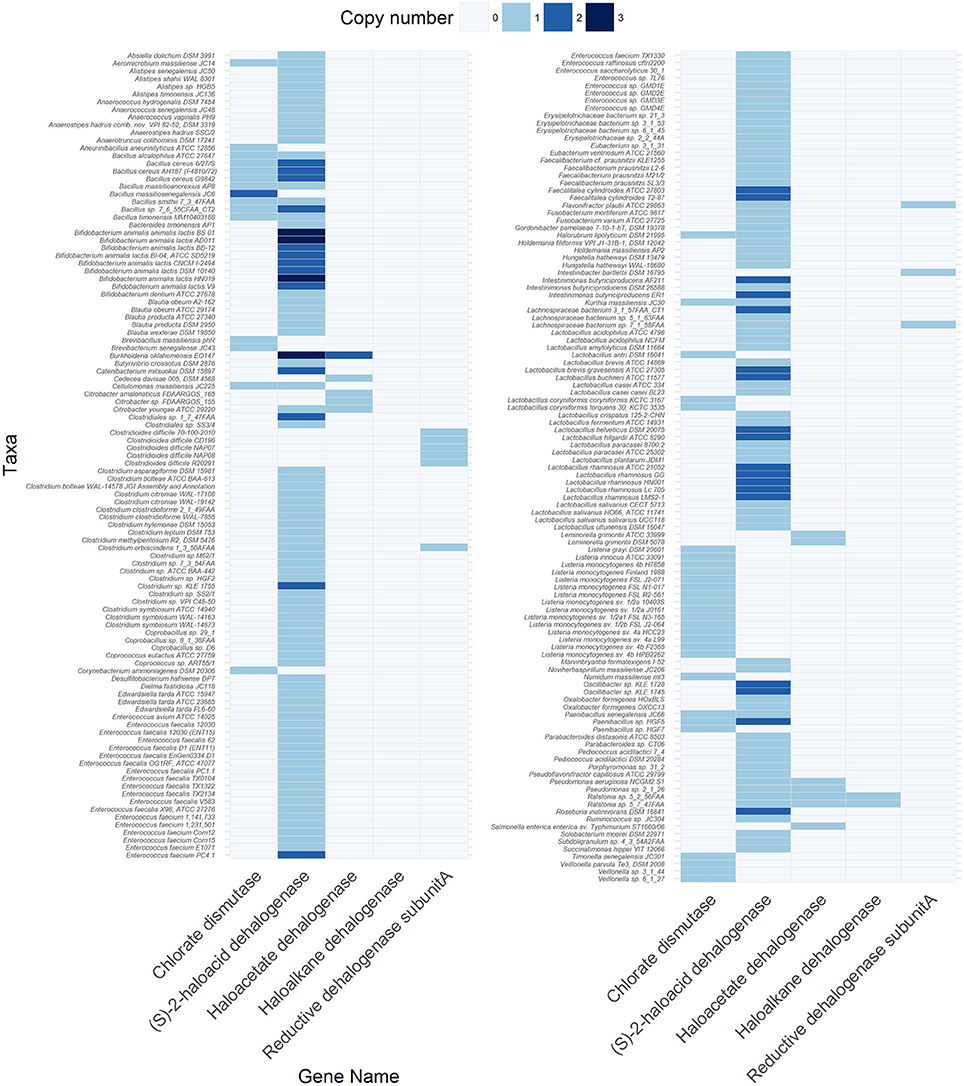
Figure 4. Genes encoding different dehalogenases found in 216 out of 670 bacterial and archaeal genomes of gut origin. The code to reproduce the figure is available at https://github.com/mibwurrepo/Atashgahi-et-al.-XenobioticReview2018.
A gene for (S)-2-haloacid dehalogenase was the most abundant in both the genome (214 genes) and metagenome (2612 genes in human and 4549 genes in non-human) datasets indicating exposure of gut microbiota to haloacids that are common DBPs (Richardson et al., 2007). Many of the known core gut genera, such as Faecalibacterium, Blautia, Roseburia, Alistipes, Eubacterium (Shetty et al., 2017) harbor a (S)-2-haloacid dehalogenase gene in their genome (Figure 4, Table S5). The encoded enzyme belongs to the family of hydrolases, acting specifically on halide bonds in α-substituted haloacids (Janssen et al., 2001). Interestingly, a (S)-2-haloacid dehalogenase encoding gene co-occurred with that of chlorite dismutase in 14 microbial genomes (Figure 4, Table S5). Chlorite dismutase mediates the last step in chlorate reduction splitting chlorite to chloride and oxygen (Liebensteiner et al., 2016). An environmental bacterium, Pseudomonas chloritidismutans AW-1T, which similarly harbored these two genes, was recently shown to concurrently degrade haloacids and chlorate as the electron donor and acceptor, respectively (Peng et al., 2017). This is an interesting finding considering that haloacids and chlorate are common DBPs in drinking water resources (Richardson et al., 2007) that might be degraded by the gut microbes harboring these genes. Further, the oxygen produced from chlorite dismutation can be used for degradation of haloacids (Peng et al., 2017) or other organic compounds (Oosterkamp et al., 2013; Atashgahi et al., 2018b) in an “intra-aerobic” pathway. If functional, this can be an important metabolism in the gut environment where oxygen is largely unavailable to serve as a terminal electron acceptor.
The well-studied reductive dehalogenase genes were more abundant in the non-human than in the human metagenomes (Table S7). Interestingly, of the 59 reductive dehalogenase genes found in the non-human metagenomes, 53 were from the rumen content of sheep, goat and cow. This implies that the rumen content of ruminant farm animals is an appropriate environment for the reductive dehalogenation metabolism that represents useful but largely unexplored sources for future enrichment/isolation of organohalide-respiring bacteria. These bacteria reductively dehalogenate organohalogens by replacing the halogen substitutes with hydrogen in a process known as organohalide respiration (Smidt and de Vos, 2004; Atashgahi et al., 2016). This metabolism usually reduces the toxicity of organohalogens and makes the otherwise chemically locked organohalogens available to other microbial metabolisms such as fermentative and aerobic degradation. Organohalide-respiring bacteria have only been found in pristine and contaminated environments impacted by natural or anthropogenic organohalogens (Atashgahi et al., 2018a). The activity of these microbes in host-associated ecosystems and their potential impacts on the organohalogen fate especially in the gut remains largely unknown. There is only one study showing reductive dehalogenation of polychlorinated biphenyls by the co-culture of Clostridium perfringens and C. beijerinckii as prominent species in the human gut (De et al., 2006) (Figure 3F). Interestingly, we found six reductive dehalogenase genes in the genomes of Clostridia (Figure 4, Table S5). This may point to an unrecognized reductive dehalogenation potential in the gut microbiota. Of interest, the gut isolate strain DP7, belonging to the genus Desulfitobacterium (Clostridia) that is known for its active organohalogen respiration metabolism (Kruse et al., 2017), was reported to lack reductive dehalogenation activity (van de Pas et al., 2001) and the corresponding genes (Kruse et al., 2017).
The toxicant-microbiota interaction has emerged in recent years as one of the novel concepts from the intensive research on the human microbiome. The gut microbiota constitutes a critical zone for xenobiotic (de)toxification and sequestration at the interphase between the external environment and our mucosal epithelial cells. An important class of toxicants are halogenated compounds from anthropogenic and natural sources that come into contact with the human gut and other body parts mainly by ingestion of, or exposure to contaminated food and water. Although canonical toxicological approaches using short-term high-dose exposure experiments are informative about the toxicity and impact of halogenated compounds, they do not represent scenarios of chronic exposure to low-level xenobiotic cocktails throughout life. The ingested concentrations of the halogenated xenobiotics in food and water resources are in most cases below the regulatory thresholds. However, little is known about the physiological impact, reactivity, bioaccumulation in food chains, additive/cumulative toxicity of these emerging contaminants, and their (bio)transformation products. For example, even at trace concentrations, mixtures of biocides, antibiotics, and heavy metals have the potential to contribute to the emergence, maintenance and transmission of antibiotic-resistant and disease-causing bacteria (Gullberg et al., 2014; Pal et al., 2015). Therefore, future studies are necessary to reveal the impact of halogenated (micro)pollutants on the gut microbial community membership, gene expression, physiology, metabolite profile, antibiotic resistance genes, and also parallel impacts on the host. Long-term incubations of the gut contents/isolates with environmentally relevant doses and diversity of halogenated compounds, in combination with other micropollutants, should aid in understanding the actual consequences of chronic low-dose exposures.
Even less information is available on the specific microorganisms responsible for halogenated xenobiotic metabolism, the molecular mechanisms and biotransformation pathways involved that can either diminish or enhance the toxicity. The metagenomic approach described here showed that genes involved in dehalogenation are widespread among gut bacteria, and this may impact flux, toxicity, bioavailability and fate of halogenated compounds. Future cultivation and omics experiments are necessary to test the actual metabolism of the halogenated compounds by the gut microbiota. To this end, we can immensely benefit from the wealth of knowledge gained about the metabolism of halogenated xenobiotic compounds in terrestrial and aquatic environments (Janssen et al., 2001; Smidt and de Vos, 2004; Atashgahi et al., 2018a) and high-throughput cultivation of the gut microbiota (Ingham et al., 2007; Lagier et al., 2016). These approaches should be coupled with untargeted metabolomics using high-resolution mass spectroscopy to identify xenobiotics and biotransformation products. Untargeted metabolomics has the potential to aid in determination of pathways and mechanisms of action (Warth et al., 2017).
Given the immense potential of gut microbiota to alter the chemical structure and bioactivity of xenobiotics with beneficial (Shin et al., 2013) or severely detrimental impacts (Okuda et al., 1998), assessments of xenobiotic metabolism should be an integral part of designing drugs and chemicals such as PPCPs and pesticides, informing toxicology risk assessment, improving nutrition, and guiding personalized medicine.
SA, HS and WMdV have designed the study, SAS has performed the metagenomic analysis and all authors wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research is supported by the Soehngen Institute of Anaerobic Microbiology (SIAM) Gravitation grant (024.002.002) and Spinoza Award of the Netherlands Organization for Scientific Research (NWO) to WMdV. We thank the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) for maintaining and curating the genomic and metagenomic, and making available via the IMG system.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00888/full#supplementary-material
Agarwal, V., Blanton, J. M., Podell, S., Taton, A., Schorn, M. A., Busch, J., et al. (2017). Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 13, 537–543. doi: 10.1038/nchembio.2330
Andrade, N. A., Lozano, N., McConnell, L. L., Torrents, A., Rice, C. P., and Ramirez, M. (2015). Long-term trends of PBDEs, triclosan, and triclocarban in biosolids from a wastewater treatment plant in the Mid-Atlantic region of the US. J. Haz. Mat. 282, 68–74. doi: 10.1016/j.jhazmat.2014.09.028
Arias-Estévez, M., López-Periago, E., Martínez-Carballo, E., Simal-Gándara, J., Mejuto, J.-C., and García-Río, L. (2008). The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agri. Ecosyst. Environ. 123, 247–260. doi: 10.1016/j.agee.2007.07.011
Arsenescu, V., Arsenescu, R. I., King, V., Swanson, H., and Cassis, L. A. (2008). Polychlorinated biphenyl-77 induces adipocyte differentiation and proinflammatory adipokines and promotes obesity and atherosclerosis. Environ. Health Perspect. 116, 761–768. doi: 10.1289/ehp.10554
Ashida, N., Ijichi, K., Watanabe, Y., and Machida, H. (1993). Metabolism of 5′-ether prodrugs of 1-β-d-Arabinofuranosyl-E-5-(2-bromovinyl) uracil in rats. Biochem. Pharmacol. 46, 2201–2207. doi: 10.1016/0006-2952(93)90610-9
Atashgahi, S., Häggblom, M. M., and Smidt, H. (2018a). Organohalide respiration in pristine environments: implications for the natural halogen cycle. Environ. Microbiol. 20, 934–948. doi: 10.1111/1462-2920.14016
Atashgahi, S., Hornung, B., Waals, M. J., Rocha, U. N., Hugenholtz, F., Nijsse, B., et al. (2018b). A benzene-degrading nitrate-reducing microbial consortium displays aerobic and anaerobic benzene degradation pathways. Sci. Rep. 8:4490. doi: 10.1038/s41598-018-22617-x
Atashgahi, S., Lu, Y., Ramiro-Garcia, J., Peng, P., Maphosa, F., Sipkema, D., et al. (2017). Geochemical parameters and reductive dechlorination determine aerobic cometabolic vs aerobic metabolic vinyl chloride biodegradation at oxic/anoxic interface of hyporheic zones. Environ. Sci. Technol. 51, 1626–1634. doi: 10.1021/acs.est.6b05041
Atashgahi, S., Lu, Y., and Smidt, H. (2016). “Overview of known organohalide-respiring bacteria - phylogenetic diversity and environmental distribution,” in Organohalide-Respiring Bacteria, eds L. Adrian and F. Löffler (Berlin; Heidelberg: Springer-Verlag Berlin Heidelberg), 63–105.
Atashgahi, S., Sánchez-Andrea, I., Heipieper, H. J., van der Meer, J. R., Stams, A. J. M., and Smidt, H. (2018c). Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 360, 743–746. doi: 10.1126/science.aar3778
Bahl, M. I., Bergström, A., and Licht, T. R. (2012). Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol. Lett. 329, 193–197. doi: 10.1111/j.1574-6968.2012.02523.x
Baker, N. A., Karounos, M., English, V., Fang, J., Wei, Y., Stromberg, A., et al. (2013). Coplanar polychlorinated biphenyls impair glucose homeostasis in lean C57BL/6 mice and mitigate beneficial effects of weight loss on glucose homeostasis in obese mice. Environ. Health Perspect. 121, 105–110. doi: 10.1289/ehp.1205421
Bakke, J. E., Bergman, A. L., and Larsen, G. L. (1982). Metabolism of 2, 4, 5-trichlorobiphenyl by the mercapturic acid pathway. Science 217, 645–647. doi: 10.1126/science.6806905
Benotti, M. J., Trenholm, R. A., Vanderford, B. J., Holady, J. C., Stanford, B. D., and Snyder, S. A. (2008). Pharmaceuticals and endocrine disrupting compounds in US drinking water. Environ. Sci. Technol. 43, 597–603. doi: 10.1021/es801845a
Besis, A., and Samara, C. (2012). Polybrominated diphenyl ethers (PBDEs) in the indoor and outdoor environments–a review on occurrence and human exposure. Environ. Pollut. 169, 217–229. doi: 10.1016/j.envpol.2012.04.009
Boxall, A. B., Johnson, P., Smith, E. J., Sinclair, C. J., Stutt, E., and Levy, L. S. (2006). Uptake of veterinary medicines from soils into plants. J. Agr. Food Chem. 54, 2288–2297. doi: 10.1021/jf053041t
Boxall, A. B., Rudd, M. A., Brooks, B. W., Caldwell, D. J., Choi, K., Hickmann, S., et al. (2012). Pharmaceuticals and personal care products in the environment: what are the big questions? Environ. Health Perspect. 120, 1221–1229. doi: 10.1289/ehp.1104477
Bradberry, S. M., Watt, B. E., Proudfoot, A. T., and Vale, J. A. (2000). Mechanisms of toxicity, clinical features, and management of acute chlorophenoxy herbicide poisoning: a review. J. Toxicol. Clin. Toxicol. 38, 111–122. doi: 10.1081/CLT-100100925
Brandt, I., Klasson-Wehler, E., Rafter, J., and Bergman, A. (1982). Metabolism of 2, 4′, 5-trichlorobiphenyl: tissue concentrations of methylsulphonyl-2, 4′, 5-trichlorobiphenyl in germfree and conventional mice. Toxicol. Lett. 12, 273–280. doi: 10.1016/0378-4274(82)90251-X
Calafat, A. M., Ye, X., Wong, L.-Y., Reidy, J. A., and Needham, L. L. (2008). Urinary concentrations of triclosan in the US population: 2003–2004. Environ. Health Perspect. 116, 303–307. doi: 10.1289/ehp.10768
Cho, K. M., Math, R. K., Islam, S. M., Lim, W. J., Hong, S. Y., Kim, J. M., et al. (2009). Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J. Agr. Food Chem. 57, 1882–1889. doi: 10.1021/jf803649z
Choi, J. J., Eum, S. Y., Rampersaud, E., Daunert, S., Abreu, M. T., and Toborek, M. (2013). Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 121, 725–730. doi: 10.1289/ehp.1306534
Chuanchuen, R., Beinlich, K., Hoang, T. T., Becher, A., Karkhoff-Schweizer, R. R., and Schweizer, H. P. (2001). Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45, 428–432. doi: 10.1128/AAC.45.2.428-432.2001
Claus, S. P., Guillou, H., and Ellero-Simatos, S. (2016). The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2:16003. doi: 10.1038/npjbiofilms.2016.3
Daiber, E. J., DeMarini, D. M., Ravuri, S. A., Liberatore, H. K., Cuthbertson, A. A., Thompson-Klemish, A., et al. (2016). Progressive increase in disinfection byproducts and mutagenicity from source to tap to swimming pool and spa water: impact of human inputs. Environ. Sci. Technol. 50, 6652–6662. doi: 10.1021/acs.est.6b00808
Damalas, C. A., and Eleftherohorinos, I. G. (2011). Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 8, 1402–1419. doi: 10.3390/ijerph8051402
De, S., Ghosh, S., and Dutta, S. K. (2006). Congener specific polychlorinated biphenyl metabolism by human intestinal microbe Clostridium species: Comparison with human liver cell line-HepG2. Ind. J. Microbiol. 46, 199–207.
Degnan, P. H., Taga, M. E., and Goodman, A. L. (2014). Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778. doi: 10.1016/j.cmet.2014.10.002
Dethlefsen, L., and Relman, D. A. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 108, 4554–4561. doi: 10.1073/pnas.1000087107
Dietert, R. R., and Silbergeld, E. K. (2015). Biomarkers for the 21st century: listening to the microbiome. Toxicol. Sci. 144, 208–216. doi: 10.1093/toxsci/kfv013
Duncan, S. H., Louis, P., Thomson, J. M., and Flint, H. J. (2009). The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 11, 2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x
Ebele, A. J., Abdallah, M. A.-E., and Harrad, S. (2017). Pharmaceuticals and personal care products (PPCPs) in the freshwater aquatic environment. Emerg. Contam. 3, 1–16. doi: 10.1016/j.emcon.2016.12.004
Elmer, G. W., and Remmel, R. P. (1984). Role of the intestinal microflora in clonazepam metabolism in the rat. Xenobiotica 14, 829–840. doi: 10.3109/00498258409151481
Fader, K. A., Nault, R., Ammendolia, D. A., Harkema, J. R., Williams, K. J., Crawford, R. B., et al. (2015). 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin alters lipid metabolism and depletes immune cell populations in the jejunum of C57BL/6 mice. Toxicol. Sci. 148, 567–580. doi: 10.1093/toxsci/kfv206
Fang, B., Li, J. W., Zhang, M., Ren, F. Z., and Pang, G. F. (2018). Chronic chlorpyrifos exposure elicits diet-specific effects on metabolism and the gut microbiome in rats. Food Chem. Toxicol. 111, 144–152. doi: 10.1016/j.fct.2017.11.001
Fetzner, S. (1998). Bacterial dehalogenation. Appl. Microbiol. Biotechnol. 50, 633–657. doi: 10.1007/s002530051346
Fick, J., Söderström, H., Lindberg, R. H., Phan, C., Tysklind, M., and Larsson, D. (2009). Contamination of surface, ground, and drinking water from pharmaceutical production. Environ. Toxicol. Chem. 28, 2522–2527. doi: 10.1897/09-073.1
Gao, B., Tu, P., Bian, X., Chi, L., Ru, H., and Lu, K. (2017). Profound perturbation induced by triclosan exposure in mouse gut microbiome: a less resilient microbial community with elevated antibiotic and metal resistomes. BMC Pharmacol. Toxicol. 18:46. doi: 10.1186/s40360-017-0150-9
Gonsior, M., Schmitt-Kopplin, P., Stavklint, H., Richardson, S. D., Hertkorn, N., and Bastviken, D. (2014). Changes in dissolved organic matter during the treatment processes of a drinking water plant in Sweden and formation of previously unknown disinfection byproducts. Environ. Sci. Technol. 48, 12714–12722. doi: 10.1021/es504349p
Gribble, G. W. (2010). Naturally Occurring Organohalogen Compounds - A Comprehensive Update. Vienna; New York, NY: Springer.
Guerra, P., Kim, M., Kinsman, L., Ng, T., Alaee, M., and Smyth, S. (2014). Parameters affecting the formation of perfluoroalkyl acids during wastewater treatment. J. Haz. Mat. 272, 148–154. doi: 10.1016/j.jhazmat.2014.03.016
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., and Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5, e01918–e01914. doi: 10.1128/mBio.01918-14
Häggblom, M. M., and Bossert, I. D. (2003). Dehalogenation: Microbial Processes and Environmental Applications. Boston, MA: Kluwer Academic Publisher Group.
Haiser, H. J., and Turnbaugh, P. J. (2013). Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 69, 21–31. doi: 10.1016/j.phrs.2012.07.009
Halden, R. U. (2016). Lessons learned from probing for impacts of triclosan and triclocarban on human microbiomes. mSphere 1, e00089–e00016. doi: 10.1128/mSphere.00089-16
Halden, R. U., and Paull, D. H. (2005). Co-occurrence of triclocarban and triclosan in US water resources. Environ. Sci. Technol. 39, 1420–1426. doi: 10.1021/es049071e
Harishankar, M. K., Sasikala, C., and Ramya, M. (2013). Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech 3, 137–142. doi: 10.1007/s13205-012-0078-0
Harris, B. E., Manning, B. W., Federle, T. W., and Diasio, R. B. (1986). Conversion of 5-fluorocytosine to 5-fluorouracil by human intestinal microflora. Antimicrob. Agents Chemother. 29, 44–48. doi: 10.1128/AAC.29.1.44
Hartmann, E. M., Hickey, R., Hsu, T., Betancourt Román, C. M., Chen, J., Schwager, R., et al. (2016). Antimicrobial chemicals are associated with elevated antibiotic resistance genes in the indoor dust microbiome. Environ. Sci. Technol. 50, 9807–9815. doi: 10.1021/acs.est.6b00262
Heidler, J., and Halden, R. U. (2009). Fate of organohalogens in US wastewater treatment plants and estimated chemical releases to soils nationwide from biosolids recycling. J. Environ. Monitor. 11, 2207–2215. doi: 10.1039/b914324f
Holt, R. (1967). The bacterial degradation of chloramphenicol. Lancet 289, 1259–1260. doi: 10.1016/S0140-6736(67)92720-1
Holvoet, K. M., Seuntjens, P., and Vanrolleghem, P. A. (2007). Monitoring and modeling pesticide fate in surface waters at the catchment scale. Ecol. Model. 209, 53–64. doi: 10.1016/j.ecolmodel.2007.07.030
Horemans, B., Raes, B., Vandermaesen, J., Simanjuntak, Y., Brocatus, H., T'Syen, J., et al. (2017). Biocarriers improve bioaugmentation efficiency of a rapid sand filter for the treatment of 2, 6-dichlorobenzamide-contaminated drinking water. Environ. Sci. Technol. 51, 1616–1625. doi: 10.1021/acs.est.6b05027
Hu, J., Raikhel, V., Gopalakrishnan, K., Fernandez-Hernandez, H., Lambertini, L., Manservisi, F., et al. (2016). Effect of postnatal low-dose exposure to environmental chemicals on the gut microbiome in a rodent model. Microbiome 4:26. doi: 10.1186/s40168-016-0173-2
Hu, K., and Bunce, N. J. (1999). Metabolism of polychlorinated dibenzo-p-dioxins by rat liver microsomes. J. Biochem. Mol. Toxicol. 13, 307–315. doi: 10.1002/(SICI)1099-0461(1999)13:6<307::AID-JBT4>3.0.CO;2-P
Ingham, C. J., Sprenkels, A., Bomer, J., Molenaar, D., van den Berg, A., van Hylckama Vlieg, J. E., et al. (2007). The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 104, 18217–18222. doi: 10.1073/pnas.0701693104
Janssen, D. B., Oppentocht, J. E., and Poelarends, G. J. (2001). Microbial dehalogenation. Curr. Opin. Biotechnol. 12, 254–258. doi: 10.1016/S0958-1669(00)00208-1
Jmaiff Blackstock, L. K., Wang, W., Vemula, S., Jaeger, B. T., and Li, X.-F. (2017). Sweetened swimming pools and hot tubs. Environ. Sci. Technol. 4, 149–153. doi: 10.1021/acs.estlett.7b00043
John, E. M., and Shaike, J. M. (2015). Chlorpyrifos: pollution and remediation. Environ. Chem. Lett. 13, 269–291. doi: 10.1007/s10311-015-0513-7
Joly, C., Gay-Quéheillard, J., Léké, A., Chardon, K., Delanaud, S., Bach, V., et al. (2013). Impact of chronic exposure to low doses of chlorpyrifos on the intestinal microbiota in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) and in the rat. Environ. Sci. Pollut. Res. 20, 2726–2734. doi: 10.1007/s11356-012-1283-4
Kakumanu, M. L., Reeves, A. M., Anderson, T. D., Rodrigues, R. R., and Williams, M. A. (2016). Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol. 7:1255. doi: 10.3389/fmicb.2016.01255
Kan, H., Zhao, F., Zhang, X.-X., Ren, H., and Gao, S. (2015). Correlations of gut microbial community shift with hepatic damage and growth inhibition of Carassius auratus induced by pentachlorophenol exposure. Environ. Sci. Technol. 49, 11894–11902. doi: 10.1021/acs.est.5b02990
Kennedy, R. C., Fling, R. R., Robeson, M. S., Saxton, A. M., Donnell, R. L., Darcy, J. L., et al. (2016). Temporal development of gut microbiota in triclocarban exposed pregnant and neonatal rats. Sci. Rep. 6:33430. doi: 10.1038/srep33430
Kim, M.-J., Marchand, P., Henegar, C., Antignac, J.-P., Alili, R., Poitou, C., et al. (2011). Fate and complex pathogenic effects of dioxins and polychlorinated biphenyls in obese subjects before and after drastic weight loss. Environ. Health Perspect. 119, 377–383. doi: 10.1289/ehp.1002848
Kiviranta, H., Ovaskainen, M. L., and Vartiainen, T. (2004). Market basket study on dietary intake of PCDD/Fs, PCBs, and PBDEs in Finland. Environ. Int. 30, 923–932. doi: 10.1016/j.envint.2004.03.002
Knutie, S. A., Gabor, C. R., Kohl, K. D., and Rohr, J. R. (2018). Do host-associated gut microbiota mediate the effect of an herbicide on disease risk in frogs? J. Animal Ecol. 87, 489–499. doi: 10.1111/1365-2656.12769
Kolmeder, C. A., Ritari, J., Verdam, F. J., Muth, T., Keskitalo, S., Varjosalo, M., et al. (2015). Colonic metaproteomic signatures of active bacteria and the host in obesity. Proteomics 15, 3544–3552. doi: 10.1002/pmic.201500049
Kolpin, D. W., Furlong, E. T., Meyer, M. T., Thurman, E. M., Zaugg, S. D., Barber, L. B., et al. (2002). Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol. 36, 1202–1211. doi: 10.1021/es011055j
Koppel, N., Rekdal, V., and Balskus, E. P. (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science 356:eaag2770. doi: 10.1126/science.aag2770
Kruse, T., Goris, T., Maillard, J., Woyke, T., Lechner, U., de Vos, W., et al. (2017). Comparative genomics of the genus Desulfitobacterium. FEMS Microbiol. Ecol. 93:fix135. doi: 10.1093/femsec/fix135
Lagier, J. C., Khelaifia, S., Alou, M. T., Ndongo, S., Dione, N., Hugon, P., et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1:16203. doi: 10.1038/nmicrobiol.2016.203
Larsson, D. G., de Pedro, C., and Paxeus, N. (2007). Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Haz. Mat. 148, 751–755. doi: 10.1016/j.jhazmat.2007.07.008
Lee, D.-H., Porta, M., Jacobs Jr, D. R., and Vandenberg, L. N. (2014). Chlorinated persistent organic pollutants, obesity, and type 2 diabetes. Endocrine Rev. 35, 557–601. doi: 10.1210/er.2013-1084
Leemann, T., Transon, C., and Dayer, P. (1993). Cytochrome P450TB (CYP2C): a major monooxygenase catalyzing diclofenac 4′-hydroxylation in human liver. Life Sci. 52, 29–34. doi: 10.1016/0024-3205(93)90285-B
Lefever, D. E., Xu, J., Chen, Y., Huang, G., Tamas, N., and Guo, T. L. (2016). TCDD modulation of gut microbiome correlated with liver and immune toxicity in streptozotocin (STZ)-induced hyperglycemic mice. Toxical. Appl. Pharmacol. 304, 48–58. doi: 10.1016/j.taap.2016.05.016
Leung, A. O., Luksemburg, W. J., Wong, A. S., and Wong, M. H. (2007). Spatial distribution of polybrominated diphenyl ethers and polychlorinated dibenzo-p-dioxins and dibenzofurans in soil and combusted residue at Guiyu, an electronic waste recycling site in southeast China. Environ. Sci. Technol. 41, 2730–2737. doi: 10.1021/es0625935
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Li, D., Zeng, S., He, M., and Gu, A. Z. (2016). Water disinfection byproducts induce antibiotic resistance-role of environmental pollutants in resistance phenomena. Environ. Sci. Technol. 50, 3193–3201. doi: 10.1021/acs.est.5b05113
Li, J., Jia, H., Cai, X., Zhong, H., Feng, Q., Sunagawa, S., et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841. doi: 10.1038/nbt.2942
Liebensteiner, M. G., Oosterkamp, M. J., and Stams, A. J. (2016). Microbial respiration with chlorine oxyanions: diversity and physiological and biochemical properties of chlorate- and perchlorate-reducing microorganisms. Ann. N. Y. Acad. Sci. 1365, 59–72. doi: 10.1111/nyas.12806
Lindström, A., Buerge, I. J., Poiger, T., Bergqvist, P.-A., Müller, M. D., and Buser, H.-R. (2002). Occurrence and environmental behavior of the bactericide triclosan and its methyl derivative in surface waters and in wastewater. Environ. Sci. Technol. 36, 2322–2329. doi: 10.1021/es0114254
Lu, K., Mahbub, R., and Fox, J. G. (2015). Xenobiotics: interaction with the intestinal microflora. ILAR J. 56, 218–227. doi: 10.1093/ilar/ilv018
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., et al. (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129. doi: 10.1016/j.chom.2007.06.010
Maier, L., Pruteanu, M., Kuhn, M., Zeller, G., Telzerow, A., Anderson, E. E., et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628. doi: 10.1038/nature25979
Mandelbaum, R. T., Allan, D. L., and Wackett, L. P. (1995). Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61, 1451–1457.
Martínez-Carballo, E., González-Barreiro, C., Scharf, S., and Gans, O. (2007). Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 148, 570–579. doi: 10.1016/j.envpol.2006.11.035
Matsumoto, M., Inoue, R., Tsukahara, T., Ushida, K., Chiji, H., Matsubara, N., et al. (2008). Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72, 572–576. doi: 10.1271/bbb.70474
Maurice, C. F., Haiser, H. J., and Turnbaugh, P. J. (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50. doi: 10.1016/j.cell.2012.10.052
Meckenstock, R. U., Elsner, M., Griebler, C., Lueders, T., Stumpp, C., Aamand, J., et al. (2015). Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 49, 7073–7081. doi: 10.1021/acs.est.5b00715
Mendel, J. L., and Walton, M. S. (1966). Conversion of p, p'-DDT to p, p'-DDD by intestinal flora of the rat. Science 151, 1527–1528. doi: 10.1126/science.151.3717.1527
Monteleone, I., MacDonald, T. T., Pallone, F., and Monteleone, G. (2012). The aryl hydrocarbon receptor in inflammatory bowel disease: linking the environment to disease pathogenesis. Curr. Opin. Gastroenterol. 28, 310–313. doi: 10.1097/MOG.0b013e328352ad69
Myre, M., and Imbeault, P. (2014). Persistent organic pollutants meet adipose tissue hypoxia: does cross-talk contribute to inflammation during obesity? Obes. Rev. 15, 19–28. doi: 10.1111/obr.12086
Nakayama, H., Kinouchi, T., Kataoka, K., Akimoto, S., Matsuda, Y., and Ohnishi, Y. (1997). Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl) uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics 7, 35–43. doi: 10.1097/00008571-199702000-00005
Nelson, G. M., Swank, A. E., Brooks, L. R., Bailey, K. C., and George, S. E. (2001). Metabolism, microflora effects, and genotoxicity in haloacetic acid-treated cultures of rat cecal microbiota. Toxicol. Sci. 60, 232–241. doi: 10.1093/toxsci/60.2.232
Nielsen, H. B., Almeida, M., Juncker, A. S., Rasmussen, S., Li, J., Sunagawa, S., et al. (2014). Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828. doi: 10.1038/nbt.2939
Noguera-Oviedo, K., and Aga, D. S. (2016). Lessons learned from more than two decades of research on emerging contaminants in the environment. J. Haz. Mat. 316, 242–251. doi: 10.1016/j.jhazmat.2016.04.058
Okuda, H., Ogura, K., Kato, A., Takubo, H., and Watabe, T. (1998). A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J. Pharmacol. Exp. Ther. 287, 791–799.
Oosterkamp, M. J., Veuskens, T., Talarico Saia, F., Weelink, S. A., Goodwin, L. A., Daligault, H. E., et al. (2013). Genome analysis and physiological comparison of Alicycliphilus denitrificans strains BC and K601T. PLoS ONE 8:e66971. doi: 10.1371/journal.pone.0066971
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. J. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5
Peng, H., Chen, C., Saunders, D. M., Sun, J., Tang, S., Codling, G., et al. (2015). Untargeted identification of organo-bromine compounds in lake sediments by ultrahigh-resolution mass spectrometry with the data-independent precursor isolation and characteristic fragment method. Anal. Chem. 87, 10237–10246. doi: 10.1021/acs.analchem.5b01435
Peng, P., Zheng, Y., Koehorst, J. J., Schaap, P. J., Stams, A. J., Smidt, H., et al. (2017). Concurrent haloalkanoate degradation and chlorate reduction by Pseudomonas chloritidismutans AW-1T. Appl. Environ. Microbiol. 83, e00325–e00317. doi: 10.1128/AEM.00325-17
Poole, A. C., Pischel, L., Ley, C., Suh, G., Goodrich, J. K., Haggerty, T. D., et al. (2016). Crossover control study of the effect of personal care products containing triclosan on the microbiome. mSphere 1, e00056–e00015. doi: 10.1128/mSphere.00056-15
Postigo, C., and Richardson, S. D. (2014). Transformation of pharmaceuticals during oxidation/disinfection processes in drinking water treatment. J. Haz. Mat. 279, 461–475. doi: 10.1016/j.jhazmat.2014.07.029
Pycke, B. F., Geer, L. A., Dalloul, M., Abulafia, O., Jenck, A. M., and Halden, R. U. (2014). Human fetal exposure to triclosan and triclocarban in an urban population from Brooklyn, New York. Environ. Sci. Technol. 48, 8831–8838. doi: 10.1021/es501100w
Ribado, J. V., Ley, C., Haggerty, T. D., Tkachenko, E., Bhatt, A. S., and Parsonnet, J. (2017). Household triclosan and triclocarban effects on the infant and maternal microbiome. EMBO Mol. Med. 9, 1732–1741. doi: 10.15252/emmm.201707882
Richardson, S. D., Plewa, M. J., Wagner, E. D., Schoeny, R., and DeMarini, D. M. (2007). Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: a review and roadmap for research. Mutat. Res. 636, 178–242. doi: 10.1016/j.mrrev.2007.09.001
Richardson, S. D., and Ternes, T. A. (2018). Water analysis: emerging contaminants and current issues. Anal. Chem. 90, 398–428. doi: 10.1021/acs.analchem.7b04577
Rickert, D. E., Long, R. M., Krakowka, S., and Dent, J. G. (1981). Metabolism and excretion of 2, 4-[14C] dinitrotoluene in conventional and axenic Fischer-344 rats. Toxical. Appl. Pharmacol. 59, 574–579. doi: 10.1016/0041-008X(81)90312-4
Salgado, R., Oehmen, A., Carvalho, G., Noronha, J. P., and Reis, M. A. (2012). Biodegradation of clofibric acid and identification of its metabolites. J. Haz. Mat. 241, 182–189. doi: 10.1016/j.jhazmat.2012.09.029
Schecter, A., Haffner, D., Colacino, J., Patel, K., Päpke, O., Opel, M., et al. (2010). Polybrominated diphenyl ethers (PBDEs) and hexabromocyclodecane (HBCD) in composite US food samples. Environ. Health Perspect. 118, 357–362. doi: 10.1289/ehp.0901345
Schneidewind, U., Haest, P. J., Atashgahi, S., Maphosa, F., Hamonts, K., Maesen, M., et al. (2014). Kinetics of dechlorination by Dehalococcoides mccartyi using different carbon sources. J. Contam. Hydrol. 157, 25–36. doi: 10.1016/j.jconhyd.2013.10.006
Schwarzenbach, R. P., Egli, T., Hofstetter, T. B., Von Gunten, U., and Wehrli, B. (2010). Global water pollution and human health. Annu. Rev. Environ. Resour. 35, 109–136. doi: 10.1146/annurev-environ-100809-125342
Schwarzenbach, R. P., Escher, B. I., Fenner, K., Hofstetter, T. B., Johnson, C. A., Von Gunten, U., et al. (2006). The challenge of micropollutants in aquatic systems. Science 313, 1072–1077. doi: 10.1126/science.1127291
Schwiertz, A., Taras, D., Schafer, K., Beijer, S., Bos, N. A., Donus, C., et al. (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195. doi: 10.1038/oby.2009.167
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infection and immunity 76, 4726–4736. doi: 10.1128/IAI.00319-08
Shaul, N. J., Dodder, N. G., Aluwihare, L. I., Mackintosh, S. A., Maruya, K. A., Chivers, S. J., et al. (2015). Nontargeted biomonitoring of halogenated organic compounds in two ecotypes of bottlenose dolphins (Tursiops truncatus) from the Southern California Bight. Environ. Sci. Technol. 49, 1328–1338. doi: 10.1021/es505156q
Shetty, S. A., Hugenholtz, F., Lahti, L., Smidt, H., and de Vos, W. M. (2017). Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol. Rev. 41, 182–199. doi: 10.1093/femsre/fuw045
Shigematsu, N., Ishimaru, S., Saito, R., Ikeda, T., Matsuba, K., Sugiyama, K., et al. (1978). Respiratory involvement in polychlorinated biphenyls poisoning. Environ. Res. 16, 92–100. doi: 10.1016/0013-9351(78)90146-9
Shin, N.-R., Lee, J.-C., Lee, H.-Y., Kim, M.-S., Whon, T. W., Lee, M.-S., et al. (2013). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 706–707. doi: 10.1136/gutjnl-2013-305370
Singer, H., Müller, S., Tixier, C., and Pillonel, L. (2002). Triclosan: occurrence and fate of a widely used biocide in the aquatic environment: field measurements in wastewater treatment plants, surface waters, and lake sediments. Environ. Sci. Technol. 36, 4998–5004. doi: 10.1021/es025750i
Smidt, H., and de Vos, W. M. (2004). Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58, 43–73. doi: 10.1146/annurev.micro.58.030603.123600
Sousa, T., Paterson, R., Moore, V., Carlsson, A., Abrahamsson, B., and Basit, A. W. (2008). The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 363, 1–25. doi: 10.1016/j.ijpharm.2008.07.009
Spanogiannopoulos, P., Bess, E. N., Carmody, R. N., and Turnbaugh, P. J. (2016). The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. doi: 10.1038/nrmicro.2016.17
Stedtfeld, R. D., Stedtfeld, T. M., Fader, K. A., Williams, M. R., Bhaduri, P., Quensen, J., et al. (2017). TCDD influences reservoir of antibiotic resistance genes in murine gut microbiome. FEMS Microbiol. Ecol. 93:fix058. doi: 10.1093/femsec/fix058
Sutton, N. B., Atashgahi, S., van der Wal, J., Wijn, G., Grotenhuis, T., Smidt, H., et al. (2015). Microbial dynamics during and after in situ chemical oxidation of chlorinated solvents. Groundwater 53, 261–270. doi: 10.1111/gwat.12209
Syed, A. K., Ghosh, S., Love, N. G., and Boles, B. R. (2014). Triclosan promotes Staphylococcus aureus nasal colonization. MBio 5, e01015–e01013. doi: 10.1128/mBio.01015-13
Taylor, K. W., Novak, R. F., Anderson, H. A., Birnbaum, L. S., Blystone, C., DeVito, M., et al. (2013). Evaluation of the association between persistent organic pollutants (POPs) and diabetes in epidemiological studies: a national toxicology program workshop review. Environ. Health Perspect. 121, 774–783. doi: 10.1289/ehp.1205502
Teuten, E. L., and Reddy, C. M. (2007). Halogenated organic compounds in archived whale oil: a pre-industrial record. Environ. Pollut. 145, 668–671. doi: 10.1016/j.envpol.2006.08.022
Unson, M., Holland, N., and Faulkner, D. (1994). A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119, 1–11. doi: 10.1007/BF00350100
van de Pas, B. A., Harmsen, H. J., Raangs, G. C., de Vos, W. M., Schraa, G., and Stams, A. J. (2001). A Desulfitobacterium strain isolated from human feces that does not dechlorinate chloroethenes or chlorophenols. Arch. Microbiol. 175, 389–394. doi: 10.1007/s002030100276
Vandermaesen, J., Horemans, B., Bers, K., Vandermeeren, P., Herrmann, S., Sekhar, A., et al. (2016). Application of biodegradation in mitigating and remediating pesticide contamination of freshwater resources: state of the art and challenges for optimization. Appl. Microbiol. Biotechnol. 100, 7361–7376. doi: 10.1007/s00253-016-7709-z
van Pée, K. H. (1996). Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. Microbiol. 50, 375–399. doi: 10.1146/annurev.micro.50.1.375
Van Pée, K. H., and Unversucht, S. (2003). Biological dehalogenation and halogenation reactions. Chemosphere 52, 299–312. doi: 10.1016/S0045-6535(03)00204-2
Vermes, A., Guchelaar, H. J., and Dankert, J. (2000). Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46, 171–179. doi: 10.1093/jac/46.2.171
Vermes, A., Kuijper, E. J., Guchelaar, H. J., and Dankert, J. (2003). An in vitro study on the active conversion of flucytosine to fluorouracil by microorganisms in the human intestinal microflora. Chemotherapy 49, 17–23. doi: 10.1159/000069784
Vetter, W., Stoll, E., Garson, M. J., Fahey, S. J., Gaus, C., and Müller, J. F. (2002). Sponge halogenated natural products found at parts-per-million levels in marine mammals. Environ. Toxicol. Chem. 21, 2014–2019. doi: 10.1002/etc.5620211002
Villanueva, C. M., Cantor, K. P., Grimalt, J. O., Castaño-Vinyals, G., Malats, N., Silverman, D., et al. (2006a). Assessment of lifetime exposure to trihalomethanes through different routes. Occup. Environ. Med. 63, 273–277. doi: 10.1136/oem.2005.023069
Villanueva, C. M., Cantor, K. P., Grimalt, J. O., Malats, N., Silverman, D., Tardon, A., et al. (2006b). Bladder cancer and exposure to water disinfection by-products through ingestion, bathing, showering, and swimming in pools. Am. J. Epidemiol. 165, 148–156. doi: 10.1093/aje/kwj364
Vrieze, A., Out, C., Fuentes, S., Jonker, L., Reuling, I., Kootte, R. S., et al. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 60, 824–831. doi: 10.1016/j.jhep.2013.11.034
Wan, Y., Choi, K., Kim, S., Ji, K., Chang, H., Wiseman, S., et al. (2010). Hydroxylated polybrominated diphenyl ethers and bisphenol A in pregnant women and their matching fetuses: placental transfer and potential risks. Environ. Sci. Technol. 44, 5233–5239. doi: 10.1021/es1002764
Wang, H. S., Chen, Z. J., Ho, K. L., Ge, L. C., Du, J., Lam, M. H. W., et al. (2012). Hydroxylated and methoxylated polybrominated diphenyl ethers in blood plasma of humans in Hong Kong. Environ. Int. 47, 66–72. doi: 10.1016/j.envint.2012.06.004
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., DuGar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Warth, B., Spangler, S., Fang, M., Johnson, C. H., Forsberg, E. M., Granados, A., et al. (2017). Exposome-scale investigations guided by global metabolomics, pathway analysis, and cognitive computing. Anal. Chem. 89, 11505–11513. doi: 10.1021/acs.analchem.7b02759
Weatherill, J. J., Atashgahi, S., Schneidewind, U., Krause, S., Ullah, S., Cassidy, N., et al. (2018). Natural attenuation of chlorinated ethenes in hyporheic zones: a review of key biogeochemical processes and in-situ transformation potential. Water Res. 128, 362–382. doi: 10.1016/j.watres.2017.10.059
Wesgate, R., Grasha, P., and Maillard, J. Y. (2016). Use of a predictive protocol to measure the antimicrobial resistance risks associated with biocidal product usage. Am. J. Infect. Control 44, 458–464. doi: 10.1016/j.ajic.2015.11.009
Xiao, Y., Chang, H., Jia, A., and Hu, J. (2008). Trace analysis of quinolone and fluoroquinolone antibiotics from wastewaters by liquid chromatography–electrospray tandem mass spectrometry. J. Chromatogr. A 1214, 100–108. doi: 10.1016/j.chroma.2008.10.090
Xu, F., Tay, J. H., Covaci, A., Padilla-Sanchez, J. A., Papadopoulou, E., Haug, L. S., et al. (2017). Assessment of dietary exposure to organohalogen contaminants, legacy and emerging flame retardants in a Norwegian cohort. Environ. Int. 102, 236–243. doi: 10.1016/j.envint.2017.03.009
Xun, L., Topp, E., and Orser, C. (1992). Glutathione is the reducing agent for the reductive dehalogenation of tetrachloro-p-hydroquinone by extracts from a Flavobacterium sp. Biochem. Biophys. Res. Commun. 182, 361–366. doi: 10.1016/S0006-291X(05)80153-6
Yamashita, N., Kannan, K., Imagawa, T., Villeneuve, D. L., Hashimoto, S., Miyazaki, A., et al. (2000). Vertical profile of polychlorinated dibenzo-p-dioxins, dibenzofurans, naphthalenes, biphenyls, polycyclic aromatic hydrocarbons, and alkylphenols in a sediment core from Tokyo Bay, Japan. Environ. Sci. Technol. 34, 3560–3567. doi: 10.1021/es001043i
Yim, Y. J., Seo, J., Kang, S. I., Ahn, J. H., and Hur, H. G. (2008). Reductive dechlorination of methoxychlor and DDT by human intestinal bacterium Eubacterium limosum under anaerobic conditions. Arch. Environ. Contam. Toxicol. 54, 406–411. doi: 10.1007/s00244-007-9044-y
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Therapeut. 138, 103–141. doi: 10.1016/j.pharmthera.2012.12.007
Zanger, U. M., Turpeinen, M., Klein, K., and Schwab, M. (2008). Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 392, 1093–1108. doi: 10.1007/s00216-008-2291-6
Zhang, J., Jiang, Y., Zhou, J., Wu, B., Liang, Y., Peng, Z., et al. (2010). Elevated body burdens of PBDEs, dioxins, and PCBs on thyroid hormone homeostasis at an electronic waste recycling site in China. Environ. Sci. Technol. 44, 3956–3962. doi: 10.1021/es902883a
Zhang, L., Nichols, R. G., Correll, J., Murray, I. A., Tanaka, N., Smith, P. B., et al. (2015). Persistent organic pollutants modify gut microbiota–host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect. 123, 679–688. doi: 10.1289/ehp.1409055
Zhang, Y., Gu, A. Z., He, M., Li, D., and Chen, J. (2016). Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 51, 570–580. doi: 10.1021/acs.est.6b03132
Zhang, Y., Zhang, Z., Zhao, Y., Cheng, S., and Ren, H. (2013). Identifying health effects of exposure to trichloroacetamide using transcriptomics and metabonomics in mice (Mus musculus). Environ. Sci. Technol. 47, 2918–2924. doi: 10.1021/es3048976
Zhang, Y., Zhao, F., Deng, Y., Zhao, Y., and Ren, H. (2015). Metagenomic and metabolomic analysis of the toxic effects of trichloroacetamide-induced gut microbiome and urine metabolome perturbations in mice. J. Proteome Res. 14, 1752–1761. doi: 10.1021/pr5011263
Zhao, X.-H., and Wang, J. (2012). A brief study on the degradation kinetics of seven organophosphorus pesticides in skimmed milk cultured with Lactobacillus spp. at 42°C. Food Chem. 131, 300–304. doi: 10.1016/j.foodchem.2011.08.046
Keywords: xenobiotics, halogenated compounds, gut microbiota, xenobiotic-microbiota interaction, dehalogenation genes, metagenomics
Citation: Atashgahi S, Shetty SA, Smidt H and de Vos WM (2018) Flux, Impact, and Fate of Halogenated Xenobiotic Compounds in the Gut. Front. Physiol. 9:888. doi: 10.3389/fphys.2018.00888
Received: 29 April 2018; Accepted: 20 June 2018;
Published: 10 July 2018.
Edited by:
Zhaoping Li, Ronald Reagan UCLA Medical Center, United StatesReviewed by:
Manlio Vinciguerra, International Clinical Research Center (FNUSA-ICRC), CzechiaCopyright © 2018 Atashgahi, Shetty, Smidt and de Vos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Willem M. de Vos, d2lsbGVtLmRldm9zQHd1ci5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.