- National-Local Joint Engineering Laboratory of Druggability and New Drug Evaluation, National Engineering Research Center for New Drug and Druggability (cultivation), Guangdong Provincial Key Laboratory of New Drug Design and Evaluation, School of Pharmaceutical Sciences, Sun Yat-Sen University, Guangzhou, China
In December 2019, a pneumonia outbreak termed COVID-19 by WHO occurred in Wuhan, Hubei province, China (Chan et al., 2020). By 30 May 2020, the pandemic had caused over 6,000,000 global laboratory-confirmed infections and 364,459 fatal cases. COVID-19 is caused by SARS-CoV-2, a single-stranded positive sense RNA virus of betacoronavirus, and can result in severe respiratory diseases, such as acute respiratory distress syndrome (ARDS) (Zhu et al., 2020). The enormous economic and social impact of the SARS-CoV-2 infection makes it paramount to develop viable vaccines and antiviral drugs.
SARS-CoV-2 is an enveloped unsegmented single-stranded positive sense RNA virus, derived from the Coronaviridae family Nidovirales. We performed multiple sequence alignments of S proteins from SARS-CoV-2 and other human coronavirus, including SARS, OC43, MERS, NL63, 229E, and HKU1, using RaTG13, a Bat-SARS-like coronavirus, as a control and found that S protein of SARS-CoV-2 maintains the highest homology (96%) sequence with RaTG13, and 75% of homology sequence with SARS-CoV, but is significantly different from other human coronavirus (Figures 1A, B). The full-length genome sequences of SARS-CoV-2 share a 79.5% sequence identity to SARS coronavirus (SARS-CoV). It was reported that, similar to SARS-CoV, the S glycoprotein of SARS-CoV-2 binds the cellular receptor angiotensin-converting enzyme 2 (ACE2) in hosts to mediate fusion of the viral and cellular membranes (Lu et al., 2020; Wrapp et al., 2020)). The binding affinity between SSARS-CoV-2 protein and ACE was suggested to be 10 to 60 nM, 10 to 20-fold higher than SSARS-CoV (Lan et al., 2020; Yan R. et al., 2020), but the underlying mechanism remains inconclusive. Variations of crucial residues in the SSARS-CoV-2 protein and their counterpart receptors may contribute to the high transmission efficiency. SARS-CoV-2 produced an evolutionary mutation of K403R compared with SSARS-CoV-2 protein, forming an adjacent RGD motif (Arg-Gly-AsP) at the interaction surface (Luan et al., 2020; Sigrist et al., 2020; Tresoldi et al., 2020; Yan S. et al., 2020). Point mutation of the second codon of “ACA” into “AGA” at site 403 creates an Arg from Thr compared with the sequence in RaTG13, while the counterpart in SARS is “AAG” coding for Lys (Figures 1C, D).
Integrin is a transmembrane heterodimeric protein comprising of α and β subunits, many of which recognize the RGD motif displayed on the exposed loops of viral capsid proteins. The RGD motif is the cell attachment site, which can recognize integrin of various epithelial cells to promote cell adhesion and virus internalization by activating transducing pathways involving phosphatidylinositol-3 kinase (PI-3K) or mitogen-activated protein kinase (MAPK) (Ruoslahti, 1996). Meanwhile, ACE2 was previously found to bind with integrin, regulates the cardiac remodeling signaling pathway, and affects cell survival and proliferation (Lin et al., 2004; Clarke et al., 2012). Integrin β1 can regulate CCL2 levels in alveolar epithelial cells, recruiting monocytes to induce an inflammatory response (Plosa et al., 2020), suggesting that the RGD sequence of the SSARS-CoV-2 protein may be recognized by integrin in alveolar epithelial cells to accelerate the infection process.
It has been confirmed that the SSARS-CoV-2 protein adopts a similar conformation and interaction mode to that of SSARS-CoV when interacting with ACE2 (Xu et al., 2020). The spatial structure of the RGD motif (403-405) is located outside of the S protein and adjacent to its interaction interface with ACE2 (Figure 1E), defining a small loop between a β-strand and an α-helix. Previous studies reported that the S protein processes a dynamic prefusion conformation during fusion into the host cell membrane (Li, 2016; Wrapp et al., 2020). When the receptor binding domain (RBD) of S1 subunit undergoes hinge-like conformational shifts, the change exposes or hides the key region of binding domain to access ACE2 by controlling the “up” and “down” conformation, exposing the RGD motif to the surface of the host cell membrane in conjunction with the key binding region. Once interacting with integrin, ACE2 may be recruited to the binding complex, facilitating the invasion of the virus. Another possible mechanism is proposed that the RGD motif may bind to integrins parallelly or sequentially in an ACE2-independent manner, which is supported by the role of ACE2 serving as a cell adhesion substrate and regulating integrin signaling (Clarke et al., 2012). However, Luan et al. held a contrary view that integrin can inhibit receptor targeting of S proteins from SARS-CoV-2 by shielding both S protein and ACE2, since there would be no space for ACE2 to contact with S if associated with integrin. In general, it is hypothesized that the RGD motif may play an important role in promoting rapid transmission in SARS-CoV-2.
Though several articles have reported the RGD motif and its potential role, no drugs have been investigated in preclinical studies or clinical trials (Sigrist et al., 2020; Tresoldi et al., 2020). Based on this rationale, high-throughput virtual screening searching for potential therapeutic drugs targeting interaction of SSARS-CoV-2 protein with both ACE2 and integrin were performed according to the hypothesis. Our compound libraries include FDA-approved drug entities (2040 species), our own medicine food homology natural products derivatives entities (1500 species), and cyclic peptides entities (230 species), along with virtual bioactive and natural products libraries (Figure 1F). The SSARS-CoV-2 structure extracted from the recent crystal structure of S protein/human ACE2 complex (NMDCS0000001) were adopted, choosing the key residues in the S protein interface (Q493, Y495, Q498, N501, and Y505) and R403, D405 as the potential binding site (Figure 1G) for virtual screening by Sybyl X using the Surflex-Dock Geom (SFXC) approach.
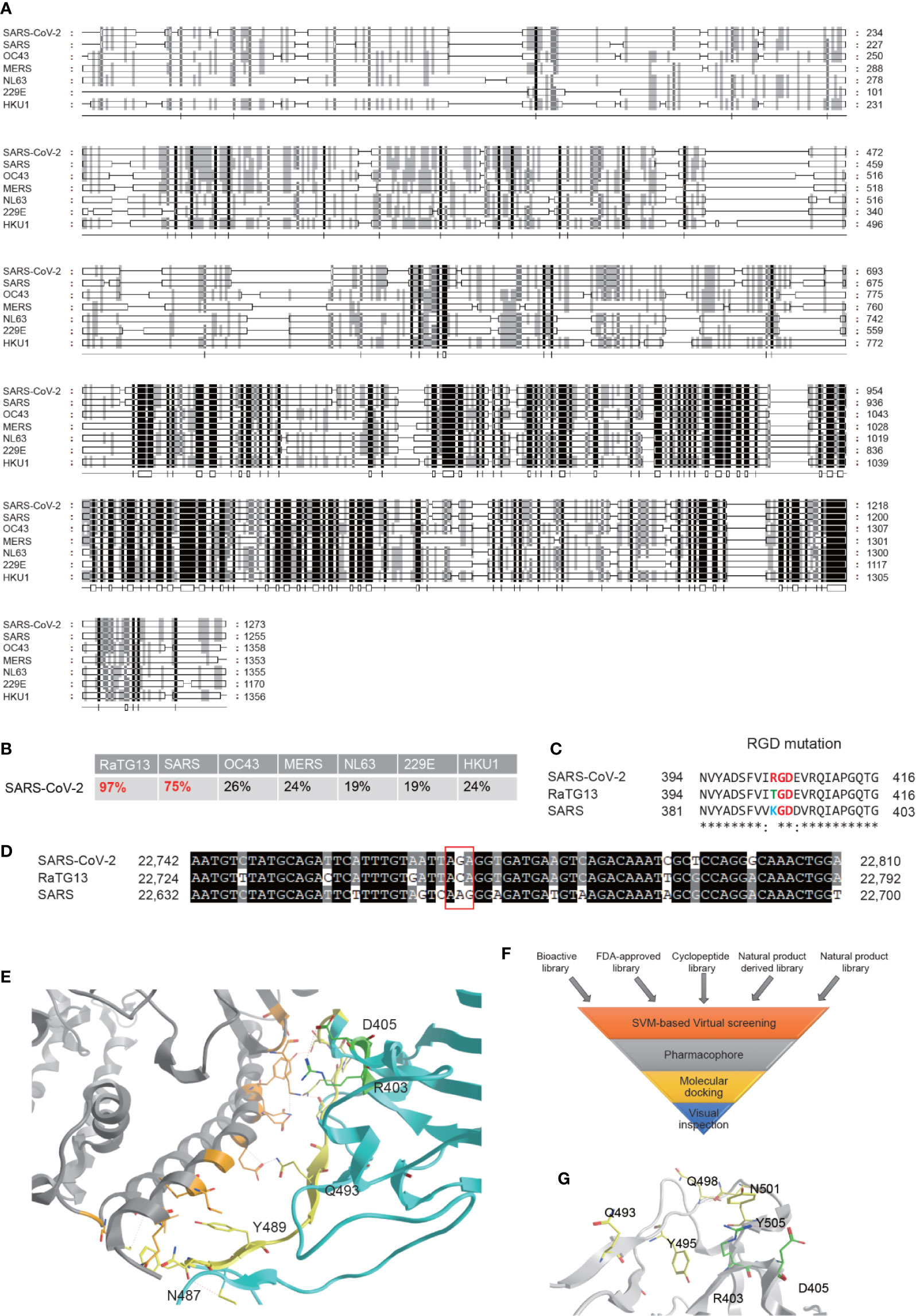
Figure 1 Bioinformatics analysis of the S protein of the SARS-CoV-2 and identification of potential inhibitors for its interaction with ACE2. (A) Amino acid sequence alignment of the S proteins in the coronaviruses infecting humans, including SARS-CoV-2, SARS, OC43, MERS, NL63, 229E, and HKU1 strains. (B) Homology of SARS-CoV-2 to other coronaviruses based on the S protein alignment. RaTG13, a Bat-SARS-like coronavirus, is used as a control. (C) Evolution of the RGD motif in the S protein of SARS-CoV-2 compared with the S protein of SARS-CoV. (D) Nucleotide mutation produces RGD in SARS-CoV-2. (E) ACE2 binding interface regions of the S protein of SARS-CoV2. Location of the RGD motif was shown in green, and the yellow residues are the interaction surfaces binding with ACE2. (F) Flowchart of interaction surface structure-based virtual high-throughput screening. (G) The key residues in the S protein interface and the RGD motif were chosen as the potential binding site to generate the protomol for virtual screening by using Surflex-Dock Geom (SFXC) approach.
The representative agents hit included: Nadide, Losartan, and Adenosine phosphate from the FDA-approved drug library; Difludionone-119 and Methyl-benzyloxychadone-844 from the natural products derivatives library; GR8-1 [Cyclo (f-P-V-R-f-P-R-L-)], MGR-7 [Cyclo (L-f-P-V-R-L-f-P0-V-R-)], and GR6-2 [(Cyclo (R-f-P-R-f-P-)) from our own cyclic peptide library; and S-9′″-Methyllithospermate B and the S-Leonurine from the bioactive and natural product library respectively(Yan S. et al., 2020). These compounds were well docked into the pocket formed by the selected key residues and RGD motif by Hydrogen bonds, and/or π-π/p-π interactions respectively, providing potential antiviral drug candidates for COVID-19.
Particularly, Nadide was scored with a high grade (10.7719), which is superior to other hits, implying it may serve as a promising drug candidate for COVID-19. Nadide is a dinucleotide of adenine and nicotinamide and has coenzyme activity in redox reactions, acting as a donor of ADP-ribose moieties (Bertoldo et al., 2020). It was postulated that supplement of nicotinamide may resist viral infection through innate immunity (Heer et al., 2020). SARS-CoV-2 infection can strikingly dysregulate the nicotinamide adenine dinucleotide (NAD) gene set by inducing a set of poly ADP-ribose polymerase (PARP) family enzymes required for the innate immune response. Overexpression of PARP10 induces a significant decrease in host cell NAD while boosting NAD through the nicotinamide and nicotinamide riboside kinase pathways, can restore antiviral PARP functions to support innate immunity to SARS-CoV-2, which provides a clue that Nadide may play a role in preventing COVID-19. Further in vivo study is needed to validate the effects of Nadide to block the interaction between the RGD motif and ACE2 protein.
Losartan is another promising potential drug candidate for COVID-19. ACE2 is a carboxypeptidase, negatively regulating Ang II production and counterbalancing the function of ACE. Losartan, a selective and competitive nonpeptide Ang II receptor antagonist, was known to block the vasoconstrictor and aldosterone-secreting effects of Ang II and interact reversibly with AT1 and AT2 receptors. It was postulated that SARS-CoV may promote severe acute lung injury pathogenesis through increased AngII production and functional alterations of the renin-angiotensin system, and the lung failure can be rescued by inhibition of AT1R (Kuba et al., 2005). Therefore, it is reasonable to presume that SARS-CoV-2 Spike may also exaggerate acute lung failure through the allied mechanism of SARS-CoV which deregulates the renin-angiotensin system, and can be rescued by inhibition of AT1R. Since losartan is a commonly used antihypertensive drug in clinical practice, its toxicological and pharmacokinetic properties have been fully studied and confirmed by a large amount of clinical data. If proven effective against the SARS-CoV-2 infection, it could be reassessed as an antiviral drug and significantly shorten the research cycle for drugs.
Since coronaviruses are under extensive mutagenesis and the mutation in key proteins are crucial to the virus, the potential clinical significance of the S protein harboring the RGD motif in SARS-CoV-2 is notable. Compared with SARS-CoV, SARS-CoV-2 has comparable, even higher, transmissibility that urges us to uncover its infection mechanism and to develop specific drugs against SARS-CoV-2 to alleviate the current pandemic (Chen et al., 2020; Li et al., 2020). It was hypothesized that the RGD motif on the S glycoprotein may bind to the integrin on the surface of host cells, resulting in higher affinity with the host cells in comparison with SARS-CoV. Further investigations are needed to verify and determine the specific subtype of integrins to interact with the SSARS-CoV-2 protein (Stewart and Nemerow, 2007). Meanwhile, infection blockers can be designed to be highly compatible with S protein to block either ACE2 binding or integrin binding. Griffithsin, for example, has been previously reported to bind to oligosaccharides of various viral glycoproteins, which can be reassessed as a treatment (Zumla et al., 2016; Lee, 2019). Multiple RGD-related peptides are currently being used in clinical trials; for example, 18F-αvβ6-BP is currently used to detect lung damage where αvβ6 is a RGD-recognizing integrin (NCT04376593). In our screening we also show several potential cyclic peptides as transmission blockers for SARS-CoV-2, but their effects are currently under investigation (Yan S. et al., 2020). Collectively, in our screen, Nadide was shown to block the interaction of the RGD motif and its unknown integrin counterpart simultaneously, serving as a promising potential drug candidate for COVID-19. Integrin-targeted drugs might modulate virus-ligand affinity and signaling of SARS-CoV-2, and provide a new strategy in controlling COVID-19.
Author Contributions
GW and XB initiated the concept and design of the study. XB, GW, SY, and HS performed the analysis. GW and SY wrote the draft. GW and XB reviewed and revised the manuscript. GW and XB supervised the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at preprints.org (Yan S. et al., 2020). This work is supported in part by grants from the National Natural Science Foundation of China 31701114 (GW), 21672266 (XB); Guangdong Basic and Applied Basic Research Foundation (2019A050510019); Science and Technology Planning Project of Guangdong Province (2017B030314030); Guangzhou Science and Technology Planning Program (202002020051); National Engineering Research Center for New Drug and Druggability Evaluation, Seed Program of Guangdong Province (2017B090903004).
References
Bertoldo, M. J., Listijono, D. R., Ho, W. J., Riepsamen, A. H., Goss, D. M., Richani, D., et al. (2020). NAD(+) Repletion Rescues Female Fertility during Reproductive Aging. Cell Rep. 30, 1670–1681 e1677. doi: 10.1016/j.celrep.2020.01.058
Chan, J. F., Yuan, S., Kok, K. H., To, K. K., Chu, H., Yang, J., et al. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523. doi: 10.1016/S0140-6736(20)30154-9
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513. doi: 10.1016/S0140-6736(20)30211-7
Clarke, N. E., Fisher, M. J., Porter, K. E., Lambert, D. W., Turner, A. J. (2012). Angiotensin converting enzyme (ACE) and ACE2 bind integrins and ACE2 regulates integrin signalling. PloS One 7, e34747. doi: 10.1371/journal.pone.0034747
Heer, C. D., Sanderson, D. J., Alhammad, Y. M. O., Schmidt, M. S., Trammell, S. A. J., Perlman, S., et al. (2020). Coronavirus and PARP expression dysregulate the NAD Metabolome: a potentially actionable component of innate immunity. bioRxiv. doi: 10.1101/2020.1104.1117.047480
Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., et al. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879. doi: 10.1038/nm1267
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi: 10.1038/s41586-020-2180-5
Lee, C. (2019). Griffithsin, a Highly Potent Broad-Spectrum Antiviral Lectin from Red Algae: From Discovery to Clinical Application. Mar. Drugs 17, 567. doi: 10.3390/md17100567
Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., Tong, Y., et al. (2020). Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 382, 1199–1207. doi: 10.1056/NEJMoa2001316
Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 3, 237–261. doi: 10.1146/annurev-virology-110615-042301
Lin, Q., Keller, R. S., Weaver, B., Zisman, L. S. (2004). Interaction of ACE2 and integrin beta1 in failing human heart. Biochim. Biophys. Acta 1689, 175–178. doi: 10.1016/j.bbadis.2004.05.005
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Luan, J., Lu, Y., Gao, S., Zhang, L. (2020). A potential inhibitory role for integrin in the receptor targeting of SARS-CoV-2. J. Infect. S0163–4453(0120)30181-X. doi: 10.31016/j.jinf.32020.30103.30046
Plosa, E. J., Benjamin, J. T., Sucre, J. M., Gulleman, P. M., Gleaves, L. A., Han, W., et al. (2020). beta1 Integrin regulates adult lung alveolar epithelial cell inflammation. JCI Insight 5, e129259. doi: 10.1172/jci.insight.129259
Ruoslahti, E. (1996). RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12, 697–715. doi: 10.1146/annurev.cellbio.12.1.697
Sigrist, C. J., Bridge, A., Le Mercier, P. (2020). A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 177, 104759. doi: 10.1016/j.antiviral.2020.104759
Stewart, P. L., Nemerow, G. R. (2007). Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15, 500–507. doi: 10.1016/j.tim.2007.10.001
Tresoldi, I., Sangiuolo, C. F., Manzari, V., Modesti, A. (2020). SARS-COV-2 and infectivity: Possible increase in infectivity associated to integrin motif expression. J. Med. Virol. doi: 10.21002/jmv.25831
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507
Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., et al. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63, 457–460. doi: 10.1007/s11427-020-1637-5
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Yan, S., Sun, H., Bu, X., Wan, G. (2020). An Evolutionary RGD Motif in the Spike Protein of SARS-CoV-2 may Serve as a Potential High Risk Factor for Virus Infection? Preprints 2020020447. doi: 10.2020020944/preprints2020202002.2020020447.v2020020441)
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Keywords: SARS-CoV-2, pandemic, COVID-19, drug screening, RGD, integrin
Citation: Yan S, Sun H, Bu X and Wan G (2020) New Strategy for COVID-19: An Evolutionary Role for RGD Motif in SARS-CoV-2 and Potential Inhibitors for Virus Infection. Front. Pharmacol. 11:912. doi: 10.3389/fphar.2020.00912
Received: 22 April 2020; Accepted: 04 June 2020;
Published: 12 June 2020.
Edited by:
Alfonso Pompella, University of Pisa, ItalyReviewed by:
Carmelo Bernabeu, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2020 Yan, Sun, Bu and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianzhang Bu, cGhzYnh6aEBtYWlsLnN5c3UuZWR1LmNu; Guohui Wan, d2FuZ3VvaEBtYWlsLnN5c3UuZWR1LmNu
 Shijia Yan
Shijia Yan Haixia Sun
Haixia Sun Xianzhang Bu
Xianzhang Bu Guohui Wan
Guohui Wan