- 1Department of Biotechnology, School of Life Science & Biotechnology, Adamas University, Kolkata, India
- 2Institute for Skeletal Aging & Orthopedic Surgery, Hallym University-Chuncheon Sacred Heart Hospital, Chuncheon-si, South Korea
As the COVID-19 is still growing throughout the globe, a thorough investigation into the specific immunopathology of SARS-CoV-2, its interaction with the host immune system and pathogen evasion mechanism may provide a clear picture of how the pathogen can breach the host immune defenses in elderly patients and patients with comorbid conditions. Such studies will also reveal the underlying mechanism of how children and young patients can withstand the disease better. The study of the immune defense mechanisms and the prolonged immune memory from patients population with convalescent plasma may help in designing a suitable vaccine candidate not only for the current outbreak but also for similar outbreaks in the future. The vital drug candidates, which are being tested as potential vaccines or therapeutics against COVID-19, include live attenuated vaccine, inactivated or killed vaccine, subunit vaccine, antibodies, interferon treatment, repurposing existing drugs, and nucleic acid-based vaccines. Several organizations around the world have fast-tracked the development of a COVID-19 vaccine, and some drugs already went to phase III of clinical trials. Hence, here, we have tried to take a quick glimpse of the development stages of vaccines or therapeutic approaches to treat this deadly disease.
Introduction
Severe Acute Respiratory Syndrome (SARS) caused by SARS Coronavirus (SARS‐CoV) initially occurred in China (November 2002) and then quickly spread to 29 countries, resulted in 8,096 cases with 774 fatalities (mortality rate 9.6%). SARS was officially contained in July 2003, about eight months since its first outbreak (WHO, 2003; Peiris et al., 2004). MERS (Middle East Respiratory Syndrome) caused by MERS‐CoV (MERS Coronavirus) has resulted in a similar outbreak by spreading into 26 countries with 2519 infected cases and 866 deaths (mortality rate 34.4%) after its first report on June 2012 in Saudi Arabia (Assiri et al., 2013; World Health Organization, 2019). The current outbreak of COVID-19 (Coronavirus Disease 2019) caused by SARS-CoV-2, which was first reported in the Wuhan (China) on December 2019 (Hubei province), now gradually spilled over 213 countries and territories resulted in over 16.3 million infected cases with and more than 650,000 deaths (4% mortality rate) as of July 26, 2020 (Wang et al., 2020a). On January 30, 2020, WHO announced the current coronavirus outbreak as a world health emergency, and on March 11, 2020, reclassified it as a pandemic (World Health Organization, 2005; Chakraborty et al., 2020c; WHO, 2020). The virus was initially named Novel Coronavirus 2019 (2019-nCoV), and later it was changed to SARS-CoV-2 (Gorbalenya, 2020). The WHO entitled the disease as COVID-19 on February 11, 2020 (World Health Organization, 2020). The SARS-CoV-2 was found to be infectious as it spreads via respiratory droplets and aerosols when an infected individual comes in contact with a healthy person (Chan et al., 2020b; Liu Y. et al., 2020). The virus incubates for about 2–14 days within humans and subsequently resulted in various mild to severe symptoms like fever, dry cough, dyspnea, severe respiratory issues, pneumonia, etc (Chakraborty et al., 2020a; Chan et al., 2020b; Huang et al., 2020; Lauer et al., 2020; Zu et al., 2020).
Coronaviruses are ssRNA (positive-sense) virus and enveloped with a diameter of 80–120 nm (Sipulwa et al., 2016). This virus (SARS-CoV-2) under the beta-coronavirus genus of the Coronaviridae family comprises four genera—α-CoV, β-CoV, γ-CoV, and δ-CoV (Chan et al., 2013). Like SARS-CoV-2, MERS-CoV and SARS-CoV are also belonged to the genus β-CoV (Chan et al., 2013). Further, four HCoVs that cause mild symptoms, i.e., common cold, belong to the genera α-CoV (HCoV-NL63 and HCoV-229E) and β-CoV (HCoV-OC43 and HCoV-HKU) (Rabi et al., 2020). The size of the SARS-CoV-2 genome was found to be about 29.9 kb (GenBank Accession Number: MN908947.3) (Wu F. et al., 2020). Preliminary studies suggested that the genome of SARS‐CoV‐2 is closer to SARS‐CoV than MERS-CoV depending on the percentage similarity, although the highest genome similarity was found with the RaTG13 virus found in bats which indicated a plausible origin of SARS-CoV-2 (bat) (Chakraborty et al., 2020b; Lu et al., 2020; Zhou et al., 2020). Both SARS-CoV-2 and SARS-CoV uses the human ACE2 as a receptor for their entrance in the cell (Ge et al., 2013; Wan et al., 2020; Wrapp et al., 2020).
The cell membrane attached ACE2 converts the vasoconstrictor peptide angiotensin II to angiotensin 1–7 (vasodilator peptide), and it protects the heart and blood vessels (Jiang et al., 2014). ACE2 is found in the heart, lung, kidney, endothelium, etc. and known to reduce the adverse effects of other RAS (Renin-Angiotensin System) components by reducing the concentration of angiotensin II and increasing the concentration of angiotensin 1–7 and regulates the blood pressure in the body. ACE2 also found to express in intestinal epithelial cells where it helps to absorb nutrients from the food particles and was predicted as one of the entry sites that may have been used initially by SARS-CoV-2 upon the consumption of contaminated food from Wuhan seafood market (Hashimoto et al., 2012; Zhang et al., 2020a). Similarly, ACE2 is also found to express on the mucosa of the oral cavity and the epithelial cell of the tongue, making these other entry routes for SARS-CoV-2 (Xu et al., 2020). Interestingly, a small subset of type II alveolar cells (AT2) was found to express the ACE2 receptor and several other genes that positively regulate viral reproduction and transmission, making the lung more susceptible to the virus. The ACE2 expressing cells in the lung triggers an immune response, which may overreact to damage the lung cells by filling up the air sacs with fluid instead of gas, causing pneumonia. Patients with a severely damaged lung can develop acute respiratory distress syndrome (ARDS), where breathing becomes difficult (Li et al., 2020). As ACE2 expresses in an array of organs, SARS-CoV-2 can attack several organs, which results in multi-organ failure often observed in patients who died of COVID-19 (Wang T. et al., 2020). Patients with chronic cardiovascular diseases often take drugs that block the angiotensin receptor or inhibit the angiotensin-converting enzyme, which in turn increases the expression of ACE2 receptors in cells. Therefore, COVID-19 patients who regularly take these medications might have an increased hazard of SARS-CoV-2 infection (Diaz, 2020).
Like other coronaviruses, SARS‐CoV‐2 also consists of two types of protein structural proteins and non-structural. Structural proteins comprise of E (envelope) protein, S (spike) protein, M (membrane) protein, and N (nucleocapsid) protein (Wu A. et al., 2020). The spike protein (S) of SARS‐CoV‐2 is a trimeric class I type of fusion protein that helps the virus to enter host cells (Bosch et al., 2003; Walls et al., 2020). The spike protein has two subunits, S1 (required for receptor recognition) and S2 (required for membrane fusion). The C-terminal RBD (receptor-binding domain) of the first subunit (S1 subunit) of spike protein directly interacts with the ACE2 receptor (Yuan et al., 2020). Upon the fusion of the S protein, which exists in a metastable prefusion state, with the ACE2 receptor, the S protein undergoes a conformational rearrangement. The binding to the ACE2 destabilizes the prefusion trimer, which results in the discharge of the S1 subunit. This allows the transition of the S2 subunit of S protein to a steady postfusion state (de Wilde et al., 2017). A cellular serine protease TMPRSS2 plays a pivotal role in this S protein priming (Hoffmann et al., 2020; Wrapp et al., 2020). The host cell-mediated S protein priming is an essential step for the virus to move into the host cells (Hoffmann et al., 2018). Once inside of the host cell, SARS-CoV-2 follows the typical life cycle of a positive-sense RNA virus as was found with MERS-CoV and SARS-CoV (Figure 1) (Fehr and Perlman, 2015).
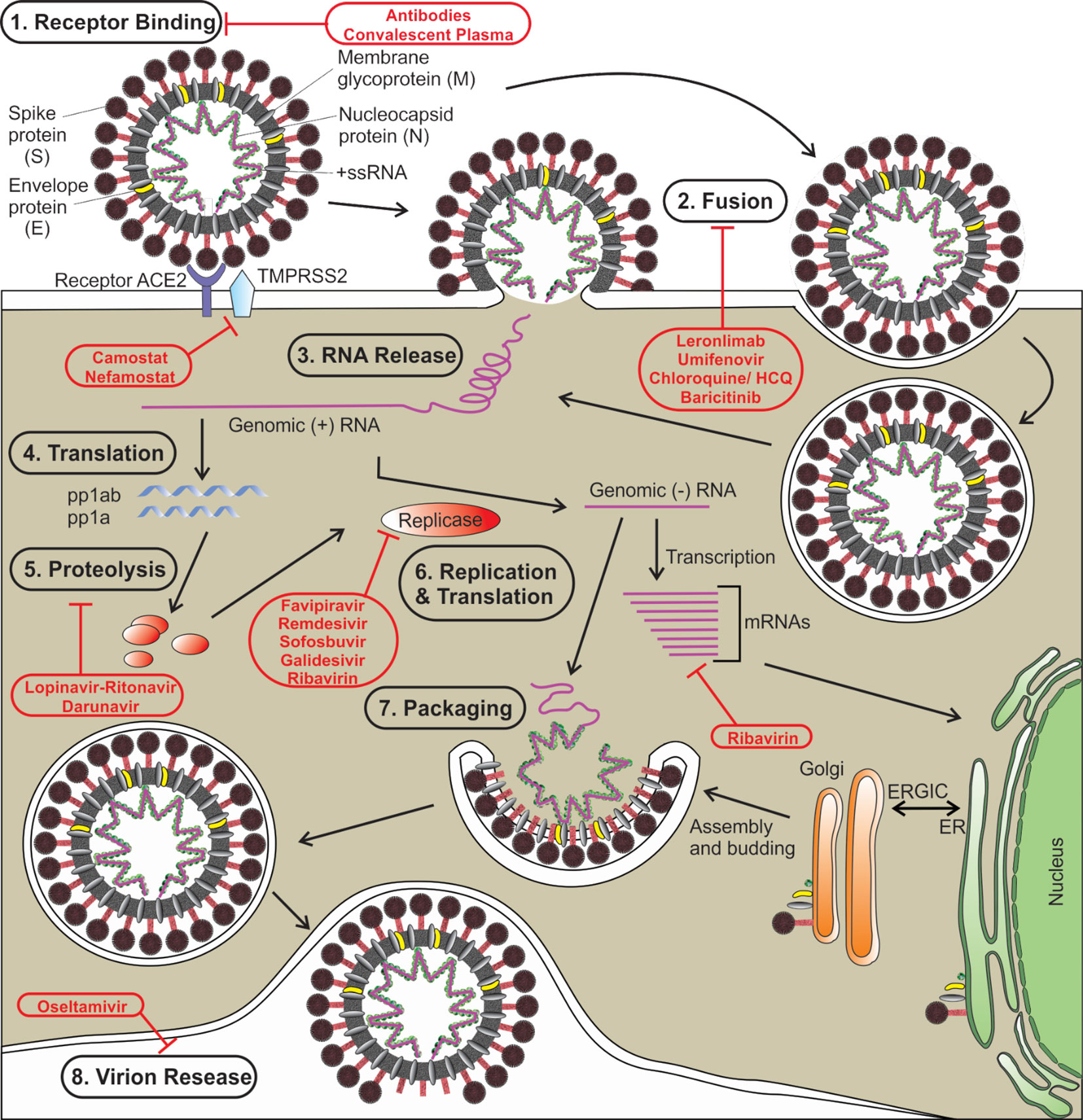
Figure 1 The life cycle of SARS-CoV-2 is shown. Various steps in the life cycle are mentioned—receptor binding of the virus, fusion with the host membrane, viral RNA release, translation of viral RNA, proteolysis of the proproteins, replication and translation, packaging of viral particles, and virion release. Possible targets of various antiviral drugs that are being repurposed/investigated for COVID-19 are indicated. S, spike protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein; HCQ, hydroxychloroquine, ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment.
According to some mathematical models, the transmission of the disease may quickly rebound if we relax measures like lockdown and social distancing (Yamey et al., 2020). In the absence of effective prophylactic treatment, such eruptions may leave the health system overburdened. The absence of a potential drug or vaccine against SARS-CoV-2 has already resulted in a pandemic situation (Wang D. et al., 2020). The designing and development of the COVID-19 vaccine that can be used globally is, therefore, the utmost priority for ending the current pandemic (Prompetchara et al., 2020). It was observed that both SARS‐CoV-2 and SARS‐CoV use the same mechanism to enter target cells has vital significance for our understanding of the SARS‐CoV‐2 pathogenesis and transmissibility. To fight this pandemic, various government and private organizations have sped up their development of vaccines and treatment procedures. In this review article, we have discussed the testing of various existing drugs that are now being repurposed and targets against which various vaccine developments are going on for COVID-19.
Host Immune Response to Viral Infections
Upon viral infection, the host cell initially activates the innate immune response via PRRs (pattern-recognition receptors) that recognizes viral particles (Takeuchi and Akira, 2009). Host cells release a group of signaling proteins called Interferons (IFNs) that play a significant role in host antiviral defense. INFs belong to a group of peptides and proteins called cytokines responsible for transferring signals by binding to the receptors on the surface of appropriate immune cells for triggering host immune response against pathogens. INFs are triggered by the activation of host PRRs. Four types of PRRs are known—TLRs, RLRs, NLRs, and CLRs although during viral infection mainly three types of PRRs are activated—RLRs, TLRs, and NLRs (González-Navajas et al., 2012; Fehr and Perlman, 2015; Nan et al., 2018; Zhang et al., 2020b). PRRs recognize several viral components including DNA, ssRNA, dsRNA, RNA with 5′-triphosphate ends, and proteins. Detection of viral particles by PRRs activates signaling pathways that release type I INFs, different types of cytokines such as proinflammatory cytokines (primarily IL-1, IL-6, TNF-α), chemokines, and co-stimulatory molecules like CD40, CD80, and CD86 that results in inflammation and subsequent engagement of innate and acquired immune cells to eliminate viral infection (González-Navajas et al., 2012; Khan et al., 2012; Nan et al., 2018).
Three types of INFs have been characterized till now—type I IFNs (mainly IFN-α/β), type II IFNs (IFN-γ), and type III (INF-λ) (Stanifer et al., 2019). INFs-α/β is secreted by all viral-infected cells including pDCs (plasmacytoid dendritic cells) which is a vital cell type for INF-α secretion during viral infection. IFN-γ, secreted by NK (natural killer) cells and immune cell-like T cells, plays a vital role in host adaptive and innate immunity. It also regulates the expression of several genes that are affected by type I IFNs. INF-λ is mainly secreted by epithelial cells in response to the viral infection at mucosal sites (Zanoni et al., 2017). INFs protect host cells by activating signaling pathways, mainly the JAK/STAT pathway (Schindler et al., 1992; Darnell et al., 1994), which subsequently trigger the expression of ISGs (IFN-stimulated genes) that controls the viral infection (Katze et al., 2002). The activated STAT proteins (STAT1, STAT2, and STAT3) in response to INF stimulation are vital for transferring signals that subsequently activate ISGs (Levy and Darnell, 2002; Tsai et al., 2019). Type I INFs induced during innate immune response also upregulate several ISGs whose expression restricts viral replication (Kane et al., 2016).
Activation of the innate immune cells is critical for setting up adaptive immune responses during the re-infection by the same virus. Activation of adaptive immunity takes a few days to weeks to become established. APCs (antigen-presenting cells; e.g., dendritic cells, B cells and macrophages), that live at the site of viral infection, binds to viral particles (antigens) and present them on major histocompatibility complex (MHC) class II to be recognized by the T cell receptor on CD4+ T cells in presence of co-stimulatory molecules (Rosendahl Huber et al., 2014). The activated CD4+ T cells release a wide range of cytokines and chemokines that helps to differentiate CD4+ T cells into several cell subtypes, mainly T helper cells (such as Th1, Th2, Tfh, etc.) as well as regulatory T cells (Treg). Th1 and Th2 cells release several cytokines (Th1-INF-γ; Th2- IL-4, IL-13, IL-5, etc.) to trigger B cell differentiation and activate macrophages (Rosendahl Huber et al., 2014). T follicular helper cells (Tfhs) also helps to activate B cells to produce specific antibodies against foreign pathogens (Crotty, 2014). Treg cells do several regulatory functions, especially controlling immunopathology (Crotty, 2014). Activated CD4+ T cells by its interaction with the APCs through CD40-CD40L upregulate expression of CD80/CD86 markers on APCs which interacts with the CD28 on the CD8+ T cells. The APCs presents viral particles on the MHC class I molecules that bind to the TCRs on the CD8+ T cells through the CD80/CD86-CD28 interactions and activate CD8+ T cells. The activated cells proliferate and differentiate into CTLs (cytotoxic T lymphocytes) which releases cytotoxic molecules, and activates the production of cytokines (e.g., TNF-α, IL-2, IFN-γ, etc.) that promotes apoptosis of virally infected cells (Crotty, 2014).
Both innate and adaptive immunity (humoral and cell-mediated) are equally important to control viral infections. Innate immunity mounts host defenses to control viral infection at the early phases by releasing proinflammatory molecules and also activates adaptive immunity by upregulating co-stimulatory molecules. In adaptive immunity, B cells (humoral immunity) and T cells (cell-mediated immunity) are activated that prevent further viral infections. Immunoglobulins (IgG, IgM, and IgA) produced by activated B lymphocytes bind to viruses to block viral spread and also eliminate virus-infected cells via ADCC (antibody-dependent cytotoxic cells) or complement-mediated pathways. CTLs differentiated from activated CD8+ T cells kill the virus-infected cells by releasing cytotoxic cytokines that trigger apoptosis of the target cells. Some of these immune cells (T cells and B cells) are converted into memory cells that prevent further infections and provide long-term immunity (Klimpel, 1996).
SARS-CoV and other coronaviruses are sensitive to IFN-α/β. Some of these viruses are also very pathogenic. It might be attributed to their ability to modulate an effective host immune response. The nucleocapsid protein of SARS-CoV can evade host interferon responses (Spiegel et al., 2005; Kopecky-Bromberg et al., 2007; Lu et al., 2011). It was reported that EV71 (Liu et al., 2014) and Ebola virus infections can downregulate the JAK-STAT pathway mediated by type-I IFNs, and promote viral replication and proliferation within the host (Okumura et al., 2010). Several antibodies, for example, MCA1, CSCC5, CDC-C2, CDC-A10, CDC-A2, MERS-GD27, etc., isolated from recovered MERS-CoV-infected patients have been found useful in controlling the disease (Chen et al., 2017; Niu et al., 2018a; Niu et al., 2018b). Recognition mechanisms involving the surface proteins of virus and the receptors of host are vital for an understanding of the cross-species transmission and host tropism to establish animal models for effective vaccine development (Ahn et al., 2020).
Some COVID-19 patients with severe symptoms experience a sudden surge of cytokines in the body, released by the immune cells in response to the viral infection, commonly referred to as ‘cytokine storm’ (Huang et al., 2020). The excessive release of the cytokines or cytokine release syndrome (CRS) is a major determinant in inducing ARDS in COVID-19 patients. The excessive secretion of proinflammatory cytokines (e.g., IL-6, IL-1, TNF-α, etc.) with the help of the innate immune system within the body leads to several lung complications like pneumonitis and ARDS which can cause multi-organ failure and death (Nicholls et al., 2003; Mahallawi et al., 2018; Ragab et al., 2020). Among various proinflammatory cytokines, IL-6 plays a major role in inducing ARDS as an increase in the concentration of IL-6 in the plasma was found to be linked with ARDS in COVID-19 patients (Ragab et al., 2020). Association of IL-6 to mIL-6R (membrane-bound IL-6 receptor) and gp130 activates the JAK-STAT3 pathway which contributes toward CRS. Besides, at high concentrations, IL-6 binds to sIL-6R (soluble form of IL-6 receptor) and gp130, and activates JAK-STAT3 pathway in cells that do not express mIL-6R which again induces cytokine storm by releasing several cytokines and chemokines (e.g., VEGF, IL-6, MCP-1/CCL2, IL-8, etc.), and by reducing E-cadherin production that leads to ARDS (Magro, 2020; Ragab et al., 2020). Therefore, preventing the occurrence of cytokine storm by drugs that inhibits the release of cytokines may help in alleviating severe COVID-19 symptoms.
Viral and Host Protein Targets
Vaccines
SARS-CoV-2 expresses four structural proteins, N (nucleocapsid), E (envelope), S (spike) protein, and M (membrane) similar to SARS-CoV. These proteins are potential antigens to induce nAbs (neutralizing antibodies) and provide protective functions (Bhattacharya et al., 2020a; Chan et al., 2020a; Shang et al., 2020). So, the finding of a protein that has the dominant neutralizing epitopes should be the first step of the investigation. Before this identification, the inactivated virus can also be used as a first-generation vaccine because it is probably easier to generate than the whole-killed virus particles. Whole-cell killed or live-attenuated vaccines represent all the antigens present in a pathogen like proteins, nucleic acids, polysaccharides, lipids, and some other components capable of inducing a potent immune response (Sharma et al., 2011). Several studies have shown that SARS-CoV inactivated through an agent such as formaldehyde, β-propiolactone and UV light can also instigate virus-neutralizing antibodies in immunized animals (He et al., 2004; Xiong et al., 2004; Jiang et al., 2005; Qu et al., 2005; Te-hui et al., 2005). So in principle, inactivated SARS-CoV-2–based vaccines can also be used. However, upon identification of the neutralizing epitopes, the vaccines that are made based on fragments containing neutralizing epitopes should be used, as they are safer and more effective than the inactivated virus vaccine. Several organizations are using viral deoptimization techniques to synthesize more effective vaccines such as live-attenuated vaccines (Zhang J. et al., 2020). Though, attenuated vaccine mimics the natural course of infection to stimulate the toll-like receptors e.g. (TLR-3, TLR-4, TLR-7, TLR-8, and TLR-9) and provide long-term immunity, ensuring low or no pathogenicity is always a major concern (Chakraborty et al., 2020d). Also, killed vaccines show difficulty in maintaining consistency in quality (Chen W. H. et al., 2020).
Most of the subunit vaccines against coronaviruses depend on mounting immune responses against the spike protein by preventing its binding to the host ACE2 receptor (Jiang et al., 2012). One way to block access to the entry receptor, i.e., human ACE2 receptor is to use the spike protein RBD (receptor-binding domain) of SARS-CoV-2 that has been shown to attach to the ACE2 receptor (Lan et al., 2020). Spike protein’s RBD from SARS-CoV has been shown to block the virus from accessing the ACE2 receptor in cell culture (Wong et al., 2004). Besides, the RBDs of spike proteins in both SARS-CoV-2 and SARS-CoV were found to interact similarly with the ACE2 receptor (Lan et al., 2020). Other researchers have proposed that the RBDs on the spike proteins of other coronaviruses like MHV (mouse hepatitis virus), TGEV (transmissible gastroenteritis virus), HCoV-229E, SARS-CoV, etc. contain key antigenic determinants that can induce production of neutralizing antibodies (Godet et al., 1994; Kubo et al., 1994; Bonavia et al., 2003; He et al., 2004). As spike proteins of coronaviruses are the most important antigenic determinants known to trigger neutralizing antibodies, spike proteins can be used as antigens for developing vaccines (Saif, 1993; Schmidt et al., 2006; Bhattacharya et al., 2020a; Bhattacharya et al., 2020b). Spike protein RBD sequences are relatively conserved. So, this may possible to find the neutralizing epitopes present into the SARS-CoV-2 spike protein for designing and developing of effective, safe vaccine against this virus. How spike protein RBD can activate extremely effective neutralizing antibodies against this virus has been elucidated by the mAbs (monoclonal antibodies) which was isolated from the inactivated virus-immunized human and mice antibody libraries (Sui et al., 2004; He et al., 2005). Thus, the RBD of this virus S protein is not only a functionally important domain for receptor binding of this virus but also a significant neutralization determinant element of SARS-CoV-2. So, the proteins that contain the RBD region or vectors encoding the spike protein RBD can be utilized for developing a highly effective vaccine candidate (Table 1). Therefore, the RBD alone could block access to ACE2 for SARS-CoV-2. Alternatively, single-domain antibodies (sdAbs) or nanobodies based on the RBD can also block the ACE2 receptor effectively (Arbabi-Ghahroudi, 2017). Researchers are developing virus-like nanoparticles based on the expression of recombinant spike protein, which can act as a potent immunogen. Others have developed subunit vaccines consisting of the RBD from SARS-CoV S protein (Chen W. H. et al., 2020). However, certain limitations of subunit vaccines exist, for example, the requirement of multiple booster shots and suitable adjuvants (Shang et al., 2020).
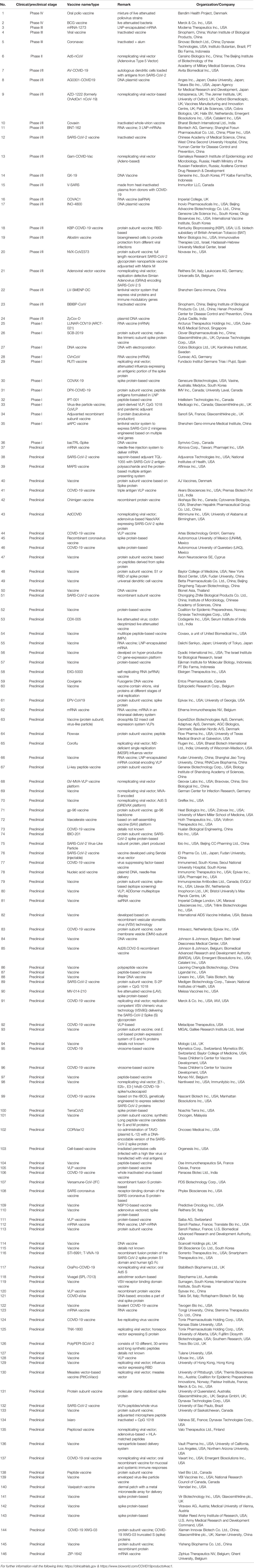
Table 1 Ongoing vaccine development initiatives against COVID-19 by different organizations that are at different phases of clinical and preclinical trials (updated on July 25, 2020).
During the vaccine candidate development against SARS-CoV-2, one may have to consider the possibility of antibody-dependent enhancement (ADE) triggering in vaccinated individuals where instead of mounting protection against the virus infection the virus-bound antibody bind to the host cell receptors to facilitate the cellular entry of the virus. Activation of ADE has been observed in vaccines against several diseases, e.g., Ebola, HIV, Dengue, feline coronavirus, etc (Takada and Kawaoka, 2003; Halstead, 2017; Takano et al., 2019). Human and rodent antibodies produced against the SARS-CoV S protein also shown to induce ADE in vitro (Liu et al., 2019). However, ADE was not observed in several pre-clinical studies done in rhesus monkeys using a SARS-CoV vaccine (Luo et al., 2018). Besides, in a pre-clinical study using an inactivated SARS-CoV-2 vaccine did not show any evidence of ADE (Gao Q. et al., 2020).
Therapeutics
SARS‐CoV‐2 does not use receptors that are utilized by other coronaviruses, for example, APN (aminopeptidase N; used by HCoV-229E), DPP4 (dipeptidyl peptidase 4; used by MERS-CoV), or O-acetylated sialic acid receptor (used by HCoV-OC43 and HCoV-HKU1) (Yeager et al., 1992; Krempl et al., 1995; Raj et al., 2013; Huang et al., 2015). It uses the human ACE2 cell receptor to enter the host cell, similar to SARS-CoV and HCoV-NL63 (Hofmann et al., 2005; Ge et al., 2013; Wrapp et al., 2020). So, soluble human ACE2 protein can also be a potential competitor for the ACE2 cell surface receptor, but it can only be achieved when the gene expression of soluble ACE2 is higher than the gene expression of cell surface ACE2 receptor. However, an increase in the concentration of soluble ACE2 in blood found to be associated with chronic cardiac dysfunction (Epelman et al., 2008; Epelman et al., 2009; Ortiz-Pérez et al., 2013). SARS-CoV was found to downregulate ACE2 by binding to it by its spike protein and inflicting severe lung damage (Kuba et al., 2005). Therefore, overexpressed soluble ACE2 may help in neutralizing SARS-CoV-2 by competitively binding to it and free the cellular ACE2 to perform its normal function. A recombinant human ACE2 (APN01) was found to decrease the levels of angiotensin II and plasma IL-6 in different patients diagnosed with ARDS (acute respiratory distress syndrome) may also be utilized for inhibiting SARS-CoV-2 from accessing cellular ACE2 receptor (Zhang et al., 2020a). Soluble human ACE2 protein was shown to bind SARS-CoV with an affinity close to the affinities of monoclonal antibodies and blocks the virus from accessing cellular ACE2 receptor in cell culture (Li et al., 2003; Sui et al., 2004). Interestingly, membrane-anchored metalloproteinase ADAM17 cleaves ACE2 to release the soluble ACE2 domain, which was predicted to have some adverse effects on the heart (Jiang et al., 2014).
Another strategy is to develop anti-ACE2 antibodies that would bind to the human ACE2 protein and block this viral entry, as was shown in SARS-CoV (Li et al., 2003). Unfortunately, there are problems with generating antibodies or protein fragments against the cellular ACE2 as it plays several important roles in controlling cardiovascular diseases including heart attack, diabetes, kidney problems, high blood pressure, etc. Therefore, inactivating the cellular ACE2 receptor is probably not a viable solution.
Alternatively, an ACE2-Fc fusion protein can also increase the lifespan of the soluble ACE2 protein in circulation and inhibit the virus from accessing the cellular ACE2 receptor. Similarly, in a study, the extracellular ACE2 domain fused to the human IgG1 domain was shown to neutralize the SARS-CoV in vitro (Gu et al., 2016), which shows that the use of ACE2-Fc could be a viable solution to block SARS-CoV-2 from infecting human cells. However, this strategy may induce ADE and therefore a thorough investigation is needed to eliminate any adverse effects. The spike protein RBD could also be attached to a human IgG Fc fragment to increase its immunogenicity and stability (Zhang et al., 2009; Li et al., 2011; Du et al., 2013b), as was done in MERS-CoV (Du et al., 2013a). The MERS-CoV spike protein RBD-Fc fusion was found useful in blocking viral cell surface receptor from accessing it by the virus and also stimulated the host immune response against the viral protein domain in mice (Du et al., 2013a). Here one has to consider the mutation of the Fc domain that eliminates its cellular Fc receptor (FcγR) binding ability and triggering of cytotoxic effects (Wang et al., 2018; Kang and Jung, 2019). The binding of the Fc region to FcγR would activate immune cells to trigger the ADCC pathway and release proinflammatory cytokines, which may lead to cytokine storm (Wang et al., 2018). Therefore, the Fc fusion strategy requires a thorough investigation of toxicity and efficacy, followed by the engineering of the Fc fragment for immune silencing and increasing effectiveness (Kang and Jung, 2019).
The other alternative strategy would be to generate antibodies or protein-fragments that would bind to the virus itself and protect the cellular ACE2 receptor from binding the virus (Jiang et al., 2020). If a protein or peptide fragment that can mimic the binding domain of ACE2 cell receptor and induce similar changes in conformation, as the receptor likely does, then also it can compete with the ACE2 cell receptor. Recently a 23-mer peptide designed from the ACE2 α1 helix has shown a specific binding affinity toward RBD of S protein from SARS-CoV-2, which shows that the development of a peptide-based therapeutics is possible that blocks of this virus interaction with human ACE2 and protecting the cell from virus entry (Zhang G. et al., 2020).
A recent report has shown that murine polyclonal antibodies generated against SARS-CoV spike protein were capable enough to inhibit spike protein-mediated cellular entry of SARS-CoV-2 (Walls et al., 2020). Also, a human monoclonal antibody (47D11), which interacts with a conserved epitope on RBD of spike protein, was found to cross-neutralize with both SARS-CoV-2 and SARS-CoV (Wang et al., 2020b). Another antibody having neutralizing property (antibody CR3022) previously isolated from the SARS-CoV infected patient was found to interact with the S protein RBD of SARS-CoV-2 at a site different from the ACE2 binding site indicating cross-reactivity of the antibody for having similar structural regions on the spike proteins of both the viruses (Yuan et al., 2020).
SARS-CoV-2 nucleocapsid protein (N) is another vital protein having several critical roles, including viral genome replication, transcription, etc., and therefore is an attractive drug target. Recently a 3D structure (x-ray crystallography) of the amino-terminal RNA-binding domain of this virus N protein has been elucidated, indicating drug targets (Kang et al., 2020). Broad-spectrum antiparasitic drug nitazoxanide has been shown to inhibit the expression of nucleocapsid protein in MERS-CoV and other coronaviruses (Rossignol, 2016). Nitazoxanide also found to suppress proinflammatory cytokines, including IL-6 in mice (Rossignol, 2016). The viral M protein is also highly conserved in evolution among different species (Neuman et al., 2011), and hence, may also be used as a candidate for developing the SARS-CoV-2 therapeutics (Table 2).

Table 2 Ongoing repurposed drug/therapeutic molecule development by different organizations against COVID-19 that are at different phases of clinical trials (updated on July 25, 2020).
Human monoclonal antibody-based drug sarilumab which inhibits IL-6 receptor is now being tested against COVID-19 (Lamb and Deeks, 2018). Monoclonal antibody-based rheumatoid arthritis drug tocilizumab which is also an inhibitor of IL-6 receptor found to be effective in critically ill COVID-19 patients with cytokine storms and elevated IL-6 levels (Venkiteshwaran, 2009; Chakraborty et al., 2020e; Luo et al., 2020; Saha et al., 2020b). Another monoclonal antibody-based drug leronlimab (PRO 140) known to bind to the CCR5 receptor on the CD4+ T lymphocytes is now being tested in COVID-19 clinical trials (Pugach et al., 2008). The proinflammatory chemokine such as C-C motif chemokine ligand 5 (CCL5) also recognized as regulated through activation, normal T cell expression, and secretion (RANTES), binds to its receptor C-C chemokine receptor type 5 (CCR5) and activates inflammatory responses by directing immune cells to the inflammation site (Vangelista and Vento, 2018). Blocking of CCR5 by leronlimab found to reduce serum IL-6 levels, which is linked with cytokine storm, in critical COVID-19 patients (Patterson et al., 2020). Interleukin-6 (IL-6) plays a vital role in inducing cytokine storm in critical COVID-19 patients and a reduction in IL-6 levels by anti-inflammatory drugs is expected to ease CRS and reduce viral loads (Zhang C. et al., 2020).
Anti-inflammatory corticosteroid drug dexamethasone has been suggested recently to treat severe COVID-19 patients with CRS. Dexamethasone reduces the production of cytokines but is also known to inhibit the protective functions of T cells and B cells. Therefore, the drug may be used selectively in some severe COVID-19 cases, but its general usage in other COVID-19 patients may cause more harm by increasing the viral load in patients due to the inhibition of protective antibody production (Lee et al., 2004; Russell et al., 2020). A recent clinical trial has shown that dexamethasone reduced the death rate among severe COVID-19 patients who needed oxygen support (Table 2). A recent study with severe COVID-19 patients found a direct link between C-reactive protein (CRP) and inflammation where higher CRP levels in the blood show greater inflammation. The study also showed that dexamethasone should only be used in severe COVID-19 patients with CRP levels above 20 mg per deciliter of blood, and the use of dexamethasone should be avoided in COVID-19 patients (under ventilator support) with CRP level below 10 as it may turn out to be fatal (Keller et al., 2020).
Anti-inflammatory rheumatoid arthritis drug baricitinib was found to reduce the levels of cytokines, including IFN-γ in severe COVID-19 patients (Huang et al., 2020). High levels of proinflammatory cytokines and chemokines including INF-γ in the plasma causes inflammatory cytokine storm that may lead to the occurrence of ARDS in virus-infected patients, therefore use of anti-inflammatory drugs in COVID-19 may help in the reduction of severe symptoms (Ye et al., 2020). Another rheumatoid arthritis drug anakinra is known to block the IL-1 receptor and reduce the inflammatory effects of IL-1. Survival rate within patients with hyperinflammatory conditions was found to increase when treated with anakinra (Shakoory et al., 2016).
Convalescent Plasma
Therapeutics
Convalescent plasma (CP) therapy is another procedure now being tested for COVID-19. This therapy is very simple yet effective, where the serum from the COVID-19 recovered persons can treat new patients (Mire et al., 2016). Recovered patients who have suffered from COVID-19 should have an elevated amount of polyclonal antibodies raised by the immune system to prevent new rounds of infection by SARS-CoV-2. Therefore, the plasma harvested from the recovered patients can be transfused to the patients who have contacted the virus (Marano et al., 2016). As the application of convalescent plasma is a well-known procedure and has been utilized before by medical practitioners, it should not be too difficult to apply this procedure to SARS-CoV-2 infected patients. Convalescent plasma has been used previously during the Ebola outbreak in 2014 and was found to be effective in treating Ebola patients (Kraft et al., 2015). A recent report has shown that CP acquired from recovered patients was effective in treating new COVID-19 infected persons (Duan et al., 2020). One problem using CP therapy is the significant variability of potency that has been found in the sera of recovered patients in neutralizing the antigen, making it a less viable option in the treatment of patients (Marano et al., 2016). Also, if the number of infected patients is much higher than the recovered patients, it would be tough to get enough CP for transfusion. Although CP therapy is being considered or used for the COVID-19 treatment, ultimately, it has limited scope in controlling the outbreak at present.
Interferon Therapy
Therapeutics
Type I interferons (IFN-I) stimulate the immune system upon viral infection by activating macrophages, natural killer cells, etc. and are expected to hinder SARS-CoV-2 infection (Samuel, 2001; Belhadi et al., 2020; Martinez, 2020). IFN-I is secreted by several cells when the pattern recognition receptors (PRRs) binds viral particles (Liu, 2005). IFN-I is recognized by the interferon-α/β receptor (IFNAR) in the plasma membrane. Upon binding of IFN-I, IFNAR induces the phosphorylation of several transcriptional factors, including STAT1. Once localized in the nucleus, STAT1 activates interferon-stimulated genes (ISGs), including PRRs, which further helps in decreasing membrane fluidity that inhibits viral entry through the membrane (Totura and Baric, 2012; Schneider et al., 2014). Although interferon treatment against SARS-CoV and MERS-CoV has shown variable efficiency (Stockman et al., 2006), the IFNβ subtype appears to work well in COVID-19 treatment if administered in the early stages of infection (Sallard et al., 2020). The side effects of interferon treatment could be toxic to a patient, especially when the patient is at critical stages of infection. Therefore, it is recommended to use this therapy in the early stages of infection.
Membrane Fusion Inhibitors
Therapeutics
Well-known antimalarial drugs chloroquine and its less toxic derivative hydroxychloroquine, both known to elevate the pH of endosomes/lysosomes that blocks membrane fusion and inhibits viral infection (Mauthe et al., 2018). Also, chloroquine found to impede glycosylation of the ACE2 receptor, which may inhibit the virus from receptor binding (Vincent et al., 2005). Both of these drugs helped inhibit this virus in the in vitro assays (Liu J. et al., 2020; Wang M. et al., 2020). However, some studies have raised concerns about the effectiveness of chloroquine/hydroxychloroquine in treating COVID-19 patients as these repurposed drugs were found to possess several side effects (Chary et al., 2020; Chen J. et al., 2020; Gautret et al., 2020; Kamp et al., 2020).
Current reports suggested that the influenza drug umifenovir is effective in reducing symptoms of COVID-19 (Zhang J. N. et al., 2020). Umifenovir (Arbidol) intercalates with the membrane lipids to inhibit the fusion between the virus particle and host membrane, which blocks the entrance point of the virus inside the host cell (Villalaín, 2010; Blaising et al., 2014). Another influenza drug oseltamivir, which reduces infection in the respiratory system by blocking viral neuraminidase and inhibits viral particles from escaping host cells, was found to be effective in the COVID-19 outbreak in China (Uyeki, 2018; Wang D. et al., 2020).
Coronaviruses use several modes of endocytosis (clathrin‐ or caveolin-mediated, or by the formation of lipid rafts) depending on the virus and cell type, and therefore, blocking of the endocytic pathways could be a promising strategy for the development of antiviral drugs (Glebov, 2020; Yang and Shen, 2020). Several anti-endocytotic drugs (e.g., chlorpromazine, bafilomycin, etc.) that are known to inhibit clathrin-or caveolin-mediated endocytosis proposed to have therapeutic activities against coronaviruses including SARS-CoV-2 (Yang and Shen, 2020). In lung AT2, alveolar epithelial cells, AAK1 regulates endocytosis, and baricitinib inhibits AAK1 with high affinity. Therefore, researchers argue that baricitinib could be one of the potential drugs against COVID-19 (Richardson et al., 2020). However, others argue that baricitinib also inhibits the JAK-STAT mediated signaling pathway which affects the interferon-mediated immune response. It might have a fatal effect on COVID-19 patients (Favalli et al., 2020). Clinical trials are currently underway to find out whether the drug has any positive effect in treating COVID-19 patients.
Protease Inhibitors
Human Protease Inhibitors (Therapeutics)
Proprotein convertases (PCs) are essential for turning precursor proteins into their active forms, e.g., furin and other proteases that control viral host cell entry and infectivity (Yamada et al., 2018; Izaguirre, 2019). Host proteases cleaved the coronavirus S proteins, including furin, TMPRSS2 (transmembrane protease serine protease 2), trypsin, cathepsin, etc., and the availability of these proteases in the infected cells are important for subsequent host cell entry (Ou et al., 2020). Furin or trypsin dependent proteolytic cleavage of the viral (SARS-CoV) S protein at two distinct sites was found to be essential for priming and subsequent membrane fusion with the host cell (Belouzard et al., 2009). MERS-CoV spike protein was also found to be activated by furin cleavage (Millet and Whittaker, 2014). Similarly, the S protein of SARS-CoV-2 has a putative cleavage site (furin) between S1 and S2 subunits, but whether it is cleaved during the priming event remains elusive (Ou et al., 2020). Another serine protease TMPRSS2 was found to be crucial for S protein priming in both SARS-CoV-2 and SARS-CoV (Matsuyama et al., 2010; Shulla et al., 2011; Iwata-Yoshikawa et al., 2019; Hoffmann et al., 2020). For SARS-CoV, it is the availability of specific proteases that appears to be the determinant factor to choose whether it enters the host cell via the cell surface or by using the endosomal cathepsin L-mediated pathway for viral entry. So, non-appearance of the host proteases within the cell surface, SARS-CoV invade host cells though a pathway (endosomal pathway) where cathepsin L activates the spike protein, allowing the association of the viras particle and endosome membranes (Simmons et al., 2004; Kam et al., 2009; Chan et al., 2013).
Previous studies have shown that the dual treatment of an inhibitor of TMPRSS2- camostat mesylate and an inhibitor of cathepsin L efficiently blocked host cell entry of SARS-CoV. This competent inhibition could be attributed to the double barrier of entry for SARS-CoV from the surface of a cell and through the endosomal pathway (Kawase et al., 2012). Serine protease inhibitor camostat mesylate was found to block TMPRSS2-mediated priming of spike protein and inhibits COVID-19 infection in lung cells in vitro (Hoffmann et al., 2020). Another TMPRSS2 inhibitor drug nafamostat mesylate was found to inhibit the membrane fusion of MERS-CoV and expected to have similar effects on this virus (Yamamoto et al., 2016; Hoffmann et al., 2020). These observations suggest that this protease inhibitor, camostat mesylate, and a cathepsin inhibitor can be used as antiviral drugs to prevent cathepsin L and TMPRSS2 -mediated SARS-CoV-2 infection.
One problem with using human protease inhibitors as antiviral drugs is that they might affect the normal physiological processes in the human cells, which may lead to further complications or side effects. Therefore, human protease inhibitors may be used in combinatorial therapies with other antiviral drugs which would allow using a less concentration of protease inhibitors to minimize side effects while keeping stronger efficacy. However, no human protease inhibitor has been approved as of now to use in treating viral infections despite having several experimental reports on their effectiveness as antiviral drugs (Steinmetzer and Hardes, 2018).
Viral Protease Inhibitors (Therapeutics)
In coronavirus, chymotrypsin-like protease (3CLpro or Mpro) is the main protease, and along with papain-like protease (PLpro) it processes the polyproteins pp1ab and pp1a (Brierley et al., 1989; Gorbalenya et al., 2006). These two proteases are attractive targets for designing drugs to inhibit cleavage functions and render the virus non-functional (Anand et al., 2003; Yang et al., 2003; Ratia et al., 2008; Hilgenfeld, 2014; Arya et al., 2020; Wu C. et al., 2020). The structures of Mpro from SARS-CoV-2 and SARS-CoV are known. Hence, the designing of drugs to inhibit the protease has been accelerated (Xue et al., 2008; Zhang L. et al., 2020). An α-ketoamide inhibitor has been identified that blocks SARS-CoV-2 Mpro from performing its functions shown in mice (Zhang L. et al., 2020). HIV protease inhibitor drug lopinavir/ritonavir was found to be useful in decreasing viral loads in COVID-19 patients (Lim et al., 2020). However, in clinical trials on COVID-19 patients, the HIV drug was found to be ineffective (Cao et al., 2020). Another HIV protease inhibitor darunavir is also under clinical trials to find out its efficacy in treating COVID-19 (Santos et al., 2019). In vitro studies have shown that several other antiretroviral protease inhibitors (e.g., nelfinavir, etc.) were highly effective in inhibiting coronaviruses (Yamamoto et al., 2004). However, the failure of Kaletra (lopinavir/ritonavir) has shown that protease inhibitors optimized for HIV are unlikely to be effective against SARS-CoV-2 as the proteases expressed by these two viruses are structurally different. Nonetheless, some efficacy against SARS-CoV-2 has been shown by HIV protease inhibitors under in vitro conditions and some of these inhibitors are also under various clinical trials to confirm their effectiveness against COVID-19 (Table 2). However, protease inhibitors specific for HIV protease (e.g., darunavir, etc.) are doubtful to be effective against SARS-CoV-2 protease because of the structural dissimilarities between them.
Replicase Inhibitors
Therapeutics
Another attractive target for drug development is the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), as this is the main molecule for the replication/transcription complex in coronaviruses. The cryo-EM structure of SARS-CoV-2 RdRp (nsp12) has been elucidated recently, along with cofactors nsp7 and nsp8 (Gao Y. et al., 2020). The structure derived using cryo-EM methodology also explained how the drug remdesivir binds to the RdRp (Gao Y. et al., 2020). The nucleotide analog remdesivir has been shown to inhibit RdRp in SARS-CoV (Agostini et al., 2018; Saha et al., 2020a), MERS-CoV (Gordon et al., 2020), and SARS-CoV-2 (Holshue et al., 2020; Wang M. et al., 2020). In a recent study, remdesivir was found to provide benefit to the majority of COVID-19 patients who needed oxygen support (Grein et al., 2020). European Medicines Agency (EMA) has given conditional marketing approval to Veklury (remdesivir) for the therapy of critical COVID-19 patients (12 years of age or higher) with pneumonia and under oxygen support. Remdesivir is the first drug to get the required authorization to use in the EU for the treatment of COVID-19 (Table 2).
Other nucleotide/nucleoside analogs, e.g., sofosbuvir (Gane et al., 2013; Appleby et al., 2015; Ju et al., 2020), and ribavirin (Elfiky, 2020), were also found to be effective in inhibiting RdRp. Favipiravir, which has a structural similarity with nucleoside analogs, found to be effective in COVID-19 clinical trials (Chen C. et al., 2020). Another nucleoside analog galidesivir (BCX-4430) was found effective in several infectious diseases, including Ebola, Zika, etc., and maybe useful in COVID-19, too (Taylor et al., 2016; Eyer et al., 2019). Similar antiviral drugs, triphosphate forms of AZT (3’-azido-3’-deoxythymidine triphosphate), and alovudine (3’-fluoro-3’-deoxythymidine triphosphate) were also predicted to inhibit SARS-CoV-2 RdRp (Ju et al., 2020). The guanosine analog ribavirin not only inhibits viral RdRp by directly interfering with it but also interferes with the RNA capping by inhibiting inosine monophosphate dehydrogenase enzyme to impede guanosine production in the host cell (Graci and Cameron, 2006; Khalili et al., 2020). Interestingly, antiparasitic drug ivermectin was found to suppress SARS-CoV-2 replication in cell culture efficiently (Caly et al., 2020). Ivermectin was predicted to inhibit the maturation of viral proteins by blocking IMPα/β1-mediated nuclear import (Wagstaff et al., 2012; Yang et al., 2020).
Nucleic Acid–Based Solutions
Vaccines
The advantages of nucleic acid-based vaccines are that they can be quickly constructed and can induce strong cell-mediated and humoral immune responses even in the absence of an adjuvant (Du et al., 2009). During the Zika virus outbreak, DNA vaccines were the first to enter clinical trials (Prompetchara et al., 2020). A DNA vaccine is a new and innovative mode of vaccination involved in direct injection of a plasmid encoding the antigens (Shang et al., 2020). Certain advancements like the use of electroporation for delivering the plasmid and use of adjuvant further increases the efficacy by invoking better immune response. Several organizations are working for pre-clinical trials of DNA vaccines against COVID-19 (Liu, 2019) (Table 1). DNA vaccines against COVID-19 mainly encode different forms of the SARS-CoV-2 S protein that was found to stimulate both cellular and humoral immune responses in mice, guinea pigs, and rhesus macaques (Amanat and Krammer, 2020; Smith et al., 2020; Yu et al., 2020). However, there is a risk of integration and mutation of DNA vaccines within the host genome. Being safer, mRNA vaccines stand as a promising alternative to DNA and other conventional vaccine approaches because of its safety and quick development (Liu, 2019). So far, several organizations are working on developing an mRNA-based vaccine for SARS-CoV-2. Small interfering RNA (siRNA) based vaccines are also being developed targeting conserved regions on the SARS-CoV-2 genome, especially 3CLpro, RdRp, and spike protein, to degrade viral mRNAs resulting inhibition of translation (Liu C. et al., 2020).
Therapeutics
Double-stranded RNA drug rintatolimod is now being tested for COVID-19, which stimulates the innate immune system by binding to one of the PRRs named TLR-3 found in the endosomal membrane. Once rintatolimod binds to TLR-3, the host cell gets a signal to produce interferons, which lead to various protective systems against pathogenic viruses or bacteria. Rintatolimod predicted to stimulate RNase L enzyme production, which degrades pathogenic RNAs of viruses (Gowen et al., 2007; Pardi et al., 2018).
Conclusions
There are several new vaccines and novel therapeutic molecules which are currently under development against COVID-19 (Tables 1 and 2). The finding of a safe and attractive target for vaccine development is of utmost importance at this point to prevent further spread of this virus. Unfortunately, the way SARS-CoV-2 is spreading around the world and infected cases increasing exponentially, we may have to witness much bigger devastation before a cure is found. Several promising drug targets have been identified, and several organizations are working relentlessly to develop vaccines against these targets (Table 1). Different available antiviral drugs (repurposed) are being tested for COVID-19 in large clinical trials, as they have shown some positive effects in initial phases (Table 2; Figure 1). Contradictory reports are also started to pouring in against some antiviral therapies targeted at COVID-19, where although initial reports suggested positive effects, later others showed no effect. For example, hydroxychloroquine treatment, along with azithromycin, has shown a significant reduction of viral load in COVID-19 infected patients (Gautret et al., 2020), but subsequent report refutes that claim and showed no benefit in severe COVID-19 patients by this treatment (Molina et al., 2020). Repurposing existing antiviral drugs against COVID-19 has shown some positive effects, but further scientific results are necessary to prove whether these affect COVID-19 treatment, or we are just looking at the placebo effect which can be dangerous for patients.
Recently, some unproven theories are spreading like wildfires, which may also hinder the actual progress on the vaccine development against COVID-19. One example is the use of the BCG vaccine, which is being advocated as a potential cure for COVID-19. Countries, where people have taken the anti-tuberculosis Bacillus Calmette-Guerin (BCG) vaccine, appear to be immune from COVID-19 compared to countries where BCG vaccination is not a norm, as per some recent non-peer-reviewed reports (Hegarty et al., 2020; Miller et al., 2020). Research organizations have already started clinical trials to test the efficacy of the BCG vaccine in COVID-19. It is not clear at this point how and whether BCG vaccination helps in preventing COVID-19 at all; therefore, further research is necessary to find the link between these two.
Several vaccine clinical and pre-clinical trials are currently ongoing (Table 1), and even if some trials finally become successful, a preventive vaccine may not be widely available for at least another 12–18 months. For a vaccine to be successful, much time is needed to conduct proper clinical trials, especially phase III and phase IV trials where the control group is large enough to get a conclusive report (Green, 2020). Therefore, fast-tracking of any clinical trial could be potentially dangerous, and comprehensive safety tests are necessary before a vaccine can be marketed. It applies the same to any repurposed drugs that show positive effects in the initial phases of clinical trials. The catastrophic failure of the respiratory syncytial virus (RSV) vaccine in 1966 showed the importance of a proper clinical trial and advocating for fast-tracking any SARS-CoV-2 clinical trials should be avoided at this stage. The RSV vaccine failed due to the lack of antibody affinity maturation, the possibility of which should be thoroughly checked to avoid a similar situation in COVID-19 (Glezen et al., 1986).
Due to the high genome mutation rates in RNA viruses as the viral RNA polymerase (e.g., influenza virus) or reverse transcriptase (e.g., HIV) lacks proofreading activity, and therefore, it is difficult to make an effective vaccine against RNA viruses (Boutwell et al., 2010; Sanjuán et al., 2010). Although the excessive mutation rate in RNA viruses helps them to adapt quickly to the variable environmental conditions, it also makes them vulnerable because of the accumulation of lethal mutations in the essential genes. Interestingly, in SARS-CoV, the nsp14 protein found to contain an exoribonuclease domain (ExoN) that provides proofreading activity and the deletion of the gene results in a reduction of virulence (Hofer, 2013; Pachetti et al., 2020). This information is important as SARS-CoV-2 also contains a similar gene on its genome, and any proofreading activity would ensure low mutational rates during the synthesis of the viral genome, which would be helpful to design and to develop a vaccine candidate against the SARS-CoV-2 virus.
Coronaviruses are known for a long time and an extensive amount of knowledge has been gathered on SARS-CoV, despite that we still do not have a vaccine against it. We still do not have an effective vaccine against HIV or malaria, for example, although these pathogens are known to us for a long time (Boutwell et al., 2010; Rts, 2015; King, 2019). Challenges posed by these pathogens are far more complex and require an extensive investigation that may take several years to complete. Therefore, extensive safety trials in humans with sizable groups of people are needed even if data from the initial phases are encouraging. Any rush at these stages may be catastrophic if upon vaccination to people who never exposed to the virus develop serious side effects.
Reports from the recent clinical trials of two COVID-19 vaccine candidates have shown promise as they were found to be safe for human use and also induced strong immune response against SARS-CoV-2 (Beyrer et al., 2012; Zhu et al., 2020). The vaccine AZD1222 (ChAdOx1 nCoV-19) developed jointly by Oxford University and AstraZeneca provides double protection against COVID-19 by producing both antibodies and T-cells that directly kill infected cells (Beyrer et al., 2012). Another vaccine (Ad5-nCOV) developed by CanSino Biologics, China, also shown to provide protection against SARS-CoV-2 (Zhu et al., 2020). These reports instill faith that a protective vaccine would be available soon to ease the suffering that the world is facing today because of COVID-19.
The virus has locked up several parts of the world from social and economic activities, and we have no other option but to wait for the development of a vaccine against COVID-19. This situation was envisaged by several scientists earlier, but no one thought we have to witness this disaster in our lifetime. Humanity always prevailed under challenging conditions and the way many research organizations are trying to find a cure one can only hope that we could get a vaccine against COVID-19 sooner than later, but until then social distancing, rigorous testing, and isolation of infected persons in COVID-19 appears to be a potent strategy to contain the spread of the virus.
Author Contributions
Writing—original draft: RS and ARS. Writing—review and editing: MKS, SS, SB, SM, and MB. Revising and supervising and funding acquisition: CC, ARS, and SSL.
Funding
This research was supported by Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A2B4012944 & NRF-2020R1C1C1008694).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Adamas University management for their kind support.
Abbreviations
HCoV, Human Coronavirus; HIV, Human Immunodeficiency Virus; SARS, Severe Acute Respiratory Syndrome; MERS, Middle East Respiratory Syndrome; WHO, World Health Organization; RAS, Renin-Angiotensin System; TLR, Toll-like receptors; STAT, Signal Transducer and Activator of Transcription, ACE2, Angiotensin-converting enzyme 2; AAK1, Adaptor-associated protein kinase 1; JAK-STAT, Janus kinases (JAKs), signal transducer and activator of transcription proteins; ADCC, Antibody-dependent cellular cytotoxicity; RdRp, RNA-dependent RNA polymerase; RSV, Respiratory syncytial virus; PLpro, papain-like protease; PRRs, pattern recognition receptors; ADE, antibody-dependent enhancement; ARDS, Acute Respiratory Distress Syndrome; CCL5, C-C motif Chemokine Ligand 5; BCG, Bacillus Calmette–Guérin; RBD, Receptor-binding domain.
References
Agostini, M. L., Andres, E. L., Sims, A. C., Graham, R. L., Sheahan, T. P., Lu, X., et al. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, e00221–e00218. doi: 10.1128/mBio.00221-00218
Ahn, D.-G., Shin, H.-J., Kim, M.-H., Lee, S., Kim, H.-S., Myoung, J., et al. (2020). Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J. Microbiol. Biotechnol. 30 (3), 313–324. doi: 10.4014/jmb.2003.03011
Amanat, F., Krammer, F. (2020). SARS-CoV-2 vaccines: status report. Immunity 52 (4), 583–589. doi: 10.1016/j.immuni.2020.03.007
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300 (5626), 1763–1767. doi: 10.1126/science.1085658
Appleby, T. C., Perry, J. K., Murakami, E., Barauskas, O., Feng, J., Cho, A., et al. (2015). Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347 (6223), 771–775. doi: 10.1126/science.1259210
Arbabi-Ghahroudi, M. (2017). Camelid single-domain antibodies: Historical perspective and future outlook. Front. Immunol. 8, 1589. doi: 10.3389/fimmu.2017.01589
Arya, R., Das, A., Prashar, V., Kumar, M. (2020). Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. chemRxiv 1–8. doi: 10.26434/chemrxiv.11860011.v2
Assiri, A., McGeer, A., Perl, T. M., Price, C. S., Al Rabeeah, A. A., Cummings, D. A., et al. (2013). Hospital outbreak of Middle East respiratory syndrome coronavirus. New Engl. J. Med. 369 (5), 407–416. doi: 10.1056/NEJMoa1306742
Belhadi, D., Peiffer-Smadja, N., Lescure, F.-X., Yazdanpanah, Y., Mentré, F., Laouénan, C. (2020). A brief review of antiviral drugs evaluated in registered clinical trials for COVID-19. medRxiv 2020.2003.2018.20038190. doi: 10.1101/2020.03.18.20038190
Belouzard, S., Chu, V. C., Whittaker, G. R. (2009). Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. 106 (14), 5871–5876. doi: 10.1073/pnas.0809524106
Beyrer, C., Baral, S. D., Van Griensven, F., Goodreau, S. M., Chariyalertsak, S., Wirtz, A. L., et al. (2012). Global epidemiology of HIV infection in men who have sex with men. Lancet 380 (9839), 367–377. doi: 10.1016/S0140-736(12)60821-6
Bhattacharya, M., Sharma, A. R., Patra, P., Ghosh, P., Sharma, G., Patra, B. C., et al. (2020a). Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 92 (6), 618–631. doi: 10.1002/jmv.25736
Bhattacharya, M., Sharma, A. R., Patra, P., Ghosh, P., Sharma, G., Patra, B. C., et al. (2020b). A SARS-CoV-2 vaccine candidate: In silico cloning and validation. Inf. Med. Unlocked 20, 100394. doi: 10.1016/j.imu.2020.100394
Blaising, J., Polyak, S. J., Pécheur, E.-I. (2014). Arbidol as a broad-spectrum antiviral: an update. Antiviral Res. 107, 84–94. doi: 10.1016/j.antiviral.2014.04.006
Bonavia, A., Zelus, B. D., Wentworth, D. E., Talbot, P. J., Holmes, K. V. (2003). Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77 (4), 2530–2538. doi: 10.1128/jvi.77.4.2530-2538.2003
Bosch, B. J., van der Zee, R., de Haan, C. A., Rottier, P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77 (16), 8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003
Boutwell, C. L., Rolland, M. M., Herbeck, J. T., Mullins, J. I., Allen, T. M. (2010). Viral evolution and escape during acute HIV-1 infection. J. Infect. Dis. 202 (Suppl 2), S309. doi: 10.1086/655653
Brierley, I., Digard, P., Inglis, S. C. (1989). Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57 (4), 537–547. doi: 10.1016/0092-8674(89)90124-4
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., Wagstaff, K. M. (2020). The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 178, 104787. doi: 10.1016/j.antiviral.2020.104787
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al. (2020). A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. New Engl. J. Med. 382 (19), 1787–1799. doi: 10.1056/NEJMoa2001282
Chakraborty, C., Sharma, A., Sharma, G., Bhattacharya, M., And Lee, S. (2020a). SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 24, 4016–4026. doi: 10.26355/eurrev_202004_20871
Chakraborty, C., Sharma, A. R., Bhattacharya, M., Sharma, G., Lee, S.-S. (2020b). The 2019 novel coronavirus disease (COVID-19) pandemic: A zoonotic prospective. Asian Pacific J. Trop. Med. 13 (6), 242. doi: 10.4103/1995-7645.281613
Chakraborty, C., Sharma, A. R., Sharma, G., Saha, R. P., Lee, S.-S. (2020c). Extensive partnership, collaboration, and teamwork is required to stop the COVID-19 outbreak. Arch. Med. Res. S0188-4409 (20), 30849-3. doi: 10.1016/j.arcmed.2020.05.021
Chakraborty, C., Sharma, A. R., Bhattacharya, M., Sharma, G., Lee, S. S., Agoramoorthy, G. (2020d). Consider TLR5 for new therapeutic development against COVID-19. J. Med. Virol. doi: 10.1002/jmv.25997
Chakraborty, C., Sharma, A. R., Bhattacharya, M., Sharma, G., Lee, S. S., Agoramoorthy, G. (2020e). COVID-19: Consider IL6 receptor antagonist for the therapy of cytokine storm syndrome in SARS-CoV-2 infected patients. J. Med. Virol. doi: 10.1002/jmv.26078
Chan, J. F.-W., To, K. K.-W., Tse, H., Jin, D.-Y., Yuen, K.-Y. (2013). Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 21 (10), 544–555. doi: 10.1016/j.tim.2013.05.005
Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., et al. (2020a). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9 (1), 221–236. doi: 10.1080/22221751.2020.1719902
Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., et al. (2020b). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395 (10223), 514–523. doi: 10.1016/S0140-6736(20)30154-9
Chary, M. A., Barbuto, A. F., Izadmehr, S., Hayes, B. D., Burns, M. M. (2020). COVID-19: Therapeutics and their toxicities. J. Med. Toxicol. 16 (3), 10.1007. doi: 10.1007/s13181-020-00777-5
Chen, Z., Bao, L., Chen, C., Zou, T., Xue, Y., Li, F., et al. (2017). Human neutralizing monoclonal antibody inhibition of middle east respiratory syndrome coronavirus replication in the common marmoset. J. Infect. Dis. 215 (12), 1807–1815. doi: 10.1093/infdis/jix209
Chen, C., Huang, J., Cheng, Z., Wu, J., Chen, S., Zhang, Y., et al. (2020). Favipiravir versus Arbidol for COVID-19: a randomized clinical trial. MedRxiv. 1–29. doi: 10.1101/2020.03.17.20037432
Chen, J., Liu, D., Liu, L., Liu, P., Xu, Q., Xia, L., et al. (2020). A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). J. Zhejiang Univ. (Med. Sci.) 49 (1), 0–0. doi: 10.3785/j.issn.1008-9292.2020.03.03
Chen, W.-H., Strych, U., Hotez, P. J., Bottazzi, M. E. (2020). The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 7, 1–4. doi: 10.1007/s40475-020-00201-6
Crotty, S. (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41 (4), 529–542. doi: 10.1016/j.immuni.2014.10.004
Darnell, J. E., Kerr, I. M., Stark, G. R. (1994). Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264 (5164), 1415–1421. doi: 10.1126/science.8197455
de Wilde, A. H., Snijder, E. J., Kikkert, M., van Hemert, M. J. (2017). “Host factors in coronavirus replication” in Roles of Host Gene and Non-coding RNA Expression in Virus Infection (Cham, Switzerland: Springer), 1–42.
Diaz, J. H. (2020). Hypothesis: angiotensin-converting enzyme inhibitors and angiotensin receptor blockers may increase the risk of severe COVID-19. J. Travel Med. 27 (3), (1–2). doi: 10.1093/jtm/taaa041
Du, L., He, Y., Zhou, Y., Liu, S., Zheng, B.-J., Jiang, S. (2009). The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7 (3), 226–236. doi: 10.1038/nrmicro2090
Du, L., Kou, Z., Ma, C., Tao, X., Wang, L., Zhao, G., et al. (2013a). A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PloS One 8 (12), e81587. doi: 10.1371/journal.pone.0081587
Du, L., Zhao, G., Kou, Z., Ma, C., Sun, S., Poon, V. K., et al. (2013b). Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 87 (17), 9939–9942. doi: 10.1128/JVI.01048-13
Duan, K., Liu, B., Li, C., Zhang, H., Yu, T., Qu, J., et al. (2020). Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 117 (17), 9490–9496. doi: 10.1073/pnas.2004168117
Elfiky, A. A. (2020). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 248, 117477. doi: 10.1016/j.lfs.2020.117477
Epelman, S., Tang, W. W., Chen, S. Y., Van Lente, F., Francis, G. S., Sen, S. (2008). Detection of soluble angiotensin-converting enzyme 2 in heart failure: insights into the endogenous counter-regulatory pathway of the renin-angiotensin-aldosterone system. J. Am. Coll. Cardiol. 52 (9), 750–754. doi: 10.1016/j.jacc.2008.02.088
Epelman, S., Shrestha, K., Troughton, R. W., Francis, G. S., Sen, S., Klein, A. L., et al. (2009). Soluble angiotensin-converting enzyme 2 in human heart failure: relation with myocardial function and clinical outcomes. J. Cardiac Failure 15 (7), 565–571. doi: 10.1016/j.cardfail.2009.01.014
Eyer, L., Nougairède, A., Uhlířová, M., Driouich, J.-S., Zouharová, D., Valdés, J. J., et al. (2019). An E460D substitution in the NS5 protein of tick-borne encephalitis virus confers resistance to the inhibitor Galidesivir (BCX4430) and also attenuates the virus for mice. J. Virol. 93 (16), e00367–e00319. doi: 10.1128/JVI.00367-19
Favalli, E. G., Biggioggero, M., Maioli, G., Caporali, R. (2020). Baricitinib for COVID-19: a suitable treatment? Lancet Infect. Dis. doi: 10.1016/S1473-3099(20)30262-0
Fehr, A. R., Perlman, S. (2015). “Coronaviruses: an overview of their replication and pathogenesis” in Coronaviruses (New York, NY: Springer), 1–23.
Gane, E. J., Stedman, C. A., Hyland, R. H., Ding, X., Svarovskaia, E., Symonds, W. T., et al. (2013). Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. New Engl. J. Med. 368 (1), 34–44. doi: 10.1056/NEJMoa1208953
Gao, Q., Bao, L., Mao, H., Wang, L., Xu, K., Li, Y., et al. (2020). Rapid development of an inactivated vaccine for SARS-CoV-2. BioRxiv 1–29. doi: 10.1101/2020.04.17.046375
Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., et al. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368 (6492), 779–782. doi: 10.1126/science.abb7498
Gautret, P., Lagier, J.-C., Parola, P., Meddeb, L., Mailhe, M., Doudier, B., et al. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 56 (1), 105949. doi: 10.1016/j.ijantimicag.2020.105949
Ge, X.-Y., Li, J.-L., Yang, X.-L., Chmura, A. A., Zhu, G., Epstein, J. H., et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503 (7477), 535–538. doi: 10.1038/nature12711
Glebov, O. O. (2020). Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. doi: 10.1111/febs.15369
Glezen, W. P., Taber, L. H., Frank, A. L., Kasel, J. A. (1986). Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Children 140 (6), 543–546. doi: 10.1001/archpedi.1986.02140200053026
Godet, M., Grosclaude, J., Delmas, B., Laude, H. (1994). Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68 (12), 8008–8016. doi: 10.1128/JVI.68.12.8008-8016.1994
González-Navajas, J. M., Lee, J., David, M., Raz, E. (2012). Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12 (2), 125–135. doi: 10.1038/nri3133
Gorbalenya, A. E., Enjuanes, L., Ziebuhr, J., Snijder, E. J. (2006). Nidovirales: evolving the largest RNA virus genome. Virus Res. 117 (1), 17–37. doi: 10.1016/j.virusres.2006.01.017
Gorbalenya, A. E. (2020). Severe acute respiratory syndrome-related coronavirus–The species and its viruses, a statement of the Coronavirus Study Group. BioRxiv 117 (1), 17–37. doi: 10.1101/2020.02.07.937862
Gordon, C. J., Tchesnokov, E. P., Feng, J. Y., Porter, D. P., Götte, M. (2020). The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295 (15), 4773–4779. doi: 10.1074/jbc.AC120.013056
Gowen, B. B., Wong, M.-H., Jung, K.-H., Sanders, A. B., Mitchell, W. M., Alexopoulou, L., et al. (2007). TLR3 is essential for the induction of protective immunity against Punta Toro Virus infection by the double-stranded RNA (dsRNA), poly (I: C12U), but not Poly (I: C): differential recognition of synthetic dsRNA molecules. J. Immunol. 178 (8), 5200–5208. doi: 10.4049/jimmunol.178.8.5200
Graci, J. D., Cameron, C. E. (2006). Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 16 (1), 37–48. doi: 10.1002/rmv.483
Green, D. R. (2020). SARS-CoV2 vaccines: Slow is fast (Washington, D.C., United States: American Association for the Advancement of Science).
Grein, J., Ohmagari, N., Shin, D., Diaz, G., Asperges, E., Castagna, A., et al. (2020). Compassionate Use of Remdesivir for Patients with Severe Covid-19. N Engl. J. Med. 382 (24), 2327–2336. doi: 10.1056/NEJMoa2007016
Gu, H., Xie, Z., Li, T., Zhang, S., Lai, C., Zhu, P., et al. (2016). Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 6, 19840. doi: 10.1038/srep19840
Halstead, S. B. (2017). Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 35 (47), 6355–6358. doi: 10.1016/j.vaccine.2017.09.089
Hashimoto, T., Perlot, T., Rehman, A., Trichereau, J., Ishiguro, H., Paolino, M., et al. (2012). ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487 (7408), 477–481. doi: 10.1038/nature11228
He, Y., Zhou, Y., Siddiqui, P., Jiang, S. (2004). Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325 (2), 445–452. doi: 10.1016/j.bbrc.2004.10.052
He, Y., Zhu, Q., Liu, S., Zhou, Y., Yang, B., Li, J., et al. (2005). Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology 334 (1), 74–82. doi: 10.1016/j.virol.2005.01.034
Hegarty, P. K., Kamat, A. M., Zafirakis, H., Dinardo, A. (2020). BCG vaccination may be protective against Covid-19. Preprint doi: 10.13140/RG.2.2.35948.10880
Hilgenfeld, R. (2014). From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 281 (18), 4085–4096. doi: 10.1111/febs.12936
Hofer, U. (2013). Viral evolution: fooling the coronavirus proofreading machinery. Nat. Rev. Microbiol. 11 (10), 662. doi: 10.1038/nrmicro3125
Hoffmann, M., Hofmann-Winkler, H., Pöhlmann, S. (2018). “Priming time: How cellular proteases arm coronavirus spike proteins,” in Activation of Viruses by Host Proteases (Cham, Switzerland: Springer), 71–98.
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 (2), 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Hofmann, H., Pyrc, K., van der Hoek, L., Geier, M., Berkhout, B., Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. 102 (22), 7988–7993. doi: 10.1073/pnas.0409465102
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. New Engl. J. Med. 382 (10), 929–936. doi: 10.1056/NEJMoa2001191
Huang, X., Dong, W., Milewska, A., Golda, A., Qi, Y., Zhu, Q. K., et al. (2015). Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 89 (14), 7202–7213. doi: 10.1128/JVI.00854-15
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395 (10223), 497–506. doi: 10.1016/S0140-6736(20)30183-5
Iwata-Yoshikawa, N., Okamura, T., Shimizu, Y., Hasegawa, H., Takeda, M., Nagata, N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 93 (6), e01815–e01818. doi: 10.1128/JVI.01815-18
Izaguirre, G. (2019). The Proteolytic Regulation of Virus Cell Entry by Furin and Other Proprotein Convertases. Viruses 11 (9), 837. doi: 10.3390/v11090837
Jiang, S., He, Y., Liu, S. (2005). SARS vaccine development. Emerg. Infect. Dis. 11 (7), 1016–1020. doi: 10.3201/1107.050219
Jiang, S., Bottazzi, M. E., Du, L., Lustigman, S., Tseng, C.-T. K., Curti, E., et al. (2012). Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev. Vacc. 11 (12), 1405–1413. doi: 10.1586/erv.12.126
Jiang, F., Yang, J., Zhang, Y., Dong, M., Wang, S., Zhang, Q., et al. (2014). Angiotensin-converting enzyme 2 and angiotensin 1–7: novel therapeutic targets. Nat. Rev. Cardiol. 11 (7), 413. doi: 10.1038/nrcardio.2014.59
Jiang, S., Hillyer, C., Du, L. (2020). Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 41 (5), 355–359. doi: 10.1016/j.it.2020.03.007
Ju, J., Li, X., Kumar, S., Jockusch, S., Chien, M., Tao, C., et al. (2020). Nucleotide Analogues as Inhibitors of SARS-CoV Polymerase. BioRxiv 1–8. doi: 10.1101/2020.03.12.989186
Kam, Y. W., Okumura, Y., Kido, H., Ng, L. F., Bruzzone, R., Altmeyer, R. (2009). Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PloS One 4 (11), e7870. doi: 10.1371/journal.pone.0007870
Kamp, T. J., Hamdan, M. H., January, C. T. (2020). Chloroquine or Hydroxychloroquine for COVID-19: Is Cardiotoxicity a Concern? J. Am. Heart Assoc. 9 (12), e016887. doi: 10.1161/JAHA.120.016887
Kane, M., Zang, T. M., Rihn, S. J., Zhang, F., Kueck, T., Alim, M., et al. (2016). Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 20 (3), 392–405. doi: 10.1016/j.chom.2016.08.005
Kang, T. H., Jung, S. T. (2019). Boosting therapeutic potency of antibodies by taming Fc domain functions. Exp. Mol. Med. 51 (11), 1–9. doi: 10.1038/s12276-019-0345-9
Kang, S., Yang, M., Hong, Z., Zhang, L., Huang, Z., Chen, X., et al. (2020). Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. BioRxiv 2020, 2003.2006.977876. doi: 10.1101/2020.03.06.977876
Katze, M. G., He, Y., Gale, M. (2002). Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2 (9), 675–687. doi: 10.1038/nri888
Kawase, M., Shirato, K., van der Hoek, L., Taguchi, F., Matsuyama, S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86 (12), 6537–6545. doi: 10.1128/JVI.00094-12
Keller, M., Kitsis, E., Arora, S., Chen, J.-T., Agarwal, S., Ross, M., et al. (2020). Effect of Systemic Glucocorticoids on Mortality or Mechanical Ventilation in Patients With COVID-19. J. Hosp. Med. 8, 489–493. doi: 10.12788/jhm.3497
Khalili, J. S., Zhu, H., Mak, A., Yan, Y., Zhu, Y. (2020). Novel coronavirus treatment with ribavirin: Groundwork for evaluation concerning COVID-19. J. Med. Virol. 92 (7), 740–746. doi: 10.1002/jmv.25798
Khan, N., Gowthaman, U., Pahari, S., Agrewala, J. N. (2012). Manipulation of costimulatory molecules by intracellular pathogens: veni, vidi, vici!! PloS Pathog. 8 (6), e1002676. doi: 10.1371/journal.ppat.1002676
King, A. (2019). Building a better malaria vaccine. Nature 575 (7784), S51. doi: 10.1038/d41586-019-03639-5
Klimpel, G. R. (1996). “Immune defenses,” in Medical Microbiology, 4th edition (Texas, United States: University of Texas Medical Branch at Galveston).
Kopecky-Bromberg, S. A., Martínez-Sobrido, L., Frieman, M., Baric, R. A., Palese, P. (2007). Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81 (2), 548–557. doi: 10.1128/JVI.01782-06
Kraft, C. S., Hewlett, A. L., Koepsell, S., Winkler, A. M., Kratochvil, C. J., Larson, L., et al. (2015). The use of TKM-100802 and Convalescent Plasma in 2 patients with ebola virus disease in the United States. Clin. Infect. Dis. 61 (4), 496–502. doi: 10.1093/cid/civ334
Krempl, C., Schultze, B., Herrler, G. (1995). “Analysis of cellular receptors for human coronavirus OC43,” in Corona-and Related Viruses (Boston, MA: Springer), 371–374.
Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., et al. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 11 (8), 875–879. doi: 10.1038/nm1267
Kubo, H., Yamada, Y. K., Taguchi, F. (1994). Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68 (9), 5403–5410. doi: 10.1128/JVI.68.9.5403-5410.1994
Lamb, Y. N., Deeks, E. D. (2018). Sarilumab: a review in moderate to severe rheumatoid arthritis. Drugs 78 (9), 929–940. doi: 10.1007/s40265-018-0929-z
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581 (7807), 1–9. doi: 10.1038/s41586-020-2180-5
Lauer, S. A., Grantz, K. H., Bi, Q., Jones, F. K., Zheng, Q., Meredith, H. R., et al. (2020). The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Internal Med. 172 (9), 577–582. doi: 10.7326/M20-0504
Lee, N., Chan, K. A., Hui, D. S., Ng, E. K., Wu, A., Chiu, R. W., et al. (2004). Effects of early corticosteroid treatment on plasma SARS-associated Coronavirus RNA concentrations in adult patients. J. Clin. Virol. 31 (4), 304–309. doi: 10.1016/j.jcv.2004.07.006
Levy, D. E., Darnell, J. (2002). Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3 (9), 651–662. doi: 10.1038/nrm909
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426 (6965), 450–454. doi: 10.1038/nature02145
Li, Z., Palaniyandi, S., Zeng, R., Tuo, W., Roopenian, D. C., Zhu, X. (2011). Transfer of IgG in the female genital tract by MHC class I-related neonatal Fc receptor (FcRn) confers protective immunity to vaginal infection. Proc. Natl. Acad. Sci. 108 (11), 4388–4393. doi: 10.1073/pnas.1012861108
Li, G., Hu, R., Zhang, X. (2020). Antihypertensive treatment with ACEI/ARB of patients with COVID-19 complicated by hypertension. Hypertension Res. 43 (6), 1–3. doi: 10.1038/s41440-020-0433-1
Lim, J., Jeon, S., Shin, H.-Y., Kim, M. J., Seong, Y. M., Lee, W. J., et al. (2020). Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 35 (6), 1–6. doi: 10.3346/jkms.2020.35.e79
Liu, Y., Zhang, Z., Zhao, X., Yu, R., Zhang, X., Wu, S., et al. (2014). Enterovirus 71 inhibits cellular type I interferon signaling by downregulating JAK1 protein expression. Viral Immunol. 27 (6), 267–276. doi: 10.1089/vim.2013.0127
Liu, L., Wei, Q., Lin, Q., Fang, J., Wang, H., Kwok, H., et al. (2019). Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4 (4), 1–20. doi: 10.1172/jci.insight.123158
Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., et al. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases (Washington, D.C., United States: ACS Publications).
Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., et al. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery 6, 16. doi: 10.1038/s41421-020-0156-0
Liu, Y., Gayle, A. A., Wilder-Smith, A., Rocklöv, J. (2020). The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 27 (2), taaa021. doi: 10.1093/jtm/taaa021
Liu, Y. J. (2005). IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23, 275–306. doi: 10.1146/annurev.immunol.23.021704.115633
Liu, M. A. (2019). A comparison of plasmid DNA and mrna as vaccine technologies. Vaccines 7 (2), 37. doi: 10.3390/vaccines7020037
Lu, X., Pan, J. A., Tao, J., Guo, D. (2011). SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42 (1), 37–45. doi: 10.1007/s11262-010-0544-x
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 (10224), 565–574. doi: 10.1016/S0140-6736(20)30251-8
Luo, F., Liao, F.-L., Wang, H., Tang, H.-B., Yang, Z.-Q., Hou, W. (2018). Evaluation of antibody-dependent enhancement of SARS-CoV infection in rhesus macaques immunized with an inactivated SARS-CoV vaccine. Virol. Sin. 33 (2), 201–204. doi: 10.1007/s12250-018-0009-2
Luo, P., Liu, Y., Qiu, L., Liu, X., Liu, D., Li, J. (2020). Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 92 (7), 814–818. doi: 10.1002/jmv.25801
Magro, G. (2020). SARS-CoV-2 and COVID-19: is interleukin-6 (IL-6) the’culprit lesion’of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine: X 2 (2), 100029. doi: 10.1016/j.cytox.2020.100029
Mahallawi, W. H., Khabour, O. F., Zhang, Q., Makhdoum, H. M., Suliman, B. A. (2018). MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 104, 8–13. doi: 10.1016/j.cyto.2018.01.025
Marano, G., Vaglio, S., Pupella, S., Facco, G., Catalano, L., Liumbruno, G. M., et al. (2016). Convalescent plasma: new evidence for an old therapeutic tool? Blood Transfus. 14 (2), 152–157. doi: 10.2450/2015.0131-15
Martinez, M. A. (2020). Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob. Agents Chemother. 64 (5), e00399–20. doi: 10.1128/AAC.00399-20
Matsuyama, S., Nagata, N., Shirato, K., Kawase, M., Takeda, M., Taguchi, F. (2010). Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84 (24), 12658–12664. doi: 10.1128/JVI.01542-10
Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K. J., et al. (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14 (8), 1435–1455. doi: 10.1080/15548627.2018.1474314
Miller, A., Reandelar, M. J., Fasciglione, K., Roumenova, V., Li, Y., Otazu, G. H. (2020). Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. MedRxiv 1–9. doi: 10.1101/2020.03.24.20042937
Millet, J. K., Whittaker, G. R. (2014). Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. 111 (42), 15214–15219. doi: 10.1073/pnas.1407087111
Mire, C. E., Geisbert, J. B., Agans, K. N., Thi, E. P., Lee, A. C., Fenton, K. A., et al. (2016). Passive Immunotherapy: Assessment of Convalescent Serum Against Ebola Virus Makona Infection in Nonhuman Primates. J. Infect. Dis. 214 (suppl 3), S367–S374. doi: 10.1093/infdis/jiw333
Molina, J. M., Delaugerre, C., Goff, J., Mela-Lima, B., Ponscarme, D., Goldwirt, L., et al. (2020). No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 50 (4), 30085–30088. doi: 10.1016/j.medmal.2020.03.006
Nan, Y., Wu, C., Zhang, Y.-J. (2018). Interferon independent non-canonical STAT Activation and virus induced inflammation. Viruses 10 (4), 196. doi: 10.3390/v10040196
Neuman, B. W., Kiss, G., Kunding, A. H., Bhella, D., Baksh, M. F., Connelly, S., et al. (2011). A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174 (1), 11–22. doi: 10.1016/j.jsb.2010.11.021
Nicholls, J. M., Poon, L. L., Lee, K. C., Ng, W. F., Lai, S. T., Leung, C. Y., et al. (2003). Lung pathology of fatal severe acute respiratory syndrome. Lancet 361 (9371), 1773–1778. doi: 10.1016/s0140-6736(03)13413-7
Niu, P., Zhang, S., Zhou, P., Huang, B., Deng, Y., Qin, K., et al. (2018a). Ultrapotent human neutralizing antibody repertoires against Middle East respiratory syndrome coronavirus from a recovered patient. J. Infect. Dis. 218 (8), 1249–1260. doi: 10.1093/infdis/jiy311
Niu, P., Zhao, G., Deng, Y., Sun, S., Wang, W., Zhou, Y., et al. (2018b). A novel human mAb (MERS-GD27) provides prophylactic and postexposure efficacy in MERS-CoV susceptible mice. Sci. China Life Sci. 61 (10), 1280–1282. doi: 10.1007/s11427-018-9343-8
Okumura, A., Pitha, P. M., Yoshimura, A., Harty, R. N. (2010). Interaction between Ebola virus glycoprotein and host toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J. Virol. 84 (1), 27–33. doi: 10.1128/JVI.01462-09
Ortiz-Pérez, J. T., Riera, M., Bosch, X., De Caralt, T. M., Perea, R. J., Pascual, J., et al. (2013). Role of circulating angiotensin converting enzyme 2 in left ventricular remodeling following myocardial infarction: a prospective controlled study. PloS One 8 (4), e61695. doi: 10.1371/journal.pone.0061695
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11 (1), 1–12. doi: 10.1038/s41467-020-15562-9
Pachetti, M., Marini, B., Benedetti, F., Giudici, F., Mauro, E., Storici, P., et al. (2020). Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Trans. Med. 18, 1–9. doi: 10.1186/s12967-020-02344-6
Pardi, N., Hogan, M. J., Porter, F. W., Weissman, D. (2018). mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discovery 17 (4), 261. doi: 10.1038/nrd.2017.243
Patterson, B. K., Seethamraju, H., Dhody, K., Corley, M. J., Kazempour, K., Lalezari, J. P., et al. (2020). Disruption of the CCL5/RANTES-CCR5 Pathway Restores Immune Homeostasis and Reduces Plasma Viral Load in Critical COVID-19. MedRxiv 1–24. doi: 10.1101/2020.05.02.20084673
Peiris, J., Guan, Y., Yuen, K. (2004). Severe acute respiratory syndrome. Nat. Med. 10 (12), S88–S97. doi: 10.1038/nm1143
Prompetchara, E., Ketloy, C., Palaga, T. (2020). Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 38 (1), 1–9. doi: 10.12932/AP-200220-0772
Pugach, P., Ketas, T. J., Michael, E., Moore, J. P. (2008). Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology 377 (2), 401–407. doi: 10.1016/j.virol.2008.04.032
Qu, D., Zheng, B., Yao, X., Guan, Y., Yuan, Z.-H., Zhong, N.-S., et al. (2005). Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine 23 (7), 924–931. doi: 10.1016/j.vaccine.2004.07.031
Rabi, F. A., Al Zoubi, M. S., Kasasbeh, G. A., Salameh, D. M., Al-Nasser, A. D. (2020). SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. Pathogens 9 (3), 1–14. doi: 10.3390/pathogens9030231
Ragab, D., Salah Eldin, H., Taeimah, M., Khattab, R., Salem, R. (2020). The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 11, 1446. doi: 10.3389/fimmu.2020.01446
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495 (7440), 251–254. doi: 10.1038/nature12005
Ratia, K., Pegan, S., Takayama, J., Sleeman, K., Coughlin, M., Baliji, S., et al. (2008). A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. 105 (42), 16119–16124. doi: 10.1073/pnas.0805240105
Richardson, P., Griffin, I., Tucker, C., Smith, D., Oechsle, O., Phelan, A., et al. (2020). Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (Lond. Engl.) 395 (10223), e30. doi: 10.1016/S0140-6736(20)30304-4
Rosendahl Huber, S., van Beek, J., de Jonge, J., Luytjes, W., van Baarle, D. (2014). T cell responses to viral infections–opportunities for peptide vaccination. Front. Immunol. 5, 171. doi: 10.3389/fimmu.2014.00171
Rossignol, J. F. (2016). Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health 9 (3), 227–230. doi: 10.1016/j.jiph.2016.04.001
Rts, S. (2015). Clinical Trials Partnership. Efficacy and safety of RTS, S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet 386 (9988), 31–45. doi: 10.1016/S0140-6736(15)60721-8
Russell, C. D., Millar, J. E., Baillie, J. K. (2020). Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 395 (10223), 473–475. doi: 10.1016/S0140-6736(20)30317-2
Saha, A., Sharma, A. R., Bhattacharya, M., Sharma, G., Lee, S.-S., Chakraborty, C. (2020a). Probable Molecular Mechanism of Remdesivir for the Treatment of COVID-19: Need to Know More. Arch. Med. Res. S0188-4409(20)30699-8. doi: 10.1016/j.arcmed.2020.05.001
Saha, A., Sharma, A. R., Bhattacharya, M., Sharma, G., Lee, S.-S., Chakraborty, C. (2020b). Tocilizumab: A therapeutic option for the treatment of cytokine storm syndrome in COVID-19. Arch. Med. Res. S0188-4409(20)30782-7. doi: 10.1016/j.arcmed.2020.05.009
Saif, L. J. (1993). Coronavirus immunogens. Vet Microbiol. 37 (3-4), 285–297. doi: 10.1016/0378-1135(93)90030-b
Sallard, E., Lescure, F. X., Yazdanpanah, Y., Mentre, F., Peiffer-Smadja, N. (2020). Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 178:104791. doi: 10.1016/j.antiviral.2020.104791
Samuel, C. E. (2001). Antiviral actions of interferons. Clin. Microbiol. Rev. 14 (4), 778–809. doi: 10.1128/CMR.14.4.778-809.2001
Sanjuán, R., Nebot, M. R., Chirico, N., Mansky, L. M., Belshaw, R. (2010). Viral mutation rates. J. Virol. 84 (19), 9733–9748. doi: 10.1128/JVI.00694-10
Santos, J. R., Curran, A., Navarro-Mercade, J., Ampuero, M. F., Pelaez, P., Pérez-Alvarez, N., et al. (2019). Simplification of antiretroviral treatment from darunavir/ritonavir monotherapy to darunavir/cobicistat monotherapy: effectiveness and safety in routine clinical practice. AIDS Res. Hum. Retroviruses 35 (6), 513–518. doi: 10.1089/aid.2018.0178
Schindler, C., Shuai, K., Prezioso, V. R., Darnell, J. E. (1992). Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257 (5071), 809–813. doi: 10.1126/science.1496401
Schmidt, A., Wolff, M. H., Weber, O. (2006). Coronaviruses with Special Emphasis on First Insights Concerning SARS (Basel, Switzerland: Birkhäuser Basel).
Schneider, W. M., Chevillotte, M. D., Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Shakoory, B., Carcillo, J. A., Chatham, W. W., Amdur, R. L., Zhao, H., Dinarello, C. A., et al. (2016). Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of the macrophage activation syndrome: Re-analysis of a prior Phase III trial. Crit. Care Med. 44 (2), 275. doi: 10.1097/CCM.0000000000001402
Shang, W., Yang, Y., Rao, Y., Rao, X. (2020). The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vacc. 5 (1), 1–3. doi: 10.1038/s41541-020-0170-0
Sharma, A., Krause, A., Worgall, S. (2011). Recent developments for Pseudomonas vaccines. Hum. Vacc. 7 (10), 999–1011. doi: 10.4161/hv.7.10.16369
Shulla, A., Heald-Sargent, T., Subramanya, G., Zhao, J., Perlman, S., Gallagher, T. (2011). A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85 (2), 873–882. doi: 10.1128/JVI.02062-10
Simmons, G., Reeves, J. D., Rennekamp, A. J., Amberg, S. M., Piefer, A. J., Bates, P. (2004). Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 101 (12), 4240–4245. doi: 10.1073/pnas.0306446101
Sipulwa, L. A., Ongus, J. R., Coldren, R. L., Bulimo, W. D. (2016). Molecular characterization of human coronaviruses and their circulation dynamics in Keny–2012. Virol. J. 13 (1), 18. doi: 10.1186/s12985-016-0474-x
Smith, T. R., Patel, A., Ramos, S., Elwood, D., Zhu, X., Yan, J., et al. (2020). Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 11 (1), 1–13. doi: 10.1038/s41467-020-16505-0
Spiegel, M., Pichlmair, A., Martínez-Sobrido, L., Cros, J., García-Sastre, A., Haller, O., et al. (2005). Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79 (4), 2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005
Stanifer, M. L., Pervolaraki, K., Boulant, S. (2019). Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 20 (6), 1445. doi: 10.3390/ijms20061445
Steinmetzer, T., Hardes, K. (2018). “The antiviral potential of host protease inhibitors,” in Activation of Viruses by Host Proteases (Cham, Switzerland: Springer), 279–325.
Stockman, L. J., Bellamy, R., Garner, P. (2006). SARS: systematic review of treatment effects. PloS Med. 3 (9), e343. doi: 10.1371/journal.pmed.0030343
Sui, J., Li, W., Murakami, A., Tamin, A., Matthews, L. J., Wong, S. K., et al. (2004). Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. 101 (8), 2536–2541. doi: 10.1073/pnas.0307140101
Takada, A., Kawaoka, Y. (2003). Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev. Med. Virol. 13 (6), 387–398. doi: 10.1002/rmv.405
Takano, T., Yamada, S., Doki, T., Hohdatsu, T. (2019). Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 81 (6), 18–0702. doi: 10.1292/jvms.18-0702
Takeuchi, O., Akira, S. (2009). Innate immunity to virus infection. Immunol. Rev. 227 (1), 75–86. doi: 10.1111/j.1600-065X.2008.00737.x
Taylor, R., Kotian, P., Warren, T., Panchal, R., Bavari, S., Julander, J., et al. (2016). BCX4430–a broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J. Infect. Public Health 9 (3), 220–226. doi: 10.1016/j.jiph.2016.04.002
Te-hui, W. C., Wang, S., Sakhatskyy, P. V., Mboudoudjeck, I., Lawrence, J. M., Huang, S., et al. (2005). Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Virology 334 (1), 134–143. doi: 10.1016/j.virol.2005.01.035
Totura, A. L., Baric, R. S. (2012). SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2 (3), 264–275. doi: 10.1016/j.coviro.2012.04.004
Tsai, M.-H., Pai, L.-M., Lee, C.-K. (2019). Fine-tuning of the type I interferon response by STAT3. Front. Immunol. 10, 1448. doi: 10.3389/fimmu.2019.01448
Uyeki, T. M. (2018). “Oseltamivir treatment of influenza in children” (Arlington, Virginia, United States: Oxford University Press US).
Vangelista, L., Vento, S. (2018). The expanding therapeutic perspective of CCR5 blockade. Front. Immunol. 8, 1981. doi: 10.3389/fimmu.2017.01981
Villalaín, J. (2010). Membranotropic effects of arbidol, a broad anti-viral molecule, on phospholipid model membranes. J. Phys. Chem. B 114 (25), 8544–8554. doi: 10.1021/jp102619w
Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., et al. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2, 69. doi: 10.1186/1743-422X-2-69
Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443 (3), 851–856. doi: 10.1042/BJ20120150
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181 (2), 281–292.e6. doi: 10.1016/j.cell.2020.02.058
Wan, Y., Shang, J., Graham, R., Baric, R. S., Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94 (7), e00127–20. doi: 10.1128/JVI.00127-20
Wang, X., Mathieu, M., Brezski, R. J. (2018). IgG Fc engineering to modulate antibody effector functions. Protein Cell 9 (1), 63–73. doi: 10.1007/s13238-017-0473-8
Wang, C., Horby, P. W., Hayden, F. G., Gao, G. F. (2020a). A novel coronavirus outbreak of global health concern. Lancet 395 (10223), 470–473. doi: 10.1016/S0140-6736(20)30185-9
Wang, C., Li, W., Drabek, D., Okba, N. M. A., van Haperen, R., Osterhaus, A. D. M. E., et al. (2020b). A human monoclonal antibody blocking SARS-CoV-2 infection. BioRxiv 2020.2003.2011.987958, 1–2. doi: 10.1101/2020.03.11.987958
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama 323 (11), 1061–1069. doi: 10.1001/jama.2020.1585
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronaviru-nCoV) in vitro. Cell Res. 30 (3), 269–271. doi: 10.1038/s41422-020-0282-0
Wang, T., Du, Z., Zhu, F., Cao, Z., An, Y., Gao, Y., et al. (2020). Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 395 (10228), e52. doi: 10.1016/S0140-6736(20)30558-4
WHO (2020). WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020. (Geneva: World Health Organization).
Wong, S. K., Li, W., Moore, M. J., Choe, H., Farzan, M. (2004). A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279 (5), 3197–3201. doi: 10.1074/jbc.C300520200
World Health Organization (2005). “Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV)”.
World Health Organization (2020). Naming the coronavirus disease (COVID-19) and the virus that causes it. World Health Organization.
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (6483), 1260–1263. doi: 10.1126/science.abb2507
Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27 (3), 0325–328. doi: 10.1016/j.chom.2020.02.001
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 10 (5), 766–788. doi: 10.1016/j.apsb.2020.02.008
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579 (7798), 265–269. doi: 10.1038/s41586-020-2008-3
Xiong, S., Wang, Y.-F., Zhang, M.-Y., Liu, X.-J., Zhang, C.-H., Liu, S.-S., et al. (2004). Immunogenicity of SARS inactivated vaccine in BALB/c mice. Immunol. Lett. 95 (2), 139–143. doi: 10.1016/j.imlet.2004.06.014
Xu, H., Zhong, L., Deng, J., Peng, J., Dan, H., Zeng, X., et al. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 12 (1), 1–5. doi: 10.1038/s41368-020-0074-x
Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., et al. (2008). Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82 (5), 2515–2527. doi: 10.1128/JVI.02114-07
Yamada, M., Hayashi, H., Yuuki, M., Matsushima, N., Yuan, B., Takagi, N. (2018). Furin inhibitor protects against neuronal cell death induced by activated NMDA receptors. Sci. Rep. 8 (1), 1–9. doi: 10.1038/s41598-018-23567-0
Yamamoto, N., Yang, R., Yoshinaka, Y., Amari, S., Nakano, T., Cinatl, J., et al. (2004). HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 318 (3), 719–725. doi: 10.1016/j.bbrc.2004.04.083
Yamamoto, M., Matsuyama, S., Li, X., Takeda, M., Kawaguchi, Y., Inoue, J. I., et al. (2016). Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrob. Agents Chemother. 60 (11), 6532–6539. doi: 10.1128/AAC.01043-16
Yamey, G., Schäferhoff, M., Hatchett, R., Pate, M., Zhao, F., McDade, K. K. (2020). Ensuring global access to COVID-19 vaccines. Lancet 395 (10234), 1405–1406. doi: 10.1016/S0140-6736(20)30763-7
Yang, N., Shen, H.-M. (2020). Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 16 (10), 1724. doi: 10.7150/ijbs.45498
Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., et al. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 100 (23), 13190–13195. doi: 10.1073/pnas.1835675100
Yang, S. N. Y., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A., et al. (2020). The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 177, S0140-6736(20)31605-6.–104760. doi: 10.1016/j.antiviral.2020.104760
Ye, Q., Wang, B., Mao, J. (2020). The pathogenesis and treatment of theCytokine Storm’in COVID-19. J. Infect. 80 (6), 607–613. doi: 10.1016/j.jinf.2020.03.037
Yeager, C. L., Ashmun, R. A., Williams, R. K., Cardellichio, C. B., Shapiro, L. H., Look, A. T., et al. (1992). Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357 (6377), 420–422. doi: 10.1038/357420a0
Yu, J., Tostanoski, L. H., Peter, L., Mercado, N. B., McMahan, K., Mahrokhian, S. H., et al. (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. doi: 10.1126/science.abc6284
Yuan, M., Wu, N. C., Zhu, X., Lee, C.-C. D., So, R. T., Lv, H., et al. (2020). A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 368 (6491), 630–633. doi: 10.1126/science.abb7269
Zanoni, I., Granucci, F., Broggi, A. (2017). Interferon (IFN)-λ takes the helm: immunomodulatory roles of type III IFNs. Front. Immunol. 8, 1661. doi: 10.3389/fimmu.2017.01661
Zhang, M.-Y., Wang, Y., Mankowski, M. K., Ptak, R. G., Dimitrov, D. S. (2009). Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: possible role of the prolonged half-life of the immunogen. Vaccine 27 (6), 857–863. doi: 10.1016/j.vaccine.2008.11.083
Zhang, C., Wu, Z., Li, J.-W., Zhao, H., Wang, G.-Q. (2020). The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents 55 (5), 105954. doi: 10.1016/j.ijantimicag.2020.105954
Zhang, G., Pomplun, S., Loftis, A. R., Loas, A., Pentelute, B. L. (2020). The first-in-class peptide binder to the SARS-CoV-2 spike protein. BioRxiv 1–15. 2020.2003.2019.999318. doi: 10.1101/2020.03.19.999318
Zhang, H., Penninger, J. M., Li, Y., Zhong, N., Slutsky, A. S. (2020a). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46 (4), 1–5. doi: 10.1007/s00134-020-05985-9
Zhang, H., Penninger, J. M., Li, Y., Zhong, N., Slutsky, A. S. (2020b). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46 (4), 586–590. doi: 10.1007/s00134-020-05985-9
Zhang, J., Zeng, H., Gu, J., Li, H., Zheng, L., Zou, Q. (2020). Progress and Prospects on Vaccine Development against SARS-CoV-2. Vaccines 8 (2), 153. doi: 10.3390/vaccines8020153
Zhang, J. N., Wang, W. J., Peng, B., Peng, W., Zhang, Y. S., Wang, Y. L., et al. (2020). Potential of Arbidol for Post-exposure Prophylaxis of COVID-19 Transmission: A Preliminary Report of a Retrospective Cohort Study. Curr. Med. Sci. 40 (3), 480–485. doi: 10.1007/s11596-020-2203-3
Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368 (6489), 409–412. doi: 10.1126/science.abb3405
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (7798), 270–273. doi: 10.1038/s41586-020-2012-7
Zhu, F.-C., Guan, X.-H., Li, Y.-H., Huang, J.-Y., Jiang, T., Hou, L.-H., et al. (2020). Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet S0140-6736(20)31605-6. doi: 10.1016/S0140-6736(20)31605-6
Keywords: coronavirus, vaccine development, SARS-CoV-2, repurposed drug, antiviral treatment, COVID-19
Citation: Saha RP, Sharma AR, Singh MK, Samanta S, Bhakta S, Mandal S, Bhattacharya M, Lee S-S and Chakraborty C (2020) Repurposing Drugs, Ongoing Vaccine, and New Therapeutic Development Initiatives Against COVID-19. Front. Pharmacol. 11:1258. doi: 10.3389/fphar.2020.01258
Received: 24 May 2020; Accepted: 30 July 2020;
Published: 19 August 2020.
Edited by:
Rosario Pignatello, University of Catania, ItalyReviewed by:
Mariusz Skwarczynski, The University of Queensland, AustraliaLucia Gozzo, Gaspare Rodolico Hospital, Italy
Copyright © 2020 Saha, Sharma, Singh, Samanta, Bhakta, Mandal, Bhattacharya, Lee and Chakraborty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rudra P. Saha, cnVkcmFwcmFzYWQuc2FoYUBhZGFtYXN1bml2ZXJzaXR5LmFjLmlu; cnVkcmFzYWhhQGdtYWlsLmNvbQ==; Sang-Soo Lee, MTIzc3NsZWVAZ21haWwuY29t; Chiranjib Chakraborty, ZHJjaGlyYW5qaWJAeWFob28uY29t
†These authors have contributed equally to this work
 Rudra P. Saha
Rudra P. Saha Ashish Ranjan Sharma
Ashish Ranjan Sharma Manoj K. Singh
Manoj K. Singh Saikat Samanta
Saikat Samanta Swarnav Bhakta
Swarnav Bhakta Snehasish Mandal1
Snehasish Mandal1 Manojit Bhattacharya
Manojit Bhattacharya Sang-Soo Lee
Sang-Soo Lee Chiranjib Chakraborty
Chiranjib Chakraborty