- 1Department of Endocrinology, Taihe Hospital, Hubei University of Medicine, Shiyan, China
- 2Department of Oral and Maxillary Surgery, Gui Zhou Provincial People's Hospital, Guiyang, China
- 3Trade Union, Taihe Hospital, Hubei University of Medicine, Shiyan, China
- 4Department of Stomatology, Center for Evidence-Based Medicine, Taihe Hospital, Hubei University of Medicine, Shiyan, China
Previous epidemiologic studies have revealed a possible association between microRNA-608 rs4919510 G>C polymorphism and digestive system cancers (DSCs) risk, but the results were not consistent. We therefore performed an updated meta-analysis to explore the association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk. Crude odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess the relationship between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk. Heterogeneity, cumulative analyses, sensitivity analyses, and publication bias were also conducted to examine the statistical power. Eight published articles with nine independent case-control studies involving 10,836 individuals were included in this meta-analysis. Overall, no significant association was found between microRNA-608 rs4919510 G>C polymorphism and DSCs risk in general populations. But some significant protective effects were observed in the subgroup of Caucasian population group in three genetic models (C vs. G: OR = 0.82, 95% CI, 0.68–0.99, P = 0.03, I2 = 0%; CC vs. GG: OR = 0.59, 95% CI = 0.36–0.97, P = 0.04, I2 = 0%; GC+CC vs. GG: OR = 0.61, 95% CI = 0.37–0.99, P = 0.05, I2 = 0%). In summary, current evidence indicates that the microRNA-608 rs4919510 G>C polymorphism maybe an important factor of DSCs susceptibility, especially in Caucasian population.
Introduction
Digestive system cancers (DSCs), which comprise esophageal cancer (EC), gastric cancer (GC), hepatocellular cancer (HC), pancreatic cancer (PC) colorectal cancer (CRC) and other solid carcinoma, is one of the most common malignancies with increasing incidence and mortality worldwide (Torre et al., 2015). According to the recent published cancer statistics, there were approximately 310,440 new DSCs cases and 157,700 DSCs-related deaths in United States in 2017 (Siegel et al., 2017). The treatment and care of patients with DSCs result in a heavy economic and psychological burden to both society and the patient's family (Hsu et al., 2017; Jinjuvadia et al., 2017). Surgery, radiotherapy, chemotherapy and other treatments may cause extensive damage to patients' tissues and organs and may result in various complications that lead to local dysfunction and seriously decrease the quality of life of patients (Nederlof et al., 2016; Bosch et al., 2017; Motoyama et al., 2017). Regrettably, no clear explanation of the mechanism underlying DSCs development and the susceptibility of different patients exist. Some researches indicate that unhealthy life styles, cigarette and alcohol use, viral infection, local inflammation and stress can trigger DSCs development (Erren et al., 2016; Gao et al., 2017; Jarzynski et al., 2017; Jayasekara et al., 2018).
Evidences suggested that abnormal gene expression and an alteration of amino acid structure can lead to abnormal biologic activities that result in an imbalance of normal physiologic function and induce the formation of tumors. MicroRNAs were discovered in 1993 and were defined as small noncoding RNA molecules with approximately 21–25 nucleotides in length and a characteristic double-stranded structure (Lee et al., 1993; Ambros, 2004). MicroRNAs always originate from endogenous transcripts and bind to imperfect complementary sequences of the 3′-untranslated regions (3'-UTR) of target mRNAs to regulate their post-transcriptional repression (Bartel, 2004). Some studies indicated that abnormal microRNA expression could result in tumor occurrence (Valeri et al., 2010; Tessitore et al., 2014). MicroRNA-608 is a newly discovered microRNA, as reported by Wang et al., who indicated that the expression of microRNA-608 was significantly reduced in hepatocellular cancer and its expression levels were associated with tumor size, differentiation, clinical stage, and overall prognosis (Wang K. et al., 2016). Further, other researchers found that the expression of microRNA-608 was significantly down-regulated in glioma stem cells (GSCs). And the up-regulation of microRNA-608 expression would inhibit the proliferation, migration, and invasion of GSCs and promotes their apoptosis (Wang Z. et al., 2016).
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation in people. SNPs alter gene function and/or expression, consequently affecting downstream biologic pathways and increasing cancer risk (Martini et al., 2013; Niu et al., 2015). In microRNA-608, rs4919510 G>C is the most common locus and has attracted increasing attention. Ye et al. conducted the first case-control study in 2008 and did not find any significant association between the microRNA-608 rs4919510 G>C polymorphism and esophageal cancer (Ye et al., 2008). Recently, a published meta-analysis addressed the association between microRNA-608 rs4919510 G>C polymorphism and cancer risk (Liu et al., 2016), but the results were not consistent with subsequent studies, especially in DSCs. Therefore, we conducted the present meta-analysis to more precisely and comprehensively assess of the association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk.
Materials and Methods
This present meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Moher et al., 2009). All included data were collected from published studies, and no ethical issues were involved.
Search Strategy
Five electronic English databases (PubMed, Embase, Web of Science, CNKI and Wanfang) were searched for relevant studies that focused on the association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk from inception up to January 1, 2018. Only studies that were written in English and Chinese were included. Moreover, the bibliographies of the collected studies and relevant reviews were retrospected to identify additional articles. The following search terms and strategy was used (e.g., in PubMed):
#1 microRNA 608
#2 microRNA-608
#3 mir 608
#4 mir-608
#5 rs4919510
#6 #1 OR #2 OR #3 OR #4 OR #5
#7 polymorphism
#8 variant
#9 mutation
#10 #7 OR #8 OR #9
#11 cancer
#12 tumor
#13 neoplasm
#14 #11 OR #12 OR #13
#15 #6 AND #10 AND #14
Eligibility Criteria
Studies were included based on the following criteria: (1) only case-control studies that focusing on the association between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk; and (2) studies had to provided sufficient frequency data on the genotype distribution to evaluate the crude odds ratios (ORs) and 95% confidence intervals (CIs); and; (3) studies had to be published only in English and Chinese; and (4) only publications with the largest or most recently updated sample data were included when there were some overlapping or duplicate publications on the same theme.
Data Extraction and Quality Evaluation
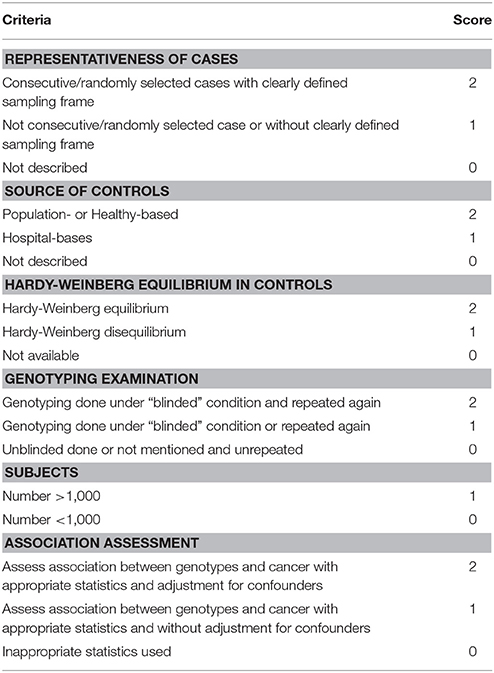
Two researchers (Li and Song) independently reviewed and extracted relevant information from all included studies, including name of the first author, publishing date, country or region where the study was conducted, race, control design, sample sizes of the cases and controls, frequency data of genotype distribution, genotyping method, Hardy-Weinberg equilibrium (HWE) assessment in controls, and cancer type. Quality evaluation of the included studies was performed by the two researchers using the modified Newcastle-Ottawa scale (NOS). The scores ranged from 0 points (worst) to 11 points (best) (Table 4); studies with a score of 9 or higher were classified as high quality.
Statistical Analysis
We calculated ORs and 95% CIs to assess the association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk. All pooled genetic models were examined, including the allele contrast (C vs. G), co-dominant models (GC vs. GG and CC vs. GG), dominant model (GC+CC vs. GG), and recessive model (CC vs. GG+GC). Heterogeneity between the included studies was calculated using Cochran's Q-test and I2 statistical method (Huedo-Medina et al., 2006). The fixed-effect model was used when the I2-value was less than 40%; otherwise, a random-effects model was used (Mantel and Haenszel, 1959; DerSimonian, 1996). Subgroup analyses were performed according to race (Asian, Caucasian, and African), control design (population base, hospital base), cancer type, subject number, and NOS evaluation. Moreover, meta-regression was performed to interpret the between-group heterogeneity. Cumulative meta-analyses were performed to assess the continuous tendency in the results. Furthermore, sensitivity analyses were conducted to examine the stability of the results by removing each study one by one. Potential publication biases were examined using Egger's linear regression and Begg's funnel plots. Statistical analyses were performed using STATA version 14.0 (Stata Corporation, College Station, TX, USA) (Begg and Mazumdar, 1994; Egger et al., 1997). A two-sided P-value less than 0.05 was considered statistically significant.
Results
Study Characteristics
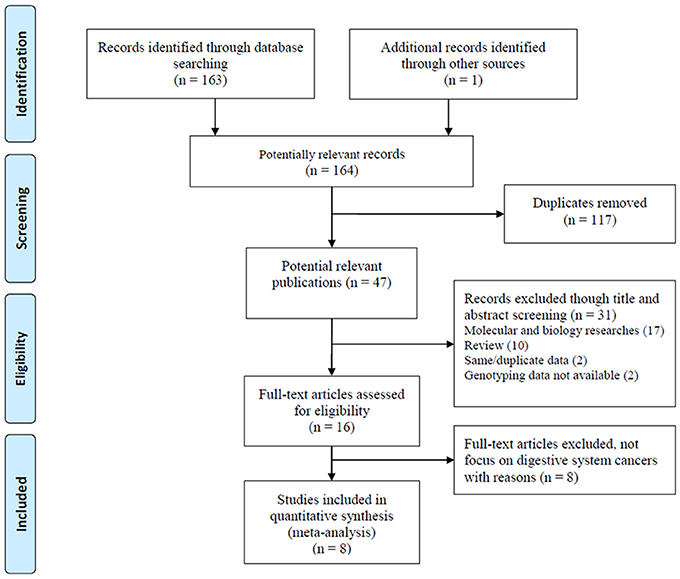
A total of 164 articles were collected. The study selection process was presented in Figure 1. One hundred and fifty-six articles were excluded during the comprehensive screening procedures based on the article titles, abstracts, and full texts. Thus, eight articles including nine independent case-control studies involving 5,224 patients and 5,612 controls were identified and included in this meta-analysis according to our inclusion criteria (Ryan et al., 2012; Zhang, 2012; Kupcinskas et al., 2014a,b; Wang et al., 2014; Zhang et al., 2015; Jiang et al., 2016; Ying et al., 2016). Five case-control studies included 4,430 cases and 4,403 controls in Asian populations(Zhang, 2012; Wang et al., 2014; Zhang et al., 2015; Jiang et al., 2016; Ying et al., 2016), thee case-control studies involved 700 cases and 1,024 controls in Caucasian populations (Ryan et al., 2012; Kupcinskas et al., 2014a,b), and one case-control study involved 94 cases and 185 controls in an African American population (Ryan et al., 2012). For genotyping, four studies used the Taqman method (Ryan et al., 2012; Kupcinskas et al., 2014a,b), three studies used the MassARRAY method (Wang et al., 2014; Jiang et al., 2016; Ying et al., 2016), and the remaining two studies used other methods (SNaPshot, PCR-RFLP) (Zhang, 2012; Zhang et al., 2015). Moreover, five studies focused on colorectal cancer(Ryan et al., 2012; Zhang, 2012; Kupcinskas et al., 2014a; Ying et al., 2016), two studies focused on gastric cancer (Kupcinskas et al., 2014b; Jiang et al., 2016), one study focused on hepatocellular cancer (Wang et al., 2014) and other focused on esophageal cancer (Zhang et al., 2015). The genotype distributions in all control groups were satisfied with the HWE. The characteristics of the included studies are presented in Table 1.
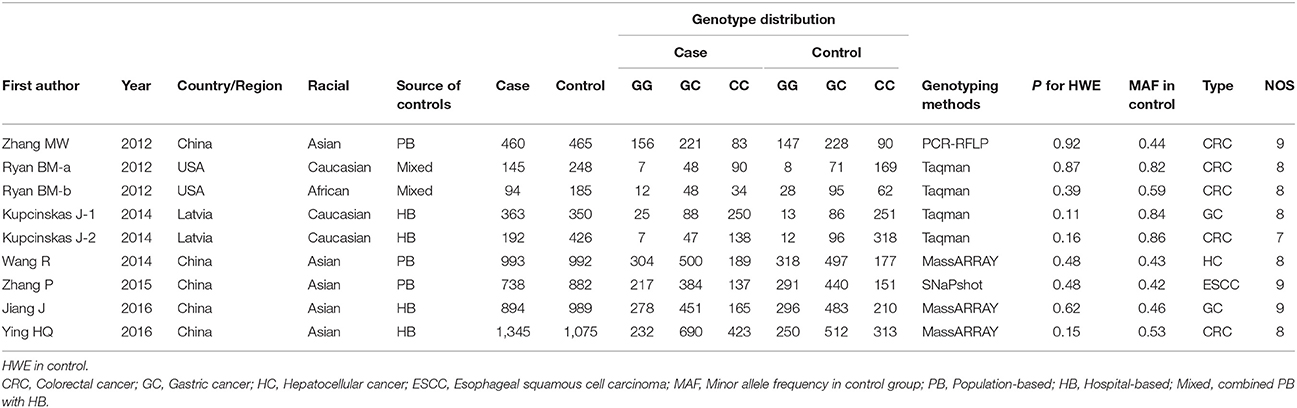
Table 1. Characteristics of included studies on MicroRNA-608 rs4919510 G>C polymorphisms and digestive system cancers risk.
Meta-Analysis
Overall, no significant association between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk was observed with the included studies (C vs. G: OR = 1.00, 95% CI = 0.91–1.10, P = 0.98, I2 = 53.4%; GC vs. GG: OR = 1.07, 95% CI = 0.93–1.24, P = 0.35, I2 = 43.0%; CC vs. GG: OR = 1.01, 95% CI = 0.83–1.24, P = 0.90, I2 = 56.4%; GC+CC vs. GG: OR = 1.05, 95% CI = 0.89–1.23, P = 0.58, I2 = 56.4%, (Figure 2); CC vs. GG+GC: OR = 0.99, 95% CI = 0.91–1.09, P = 0.89, I2 = 0%) (Supplementary Figure S1).
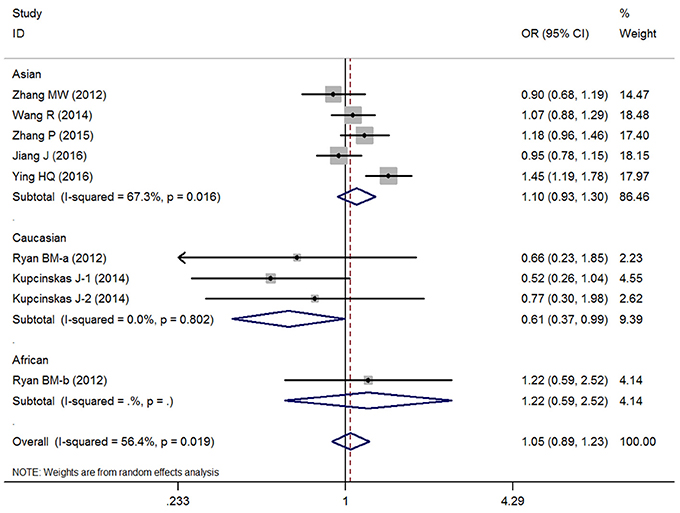
Figure 2. OR and 95% CIs of the associations between microRNA-608 rs4919510 G>C polymorphism and digestive system cancer risk in GC+CC vs. GG model.
Subgroup and Meta-Regression Analysis
The results of race diversity demonstrated some significant protective effects between microRNA-608 rs4919510 G>C polymorphism and DSCs risk in Caucasian populations (C vs. G: OR = 0.82, 95% CI = 0.68–0.99, P = 0.03, I2 = 0%; CC vs. GG: OR = 0.59, 95% CI = 0.36–0.97, P = 0.04, I2 = 0%; GC+CC vs. GG: OR = 0.61, 95% CI = 0.37–0.99, P = 0.05, I2 = 0%, Figure 2). But the other Subgroup analysis based on control design, cancer type, subject number, and NOS evaluation did not present any significant association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk. All these statistical results were presented in Table 2. Moreover, meta-regression were conducted due to the slight heterogeneity between included studies existed, but the statistical test did not find any remarkable factors that contributed to the current heterogeneity (Table 3).
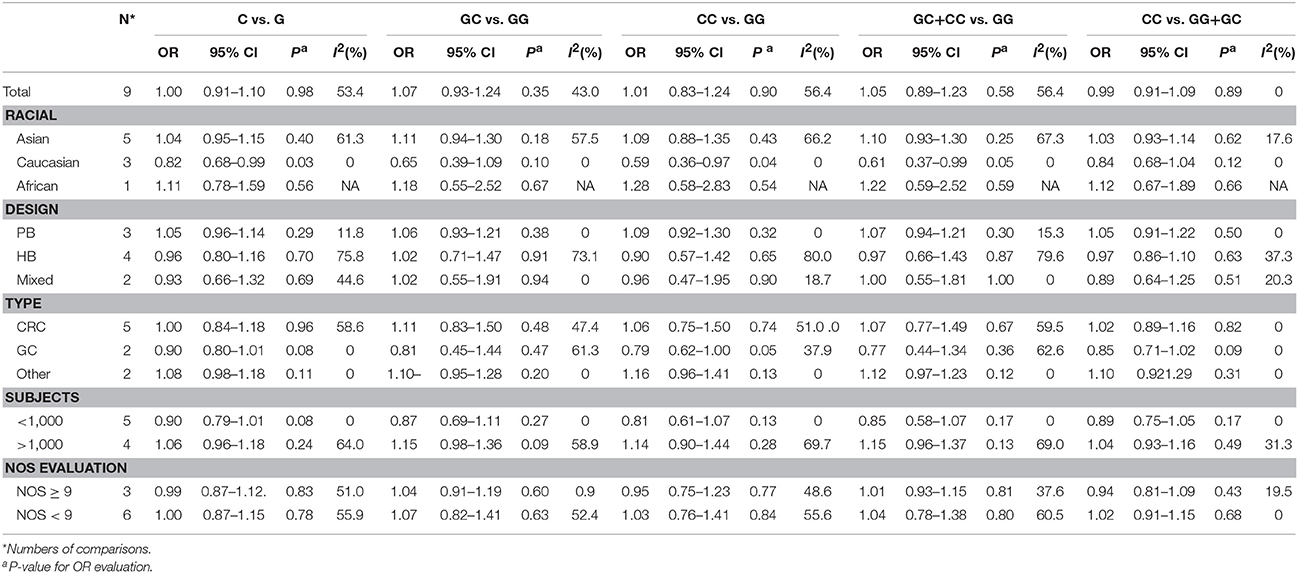
Table 2. Summary ORs and 95% CI of MicroRNA-608 rs4919510 G>C polymorphisms and digestive system cancers risk.
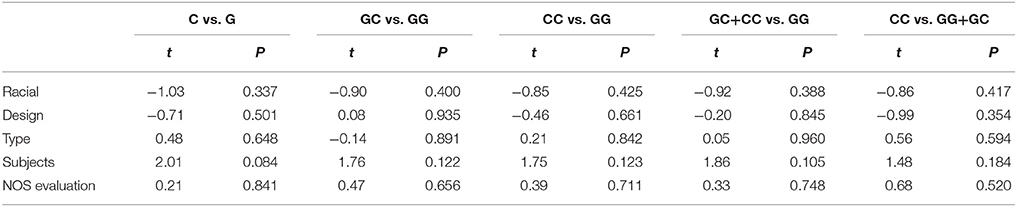
Table 3. Characteristics of included covariate in meta-regression analysis in all five genetic models.
Cumulative and Sensitivity Analyses
Cumulative analysis indicated a consistent tendency accompanied with the continuously published studies (Figure 3 for GC+CC vs. GG model) (Supplementary Figure S2). Sensitive analyses were performed by omitting each study one at a time, and the results did not indicate any significant changes (Figure 4 for GC+CC vs. GG model) (Supplementary Figure S3).
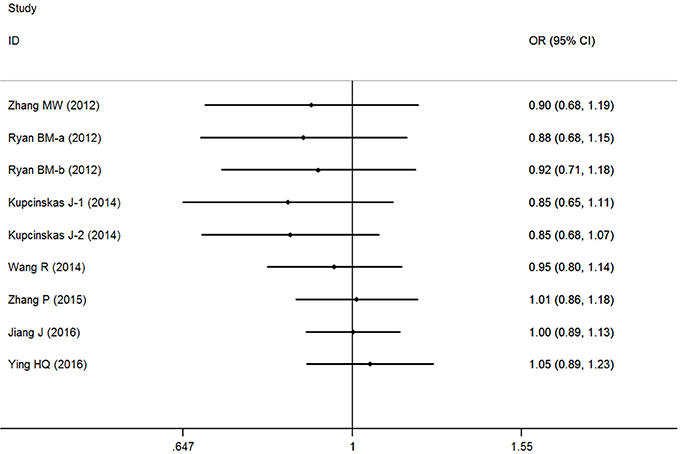
Figure 3. Cumulative meta-analyses according to publication year in GC+CC vs. GG model of microRNA-608 rs4919510 G>C polymorphism.
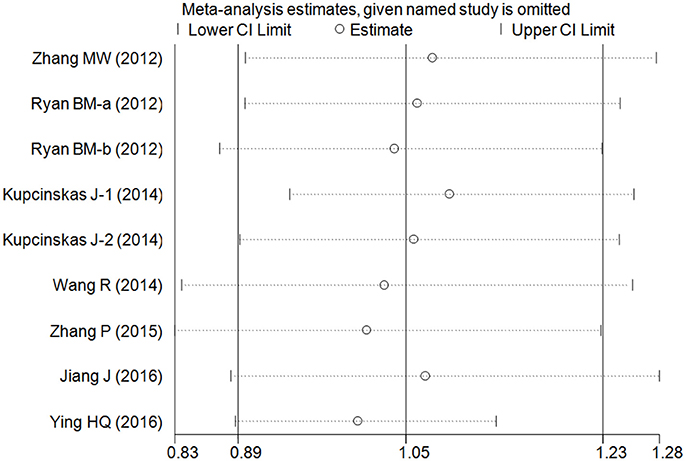
Figure 4. Sensitivity analysis involving deletion of each study to reflect the influence of the individual dataset to the pooled ORs in GC+CC vs. GG model of microRNA-608 rs4919510 G>C polymorphism.
Publication Bias
Publication bias was examined with Begg's test and no apparent asymmetry of the funnel plot was found (Figure 5 for GC+CC vs. GG model) (Supplementary Figure S4). These results were confirmed with Egger's test (C vs. G: P = 0.11; GC vs. GG: P = 0.24; CC vs. GG: P = 0.16; GC+CC vs. GG: P = 0.22; CC vs. GG+GC: P = 0.19).
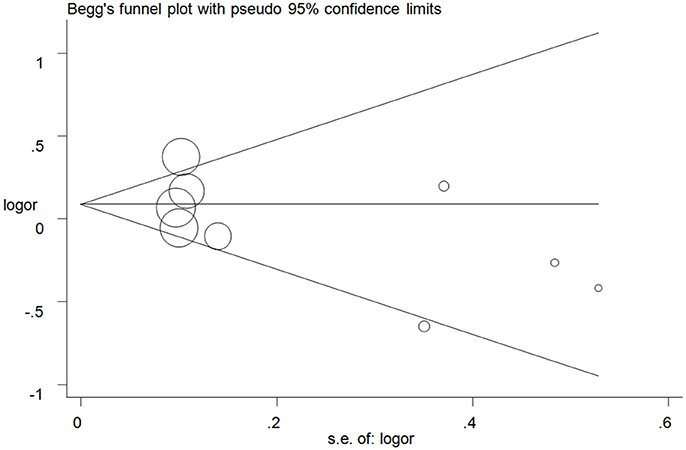
Figure 5. Funnel plot analysis to detect publication bias for GC+CC vs. GG model of microRNA-608 rs4919510 G>C polymorphism. Circles represent the weight of the studies.
Discussion
MicroRNAs is a family of small, noncoding, evolutionarily conserved endogenous RNAs consisting of 22 nucleotides that can regulate gene expression through a post-transcriptional pathway binding to the 3′-UTR of target mRNAs (Lagos-Quintana et al., 2001). MicroRNAs take part in numerous biologic processes, including cell proliferation, differentiation and apoptosis (Ambros, 2003). Evidences have shown that microRNAs play a composite function for oncogenes and tumor suppressor genes. Some mutations, such as SNPs in microRNAs or their binding site, may alter cancer susceptibility in various tumor types and different populations (Niu et al., 2014, 2015).
MicroRNA-608 is novel small RNA that has been shown to be significantly associated with cancer development, metastasis and recurrence. Wang et al. had found that the expression of miR-608 expression was significantly reduced in hepatocellular carcinoma and presented a negative correlation of the tumor size, differentiation, and so on (Wang K. et al., 2016). Yang et al. reported that the miR-608 could suppress the tumorigenesis of colon cancer cells by targeting NAA10 mRNA for degradation. And the overexpression of miR-608 could decrease NAA10 mRNA and protein levels, and thereby suppressed cell proliferation, migration, and cell-cycle progression (Yang et al., 2016).
Rs4919510 G>C is the most common SNP locus in microRNA-608 and might contribute a different free energy to its binding site of target genes, including INSR,CD4, GHR and RXRB (Landi et al., 2008). According the research of Huang et al., the variant genotypes (CG/GG) and variant G-allele were specifically associated with increased risk of HER2-positive subtype in breast cancer patients. HSF1 is a predicted target of miR-608 and the C-to-G substitution of rs4919510 might weaken the suppression of HSF1 mRNA by miR-608, leading to relatively high expression of HSF1 protein, which could up-regulate the expression of HER2 and facilitated tumorigenesis. The above evidence indicated that the alteration of free energy caused by gene mutation between miR-608 and targeted mRNA would influence the protein expression and change the cancer susceptibility (Huang et al., 2012). Since 2008, a series of case-control studies focused on microRNA-608 rs4919510 G>C polymorphism and DSCs risk were published, but the conclusions were inconsistent. Kupcinskas et al. explored the association between microRNA-608 rs4919510 G>C polymorphism and gastric cancer and found an apparently increased risk of gastric cancer in a Latvian population with GG genotype (GG vs. CC, OR = 2.34, 95% CI, 1.08–5.04) (Kupcinskas et al., 2014b). Conversely, Ying et al. evaluated the microRNA-608 rs4919510 G>C polymorphism in Chinese individuals and concluded that the rs4919510 G-allele and genotype GG were protective factors for stage 0 to II colorectal cancer (CRC) (GG vs. CC, OR = 0.70, 95% CI = 0.55–0.88) (Ying et al., 2016). However, other studies did not find any significant relationship between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk, including these studies by Wang et al. (2014) and Zhang et al. (2015). How can we conduct a more precise assessment of the association between microRNA-608 rs4919510 G>C polymorphism and DSC risk with these inconsistent results? As we know, few studies and small sample size maybe the important reasons due to the confusing results. Therefore, we conducted the meta-analysis with nine published case-control studies to investigate the association between the microRNA-608 rs4919510 G>C polymorphism and DSCs susceptibility. Overall, our meta-analysis did not identify any significant association in all of the genetic models. To our knowledge, the potential real results might be confounded or even conceal by some subgroup factors, such as race diversity, control design, cancer type, subject number, and NOS evaluation. So, we subsequently conducted stratified analyses by these subgroup factors. The results indicate that the microRNA-608 rs4919510 G>C polymorphism play an important protective effect against DSCs development in Caucasian population only, but not in other subgroup analysis. It was supposed that the racial diversity maybe the critical factor that contributed to the different result. However, according to current results in general population and subgroups, the precise biological mechanism of the association between microRNA-608 rs4919510 G>C polymorphism and DSCs risk were remains unclear. To our minds, the possible explanation was that the microRNA-608 rs4919510 G>C polymorphism not participated in cancer susceptibility directly, but play a connected and synergism role during the development of cancer. All above results and speculations were needed to confirm with new molecular and epidemiological studies in various races.
In 2017, Liu et al. (2016) and Wu et al. (2017) published two meta-analysis assessing the association between the microRNA-608 rs4919510 G>C polymorphism and cancer risk. Their researches included 10 and 18 case-control studies respectively, which consisted of thyroid cancer, breast cancer, lung cancer and others. Liu et al. indicated that the presence of CC genotypes might be protective against tumorigenesis, especially in Caucasians. And Wu et al. suggested that CG genotypes would increase the cancer risk in the Chinese populations. However, the results of their meta-analysis focused on the general cancer, and the association between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk was not conducted. To our knowledge, this is the first meta-analysis to explore the association between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk. Compared with previous meta-analyses, the present meta-analysis used a more scientific retrieval strategy and a larger sample size. Additionally, more rigorous methodology, including cumulative and sensitivity analyses and quality evaluation with modified NOS, were used to estimate the genetic effects of the microRNA-608 rs4919510 G>C polymorphism on carcinogenesis. This provided a more accurate evaluation of the association between the microRNA-608 rs4919510 G>C polymorphism and DSCs risk.
However, some limitations of this analysis could not be avoided and should be addressed. First, some heterogeneity was apparent among the included studies, which might distort the results in the current meta-analysis. The meta-regression was conducted but not found any distinct factor that contributes to the current heterogeneity. Fortunately, the heterogeneity was partly alleviated in the subsequent stratified analysis, such as race diversity and cancer type. Second, this meta-analysis only focused on one SNP locus (microRNA-608 rs4919510 G>C), and the results were calculated without gene-gene and gene-environment risk factors, leading to a failure to interpret the potential interaction mechanisms. Third, almost all of the included studies focused on Asian and Caucasian populations, which would restrict the application of our results to other populations.
In summary, our meta-analysis demonstrated that the microRNA-608 rs4919510 G>C polymorphism might play an important role in DSCs susceptibility. Further studies in different races and regions with larger population sizes are needed to confirm our findings.
Author Contributions
X-FL, J-KS, and Y-MN conceived the study. X-FL, J-KS, and J-WC searched the databases and extracted the data. J-KS, Y-QZ, and ML analyzed the data. X-FL, J-KS, and J-WC wrote the draft of the paper. JZ and Y-MN reviewed the manuscript. All the authors approved the final manuscript.
Funding
This study was supported by the Foundations of the Science and Technology Department of Hubei Province (No. 2016CFB567, 2014CFB364), and the Hubei Province health and family planning scientific research project (No. WJ2017F069, WJ2015Q041) and Taihe Hospital (2016BSQD02). The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00705/full#supplementary-material
References
Ambros, V. (2003). MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113, 673–676. doi: 10.1016/S0092-8674(03)00428-8
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Begg, C. B., and Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101. doi: 10.2307/2533446
Bosch, S. L., van Rooijen, S. J., Bokkerink, G. M., Braam, H. J., Derikx, L. A., Poortmans, P., et al. (2017). Acute toxicity and surgical complications after preoperative (chemo)radiation therapy for rectal cancer in patients with inflammatory bowel disease. Radiother. Oncol. 123, 147–153. doi: 10.1016/j.radonc.2017.02.009
DerSimonian, R. (1996). Meta-analysis in the design and monitoring of clinical trials. Stat. Med. 15, 1237-1248. doi: 10.1002/(SICI)1097-0258(19960630)15:12<1237::AID-SIM301>3.0.CO;2-N
Egger, M., Davey Smith, G., Schneider, M., and Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634. doi: 10.1136/bmj.315.7109.629
Erren, T. C., Morfeld, P., Foster, R. G., Reiter, R. J., Gross, J. V., and Westermann, I. K. (2016). Sleep and cancer: synthesis of experimental data and meta-analyses of cancer incidence among some 1,500,000 study individuals in 13 countries. Chronobiol. Int. 33, 325–350. doi: 10.3109/07420528.2016.1149486
Gao, C., Ganesh, B. P., Shi, Z., Shah, R. R., Fultz, R., Major, A., et al. (2017). Gut microbe-mediated suppression of inflammation-associated colon carcinogenesis by luminal histamine production. Am. J. Pathol. 187, 2323–2336. doi: 10.1016/j.ajpath.2017.06.011
Hsu, C. Y., Liu, P. H., Ho, S. Y., Huang, Y. H., Lee, Y. H., Chiou, Y. Y., et al. (2017). Impact of tumor burden on prognostic prediction for patients with terminal stage hepatocellular carcinoma: a nomogram study. PLoS ONE 12:e0188031. doi: 10.1371/journal.pone.0188031
Huang, A. J., Yu, K. D., Li, J., Fan, L., and Shao, Z. M. (2012). Polymorphism rs4919510:C>G in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PLoS ONE 7:e35252. doi: 10.1371/journal.pone.0035252
Huedo-Medina, T. B., Sanchez-Meca, J., Marin-Martinez, F., and Botella, J. (2006). Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol. Methods 11, 193–206.doi: 10.1037/1082-989X.11.2.193
Jarzynski, A., Zajac, P., Zebrowski, R., Boguszewska, A., and Polz-Dacewicz, M. (2017). Occurrence of BK virus and human papilloma virus in colorectal cancer. Ann. Agric. Environ. Med. 24, 440–445. doi: 10.26444/aaem/74648
Jayasekara, H., English, D. R., Haydon, A., Hodge, A. M., Lynch, B. M., Rosty, C., et al. (2018). Associations of alcohol intake, smoking, physical activity and obesity with survival following colorectal cancer diagnosis by stage, anatomic site and tumor molecular subtype. Int. J. Cancer 142, 238–250. doi: 10.1002/ijc.31049
Jiang, J., Jia, Z. F., Cao, D. H., Wu, Y. H., Sun, Z. W., and Cao, X. Y. (2016). Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 12, 2215–2226. doi: 10.2217/fon-2016-0224
Jinjuvadia, R., Salami, A., Lenhart, A., Jinjuvadia, K., Liangpunsakul, S., and Salgia, R. (2017). Hepatocellular carcinoma: a decade of hospitalizations and financial burden in the united states. Am. J. Med. Sci. 354, 362–369. doi: 10.1016/j.amjms.2017.05.016
Kupcinskas, J., Bruzaite, I., Juzenas, S., Gyvyte, U., Jonaitis, L., Kiudelis, G., et al. (2014a). Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci. Rep. 4:5993. doi: 10.1038/srep05993
Kupcinskas, J., Wex, T., Link, A., Leja, M., Bruzaite, I., Steponaitiene, R., et al. (2014b). Gene polymorphisms of micrornas in helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLoS ONE 9:e87467. doi: 10.1371/journal.pone.0087467
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Landi, D., Gemignani, F., Barale, R., and Landi, S. (2008). A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 27, 35–43. doi: 10.1089/dna.2007.0650
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Liu, H., Zhou, Y., Liu, Q., Xiao, G., Wang, B., Li, W., et al. (2016). Association of miR-608 rs4919510 polymorphism and cancer risk: a meta-analysis based on 13,664 subjects. Oncotarget. 8, 37023–37031. doi: 10.18632/oncotarget.9509
Mantel, N., and Haenszel, W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748.
Martini, S., Nair, V., Patel, S. R., Eichinger, F., Nelson, R. G., Weil, E. J., et al. (2013). From single nucleotide polymorphism to transcriptional mechanism: a model for FRMD3 in diabetic nephropathy. Diabetes 62, 2605–2612. doi: 10.2337/db12-1416
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339:b2535. doi: 10.1136/bmj.b2535
Motoyama, S., Sato, Y., Sasaki, T., Wakita, A., Kawakita, Y., Liu, J., et al. (2017). Efficacy and safety of neoadjuvant chemoradiotherapy following esophagectomy with japanese-style extended 3-field lymphadenectomy for thoracic esophageal cancer. Anticancer Res. 37, 5837–5843. doi: 10.21873/anticanres.12027
Nederlof, N., Slaman, A. E., van Hagen, P., van der Gaast, A., Slankamenac, K., Gisbertz, S. S., et al. (2016). Using the comprehensive complication index to assess the impact of neoadjuvant chemoradiotherapy on complication severity after esophagectomy for cancer. Ann. Surg. Oncol. 23, 3964–3971. doi: 10.1245/s10434-016-5291-3
Niu, Y. M., Du, X. Y., Lu, M. Y., Xu, Q. L., Luo, J., and Shen, M. (2015). Significant association between functional microRNA polymorphisms and head and neck cancer susceptibility: a comprehensive meta-analysis. Sci. Rep. 5:12972. doi: 10.1038/srep12972
Niu, Y., Yuan, H., Shen, M., Li, H., Hu, Y., and Chen, N. (2014). Association between cyclooxygenase-2 gene polymorphisms and head and neck squamous cell carcinoma risk. J. Craniofac. Surg. 25, 333–337. doi: 10.1097/SCS.0000000000000372
Ryan, B. M., McClary, A. C., Valeri, N., Robinson, D., Paone, A., Bowman, E. D., et al. (2012). rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer. PLoS ONE 7:e36306. doi: 10.1371/journal.pone.0036306
Siegel, R. L., Miller, K. D., and Jemal, A. (2017). Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30. doi: 10.3322/caac.21387
Tessitore, A., Cicciarelli, G., Del Vecchio, F., Gaggiano, A., Verzella, D., Fischietti, M., et al. (2014). MicroRNAs in the DNA damage/repair network and cancer. Int. J. Genomics 2014:820248. doi: 10.1155/2014/820248
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Valeri, N., Gasparini, P., Fabbri, M., Braconi, C., Veronese, A., Lovat, F., et al. (2010). Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. U.S.A. 107, 6982–6987. doi: 10.1073/pnas.1002472107
Wang, K., Liang, Q., Wei, L., Zhang, W., and Zhu, P. (2016). MicroRNA-608 acts as a prognostic marker and inhibits the cell proliferation in hepatocellular carcinoma by macrophage migration inhibitory factor. Tumour Biol. 37, 3823–3830. doi: 10.1007/s13277-015-4213-5
Wang, R., Zhang, J., Ma, Y., Chen, L., Guo, S., Zhang, X., et al. (2014). Association study of miR149 rs2292832 and miR608 rs4919510 and the risk of hepatocellular carcinoma in a largescale population. Mol. Med. Rep. 10, 2736–2744. doi: 10.3892/mmr.2014.2536
Wang, Z., Xue, Y., Wang, P., Zhu, J., and Ma, J. (2016). MiR-608 inhibits the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor. Oncol. Rep. 35, 2733–2742. doi: 10.3892/or.2016.4652
Wu, S., Yuan, W., Shen, Y., Lu, X., Li, Y., Tian, T., et al. (2017). The miR-608 rs4919510 polymorphism may modify cancer susceptibility based on type. Tumour Biol. 39:1010428317703819. doi: 10.1177/1010428317703819
Yang, H., Li, Q., Niu, J., Li, B., Jiang, D., Wan, Z., et al. (2016). microRNA-342-5p and miR-608 inhibit colon cancer tumorigenesis by targeting NAA10. Oncotarget 7, 2709–2720. doi: 10.18632/oncotarget.6458
Ye, Y., Wang, K. K., Gu, J., Yang, H., Lin, J., Ajani, J. A., et al. (2008). Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila). 1, 460–469. doi: 10.1158/1940-6207.CAPR-08-0135
Ying, H. Q., Peng, H. X., He, B. S., Pan, Y. Q., Wang, F., Sun, H. L., et al. (2016). MiR-608, pre-miR-124-1 and pre-miR26a-1 polymorphisms modify susceptibility and recurrence-free survival in surgically resected CRC individuals. Oncotarget 7, 75865–75873. doi: 10.18632/oncotarget.12422
Zhang, M. W. (2012). Study on the Associations of Lifestyle-Related Factors, Genetic Variants in miRNA Encoding Regions and miRNA Binding Sites With Colorectal Cancer Risk. Zhejiang: Zhejiang University.
Keywords: microRNA-608, rs4919510, cancer, polymorphism, susceptibility
Citation: Li X-F, Song J-K, Cai J-W, Zeng Y-Q, Li M, Zhu J and Niu Y-M (2018) No Association Between MicroRNA-608 rs4919510 G>C Polymorphism and Digestive System Cancers Susceptibility: A Meta-Analysis Based on 10,836 Individuals. Front. Physiol. 9:705. doi: 10.3389/fphys.2018.00705
Received: 05 March 2018; Accepted: 22 May 2018;
Published: 07 June 2018.
Edited by:
Brian James Morris, University of Sydney, AustraliaReviewed by:
Hector A. Cabrera-Fuentes, Justus Liebig Universität Gießen, GermanyAndrey Turchinovich, Deutsches Krebsforschungszentrum (DKFZ), Germany
Copyright © 2018 Li, Song, Cai, Zeng, Li, Zhu and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Ming Niu, bjRvbmVvbmVAMTI2LmNvbQ==
†These authors have contributed equally to this work.
 Xue-Feng Li1†
Xue-Feng Li1† Ju-Kun Song
Ju-Kun Song Yu-Ming Niu
Yu-Ming Niu