
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 08 May 2020
Sec. Drug Metabolism and Transport
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.00664
 Dehua Liao1,2
Dehua Liao1,2 Zhigang Liu3
Zhigang Liu3 Yongchang Zhang4
Yongchang Zhang4 Ni Liu2
Ni Liu2 Dunwu Yao2
Dunwu Yao2 Lizhi Cao2
Lizhi Cao2 Yun Chen2
Yun Chen2 Yilan Fu2
Yilan Fu2 Nong Yang4*
Nong Yang4* Daxiong Xiang1*
Daxiong Xiang1*Background: Erlotinib is presently the first line treatment for non-small cell lung cancer (NSCLC) with epidermal growth factor receptor (EGFR) active mutation. An increasing number of evidences show that the treatment efficacy and toxicities are considerably heterogeneous among individuals. Hence, it is necessary to find biological predictors for further individualized treatment of erlotinib in NSCLC patients.
Methods: Our present study enrolled 87 cases of NSCLC patients who had been administrated erlotinib with a fixed dose (150 mg/d). Eleven polymorphisms in seven genes of drug-metabolizing enzymes and transporters were genotyped and the steady state trough concentrations were also determined.
Results: There were significant variances in the steady-state erlotinib trough plasma concentrations, ranging from 315.6 ng/ml to 4479.83 ng/ml. Erlotinib steady state trough concentration was remarkably lower in current smoking patients. The steady state trough concentration of GG in rs1048943 of CYP1A1 was significantly higher than that of AA allele carriers. The polymorphism of CYP1A2 was significantly associated with the severity of skin rash, and the development of diarrhea was associated with SNPs in ABCB1 and CYP3A5. We also observed that GG allele in CYP1A1 was accompanied with a longer PFS in our study.
Conclusion: A large variability of erlotinib steady state trough concentration was found among Chinese Han population. SNPs in CYP1A1 appeared to influence the steady state trough concentration of erlotinib. Correlation between CYP1A2 polymorphisms and severity of skin rash was observed, together with the correlation between the development of diarrhea and SNPs in ABCB1 and CYP3A5.
Non-small cell lung cancer (NSCLC) is a very common carcinoma, accounting for 80%–85% of lung cancer cases, and also the leading cause of cancer-related deaths worldwide. (Maemondo et al., 2010; Novello et al., 2016). The management of treatment for NSCLC could be optimized by using pharmacogenetic testing (Hamada et al., 2012). Numerous studies have showed that aberrant epidermal growth factor receptor (EGFR) pathway is a driving gene of NSCLC cancerogenesis, playing a considerable role in the process of malignancies, such as cell proliferation, differentiation and migration (Doerks et al., 2002; Bardelli et al., 2003). It was reported that the EGFR tyrosine kinase inhibitors (TKIs) were effective in patients with EGFR sensitive mutations (Mok et al., 2009; Mitsudomi et al., 2010).
Erlotinib is an orally administered, reversible TKI indicated as first-line treatment for advanced NSCLC in patients with activating EGFR mutations and as second- and third-line treatment in patients with EGFR wild-type NSCLC (Timmers et al., 2015). The most common activating mutations in EGFR are the exon 19 deletion and the exon 21-point mutation L858R (Tournier et al., 2017). Compared with standard platinum doublet therapy, erlotinib showed dramatically improved the long-term survival rate and clinical response of EGFR sensitive mutation NSCLC. Despite of its striking efficacy, however, real-world clinical evidence demonstrated considerable individual variability in both therapeutic efficacy and toxicity (Riely et al., 2009; Gainor and Shaw, 2013). Suboptimal response and response failure to the treatment are also observed in clinical treatment. In addition, most of the patients experienced drug-related adverse events (such as skin rash and diarrhea) with fixed dose of erlotinib during their treatment course (Endo-Tsukude et al., 2018).
Research in advanced NSCLC has since focused on finding biological predictors of response and adverse events allowing for treatment optimization. More evidences showed that inter-patient variability in drug response and adverse events which reflect the systemic levels or intracellular concentrations of the drug (Decosterd et al., 2015). Absorption, distribution, and metabolism are important pharmacokinetics parameters for a drug, which are all significantly associated with the exposure level. Statistical correlations between these pharmacokinetic parameters and treatment outcomes and adverse events were observed. Thus, inter-individual variations in erlotinib pharmacokinetics may present important clinical consequences.
Increasing body of evidences recognized that genetic polymorphisms of drug-metabolizing enzymes and transporters may significantly influence the inter-individual variations in drug reaction and disposition (Evans and McLeod, 2003). As an orally administrated small molecule TKI, erlotinib is mainly metabolized in the liver by cytochrome P450 CYP1A1, 1A2, 3A4, 3A5, and 2D6. Activities of these drug-metabolizing enzymes determine pharmacokinetics of the agents and therefore influence adverse events and therapeutic response rates. Nie et al. have reported that the polymorphism of CYP1A1 is a predictor for clinical outcome in NSCLC patients treated with EGFR-TKI therapy (Nie et al., 2011b). Guillen et al. have also reported that determination of CYP3A4 and CYP1A2 phenotyping in plasma and dried blood spots could predict the pharmacokinetics and toxicity of erlotinib in NSCLC patients (Parra-Guillen et al., 2017). ATP binding cassette subfamily B member 1 (ABCB1) and ATP binding cassette subfamily G member 2 (ABCG2) are important transporter of several drugs, the expression of which appear to affect the pharmacokinetics of erlotinib (Shi et al., 2007; Marchetti et al., 2008; Li et al., 2014). Hamada et al. have reported that ABCB1 1236TT–2677TT–3435TT genotype is associated with higher plasma concentration and the risk of higher toxicity in patients treated with erlotinib (Hamada et al., 2012). Neul et al. have reported that transporter genetic variants are promising novel factors to explain inter-individual variability in the response to TKI therapy (Neul et al., 2016). Chihiro et al. have also reported that the ABCB1 genotype is a potential predictor for erlotinib adverse events (Endo-Tsukude et al., 2018). Although the relationship between SNPs and treatment outcomes and toxicities were widely investigated, the results were inconsistent and none of those analyses clearly explains the determinants of the large inter-individual variability in the treatment outcomes and toxicity of erlotinib treatment.
Accordingly, genetic polymorphisms of the metabolizing enzymes and transporters could affect the expression of corresponding proteins, thus, may predict differences in erlotinib response and their adverse events. To validate this hypothesis, we determined to measure the steady state trough concentration of erlotinb, and analyzed the SNPs in metabolizing enzymes (P450) and transporters (ABC) in NSCLC patients treated with fixed dose erlotinib. We also investigated the correlations between the polymorphisms of these genes with the treatment outcomes and toxicities, respectively.
The study was carried out at Hunan Cancer Hospital, Hunan, China, between April 2016 and October 2018. Patients with advanced NSCLC who were treated with oral erlotinib at a standard dose of 150 mg were enrolled in our study. Inclusion criteria were as follows: 1) histologically confirmed NSCLC with EGFR sensitive mutation; 2) unmodified dosage of erlotinib during the treatment period. This study was designed and implemented by following the guidelines dictated in the Declaration of Helsinki. Ethics approval was obtained from the Independent Ethics Committee of Hunan Cancer Hospital and all patients were provided written informed consent.
Blood samples were collected 24 ± 3 h after the last drug administration but before the next administration at day 26-30 after the first dose of erlotinib, which aimed to ensure that the blood samples were steady state trough concentration samples. Blood samples (3 ml each) were collected into EDTA polypropylene tubes, centrifuged at 3000 r/min for 5 min. The remaining samples were prepared for germline mutation detection, while the supernatant in the centrifuged tubes were collected and stored at -80 ℃ until to be analyzed. The plasma concentration of erlotinib was determined by an on-line column extraction HPLC (Shimadzu Corporation, Kyoto, Japan) coupled with binary peak focusing system that we developed. Chromatography was performed as previously described (Liao et al., 2018). The lower limit of quantification was 50 ng/ml for erlotinib.
The DNA of the patients for genotyping was extracted from peripheral blood according to the method described previously. The genetic polymorphisms of CYP3A4 rs2242480, CYP3A5 rs 776746, CYP2D6 rs 1065852, CYP1A1 rs1048943, CYP1A2 rs762551, rs2470890, ABCB1 rs1128503, rs1045642, ABCG2 rs2231142, rs2622604, rs7699188 were determined by multiple SNP typing techniques. All samples were analyzed in duplicates and negative controls were included to ensure authenticity of the results.
As typical adverse events, the development of skin rash and diarrhea in all patients were monitored in our study, which were rated according to the National Cancer Institute Common Terminology Criteria for Adverse Events version 4.0 (NCI CTCAE v4.0). Since the median onset of erlotinib induced skin rash and diarrhea are typical within 1–2 weeks of therapy initiation, and toxicities might be confounded by the number of treatment periods, the grade of erlotinib-induced toxicity was determined during the first 30 days. According to NCI CTCAE v4.0, the severity of skin rash was graded as follows: Grade 1: Papules and/or pustules covering <10% body surface area (BSA), which may or may not be associated with symptoms of pruritus or tenderness. Grade 2: Papules and/or pustules covering 10%–30% BSA, which may or may not be associated with symptoms of pruritus or tenderness; associated with psychosocial impact; limiting instrumental activities of daily living (ADL). Grade 3: Papules and/or pustules covering >30% BSA, which may or may not be associated with symptoms of pruritus or tenderness; limiting self care ADL; associated with local superinfection with oral antibiotics indicated. Grade 4: Papules and/or pustules covering any % BSA, which may or may not be associated with symptoms of pruritus or tenderness and are associated with extensive superinfection with IV antibiotics indicated; life threatening consequences. Grade 5: Death. The severity of diarrhea was graded as follows: Grade 1: Increase of <4 stools per day over baseline; mild increase in ostomy output compared to baseline. Grade 2: Increase of 4–6 stools per day over baseline; moderate increase in ostomy output compared to baseline. Grade 3: Increase of ≥7 stools per day over baseline; incontinence; hospitalization indicated; severe increase in ostomy output compared to baseline; limiting self care ADL. Grade 4: Life-threatening consequences; urgent intervention indicated. Grade 5: Death.
Patient characteristic information, such as sex, height, weight, performance status (PS), disease stage (based on the TNM classification), smoking status, mutational status were obtained from their medical records, and progression free survival (PFS) and AEs were strictly assessed and collected by the members in our study after each follow-up visit. Chest computed tomography (CT) was performed just before treatment, besides, chest CT, a complete blood count, and blood chemistry studies were repeated at least once a month until disease progression. Treatment response was assessed using the Response Evaluation Criteria in Solid Tumors version 1.1, and response of treatment was defined as complete response (CR: Disappearance of all target lesions. Any pathological lymph nodes must have reduction in short axis to <10 mm), Partial Response (PR: At least a 30% decrease in the sum of the diameters of target lesions, taking as reference the baseline sum diameters), Progressive Disease (PD: At least a 20% increase in the sum of the diameters of target lesions, taking as reference the smallest sum on study). Stable Disease (SD: Neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for PD, taking as reference the smallest sum diameters while on study). In addition to the relative increase of 20%, the sum must also demonstrate an absolute increase of at least 5 mm. Follow-up was considered as the duration of treatment under erlotinib. PFS was defined as from the date of erlotinib treatment initiation to the date of first documented progression or death. PFS and adverse event were included in the present analysis to correlate with the mean trough level concentration at steady state and pharmacogenetic factors.
Data analysis were carried by SPSS version 13.0 software (SPSS Inc., Chicago, IL, USA) in our study and all values were presented as means ± SD. The influence of different genotypes on erlotinib trough levels was analyzed statistically by one-way analysis of variance (ANOVA) with dunnet t test. Fisher's exact tests were used to analyze the association between genetic polymorphisms and toxicity. The patient characteristic factors were tested univariately with Cox regression or Kaplane-Meier analysis. The association between SNPs and PFS was univariately tested with Kaplan-Meier analysis. The SNPs were added to the combined clinical characteristic factors to determine the impact of SNPs on PFS. Statistical significance was assumed for p values < 0.05.
Patient characteristics at the analysis of the erlotinib plasma concentration were summarized in Table 1. A total of 87 advanced NSCLC patients were enrolled in our study, 52 (59.8%) male and 35 (40.2%) female, with a median age of 57 years (29-77). The median body weight was 57 kg (37.0–82.5) and the median body surface area was 1.59 m2 (1.24–1.92). All the patients in our study were given a fixed dose (150 mg/d) of erlotinib and had good PS, without dose adjustment. The cohort was consisted of 57 non-smokers, 16 patients with smoking history, and 14 currently-smoking patients. All of the patients had major EGFR mutations, with exon 19 deletions (66.7%) and exon 21 L858R (33.3%) respectively.
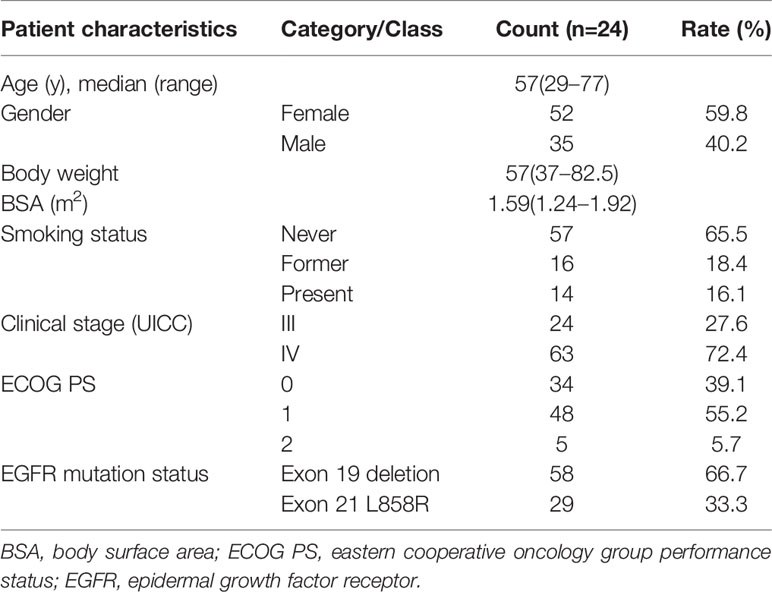
Table 1 Baseline clinical and pathological characteristics of the 87 patients enrolled in the study.
The genotype status of the drug-metabolizing enzymes and transporters were determined for all 87 patients. The genotype and allele frequency in the study population were presented in Table 2.
Erlotinib trough concentration at steady state was analyzed, and the results were available for 87 patients. The studied NSCLC patients had a wide inter-individual variation in erlotinb trough plasma concentrations, and the mean erlotinib trough plasma concentration was 1354.5 ± 700.3 ng/ml, ranging from 315.6 ng/ml to 4479.83 ng/ml. Trough plasma erlotinib concentration of 8.05% patients were under the target value (500 ng/ml) (Hidalgo et al., 2001), which indicates a high potential risk of disease progression and treatment failure.
Previous study has reported that erlotinib pharmacokinetic was related to smoking status (Hamilton et al., 2006). In our present study, the smoking status for 14 patients was concurrent smoking. The relationship between smoking status and erlotinib steady state trough concentration was investigated, and the results showed that the mean erlotinib trough plasma concentration for concurrent smokers was significantly lower than those without smoking history (p=0.021). The mean erlotinib trough plasma concentration in concurrent smokers was also lower than former smokers, but without statistical difference (Figure 1).
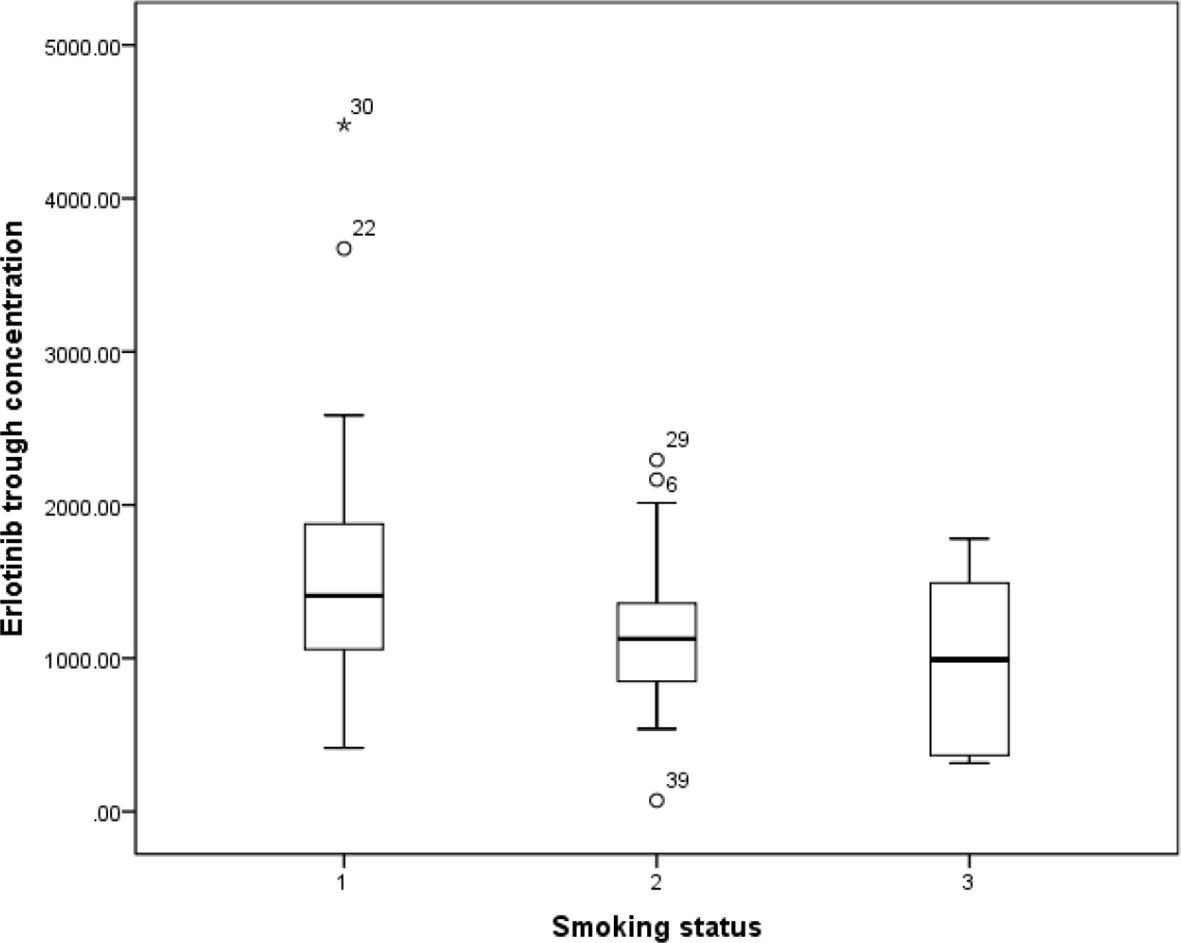
Figure 1 The relationship between erlotinib steady state trough plasma concentrations and smoking status.
Effects of genetic polymorphisms of drug-metabolizing enzymes and transporters on erlotinib trough level were investigated in our study. For CYP1A1rs1048943 (Figure 2A), the mean steady state erlotinib trough plasma concentration in wild-types (AA) was significantly lower than that of mutant-types (GG) (p=0.007). For CYP1A2rs762551 (Figure 2B), rs2470890 (Figure 2C), CYP2D6rs1065852 (Figure 2D), CYP3A4rs2242480 (Figure 2E), CYP3A5rs776746 (Figure 2F), no significant difference in the mean steady state erlotinib trough plasma concentration was found in observed genotypes (p > 0.05). In addition, there was no significant difference among the mean trough concentration of patients stratified based on genotypes of all the drug transporters (ABCB1rs1045642, rs1128503, ABCG2rs2231142, rs2622604, rs7699188) (Figure 3).
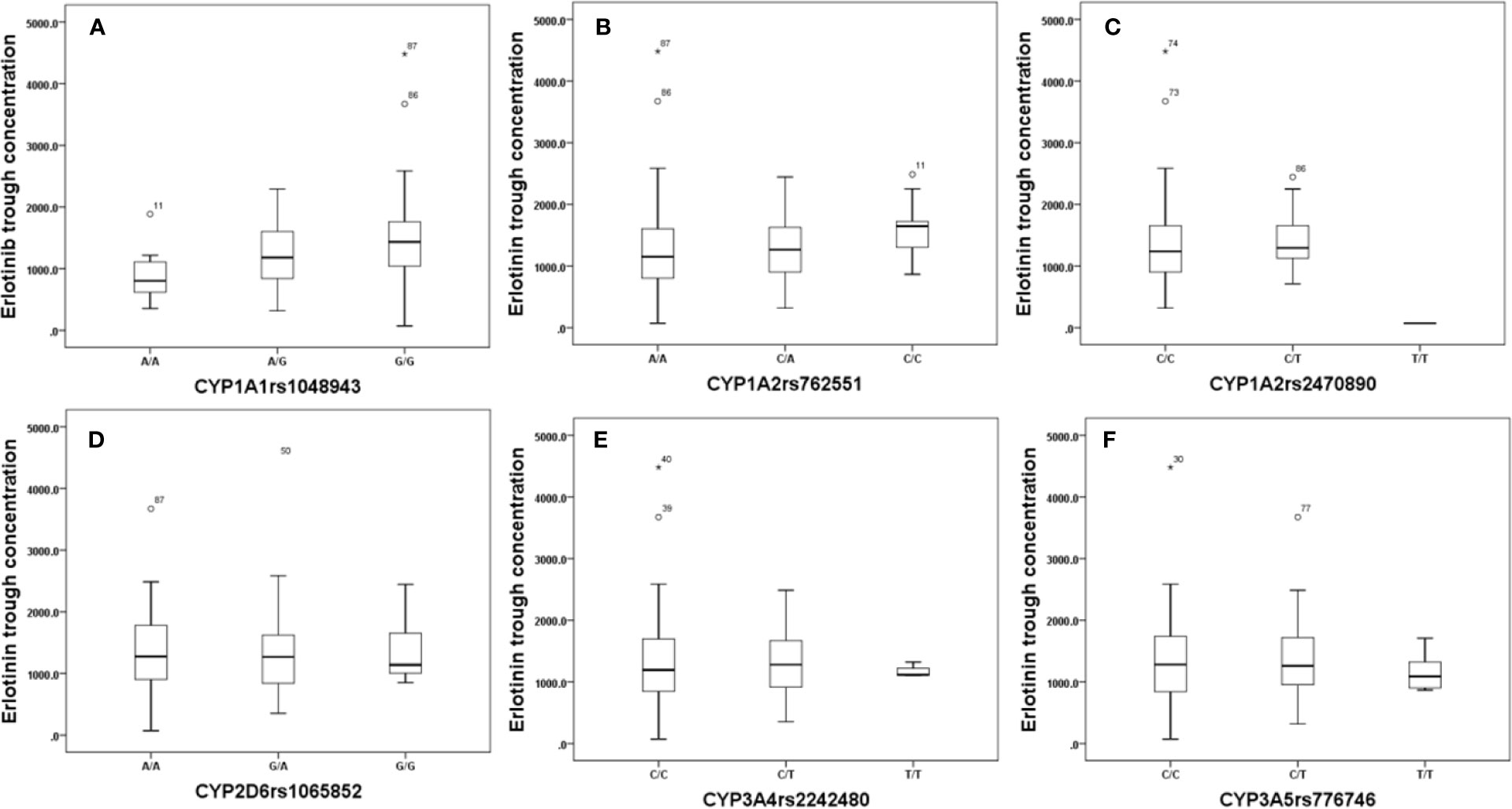
Figure 2 The relationship between erlotinib steady state trough plasma concentrations and CYP1A1rs1048943 (A), CYP1A2rs762551(B), rs2470890 (C), CYP2D6rs1065852 (D), CYP3A4rs2242480 (E), CYP3A5rs776746 (F).
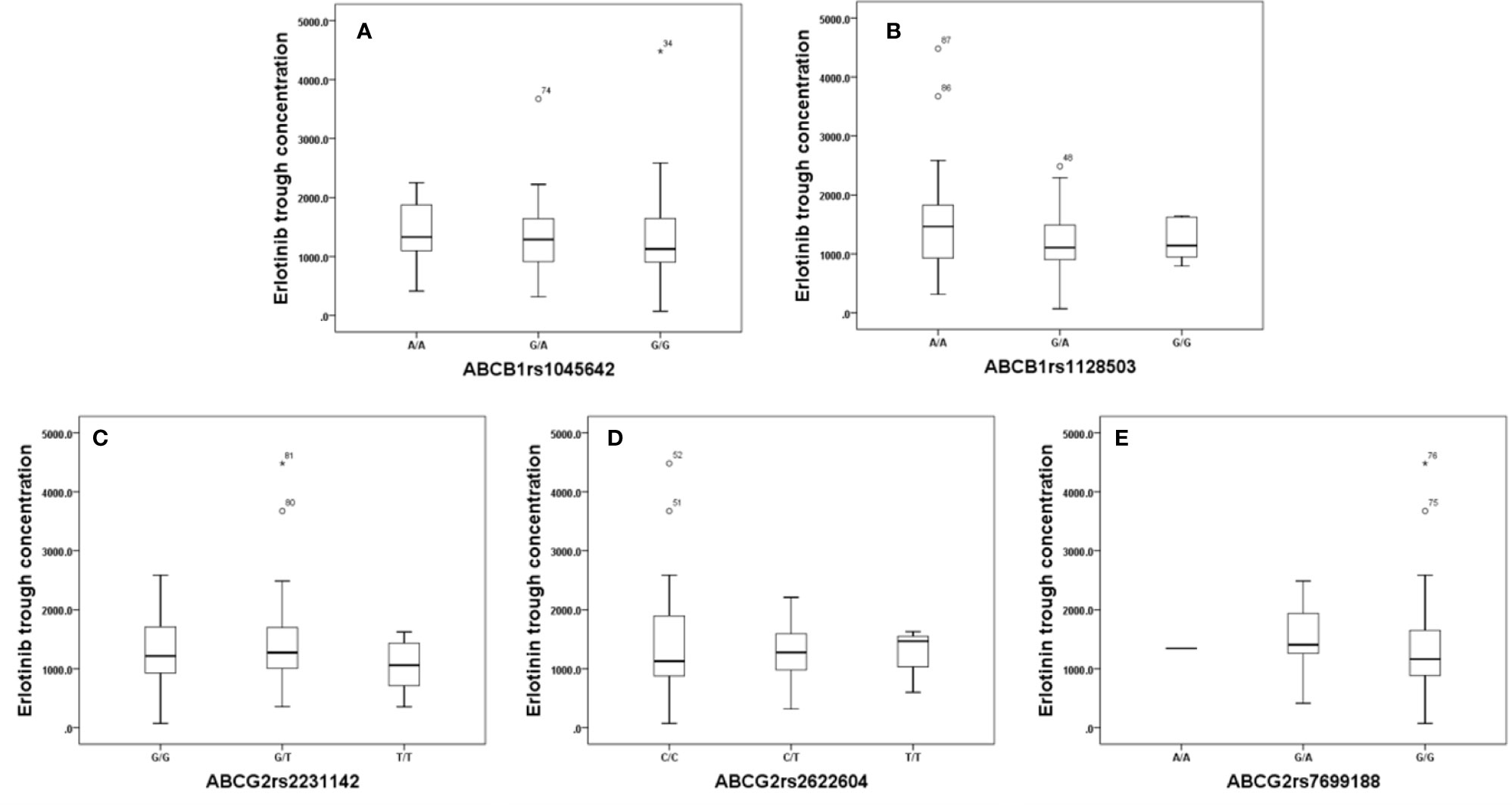
Figure 3 The relationship between erlotinib steady state trough plasma concentrations and ABCB1rs1045642 (A), rs1128503 (B), ABCG2rs2231142 (C), rs2622604 (D), rs7699188 (E).
As the most frequently observed adverse reactions, the development and severity of skin rash and diarrhea were recorded in our study. As shown in Table 3, the severity of skin rash was only associated with the polymorphism of rs762551 in CYP1A2 (p=0.029). For diarrhea, the polymorphisms of rs1045642 (p=0.005) and rs1128503 (p=0.007) in ABCB1 and rs776746 (p=0.048) in CYP3A5 was significantly associated with its development, but not the severity grade. For other metabolizing enzymes and transporters, no significant association was observed between polymorphisms and the development and severity of adverse reactions (p > 0.05).
The relationship between pharmacogenetic factors and PFS was analyzed in our study. In the univariate analysis of PFS, the SNP of rs1048943 in CYP1A1 (AA vs. AG vs. GG, p=0.030) relating to the pharmacodynamics of erlotinib showed an association with survival (Table 4). As to the multivariate analysis, the characteristic factors of patient body surface area and smoking status (HR 1.042, P=0.008, HR 5.317, P=0.029, respectively) were associated with shorter PFS. When the SNPs of metabolizing enzymes and transporters were added to this model, it can be observed that the CT allele in rs1048943 was associated with longer PFS (HR 0.226, p=0.038).
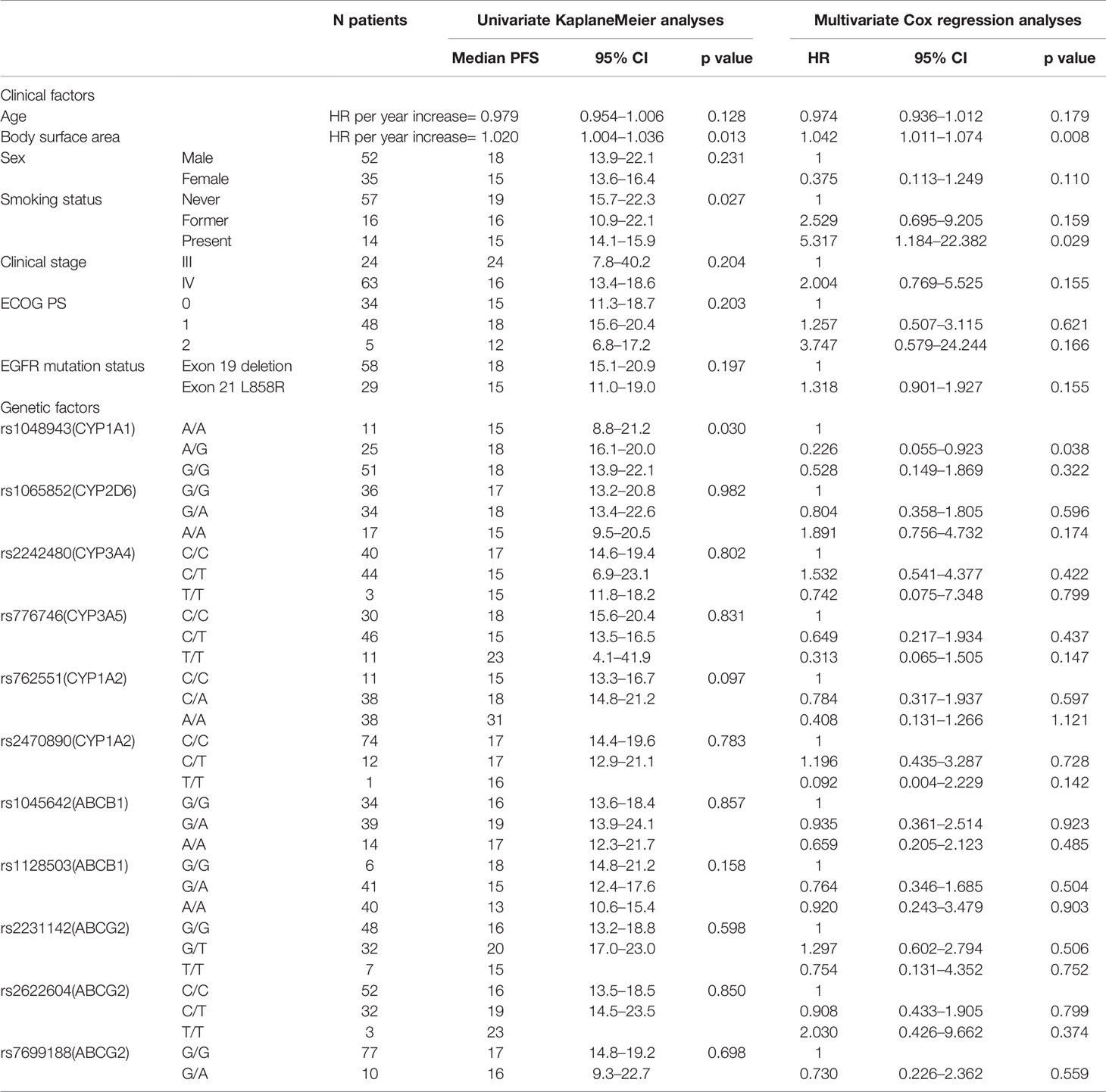
Table 4 Univariate and multivariate analyses of progression-free survival of EGFR mutated NSCLC with 150 mg erlotinib.
Pharmacokinetics is a potential source of biomarkers and could partially explain the inter-individual variability in erlotinib response in previous studies. In our present study, significant variation of erlotinib steady state trough plasma concentration was observed. The influence of polymorphism of drug metabolizing enzymes and transporters on trough plasma concentration, PFS and adverse events was also observed.
The average erlotinib trough plasma concentration was 1354.5 ± 700.3 ng/ml, ranging from 315.6 ng/ml to 4479.8 ng/ml, with a nearly 14.2 fold variance. Results from our research were consistent with previous studies. Hamada et al. has reported that the mean trough plasma concentration of erlotinib was 1530 ± 780 ng/ml in Japanese NSCLC patient, also with a wide variation (range: 320–3390 ng/ml) (Hamada et al., 2012). Similarly, trough concentration of erlotinib has also been monitored in the study by Nienke and co-workers, in which the mean trough plasma concentrations for erlotinib was 1010 ng/ml, with a nearly 74.0 fold variance, which was larger than our study (Lankheet et al., 2014). 500 ng/ml was recognized as the targeting trough concentration of erlotinib in the treatment of NSCLC (Hidalgo et al., 2001). However, 8.05% of patients have not reached this threshold in our study population, which may lead to a higher risk of disease progression and treatment failure.
The relationship between genetic polymorphisms and erlotinib trough level was also investigated in our study, and the results showed that the mean steady state erlotinib trough plasma concentration in wild-types (AA) of CYP1A1rs1048943 was significantly lower than that of mutant-types (GG) (p=0.007). CYP1A1 is a polymorphic gene which codes for the important phase-I XME aryl hydrocarbon hydroxylase and involves in drug metabolism (Indulski and Lutz, 2000). In addition, rs1048943 is one of the common genetic mutant sites in CYP1A1, which is strongly associated with susceptibility to various cancers (Abd El et al., 2017; Jain et al., 2017). Previous study showed that the polymorphism of CYP1A1 was associated with the treatment outcome of EGFR-TKI in advanced lung cancer patients (Nie et al., 2011b). Samyuktha et al. have also reported that a longer survival rate was observed in those patients who carry the mutant-type gene of CYP1A1 in imatinib treated chronic myeloid leukemia patients (Lakkireddy et al., 2015). The response of imatinib was positively associated with its exposure, which may accounts for the fact that a higher steady state concentration of imatinib was achieved in rs1048943 mutant-type gene patients. Mutant allele GG in rs1048943 is thought to be a risk factor for the development of adverse event, while allele AA in rs1048943 is considered as a risk factor for predicting inadequate clinical efficacy of erlotinib treatment. Therefore, for erlotinib treatment, dose adjustment and drug monitoring are highly suggested in patients who carry allele GG in rs1048943.
The effect of smoking on oral erlotinib pharmacokinetics has been widely studied. In our present study, the steady state trough concentration of erlotinb in concurrent smoking patients was significantly lower than that of those without smoking history, which was consistent with previous studies. Hamilton et al. (2006) have reported that smokers required 300mg of erlotinib to achieve similar Cmax and area under the curve to those of non-smokers only given a 150 mg dose. Clark et al. (2006) have also reported that steady-state Ctrough values were about 2-fold lower and the CL/F of erlotinib was about 24% increased in current smokers compared with the values in non-smokers. Investigations into the contribution of CYP1A2 to the metabolism of erlotinib and their induction by cigarette smoking have been widely reported (Hamilton et al., 2006; Anderson and Chan, 2016). Parra-Guillen et al. (2017) have reported that higher CYP1A2 activity was observed in current smoking NSCLC patients. Efficacy of erlotinib in smoking patients following administration of standard doses diminished possibly due to CYP1A2 mediated increased metabolism of the drug in the liver and tumor tissue (Li et al., 2007). However, the effect of smoking on the activity of CYP1A2 was not investigated in our present study, which could be a limitation. In addition, considering that the exact recommendations for the erlotinib dose modification have not been established yet (Hughes et al., 2009), traditional fixed dose (150 mg/d) was chosen as the dosage of erlotinib for concurrent smoking patients, not the increased 300 mg/d in our present study.
It is well established that in vivo drug-related therapeutic efficacy and toxicity are strongly associated with genetic factors (Scott, 2011; Patel, 2016). However, few pharmacogenomic biomarkers were used as predictive factor of toxicity for target therapy. To our knowledge, the incidence rates of skin rash and diarrhea were 81.6% and 35.5%, respectively. Findings from our present study showed that the polymorphism of CYP1A2 was the only statically significant covariate responsible for erlotinib induced skin rash (p = 0.029). As previous study showed, CYP1A2 was considerably involved in erlotinib metabolism (Li et al., 2007) and may be of value in predicting the individual metabolizer status of erlotinib (Parra-Guillen et al., 2017). The polymorphism of CYP1A2 may have a significant influence on the pharmacokinetic of erlotinib, which resulted in the variation of the severity of skin rash in our study. However, Chihiro et al. has pointed out that the SNPs in CYP1A2 have no effect on skin rash of erlotinib in Japanese patients, which was contrary to our study (Endo-Tsukude et al., 2018). Other factors additional to CYP1A2 activity may also influence erlotinib induced skin rash. Our present study showed that the occurrence of diarrhea was significantly associated with the tested polymorphisms of the selected genes (ABCB1 and CYP3A5). As a common transporter of erlotinib, ABCB1 was expressed in various organs, especially highly in the entire intestine. Polymorphism of ABCB1 can induce the change in transporter activity, and lower activity leads to higher absorption. Hamada et al. has previously reported that polymorphisms of ABCB1 was prominently related with erlotinib pharmacokinetics and toxicity in NSCLC patients (Hamada et al., 2012). Ma et al. has also reported that several SNPs in ABCB1 were associated with diarrhea in NSCLC patients treated with gefitinib (Ma et al., 2017). Consistent with the studies mentioned above, our present study also observed that SNPs in CYP3A5 were statistically associated with the development of diarrhea. Erlotinib is a substrate of CYP3A5 which is highly and polymorphically expressed (Wojnowski, 2004; Thorn et al., 2005). Charles et al. has also reported that polymorphism of CYP3A5 were accompanied with a higher risk of diarrhea in erlotinib treated NSCLC patients (Rudin et al., 2008). However, the relationship between SNPs in CYP3A5 and diarrhea after administration of erlotinib was rarely reported in other study.
The polymorphisms of CYP1A1 were found to be strongly associated with susceptibility to various cancers (Abd El et al., 2017; Jain et al., 2017). However, little information is known available on the polymorphism of CYP1A1 in relation to the clinical outcomes of NSCLC patients undergoing TKI therapy. Our pharmacogenetic study showed that SNPs in the genes encoding for CYP1A1 were associated with PFS in EGFR sensitive mutation patients who were treated with erlotinib 150 mg/d. The rs1048943 in CYP1A1 were significantly associated with a shorter PFS of patients in the present study. We found that the steady state trough level of erlotinib was significantly increased with this SNP mutation, and previous studies have reported that PFS are positively related with trough concentration in NSCLC patients. Hence, the longer PFS may be induced by the decreased metabolic capacity of CYP1A1 in mutant patients. Nie et al. previously reported that the polymorphism of CYP1A1 has a linkage with clinical outcome of NSCLC patients treated with EGFR-TKI (Nie et al., 2011a; Nie et al., 2011b). Previous study also showed that CYP1A1 genetic polymorphism was a promising predictor to improve chemotherapy effects in patients with metastatic breast cancer (Zhou et al., 2018).
In our present study, SNPs in the pharmacokinetic genes encoding for CYP3A4, CYP2D6 and ABCG2 were not significant associated with erlotinib steady state trough concentration. Furthermore, the same results were also observed for toxicity and survival. Our results were consistent with previous studies (Rudin et al., 2008; Parra-Guillen et al., 2017; Endo-Tsukude et al., 2018), however, it should be pointed out that some results in our study were conflicted with other studies (Rudin et al., 2008; Endo-Tsukude et al., 2018). For the relationship between polymorphisms of CYP3A4, CYP2D6, ABCG2, and survival, a hypothetical explanation would be that most patients had an erlotinib steady state serum level higher than the threshold (500ng/ml) needed for clinical activity, negating any effects that these SNPs may have on the actual serum level above this threshold.
Still, several limitations exist in our study. Firstly, we have only investigated the predictive value of polymorphism of common metabolize enzymes and transporters, the other metabolize enzymes and transporters (such as: organic cation transporters) possibly influencing erlotinib treatment outcomes and toxicity to be explored yet. Secondly, the influence of smoking status on the activity of CYP1A2 was not investigated. Thirdly, the concomitant medication factors with potential PK or pharmacodynamic interactions were not taken into account in our study, which is a limitation of the present study. According to the recorded information of patients in our present study, the incidence rate of concomitantly administered drug was low, which could justify our decision that concomitant medication factors were not considered in the study. However, the effect of concomitant medication factors with potential PK or pharmacodynamic interactions will be considered and investigated in our future study. Last but not the least, a larger sample study is needed to confirm our findings.
In summary, our results demonstrated that a large variability of erlotinib steady state trough concentration exists in NSCLC patients. It is also observed that SNPs in CYP1A1 may influence the steady state trough concentration of erlotinib and PFS. The polymorphisms of CYP1A2 plays an important role in the severity of skin rash, and the development of diarrhea was associated with SNPs in ABCB1 and CYP3A5. On the basis of these findings, with the aim of improving treatment efficacy and reducing toxicity, it is highly recommended that TDM (therapeutic drug monitoring) and genotyping should be taken into account in an attempt to further individualize treatment of erlotinib in NSCLC.
The data generated for this study is available to download here: https://www.ebi.ac.uk/ena/data/view/PRJEB37674.
The studies involving human participants were reviewed and approved by Ethics approval was obtained from the Independent Ethics Committee of Hunan Cancer Hospital. The patients/participants provided their written informed consent to participate in this study.
NY, DX, and DL designed the study and wrote the protocol. DL, ZL, YZ, DY, NL, and LC performed the experiments and analyzed the data. YC, YF contributed to the reagents and materials. DL drafted the manuscript. NY and DL revised the manuscript content. All authors read and approved the final manuscript.
This study was supported by National Natural Science Foundation of China (NSFC: 81603206), Hunan Provincial Pharmaceutical Association Fund (NO: hn2017005) and Health and Family Planning Commission Foundation of Hunan Province (grant number B20180252).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abd El, W. N., Shafik, N. F., Shafik, R. E., Taha Sh, A., Shafik, H. E., Darwish, A. D. (2017). Association of CYP3A5*3 and CYP1A1*2C Polymorphism with Development of Acute Myeloid Leukemia in Egyptian Patients. Asian Pac. J. Cancer Prev. 18 (3), 747–752. doi: 10.22034/APJCP.2017.18.3.747
Anderson, G. D., Chan, L. N. (2016). Pharmacokinetic Drug Interactions with Tobacco, Cannabinoids and Smoking Cessation Products. Clin. Pharmacokinet. 55 (11), 1353–1368. doi: 10.1007/s40262-016-0400-9
Bardelli, A., Parsons, D. W., Silliman, N., Ptak, J., Szabo, S., Saha, S., et al. (2003). Mutational Analysis of the Tyrosine Kinome in Colorectal Cancers. Science 300 (5621), 949. doi: 10.1126/science.1082596
Clark, G. M., Zborowski, D. M., Santabárbara, P., Ding, K., Whitehead, M., Seymour, L., et al. (2006). Smoking History and Epidermal Growth Factor Receptor Expression as Predictors of Survival Benefit from Erlotinib for Patients with Non–Small-Cell Lung Cancer in the National Cancer Institute of Canada Clinical Trials Group Study BR.21. Clin. Lung Cancer 7 (6), 389–394. doi: 10.3816/CLC.2006.n.022
Decosterd, L. A., Widmer, N., Zaman, K., Cardoso, E., Buclin, T., Csajka, C. (2015). Therapeutic drug monitoring of targeted anticancer therapy. Biomark Med. 9 (9), 6. doi: 10.2217/bmm.15.78
Doerks, T., Copley, R. R., Schultz, J., Ponting, C. P., Bork, P. (2002). Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genome Res. 12 (1), 47–56. doi: 10.1101/gr.203201
Endo-Tsukude, C., Sasaki, J. I., Saeki, S., Iwamoto, N., Inaba, M., Ushijima, S., et al. (2018). Population Pharmacokinetics and Adverse Events of Erlotinib in Japanese Patients with Non-small-cell Lung Cancer Impact of Genetic Polymorphisms in Metabolizing Enzymes and Transporters. Biol. Pharm. Bull. 41 (1), 9. doi: 10.1248/bpb.b17-00521
Evans, W. E., McLeod, H. L. (2003). Pharmacogenomics-Drug Disposition,Drug Targets, and Side Effects. N. Engl. J. Med. 348 (6), 11. doi: 10.1056/NEJMra020526
Gainor, J. F., Shaw, A. T. (2013). Novel Targets in Non-Small Cell Lung Cancer ROS1 and RET Fusions. Oncologist 18 (7), 10. doi: 10.1634/theoncologist.2013-0095
Hamada, A., Sasaki, J., Saeki, S., Iwamoto, N., Inaba, M., Ushijima, S., et al. (2012). Association of ABCB1 polymorphisms with erlotinib pharmacokinetics and toxicity in Japanese patients with non-small-cell lung cancer. Pharmacogenomics 13 (5), 9. doi: 10.2217/PGS.11.176
Hamilton, M., Wolf, J. L., Rusk, J., Beard, S. E., Clark, G. M., Witt, K., et al. (2006). Effects of smoking on the pharmacokinetics of erlotinib. Clin. Cancer Res. 12 (7 Pt 1), 2166–2171. doi: 10.1158/1078-0432.CCR-05-2235
Hidalgo, M., Siu, L. L., Nemunaitis, J., Rizzo, J., Hammond, L. A., Takimoto, C., et al. (2001). Phase I and Pharmacologic Study of OSI-774, an Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor, in Patients With Advanced Solid Malignancies. J. Clin. Oncol. 19 (13), 12. doi: 10.1200/JCO.2001.19.13.3267
Hughes, A. N., O'Brien, M. E., Petty, W. J., Chick, J. B., Rankin, E., Woll, P. J., et al. (2009). Overcoming CYP1A1/1A2 mediated induction of metabolism by escalating erlotinib dose in current smokers. J. Clin. Oncol. 27 (8), 1220–1226. doi: 10.1200/JCO.2008.19.3995
Indulski, J. A., Lutz, W. (2000). Metabolic genotype in relation to individual susceptibility to environmental carcinogens. Int. Arch. Occup. Environ. Health 73 (2), 14. doi: 10.1007/PL00007942
Jain, V., Ratre, Y. K., Amle, D., Mishra, P. K., Patra, P. K. (2017). Polymorphism of CYP1A1 gene variants rs4646903 and rs1048943 relation to the incidence of cervical cancer in Chhattisgarh. Environ. Toxicol. Pharmacol. 52, 188–192. doi: 10.1016/j.etap.2017.04.009
Lakkireddy, S., Aula, S., Avn, S., Kapley, A., Rao Digumarti, R., Jamil, K. (2015). Association of The Common CYP1A1 2C Variant (Ile462Val Polymorphism) with Chronic Myeloid Leukemia (CML) in Patients Undergoing Imatinib Therapy. Cell J. 17 (3), 9. doi: 10.22074/cellj.2015.11
Lankheet, N. A. G., Knapen, L. M., Schellens, J. H. M., Beijnen, J. H., Steeghs, N., Huitema, A. D. R. (2014). Plasma Concentrations of Tyrosine Kinase Inhibitors Imatinib, Erlotinib, and Sunitinib in Routine Clinical Outpatient Cancer Care. Ther. Drug Monit. 36 (3), 8. doi: 10.1097/FTD.0000000000000004
Li, J., Zhao, M., He, P., Hidalgo, M., Baker, S. D. (2007). Differential Metabolism of Gefitinib and Erlotinib by Human Cytochrome P450 Enzymes. Clin. Cancer Res. 13 (12), 3731–3737. doi: 10.1158/1078-0432.ccr-07-0088
Li, J., Cusatis, G., Brahmer, J., Sparreboom, A., Robey, R. W., Bates, S. E., et al. (2014). Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol. Ther. 6 (3), 432–438. doi: 10.4161/cbt.6.3.3763
Liao, D., Yao, D., Liu, N., Cao, L., Xiang, D., Yang, N., et al. (2018). Correlation of plasma erlotinib trough concentration with skin rash in Chinese NSCLC patients harboring exon 19 deletion mutation. Cancer Chemother. Pharmacol. 82 (3), 551–559. doi: 10.1007/s00280-018-3642-4
Ma, Y., Xin, S., Huang, M., Yang, Y., Zhu, C., Zhao, H., et al. (2017). Determinants of Gefitinib toxicity in advanced non-small cell lung cancer (NSCLC): a pharmacogenomic study of metabolic enzymes and transporters. Pharmacogenom. J. 17 (4), 325–330. doi: 10.1038/tpj.2016.31
Maemondo, M., Inoue, A., Kobayashi, K., Sugawara, S., Oizumi, S., Isobe, H., et al. (2010). Gefitinib or Chemotherapy for Non–Small Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 362 (25), 8. doi: 10.1056/NEJMoa0909530
Marchetti, S., de Vries, N. A., Buckle, T., Bolijn, M. J., van Eijndhoven, M. A., Beijnen, J. H., et al. (2008). Effect of the ATP-binding cassette drug transporters ABCB1, ABCG2, and ABCC2 on erlotinib hydrochloride (Tarceva) disposition in in vitro and in vivo pharmacokinetic studies employing Bcrp1-/-/Mdr1a/1b-/- (triple-knockout) and wild-type mice. Mol. Cancer Ther. 7 (8), 2280–2287. doi: 10.1158/1535-7163.MCT-07-2250
Mitsudomi, T., Morita, S., Yatabe, Y., Negoro, S., Okamoto, I., Tsurutani, J., et al. (2010). Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 11 (2), 8. doi: 10.1016/s14702045(09)70364-x
Mok, T. S., Wu, Y. L., Thongprasert, S., Yang, C. H., Chu, D. T., Saijo, N., et al. (2009). Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 361 (10), 10. doi: 10.1056/NEJMoa0810699
Neul, C., Schaeffeler, E., Sparreboom, A., Laufer, S., Schwab, M., Nies, A. T. (2016). Impact of Membrane Drug Transporters on Resistance to Small-Molecule Tyrosine Kinase Inhibitors. Trends Pharmacol. Sci. 37 (11), 904–932. doi: 10.1016/j.tips.2016.08.003
Nie, Q., An, S., Yang, X., Zhong, W., Liao, R., Yang, J., et al. (2011a). CYP1A12A polymorphism as a prognostic factor for the advanced lung cancer patients treated with EGFR-TKI and its correlation with EGFR mutation. J. Clin. Oncol. 29 (15), e13098. doi: 10.1200/jco.2011.29.15_suppl.e13098
Nie, Q., Yang, X. N., An, S. J., Zhang, X. C., Yang, J. J., Zhong, W. Z., et al. (2011b). CYP1A1*2A polymorphism as a predictor of clinical outcome in advanced lung cancer patients treated with EGFR-TKI and its combined effects with EGFR intron 1 (CA)n polymorphism. Eur. J. Cancer 47 (13), 1962–1970. doi: 10.1016/j.ejca.2011.04.018
Novello, S., Barlesi, F., Califano, R., Cufer, T., Ekman, S., Levra, M. G., et al. (2016). Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 27 (suppl 5), v1–v27. doi: 10.1093/annonc/mdw326
Parra-Guillen, Z. P., Berger, P. B., Haschke, M., Donzelli, M., Winogradova, D., Pfister, B., et al. (2017). Role of Cytochrome P450 3A4 and 1A2 Phenotyping in Patients with Advanced Non-small-Cell Lung Cancer Receiving Erlotinib Treatment. Basic Clin. Pharmacol. Toxicol. 121 (4), 309–315. doi: 10.1111/bcpt.12801
Patel, J. N. (2016). Cancer pharmacogenomics, challenges in implementation, and patient-focused perspectives. Pharmgenom. Pers. Med. 9, 12. doi: 10.2147/PGPM.S62918
Riely, G. J., Marks, J., Pao, W. (2009). KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 6 (2), 201–205. doi: 10.1513/pats.200809-107LC
Rudin, C. M., Liu, W., Desai, A., Karrison, T., Jiang, X., Janisch, L., et al. (2008). Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J. Clin. Oncol. 26 (7), 1119–1127. doi: 10.1200/JCO.2007.13.1128
Scott, S. A. (2011). Personalizing medicine with clinical pharmacogenetics. Genet. Med. 13 (12), 987–995. doi: 10.1097/GIM.0b013e318238b38c
Shi, Z., Peng, X. X., Kim, I. W., Shukla, S., Si, Q. S., Robey, R. W., et al. (2007). Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 67 (22), 11012–11020. doi: 10.1158/0008-5472.CAN-07-2686
Thorn, M., Finnstrom, N., Lundgren, S., Rane, A., Loof, L. (2005). Cytochromes P450 and MDR1 mRNA expression along the human gastrointestinal tract. Br. J. Clin. Pharmacol. 60 (1), 54–60. doi: 10.1111/j.1365-2125.2005.02389.x
Timmers, L., Boons, C. C., Moes-Ten Hove, J., Smit, E. F., van de Ven, P. M., Aerts, J. G., et al. (2015). Adherence, exposure and patients' experiences with the use of erlotinib in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 141 (8), 1481–1491. doi: 10.1007/s00432-015-1935-0
Tournier, N., Goutal, S., Auvity, S., Traxl, A., Mairinger, S., Wanek, T., et al. (2017). Strategies to Inhibit ABCB1- and ABCG2-Mediated Efflux Transport of Erlotinib at the Blood-Brain Barrier: A PET Study on Nonhuman Primates. J. Nucl. Med. 58 (1), 117–122. doi: 10.2967/jnumed.116.178665
Wojnowski, L. (2004). Genetics of the Variable Expression of CYP3A in Humans. Ther. Drug Monit. 26 (2), 7. doi: 10.1097/00007691-200404000-00019
Zhou, X., Qiao, G., Wang, X., Song, Q., Morse, M. A., Hobeika, A., et al. (2018). CYP1A1 genetic polymorphism is a promising predictor to improve chemotherapy effects in patients with metastatic breast cancer treated with docetaxel plus thiotepa vs. docetaxel plus capecitabine. Cancer Chemother. Pharmacol. 81 (2), 365–372. doi: 10.1007/s00280-017-3500-9
Keywords: erlotinib, non-small cell lung cancer, polymorphism, drug-metabolizing enzymes, transporters, steady state trough concentration
Citation: Liao D, Liu Z, Zhang Y, Liu N, Yao D, Cao L, Chen Y, Fu Y, Yang N and Xiang D (2020) Polymorphisms of Drug-Metabolizing Enzymes and Transporters Contribute to the Individual Variations of Erlotinib Steady State Trough Concentration, Treatment Outcomes, and Adverse Reactions in Epidermal Growth Factor Receptor–Mutated Non-Small Cell Lung Cancer Patients. Front. Pharmacol. 11:664. doi: 10.3389/fphar.2020.00664
Received: 02 August 2019; Accepted: 23 April 2020;
Published: 08 May 2020.
Edited by:
Olavi R. Pelkonen, University of Oulu, FinlandReviewed by:
Snehal Samant, University of Florida, United StatesCopyright © 2020 Liao, Liu, Zhang, Liu, Yao, Cao, Chen, Fu, Yang and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nong Yang, eWFuZ25vbmczMkAxNjMuY29t; Daxiong Xiang, eGlhbmdkYXhpb25nQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.