- 1Hainan Provincial Key Laboratory of Resources Conservation and Development of Southern Medicine, Hainan Branch of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Haikou, China
- 2Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 4National Center of Traditional Medicine, Ministry of Health of Cambodia, Phnom Penh, Cambodia
In Cambodia, medicinal plants are often used to treat various illnesses. However, the identities of many medicinal plants remain unknown. In this study, we collected 50 types of traditional Cambodian medicinal plants that could not be identified by their appearance from a domestic market. We utilized the DNA barcoding technique, combined with the literature survey, to trace their identities. In the end, 33 species were identified at the species level and 7 species were identified at the genus level. The ethnopharmacological information of 33 medicinal plants was documented. The DNA barcoding technique is useful in the identification of medicinal plants with no previous information.
Introduction
Cambodia is located in the Indo-China Peninsula, where it borders Thailand, Vietnam, and Lao PDR in Southeast Asia. Although it does not exceed 4% (181,035 km2) of the total area of Southeast Asia, Cambodia is well-known for its rich biodiversity, overlapping with four of the 25 “biodiversity hotspots” and maintaining rich natural resources and a unique ecosystem. It is estimated that the country has more than 3,000 vascular plant species (Chassagne et al., 2016). Approximately 1,200 medicinal plants are used to treat diseases (Xu, 2008; Walker, 2017). Traditional medicine plays an important role in the lives of most Cambodians. In the face of disease, 70–80% of Cambodians opt for traditional medicinal methods (Walker, 2017; Yao et al., 2017) with approximately 40–50% of the population using medicinal plants daily (Chassagne et al., 2017).
Traditional Cambodian medicine involves several cultural and regional traditions derived from Theravada Buddhism, Ayurveda, traditional Chinese medicine, and French pharmaceutical traditions (Chassagne et al., 2017). Among these, Chinese and Ayurvedic medicines are the two oldest and most comprehensive medical systems based on natural medicinal agents. Consequently, the importance of traditional medicinal plant research in Cambodia is relatively high. However, Cambodia still does not have a national ethnopharmacopoeia (World Health Organization [WHO], 2005). Furthermore, there are few curricula teaching traditional Cambodian medicine, and books offer inconsistent and confusing information (Richman et al., 2010). Due to climate change and agro-industrial development in Cambodia, the local ecological environment is threatened (Chassagne et al., 2016). At the same time, Western medicine is being promoted, and knowledge of medicinal plants is being lost (Xu, 2008). Therefore, the study of medicinal plants in this country is very important. The medicinal plant market is not only a place for the sale of natural therapies but also a place for people to exchange information on medicinal plants, which preserves the knowledge as the information is passed from one generation to the next (Lee et al., 2008; de Carvalho Nilo Bitu et al., 2015; Jin et al., 2018). Therefore, we chose to conduct our study at a traditional market in Phnom Penh, the Cambodian capital.
We performed this study in August 2016, December 2016, and November 2017 in Orussey Market, which is one of the largest traditional markets for Chinese merchants in Phnom Penh carrying a wide variety of medicinal plants. The medicinal materials market that we surveyed represented only a small part of the entire Orussey Market (Figure 1). Only about 10 merchants were selling herbal medicines. The business model of the Cambodian medicinal plant market is mainly retail sales. Each store was small and independently run, with its own shop name. The medicinal plants were stored outside of the shops for customers to browse and purchase. The herbs had no fixed specifications, and they were derived from plants in the region. There were various types of herbs that included roots, stems, leaves, fruits, and whole plants. Most medicinal plants were previously dried, and a few fresh medicinal plants were formulated into medicines or single-flavored products. Due to the large number of Chinese customers, the shops also sold commonly used herbs in China, such as red dates, pepper, and Atractylodes macrocephalae rhizome (Atractylodes macrocephala Koidz.). The quality and specifications of the medicinal plants were not significantly different from store to store, and the price was similar across different shops. We collected samples from a total of 118 medicinal plants, of which 68 could be identified by morphology, whereas the remaining 50 species could not be morphologically distinguished. The main objective of this study was to clarify the original species of these medicinal plants.
To identify medicinal plants, morphological, microscopical, and physical and chemical identification methods are commonly used, but they require experienced investigators that are knowledgeable in the field. A shortage of experienced investigators has led to difficulties in identifying unknown plants. With advances in science and technology, such as chromatography, spectroscopy, and X-ray diffraction, new methods have been used to study medicinal plants (Chen et al., 2012). Although most methods are not useful when combined with the starting material of unknown origin, they can provide indirect evidence for the authenticity of the material (Han et al., 2016).
The DNA barcoding technique is an effective tool that can identify unknown medicinal plants with no background information (Chen S. et al., 2013). It uses a short DNA sequence from a standard and agreed-upon position in the genome to identify the species rapidly and accurately (de Vere et al., 2015; Li et al., 2015). The experimental method is fast, standardized, and simple. It generates a large experimental throughput and easily identifies the species (Chen et al., 2011). The common DNA barcodes for plants are rbcL, matK, ITS2, and psbA-trnH; however, matK is difficult to amplify with commonly used primers (Li et al., 2015); therefore, different taxonomic groups require different sets of primers (Hollingsworth, 2008). Furthermore, matK sequences evolve slowly, and this locus has by far the lowest divergence among plastid genes in flowering plants (Kress et al., 2005). Due to its modest discriminatory ability, it is not recommended for studies at the species level. Presently, psbA-trnH is the most widely used plastid barcode for species identification, as its universal primers can amplify nearly all angiosperms (Shaw et al., 2007). Internal transcribed spacer 2 (ITS2), a part of the nuclear DNA, is another ideal barcode because of its short length, easy amplification with a single primer pair, high sequencing efficiency, and high variation between species (Yu et al., 2017).
Internal transcribed spacer 2 and psbA-trnH represent the universal barcode for the reliable identification of medicinal plants (Chen et al., 2010; Chen S. et al., 2013; Yao et al., 2010). The ITS2 sequence can accurately identify Solani nigri (Solanum nigrum L.) and its sibling species (Chen et al., 2017) Scutellaria barbata D.Don and its adulterants (Guo et al., 2016), and Lycium barbarum L. and its adulterants (Xin et al., 2013), as well as the origin of various ginseng plants based on SNP barcodes (Chen X. et al., 2013). This technique can also be used to classify and identify medicinal plants from family Orchidaceae (Tang et al., 2017). Furthermore, the ITS2 region has been used to supervise the proportions and varieties of adulterant species (Yu et al., 2017). Presently, DNA barcoding is widely used, as it has been applied in the authentication and identification of small berry fruits (Wu et al., 2018), as well as in the study of Li minority medicine (Cui et al., 2019) and various animal species (Yang et al., 2018). The technique has also been used to identify Sida L. herbal products (Santhosh Kumar et al., 2015), traditional Dai medicines, and laxative producing plants (Seethapathy et al., 2015). Han et al. (2016) identified unknown herbal plants in common markets and reported the adulterant rate. In this study, we used the DNA barcoding technique to identify the 50 unknown medicinal plants obtained from the Orussey Market, one of the largest Cambodian traditional markets.
Results and Discussion
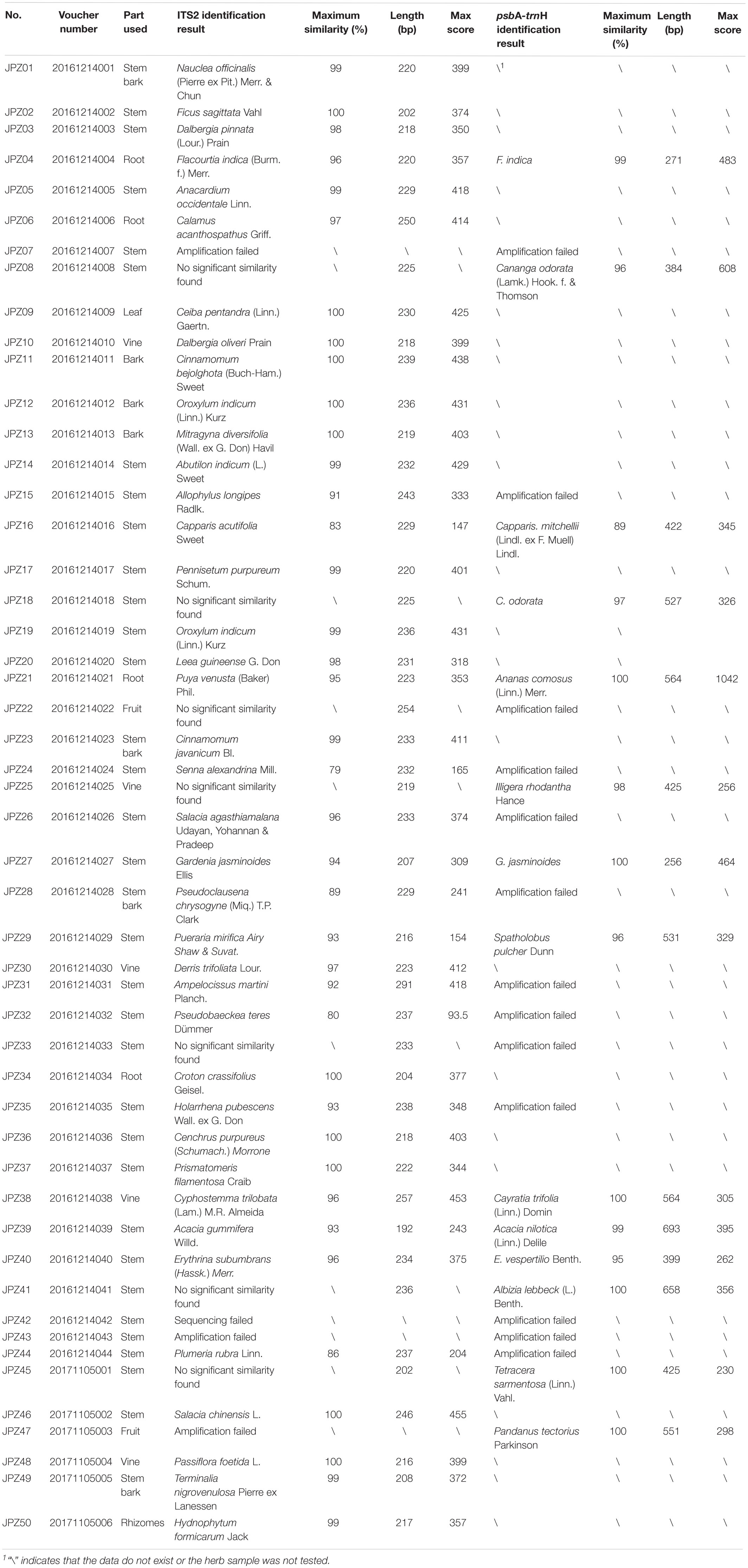
In total, we collected samples from 118 medicinal plants, of which 68 could be morphologically identified, the remaining 50 were randomly selected for DNA barcoding analysis. According to Table 1, 42 of the 50 samples were derived from the stem, bark, and vine, except for two fruits, four roots, one leaf, and one rhizome. Based on their morphological appearance, it was difficult to confirm their identity. The ITS2 amplification success rate was 94% (47/50), and the sequencing success rate was 98% (46/47). According to the results of ITS2 and psbA-trnH experiments, 33 plants were identified at the species level. Seven were identified at the genus level. Ten plants were unidentifiable (five were due to either amplification failures, sequencing failures or no matched results, and five were due to the low maximum similarity). There were 27 medicinal plants with a maximum similarity <97%, and the psbA-trnH region was amplified in these samples. The psbA-trnH amplification success rate was 52% (14/27), and the sequencing success rate was 100% (14/14). All the identification results (matched species, maximum similarity, length, and maximum score) are shown in Table 1. The maximum similarity range of ITS2 was 83–100%, and that for psbA-trnH was 89–100%. In this study, the ITS2 amplification of three samples and psbA-trnH amplification of 13 samples were unsuccessful, which might have been due to impure or degraded DNA of processed medicinal materials. This shows the boundedness of DNA barcoding due to the instability of DNA. This result also reflects the limitations of Sanger sequencing, such as the unspecific amplification of non-target DNA when target DNA is degraded (Pawar et al., 2017). In the method part, we have taken some measures to avoid these problems. If mixed sequencing signals were present, we would clone the PCR products and sequence the single colonies to identify target amplicons. To overcome the limitations of Sanger sequencing, we can employ next-generation sequencing (NGS) approach technology to simultaneously detect plant and fungal DNAs (Pawar et al., 2017). And it is also possible that the samples were not suitable for ITS2 or psbA-trnH amplification. To tackle this issue, specific primers (Zhao et al., 2018) or other types of barcodes, such as mini-barcodes (Song et al., 2017; Liu, 2018) or plastid super-barcodes (Krawczyk et al., 2018), can be used. However, this method also has limitations, according to our data, seven medicinal species were identified at the genus level, five exhibited low maximum similarity (<90%), and several could not be matched to any of the existing medicinal species, presumably due to the low species-level resolution of many plant genera and insufficient database information of GenBank. Therefore, the analyses with multiple genetic loci (e.g., single-nucleotide polymorphisms, SNPs) and other analytical methods, such as infrared spectroscopy and X-ray diffraction, must be employed to achieve high resolution for species differentiation (Chen et al., 2012).
Seventeen herbs could not be identified at the species level; besides the limitations of DNA barcoding, one plausible explanation can be that incomplete database information. In this context, accurate species identification by botanists is key to improve the NCBI database for identifying unknown medicinal species.
Interestingly, Chhke sreng, a pricy panacea in traditional medicine in Cambodia, was identified as Cananga brandisiana (Pierre) Saff. [syn. Cananga latifolia (Hook.f. & Thomson) Finet & Gagnep.] but not Cananga odorata (Lamk.) Hook. f. & Thomson (also called “Kdang nie”) by plant taxonomists (Dy Phon, 2000).
However, C. odorata and Chhke sreng are related to each other as indicated by several references, presumably owing to the mismatch of local and Latin names of some medicinal species or the adulteration and mis-authentication of C. odorata in Cambodian markets. Thus, DNA barcoding plays a key role in ensuring medicinal safety in Cambodia and it would be better when DNA barcoding combines chemical information.
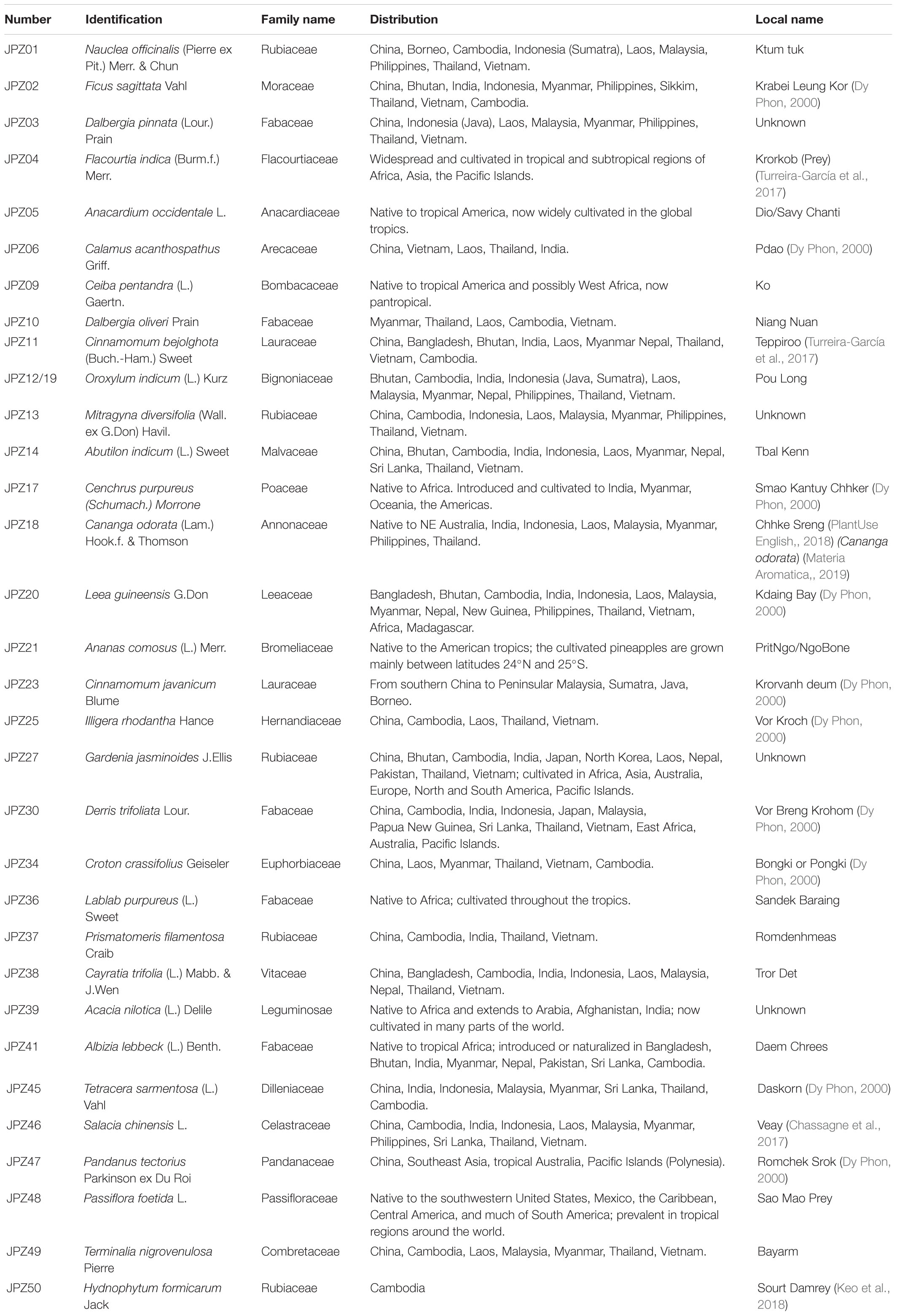
The ethnopharmacological information (family name, distribution, local name) of 33 plants is shown in Table 2. Among these, there were five Rubiaceae and eight Fabaceae plants. The pictures of five representative medicines are shown in Figure 2. Furthermore, legumes were the most cited plants in studies from Cambodia, Thailand, and Laos (Chassagne et al., 2017).
Importantly, 19 out of 33 medicinal plants were also used as Chinese medicines, and they were Nauclea officinalis, Ficus sagittata, Dalbergia pinnata, Flacourtia indica, Anacardium occidentale, Dalbergia oliveri, Cinnamomum bejolghota, Oroxylum indicum, Abutilon indicum, Illigera rhodantha, Gardenia jasminoides, Derris trifoliata, Croton crassifolius, Lablab purpureus, Cayratia trifolia, Acacia nilotica, Pandanus tectorius, Passiflora foetida, and Terminalia nigrovenulosa. Among these, O. indicum, P. tectorius, and N. officinalis were the typical traditional Chinese herbs with a long history of use. N. officinalis, also known as Li minority medicine from which several new drugs have been developed, can lower body temperature, detoxify the body, reduce swelling, and relieve pain. It is often used in the treatment of colds, fevers, acute tonsillitis, pharyngitis, pneumonia, urinary system infections, enteritis, dysentery, carbuncle, and other diseases. Oroxylum indicum has been included in the 2015 Chinese Pharmacopoeia for treating fevers, coughs, sore throats, lung phlegm, liver diseases, and stomach pain (Chinese Pharmacopoeia Comission, 2015). The flowers, leaves, fruits, roots, and rhizomes of P. tectorius have high medicinal value in the remedy of colds, fevers, hepatitis, dysentery, hemorrhoids, orchitis, syphilis, and diabetes mellitus (Zhan, 2013). Therefore, studies are needed to further characterize these plants of medicinal value. According to the results shown in Table 2, there were 19 species distributed in Guangdong, 17 species in Guangxi, and 15 species in Yunnan. Therefore, the medicinal plants in Cambodia are similar to the three aforementioned provinces in China.
Materials and Methods
Study Area and Materials
In Orussey Market, we interviewed a total of 10 medicine retailors, all sellers were briefed on the purpose and details of the investigation, they were also informed that the investigation could be terminated any time as needed. As shown in Tables 1, 2, we collected medicinal plant samples and recorded the local names of the medicinal plants. Some local names of these plants were provided by the sellers, others were obtained by reviewing the literature. Ethnopharmacological information including family name and distribution of medicinal species were also obtained from the literature. The medicinal material samples were dried and stored in Herbarium of Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences (IMD). Voucher numbers are listed in Table 1.
DNA Extraction
Approximately 30 mg of each sample was ground for 2 min (40 Hz) using a high-throughput tissue grinder (Scientz-48, Ningbo, China). Total genomic DNA was extracted using a plant genomic DNA extraction kit (Tiangen Biotech Co., China). Occasionally, alien DNA sequences from other species – such as fungi and algae – or mixed sequence signals were repeatedly detected if the primers were not specific. To prevent non-specific PCR amplification, we washed the samples of medicinal materials using 75% alcohol to remove fungi and other plant powder contaminations.
PCR Amplification and Sequencing
The primers used for amplification and sequencing were as follows: ITS2 (the second ITS) (forward, 5′-GCGATACTTG GTGTGAAT-3′; reverse, 5′-GACGCTTCTCCAGACTACA AT-3′) (Chen et al., 2010) and psbA-trnH intergenic spacer [forward, 5′-GTTATGCATGAACGTAATGCTC-3′ (Sang et al., 1997); reverse, 5′-CGCGCATG GTGGATTCACAATCC-3′ (Tate and Simpson, 2003)]. Primers were synthesized by Shanghai Shenggong Bioengineering Co., Ltd. The 25 μL PCR reaction contained 12.5 μL of 2× Taq PCR Mix, 1.0 μL each of the forward and reverse primers (2.5 μmol L–1), 8.5 μL of double distilled water, and 2.0 μL of the template (genomic DNA < 0.1 ng). The PCR amplification procedure for ITS2 was as follows: denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 45 s. The PCR amplification procedure for psbA-trnH was as follows: denaturation at 95°C for 4 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min. A final extension was performed at 72°C for 10 min for both PCR amplification procedures. The PCR was conducted in a thermal cycler (model 2720; Thermo Fisher Scientific). Bi-directional sequencing of the PCR products was performed by Beijing Qingke New Industry Biotechnology Co., Ltd.
Data Analysis
Codon Code Aligner V 7.0.1 (CodonCode Co., United States) was used to assemble and cut the contigs and to generate the ITS2 and psbA-trnH sequences. The sequences were submitted to the National Center for Biotechnology Information (NCBI) database to search for other similar sequences, which have been taxonomically validated from published literatures. To identify the species of each medicinal plant, each species was searched in the literature in descending order of similarity. The maximum score was used to determine if the medicinal plant distributed in Cambodia. If its origin was from Cambodia and its similarity was ≥97% [97% was used as the DNA barcoding identification similarity threshold for medicinal plants (Chen et al., 2012; Gu et al., 2015)], the species identified were considered to be the final. If the similarity was ≤97%, the psbA-trnH sequence was amplified. The final identification results of similarities ≥97% were determined according to the aforementioned ITS2 method. If the similarity was between 90 and 97%, the results revealed the genus level. In cases of inconsistent ITS2 and psbA-trnH results, we choose the one with the higher similarity.
Ethics
We plan to work with the National Center of Traditional Medicine (NCTM), Ministry of Health of Cambodia, to publish Handbook of medicinal plants in Cambodia, which will be made available to the Cambodian sellers we interviewed after its publication. Meanwhile, we also collaborate with NCTM on the Sino-Cambodian International Exchange Project to promote educational and academic communications between China and Cambodia; this project will also provide Cambodia with technical and theoretical supports in the identification and marker-assisted selection of medicinal plants.
Conclusion
Cambodia has a history of nearly 2000 years, and for a long time, it has suffered from civil wars and wars of aggression. In light of the extremely poor conditions, the Cambodian people have relied on their own practices to identify medicinal plants to fight diseases. Although there is some information on various medicinal plants, there is no pharmacopoeia and no readily available body of medicinal plant literature, which has hindered the application of medicinal plants and the dissemination of results from investigators of other countries. Therefore, the current study of medicinal plants in Cambodia is incomplete, and there remain many gaps in knowledge. Furthermore, many plants have become endangered due to industrialization and environmental pollution (Chassagne et al., 2016). In light of this, the DNA barcoding technique can provide useful information on the species of various medicinal plants in Cambodia. This will not only preserve plant knowledge in Cambodia, but also help develop an ethnopharmacopia and provide new insights on the development of new drugs in the future. At the same time, the use of DNA barcoding is one step in supporting the improvement in the quality control of plants being sold for medicinal use in Cambodia and it will emphasize the importance of protecting endangered species. This approach can also be used in other countries or regions with relatively backward economies and underdeveloped research practices. Through DNA barcoding, commonly used medicinal plants can be completely characterized.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XC, XZ, WL, YQ, TH, RL, and YY: investigation. WS, XZ, YS, XM, and YM: methodology. JW and XZ: project administration. XZ, WS, and WL: resources. JW: supervision. XC: writing – original draft. XZ and WS: writing – review and editing.
Funding
This research was funded by the CAMS Innovation Fund for Medical Sciences (CIFMS), grant number 2016-I2M-2-003.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the locals who shared traditional knowledge and helped us during the market research process. we also thank Professor Bengang Zhang, Associate Researcher Haitao Li, Liwei Jia, Danqi Chen, Shu Pan, and others for their help during the investigation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00244/full#supplementary-material
References
Chassagne, F., Deharo, E., Punley, H., and Bourdy, G. (2017). Treatment and management of liver diseases by Khmer traditional healers practicing in Phnom Penh area, Cambodia. J. Ethnopharmacol. 202, 38–53. doi: 10.1016/j.jep.2017.03.002
Chassagne, F., Hul, S., Deharo, E., and Bourdy, G. (2016). Natural remedies used by Bunong people in Mondulkiri province (Northeast Cambodia) with special reference to the treatment of 11 most common ailments. J. Ethnopharmacol. 191, 41–70. doi: 10.1016/j.jep.2016.06.003
Chen, S., Guo, B., Zhang, G., Yan, Z., Luo, G., Sun, S., et al. (2012). Advances of studies on new technology and method for identifying traditional Chinese medicinal materials. China J. Chin. Mater. Med. 37, 1043–1055.
Chen, S., Pang, X., Yao, H., Han, J., and Luo, K. (2011). Identification system and perspective for DNA barcoding traditional Chinese materia medica. World Sci. Technol. 5, 747–754.
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613. doi: 10.1371/journal.pone.0008613
Chen, S., Yao, H., Han, J., Xin, T., Pang, X., Shi, L., et al. (2013). Principles for molecular identification of traditional chinese materia medica using DNA barcoding. China J. Chin. Mater. Med. 38, 141–148.
Chen, X., Bai, N., Xiang, L., Sun, W., and Mei, Q. (2017). Molecular identification of Solani Nigri Herba and its sibling species based on DNA barcoding. J. Chin. Med. Mater. 40, 290–294.
Chen, X., Liao, B., Song, J., Pang, X., Han, J., and Chen, S. (2013). A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene 530, 39–43. doi: 10.1016/j.gene.2013.07.097
Chinese Pharmacopoeia Comission, (2015). Pharmacopoeia of the People Republic of China, Vol. 1. Beijing: China Medical Science Press.
Cui, X., Sun, W., Xiong, C., Meng, X., Shi, Y., Wu, L., et al. (2019). Identification of 23 unknown Li medicinal plants based on DNA barcoding. China J. Chin. Mater. Med. 44, 357–366. doi: 10.19540/j.cnki.cjcmm.20181106.002
de Carvalho Nilo Bitu, V., de Carvalho Nilo Bitu, V., Matias, E. F. F., de Lima, W. P., da Costa Portelo, A., Coutinho, H. D. M., et al. (2015). Ethnopharmacological study of plants sold for therapeutic purposes in public markets in Northeast Brazil. J. Ethnopharmacol. 172, 265–272. doi: 10.1016/j.jep.2015.06.022
de Vere, N., Rich, T. C. G., Trinder, S. A., and Long, C. (2015). DNA barcoding for plants. Methods Mol. Biol. 1245, 101–118. doi: 10.1007/978-1-4939-1966-6_8
Gu, X., Li, Y.-p., Cui, X.-m., Wang, J., Han, L., Zhao, B., et al. (2015). The pharmaceutical research on Brazilian herb CENTELHA ASIATICA (Centella asiatica) based on a combined analysis of DNA barcoding-origin-morphology. Glob. Tradit. Chinese Med. 8, 1294–1297. doi: 10.3969/j.issn.1674-1749.2015.11.005
Guo, M., Ren, L., Chen, X., and Pang, X. (2016). Identification of Scutellaria barbata and its adulterants using ITS2 barcode. World Chin. Med. 5, 796–800.
Han, J., Pang, X., Liao, B., Yao, H., Song, J., and Chen, S. (2016). An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 6:18723. doi: 10.1038/srep18723
Hollingsworth, P. M. (2008). DNA barcoding plants in biodiversity hot spots: progress and outstanding questions. Heredity (Edinb.) 101, 1–2. doi: 10.1038/hdy.2008.16
Jin, B., Liu, Y., Xie, J., Luo, B., and Long, C. (2018). Ethnobotanical survey of plant species for herbal tea in a Yao autonomous county (Jianghua, China): results of a 2-year study of traditional medicinal markets on the Dragon Boat Festival. J. Ethnobiol. Ethnomed. 14, 58. doi: 10.1186/s13002-018-0257-0
Keo, S., Meng, C., Chhea, S., and Chea, S. (2018). Ethnobotanical survey of medicinal plants used by traditional healers in Kampong Speu province, Cambodia. Asian J. Pharmacogn. 2, 21–32.
Krawczyk, K., Nobis, M., Myszczyński, K., Klichowska, E., and Sawicki, J. (2018). Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci. Rep. 8:1924. doi: 10.1038/s41598-018-20399-w
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., and Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Lee, S., Xiao, C., and Pei, S. (2008). Ethnobotanical survey of medicinal plants at periodic markets of Honghe Prefecture in Yunnan Province, SW China. J. Ethnopharmacol. 117, 362–377. doi: 10.1016/j.jep.2008.02.001
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Liu, Y. (2018). Study of TCM Nucleotide Signature Based on Mini-Barcoding Panax quinquefolius, Ophiocordyceps sinensis and Ginkgo biloba for Examples. Beijing: Peking Union Medical College.
Materia Aromatica, (2019). Available online at: https://materiaaromatica.com/oil-profiles/ylang-ylang-oil (accessed July 11, 2019).
Pawar, R. S., Handy, S. M., Cheng, R., Shyong, N., and Grundel, E. (2017). Assessment of the authenticity of herbal dietary supplements: comparison of chemical and DNA barcoding methods. Planta Med. 83, 921–936. doi: 10.1055/s-0043-107881
PlantUse English, (2018). Plant Use. Available online at: https://uses.plantnet-project.org/en/Cananga_odorata_(PROSEA) (accessed July 10, 2019).
Richman, M. J., Nawabi, S., Patty, L., and Ziment, I. (2010). Traditional Cambodian medicine. J. Complement. Integr. Med. 7, 1–14. doi: 10.2202/1553-3840.1194
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi: 10.2307/2446155
Santhosh Kumar, J. U., Krishna, V., Seethapathy, G. S., Senthilkumar, U., Ragupathy, S., Ganeshaiah, K. N., et al. (2015). DNA barcoding to assess species adulteration in raw drug trade of “Bala” (Genus: Sida L.) herbal products in South India. Biochem. Syst. Ecol. 61, 501–509. doi: 10.1016/j.bse.2015.07.024
Seethapathy, G. S., Ganesh, D., Santhosh Kumar, J. U., Senthilkumar, U., Newmaster, S. G., Ragupathy, S., et al. (2015). Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int. J. Legal Med. 129, 693–700. doi: 10.1007/s00414-014-1120-z
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Song, M., Dong, G., Zhang, Y., Liu, X., and Sun, W. (2017). Identification of processed Chinese medicinal materials using DNA mini-barcoding. Chin. J. Nat. Med. 15, 481–486. doi: 10.1016/S1875-5364(17)30073-0
Tang, H., Xiang, L., Li, X., Sun, W., Wang, M., Huang, Y., et al. (2017). DNA barcoding identification of endangered medicinal plants of Orchidaceae. China J. Chin. Mater. Med. 42, 2058–2067. doi: 10.19540/J.CNKI.CJCMM.2017.0090
Tate, J. A., and Simpson, B. B. (2003). Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 28, 723–737.
Turreira-García, N., Argyriou, D., Phourin, C., Srisanga, P., and Theilade, I. (2017). Ethnobotanical knowledge of the Kuy and Khmer people in Prey Lang, Cambodia. Cambodian J. Nat. Hist. 2017, 76–101.
Walker, T. (2017). An Examination of Medicinal Ethnobotany and Biomedicine Use in Two Villages on the Phnom Kulen plateau. thesis, Hollins University, Roanoke.
World Health Organization [WHO] (2005). National Policy on Traditional Medicine and Regulation of Herbal Medicines. Report of a WHO Global Survey, 1–168. Available online at: https://apps.who.int/iris/bitstream/handle/10665/43229/9241593237.pdf (accessed April 23, 2019).
Wu, Y., Li, M., Yang, Y., Jiang, L., Liu, M., Wang, B., et al. (2018). Authentication of small berry fruit in fruit products by DNA barcoding method. J. Food Sci. 83, 1494–1504. doi: 10.1111/1750-3841.14177
Xin, T., Yao, H., Gao, H., Zhou, X., Ma, X., Xu, C., et al. (2013). Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res. Int. 54, 1699–1704. doi: 10.1016/J.FOODRES.2013.10.007
Xu, Z. (2008). Preliminary comparison on traditional knowledge of medicinal plants used by Dai Xishuangbanna China and Khmer Cambodia. Acta Bot. Yunnan. 30, 371–377.
Yang, F., Ding, F., Chen, H., He, M., Zhu, S., Ma, X., et al. (2018). DNA Barcoding for the identification and authentication of animal species in traditional medicine. Evid. Based Complement. Altern. Med. 2018:5160254. doi: 10.1155/2018/5160254
Yao, H., Song, J., Liu, C., Luo, K., Han, J., Li, Y., et al. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 5:e13102. doi: 10.1371/journal.pone.0013102
Yao, X., Pen, S., and Meng, Y. (2017). Commonly used traditional medicinal plants in Cambodia. Mod. Chin. Med. 19, 290–294.
Yu, N., Wei, Y., Zhang, X., Zhu, N., Wang, Y., Zhu, Y., et al. (2017). Barcode ITS2: a useful tool for identifying Trachelospermum jasminoides and a good monitor for medicine market. Sci. Rep. 7:5037. doi: 10.1038/s41598-017-04674-w
Zhan, L. (2013). Study on Extraction and Separation of Active Ingredients from Pandanus tectorius Sol. Haikou: Hainan University.
Keywords: Cambodia, herb, DNA barcoding, ITS2, psbA-trnH
Citation: Cui X, Li W, Wei J, Qi Y, Li R, Yang Y, Shi Y, Meng X, Mi Y, Huot T, Sun W and Zheng X (2020) Assessing the Identity of Commercial Herbs From a Cambodian Market Using DNA Barcoding. Front. Pharmacol. 11:244. doi: 10.3389/fphar.2020.00244
Received: 09 May 2019; Accepted: 21 February 2020;
Published: 24 March 2020.
Edited by:
Rong-Rong He, Jinan University, ChinaReviewed by:
Natalia V. Ivanova, University of Guelph, CanadaFrançois Chassagne, Emory University, United States
Copyright © 2020 Cui, Li, Wei, Qi, Li, Yang, Shi, Meng, Mi, Huot, Sun and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Sun, d3N1bkBpY21tLmFjLmNu; Xilong Zheng, emhlbmd4bDIwMTJAc2luYS5jb20=
 Xinyun Cui
Xinyun Cui Weijie Li
Weijie Li Jianhe Wei1,2
Jianhe Wei1,2 Wei Sun
Wei Sun