- 1Halo Research Group, Queen’s University Belfast, Belfast, United Kingdom
- 2Centre for Experimental Medicine, School of Medicine, Dentistry and Biomedical Sciences, Queen’s University Belfast, Belfast, United Kingdom
- 3School of Pharmacy, Queen’s University Belfast, Belfast, United Kingdom
The advent of high-throughput multi-omics technologies has underpinned the expansion in lung microbiome research, increasing our understanding of the nature, complexity and significance of the polymicrobial communities harbored by people with CF (PWCF). Having established that structurally complex microbial communities exist within the airways, the focus of recent research has now widened to investigating the function and dynamics of the resident microbiota during disease as well as in health. With further refinement, multi-omics approaches present the opportunity to untangle the complex interplay between microbe–microbe and microbe–host interactions in the lung and the relationship with respiratory disease progression, offering invaluable opportunities to discover new therapeutic approaches for our management of airway infection in CF.
Introduction
In recent years, the growing and widespread application of genomics focused, culture-independent techniques for microbiological analysis has been universally acknowledged as revolutionary for the research and management of disease (Malla et al., 2019). As the field of genomics has matured, we have learned that healthy lungs are inhabited by a diverse and complex array of airborne particles and microbial life (Hilty et al., 2010; Dickson and Huffnagle, 2015) . Moreover, we have realized the extent and significance of the microbiome and its role in respiratory health and disease (Nguyen et al., 2015; Barcik et al., 2020).
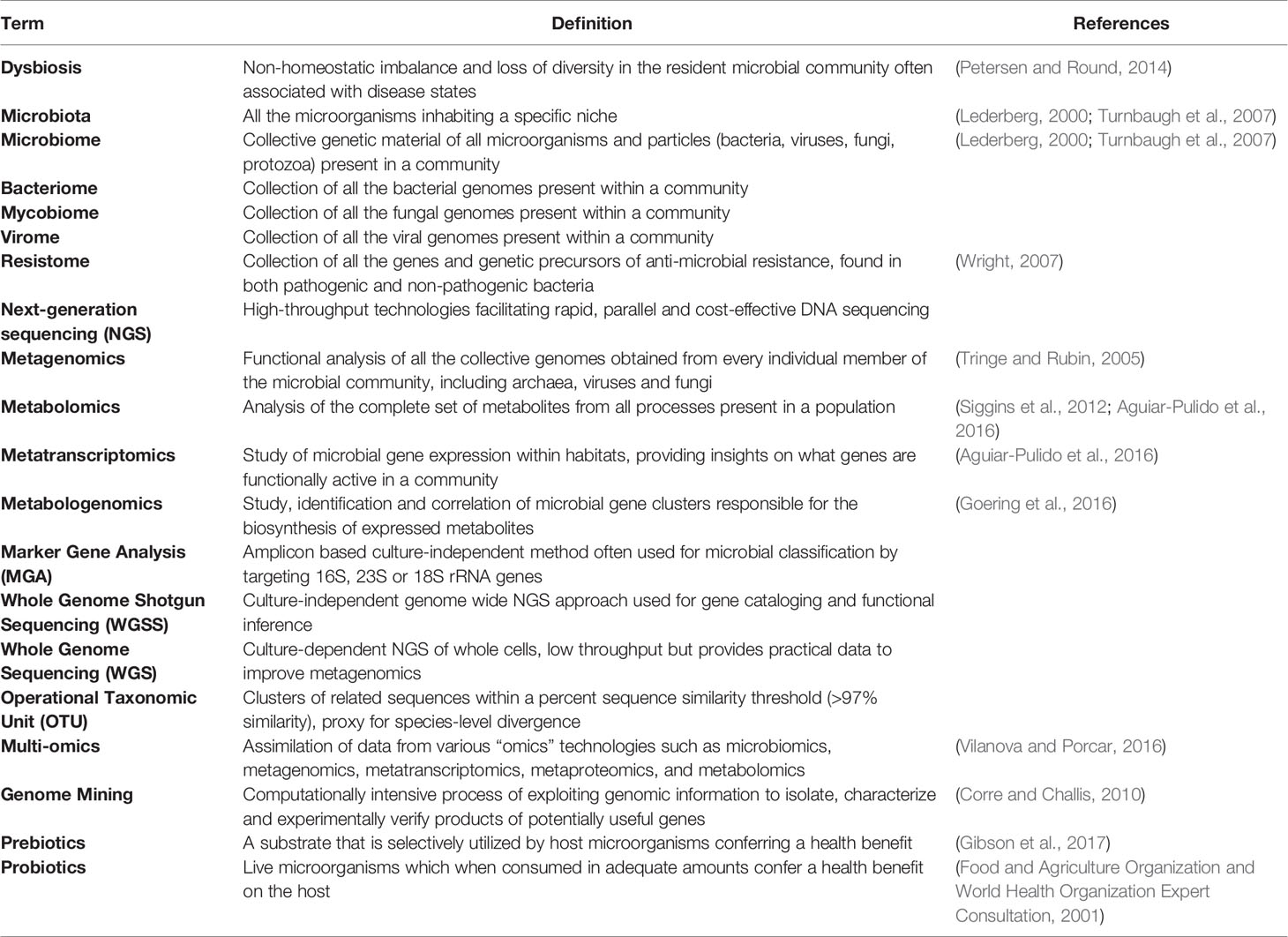
Technological advances have enabled cost-efficient, high-throughput, in-depth analysis of transcripts, proteins, and metabolites (e.g. metatranscriptomics, metaproteomics, metabolomics, and metabologenomics) exponentially increasing our understanding of our own microbiota, the vital role it plays in maintaining our health and offering opportunities to understand its capacity to contribute to disease (Table 1).
Collectively the respiratory microbiome consists of the upper (nasal and oral passages) and lower (lungs) airways. In those without respiratory disease, specific differences in the composition and load of the microbiota between these connected mucosal sites are apparent (Table 2). No firm consensus exists regarding the bacterial composition of a “typical healthy” lung; however it is now accepted that the lower airways harbor a diverse and dynamic ecosystem inhabited by a range of facultatively and obligately aerobic and anaerobic microorganisms with considerably greater inter versus intrasubject variation in composition (Erb-Downward et al., 2011; Invernizzi et al., 2020).
The Lung Microbiota During Disease
Personal health is linked to the presence of a broad and diverse microbiota, and this varies widely from individual to individual. Evidence thus far suggests that any disorder in the balance of the communities present, which ultimately leads to a loss of microbial diversity (dysbiosis) could potentially act as a catalyst for the development of illness (Malla et al., 2019). In CF, Achromobacter, Burkholderia, Pseudomonas, Staphylococcus, Stenotrophomonas, and Streptococcus spp. in the airways of PWCF has been described (Fodor et al., 2012; Feigelman et al., 2017; Frayman et al., 2017; Einarsson et al., 2019a). The presence of obligate anaerobes such as Prevotella and Veillonella in the CF lung has also been noted (Tunney et al., 2008; Field et al., 2010; Tunney et al., 2011) and such diverse taxa can potentially negatively or positively impact respiratory health depending on the species present (O’Neill et al., 2015; Sherrard et al., 2016). Moreover, it is worth noting that the prevalence of non-tuberculosus mycobacterial (NTM) respiratory infection in CF has been increasing in recent years, with those commonly isolated belonging to either Mycobacterium avium complex (MAC) or Mycobacterium abscessus group (MABS) (Salsgiver et al., 2016; Adjemian et al., 2018; Martiniano et al., 2019).
Microbial Dysbiosis and the Gut–Lung Axis
Numerous studies have shown the importance of the gut microbiome in metabolic function (O’Hara and Shanahan, 2006), immune function (Cho and Blaser, 2012), pathogen resistance (Kamada et al., 2013) and chronic inflammation (Lobionda et al., 2019; Tilg et al., 2020). Of particular interest is the proven links between gut microbiome health, intestinal dysbiosis and inflammation in people with CF (Rogers et al., 2010; Li and Somerset, 2014; Dhaliwal et al., 2015; Flass et al., 2015; Nielsen et al., 2016). This identified crosstalk occurring between the intestinal microbiota and the lungs has been termed the gut–lung axis, and there is growing interest in understanding how intestinal dysbiosis could potentially impact the progression and severity of airway disease in CF (Figure 1) (Marsland et al., 2015).
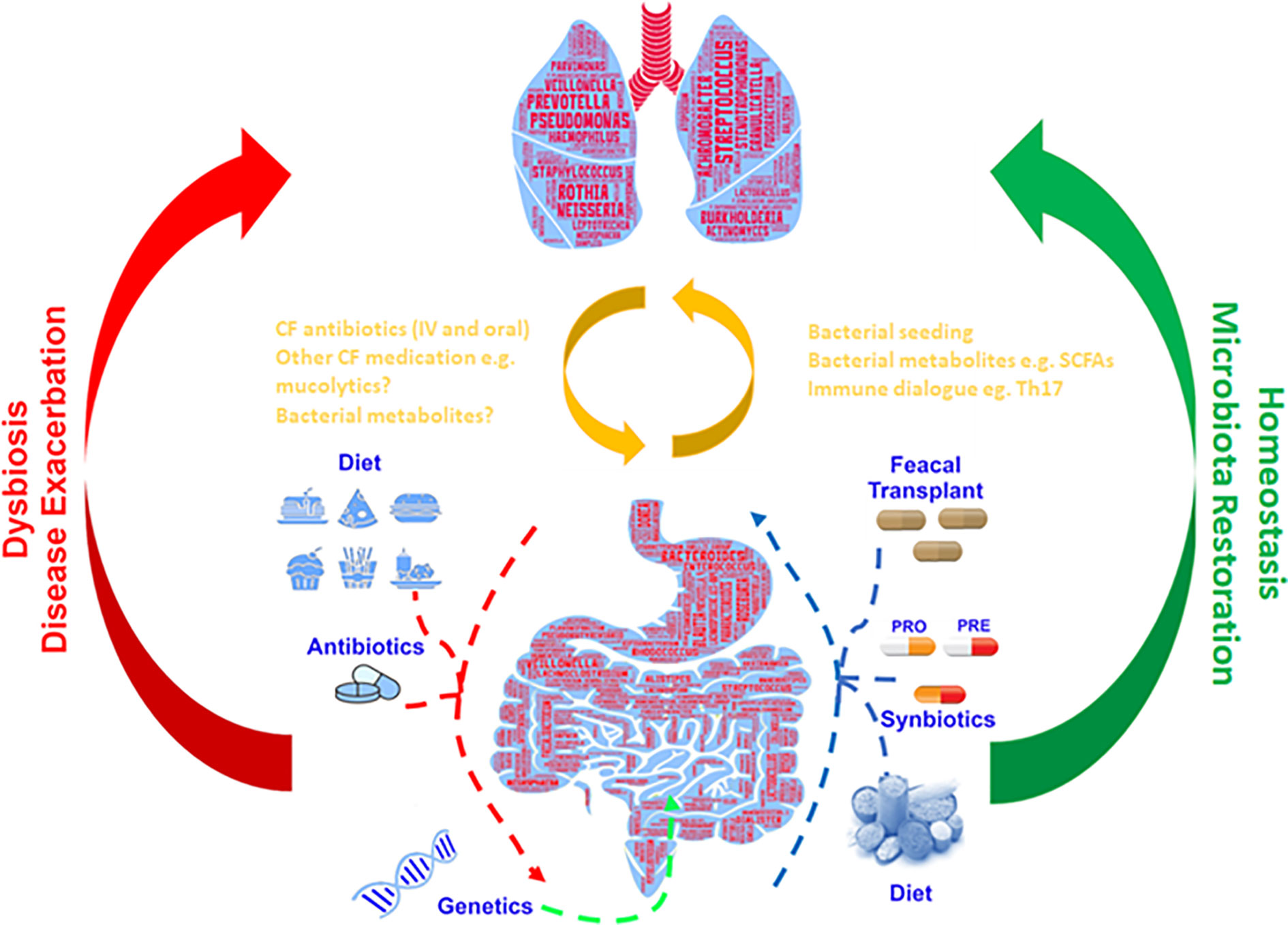
Figure 1 Gut–lung axis: Schematic depiction of the bidirectional cross-talk between the gut and lung which helps maintain microbial homeostasis. Respiratory disease, genetics and additional external factors can modulate the balance of this axis leading to the formation of a dysbiotic state.
The neonatal period has been established as a critical stage in microbiota development and immune system maturation (Pattaroni et al., 2018). CFTR dysfunction has been shown to present as gut complications from birth that affect infant development, negatively impact the establishment of a beneficial gut and airway microbiota and consequently continue throughout adulthood (Bronstein et al., 1992; Li and Somerset, 2014; Leung et al., 2015; Boutin and Dalpke, 2017; Burke et al., 2017). Intestinal dysbiosis and inflammation observed from birth (Munck, 2014; Hoen et al., 2015) results in a decrease in beneficial gut commensals that could adversely affect the airway microbiota (Scanlan et al., 2012).
Evidence of the crosstalk which occurs between the intestinal and airway microbiota has been strengthened considerably by the discovery that both the gut (Ríos-Covián et al., 2016) and lungs (Mirković et al., 2015) are dominated by bacteria producing short-chain fatty acids (SCFA) and that those found in the lung are likely to originate from the gut. The mesenteric lymphatic system connects the gut with the lungs and it is possible this is the route by which bacteria or their metabolites could “seed” the airways; this could result in modulation of the immune response, as SCFAs have been shown to promote anti-inflammatory mechanisms and maintain intestinal homeostasis (Peng et al., 2009; Invernizzi et al., 2020). It is therefore conceivable that methods to combat microbial dysbiosis in one community could positively influence the other (Budden et al., 2017).
Targeted investigation and quantification of metabolites could provide better understanding of how the metabolic activity of the lung and gut microbiota affects respiratory health. Metabolomics offers the opportunity to validate and identify those metabolites and pathways of microbial origin, discern their interaction with the host and any potential role in inflammation and immune system development (Lee-Sarwar et al., 2020). Furthermore, the establishment of distinct microbiome and metabolome signatures offers the opportunity to advance a more personalized approach to patient diagnosis and treatment; for example, metabolomic profiles of exhaled breath condensate (EBC) have been considered as a potential prognostic biomarker for individuals with chronic respiratory disorders (Maniscalco et al., 2019) and for the early detection of pulmonary exacerbations in PWCF (Zang et al., 2017).
Microbiota Diversity as an Indicator of Lung Inflammation
Inflammation of the airways is a common biological response to damage or infection and though largely beneficial it can play a key role in influencing the composition of the airway microbiome (Huffnagle et al., 2017). In the CF airways, increased secretion of proinflammatory mediators (e.g. IL-6 and IL-8) by potentially defective airway epithelium and immune cells leads to the disproportionate influx of neutrophils. Subsequent release of proteases, such as neutrophil elastase, damage structural lung proteins leading to an inevitable decline in pulmonary function.
Thus, the development of a range of microbiome-based molecular biomarkers would be a useful tool to monitor the onset or progression of airway disease, giving clinicians advanced warning of changing conditions and guiding therapeutic interventions. Rapid PCR diagnostics could be employed to determine the presence/absence of specific indicator organisms, such as known pathogens or established beneficial commensals, with a change in the load of either predicting the onset of deteriorating conditions (Harun et al., 2011; Boutin et al., 2018; Taylor et al., 2019). Pathogenic Proteobacteria possess potent inflammation enhancing pathogen-associated molecular patterns, such as lipopolysaccharides, and their association with inflammatory disorders is well known (Larsen et al., 2015; Rizzatti et al., 2017). For this reason, it has been suggested that this phylum could be used as an indicator of disease or marker of microbiota instability (Shin et al., 2015; Litvak et al., 2017). Likewise, messenger molecules involved in the host inflammatory response could also form the basis for further PCR targets (Eckrich et al., 2017).
As genomics technologies continue to advance, it is conceivable that monitoring of airway bacterial communities will become routine and so by utilizing either MGA or WGSS, the host’s respiratory microbiota could potentially function as a marker (Acosta et al., 2018; Sherrard and Bell, 2018). Comparing it to an idealized healthy benchmark may give advanced warning of unfavorable alterations in the composition of the microbiota, or potential “blooms” of pathogens, allowing for earlier intervention, especially if the patient is harboring slow growing pathogens or strains resistant to standard clinical culture (Harris et al., 2007; Woo et al., 2008). Recently Keravec et al. (2019) used a 16S MGA approach to identify prognostic biomarkers of P. aeruginosa infection in patients with CF and proposed representatives of the genus Porphyromonas as possible predictive markers.
Microbiota Directed Therapeutic Approaches
If correctly exploited, the information generated by multi-omics investigations of the complete airway biome offers the potential to develop novel methods for the treatment of CF. Microbiome-based interventions offer greater scope for controlling health outcomes and although this field is in its infancy, options on how to leverage this untapped resource are discussed below.
Antibiotic Regimen Optimization
One of the first studies to utilize culture-independent microbiota data in the selection of antimicrobial therapy for the treatment of pulmonary exacerbations in PWCF is the “Cystic Fibrosis Microbiome-determined Antibiotic Therapy Trial in Exacerbations: Results Stratified (CFMATTERS) Study” (Einarsson et al., 2017; Einarsson et al., 2019b). This study was designed to determine targeted antibacterial therapy compared to standard empirical therapy in the treatment of pulmonary exacerbations. Antibiotic treatment was selected based on the bacterial composition of a patient’s microbiota determined via NGS. Patients were randomized to either the control group where they received empirical antimicrobial therapy (ceftazidime and tobramycin/aztreonam) or the microbiome-directed treatment group where they received empirical therapy plus an additional antibiotic based on the top four most abundant taxa determined via NGS of a baseline sputum sample. However, there was no significant difference in the primary outcome, percentage change in recovery (post-exacerbation) FEV1 relative to the previous pre-exacerbation FEV1, between groups. Furthermore, in both the microbiome directed and empirical treatment arms, community composition appeared relatively stable over time, despite often showing a major disruption in community composition during the period of active antibiotic treatment. This highlights the need to further understand how bacterial community composition is affected in chronic respiratory diseases, as well as how a disruption or longer stability in the community composition impacts inflammatory processes within the airways of chronically infected PWCF.
Enhanced Culturomics
Culture-independent metagenomics methods have proven expedient for generating vast quantities of data and accelerating our understanding of microbiomes. This has led to suggestions they could supplant traditional time-consuming culture techniques despite evidence that bacterial cultivability is much higher than previously thought (Kaeberlein et al., 2002). However, these methods can be disadvantaged by imperfect data analysis or experimental design (Bilen et al., 2018). Culturomics promises to bridge gaps in our knowledge by identifying unassigned sequences and uncovering new bacterial strains missed or overlooked by NGS (Lagier et al., 2012; Browne et al., 2016; Lagier et al., 2016) and provide more comprehensive data than metagenomics can alone (Dubourg et al., 2013). Using diverse and large-scale culture conditions, often complimented with MALDI-TOF and metagenomics, it has been possible to isolate and sequence previously uncultivable strains providing functional data to improve metagenomics. To more fully explore host-microbe interactions, complete the identification of microbiomes and test possible therapies, culturing pure isolates is still desirable (Lagier et al., 2015). Recent investigation of the CF lung by these methods demonstrated that not only could the majority of OTUs identified by amplicon sequencing be cultured but many more OTUs were described by culturomics than direct sequencing (Whelan et al., 2020).
Pre-, Pro- and Syn-Biotic Therapies
The use of nutritional supplements containing substrates that stimulate the growth of beneficial microbes, live microorganisms, or combinations of both has been considered for some time as a treatment for intestinal dysbiosis (Vyas and Ranganathan, 2012; Coffey et al., 2018). The implication that microbiomes in anatomically distinct and distal sites, such as the gut and lungs, can communicate with each other presents an opportunity for direct and indirect targeted treatment of airway disease. Once such approach would be modulation of the intestinal microbiota in ways we know influence the airways beneficially, such as supporting the growth of gut commensals identified as indirectly enhancing alveolar macrophages and lung immune function (Clarke, 2014; Trompette et al., 2014; Martin et al., 2015). Moreover, directly transplanting bacteria into the lungs that increase diversity or seeding the airways with abiotic growth factors may improve respiratory health. Such therapies could possibly be used in addition to conventional therapy for treatment of chronic lung inflammation. However, they would require a high degree of personalization to ensure the combination of bacteria used produces the intended results and does not further exacerbate inflammation, a potential consequence if the bacterial constituents of the treatment are not normal members of the patients microbiota. Although limited in nature, human and murine investigations using prebiotics have shown promising effects on partially correcting intestinal dysbiosis and positively impacting colitis and ulcerative colitis symptoms (Hanai et al., 2004; Casellas et al., 2007; Koleva et al., 2012). Probiotic studies are much more varied and have focused principally on supplementation with Lactobacillus and Bifidobacteria spp. Symptom severity and improved lung function in children suffering from asthma was noted following administration of capsules containing Lactobacillus gasseri A5 (Chen et al., 2010) or a combination of Lactobacillus acidophilus, Bifidobacterium bifidum and Lactobacillus delbrueckii subsp. Bulgaricus (Gutkowski et al., 2010). However, after initially promising observations showing restoration of gut microbiota in children with CF when treated orally with Lactobacillus GG (Bruzzese et al., 2014), further investigation by the same group showed no significant clinical effect on respiratory outcomes (Bruzzese et al., 2018). The best known naturally occurring synbiotic is human breast milk (Martín et al., 2005) and the link between breastfeeding and lower incidence of asthma and associated airway inflammation is well documented (Oddy et al., 2002; Corey et al., 2019). Collectively, the use of prebiotic, probiotic or synbiotics to potentially suppress identified airway pathogens or reinstate beneficial taxa that have been eliminated during disease is promising. However substantial additional data, including in vivo studies, is required before it can be conclusively proven they positively impact the progression of CF chronic airway inflammation.
Microbiome Derived Biomolecules
Genome mining of data acquired by WGSS can identify and characterize the biosynthetic gene clusters (BGCs) present within microbiomes that encode for bacterial natural products (BNPs), presenting an opportunity to discover unique biomolecules that could deliver new treatments for reducing inflammation and combating pathogen colonization (Vakhlu et al., 2008; Bachmann et al., 2014; Cheng et al., 2019). Multi-omics approaches can be used to analyze enzymes encoded by these gene clusters and any resulting products identified experimentally (Ziemert et al., 2016). This shift in focus from using culture-independent techniques to purely describe what constitutes a microbiota to what metabolic functions the microbiota demonstrate has revealed the vast extent of BGCs present and the drug-like products they encode for (Donia et al., 2014).
Given the complex communities and competition for resources that occurs within microbiomes, an extensive range of BNPs that exhibit narrow spectrum anti-microbial effects have been identified (Arnison et al., 2013). An opportunity to identify and develop microbiome-sourced antimicrobial compounds is an exciting possibility for the treatment of respiratory diseases. The use of natural products as therapies could reduce the use of broad-spectrum antibiotics and avoid associated complications such as dysbiosis and increased AMR. Lantibiotics have been found in both commensals such as Staphylococcus epidermidis (Velásquez et al., 2011) and human pathogens like Enterococcus faecalis (Sawa et al., 2012). Microcins are potent antibacterials derived exclusively from both commensal and pathogenic enterobacteria (Duquesne et al., 2007). TOMMs (thiazole/oxazole modified microcins) are related to microcins and display prolific functional diversity with over 300 TOMM BGCs identified to date (Melby et al., 2011). Bacterial pathways that exert anti-inflammatory responses could be inferred from gene mining of shotgun sequencing data. From this, BNPs that show beneficial immunomodulatory activity, could be identified and form the basis of new treatment opportunities.
Modulate Microbiome Interactions
Deciphering the biochemical mechanisms between host cells and the microbiota offer novel avenues for therapy (Holmes et al., 2012) as it is now understood that the biotransformation of xenobiotics is commonly affected by the genetics of the host microbiome (Clayton et al., 2009; Wikoff et al., 2009) and they in turn can influence bacterial signaling and stress response pathways within the system (Maurice et al., 2013).
Metatransciptomics (Shakya et al., 2019) could be employed to examine the chemical roles bacterial members perform within the microbiome. This could provide specific information on bacterial metabolite usage, enzyme induction, regulation of inflammatory markers, secondary metabolism, the efficacy and unintended consequences of administered drugs and may reveal components of the microbiota that could be disrupted or selectively “drugged” (Wallace and Redinbo, 2013). Synthetic systems models are now being developed to study host-microbe interactions (Elzinga et al., 2019), an important step due to the growing appreciation that the gut microbiota and its metabolites can influence and modulate host immune function (Li and Somerset, 2018) and that these metabolites are present in distal organs such as the lungs (Schroeder and Bäckhed, 2016).
Synthetic Bio-Delivery
An emerging field of therapeutic research is the use of synthetic engineering and the native microbiome itself for drug delivery and modulation of disease via either genetically modified bacteria (Nicaise et al., 2008; Mimee et al., 2016; Singhvi et al., 2018), vesicles (Chellappan et al., 2019) or phages (Nobrega et al., 2015). The latter has already shown promising results for the treatment of drug-resistant M. abscessus and P. aeruginosa infections in CF (Alemayehu et al., 2012; Dedrick et al., 2019; Law et al., 2019) and against dual-species biofilms formed by both P. aeruginosa and S. aureus (Tkhilaishvili et al., 2020). Deacon et al. (2015) enhanced the effectiveness of nanoparticle encapsulated tobramycin through DNase functionalization, improving CF sputum penetration. Understanding of the microbiome using enhanced culture-dependent and independent methods will be key to maximizing the advantages provided by synthetic engineering and its use as a therapy for respiratory disease.
Future Directions
With the transformative potential that NGS technologies and metagenomics approaches promise, it is tempting to consider them a sole successor to all other current forms of testing and employ them wholesale in all areas of medicine and public health. However, considerable refinement and development of these sequencing technologies, their necessary downstream analytical pipelines, and investment in bioinformatic support is still required before widespread adoption into routine practice. Further studies utilizing these approaches are necessary to improve our understanding of the nature and composition of the lung microbiome, address the mechanisms by which microbe and host interact and ultimately the impact the respiratory microbiome in CF has on health and disease. More information resulting from such investigations will prove instrumental in improving molecular diagnostics and developing novel therapeutic approaches for the treatment of lung disease.
Author Contributions
AL conceptualized and wrote the manuscript with support from GE. GE, DG, and MT provided consultation and critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
AL and GE are funded by the Innovative Medicines Initiative Joint Undertaking – European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies in kind contribution (no. # 115721-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acosta, N., Heirali, A., Somayaji, R., Surette, M. G., Workentine, M. L., Sibley, C. D., et al. (2018). Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax 73 (11), 1016–1025. doi: 10.1136/thoraxjnl-2018-211510
Adjemian, J., Olivier, K. N., Prevots, D. R. (2018). Epidemiology of Pulmonary Nontuberculous Mycobacterial Sputum Positivity in Patients with Cystic Fibrosis in the United States, 2010-2014. Ann. Am. Thorac. Soc. 15 (7), 817–826. doi: 10.1513/AnnalsATS.201709-727OC
Aguiar-Pulido, V., Huang, W., Suarez-Ulloa, V., Cickovski, T., Mathee, K., Narasimhan, G. (2016). Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. Online 12 (Suppl 1), 5–16. doi: 10.4137/EBO.S36436
Alemayehu, D., Casey, P. G., McAuliffe, O., Guinane, C. M., Martin, J. G., Shanahan, F., et al. (2012). Bacteriophages ϕMR299-2 and ϕNH-4 Can Eliminate Pseudomonas aeruginosa in the Murine Lung and on Cystic Fibrosis Lung Airway Cells. mBio 3 (2), e00029–e00012. doi: 10.1128/mBio.00029-12
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30 (1), 108–160. doi: 10.1039/C2NP20085F
Bachmann, B. O., Van Lanen, S. G., Baltz, R. H. (2014). Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biot. 41 (2), 175–184. doi: 10.1007/s10295-013-1389-9
Barcik, W., Boutin, R. C. T., Sokolowska, M., Finlay, B. B. (2020). The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 52 (2), 241–255. doi: 10.1016/j.immuni.2020.01.007
Bassis, C. M., Tang, A. L., Young, V. B., Pynnonen, M. A. (2014). The nasal cavity microbiota of healthy adults. Microbiome 2 (1), 27. doi: 10.1186/2049-2618-2-27
Bilen, M., Dufour, J.-C., Lagier, J.-C., Cadoret, F., Daoud, Z., Dubourg, G., et al. (2018). The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome 6 (1), 94. doi: 10.1186/s40168-018-0485-5
Boutin, S., Dalpke, A. H. (2017). Acquisition and adaptation of the airway microbiota in the early life of cystic fibrosis patients. Mol. Cell Pediatr. 4 (1), 1–1. doi: 10.1186/s40348-016-0067-1
Boutin, S., Weitnauer, M., Hassel, S., Graeber, S. Y., Stahl, M., Dittrich, A. S., et al. (2018). One-time quantitative PCR detection of Pseudomonas aeruginosa to discriminate intermittent from chronic infection in cystic fibrosis. J. Cyst. Fibros 17 (3), 348–355. doi: 10.1016/j.jcf.2017.12.007
Bronstein, M. N., Sokol, R. J., Abman, S. H., Chatfield, B. A., Hammond, K. B., Hambidge, K. M., et al. (1992). Pancreatic insufficiency, growth, and nutrition in infants identified by newborn screening as having cystic fibrosis. J. Pediatr. 120 (4, Part 1), 533–540. doi: 10.1016/S0022-3476(05)82478-3
Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., et al. (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533 (7604), 543–546. doi: 10.1038/nature17645
Bruzzese, E., Callegari, M. L., Raia, V., Viscovo, S., Scotto, R., Ferrari, S., et al. (2014). Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One 9 (2), e87796. doi: 10.1371/journal.pone.0087796
Bruzzese, E., Raia, V., Ruberto, E., Scotto, R., Giannattasio, A., Bruzzese, D., et al. (2018). Lack of efficacy of Lactobacillus GG in reducing pulmonary exacerbations and hospital admissions in children with cystic fibrosis: A randomised placebo controlled trial. J. Cyst. Fibros 17 (3), 375–382. doi: 10.1016/j.jcf.2017.10.014
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15 (1), 55–63. doi: 10.1038/nrmicro.2016.142
Burke, D. G., Fouhy, F., Harrison, M. J., Rea, M. C., Cotter, P. D., O’Sullivan, O., et al. (2017). The altered gut microbiota in adults with cystic fibrosis. BMC Microbiol. 17 (1), 58. doi: 10.1186/s12866-017-0968-8
Casellas, F., Borruel, N., Torrejón, A., Varela, E., Antolin, M., Guarner, F., et al. (2007). Oral oligofructose-enriched inulin supplementation in acute ulcerative colitis is well tolerated and associated with lowered faecal calprotectin. Aliment. Pharmacol. Ther. 25 (9), 1061–1067. doi: 10.1111/j.1365-2036.2007.03288.x
Charlson, E. S., Bittinger, K., Haas, A. R., Fitzgerald, A. S., Frank, I., Yadav, A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184 (8), 957–963. doi: 10.1164/rccm.201104-0655OC
Chellappan, D. K., Sze Ning, Q. L., Su Min, S. K., Bin, S. Y., Chern, P. J., Shi, T. P., et al. (2019). Interactions between microbiome and lungs: Paving new paths for microbiome based bio-engineered drug delivery systems in chronic respiratory diseases. Chem-Biol Interact. 310. (no pagination). doi: 10.1016/j.cbi.2019.108732
Chen, Y. S., Jan, R. L., Lin, Y. L., Chen, H. H., Wang, J. Y. (2010). Randomized placebo-controlled trial of lactobacillus on asthmatic children with allergic rhinitis. Pediatr. Pulmonol. 45 (11), 1111–1120. doi: 10.1002/ppul.21296
Cheng, M., Cao, L., Ning, K. (2019). Microbiome Big-Data Mining and Applications Using Single-Cell Technologies and Metagenomics Approaches Toward Precision Medicine. Front. Genet. 10, 109–118. doi: 10.3389/fgene.2019.00972
Cho, I., Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13 (4), 260–270. doi: 10.1038/nrg3182
Clarke, T. B. (2014). Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 82 (11), 4596–4606. doi: 10.1128/IAI.02212-14
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R., Nicholson, J. K. (2009). Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U. S. A. 106 (34), 14728–14733. doi: 10.1073/pnas.0904489106
Coffey, M. J., Garg, M., Homaira, N., Jaffe, A., Ooi, C. Y. (2018). Probiotics for people with cystic fibrosis. Cochrane Db Syst. Rev. 2, 1–13. doi: 10.1002/14651858.CD012949
Corey, S., Kvederis, L., Kingsbury, C., Bonsack, B., Sanberg, P. R., Castelli, V., et al. (2019). Gut microbiome: Lactation, childbirth, lung dysbiosis, animal modeling, stem cell treatment, and CNS disorders. CNS Neuro. Disord. - Drug Targets 18 (9), 687–694. doi: 10.2174/1871527318666191021145252
Corre, C., Challis, G. L. (2010). “2.12 - Exploiting Genomics for New Natural Product Discovery in Prokaryotes,” in Comp Nat Prod II. Eds. Liu, H.-W., Mander, L. (Oxford: Elsevier), 429–453.
de Steenhuijsen Piters, W. A. A., Sanders, E. A. M., Bogaert, D. (2015). The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 370 (1675), 20140294. doi: 10.1098/rstb.2014.0294
Deacon, J., Abdelghany, S. M., Quinn, D. J., Schmid, D., Megaw, J., Donnelly, R. F., et al. (2015). Antimicrobial efficacy of tobramycin polymeric nanoparticles for Pseudomonas aeruginosa infections in cystic fibrosis: formulation, characterisation and functionalisation with dornase alfa (DNase). J. Control Release 198, 55–61. doi: 10.1016/j.jconrel.2014.11.022
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Russell, D. A., Ford, K., Harris, K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25 (5), 730–733. doi: 10.1038/s41591-019-0437-z
Deo, P. N., Deshmukh, R. (2019). Oral microbiome: Unveiling the fundamentals. J. Oral. Maxillofacial Pathol. JOMFP 23 (1), 122–128. doi: 10.4103/jomfp.JOMFP_304_18
Dhaliwal, J., Leach, S. T., Katz, T., Nahidi, L., Pang, T., Lee, J. M., et al. (2015). Intestinal inflammation and impact on growth in children with cystic fibrosis. J. Pediatr. Gastr Nutr. 60 (4), 521–526. doi: 10.1097/MPG.0000000000000683
Dickson, R. P., Huffnagle, G. B. (2015). The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLoS Pathog. 11 (7), e1004923–e1004923. doi: 10.1371/journal.ppat.1004923
Dimitri-Pinheiro, S., Soares, R., Barata, P. (2020). The Microbiome of the Nose—Friend or Foe? Allergy Rh 11, 2152656720911605. doi: 10.1177/2152656720911605
Donia, M. S., Cimermancic, P., Schulze, C. J., Wieland Brown, L. C., Martin, J., Mitreva, M., et al. (2014). A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158 (6), 1402–1414. doi: 10.1016/j.cell.2014.08.032
Dubourg, G., Lagier, J. C., Armougom, F., Robert, C., Hamad, I., Brouqui, P., et al. (2013). The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 32 (5), 637–645. doi: 10.1007/s10096-012-1787-3
Duquesne, S., Petit, V., Peduzzi, J., Rebuffat, S. (2007). Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J. Mol. Microbiol. Biotechnol. 13 (4), 200–209. doi: 10.1159/000104748
Eckrich, J., Zissler, U. M., Serve, F., Leutz, P., Smaczny, C., Schmitt-Grohé, S., et al. (2017). Airway inflammation in mild cystic fibrosis. J. Cyst. Fibros 16 (1), 107–115. doi: 10.1016/j.jcf.2016.05.016
Einarsson, G. G., Comer, D. M., McIlreavey, L., Parkhill, J., Ennis, M., Tunney, M. M., et al. (2016). Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 71 (9), 795–803. doi: 10.1136/thoraxjnl-2015-207235
Einarsson, G., Flanagan, E., Lee, A., Elborn, J. S., Tunney, M., Plant, B. J. (2017). WS03. 1 Longitudinal airway microbiota profiling in cystic fibrosis patients enrolled in the CFMATTERS clinical trial. J. Cyst. Fibros 16, S4. doi: 10.1016/S1569-1993(17)30168-6
Einarsson, G. G., Zhao, J., LiPuma, J. J., Downey, D. G., Tunney, M. M., Elborn, J. S. (2019a). Community analysis and co-occurrence patterns in airway microbial communities during health and disease. ERJ Open Res. 5 (3), 00128–02017. doi: 10.1183/23120541.00128-2017
Einarsson, G. G., Flanagan, E., Lee, A. J., McGettigan, C., Smith, C., Gilpin, D. F., et al. (2019b). WS19-3 Microbiota profiling during 1-year of clinical stability in people with cystic fibrosis-CFMATTERS Consortium. J. Cyst. Fibros 18, S35. doi: 10.1016/S1569-1993(19)30230-9
Elzinga, J., van der Oost, J., de Vos, W. M., Smidt, H. (2019). The Use of Defined Microbial Communities To Model Host-Microbe Interactions in the Human Gut. Microbiol. Mol. Biol. R 83 (2), e00054–e00018. doi: 10.1128/MMBR.00054-18
Erb-Downward, J. R., Thompson, D. L., Han, M. K., Freeman, C. M., McCloskey, L., Schmidt, L. A., et al. (2011). Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One 6 (2), e16384–e16384. doi: 10.1371/journal.pone.0016384
Feigelman, R., Kahlert, C. R., Baty, F., Rassouli, F., Kleiner, R. L., Kohler, P., et al. (2017). Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome 5 (1), 20. doi: 10.1186/s40168-017-0234-1
Field, T. R., Sibley, C. D., Parkins, M. D., Rabin, H. R., Surette, M. G. (2010). The genus Prevotella in cystic fibrosis airways. Anaerobe 16 (4), 337–344. doi: 10.1016/j.anaerobe.2010.04.002
Flass, T., Tong, S., Frank, D. N., Wagner, B. D., Robertson, C. E., Kotter, C. V., et al. (2015). Intestinal lesions are associated with altered intestinal microbiome and are more frequent in children and young adults with cystic fibrosis and cirrhosis. PLoS One 10 (2), e0116967–e0116967. doi: 10.1371/journal.pone.0116967
Fodor, A. A., Klem, E. R., Gilpin, D. F., Elborn, J. S., Boucher, R. C., Tunney, M. M., et al. (2012). The Adult Cystic Fibrosis Airway Microbiota Is Stable over Time and Infection Type, and Highly Resilient to Antibiotic Treatment of Exacerbations. PLoS One 7 (9), e45001. doi: 10.1371/journal.pone.0045001
Food and Agriculture Organization and World Health Organization Expert Consultation (2001). Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria (Córdoba, Argentina: Food and Agriculture Organization of the United Nations and World Health Organization).
Frayman, K. B., Armstrong, D. S., Carzino, R., Ferkol, T. W., Grimwood, K., Storch, G. A., et al. (2017). The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 72 (12), 1104–1112. doi: 10.1136/thoraxjnl-2016-209279
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14 (8), 491–502. doi: 10.1038/nrgastro.2017.75
Goering, A. W., McClure, R. A., Doroghazi, J. R., Albright, J. C., Haverland, N. A., Zhang, Y., et al. (2016). Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. ACS Cent. Sci. 2 (2), 99–108. doi: 10.1021/acscentsci.5b00331
Gutkowski, P., Grek, M., Dmeńska, H., Syczewska, M., Michalkiewicz, J. (2010). Effect of orally administered probiotic strains Lactobacillus and Bifidobacterium in children with atopic asthma. Cent. Eur. J. Immunol. 35, 233–238.
Hanai, H., Kanauchi, O., Mitsuyama, K., Andoh, A., Takeuchi, K., Takayuki, I., et al. (2004). Germinated barley foodstuff prolongs remission in patients with ulcerative colitis. Int. J. Mol. Med. 13 (5), 643–647. doi: 10.3892/ijmm.13.5.643
Harris, J. K., De Groote, M. A., Sagel, S. D., Zemanick, E. T., Kapsner, R., Penvari, C., et al. (2007). Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. 104 (51), 20529–20533. doi: 10.1073/pnas.0709804104
Harun, A., Blyth, C. C., Gilgado, F., Middleton, P., Chen, S. C.-A., Meyer, W. (2011). Development and Validation of a Multiplex PCR for Detection of Scedosporium spp. in Respiratory Tract Specimens from Patients with Cystic Fibrosis. J. Clin. Microbiol. 49 (4), 1508–1512. doi: 10.1128/jcm.01810-10
Hilty, M. B. C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., Davies, J., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS One 5. doi: 10.1371/journal.pone.0008578
Hoen, A. G., Li, J., Moulton, L. A., O’Toole, G. A., Housman, M. L., Koestler, D. C., et al. (2015). Associations between Gut Microbial Colonization in Early Life and Respiratory Outcomes in Cystic Fibrosis. J. Pediatr. 167 (1), 138–147.e131-133. doi: 10.1016/j.jpeds.2015.02.049
Holmes, E., Li, J. V., Marchesi, J. R., Nicholson, J. K. (2012). Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 16 (5), 559–564. doi: 10.1016/j.cmet.2012.10.007
Huffnagle, G. B., Dickson, R. P., Lukacs, N. W. (2017). The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10 (2), 299–306. doi: 10.1038/mi.2016.108
Invernizzi, R., Lloyd, C. M., Molyneaux, P. L. (2020). Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology 160 (2), 171–182. doi: 10.1111/imm.13195
Kaeberlein, T., Lewis, K., Epstein, S. S. (2002). Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science 296 (5570), 1127–1129. doi: 10.1126/science.1070633
Kamada, N., Chen, G. Y., Inohara, N., Núñez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14 (7), 685–690. doi: 10.1038/ni.2608
Keravec, M., Mounier, J., Guilloux, C.-A., Fangous, M.-S., Mondot, S., Vallet, S., et al. (2019). Porphyromonas, a potential predictive biomarker of Pseudomonas aeruginosa pulmonary infection in cystic fibrosis. BMJ Open Respir. Res. 6 (1), e000374–e000374. doi: 10.1136/bmjresp-2018-000374
Koleva, P. T., Valcheva, R. S., Sun, X., Gänzle, M. G., Dieleman, L. A. (2012). Inulin and fructo-oligosaccharides have divergent effects on colitis and commensal microbiota in HLA-B27 transgenic rats. Brit J. Nutr. 108 (9), 1633–1643. doi: 10.1017/S0007114511007203
Kumpitsch, C., Koskinen, K., Schöpf, V., Moissl-Eichinger, C. (2019). The microbiome of the upper respiratory tract in health and disease. BMC Biol. 17 (1), 87. doi: 10.1186/s12915-019-0703-z
Lagier, J. C., Armougom, F., Million, M., Hugon, P., Pagnier, I., Robert, C., et al. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infec. 18 (12), 1185–1193. doi: 10.1111/1469-0691.12023
Lagier, J. C., Hugon, P., Khelaifia, S., Fournier, P. E., La Scola, B., Raoult, D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28 (1), 237–264. doi: 10.1128/CMR.00014-14
Lagier, J.-C., Khelaifia, S., Alou, M. T., Ndongo, S., Dione, N., Hugon, P., et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1 (12), 16203. doi: 10.1038/nmicrobiol.2016.203
Larsen, J. M., Musavian, H. S., Butt, T. M., Ingvorsen, C., Thysen, A. H., Brix, S. (2015). Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 144 (2), 333–342. doi: 10.1111/imm.12376
Law, N., Logan, C., Yung, G., Furr, C.-L. L., Lehman, S. M., Morales, S., et al. (2019). Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 47 (4), 665–668. doi: 10.1007/s15010-019-01319-0
Lederberg, J. (2000). Infectious History. Science 288 (5464), 287–293. doi: 10.1126/science.288.5464.287
Lee-Sarwar, K. A., Lasky-Su, J., Kelly, R. S., Litonjua, A. A., Weiss, S. T. (2020). Gut Microbial-Derived Metabolomics of Asthma. Metabolites 10 (3), 97. doi: 10.3390/metabo10030097
Lemon, K. P., Klepac-Ceraj, V., Schiffer, H. K., Brodie, E. L., Lynch, S. V., Kolter, R. (2010). Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1 (3), e00129–e00110. doi: 10.1128/mBio.00129-10
Leung, D. H., Borowitz, D., Heltshe, S. L., Kloster, M., Gelfond, D., Heubi, J. E., et al. (2015). CF infant growth and pulmonary status in the first year of life: The bonus study. Pediatr. Pulm. 50, 158–159. doi: 10.1002/ppul.23296
Li, L., Somerset, S. (2014). The clinical significance of the gut microbiota in cystic fibrosis and the potential for dietary therapies. Clin. Nutr. 33 (4), 571–580. doi: 10.1016/j.clnu.2014.04.004
Li, L., Somerset, S. (2018). Associations between Flavonoid Intakes and Gut Microbiota in a Group of Adults with Cystic Fibrosis. Nutrients 10 (9), 1264. doi: 10.3390/nu10091264
Litvak, Y., Byndloss, M. X., Tsolis, R. M., Bäumler, A. J. (2017). Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6. doi: 10.1016/j.mib.2017.07.003
Lobionda, S., Sittipo, P., Kwon, H. Y., Lee, Y. K. (2019). The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms 7 (8), 271–287. doi: 10.3390/microorganisms7080271
Malla, M. A., Dubey, A., Kumar, A., Yadav, S., Hashem, A., Abd-Allah, E. F. (2019). Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 9, 2868. doi: 10.3389/fimmu.2018.02868
Mammen, M. J., Sethi, S. (2016). COPD and the microbiome. Respirology 21 (4), 590–599. doi: 10.1111/resp.12732
Maniscalco, M., Fuschillo, S., Paris, D., Cutignano, A., Sanduzzi, A., Motta, A. (2019). “Chapter Five - Clinical metabolomics of exhaled breath condensate in chronic respiratory diseases,” in Advances in Clinical Chemistry, vol. 88 . Ed. Makowski, G. S. (Elsevier), 121–149.
Marsland, B. J., Trompette, A., Gollwitzer, E. S. (2015). The Gut–Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 12 (Supplement 2), S150–S156. doi: 10.1513/AnnalsATS.201503-133AW
Martín, R., Olivares, M., Marín, M. L., Fernández, L., Xaus, J., Rodríguez, J. M. (2005). Probiotic potential of 3 Lactobacilli strains isolated from breast milk. J. Hum. Lact. 21 (1), 8–17. doi: 10.1177/0890334404272393
Martin, C., Burgel, P.-R., Lepage, P., Andréjak, C., d. Blic, J., Bourdin, A., et al. (2015). Host–microbe interactions in distal airways: relevance to chronic airway diseases. Eur. Respir. Rev. 24 (135), 78–91. doi: 10.1183/09059180.00011614
Martiniano, S. L., Nick, J. A., Daley, C. L. (2019). Nontuberculous Mycobacterial Infections in Cystic Fibrosis. Thorac. Surg. Clin. 29 (1), 95–108. doi: 10.1016/j.thorsurg.2018.09.008
Maurice, C. F., Haiser, H. J., Turnbaugh, P. J. (2013). Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Cell 152 (1), 39–50. doi: 10.1016/j.cell.2012.10.052
McLean, J. S. (2014). Advancements toward a systems level understanding of the human oral microbiome. Front. Cell Infect. Mi 4 (98), 52–64. doi: 10.3389/fcimb.2014.00098
Melby, J., Nard, N., Mitchell, D. (2011). Thiazole/oxazole-modified microcins: Complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 15, 369–378. doi: 10.1016/j.cbpa.2011.02.027
Mendez, R., Bhattacharya, S. K., Banerjee, S. (2019). Lung inflammation and disease: A perspective on microbial homeostasis and metabolism. IUBMB Life 71 (2), 152–165. doi: 10.1002/iub.1969
Mimee, M., Citorik, R. J., Lu, T. K. (2016). Microbiome therapeutics — Advances and challenges. Adv. Drug Deliver Rev. 105, 44–54. doi: 10.1016/j.addr.2016.04.032
Mirković, B., Murray, M. A., Lavelle, G. M., Molloy, K., Azim, A. A., Gunaratnam, C., et al. (2015). The Role of Short-Chain Fatty Acids, Produced by Anaerobic Bacteria, in the Cystic Fibrosis Airway. Am. J. Resp. Crit. Care 192 (11), 1314–1324. doi: 10.1164/rccm.201505-0943OC
Moffatt, M. F., Cookson, W. O. (2017). The lung microbiome in health and disease. Clin. Med. (Lond.) 17 (6), 525–529. doi: 10.7861/clinmedicine.17-6-525
Munck, A. (2014). Cystic fibrosis: evidence for gut inflammation. Int. J. Biochem. Cell Biol. 52, 180–183. doi: 10.1016/j.biocel.2014.02.005
Nguyen, L. D. N., Viscogliosi, E., Delhaes, L. (2015). The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00089
Nicaise, C., Prozzi, D., Viaene, E., Moreno, C., Gustot, T., Quertinmont, E., et al. (2008). Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology 48 (4), 1184–1192. doi: 10.1002/hep.22445
Nielsen, S., Needham, B., Leach, S. T., Day, A. S., Jaffe, A., Thomas, T., et al. (2016). Disrupted progression of the intestinal microbiota with age in children with cystic fibrosis. Sci. Rep. 6 (1), 24857. doi: 10.1038/srep24857
Nobrega, F. L., Costa, A. R., Kluskens, L. D., Azeredo, J. (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23 (4), 185–191. doi: 10.1016/j.tim.2015.01.006
Oddy, W. H., de Klerk, N. H., Sly, P. D., Holt, P. G. (2002). The effects of respiratory infections, atopy, and breastfeeding on childhood asthma. Eur. Respir. J. 19 (5), 899–905. doi: 10.1183/09031936.02.00103602
O’Hara, A. M., Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7 (7), 688–693. doi: 10.1038/sj.embor.7400731
O’Neill, K., Bradley, J. M., Johnston, E., McGrath, S., McIlreavey, L., Rowan, S., et al. (2015). Reduced bacterial colony count of anaerobic bacteria is associated with a worsening in lung clearance index and inflammation in cystic fibrosis. PLoS One 10 (5), e0126980. doi: 10.1371/journal.pone.0126980
Pattaroni, C., Watzenboeck, M. L., Schneidegger, S., Kieser, S., Wong, N. C., Bernasconi, E., et al. (2018). Early-Life Formation of the Microbial and Immunological Environment of the Human Airways. Cell Host Microbe 24 (6), 857–865.e854. doi: 10.1016/j.chom.2018.10.019
Peng, L., Li, Z. R., Green, R. S., Holzman, I. R., Lin, J. (2009). Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139 (9), 1619–1625. doi: 10.3945/jn.109.104638
Petersen, C., Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 16 (7), 1024–1033. doi: 10.1111/cmi.12308
Ramakrishnan, V. R., Hauser, L. J., Frank, D. N. (2016). The sinonasal bacterial microbiome in health and disease. Curr. Opin. Otolaryngo 24 (1), 20–25. doi: 10.1097/MOO.0000000000000221
Ríos-Covián, D., Ruas-Madiedo, P., Margolles, A., Gueimonde, M., de los Reyes-Gavilán, C. G., Salazar, N. (2016). Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 7 (185). doi: 10.3389/fmicb.2016.00185
Rizzatti, G., Lopetuso, L. R., Gibiino, G., Binda, C., Gasbarrini, A. (2017). Proteobacteria: A Common Factor in Human Diseases. BioMed. Res. Int. 2017, 9351507. doi: 10.1155/2017/9351507
Rogers, G. B., Carroll, M., Hoffman, L., Walker, A., Fine, D., Bruce, K. (2010). Comparing the microbiota of the cystic fibrosis lung and human gut. Gut Microbes 1 (2), 85–93. doi: 10.4161/gmic.1.2.11350
Salsgiver, E. L., Fink, A. K., Knapp, E. A., LiPuma, J. J., Olivier, K. N., Marshall, B. C., et al. (2016). Changing Epidemiology of the Respiratory Bacteriology of Patients With Cystic Fibrosis. Chest 149 (2), 390–400. doi: 10.1378/chest.15-0676
Sawa, N., Wilaipun, P., Kinoshita, S., Zendo, T., Leelawatcharamas, V., Nakayama, J., et al. (2012). Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1. Appl. Environ. Microbiol. 78 (3), 900–903. doi: 10.1128/AEM.06497-11
Scanlan, P. D., Buckling, A., Kong, W., Wild, Y., Lynch, S. V., Harrison, F. (2012). Gut dysbiosis in cystic fibrosis. J. Cyst. Fibros 11 (5), 454–455. doi: 10.1016/j.jcf.2012.03.007
Schroeder, B. O., Bäckhed, F. (2016). Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22 (10), 1079–1089. doi: 10.1038/nm.4185
Segal, L. N., Alekseyenko, A., Wu, B., Rom, W., Blaser, M., Weiden, M. (2013). Enrichment of lung microbiome with supraglotic microbes is a risk factor for increased pulmonary inflammation. Clin. Transl. Sci. 6 (2), 139. doi: 10.1111/cts.12047
Shakya, M., Lo, C.-C., Chain, P. S. G. (2019). Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 10, 904–904. doi: 10.3389/fgene.2019.00904
Sharma, N., Bhatia, S., Sodhi, A. S., Batra, N. (2018). Oral microbiome and health. AIMS Microbiol. 4 (1), 42–66. doi: 10.3934/microbiol.2018.1.42
Sherrard, L. J., Bell, S. C. (2018). Lower airway microbiota for ‘biomarker’ measurements of cystic fibrosis disease progression? Thorax 73 (11), 1001–1003. doi: 10.1136/thoraxjnl-2018-212165
Sherrard, L. J., Bell, S. C., Tunney, M. M. (2016). The role of anaerobic bacteria in the cystic fibrosis airway. Curr. Opin. Pulm. Med. 22 (6), 637–643. doi: 10.1097/MCP.0000000000000299
Shin, N.-R., Whon, T. W., Bae, J.-W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33 (9), 496–503. doi: 10.1016/j.tibtech.2015.06.011
Siggins, A., Gunnigle, E., Abram, F. (2012). Exploring mixed microbial community functioning: recent advances in metaproteomics. FEMS Microbiol. Ecol. 80 (2), 265–280. doi: 10.1111/j.1574-6941.2011.01284.x
Singhvi, G., Girdhar, V., Patil, S., Gupta, G., Hansbro, P. M., Dua, K. (2018). Microbiome as therapeutics in vesicular delivery. BioMed. Pharmacother. 104, 738–741. doi: 10.1016/j.biopha.2018.05.099
Sultan, A. S., Kong, E. F., Rizk, A. M., Jabra-Rizk, M. A. (2018). The oral microbiome: A Lesson in coexistence. PLoS Pathog. 14 (1), e1006719–e1006719. doi: 10.1371/journal.ppat.1006719
Taylor, S. L., O’Farrell, H. E., Simpson, J. L., Yang, I. A., Rogers, G. B. (2019). The contribution of respiratory microbiome analysis to a treatable traits model of care. Respirology 24 (1), 19–28. doi: 10.1111/resp.13411
Tilg, H., Zmora, N., Adolph, T. E., Elinav, E. (2020). The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 20 (1), 40–54. doi: 10.1038/s41577-019-0198-4
Tkhilaishvili, T., Wang, L., Perka, C., Trampuz, A., Gonzalez Moreno, M. (2020). Using Bacteriophages as a Trojan Horse to the Killing of Dual-Species Biofilm Formed by Pseudomonas aeruginosa and Methicillin Resistant Staphylococcus aureus. Front. Microbiol. 11, 695–695. doi: 10.3389/fmicb.2020.00695
Tringe, S. G., Rubin, E. M. (2005). Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6 (11), 805–814. doi: 10.1038/nrg1709
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20 (2), 159–166. doi: 10.1038/nm.3444
Tunney, M. M., Field, T. R., Moriarty, T. F., Patrick, S., Doering, G., Muhlebach, M. S., et al. (2008). Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 177 (9), 995–1001. doi: 10.1164/rccm.200708-1151OC
Tunney, M. M., Klem, E. R., Fodor, A. A., Gilpin, D. F., Moriarty, T. F., McGrath, S. J., et al. (2011). Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 66 (7), 579–584. doi: 10.1136/thx.2010.137281
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., Gordon, J., II (2007). The Human Microbiome Project. Nature 449 (7164), 804–810. doi: 10.1038/nature06244
Vakhlu, J., Sudan, A. K., Johri, B. N. (2008). Metagenomics: Future of microbial gene mining. Indian J. Microbiol. 48 (2), 202–215. doi: 10.1007/s12088-008-0033-2
Velásquez, J. E., Zhang, X., van der Donk, W. A. (2011). Biosynthesis of the antimicrobial peptide epilancin 15X and its N-terminal lactate. Chem. Biol. 18 (7), 857–867. doi: 10.1016/j.chembiol.2011.05.007
Vilanova, C., Porcar, M. (2016). Are multi-omics enough? Nat. Microbiol. 1 (8), 16101. doi: 10.1038/nmicrobiol.2016.101
Vyas, U., Ranganathan, N. (2012). Probiotics, prebiotics, and synbiotics: Gut and beyond. Gastroent. Res. Pract. 2012. doi: 10.1155/2012/872716
Wallace, B. D., Redinbo, M. R. (2013). The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol. 17 (3), 379–384. doi: 10.1016/j.cbpa.2013.04.011
Whelan, F. J., Waddell, B., Syed, S. A., Shekarriz, S., Rabin, H. R., Parkins, M. D., et al. (2020). Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat. Microbiol. 5 (2), 379–390. doi: 10.1038/s41564-019-0643-y
Wikoff, W. R., Anfora, A. T., Liu, J., Schultz, P. G., Lesley, S. A., Peters, E. C., et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. 106 (10), 3698–3703. doi: 10.1073/pnas.0812874106
Woo, P. C. Y., Lau, S. K. P., Teng, J. L. L., Tse, H., Yuen, K.-Y. (2008). Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infec. 14 (10), 908–934. doi: 10.1111/j.1469-0691.2008.02070.x
Wright, G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5 (3), 175–186. doi: 10.1038/nrmicro1614
Zang, X., Monge, M. E., McCarty, N. A., Stecenko, A. A., Fernández, F. M. (2017). Feasibility of Early Detection of Cystic Fibrosis Acute Pulmonary Exacerbations by Exhaled Breath Condensate Metabolomics: A Pilot Study. J. Proteome Res. 16 (2), 550–558. doi: 10.1021/acs.jproteome.6b00675
Keywords: cystic fibrosis, inflammation, lungs, metagenomics, microbiome, microbiota, multi-omics
Citation: Lee AJ, Einarsson GG, Gilpin DF and Tunney MM (2020) Multi-Omics Approaches: The Key to Improving Respiratory Health in People With Cystic Fibrosis? Front. Pharmacol. 11:569821. doi: 10.3389/fphar.2020.569821
Received: 05 June 2020; Accepted: 17 August 2020;
Published: 03 September 2020.
Edited by:
Noel Gerard McElvaney, Royal College of Surgeons in Ireland, IrelandReviewed by:
Michal Letek, Universidad de León, SpainClaudio Ferrante, University of Studies G. d’Annunzio Chieti and Pescara, Italy
Copyright © 2020 Lee, Einarsson, Gilpin and Tunney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew J. Lee, YS5qLmxlZUBxdWIuYWMudWs=
 Andrew J. Lee
Andrew J. Lee Gisli G. Einarsson
Gisli G. Einarsson Deirdre F. Gilpin
Deirdre F. Gilpin Michael M. Tunney1,3
Michael M. Tunney1,3