- 1Department of Clinical Epidemiology, First Affiliated Hospital, China Medical University, Shenyang, China
- 2Department of Epidemiology, School of Public Health, China Medical University, Shenyang, China
- 3Key Laboratory of Cancer Etiology and Prevention, Liaoning Provincial Department of Education, China Medical University, Shenyang, China
MicroRNAs (miRNAs) are a class of endogenous, short and non-coding RNAs that may play important roles in the pathogenesis of tumor. The associations between microRNA-499 rs3746444 polymorphism and cancer risk in different systems remain inconclusive. This article is aimed to obtain more exact estimation of these relationships through a meta-analysis based on 52,456 individuals. We retrieved relevant and eligible studies from Pubmed and Embase database up to January 10, 2018. ORs and 95% CIs were used to estimate the associations between miR-499 polymorphism and cancer susceptibility in different systems. All analyses were performed using the Stata 11.0 software. A total of 65 case-control studies were retrieved using explicit inclusion and exclusion criteria. The study included 23,762 cases and 28,694 controls. Overall cancer analysis showed the association between miR-499 polymorphism and susceptibility to cancer was significant. MicroRNA-499 rs3746444 was found to be significantly associated with increased risk of cancer of the respiratory system (CC vs. TT: OR = 1.575, 95% CI = 1.268–1.955, CC vs. TC+TT: OR = 1.527, 95% CI = 1.232–1.892), digestive system (CC vs. TT: OR = 1.153, 95% CI = 1.027–1.295; TC vs. TT: OR = 1.109, 95% CI = 1.046–1.176; CC+TC vs. TT: OR = 1.112, 95% CI = 1.018–1.216; CC vs. TC+TT: OR = 1.137, 95% CI = 1.016–1.272; C vs. T: OR = 1.112, 95% CI = 1.025–1.206), urinary system (TC vs. TT: OR = 1.307, 95% CI = 1.130–1.512; CC+TC vs. TT: OR = 1.259, 95% CI = 1.097–1.446; C vs. T: OR = 1.132, 95% CI = 1.014–1.264), and gynecological system (C vs. T: OR = 1.169, 95% CI = 1.002–1.364). In the subgroup analysis by ethnicity, the result showed that significant association with an increased cancer risk was found in Asian. Subgroup analysis based on type of tumor was also performed, miR-499 rs3746444 is associated with susceptibility of cervical squamous cell carcinoma, lung cancer, prostate cancer, and hepatocellular carcinoma.
Introduction
Cancer is a chronic disease that severely affects the lives and health of people in most countries and regions worldwide. The World Health Organization's International Agency for Research on Cancer recently published cancer statistics indicating that: 14.1 million new cancer cases, 8.4 million deaths occurred in 2012 worldwide (Torre et al., 2015). There is a growing body of evidence showing that carcinogenic effects are complex processes involving multiple environmental and genetic factors, although the cause of carcinogenic effects has not yet been completely clarified.
Micro-ribonucleic acids (miRNAs) are ~19- to 24-nucleotide RNA molecules that can regulate the expression of other genes in eukaryotes (Bartel, 2004). A growing number of studies have shown that miRNAs play an important role in regulating a variety of cellular functions. Impairment of miRNA expression is usually associated with various diseases. MicroRNAs play an important regulatory role in tumorigenesis through the regulation of cancer-related target gene expression and pathways. A variety of evidence suggests that miRNAs can participate in immune responses; inflammatory responses; infections; and cellular metabolism, growth and migration (Jovanovic and Hengartner, 2006).
MiR-499 is one of the microRNAs that plays important posttranscriptional regulatory roles by modulating several genes and signaling pathways, particularly under hypoxic-ischemic conditions such as in cancer and myocardial infarction (Wilson et al., 2010; Ando et al., 2014). Liu et al. (2011) indicated that miR-499 could promote metastasis of colorectal cancer cells by targeting FOXO4 and PDCD4, and miR-499 might be regarded as a new potential therapeutic target for colorectal cancer. Li et al. also found that miR-499 functions as a tumor suppressor in non-small cell lung cancer by targeting VAV3 (Li M. et al., 2016). MiR-499 is related to several signaling pathways such as wnt/β-catenin signal pathway (Zhang et al., 2012). A study by Wang et al. (2015) suggested that downregulation of the miR-499-5p level impaired the PI3K/AKT/GSK signaling pathway and glycogen synthesis by targeting PTEN. Ando et al. (2014) reported that the tumor-targeted delivery of miR-499 was very effective so as to implement cancer therapy. Okamoto et al. (2016) found that APRPG-miR-499 may be a combination therapeutic agent for cancer treatment.
Single nucleotide genetic variants that occur in miRNAs may influence microRNA biogenesis, stability of mature microRNA molecules, efficiency of target gene regulation, as well as specificity of targets, which may involve developing susceptibility to cancer, thus, miRNAs play an important role in the occurrence and development of malignant tumors (Nikolic et al., 2015).
Accumulated evidence suggests that miR-499 rs3746444 is associated with the susceptibility to many cancers. Zhang et al. (2017) reported that the miR-499 rs3746444 polymorphism is associated with the risk of oral squamous cell cancer in Chinese individuals. Hashemi et al. (2016) found that the rs3746444 polymorphism in miR-499 can increase the risk of prostate cancer in an Iranian population. The rs3746444 single nucleotide polymorphism was also found to be clearly associated with lung cancer (Li D. et al., 2016). Wang et al. (2014) found that miR-499A>G polymorphism is associated with an increased risk of hepatocellular carcinoma in Chinese individuals. However, Zhang et al. (2016) suggested that there is no association between miR-499 variant and the risk of hepatocellular carcinoma in the co-dominant, dominant, and recessive models. These two articles similarly focus on hepatocellular carcinoma, but it led to very different results. In a Greek population, a case–control study demonstrated that the rs3746444 polymorphism in miR-499 is not associated with colorectal cancer (Dikaiakos et al., 2015). For this gene locus, many investigators have conducted exploratory studies and also obtained a number of different results, different outcomes for different types of cancer, and even the same cancer may have different results. The association between miR-499 rs3746444 and the susceptibility to cancer has not yet reached a clear consensus.
Now that increasingly more and more researchers have studied the association between cancer risk and miR-499 polymorphism, it is necessary to integrate these results to further explore the relationship between the miR-499 polymorphism and cancer risk of different systems at the level of statistics.
Materials and Methods
Literature Retrieval
We retrieved articles from PubMed and EMBASE using the following terms “microRNA-499 or miR-499 or rs3746444” and “cancer or carcinoma” published prior to January 10, 2018. In order to be more comprehensive and rigorous, we searched for the documents mentioned in the original article and the references to find more qualified article. There is no language restriction on the retrieved literature.
Study Selection and Data Extraction
All incorporated literature must clearly meet the following three criteria: (a) evaluation of the miR-499 rs3746444 polymorphism and cancer risks; (b) a case-control study; and (c) sufficient published data for the computation of odds ratios (ORs) with 95% confidence intervals (CIs). The exclusion criteria were as follows: (a) Animal studies; (b) not for cancer research; (c) meta-analyses or review articles; Two reviewers independently extract data. If the opinion is inconsistent, the third review will join the in-depth discussion to determine whether the article should be obtained. The main contents of extracted information included the name of first author, publication date, country of origin, ethnicity, cancer type, characteristics of controls, the number of cases and controls with miR-499 T/C genotypes and the total number of cases and controls. According to the source of the control, we defined controls as population-based and hospital-based.
Statistical Methods
We firstly calculated the value of Hardy-Weinberg equilibrium (HWE) of the control groups by using the chi-square goodness-of-fit test including all retrieved literature (a P < 0.05 was regarded representative of a departure from HWE). ORs corresponding to 95% CI was used to estimate the strength of association between miR-499 rs3746444 T/C and different cancer risk. The pooled ORs were conducted for five genetic comparison models: allelic model (C vs. T), homozygote comparison (CC vs. TT), heterozygote comparison (TC vs. TT), recessive model (CC vs. TC/TT), dominant model (CC/TC vs. TT). Heterogeneity among retrieved literature was used to evaluate by I2 test and Q test (a significance level of P < 0.05 and/or I2 ≥ 50%). If there was obvious heterogeneity (I2 ≥ 50%), we conducted with a random-effects model (DerSimonian and Laird), otherwise, we analyzed with a fixed-effects model (Mantel–Haenszel) to obtain summary associations between miR-499 rs3746444 T/C and different cancer risk. Then subgroup analysis was done by ethnicity, different systems, source of control, and type of tumor, HWE in controls (yes/no). Sensitivity analyses were used to evaluate the stabilization of the results. Potential effect of publication bias of literatures was estimated by funnel plots and the Egger's test. A P < 0.05 or the unsymmetrical funnel plot were regarded as statistically significant. Statistical analysis was done by Stata11.0 Software. All the P-values are two-sided.
Results
Study Characteristics
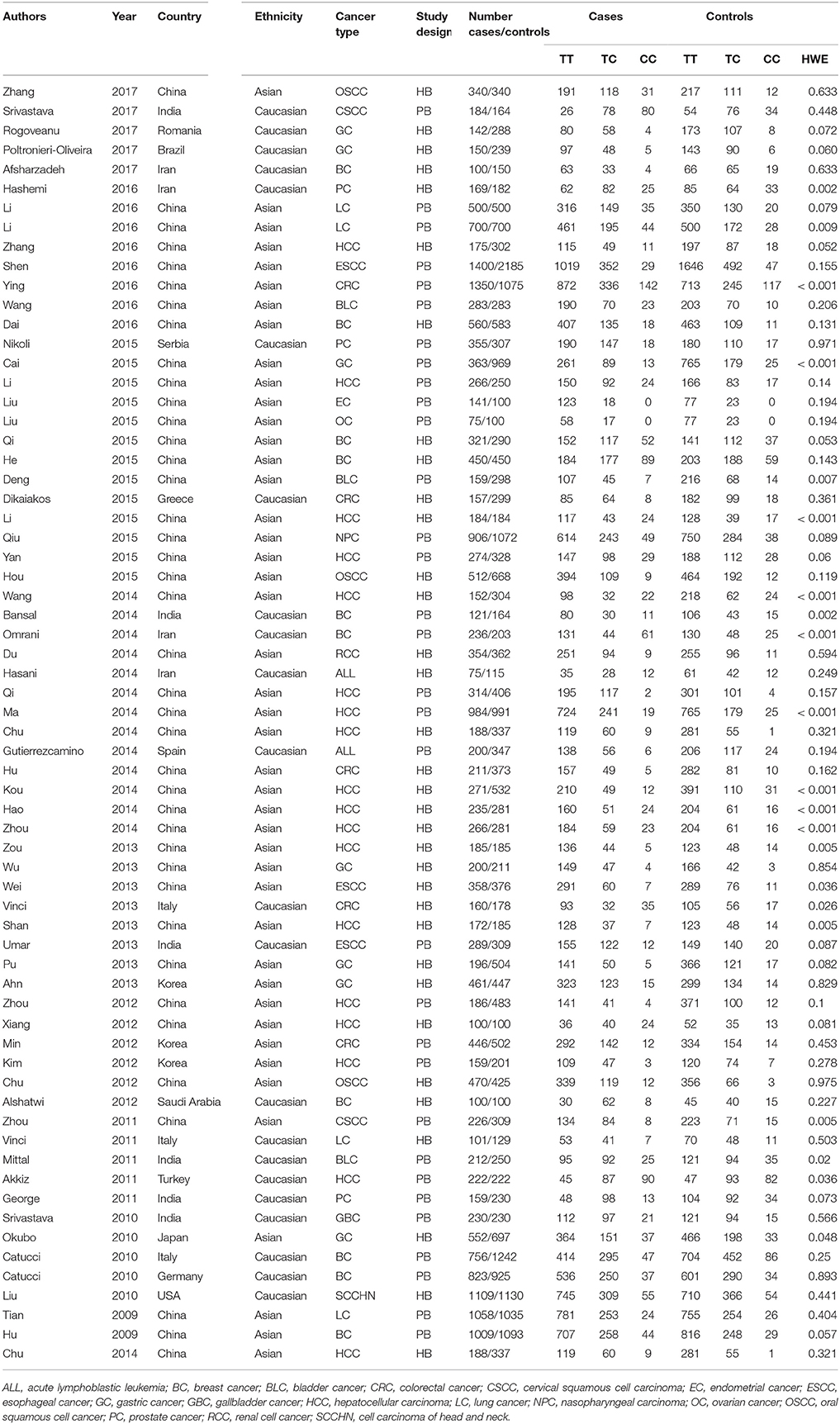
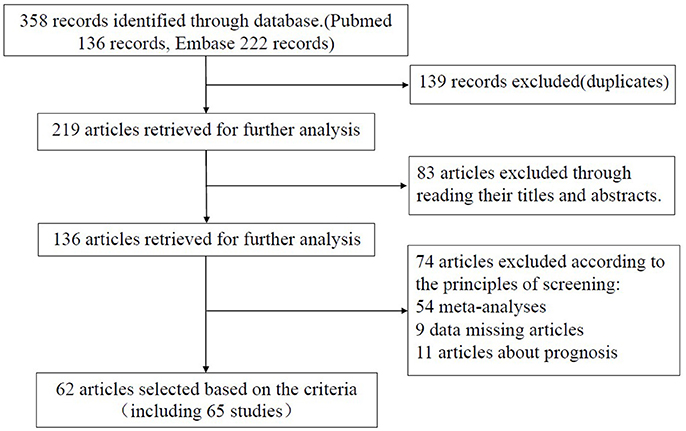
We first obtained 219 articles by using our specified search terms (Figure 1). After reading the titles and the abstracts, 83 articles were excluded. To meet the screening criteria set out earlier, we also excluded an additional 74 articles, including 54 meta-analyses, 9 articles with missing data, and 11 articles concerning prognosis. Finally, a total of 62 articles (65 studies) consisting of 23,762 cases and 28,694 controls were used for further analysis. Of the 65 studies, sample sizes ranged from 175 to 3,585, and publication year ranged from 2009 to 2017. In total, there were 2 acute lymphoblastic leukemia studies (Gutierrez-Camino et al., 2014; Hasani et al., 2014), 10 breast cancer studies (Hu et al., 2009; Catucci et al., 2010; Alshatwi et al., 2012; Bansal et al., 2014; Omrani et al., 2014; He et al., 2015; Qi et al., 2015; Dai et al., 2016; Afsharzadeh et al., 2017), 3 bladder cancer studies (Mittal et al., 2011; Deng et al., 2015; Wang et al., 2016), 5 colorectal cancer studies (Min et al., 2012; Vinci et al., 2013; Hu et al., 2014; Dikaiakos et al., 2015; Ying et al., 2016), 2 cervical squamous cell carcinoma studies (Zhou et al., 2011; Srivastava et al., 2017), 3 esophageal cancer studies (Umar et al., 2013; Wei et al., 2013; Shen et al., 2016), 7 gastric cancer studies (Okubo et al., 2010; Ahn et al., 2013; Wu et al., 2013; Pu et al., 2014; Cai et al., 2015; Poltronieri-Oliveira et al., 2017; Rogoveanu et al., 2017), 17 hepatocellular carcinoma studies (Akkiz et al., 2011; Kim et al., 2012; Xiang et al., 2012; Zhou et al., 2012, 2014; Shan et al., 2013; Zou and Zhao, 2013; Chu et al., 2014; Hao et al., 2014; Kou et al., 2014; Ma et al., 2014; Qi et al., 2014; Wang et al., 2014; Li D. et al., 2015; Li X. et al., 2015; Yan et al., 2015; Zhang et al., 2016), 4 lung cancer studies (Tian et al., 2009; Vinci et al., 2011; Li D. et al., 2016), 2 oral squamous cell cancer studies (Hou et al., 2015; Zhang et al., 2017), 3 prostate cancer (George et al., 2011; Nikolic et al., 2015; Hashemi et al., 2016), and other cancer studies [1 cervical squamous cell carcinoma study (Zhou et al., 2011), 1 endometrial cancer study (Liu et al., 2015), 1 gallbladder cancer study (Srivastava et al., 2010), 1 ovarian cancer study (Liu et al., 2015), 1 nasopharyngeal carcinoma study (Qiu et al., 2015), 1 renal cell cancer study (Du et al., 2014), 1 cell carcinoma of head and neck study (Liu et al., 2010)]. There were 22 studies on Caucasians and 43 studies on Asians. Table 1 indicates the main characteristics of each study included in this meta-analysis. We separated these studies into several categories, such as cancers of the respiratory system (8 studies), digestive system (33 studies), urinary system (7 studies), gynecological system (14 studies), and lymphatic and hematopoietic system (2 studies).
Meta-Analysis Results
As is shown in Table 2, the main analyses performed included association and heterogeneity tests. Among all studies, we found significant associations between miR-499 rs3746444 polymorphism and cancer susceptibility in five kinds of genetic models. (CC vs. TT: OR = 1.227, 95% CI = 1.084–1.388, P = 0.001; TC vs. TT: OR = 1.108, 95% CI = 1.035–1.186, P = 0.003; CC+TC vs. TT: OR = 1.136, 95% CI = 1.061–1.216, P < 0.001; CC vs. TC+TT: OR = 1.177, 95% CI = 1.045–1.326, P = 0.007; C vs. T: OR = 1.126, 95% CI = 1.059–1.197, P < 0.001).
In the respiratory system, cancer risk was found to be significantly increased in the CC genetic model as compared with those in the TT and TC+TT (recessive) models (CC vs.TT: OR = 1.575, 95% CI = 1.268–1.955, P < 0.001; CC vs. TC+TT: OR = 1.527, 95% CI = 1.232–1.892, P < 0.001) (Figure 2), especially in lung cancer. We also found significantly increased cancer risk in all five genetic models of the digestive system (CC vs.TT: OR = 1.153, 95% CI = 1.027–1.295, P = 0.016; TC vs. TT: OR = 1.109, 95% CI = 1.046–1.176, P = 0.001; CC+TC vs. TT: OR = 1.112, 95% CI = 1.018–1.216, P = 0.019; CC vs. TC+TT: OR = 1.137, 95% CI = 1.016–1.272, P = 0.025; C vs. T: OR = 1.112, 95% CI = 1.025–1.206, P = 0.011; Figure 3), particularly in hepatocellular carcinoma. Significant association was observed between rs3746444 and increased cancer risk in three genetic models in the urinary system (TC vs. TT: OR = 1.307, 95% CI = 1.130–1.512, P < 0.001; CC+TC vs. TT = 1.259, 95% CI = 1.097–1.446, P = 0.001; C vs. T: OR = 1.132, 95% CI = 1.014–1.264, P = 0.027; Figure 4), as well as for prostate cancer. Our results showed that rs3746444 could increase cancer risk of the gynecological system in the allelic model (C vs. T: OR = 1.169, 95% CI = 1.002–1.364, P = 0.047; Figure 5), particularly for cervical squamous cell carcinoma. However, studies on lymphatic and hematopoietic system cancers did not reveal any significant association between cancer risk and genetic models. In addition, no significant association between the five different genetic models and cancer risk in other types of cancers were observed.
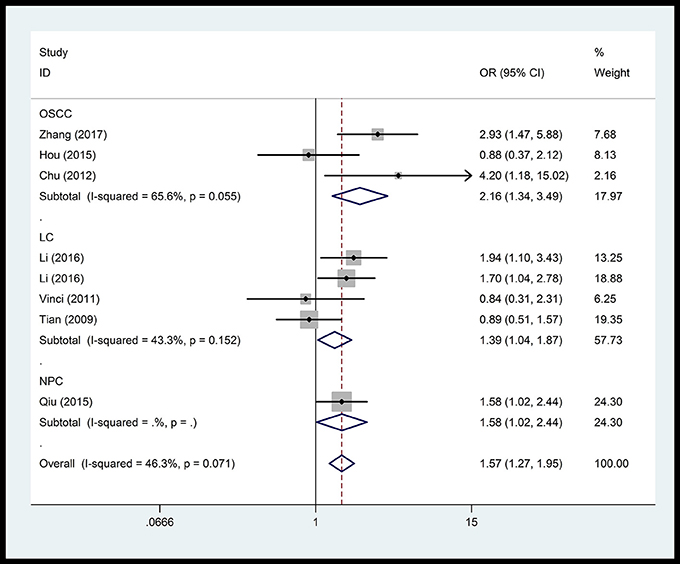
Figure 2. Forest plot of the OR and 95% CIs for association between microRNA-499 rs3746444 polymorphism and cancer risk of respiratory system in homozygote comparison model. OSCC, oral squamous cell cancer; LC, lung cancer; NPC, nasopharyngeal carcinoma.
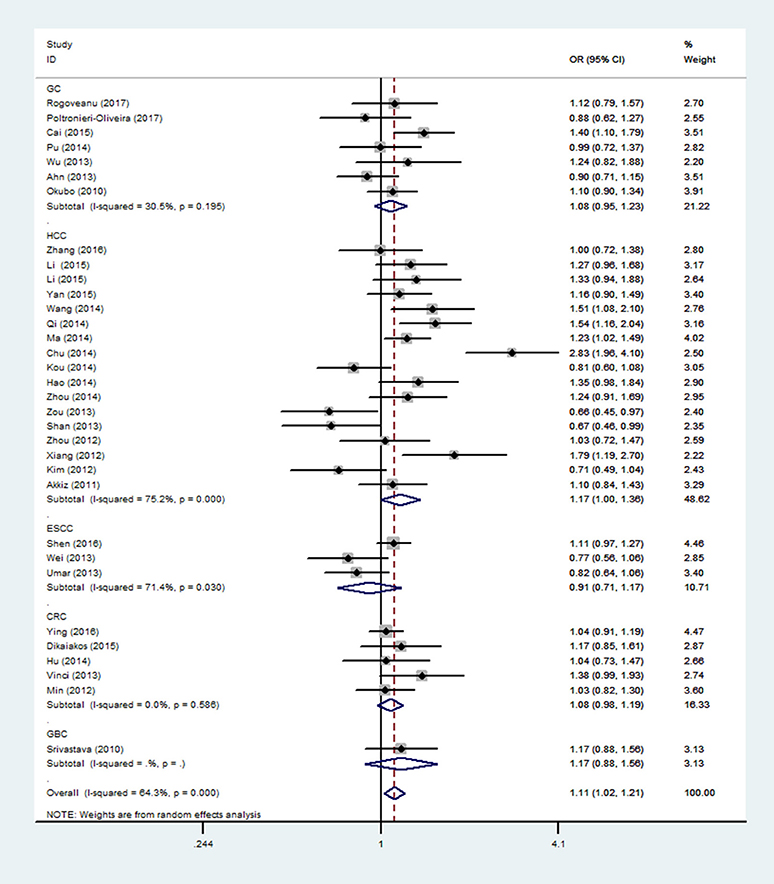
Figure 3. Forest plot of the OR and 95% CIs for association between microRNA-499 rs3746444 polymorphism and cancer risk of digestive system in allelic model. HCC, hepatocellular carcinoma; ESCC, esophageal cancer; CRC, colorectal cancer; GC, gastric cancer; GBC, gallbladder cancer.
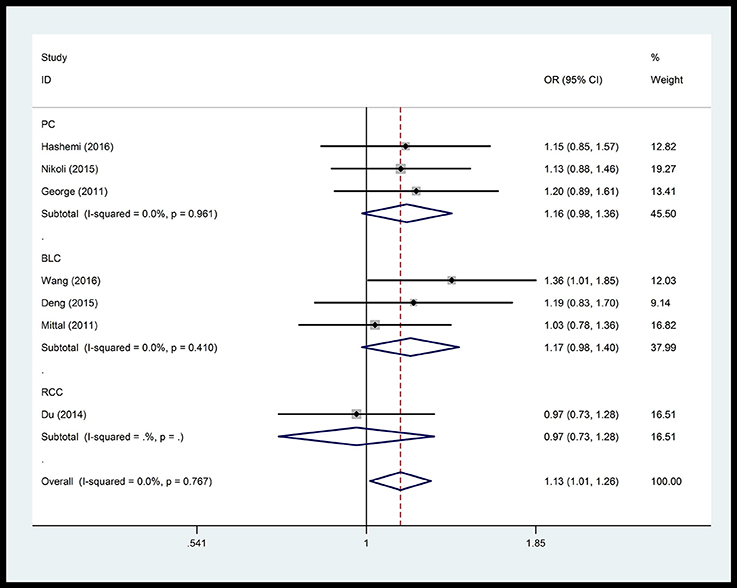
Figure 4. Forest plot of the OR and 95% CIs for association between microRNA-499 rs3746444 polymorphism and cancer risk of urinary system in allelic model. PC, prostate cancer; BLC, bladder cancer; RCC, renal cell cancer.

Figure 5. Forest plot of the OR and 95% CIs for association between microRNA-499 rs3746444 polymorphism and cancer risk of gynecological system in allelic model. BC, breast cancer; CSCC, cervical squamous cell carcinoma; EC, endometrial cancer; OC, ovarian cancer.
We then performed subgroup analysis based on ethnicity, source of control, and HWE in controls. When results were stratified by ethnicity, significant association was found between miR-499 rs3746444 polymorphism and cancer risk in the Asians; (CC vs. TT: OR = 1.301, 95% CI = 1.180–1.434, P < 0.001; TC vs. TT: OR = 1.123, 95% CI = 1.040–1.213, P = 0.003; CC+TC vs. TT: OR = 1.152, 95% CI = 1.065–1.247, P < 0.001; CC vs. TC+TT: OR = 1.267, 95% CI = 1.151–1.394, P < 0.001; C vs. T: OR = 1.136, 95% CI = 1.072–1.237, P < 0.001) this association was absent in the Caucasians. When analysis was performed on source of control, the ORs were significant in four genetic models for the population-based control. At the same time, significant associations were observed for the hospital-based control in the homozygote comparison model and the recessive model. Following the removal of 21 studies whose genotypic distributions in controls were not in accordance with HWE, the results remained the same.
Sensitivity Analysis
Sensitivity analysis was used to estimate the contribution of each study to the pooled ORs. Our results showed that the total ORs were not significantly altered following removal of studies where the controls were not in accordance with HWE. This further supported the significant association between miR-499 rs3746444 and cancer risk.
Publication Bias
In this meta-analysis, the potential effect of publication bias in literatures was estimated by funnel plots (Figure 6) and the Egger's test. Based on our analysis, no publication bias was found (P > 0.05).
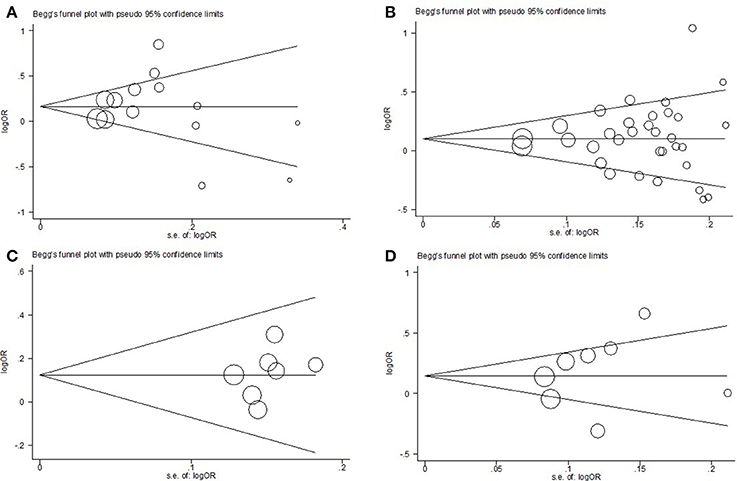
Figure 6. Funnel plot analysis to detect publication bias for allelic model. (A) respiratory system; (B) digestive system; (C) urinary system; (D) gynecological system.
Discussion
The odds of getting cancer varies widely among different individuals. Many studies have discovered that miRNAs are strongly associated with tumorigenesis and can act as oncogenes or tumor suppressor genes that affect the initiation and development of tumors (Zhou et al., 2015). In recent years, the status of SNPs in microRNAs and their potential effects on cancer risk have been extensively studied. Though several studies and meta-analyses have been carried out to evaluate the association between miR-499 rs3746444 T > C polymorphism and cancer risk, no conclusive findings have been obtained in different cancers. Hence, integrating and updating these results to explore the relationship further is important.
In this study, our results demonstrate significant associations between miR-499 rs3746444 polymorphism and cancer susceptibility for the first time, in five different genetic models from 65 case-control studies. Meta-analyses of all types of cancer risks by other groups have found either no association between the miR-499 polymorphism and cancer susceptibility or significant associations in one or two genetic models (Xu et al., 2015). Several meta-analyses have explored the association between the miR-499 rs3746444 polymorphism and cancer risk and it is difficult to judge if the analysis without studies departing from HWE would be more valid or not. An overall meta-analysis by Jiang et al. (2015) demonstrated a borderline association (TC+CC vs. TT: OR = 1.15, P = 0.04). However, the association disappeared (TC+CC vs. TT: OR = 1.18, P = 0.21) when studies departing from HWE were excluded. In our study, after the exclusion of 21 studies wherein the genotypic distributions in controls were not in accordance with HWE, the results were still significant. Therefore, though there is still no consensus on whether or not to include studies that deviate from HWE, our results basically were not affected by such studies.
The most important feature of our study is that it is the first meta-analysis to explore the associations between miR-499 polymorphism and cancer susceptibility in different systems comprehensively. We found that microRNA-499 rs3746444 was significantly associated with an increased risk of cancer of the respiratory system in two genetic models, digestive system in all five genetic models, the urinary system in three genetic models, and gynecological system in an allelic model. To the best of our knowledge, no previous meta-analysis has explored this association in cancer of the respiratory system. We also found that miR-499 rs3746444 C allele might increase the risk of lung cancer in a homozygote comparison model. Previous studies have reported that rs3746444 was significantly associated with an increased risk of lung cancer and could contribute to a poor prognosis by modulating the expression of cancer-related genes leading to tumorigenesis and resistance to chemotherapy in lung cancer (Li D. et al., 2016). Our data differs from that of Li et al. (2013), since their analysis which included 12 gastrointestinal cancer studies, demonstrated no association between miR-499 rs3746444 and gastrointestinal cancer. We included 33 gastrointestinal cancer studies in this meta-analysis. In our analysis, though the number of the studies evaluating the association between hepatocellular carcinoma and miR-499 was the largest, the results were inconsistent on account of the small sample sizes and different ethnic groups. MicroRNA-499 downregulates the expression of the ets1 proto-oncogene in HepG2 cells and plays a vital role in the pathogenesis of HCC (Wei et al., 2012). In addition, we found that miR-499 rs3746444 C allele can increase the risk of hepatocellular carcinoma in recessive and allelic models. Our findings from the allelic hepatocellular carcinoma model were consistent with those of Yu et al. (2017). We also found that that rs3746444 could increase the risk of gynecological cancers in an allelic model. At the same time, miR-499 rs3746444 was associated with susceptibility to cervical squamous cell carcinoma. Consistent with the findings of Wang et al. (2017), we also found a significant association between rs3746444 and increased risk for cancer of the urinary system in two genetic models, but not the allelic model. Our results also suggested a significant association between miR-499 rs3746444 and prostate cancer risk in the heterozygote comparison and dominant models. George et al. have shown that a heterozygous miR-499 genotype, confers an increased risk of developing prostate cancer in the North Indian population (George et al., 2011). Nikolic et al. found that rs3746444 qualifies for a genetic variant potentially associated with aggressive prostate cancer in the Serbian population (Nikolic et al., 2015). Classifying cancer into different systems provides a new perspective and an in-depth understanding of the association between the miR-499 polymorphism and cancer susceptibility.
As for subgroup analysis based on ethnicity, our results were consistent with those of other meta-analyses. We found an association between the miR-499 polymorphism and cancer risk in Asians, but not in Caucasians. Unlike the other analyses, we found an association among the five genetic models in the Asian population. Our results suggest that miR-499 rs3746444 is associated with susceptibility to cervical squamous cell carcinoma, lung cancer, prostate cancer, and hepatocellular carcinoma, but not other cancer types, indicating that rs3746444 may have different effects on different cancer types. Compared to the previous studies, the difference in results could also be due to a lack of articles on miR-499 polymorphism and cancer risk.
Although our study pooled a large number of cases and controls including 52456 individuals, limitations which might affect the objectivity of the results still exist. First, our studies included data from only Asians and Caucasians and none from the African population. Second, detailed and original patient information such as age, gender, lifestyle, family history, nutrient intake was lacking. Third, gene-gene and gene-environment interactions should have been taken into consideration, if the relevant information was available.
In conclusion, our meta-analysis found that miR-499 rs3746444 polymorphism is associated with the risk of cancer in five genetic models. MicroRNA-499 rs3746444 was found to be significantly associated with increased risk of cancer of the respiratory, digestive, urinary, and gynecological systems. The subgroup analysis by ethnicity showed a significant association with increased cancer risk in the Asian population. Our findings also suggest that the miR-499 rs3746444 C allele may increase the risk of cervical squamous cell carcinoma, lung cancer, prostate cancer, and hepatocellular carcinoma. Well-designed and large-scale studies are, therefore, necessary to further verify these findings.
Author Contributions
XY and BZ designed this study. XY and XL searched the databases and extracted the data. XY wrote the manuscript. XL and BZ reviewed the manuscript. XY, XL, and BZ approved the final manuscript.
Funding
This study is supported by grants No. 81773524 and No. 81502878 from the National Natural Science Foundation of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are very appreciated for all the participants in the present study.
References
Afsharzadeh, S. M., Mohaddes Ardebili, S. M., Seyedi, S. M., Karimian Fathi, N., and Mojarrad, M. (2017). Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene 11, 20–25. doi: 10.1016/j.mgene.2016.11.004
Ahn, D. H., Rah, H., Choi, Y. K., Jeon, Y. J., Min, K. T., Kwack, K., et al. (2013). Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol. Carcinog. 52(Suppl. 1), E39–E51. doi: 10.1002/mc.21962
Akkiz, H., Bayram, S., Bekar, A., Akgollu, E., and Uskudar, O. (2011). Genetic variation in the microRNA-499 gene and hepatocellular carcinoma risk in a Turkish population: lack of any association in a case-control study. Asian Pac. J. Cancer Prev. 12, 3107–3112.
Alshatwi, A. A., Shafi, G., Hasan, T. N., Syed, N. A., Al-Hazzani, A. A., Alsaif, M. A., et al. (2012). Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS ONE 7:e30049. doi: 10.1371/journal.pone.0030049
Ando, H., Asai, T., Koide, H., Okamoto, A., Maeda, N., Tomita, K., et al. (2014). Advanced cancer therapy by integrative antitumor actions via systemic administration of miR-499. J. Control. Release 181, 32–39. doi: 10.1016/j.jconrel.2014.02.019
Bansal, C., Sharma, K. L., Misra, S., Srivastava, A. N., Mittal, B., and Singh, U. S. (2014). Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience 8:473. doi: 10.3332/ecancer.2014.473
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Cai, M., Zhang, Y., Ma, Y., Li, W., Min, P., Qiu, J., et al. (2015). Association between microRNA-499 polymorphism and gastric cancer risk in Chinese population. Bull. Cancer 102, 973–978. doi: 10.1016/j.bulcan.2015.09.012
Catucci, I., Yang, R., Verderio, P., Pizzamiglio, S., Heesen, L., Hemminki, K., et al. (2010). Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 31, E1052–E1057. doi: 10.1002/humu.21141
Chu, Y. H., Hsieh, M. J., Chiou, H. L., Liou, Y. S., Yang, C. C., Yang, S. F., et al. (2014). MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS ONE 9:e89930. doi: 10.1371/journal.pone.0089930
Dai, Z. M., Kang, H. F., Zhang, W. G., Li, H. B., Zhang, S. Q., Ma, X. B., et al. (2016). The associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: a STROBE-compliant observational study. Medicine 95:e2826. doi: 10.1097/MD.0000000000002826
Deng, S., Wang, W., Li, X., and Zhang, P. (2015). Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J. Surg. Oncol. 13:297. doi: 10.1186/s12957-015-0683-6
Dikaiakos, P., Gazouli, M., Rizos, S., Zografos, G., and Theodoropoulos, G. E. (2015). Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biomark. 15, 157–162. doi: 10.3233/CBM-140449
Du, M., Lu, D., Wang, Q., Chu, H., Tong, N., Pan, X., et al. (2014). Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis 35, 1629–1635. doi: 10.1093/carcin/bgu082
George, G. P., Gangwar, R., Mandal, R. K., Sankhwar, S. N., and Mittal, R. D. (2011). Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol. Biol. Rep. 38, 1609–1615. doi: 10.1007/s11033-010-0270-4
Gutierrez-Camino, A., Lopez-Lopez, E., Martin-Guerrero, I., Pinan, M. A., Garcia-Miguel, P., Sanchez-Toledo, J., et al. (2014). Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 75, 767–773. doi: 10.1038/pr.2014.43
Hao, Y. X., Wang, J. P., and Zhao, L. F. (2014). Associations between three common MicroRNA polymorphisms and hepatocellular carcinoma risk in Chinese. Asian Pac. J. Cancer Prev. 14, 6601–6604. doi: 10.7314/APJCP.2013.14.11.6601
Hasani, S. S., Hashemi, M., Eskandari-Nasab, E., Naderi, M., Omrani, M., and Sheybani-Nasab, M. (2014). A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumour Biol. 35, 219–225. doi: 10.1007/s13277-013-1027-1
Hashemi, M., Moradi, N., Ziaee, S. A., Narouie, B., Soltani, M. H., Rezaei, M., et al. (2016). Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J. Adv. Res. 7, 491–498. doi: 10.1016/j.jare.2016.03.008
He, B., Pan, Y., Xu, Y., Deng, Q., Sun, H., Gao, T., et al. (2015). Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 36, 4575–4582. doi: 10.1007/s13277-015-3102-2
Hou, Y. Y., Lee, J. H., Chen, H. C., Yang, C. M., Huang, S. J., Liou, H. H., et al. (2015). The association between miR-499a polymorphism and oral squamous cell carcinoma progression. Oral Dis. 21, 195–206. doi: 10.1111/odi.12241
Hu, X., Li, L., Shang, M., Zhou, J., Song, X., Lu, X., et al. (2014). Association between microRNA genetic variants and susceptibility to colorectal cancer in Chinese population. Tumour Biol. 35, 2151–2156. doi: 10.1007/s13277-013-1285-y
Hu, Z., Liang, J., Wang, Z., Tian, T., Zhou, X., Chen, J., et al. (2009). Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 30, 79–84. doi: 10.1002/humu.20837
Jiang, S. G., Chen, L., Tang, J. H., Zhao, J. H., and Zhong, S. L. (2015). Lack of association between Hsa-Mir-499 rs3746444 polymorphism and cancer risk: meta-analysis findings. Asian Pac. J. Cancer Prev. 16, 339–344. doi: 10.7314/APJCP.2015.16.1.339
Jovanovic, M., and Hengartner, M. O. (2006). miRNAs and apoptosis: RNAs to die for. Oncogene 25, 6176–6187. doi: 10.1038/sj.onc.1209912
Kim, W. H., Min, K. T., Jeon, Y. J., Kwon, C. I., Ko, K. H., Park, P. W., et al. (2012). Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene 504, 92–97. doi: 10.1016/j.gene.2012.05.014
Kou, J. T., Fan, H., Han, D., Li, L., Li, P., Zhu, J., et al. (2014). Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol. Lett. 8, 1255–1260. doi: 10.3892/ol.2014.2257
Li, D., Peng, J. J., Tan, Y., Chen, T., Wei, D., Du, M., et al. (2015). Genetic variations in microRNA genes and susceptibility to hepatocellular carcinoma. Genet. Mol. Res. 14, 1926–1931. doi: 10.4238/2015
Li, D., Zhu, G., Di, H., Li, H., Liu, X., Zhao, M., et al. (2016). Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget 7, 41715–41724. doi: 10.18632/oncotarget.9566
Li, L., Sheng, Y., Lv, L., and Gao, J. (2013). The association between Two MicroRNA variants (miR-499, miR-149) and gastrointestinal cancer risk: a meta-analysis. PLoS ONE 8:e81967. doi: 10.1371/journal.pone.0081967
Li, M., Zhang, S., Wu, N., Wu, L., Wang, C., and Lin, Y. (2016). Overexpression of miR-499-5p inhibits non-small cell lung cancer proliferation and metastasis by targeting VAV3. Sci. Rep. 6:23100. doi: 10.1038/srep23100
Li, X., Li, K., and Wu, Z. (2015). Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 8, 9560–9566.
Liu, X., Xu, B., Li, S., Zhang, B., Mao, P., Qian, B., et al. (2015). Association of SNPs in miR-146a, miR-196a2, and miR-499 with the risk of endometrial/ovarian cancer. Acta Biochim. Biophys. Sin. 47, 564–566. doi: 10.1093/abbs/gmv042
Liu, X., Zhang, Z., Sun, L., Chai, N., Tang, S., Jin, J., et al. (2011). MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis 32, 1798–1805. doi: 10.1093/carcin/bgr213
Liu, Z., Li, G., Wei, S., Niu, J., El-Naggar, A. K., Sturgis, E. M., et al. (2010). Genetic variants in selected pre-microrna genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116, 4753–4760. doi: 10.1002/cncr.25323
Ma, Y., Wang, R., Zhang, J., Li, W., Gao, C., Liu, J., et al. (2014). Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet. Test. Mol. Biomarkers 18, 516–524. doi: 10.1089/gtmb.2013.0510
Min, K. T., Kim, J. W., Jeon, Y. J., Jang, M. J., Chong, S. Y., Oh, D., et al. (2012). Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol. Carcinog. 51(Suppl. 1), E65–E73. doi: 10.1002/mc.21849
Mittal, R. D., Gangwar, R., George, G. P., Mittal, T., and Kapoor, R. (2011). Investigative role of pre-microRNAs in bladder cancer patients: a case-control study in North India. DNA Cell Biol. 30, 401–406. doi: 10.1089/dna.2010.1159
Nikolic, Z., Savic Pavicevic, D., Vucic, N., Cidilko, S., Filipovic, N., Cerovic, S., et al. (2015). Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp. Mol. Pathol. 99, 145–150. doi: 10.1016/j.yexmp.2015.06.009
Okamoto, A., Asai, T., Ryu, S., Ando, H., Maeda, N., Dewa, T., et al. (2016). Enhanced efficacy of doxorubicin by microRNA-499-mediated improvement of tumor blood flow. J. Clin. Med. 5:10. doi: 10.3390/jcm5010010
Okubo, M., Tahara, T., Shibata, T., Yamashita, H., Nakamura, M., Yoshioka, D., et al. (2010). Association between common genetic variants in Pre-microRNAs and gastric cancer risk in japanese population. Helicobacter 15, 524–531. doi: 10.1111/j.1523-5378.2010.00806.x
Omrani, M., Hashemi, M., Eskandari-Nasab, E., Hasani, S. S., Mashhadi, M. A., Arbabi, F., et al. (2014). hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark. Med. 8, 259–267. doi: 10.2217/bmm.13.118
Poltronieri-Oliveira, A. B., Madeira, F. F., Nunes, D. B. S. M., Rodrigues, G. H., Lopes, B. C., Manoel-Caetano, F. S., et al. (2017). Polymorphisms of miR-196a2 (rs11614913) and miR-605 (rs2043556) confer susceptibility to gastric cancer. Gene Rep. 7, 154–163. doi: 10.1016/j.genrep.2017.04.006
Pu, J. Y., Dong, W., Zhang, L., Liang, W. B., Yang, Y., and Lv, M. L. (2014). No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iran. J. Basic Med. Sci. 17, 128–133. doi: 10.22038/ijbms.2014.2246
Qi, J. H., Wang, J., Chen, J., Shen, F., Huang, J. T., Sen, S., et al. (2014). High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer 14:643. doi: 10.1186/1471-2407-14-643
Qi, P., Wang, L., Zhou, B., Yao, W. J., Xu, S., Zhou, Y., et al. (2015). Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet. Mol. Res. 14, 6289–6296. doi: 10.4238/2015.June.11.2
Qiu, F., Yang, L., Zhang, L., Yang, X., Yang, R., Fang, W., et al. (2015). Polymorphism in mature microRNA-608 sequence is associated with an increased risk of nasopharyngeal carcinoma. Gene 565, 180–186. doi: 10.1016/j.gene.2015.04.008
Rogoveanu, I., Burada, F., Cucu, M. G., Vere, C. C., Ioana, M., and Cimpeanu, R. A. (2017). Association of microRNA polymorphisms with the risk of gastric cancer in a romanian population. J. Gastrointestin. Liver Dis. 26, 231–238. doi: 10.15403/jgld.2014.1121.263.rog
Shan, Y. F., Huang, Y. H., Chen, Z. K., Huang, K. T., Zhou, M. T., Shi, H. Q., et al. (2013). miR-499A>G rs3746444 and miR-146aG>C expression and hepatocellular carcinoma risk in the Chinese population. Genet. Mol. Res. 12, 5365–5371. doi: 10.4238/2013.November.7.11
Shen, F., Chen, J., Guo, S., Zhou, Y., Zheng, Y., Yang, Y., et al. (2016). Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 37, 4777–4784. doi: 10.1007/s13277-015-4268-3
Srivastava, K., Srivastava, A., and Mittal, B. (2010). Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J. Hum. Genet. 55, 495–499. doi: 10.1038/jhg.2010.54
Srivastava, S., Singh, S., Fatima, N., Mittal, B., and Srivastava, A. N. (2017). Pre-microrna gene polymorphisms and risk of cervical squamous cell carcinoma. J. Clin. Diagnos. Res. 11, GC01–GC04. doi: 10.7860/JCDR/2017/25361.10543
Tian, T., Shu, Y., Chen, J., Hu, Z., Xu, L., Jin, G., et al. (2009). A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol. Biomarkers Prev. 18, 1183–1187. doi: 10.1158/1055-9965.EPI-08-0814
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Umar, M., Upadhyay, R., Prakash, G., Kumar, S., Ghoshal, U. C., and Mittal, B. (2013). Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol. Carcinog. 52, 10–18. doi: 10.1002/mc.21931
Vinci, S., Gelmini, S., Mancini, I., Malentacchi, F., Pazzagli, M., Beltrami, C., et al. (2013). Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods 59, 138–146. doi: 10.1016/j.ymeth.2012.09.002
Vinci, S., Gelmini, S., Pratesi, N., Conti, S., Malentacchi, F., Simi, L., et al. (2011). Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin. Chem. Lab. Med. 49, 2073–2080. doi: 10.1515/CCLM.2011.708
Wang, J., Zhang, Y., Zhang, Y., and Chen, L. (2016). Correlation between miRNA-196a2 and miRNA-499 polymorphisms and bladder cancer. Int. J. Clin. Exp. Med. 9, 20484–20488.
Wang, L., Zhang, N., Pan, H. P., Wang, Z., and Cao, Z. Y. (2015). MiR-499-5p contributes to hepatic insulin resistance by suppressing PTEN. Cell. Physiol. Biochem. 36, 2357–2365. doi: 10.1159/000430198
Wang, X. H., Wang, F. R., Tang, Y. F., Zou, H. Z., and Zhao, Y. Q. (2014). Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet. Mol. Res. 13, 5048–5054. doi: 10.4238/2014.July.4.20
Wang, Y. H., Hu, H. N., Weng, H., Chen, H., Luo, C. L., Ji, J., et al. (2017). Association between polymorphisms in MicroRNAs and risk of urological cancer: a meta-analysis based on 17,019 subjects. Front. Physiol. 8:325. doi: 10.3389/fphys.2017.00975
Wei, J., Zheng, L., Liu, S., Yin, J., Wang, L., Wang, X., et al. (2013). MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum. Immunol. 74, 1199–1205. doi: 10.1016/j.humimm.2013.06.012
Wei, W., Hu, Z., Fu, H., Tie, Y., Zhang, H., Wu, Y., et al. (2012). MicroRNA-1 and microRNA-499 downregulate the expression of the ets1 proto-oncogene in HepG2 cells. Oncol. Rep. 28, 701–706. doi: 10.3892/or.2012.1850
Wilson, K. D., Hu, S., Venkatasubrahmanyam, S., Fu, J. D., Sun, N., Abilez, O. J., et al. (2010). Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ. Cardiovasc. Genet. 3, 426–435. doi: 10.1161/CIRCGENETICS.109.934281
Wu, X. J., Mi, Y. Y., Yang, H., Hu, A. K., Li, C., Li, X. D., et al. (2013). Association of the hsa-mir-499 (rs3746444) polymorphisms with gastric cancer risk in the Chinese population. Onkologie 36, 573–576. doi: 10.1159/000355518
Xiang, Y., Fan, S., Cao, J., Huang, S., and Zhang, L. P. (2012). Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol. Biol. Rep. 39, 7019–7023. doi: 10.1007/s11033-012-1532-0
Xu, Z., Zhang, E., Duan, W., Sun, C., Bai, S., and Tan, X. (2015). The association between miR-499 polymorphism and cancer susceptibility: a meta-analysis. Onco. Targets. Ther. 8, 2179–2186. doi: 10.2147/OTT.S88224
Yan, P., Xia, M., Gao, F., Tang, G., Zeng, H., Yang, S., et al. (2015). Predictive role of miR-146a rs2910164 (C>G), miR-149 rs2292832 (T>C), miR-196a2 rs11614913 (T>C) and miR-499 rs3746444 (T>C) in the development of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 8, 15177–15183.
Ying, H. Q., Peng, H. X., He, B. S., Pan, Y. Q., Wang, F., Sun, H. L., et al. (2016). MiR-608, pre-miR-124-1 and pre-miR26a-1 polymorphisms modify susceptibility and recurrence-free survival in surgically resected CRC individuals. Oncotarget 7, 75865–75873. doi: 10.18632/oncotarget.12422
Yu, J. Y., Hu, F., Du, W., Ma, X. L., and Yuan, K. (2017). Study of the association between five polymorphisms and risk of hepatocellular carcinoma: a meta-analysis. J. Chin. Med. Assoc. 80, 191–203. doi: 10.1016/j.jcma.2016.09.009
Zhang, E., Xu, Z., Duan, W., Huang, S., and Lu, L. (2017). Association between polymorphisms in pre-miRNA genes and risk of oral squamous cell cancer in a Chinese population. PLoS ONE 12:e0176044. doi: 10.1371/journal.pone.0176044
Zhang, L. H., Hao, B. B., Zhang, C. Y., Dai, X. Z., and Zhang, F. (2016). Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Genet. Mol. Res. 15:gmr.15038582. doi: 10.4238/gmr.15038582
Zhang, L. L., Liu, J. J., Liu, F., Liu, W. H., Wang, Y. S., Zhu, B., et al. (2012). MiR-499 induces cardiac differentiation of rat mesenchymal stem cells through wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 420, 875–881. doi: 10.1016/j.bbrc.2012.03.092
Zhou, B., Dong, L. P., Jing, X. Y., Li, J. S., Yang, S. J., Wang, J. P., et al. (2014). Association between miR-146aG>C and miR-196a2C>T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 35, 7775–7780. doi: 10.1007/s13277-014-2020-z
Zhou, B., Wang, K., Wang, Y., Xi, M., Zhang, Z., Song, Y., et al. (2011). Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol. Carcinog. 50, 499–505. doi: 10.1002/mc.20740
Zhou, J., Lv, R., Song, X., Li, D., Hu, X., Ying, B., et al. (2012). Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 31, 524–530. doi: 10.1089/dna.2011.1340
Zhou, X., Du, Y. L., Jin, P., and Ma, F. (2015). Bioinformatic analysis of cancer-related microRNAs and their target genes. Yi Chuan 37, 855–864. doi: 10.16288/j.yczz.14-439
Keywords: miR-499, rs3746444, cancer risk, polymorphism, meta-analysis
Citation: Yang X, Li X and Zhou B (2018) A Meta-Analysis of miR-499 rs3746444 Polymorphism for Cancer Risk of Different Systems: Evidence From 65 Case-Control Studies. Front. Physiol. 9:737. doi: 10.3389/fphys.2018.00737
Received: 30 January 2018; Accepted: 28 May 2018;
Published: 12 June 2018.
Edited by:
Brian James Morris, University of Sydney, AustraliaReviewed by:
Antonio Longo, Università degli Studi di Catania, ItalyGoran Negosava Brajušković, University of Belgrade, Serbia
Copyright © 2018 Yang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baosen Zhou, YnN6aG91QG1haWwuY211LmVkdS5jbg==
 Xianglin Yang
Xianglin Yang Xuelian Li2,3
Xuelian Li2,3