- 1Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Emory University, Atlanta, GA, United States
- 2Department of Chemistry, Emory University, Atlanta, GA, United States
The cells and receptors of the immune system are mechanically active. Single molecule force spectroscopy, traction force microscopy, and molecular tension probe measurements all point to the importance of piconewton (pN) molecular forces in immune function. For example, forces enhance the ability of a T-cell to discriminate between nearly identical antigens. The role of molecular forces at these critical immune recognition junctions is puzzling as mechanical forces generally facilitate bond dissociation, potentially increasing the difficulty for a receptor to recognize its cognate antigen. The advantage that molecular forces confer in the process of immune recognition is not clear. Why would cells expend energy to exert force on the critical, but tenuous bonds that mediate immune surveillance? Do molecular forces provide some advantage to the immune system? The premise of this review is that molecular forces provide a specificity advantage to immune cells. Inspired by the recent discovery that receptor forces regulate immune signaling in T-cell and B-cells, we dub this notion “mechanical proofreading,” akin to more classic kinetic proofreading models. During the process of mechanical proofreading, cells exert pN receptor forces on receptor-ligand interactions, deliberately increasing the energy cost of the immune recognition process in exchange for increased specificity of signaling. Here, we review the role of molecular forces in the immune system and suggest how these forces may facilitate mechanical proofreading to increase the specificity of the immune response.
Introduction
Immune cells must detect and respond to rare traces of malignancies or infection. Accordingly, the immune response must display extraordinary sensitivity and specificity. The requirements of specificity and sensitivity are often mutually exclusive: for example, if the signaling threshold required to initiate an immune response is set very high, the immune system is unlikely to make a mistake, but also more likely to miss an infection.
T-cell antigen recognition is a striking example of a vital immune recognition event that must balance both extreme sensitivity and specificity. The T-cell receptor (TCR) physically engages with peptide antigens bound to the major histocompatibility complex (pMHC). Virtually all nucleated cells present fragments of their proteome on the MHC for TCR inspection. When a TCR recognizes an antigen, TCR-pMHC binding triggers biochemical signaling leading to T-cell activation [1]. However, the origins of T-cell triggering in response to antigen binding are the subject of intense debate [2]. For example, TCR-pMHC affinity (as measured by 3D techniques such as surface plasmon resonance) is low, with dissociation constants in the range of 1–100 μM [3], one of the weakest measured affinities for receptor-ligand binding. Somehow, despite the poor TCR-pMHC affinity, single amino acid alterations in the peptide antigen can produce a 10,000-fold difference in T-cell activation [4] and some reports suggest that the TCR possesses single-molecule antigen sensitivity [5, 6]. TCR binding affinity (KD), off rates (koff), and on rates (kon) sometimes, but not always correlate with T-cell activity [7–9]. Additionally, a comparison of crystal structures of TCR-pMHC bound and unliganded TCRs, reveal only minor conformational shifts upon TCR-pMHC binding [10]. The mechanism through which TCR-pMHC binding produces a high-fidelity signal to trigger T-cell activation remains a mystery [2, 11].
Further complicating the issue of T-cell antigen recognition is the observation that T-cells are mechanically active. The pMHC-TCR interaction forms only when a T-cell physically touches target cells; thus, it is likely that the TCR-pMHC complex experiences force. In support of this notion, soluble, monovalent pMHC can bind to the TCR but fails to activate T-cells [12, 13], while pMHC attached to a planar lipid bilayer does activate T-cells [14]. Collectively, these results suggest that mechanical forces may regulate TCR triggering. A seminal optical tweezer study by Reinherz et al. demonstrated that T-cells trigger in response to forces exerted on the TCR-pMHC complex, positioning the TCR as a mechanosensor [15]. Furthermore, the TCR is not a passive recipient of external forces. Our group pioneered the development of molecular probes to map pN forces applied by cells [16–19]. In particular, DNA-based probes revealed that the TCR transmits defined piconewton forces to the pMHC, and that these forces facilitate TCR antigen discrimination (Figures 1A–D) [20, 22]. Traction force microscopy and micropillar measurements also demonstrate that T-cells transmit forces through the TCR-pMHC interaction and through the CD3 complex [23, 24]. Additionally, biomembrane force probe and optical tweezer measurements have revealed that TCR-pMHC bond lifetimes increase under force (catch bond behavior) for agonists, but decrease under force for non-agonist pMHC [21, 25–27] (Figures 1E,F). Additional studies have revealed the role molecular forces play in regulating the pore-forming capability of cytotoxic T-cells [28] and mediating the antigen recognition by CD8+ T-cells [20]. The many contexts in which receptor forces regulate immune processes have been reviewed elsewhere [29, 30]. Many mechanisms to explain T-cell antigen recognition have been proposed [2], but vital questions remain unanswered. Why would a T-cell expend energy to exert force on an already tenuous, low affinity interaction? How do molecular forces aid in immune recognition?
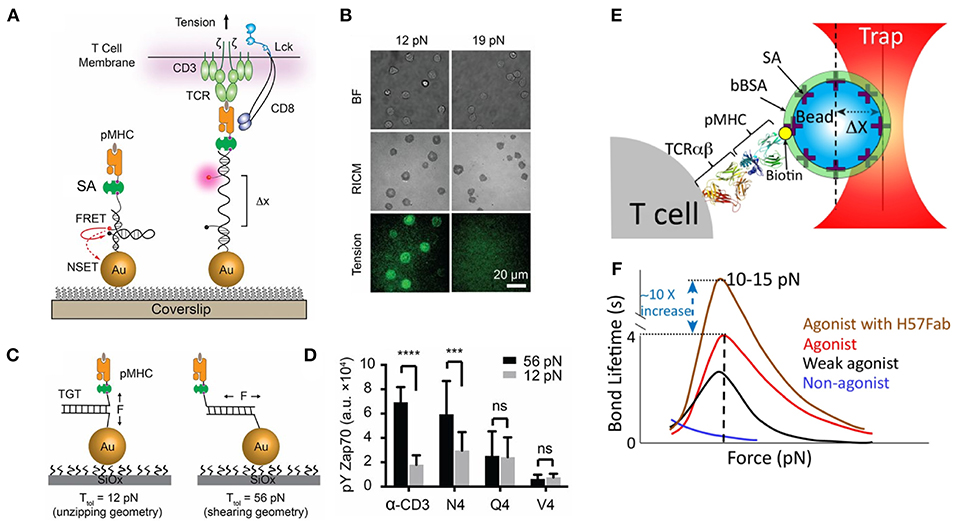
Figure 1. Piconewton (pN) molecular forces are important to TCR antigen specificity. (A) Molecular tension probes are comprised of a fluorophore-quencher pair flanking a force-extensible domain. Molecular forces extend the probe, separating the fluorophore-quencher pair. DNA-based molecular tension probes, highlighted here, produce up to a 100x increase in fluorescence under the influence of cellular forces. (B) Using DNA molecular tension probes, OT-1 CD8+ T-cells were found to exert defined (12–19pN) forces through the TCR. (C) Tension gauge tethers rupture irreversibly when receptor forces reach a threshold, capping the maximum tension a cell can exert through a receptor. (D) OT-1 CD8+ T-cells exhibit more potent activation when the pMHC-TCR complex is more mechanically stable. (E) Optical tweezers, formed by trapping a bead near the focus of a tightly focused laser, enable precise control over the movement of the bead and are useful for exerting pN forces on receptor-ligand interactions. (F) Optical tweezer manipulation of pMHC-presenting beads enabled the application of pN forces to TCR-pMHC complexes. TCR-pMHC bond lifetime increases for agonist pMHC (catch bond behavior) but decreases for non-agonist pMHC (slip bond behavior). The TCR FG loop is thought to elongate under cellular force and to be responsible for catch bond behavior. Stabilizing the FG loop with the H57Fab frag further enhances bond lifetime. (A–D) Are adapted from with permission from Liu et al. [20] while panels (E,F) are adapted from Feng et al. [21] licensed under Creative Commons (CC BY 4.0). NSET, near surface energy transfer; BF, brightfield; RICM, reflection interference contrast microscopy which displays cell-surface contact area; TGT, tension gauge tether; Ttol, the tension tolerance of a TGT, the force at which the TGT ruptures within 2 s; bBSA, biotinylated bovine serum albumin; SA, streptavidin. *** Indicates p < 0.001 and **** indicates p < 0.0001.
Receptor forces have many potential effects on receptor-ligand binding which have been excellently reviewed elsewhere [31]. Many effects of molecular forces might hinder antigen recognition (Figure 2). For example, receptor forces alter equilibrium by shifting the energy landscape of bound and unbound receptor-ligand pairs (Figure 2A). Cell-generated forces may therefore destabilize receptor-ligand complexes critical to immune recognition events, potentially dampening cellular activation. Additionally, as predicted by the Bell model, tensile forces on receptor ligand interactions typically increase the bond dissociation rate in a force-dependent manner [32], a phenomenon-termed a slip-bond (Figure 2B). The increased dissociation rate reduces the number of bound receptors and is expected to dampen signaling. Individual slip bonds do not provide a specificity benefit under constant forces for pure affinity-based discrimination. For example, if a cognate antigen displays a koff, cognate = 1 s−1 while a non-cognate antigen has koff, non−cognate = 10 s−1 then the binding error rate (assuming identical on rates) is approximately the ratio of the off rates, koff, cognate/koff, non−cognate = 1/10. The Bell model (see equation, Figure 2B) dictates that force alters the off rate of both the non-cognate and cognate antigen by the same factor of exp(FΔx/kBT), leaving the ratio koff, cognate and koff, non−cognate unchanged when both antigens experience the same cellular force and assuming Δx is similar for both antigens. This simple analysis is valid for constant forces, but dynamic forces [33] will have more complex effects that are difficult to treat in the context of this review. In rare cases, forces increase the lifetime of receptor-ligand interactions under force, a phenomenon known as a catch bond [34, 35]. Catch bonds could conceivably stabilize receptor-ligand recognition under force, facilitating signaling. Finally, pN forces alter protein energy landscapes, which may facilitate conformational shifts (Figure 2C). Force-induced conformational changes may lead to the recruitment of accessory proteins to mechanically strained receptors, amplifying the signal of mechanically strained receptor-ligand complexes. Given the many possible effects of molecular forces, it is crucial to define the precise role of pN molecular forces in the immune system.
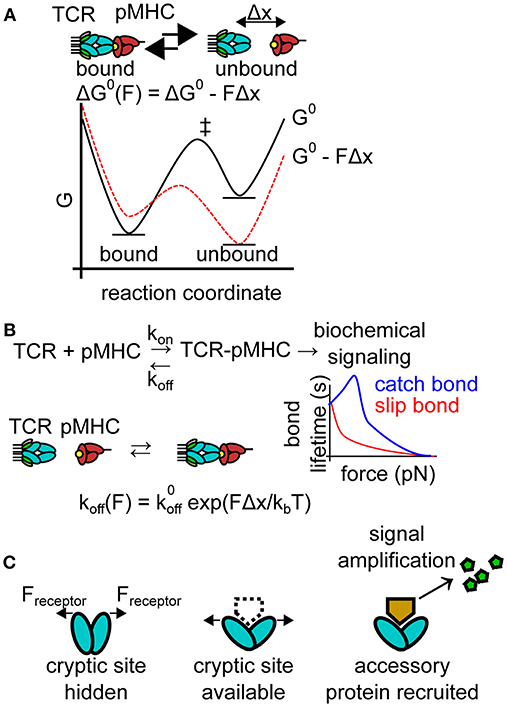
Figure 2. Effects of pN forces on receptor-ligand interactions. (A) Molecular forces may skew the energy landscape of ligand binding. Under most conditions, pN molecular forces destabilize receptor-ligand complexes, shifting thermodynamic equilibrium. The plot depicts an idealized free energy diagram for a two-state receptor-ligand interaction in the presence and absence of pN force. G0, standard free energy of the interaction pathway in the absence of force; Δx, distance to the transition state of receptor-ligand unbinding; and ‡ indicates the transition states on the free energy diagram. (B) Molecular forces also generally reduce bond lifetime (increasing koff under force). This force-dependent reduction in bond lifetime is known as a slip bond and is well-described mathematically by the Bell model. Select bonds exhibit increased bond lifetime under force, a phenomenon known as a catch bond. The plot depicts the force-bond lifetime relationship for idealized catch and slip bonds. , zero-force off rate; kB, Boltzmann constant; T, temperature. (C) Molecular forces may also alter the energy landscape of a protein, facilitating conformational shifts, exposing cryptic binding sites for accessory proteins (dotted line). Accessory proteins may produce secondary messengers, amplifying the signal produced by force-induced conformational shifts.
The purpose of this review is to suggest that pN molecular forces provide a benefit to receptor-ligand interactions by increasing the specificity of signaling. In effect, cells expend energy in the form of pN receptor forces in exchange for increased specificity. With kinetic proofreading as a hallmark example, literature precedent exists for biological systems expending energy to purchase specificity. DNA replication and protein synthesis both leverage kinetic proofreading, increasing fidelity by using far-from-equilibrium intermediate states driven by triphosphate hydrolysis. These irreversible intermediate steps enable discrimination based on the off rates of “correct” vs. “incorrect” substrates [36]. In addition to kinetic proofreading, the serial engagement model has also been widely discussed as a mechanism to facilitate TCR antigen discrimination [37, 38]. These models have been reviewed elsewhere [2, 39], and are not the focus of this review. Instead, we focus on how molecular forces open many new possibilities to explain the specificity of immune recognition events.
Here, we review evidence for the importance of molecular forces in the immune system. Inspired by the work of Tolar [40], Zhu [25], Reinherz et al. [15], Lang et al. [21], and by our recent work [20], we interpret how forces might enable a cell to expend mechanical work to increase signaling accuracy. In an analogy to kinetic proofreading, we term the concept that cells harness mechanical work to increase the specificity of signaling “mechanical proofreading.”
Mechanisms of Mechanical Proofreading
We highlighted three potential mechanisms for mechanical proofreading. First, bond lifetime may be altered by molecular forces to facilitate mechanical proofreading. Most biological bonds decrease in duration under force (slip bonds), but a subset of bonds exhibit an increased lifetime under force, a phenomenon known as catch bonds [34, 35]. The change in bond lifetime due to molecular forces may provide a potent means of mechanical proofreading (Figures 2B, 3A). Through the alteration of the bond lifetime, mechanical proofreading may enable immune receptors to select for the rare ligands that produce catch bond behavior under force, increasing the fidelity of signaling. Second, mechanical proofreading could occur via a “stress test.” Stress is defined as force per unit area. Analogously, cells may apply forces too great for a single receptor-ligand interaction to withstand. Only clusters of proteins sharing cellular forces among many bonds reduce the applied stress below a threshold level, enabling longer, more stable bonds (Figure 3B). The mechanical proofreading stress test selects for multivalent interactions and does not require catch bonds to function. A third mechanism of mechanical proofreading is a “strain test.” Strain is defined as the change in length of a material due to mechanical stress divided by the original length of the material. Analogously, if a receptor-ligand bond can withstand forces applied to it, the force may produce a conformational change (loosely analogous to strain) in one or both binding partners. This conformational change could result in adaptor protein recruitment, or exposure of cryptic binding sites, producing downstream signaling (Figure 3C). The mechanical proofreading strain test selects for, and subsequently amplifies, the signal from mechanically stable individual receptor-ligand bonds that can survive mechanical forces. While the strain test mechanism may contribute to the formation of catch bonds, the signaling outcome of the strain model is not the increased bond lifetime, but rather the activation of a mechanosensitive switch that enhances the fidelity of receptor-ligand binding.
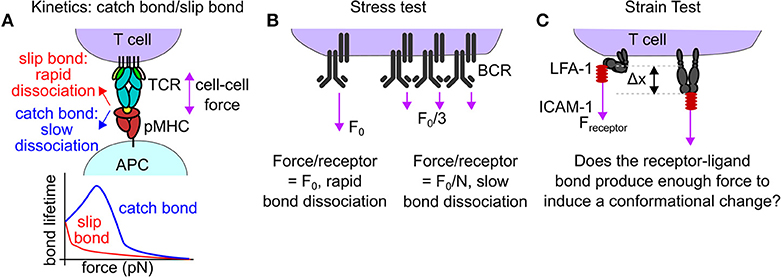
Figure 3. Mechanisms of mechanical proofreading. (A) In rare cases, forces borne by receptor-ligand interaction reduce the off rate of that interaction, a behavior known as a catch bond. The TCR-pMHC bond is known to exhibit catch bond behavior. The increased bond lifetime under force may facilitate downstream signaling, acting as a form of mechanical proofreading. (B) Mechanical proofreading may occur via a stress test if single bonds are insufficient to withstand the applied cellular force. Only when the force is spread across many bonds will the entire force-bearing structure survive to produce biological signaling. This mode of mechanical proofreading may occur in BCR-antigen internalization. (C) Mechanical proofreading may occur via a strain test. Piconewton receptor forces act to produce a conformational shift in a receptor only if the strength of the receptor-ligand bond is sufficient. A potential example of the mechanical proofreading strain test is the LFA-1:ICAM bond. LFA-1 is known to extend in response to interactions with ICAM-1 and to modulate T-cell functions.
Mode 1: Alteration of Bond Lifetime
Receptor-ligand interactions at cell-cell or cell-extracellular matrix junctions frequently experience force. These pN forces alter receptor-ligand bond kinetics (Figures 2B, 3A). Most frequently, receptor-ligand forces produce reduced bond lifetime (slip bonds) [32]. In special cases, bond lifetime increases under force, a phenomenon known as a catch bond [34]. AFM measurements provided the first direct evidence for a catch bond between P-selectin and P-selectin glycoprotein ligand-1 [35]. Since this initial observation, catch bonds have been demonstrated in selectins, integrins, the platelet glycoprotein GPIbα, E-cadherin, and recently in the TCR-pMHC interaction [25, 35, 41–43].
Qualitatively, catch bonds provide a means to spend mechanical energy (from cyotoskeletal and motor protein forces) in exchange for specificity. Most molecular bonds exhibit slip bond behavior; therefore, if a cell expends mechanical energy via applying force to a receptor-ligand bond and bond lifetime increases, the interaction is very likely to be “correct.” This enhancement of bond lifetime under force may amplify the downstream effects of cognate ligand binding events relative to the shorter bond lifetime (under force) of non-cognate ligands through the classic kinetic proof reading model. Catch bonds thus complement the kinetic proofreading model and offer a potent means for mechanical proofreading.
T-cells are mechnaosensitive, transducing TCR forces into biochemical signals, such as calcium flux [15]. Additionally, the TCR transmits piconewton pN forces to the pMHC, and these forces are important to T-cell antigen recognition [20]. 2D affinity measurement of the TCR-pMHC bond demonstrated that agonist pMHC actually has a shorter bond lifetime than weak agonist pMHC [8]. Conversely, biomembrane force probe measurements revealed that the TCR-pMHC interaction exhibits catch bond behavior, but only for agonist antigen [25] (Figure 3A). The catch bond behavior of the TCR-pMHC bond was also confirmed by optical tweezer measurements [26]. Sibner and colleagues recently interrogated TCRs that bind pMHC but are not activated. When agonist peptides to the formerly non-responsive TCRs were generated, catch bonds that correlated with CD 45 exclusion from the T-cell antigen presenting the cell contact area were identified in the new, activating interactions [44]. This finding further emphasizes the importance of catch bonds, and thus mechanical proofreading, in T-cell antigen discrimination. The accumulation of TCR-pMHC bond lifetimes may link TCR forces with the serial-engagement model of T-cell activation [38], allowing the catch bond behavior of many TCR-pMHC interactions to cumulatively trigger sustained calcium signaling during T-cell activation [25, 45]. The increase in bond lifetime for agonist pMHC could serve as an extraordinarily specific indicator that a T-cell has found agonist antigen, specificity purchased at the cost of mechanical work.
Catch bonds are not limited to the TCR-pMHC interaction. A catch bond has been demonstrated in the bond between lymphocyte function-associated antigen-1 (LFA-1) and intercellular adhesion molecule 1 (ICAM-1) [41]. Combined with the observation that dendritic cells (DC) immobilize ICAM-1 on their surface in response to inflammatory signals [46], these results raise the possibility that that the LFA-1-ICAM-1 catch bond may help T-cells recognize stimulated DCs.
Finally, catch bonds likely mediate mechanical proofreading in non-immune biological systems. For example, the platelet glycoprotein GPIbα exhibits catch bond behavior when interacting with Von Willebrand factor [42]. Bacterial adhesion under flow is regulated by a catch bond between FimH and mannose [47]. Leukocyte adhesion against the flow of blood depends on a catch bond [35]. It is likely that catch bonds are a biologically general mechanism for mechanical proofreading, ensuring that biological processes occur only at specific, desirable interfaces via extremely specific mechanically stable receptor-ligand bonds.
Mode 2: Stress Test
Most receptor-ligand interactions are not catch bonds; however, slip bonds in groups of mechanically strained receptors can also facilitate mechanical proofreading. Receptor clustering is crucial to many biological signaling pathways and may also provide a mechanical advantage to force-bearing receptors. When force-bearing receptors cluster together, the stress (defined as force per unit area) applied at a cell-cell junction may be reduced because the force is distributed over many bonds. Receptor clustering therefore offers an opportunity for a mechanical proofreading “stress test,” which selects for cellular structures composed of many force-bearing proteins. Collectively, clustered proteins may withstand forces that would rupture any single bond. Note that for the stress test mechanism to function, the cellular force-generating machinery must be connected to groups of receptors (e.g., one actin stress fiber transmitting force to many integrins).
Consider a cluster of N receptors withstanding a total force F at a cell-cell junction. Force balance dictates that each receptor is bearing a force of approximately F/N. Clustering leads to an N-fold decrease in the force experienced per ligand-receptor pair. Based on the Bell model [32], the reduction in force on each receptor-ligand pair will lead to a significant enhancement in the bond lifetime, increasing the probability that the cellular structure will survive long enough to initiate biochemical signaling. To achieve the desired specificity and sensitivity, biological systems can alter both F and N. Increasing F produces shortened bond lifetime, decreasing sensitivity. Conversely, increasing N reduces the per-receptor force, facilitating signaling under force, potentially decreasing specificity. High F and N may achieve both high sensitivity and specificity by enabling only highly multivalent interations to initiate biochemical signaling.
B cell receptor (BCR) antigen internalization is an example of a mechanical proofreading stress test. BCR signaling is intricately related to the B cell cytoskeleton [48]. Clusters of BCRs are thought to use mechanical force to internalize antigen. Natkanski et al. demonstrated that B cells internalize plasma membrane sheet bound antigen in a myosin IIA dependent manner, and that clathrin-coated structures seem to be associated with antigen internalization [40]. Interestingly, BCR microclusters were observed to resist contractile forces for 20–30 s; however, AFM measurements demonstrated that single BCR-antigen bonds do not endure long enough for the observed membrane invagination to occur [40]. The authors hypothesize that forces exerted by B cells shorten the BCR-antigen bond lifetime, ensuring that only multivalent, high-affinity antigens which collectively withstand the cellular force are internalized (Figure 3B) [40]. These findings are supported by the observation that B cells exert measurable traction forces through the B cell receptor, and that the magnitude of traction force scales with the number of clustered BCRs involved in force transmission [49]. Additionally, DNA-based molecular tension sensor measurements demonstrated that the BCR utilizes molecular forces to extract antigen from follicular dendritic cells, and that stiff follicular dendritic cells produce stronger BCR forces and more stringent antigen affinity discrimination [50]. Collectively, this evidence points toward a mechanical stress test facilitating BCR selection for high-affinity antigen.
A mechanical stress test may also be important to TCR signaling. The TCR is known to exert pN forces on the TCR-pMHC bond and also on TCR-pMHC clusters [20, 22], but the importance of nanoclusters in TCR force transmission is not known. Super-resolution imaging has suggested that TCRs form nanoclusters, and that clustered TCRs are more likley to be phosphorylated and to participate in downstream signaling [51]. TCRs are known to cluster at the surface of microvilli on the T-cell surface [52] and TCR-bearing microvilli are selectively stabilized at the T-cell antigen presenting cell interface [53]. Collectively, this evidence raises the possibility that TCR nanoclusters, and potentially microvilli, may facilitate a mechanical stress test.
Mechanical stress within other supramolecular complexes may also be important. For example, the focal adhesion has been proposed to behave like a molecular clutch, with mechanical unfolding of adaptor proteins serving to recruit more integrins to the adhesion site to share the applied load [54]. Thus, focal adhesions may sense and respond to substrate stiffness via a mechancial proofreading stress test, where specific combinations of force and adhesion site density produce focal adhesion growth and maturation.
Mode 3: Strain Test
Another mode of mechanical proofreading is a “strain test.” Strain is defined as the change in length of an object relative to its original length. Analogously, many receptors undergo conformational changes due to ligand or allosteric interactions. Some conformational shifts are facilitated by pN scale forces [55, 56]. Conformational shifts may expose cryptic binding sites, leading to protein recruitment or to phosphorylation of a previously inaccessible site. Force-reconfigurable proteins therefore offer a third potential mode of mechanical proofreading. In the mechanical proofreading strain test, a receptor-ligand bond must withstand a threshold force to produce a conformational shift before eliciting biochemical signaling. Note that for the strain test to function, a large energy threshold must prevent the receptor from spontaneously experiencing the conformational shift in the absence of a large input of mechanical work by a cell.
Integrins are well-studied examples of the mechanical proofreading strain test. They exhibit profound conformational shifts, existing in a low-affinity, bent conformation at rest, but able to adopt a high affinity extended state [57]. Integrins may be an ultrasensitive molecular switch, able to extend in response to pN forces [55].
In the context of lymphocytes, the integrin αLβ2, also known as LFA-1, binds to ICAM, and is important to immune cell-cell adhesion and to T-cell function [58]. LFA-1 undergoes outside-in activation in response to surface bound, but not soluble ICAM [59], and molecular forces increase the rate of LFA-1 extension and slow the rate of LFA-1 bending (Figure 3C) [56]. It is believed that actin forces transmitted through LFA-1 to immobilized ICAM-1 induce extension of LFA-1 and that actin is critical to the formation and maintenance of the immunological synapse [60]. Additionally, recent interference photoactivation localization microscopy (iPALM) measurements, which resolve the location of fluorophores along the microscope optical axis with ~10 nm resolution, have measured the extension of LFA-1 at a T-cell surface interface [61]. Outside-in activation of LFA-1 has functional consequences; for example, severing ICAM's cytoskeletal anchorage prevented natural killer cells from forming junctions with target cells and disrupted granule polarization [62]. Additionally, when DCs are induced to mature via treatment with lipopolysaccharide, they utilize their actin cytoskeleton to restrict ICAM-1 surface mobility. T-cells sense the change in ICAM-1 mobility, producing more extended LFA-1 on the T-cells surface [46]. Collectively, these findings demonstrate that LFA-1 exhibits force-sensitive outside-in activation, positioning LFA-1 as a potential mechanical proofreading strain sensor.
The T-cell receptor (TCR) may also utilize a mechanical strain test during T-cell antigen recognition. Optical tweezer measurements demonstrated a force dependent extension in the FG loop of the TCR [26]. The FG loop extends 8–15 nm under the influence of pN forces. Deleting the FG loop of the TCR removed the force-dependent extension behavior and reduced the ability of T-cells to respond to antigen as assessed by IL-2 production [26]. The FG loop extension of the TCR therefore presents another example of a potential mechanical proofreading strain test. Because TCR forces are between 12 and 19 pN [20], and because the FG loop extension is known [26], the height of the mechanical proofreading energy barrier that TCR forces must surmount has been calculated as ~37 kBT (assuming 10 nm displacement and 15 pN forces), or almost twice the energy of one ATP hydrolysis. This energy of discrimination could explain the remarkable specificity of T-cell antigen recognition [21]. The mechanical proofreading strain test is therefore capable of extreme specificity.
The strain test mode of mechanical proofreading is not limited to the immune system. For example, platelets action must be tightly regulated to prevent erroneous clot formation. The platelet integrin αIIbβ3 is anisotropically mechanosensitive, requiring lateral forces to undergo outside in activation to enable platelet spreading and activation on a surface [63, 64] which may explain why platelets ignore soluble fibrinogen in the blood, only binding to fibrinogen attached to other activated platelets. Additionally, the platelet glycoprotein complex GPIb-IX-V, has a mechanosensitive domain that extends several nanometers under force [65], making it another likely candidate for a strain test for mechanical proofreading.
Combinations of Mechanical Proofreading Modes
The modes of mechanical proofreading may also work together to produce increased specificity. For example, the GP1b-IX-V:von-Willebrand factor bond is a catch bond [42]. The prolonged bond lifetime under force may facilitate the opening of the mechanosensitive domain of GP1b-IX-V [66]. Similarly, the TCR exhibits catch bond behavior, an extension of its FG loop, and receptor clustering [25, 26, 51]. Various mechanisms invoked to explain the extreme sensitivity and specificity of the TCR-pMHC bond involve each of the proposed modes of mechanical proofreading [2, 11]. The molecular clutch model of the focal adhesion [54, 67] argues that talin unfolding under force (a strain test) regulates integrin recruitment to the focal adhesion (regulating stress), thus these two models may work cooperatively in focal adhesions. Elucidating which mechanisms are critical to regulating cellular decision-making will require careful experimentation to isolate the effects of each of the modes of mechanical proofreading.
Conclusion
Here, we have focused on applications of mechanical proofreading within the immune system, but mechanical proofreading is likely a biologically general mechanism. Because mechanical forces generally disrupt receptor-ligand interactions, as described by the Bell model [32], careful examination of force-exerting systems is required to determine what functional benefit is derived by expending cellular energy on force generation. Mechanical forces may be vital to biological differentiation between soluble and a surface bound ligand. Soluble ligands are not capable of resisting mechanical force. Indeed, it is conceivable that from a mechanistic point of view, mechanical forces are the principle difference between cell-cell interfaces and receptors interacting with soluble ligands.
Mechanical proofreading likely has great physiological relevance. Zhu et al. found that during thymic selection, negative selection ligands produced cooperative trimolecular catch bonds (TCR-pMHC-CD8) while positive selection ligands formed slip bonds [68]. This result strongly suggests that mechanical proofreading, by means of catch bonds, assists in the elimination of strongly self-reactive thymocytes during negative selection. Additionally, Huse et al. recently found that the exertion of force by T-cells is spatially and temporally linked to the release of cytotoxin release and target cell killing [28]. Thus, mechanical proofreading is also implicated in the killing of target cells by cytotoxic T-cells. Finally, T-cells have been demonstrated to respond to increased stiffness of a substrate presenting activating antibodies to CD3 and CD28 with increased IL-2 secretion [69]. Importantly, human primary immune cells, including macrophages and dendritic cells, modulate their stiffness in response to inflammatory signals [70]. Collectively, the ability of antigen-presenting cells to modulate their stiffness in response to inflammatory signals combined with the stiffness sensitivity of T-cells suggests that mechanical proofreading may play a role in enabling T-cells to respond appropriately to complex environmental signals.
Additionally, mechanical proofreading has the potential to resolve longstanding debates in immunology. For example, mechanical proofreading may provide insight into how adaptive immune responses are initiated. TCR binding affinity for pMHC does not always correlate with T-cell activity [7–9]. The ability of a pMHC to bind to the TCR does not guarantee that the TCR will activate [71]. Garcia et al. recently interrogated TCRs that bind pMHC but are not activated. When agonist peptides were produced to the formerly non-responsive TCRs, these new agonist pMHC-TCR bonds exhibited catch bond behavior [44]. This finding further emphasizes the importance of mechanical forces, catch bonds, and mechanical proofreading in T-cell antigen discrimination.
Finally, mechanical proofreading may have utility for cell-based immunotherapies. Kam et al. have recently demonstrated the use of polydimethylsiloxane microbeads coated in activating antibodies to CD3 and CD28 to enhance the ex vivo activation and proliferation of both CD4+ and CD8+ T-cells for immunotherapy [72]. Intriguingly, T-cell expansion was found to be enhanced in soft beads, implicating a role for mechanical forces in ex vivo T-cell expansion. It is also possible that some variety of mechanical proofreading may operate in chimeric antigen receptor (CAR) T-cell recognition of antigen. Chen and colleagues recently rewired CAR-T-cells to recognize soluble antigen [73]. However, CAR signaling in these cells required ligand-mediated CAR dimerization, which the authors believe implicates the role of mechanotransduction in CAR signaling. Interestingly, the chimeric antigen-receptor is comprised of an antibody-derived fragment for ligand binding. Antibodies exhibit a slip bond character under force [74]. Mechanotransduction during CAR signaling likely proceeds via a different mechanism than the catch bond that influences TCR-pMHC interactions.
Experiments designed to test the models outlined here may be critical in determining the mechanical origins of cellular decision-making. For example, deletion of mechanically sensitive domains in the TCR reduced IL-2 production [26]. Likewise, the mecahnosensitivity of platelets has been partially explained via a mechanosensitive domain discovered in the platelet glycoprotein GpIbα [65]. Deletion of a portion of the mechanosensitive domain produces constitutive GpIbα signaling [75]. These experiments may be facilitated by the recent development of powerful molecular tools capable of measuring both the magnitude and orientation of molecular forces [18, 20, 22, 64, 76]. We anticipate that understanding the precise role of molecular forces in receptor-ligand interactions will provide fundamental insight into the exquisite specificiy of cellular decision-making.
Author Contributions
JB and KS conceived the idea of mechanical proofreading and wrote the manuscript.
Funding
This work was supported through NIGMS R01 GM124472 (KS), NSF 1350829 (KS), NSF GRFP 1444932 (JB), and NCI-F99CA234959 (JB). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institute of Health or of the National Science Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Victor Pui-Yan Ma for comments on the manuscript and for helpful discussion.
References
1. van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. (2011) 11:47–55. doi: 10.1038/nri2887
2. Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. (2014) 15:798–807. doi: 10.1038/ni.2940
3. Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci USA. (1994) 91:12862–6.
4. Sasada T, Ghendler Y, Wang JH, Reinherz EL. Thymic selection is influenced by subtle structural variation involving the p4 residue of an MHC class I-bound peptide. Eur J Immunol. (2000) 30:1281–9. doi: 10.1002/(SICI)1521-4141(200005)30:5<1281::AID-IMMU1281>3.0.CO;2-D
5. Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity (1996) 4:565–71.
6. Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature (2002) 419:845–9. doi: 10.1038/nature01076
7. Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology (2009) 126:165–76. doi: 10.1111/j.1365-2567.2008.03015.x
8. Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature (2010) 464:932–6. doi: 10.1038/Nature08944
9. Lyons DS, Lieberman SA, Hampl J, Boniface JJ, Chien Y, Berg LJ, et al. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity (1996) 5:53–61.
10. Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. (2006) 24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658
11. Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol (2009) 27:591–619. doi: 10.1146/annurev.immunol.021908.132706
12. Boniface JJ, Rabinowitz JD, Wülfing C, Hampl J, Reich Z, Altman JD, et al. Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands [corrected]. Immunity (1998) 9:459–66
13. Cochran JR, Cameron TO, Stern LJ. The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity (2000) 12:241–50. doi: 10.1016/S1074-7613(00)80177-6
14. Watts TH, McConnell HM. High-affinity fluorescent peptide binding to I-Ad in lipid membranes. Proc Natl Acad Sci USA. (1986) 83:9660–4.
15. Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. (2009) 284:31028–37. doi: 10.1074/jbc.M109.052712
16. Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Methods (2012) 9:64–7. doi: 10.1038/Nmeth.1747
17. Jurchenko C, Salaita KS. Lighting up the force: investigating mechanisms of mechanotransduction using fluorescent tension probes. Mol Cell Biol. (2015) 35:2570–82. doi: 10.1128/MCB.00195-15
18. Liu Y, Galior K, Ma VP, Salaita K. Molecular tension probes for imaging forces at the cell surface. Acc Chem Res. (2017) 50:2915–24. doi: 10.1021/acs.accounts.7b00305
19. Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. (2014) 5:5167. doi: 10.1038/ncomms6167
20. Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA. (2016) 113:5610–5. doi: 10.1073/pnas.1600163113
21. Feng Y, Reinherz EL, Lang MJ. αβ T Cell Receptor Mechanosensing Forces out Serial Engagement. Trends Immunol. (2018) 39:596–609. doi: 10.1016/j.it.2018.05.005
22. Ma VP, Liu Y, Blanchfield L, Su H, Evavold BD, Salaita K. Ratiometric tension probes for mapping receptor forces and clustering at intermembrane junctions. Nano Lett. (2016) 16:4552–9. doi: 10.1021/acs.nanolett.6b01817
23. Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci USA. (2014) 111:2241–6. doi: 10.1073/pnas.1315606111
24. Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol Biol Cell (2015) 26:685–95. doi: 10.1091/mbc.E14-03-0830
25. Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell (2014) 157:357–68. doi: 10.1016/j.cell.2014.02.053
26. Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA. (2015) 112:1517–22. doi: 10.1073/pnas.1424829112
27. Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ. Mechanosensing drives acuity of alphabeta T-cell recognition. Proc Natl Acad Sci USA. (2017) 114:E8204–13. doi: 10.1073/pnas.1703559114
28. Basu R, Whitlock BM, Husson J, Le Floc'h A, Jin W, Oyler-Yaniv A, et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell (2016) 165:100–110. doi: 10.1016/j.cell.2016.01.021
29. Huse M. Mechanical forces in the immune system. Nat Rev Immunol. (2017) 17:679–90. doi: 10.1038/nri.2017.74
30. Chen W, Zhu C. Mechanical regulation of T-cell functions. Immunol Rev. (2013) 256:160–76. doi: 10.1111/imr.12122
31. Chen Y, Ju L, Rushdi M, Ge C, Zhu C. Receptor-mediated cell mechanosensing. Mol Biol Cell (2017) 28:3134–55. doi: 10.1091/mbc.E17-04-0228
33. Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell (2012) 151:1513–27. doi: 10.1016/j.cell.2012.11.034
34. Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. (1988) 234:55–83.
35. Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature (2003) 423:190–3. doi: 10.1038/nature01605
36. Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. (1974) 71:4135–9.
37. McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. (1995) 92:5042–46.
38. Valitutti S, Müller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature (1995) 375:148–51. doi: 10.1038/375148a0
39. Valitutti S. The serial engagement model 17 years after: from TCR triggering to immunotherapy. Front Immunol. (2012) 3:272. doi: 10.3389/fimmu.2012.00272
40. Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science (2013) 340:1587–90. doi: 10.1126/science.1237572
41. Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. (2010) 285:35967–78. doi: 10.1074/jbc.M110.155770
42. Yago T, Lou J, Wu T, Yang J, Miner JJ, Coburn L, et al. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. (2008) 118:3195–207. doi: 10.1172/JCI35754
43. Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci USA. (2012) 109:18815–20. doi: 10.1073/pnas.1208349109
44. Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell (2018) 174:672.e27–87. doi: 10.1016/j.cell.2018.06.017
45. Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C. Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J Immunol. (2014) 193:68–76. doi: 10.4049/jimmunol.1303436
46. Comrie WA, Li S, Boyle S, Burkhardt JK. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J Cell Biol. (2015) 208:457–73. doi: 10.1083/jcb.201406120
47. Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, et al. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. (2008) 283:11596–605. doi: 10.1074/jbc.M707815200
48. Tolar P. Cytoskeletal control of B cell responses to antigens. Nat Rev Immunol. (2017) 17:621–34. doi: 10.1038/nri.2017.67
49. Wang J, Lin F, Wan Z, Sun X, Lu Y, Huang J, et al. Profiling the origin, dynamics, and function of traction force in B cell activation. Sci Signal. (2018) 11:eaai9192. doi: 10.1126/scisignal.aai9192
50. Spillane KM, Tolar P. B cell antigen extraction is regulated by physical properties of antigen-presenting cells. J Cell Biol. (2017) 216:217–30. doi: 10.1083/jcb.201607064
51. Pageon SV, Tabarin T, Yamamoto Y, Ma Y, Nicovich PR, Bridgeman JS, et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc Natl Acad Sci USA. (2016) 113:E5454–63. doi: 10.1073/pnas.1607436113
52. Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci USA. (2016) 113:E5916–24. doi: 10.1073/pnas.1605399113
53. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science (2017) 356:eaal3118. doi: 10.1126/science.aal3118
54. Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, et al. Force loading explains spatial sensing of ligands by cells. Nature (2017) 552:219–24. doi: 10.1038/nature24662
55. Li J, Springer TA. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc Natl Acad Sci USA. (2017) 114:4685–90. doi: 10.1073/pnas.1704171114
56. Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol. (2012) 199:497–512. doi: 10.1083/jcb.201201091
57. Li J, Su Y, Xia W, Qin Y, Humphries MJ, Vestweber D, et al. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. (2017) 36:629–45. doi: 10.15252/embj.201695803
58. Hogg N, Patzak I, Willenbrock F. The insider's guide to leukocyte integrin signalling and function. Nat Rev Immunol. (2011) 11:416–26. doi: 10.1038/nri2986
59. Feigelson SW, Pasvolsky R, Cemerski S, Shulman Z, Grabovsky V, Ilani T, et al. Occupancy of lymphocyte LFA-1 by surface-immobilized ICAM-1 is critical for TCR- but not for chemokine-triggered LFA-1 conversion to an open headpiece high-affinity state. J Immunol. (2010) 185:7394–404. doi: 10.4049/jimmunol.1002246
60. Comrie WA, Babich A, Burkhardt JK. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J Cell Biol. (2015) 208:475–91. doi: 10.1083/jcb.201406121
61. Moore TI, Aaron J, Chew TL, Springer TA. Measuring integrin conformational change on the cell surface with super-resolution microscopy. Cell Rep. (2018) 22:1903–12. doi: 10.1016/j.celrep.2018.01.062
62. Gross CC, Brzostowski JA, Liu D, Long EO. Tethering of intercellular adhesion molecule on target cells is required for LFA-1-dependent NK cell adhesion and granule polarization. J Immunol. (2010) 185:2918–26. doi: 10.4049/jimmunol.1000761
63. Zhang Y, Qiu Y, Blanchard AT, Chang Y, Brockman JM, Ma VP, et al. Platelet integrins exhitib anisotropic mechanosensing and harness pN forces to mediate platelet aggregation. Proc Natl Acad Sci USA. (2017) 115:325–30. doi: 10.1073/pnas.1710828115
64. Brockman JM, Blanchard AT, Pui-Yan V, Derricotte WD, Zhang Y, Fay ME, et al. Mapping the 3D orientation of piconewton integrin traction forces. Nat Methods (2018) 15:115–8. doi: 10.1038/nmeth.4536
65. Zhang W, Deng W, Zhou L, Xu Y, Yang W, Liang X, et al. Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood (2015) 125:562–9. doi: 10.1182/blood-2014-07-589507
66. Cheng Z, Huang M, Chen G, Yang G, Zhou X, Chen C, et al. Cell-based assays in combination with ultra-high performance liquid chromatography-quadrupole time of flight tandem mass spectrometry for screening bioactive capilliposide C metabolites generated by rat intestinal microflora. J Pharm Biomed Anal. (2016) 119:130–8. doi: 10.1016/j.jpba.2015.11.029
67. Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. (2016) 18:540–8. doi: 10.1038/ncb3336
68. Hong J, Ge C, Jothikumar P, Yuan Z, Liu B, Bai K, et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat Immunol. (2018) 19:1379–90. doi: 10.1038/s41590-018-0259-z
69. Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. (2012) 102:L5–7. doi: 10.1016/j.bpj.2011.12.011
70. Bufi N, Saitakis M, Dogniaux S, Buschinger O, Bohineust A, Richert A, et al. Human Primary Immune Cells Exhibit Distinct Mechanical Properties that Are Modified by Inflammation. Biophys J. (2015) 108:2181–90. doi: 10.1016/j.bpj.2015.03.047
71. Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity (2011) 35:681–93. doi: 10.1016/j.immuni.2011.09.013
72. Lambert LH, Goebrecht GK, De Leo SE, O'Connor RS, Nunez-Cruz S, Li TD, et al. Improving T cell expansion with a soft touch. Nano Lett. (2017) 17:821–6. doi: 10.1021/acs.nanolett.6b04071
73. Chang ZL, Lorenzini MH, Chen X, Tran U, Bangayan NJ, Chen YY. Rewiring T-cell responses to soluble factors with chimeric antigen receptors. Nat Chem Biol. (2018) 14:317–24. doi: 10.1038/nchembio.2565
74. Schwesinger F, Ros R, Strunz T, Anselmetti D, Güntherodt HJ, Honegger A, et al. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc Natl Acad Sci USA. (2000) 97:9972–7. doi: 10.1073/pnas.97.18.9972
75. Deng W, Xu Y, Chen W, Paul DS, Syed AK, Dragovich MA, et al. Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat Commun. (2016) 7:12863. doi: 10.1038/ncomms12863
Keywords: mechanical proofreading, molecular forces, mechanobiology, immune recognition, T-cell activation, mechanotransduction
Citation: Brockman JM and Salaita K (2019) Mechanical Proofreading: A General Mechanism to Enhance the Fidelity of Information Transfer Between Cells. Front. Phys. 7:14. doi: 10.3389/fphy.2019.00014
Received: 08 November 2018; Accepted: 23 January 2019;
Published: 19 February 2019.
Edited by:
Jorge Bernardino De La Serna, United Kingdom Research and Innovation, United KingdomReviewed by:
Pavel Tolar, Francis Crick Institute, United KingdomLance Kam, Columbia University, United States
Copyright © 2019 Brockman and Salaita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khalid Salaita, ay5zYWxhaXRhQGVtb3J5LmVkdQ==
 Joshua M. Brockman
Joshua M. Brockman Khalid Salaita
Khalid Salaita