- 1Department of Pharmacognosy, Institute of Pharmaceutical Sciences, University of Graz, Graz, Austria
- 2Department of Pharmaceutical Chemistry, Institute of Pharmaceutical Sciences, University of Graz, Graz, Austria
- 3Department of Pharmacognosy, Molecular Targets, University of Vienna, Vienna, Austria
In order to identify active constituents and to gain some information regarding their mode of action, extracts from leaves of Epipremnum pinnatum were tested for their ability to inhibit inflammatory gene expression in endothelial- and monocyte-like cells (HUVECtert and THP-1, respectively). Bioactivity-guided fractionation using expression of PTGS2 (COX-2) mRNA as a readout resulted in the isolation of two C13 megastigmane glycosides, gusanlungionoside C (1) and citroside A (3), and the phenylalcohol glycoside phenylmethyl-2-O-(6-O-rhamnosyl)-ß-D-galactopyranoside (2). Further analysis identified six additional megastigmane glycosides and the aglycones β-damascenone (10), megastigmatrienone (11), 3-hydroxy-β-damascenone (12), and 3-oxo-7,8-dihydro-α-ionol (13). Pharmacological analysis demonstrated that 10 inhibits LPS-stimulated induction of mRNAs encoding for proinflammatory cytokines and leukocyte adhesion molecules, such as TNF-α, IL-1β, IL-8, COX-2, E-selectin, ICAM-1, and VCAM-1 in HUVECtert and THP-1 cells. 10 inhibited induction of inflammatory genes in HUVECtert and THP-1 cells treated with different agonists, such as TNF-α, IL-1β, and LPS. In addition to mRNA, also the upregulation of inflammatory proteins was inhibited by 10 as demonstrated by immune assays for cell surface E-selectin and secreted TNF-α. Finally, using a luciferase reporter construct, it was shown, that 10 inhibits NF-κB-dependent transcription. Therefore, we hypothesize that inhibition of NF-κB by β-damascenone (10) may represent one of the mechanisms underlying the in vitro anti-inflammatory activity of Epipremnum pinnatum extracts.
Introduction
Epipremnum pinnatum (L.) Engl. (Araceae) is a slender to gigantic liana mostly distributed in tropical Southeast Asia and the Pacific Islands (Boyce, 1998; Wu et al., 2010). It is known as a popular indoor ornamental plant in western countries with the property to remove air pollutants, such as benzene, formaldehyde, and chloroform (Liu et al., 2011; Tada et al., 2010). In traditional Chinese medicine, aqueous leaf extracts have been used for detoxification and treatment of tendonitis, fractures, burns, carbuncles, sores, redness, and cancer (Chan et al., 2008; Siew et al., 2014). In other regions, such as Papua New Guinea and Rotuman, the plant has been used against inflammation, diabetes, and malaria (McClatchey, 1996; WHO, 2009). Even today, Epipremnum is part of herbal formulations, such as plasters, ointments, and medicinal wines (Ren, 2010; Li, 2013; Zhang 2013; Xiang and Zhang, 2014; Zong, 2014; Liu, 2015; Wang, 2015; Zong, 2015).
Previous investigations of lipophilic n-hexane leaf and stem extracts demonstrated a cytotoxic effect against T-47D breast cancer cells (EC50 = 2.90 µg/ml) by upregulation of c-myc and caspase 3 mRNA through the activation of PKC and tyrosine kinases pathways (Tan et al., 2005; Lan et al., 2007). Cytotoxic activity against P388 murine lymphocytic leukemia cells, MOLT4 human leukemia cells, KB human nasopharynx cancer cells, and SW 620 human colon adenocarcinoma cells was also reported from a diethylether extract (Lan et al., 2007). Antibacterial and antifungal activity was demonstrated by an aqueous extract (Sonawane et al., 2011), while the extracts obtained using supercritical CO2 inhibited pancreatic lipase (Hamzah et al., 2015). Moreover, the EtOH extract of aerial parts of Epipremnum pinnatum exerted anti-inflammatory activity in carrageenan-induced rat paw edema model, and induced analgesic effects in mice (Linnet et al., 2010).
Phytochemical screening of Epipremnum pinnatum revealed the presence of alkaloids, flavonoids, anthraquinones, tannins, glycosides, phenols, phytosterols, carbohydrates, and terpenoids (Arulpriya and Lalitha, 2011; Das et al., 2015; Meshram and Srivastava, 2015). One unstable cytotoxic compound has been isolated and identified as 5,6-dihyroxyindole. This compound rapidly undergoes autoxidation to form a black polymeric material (Wong et al., 1996; Goh, 1998).
The nuclear factor kappa B (NF-κB) family of transcription factors is ubiquitously expressed in mammalian cells (May and Ghosh, 1998) and is activated by complex signaling cascades (Hayden and Ghosh, 2012; Hoesel and Schmid, 2013). NF-κB is involved in the regulation of cell differentiation, proliferation, and cell survival. Moreover, it is a major player within the immune system (Pahl, 1999; Hoesel and Schmid, 2013).
NF-κB can be activated by pro-inflammatory stimuli including the cytokines lL-1β and TNF-α, as well as bacterial and viral antigens (Pahl, 1999; Hayden and Ghosh, 2012). Among the wide range of target genes controlled by NF-κB are genes encoding for cytokines like TNF-α, IL-1ß, cell adhesion molecules like ICAM-1, VCAM-1, and E-selectin or genes encoding for cyclooxygenase-2. All these proteins are essentially involved within the pathogenesis and regulation of inflammation. (Pahl, 1999; Liu et al., 2017).
The aim of the present study was to identify compounds, which are contributing to the anti-inflammatory activity of E. pinnatum and to evaluate their inhibitory activity on the inflammatory response in THP-1 and human umbilical vein endothelial cells to gain information about the underlying mechanisms.
Materials and Methods
General Experimental Procedures
UV spectra were recorded on a Thermo Scientific Ultimate 3000 diode-array detector. 1D and 2D NMR spectra were measured in methanol-d4 (δH 3.31/δC 49.0) either on a Varian UnityInova 600 spectrometer (1H: 400 MHz, 13C: 100 MHz) or on a Brucker Avance III NMR spectrometer (1H: 700.0 MHz, 13C: 166.0 MHz) equipped with a cryo probe. HR-MS was conducted on a Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap equipped with electron spray ionization. ESI-MS were obtained on a LTQ XL mass detector in positive and in negative mode. Analytical HPLC was conducted on an Agilent 1100 with an Agilent 1100 diode-array detector. A RP-18 Kinetex (2.6 µm, 100 × 2.6 mm; Phenomenex) column was used for analytical separations employed with a 40-min gradient separation of 0.1% HCOOH in water-MeCN (95:5–0:100 over 30 min, 0:100 held for 10 min). Semipreparative HPLC was performed on a Shimadzu LC-20AT with a Shimadzu DPS-M20A detector on a LUNA RP-18 column (10 µm, 250 × 10 mm, Phenomenex). GC-MS was performed on an Agilent 7890A GC equipped with an Agilent 5975 VL MSD on a HP-5MS capillary column (30 m × 250 µm × 0.25 µm, Agilent). GC-MS method consisted of a 66-min temperature program with an initial temperature of 60°C held for 1 min followed by 4°C/min increase to 280°C held for 10 min. The flow rate of the carrier gas helium was 1.2 ml/min performed in splitless mode. The GC-MS system was operated in EI mode at 70 eV. The identification of the C13 aglycones was performed by comparison of measured chromatogram with available reference compounds or with library softwares (HPCH2205, NIST 08, Wiley138). Column chromatography was performed on a silica gel 60 (0.04–0.063 mm, Merck), and on a sephadex LH 20 (163 µm, GE-Healthcare). Separations were monitored using TLC 60 F254 (Merck) by staining with 5% sulfuric acid in EtOH and 1% vanilin in EtOH.
Reference Compounds
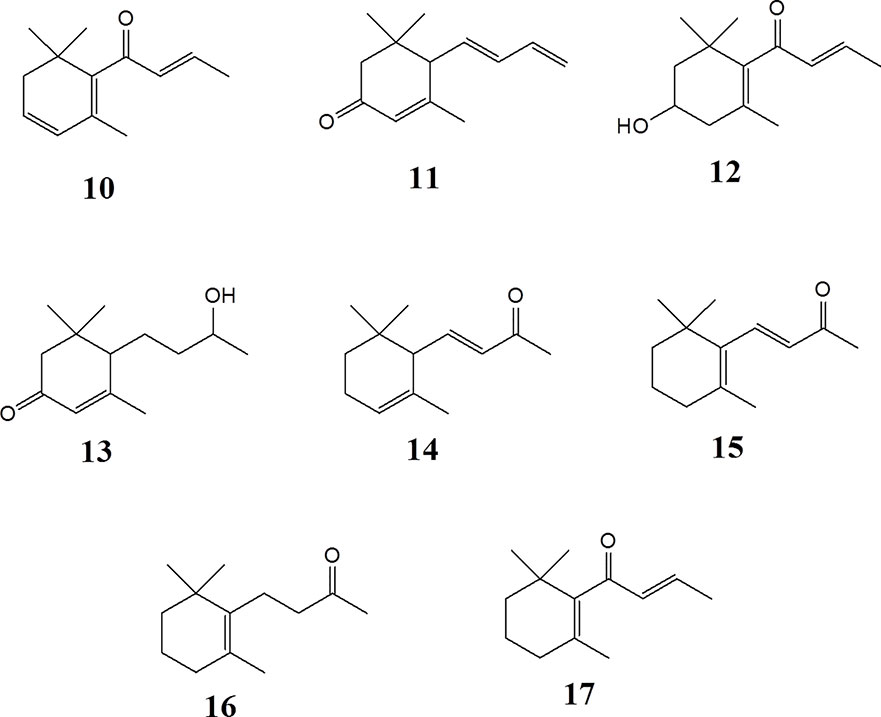
α-Ionone (> 96%) (14), β-ionone (96%) (15), dihydro-β-ionone (90%) (16), β-damascone (95%) (17), and β-damascenone (> 98%) (10), were purchased from Sigma-Aldrich. Megastigmatrienone/tabanone (mixtures of isomers) (11) was kindly provided by Lothar Streeck GmbH & Co. KG. Flavors & Fragrances (Germany, Hamburg).
Plant Material
Leaves of Epipremnum pinnatum (L.) Engl. were collected in May 2014 from cultivated stock (IPEN ID number: XX-0-GZU-98100700) in the botanical garden of Graz, Graz, Austria. The identification was conducted by Christian Berg (Institute of Plant Sciences, University of Graz, Austria). Voucher specimen (080514_EPI_Fol_tot_GZU, 080514_EPI_Fol_pulv_GZU) has been deposited at the herbarium of the Department of Pharmacognosy, University of Graz, Austria. The fresh plant material was air-dried for 3 weeks at 30°C in an air-flow chamber.
Extraction and Isolation
Dried leaves (47 g) were crushed in a blender and submitted to ASE (Dionex Accelerated Solvent Extractor-200) for successive extraction using n-hexane, dichloromethane, and MeOH as solvents. ASE conditions were set as follows: pressure, 69 bar; temperature, n-hexane: 72°C; dichloromethane: 44°C; MeOH: 98°C; static, 5 min; heat, 5 min; flush, 40%; purge 60 s; cycles, 3; total solvent volume of each solvents: 33 ml; amount of samples: 10.0 g each cell. The MeOH extract (5.59 g) was dried under N2 and chromatographed on a Silica gel column with mixtures of n-hexane-EtOAc-MeOH gradient system (from 100:0:0 to 0:0:100) as eluents. Fractions with similar compositions were combined according to TLC monitoring, producing fractions E1 to E20. Fraction E16 (758 mg), obtained with n-hexane-EtOAc-MeOH (1:1:2) was further separated on a Sephadex LH 20 column using MeOH as eluent. The collected volume of each fractions was 20 ml. Fractions were combined to 11 fractions after TLC monitoring (S1–S11). Fractions S3-S6 were subjected to semi-preparative HPLC using a RP-18 column (Phenomenex LUNA® 10 µm, 250 × 10 mm) as stationary phase and a gradient system of MeCN-water (15:85–100:0) as mobile phase (flow rate, 4.0 ml/min) over 30 min at 30°C column temperature to afford 1 (2.56 mg), and a mixture of 2 and 3 (1.93 mg). Other C13 compounds and aglycones were identified using GC-MS and HR-MS in fractions S3-S6 by comparison with available reference compounds or literature data as specified in the text.
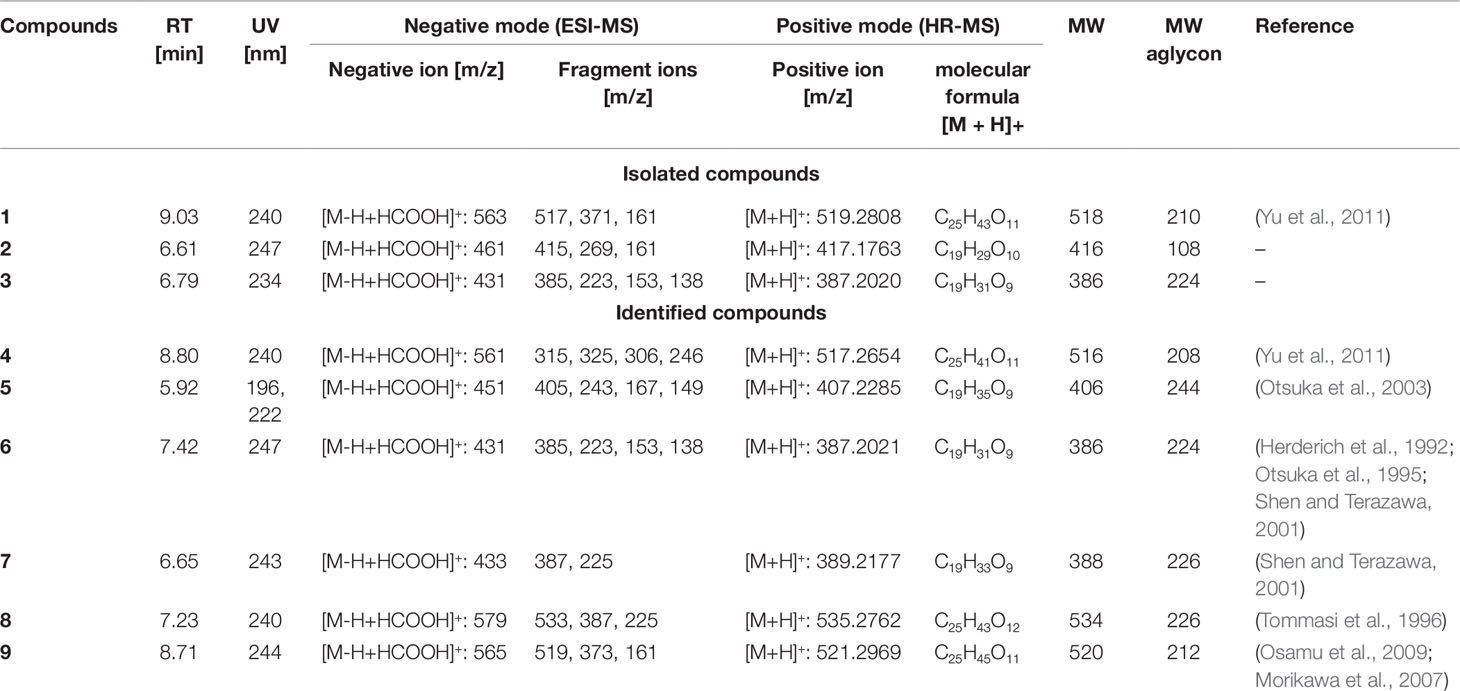
Gusanlungionoside C (1). Colorless powder; λmax 245.0 nm; 1H NMR and 13C NMR data, see Supplementary Data; HRESIMS m/z 519.2808 [M + H]+ (calc for C25H43O11, 519.2808); ESIMS positive mode m/z 519 [M + H]+, 373 [M + H–Rha]+, 211 [M + H–Rha–Glc]+; ESIMS negative mode m/z 563 [M–H + HCOOH]+, 517 [M–H]−, 371 [M–H–Rha]−, 161 [M–H–356]−.
Phenylmethyl-2-O-(6-O-rhamnosyl)-ß--galactopyranoside (2). Colorless powder; λmax 247.0 nm; 1H NMR and 13C NMR data, see Supplementary Data; HRESIMS m/z 417.1763 [M + H]+ (calc for C19H29O10, 417.1763); ESIMS negative mode m/z 461 [M–H+ HCOOH]+, 415 [M–H]−, 269 [M–H–Rha]; 161 [M–H–254]−.
Citroside A (3). Colorless powder; λmax 234.0 nm; 1H NMR and 13C NMR data, see Supplementary Data; HRESIMS m/z 387.2020 [M + H]+ (cald for C19H31O8, 387.2020); ESIMS in negative mode m/z 431 [M–H + HCOOH]+, 385 [M–H]−, 223 [M–H–Glc]−, 138 [M–H–247]−.
qPCR Analysis of COX-2 mRNA Level
The human monocytic cell line THP-1 was obtained from the European Collection of Cell Cultures (ECACC, catalogue No: 88081201) and maintained in medium RPMI 1640 (Gibco, ThermoFisher Scientific Inc., NY, USA) containing 2 mM L-glutamine, 10 mM HEPES (Gibco), 10% fetal bovine serum (FBS, Gibco), 100 units/ml penicillin (Gibco) and 100 µg/ml streptomycin (Gibco) at 5% CO2 and 37°C in humidified atmosphere. To initiate the monocyte to macrophage differentiation, THP-1 were seeded 1 × 106/ml in a 24-well plate and incubated with medium containing 12 nM phorbol-12-myristate-13-acetate (PMA, Sigma-Aldrich) for 48 h. Cells were further incubated with test compounds for 1 h followed by the stimulation with 7.5 ng/ml LPS for additional 3 h. Subsequently, total RNA was extracted with the GenElute™ mammalian total RNA miniprep kit (Sigma-Aldrich) and reverse transcribed with the high-capacity cDNA reverse transcription kit (Applied Biosystems®) according to manufacturer's instructions. The cycler conditions was set as followed: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 s. Quantitative real-time PCR (qPCR) was performed on a ABI 7300 real-time PCR Systems (Applied Biosystems®) with primers designed with Primer Express Software (Applied Biosystems®). The sequences for COX-2 were as follows: COX-2 forward: 5′-GAA-TCA-TTC-ACC-AGG-CAA-ATT-G-3′, COX-2 reverse: 5′-TCT-GTA-CTG-CGG-GTG-GAA-CA-3′, and COX-2 probe: 5′-FAM-TCC-TAC-CAC-CAG-CAA-CCC-TGC-CA-TAMRA-3′. TaqMan probe was used against endogenous control GAPDH (pre-developed TaqMan® assay, Applied Biosystems®). The mRNA expression was quantified using the ΔΔCt method. The instrument was set to 50°C for 2 min, 95°C for 10 min, followed by 40 PCR cycles of 95°C for 15 s and 60°C for 1 min (Lajter et al., 2015).
IL-8 ELISA
Human immortalized umbilical vein endothelial cells (HUVECtert) were seeded at a density of 4000 cells/well in a 96-well plate with M199 medium supplemented with 20% FCS, antibiotics (100 units/ml penicillin, 100 µg/ml streptomycine, 250 ng/ml fungizone, Lonza), 2 mM L-glutamine, and 12 µg/ml endothelial cell growth supplement/90 µg/ml heparin (PromoCell). After 24 h, the medium was replaced with medium containing 3% FCS and tested compounds. Treatment was performed in quintuplicates. After 15 min of preincubation with inhibitors the cells were stimulated with 0.3 ng/ml TNF-α or 30 ng/ml LPS (end concentration) and incubated for 6 h. Subsequently, medium supernatant was used to quantify IL-8 protein concentration with DuoSet ELISA development kit (R&D Systems) (Oskolkova et al., 2017).
Analysis of NF-κB-dependent Genes
Analysis of NF-κB-dependent genes was performed in human umbilical vein endothelial cells (HUVECtert) and the human monocytic cell line THP-1. HUVECtert cells were seeded in a 12-well plate with M199 medium supplemented with 20% FCS, antibiotics (100 units/ml penicillin, 100 µg/ml streptomycine, 250 ng/ml fungizone; Lonza), 2 mM L-glutamine, and 12 µg/ml endothelial cell growth supplement/90 µg/ml heparin (PromoCell). Cells were grown to full confluence, before they were used for experiments. THP-1 cells were cultured as described above, seeded at density of 1x106 cells/well in a 12-well plate, and used for experiments without inducing differentiation into macrophages by phorbol ester treatment. For experiment, growing medium was replaced with medium containing 3% FCS and 20 mM HEPES. Cells were pre-incubated with sample compounds for 30 min. Afterward, LPS (30 ng/ml), TNF-α (0.3 ng/ml), or IL-1β (1 ng/ml) were added and incubated for 4 h at 37°C. Subsequently, cells were lysed in RNAzol (Molecular Research Center Inc.) and total RNA was isolated according to manufacturer's instructions.
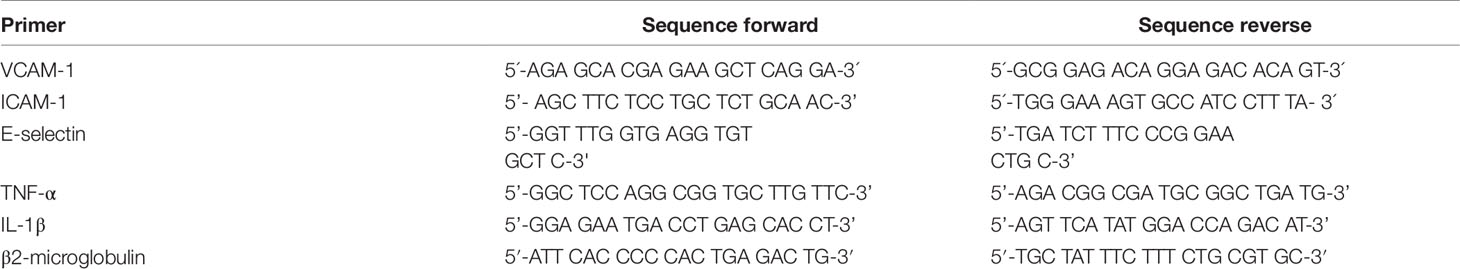
900 ng RNA were reversely transcribed using murine leukemia virus reverse transcriptase (MuLV from Applied Biosystems®) and oligo (dT)16 primers (Invitrogen). T100™ Thermal Cycler was set to following conditions: 25°C for 10 min, 42°C for 45 min, and 95°C for 5 min. Quantitative real time PCR was performed on a StepOnePlus instrument (Applied Biosystems®) using SYBRGreen master mix (PCR Biosystems Ltd). Primer sequences are presented in Table 1. mRNA expression was quantified using the ΔΔCt method. Amplification program was set as following: 30 s at 90°C, followed by 40 PCR cycles of 3 s at 95°C and 30 s at 60°C; melting point analysis in 0.1°C steps. The mRNA level of each target gene was normalized to expression of β2-microglobulin. Fold expression was defined as fold increase relative to β2-microglobulin-normalized level of expression in mock-stimulated treatment with vehicle) cells (0.1% DMSO).
TNF-α ELISA
THP-1 cells were seeded at a density of 1x105 cells per well in a 96-well plate with RPMI 1640 supplemented with 20 mM HEPES, 3% FCS and 100 units/ml penicillin (Gibco) and 100 µg/ml streptomycin. After 30 min pre-incubation with test compounds, cells were stimulated with 30 ng/ml LPS and incubated for another 4 h. Subsequently, TNF-α protein was quantified in medium supernatant by a DuoSet ELISA kit (R&D Systems).
Cell-based E-selectin ELISA
HUVECtert cells were seeded at density of 4000 cells per well in 96 well plates and incubated for 48 h before experiment. Afterward, medium was replaced with medium containing 3% FCS and 20 mM HEPES. Cells were pretreated with sample compounds for 30 min, followed by stimulation with LPS (30 ng/ml) for 4 h. Cells were fixed with 0.1% glutaraldehyde in PBS for 15 min at 4°C. Blocking was performed with 5% BSA in PBS for 1 h at 37°C. Afterward, cell-bound E-selectin was analyzed using a DuoSet ELISA development kit (R&D Systems).
NF-κB-Driven Luciferase Reporter Gene Transactivation
The stable cell line HEK 293/NF-κB-luc was obtained from Panomics (catalog RC0014). TNF-α stimulation of these cells induces the expression of the luciferase reporter gene that is regulated by multiple copies of NF-κB response elements. For the assay, cells were seeded in 96 well plates at a density of 4 × 104 cells per well in serum-free DMEM overnight. Cells were stained with 2 µM cell tracker green (CTG, Thermo Scientific) for 1 h, followed by subsequent treatment with respective compounds for 1 h and then stimulation with 2 ng/ml TNF-α for 4 h, which initiated NF-κB activation. Fluorescence of CTG-stained cells and luminescence of the luciferase product were measured on a Tecan SPARK®. The obtained luminescence values were normalized to CTG-derived fluorescence related to DMSO control. Normalized fluorescence units were used as indicator for cell number (Fakhrudin et al., 2014). To test whether β-damascenone inhibits NF-κB signaling via an electrophilic attack, 5 mM glutathione was added shortly before test compounds to all samples.
Cell Viability Assay
100.000 THP-1 cells/ml (100 µl/well) were seeded in 96-well plates and incubated with 12 ng/ml PMA for 48 h. HUVECtert cells were seeded at density of 7,000 cells/well in 96 well plates and incubated for 48 h. Afterward, medium was replaced to medium containing 3% FCS and indicated concentrations of sample compounds. After 24 h of incubation, medium was exchanged to medium with 0.2 mg/ml 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (Molecular Probes) and 5 nM phenazine methosulfate (Acros-Organic). After 1.5 h of incubation, 1 µg/ml Hoechst-33342 (final concentration) was added. Cells were incubated for 30 min. Afterward, absorbance at 450 nm (XTT), number of stained nuclei (Hoechst) and confluence were measured using the EnSight multimode plate reader equipped with Kaleido data acquisition and analysis software (PerkinElmer).
Statistical Analysis
Statistical analysis was calculated by one-way analysis of variance (ANOVA) with Bonferroni post hoc test using IBM SPSS Statistics 25 software. In graphs, data were expressed as mean values ± standard deviation (SD). Statistical significance was expressed as p values: p > 0.05 *, p > 0.01 **, p > 0.001 ***.
Results and Discussion
Out of three successively prepared extracts (n-hexane, dichloromethane, and MeOH) from E. pinnatum leaves, the MeOH extract inhibited the LPS-induced COX-2 mRNA expression most potently (inhibition at 20 µg/ml: n-hexane: 10.8 ± 10.4%, DCM: no effect, MeOH: 54.3 ± 9.2%) and was, therefore, subjected to activity-guided isolation of the active compounds. First, the MeOH extract was fractionated by silica gel column chromatography using mixtures of n-hexane, ethyl acetate, and water. After TLC comparison and combining similar fractions (175 fractions in total), 20 fractions E1 to E20 were obtained. Subsequent pharmacological testing revealed two active fractions, E16 and E20 (inhibition of LPS-induced COX-2 mRNA expression at 20 µg/ml: E16: 32.7 ± 6.7%, E20: 31.3 ± 7.1%). E16 was further separated on a size exclusion column using Sephadex LH 20 to gain 11 fractions (S1–S11). Active fractions (S3–S6) were combined (inhibition at 20 µg/ml from 20.9 to 71.8%) and compounds of interest were separated on a RP18 column by semi-preparative HPLC to yield gusanlungionoside C (1) and a mixture of phenylmethyl-2-O-(6-O-rhamnosyl)-ß-D-galactopyranoside (2) and citroside A (3). Figure 1 depicts the isolation scheme. Structure elucidation was carried out using 1-D proton NMR, 2-D NMR (DQFCOSY, HSQC, HMBC), ESI-MS, HR-MS, and GC-MS experiments. For NMR spectra and data, see Supplementary Data.
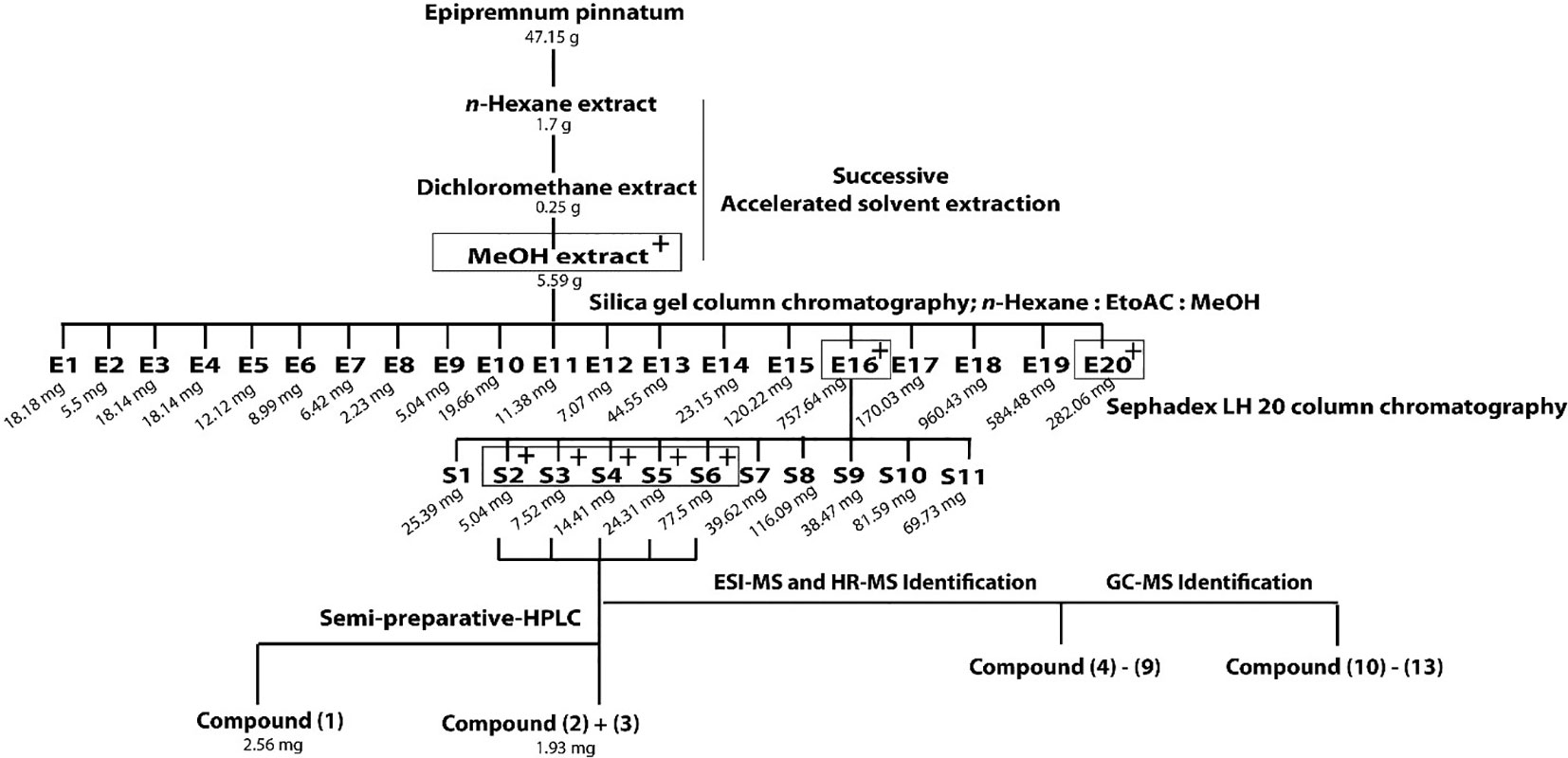
Figure 1 Extraction and isolation scheme of anti-inflammatory components from leaves of Epipremnum pinnatum. The plant material was successively extracted with n-hexane, dichloromethane and methanol. The active methanol extract was further fractionated on a silica gel column to gain E1-E20. Fraction E16 was fractionated on a Sephadex LH20 column to produce fractions S1-S11. The most active fraction (S2-S6) were combined to isolate 1 and a mixture of 2 and 3. Compounds 4-9 and 10-13 could be identified with ESI-MS, HR-MS and GC-MS. Active fractions are determined using qPCR analysis of COX-2 mRNA and are indicated with "+" in grey boxes.
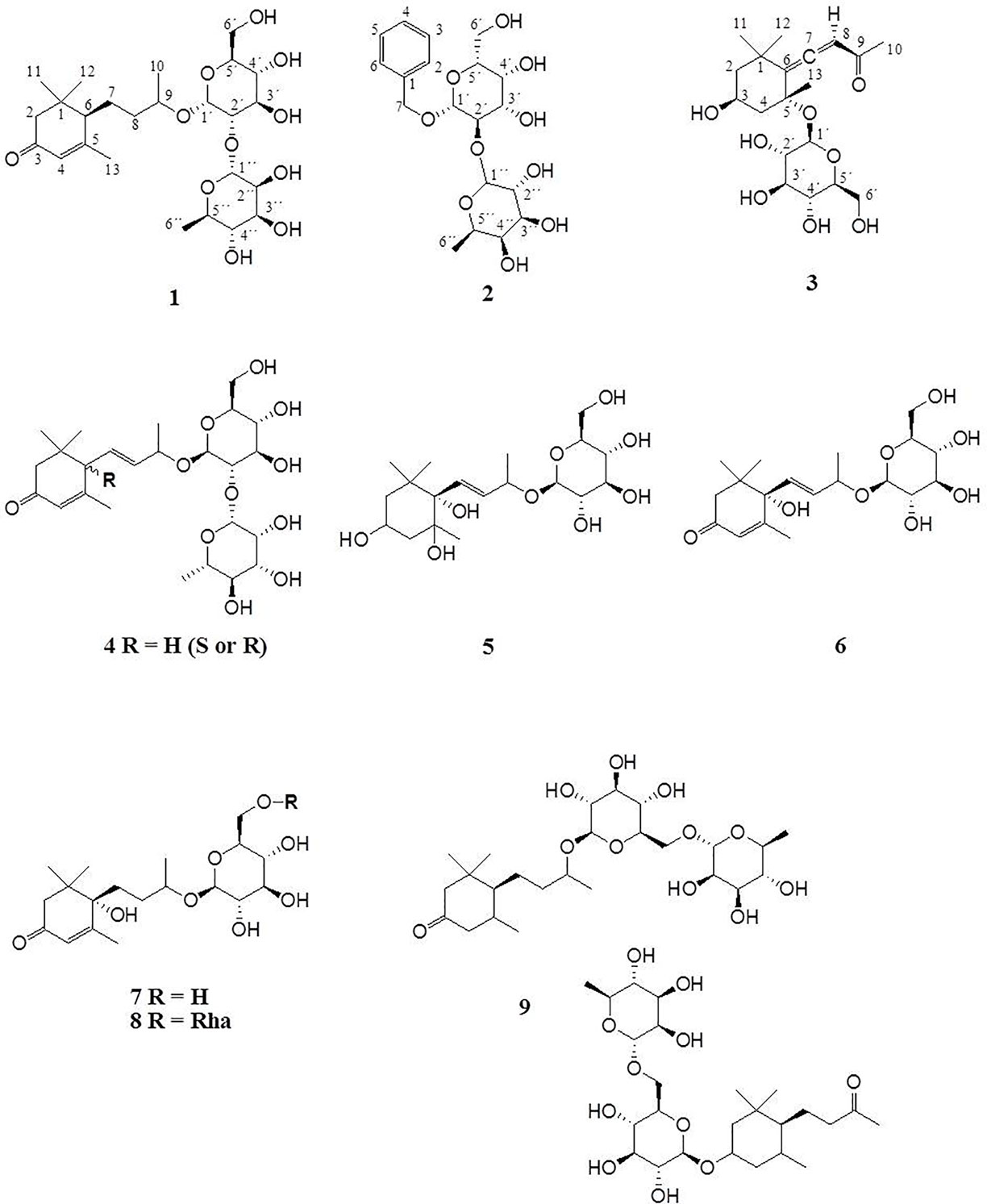
Compound 1 was isolated as colorless powder. HR-MS and NMR spectroscopic data indicated a megastigmane derivative with a trimethylcyclohexane moiety attached to a hexose and a deoxyhexose unit. HR-MS suggested a molecular formula of C25H42O11, according to a monoisotopic mass at m/z 519.2808 [M + H]+ (calculated for C25H43O11, 519.2808). ESI-MS detected a deprotonated molecule at m/z 563 [M–H + HCOOH]+, which fragmented to its molecular ion at m/z 517 [M–H]−. This compound was subsequently fragmented, losing a rhamnose molecule (m/z 371, [M–H–Rha]−) and its aglycon, with a remaining glucose molecule at m/z 161 [M–H–Rha – 210]−. The constitution of 1 was determined by complete assignment of 1H, DQF-COSY, HSQC, and HMBC spectra, which indicated the presence of a megastigmane derivative with two sugar units. The homonuclear coupling constant (H-1′, 7.8 Hz) of the glucose and the carbon chemical shift (69.5 ppm) of C-5 of rhamnose indicated the presence of a β-glucopyranose and a α-rhamnopyranose. HMBC correlation between the anomeric proton (H-1') of the glucose and the carbon at 75 ppm indicated that the glucose was attached to C-9 of the aglycon, while HMBC correlation between the anomeric proton of the rhamnose (H-1′′) and the carbon at 78.3 ppm pointed to the attachment of this sugar to C-2′ of the glucose. Hence, the constitution of the compound was determined as depicted in Figure 2. Three DBEs are provided by the ring fusions, two by the double bonds in the aglycon. The relative configurations of the stereogenic centers C-6 and C-9 of the aglycon were elucidated by comparison of the proton and carbon resonance values with literature data. The values fit well with compound 3 of the publication by Yu et al. (2011) with reported absolute configurations 6S, 9R, assigned to a compound named gusanlungionoside C.
A mixture of 2 and 3 was isolated as colorless powder. According to proton NMR, the sample comprised two major glycosidic compounds. As the aglycon parts of these components were chemically quite different, complete resonance assignments for the non-sugar portions of the compounds were possible. HR-MS measurement identified protonated molecules at m/z 417.1764 (2) and 387.2020 (3) [M + H]+ (calc for C19H29O10, 417.1764; calc for C19H31O8, 387.2020). ESI-MS of 2 showed characteristic fragment loss of a formic acid adduct at m/z 461 [M–H + HCOOH]+, with further loss at m/z 415 [M–H]−, 269 [M–H–Rha]−, and 161 [M–H–Rha – 108]−, indicating a cleavage of rhamnose and its aglycon with a remaining glucose molecule. This is in accordance with the NMR data. According to the observed resonances in proton NMR and HSQC, the disaccharide was formed by a hexopyranose in the β-form and a β-rhamnopyranose. The correlations in the HMBC spectrum between H-7 (4.94 ppm) and C-1' (101.6 ppm), and H-1' (4.42 ppm) and C-7 (71.5 ppm), respectively, indicated that the benzyl alcohol was attached to the anomeric carbon of the hexose. The three bond correlation between H-1'' (5.20 ppm) and 79.2 ppm indicated that the rhamnose was attached to C-2′. Due to severe signal overlap in the NMR spectra of the hexoses of 2 and 3, it was not possible to determine the relative configuration at C-4′, i.e. it was not possible to determine whether the β-hexose is a glucose or galactose. However, the relatively low carbon shift value of C-2' (79 ppm) points more to a galactose, because for glucose more than 82 ppm would be expected. Compound 2 has therefore been tentatively assigned as phenylmethyl-2-O-(6-O-rhamnosyl)-ß-D-galactopyranoside.
The MS of compound 3 exhibited a formate adduct at m/z 431 [M–H + HCOOH]+, with sequential fragment loss to its molecular ion at m/z 385 [M–H]−, 223 [M–H–Glc]−, and 138. According to NMR analysis, compound 3 was a C13 megastigmane derivative with a trimethylcyclohexane portion attached to a hexose. Complete analysis of HMBC correlations revealed that the aglycon was the so-called “grasshopper ketone”with a β-glucopyranose moiety attached to C-5. This compound is a cumulene with two different terminal substituents (H, acyl moiety) at C-8. This structural feature leads to a chiral axis and due to the stereogenic centers C-3 and C-5, respectively, to two possible diastereomers with different spatial orientation of the terminal groups, which are described as citroside A and citroside B in the literature. The NMR-resonance values of component A fit much better with those of citroside A (Umehara et al., 1988; Zhang et al., 2010).
HR-MS and ESI-MS Identification of Megastigmane Glycosides
ESI-MS and HR-MS analyses of the initial combined fractions S3-S6 showed six additional compounds with characteristic fragmentations of megastigmane glycosides. Since the megastigmane aglycones can vary in the degrees of saturation and hydroxylation (Gribble, 1991; Hou et al., 2016), a structural determination could be made in comparison to the isolated compounds 1 and 3. Table 2 depicts the mass fragmentation of identified compounds. They were identified as gusanlungionoside A or B (4), actinidioionoside (5), roseoside (6), 7,8-dihydroroseoside (7), blumenol B 9-O-α-L-rhamnopyranosyl-(1 6)-β-D-glucopyranoside (8), sedumoside J or G (9), as described below.
In the HR-MS spectra, 4 showed a protonated molecule at 517.2654 [M + H]+ with a calculated formula of C25H41O11. ESI-MS fragmentation exhibited a formate adduct ion at m/z 561 [M–H + HCOOH]+, with the sequential fragments at m/z 517 [M–H]−, 371 [M–H–Rha]−, 161 [M–H–Rha–210]−. Compared to 1, compound 4 differed by m/z 2, indicating a structural difference by a double bond. These measured values fit well to the stereochemical isomers assigned as gusanlungionosides A and B described by Yu et al. (Yu et al., 2011).
Compound 5 showed a protonated molecular ion at m/z 407.2285 [M + H]+ corresponding to the formula C19H35O9. ESI-MS revealed a formate adduct ion at m/z 451 [M–H + HCOOH]+. Its molecular ion at m/z 405 [M–H]− fragmented in m/z 243 [M–H–Glc]−, 167 [M–H–Glc–76]−, 149 [M–H–Glc–94]−. The aglycon with the deprotonated molecule at m/z 243 differed from the aglycon of 1 by additional m/z 34 indicating a difference of a saturated double bond and two oxygen moieties. This compound was described as actinidioionoside by Otsuka et al. (2003).
Compound 6 shares the same fragmentation pattern and molecular formula as 3. HR-MS detected a protonated molecule at m/z 387.2021 [M + H]+ (calc C19H31O9). This compound has been assigned as roseoside according to literature values (Herderich et al., 1992; Otsuka et al., 1995; Shen and Terazawa, 2001).
In HR-MS, 7 exhibited a monoisotopic mass at m/z 389.2177 [M + H]+ (calc C19H33O9). ESI-MS displayed a formate adduct ion at m/z 433 [M–H + HCOOH]+, with further fragments at m/z 387, and 225. Compared to 6, compound 7 varied by additional m/z 2 indicating a saturated double bond in its structure. 7,8-dihydroroseoside fits well to the measured and described literature values (Shen and Terazawa, 2001).
Compound 8 is a megastigmane diglucoside comprised of 7 additionally linked with a rhamnose. HR-MS exhibited a protonated molecule at m/z 535.2762 [M + H]+, indicating a molecular formula of C25H43O12. ESI-MS showed characteristic fragments at m/z 579 [M–H + HCOOH]+, 533 [M–H]−, 387 [M–H–Rha]−, and 225 [M–H–Rha–Glc]−. This compound has been previously isolated by Tommasi et al. (1996) and assigned as blumenol B 9-O-α--rhamnopyranosyl-(1 6)-β--glucopyranoside.
Compound 9 gained a molecular ion peak at m/z 521.2969 [M + H]+ (calc C25H45O11). ESI-MS fragmentation showed deprotonated fragment ions at m/z 565 [M–H + HCOOH]+, 519 [M–H]−, 373 [M–H–Rha]−, and 161 [M–H–Rha–212]−. These results differed from 1 by additional m/z 2, indicating a saturation of one double bond. Two compounds fitting to the measured values are sedumoside J or sedumoside G (Morikawa et al., 2007; Osamu et al., 2009). The position of the sugar linkage could not be determined by mass spectrometry.
Figure 2 depicts the identified megastigmane glycosides 1–9.
GC-MS Identification of Megastigmane Aglycones
C13 megastigmane derivatives represent a large group of natural products important as aromatic components in fruits and plants. In order to identify the low molecular weight volatile megastigmane aglycones, capillary GC-MS has been employed. GC-MS of the fractions S3 to S6, as shown in Figure S5 (supporting information), allowed the identification of the four megastigmane aglycones β-damascenone (10), megastigmatrienone (11), 3-hydroxy-β-damascone (12), and 3-oxo-7,8-dihydro-α-ionol (13). The identification was performed by comparing the MS fragmentation patterns with library data (HPC2205, NIST08, Wiley138) and reference compounds. The mass spectra are dispicted in the supporting information. The other peaks were not identified because they were from a fraction, which did not show activity.
In Vitro Pharmacology of Megastigmane Derivatives
Epipremnum pinnatum is used in traditional medicine for treatment of various inflammation-associated conditions. We addressed the question whether megastigmane derivatives identified in this project can inhibit inflammatory reactions in cell types relevant to inflammation, such as endothelial cells and mononuclear leukocytes. To this end, LPS-induced upregulation of the pro-inflammatory PTGS2 (COX-2) mRNA in the monocyte-like THP-1 cell line was used as a readout. In addition to the isolated compounds, α-ionone (14), β-ionone (15), 7,8-dihydro-β-ionone (16), and damascone (17), which all have similar structures, were available as reference compounds. Figure 3 depicts the megastigmane aglycones 10–17. Analysis of the relative inhibitory activity showed that out of the tested compounds, only compound 10 was able to inhibit PTGS2 (COX-2) mRNA expression (Table 3). The inhibitory effect was dose-dependent (Figure 4). Since only 10 demonstrated significant activity, further studies were focused on the pharmacological analysis of this substance.

Table 3 IC50 values of isolated and reference compounds in COX-2 gene expression assay in THP-1 macrophages. n.a. no activity was observed up to a concentration of a 20 µg/ml; b 50 µM.
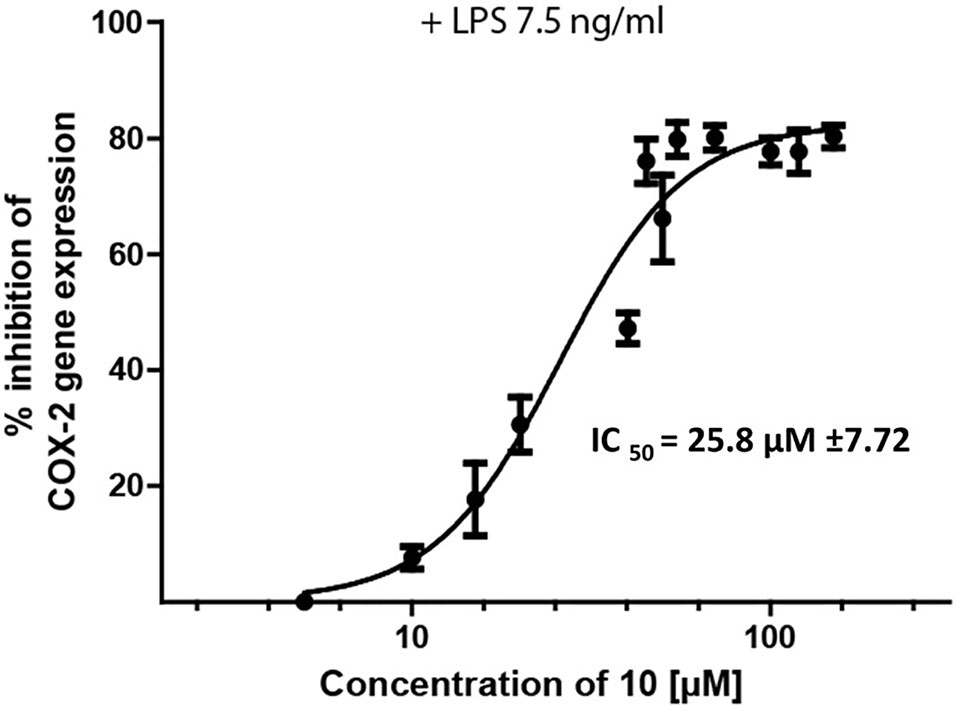
Figure 4 Inhibition of LPS-induced (7.5 ng/ml) PTSG2 (COX-2) mRNA expression by 10. Cells were pretreated with β-damascenone (10) for 1 h followed by LPS stimulation for 3 h. Total RNA was isolated and reverse transcribed to cDNA. cDNA was amplified and expression of PTSG2 (COX-2) was normalized to the expression of GAPDH mRNA. Data are presented as mean ± SE (n = 3).
To characterize the anti-inflammatory properties of 10, endothelial cells (HUVECtert) were pretreated with this compound and then stimulated with LPS for 4 h. 10 inhibited the induction of typical NFκB-regulated pro-inflammatory genes, i.e. E-selectin, ICAM-1, and VCAM-1 (Figures 5A–C), as well as IL-8 (Figure 6). Similarly, in the monocyte-like cell line THP-1, 10 inhibited the induction of the NFκB-responsive genes TNF-α and IL-1β (Figures 5D, E). The effects were not due to cytotoxicity of 10, as illustrated by the lack of cytotoxic effect after 24 h (Figure 7). The concentration dependence of the anti-inflammatory action of 10 was analyzed using E-selectin mRNA expression in HUVECtert cells as readout. The inhibition of E-selectin mRNA expression was concentration-dependent showing a residual activity of 37.9% at a concentration of 5 µM (Figure 8). Similar effects were observed for the inhibition of E-selectin protein expression on the surface of HUVECtert cells (Figure 9) and the secretion of TNFα protein by LPS-stimulated THP-1 cells (Figure 9). The data of Figure 9 show that 10 not only reduced induction of mRNA of pro-inflammatory genes, but also blocked their expression at the protein level.
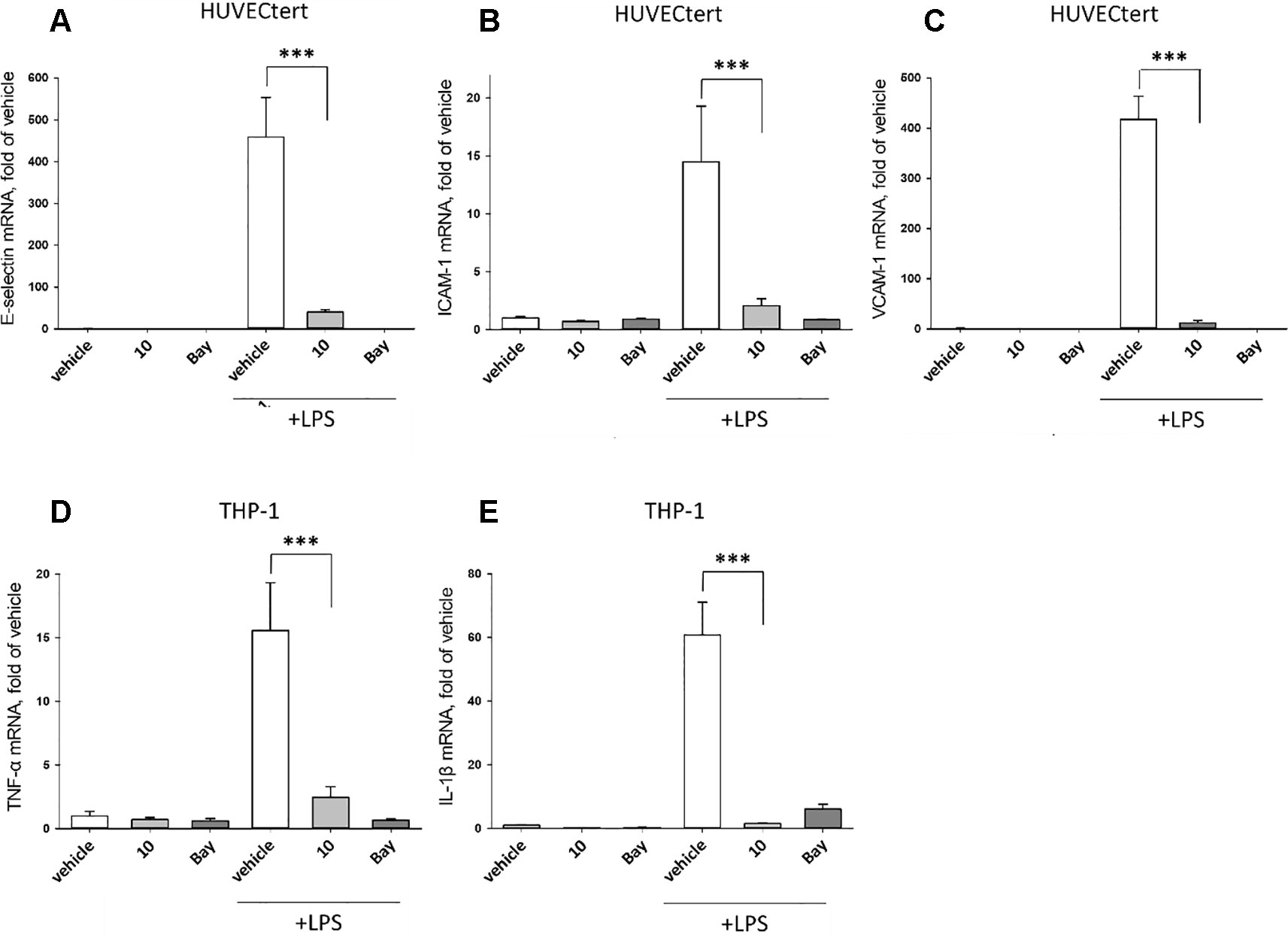
Figure 5 Inhibition of (A) E-selectin, (B) ICAM-1 and (C) VCAM-1 gene expression in HUCECtert cells, and inhibition of (D) TNF-α and (E) IL-1β gene expression in THP-1cells stimulated with LPS. Cells were pretreated with 30 µM β-damascenone (10) for 30 min followed by stimulation with LPS (30 ng/ml) for 4 hours. Basal values refer to vehicle-stimulated cells. Bay-11–7082 (5 µM) served as a positive control. Isolation of total RNA, cDNA synthesis and real-time PCR were performed as described in materials and methods section. Results are normalized to β2-microglobulin. Data are presented as mean ± SE (n=4). P-values are shown as *< 0.5, **<0.01 ***<0.001.
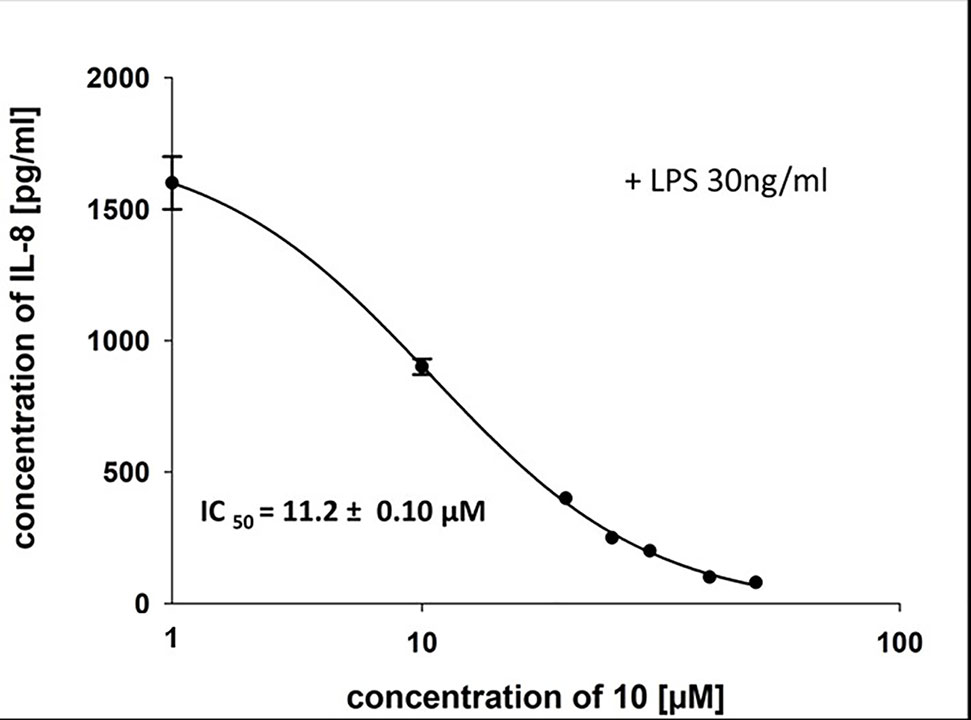
Figure 6 Inhibition of IL-8 secretion in HUVECtert by β-damascenone (10). Cells were pretreated with 10 for 15 min followed by stimulation with LPS (30 ng/ml) for 4 h. The IL-8 concentration in the medium was quantified by ELISA. Bay 11-7082 at 5 µM served as positive control. 0.1% MeOH was used as solvent control. Results are expressed as mean values ± SE.
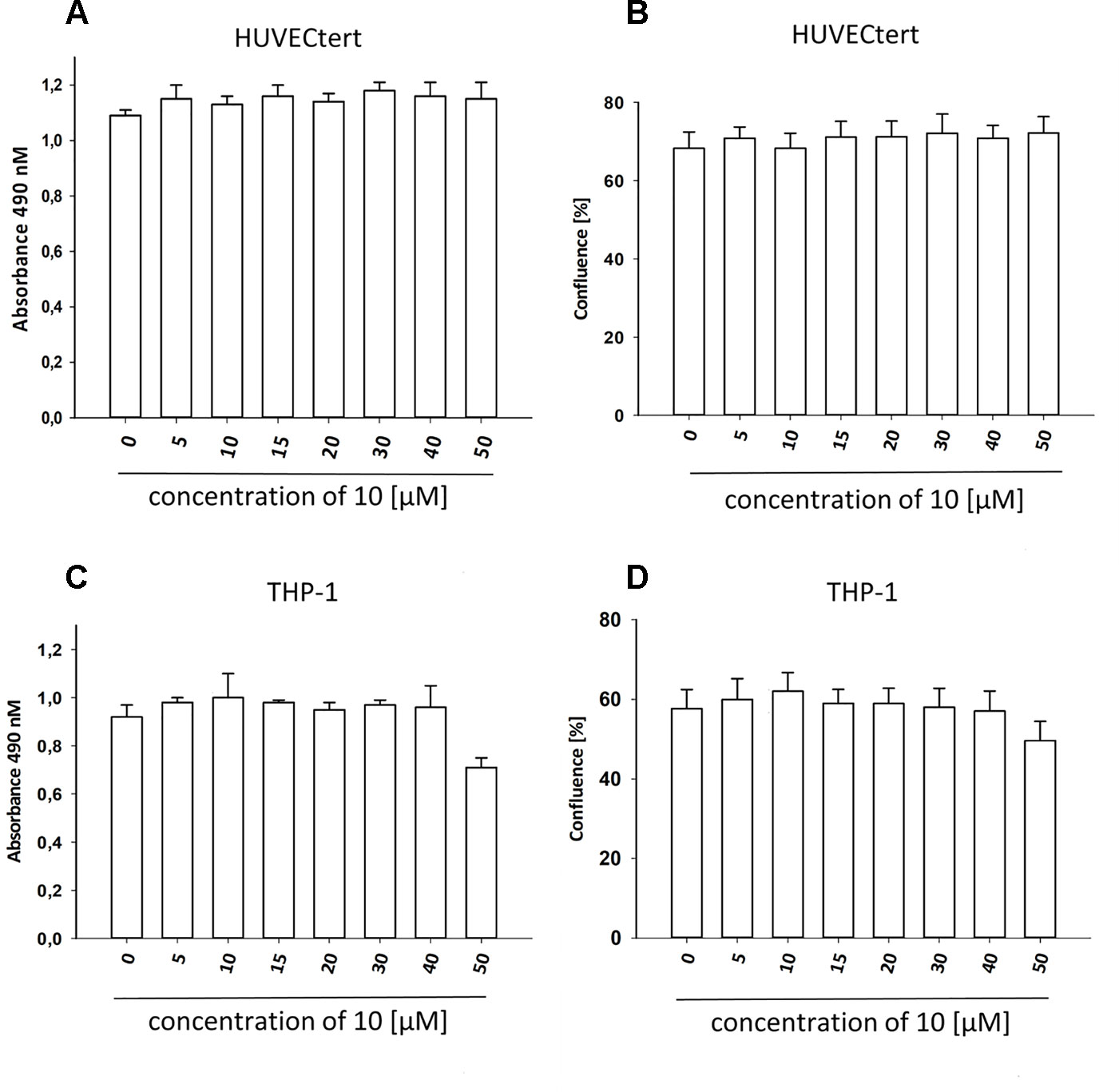
Figure 7 β-Damascenone at tested concentrations is not toxic. HUVECtert. (A, B) and differentiated THP-1 (C, D) cells were treated with different concentrations of β-damascenone. 0.2% DMSO served as vehicle control. After 24 h, confluence (B, D) and metabolic activity (XTT assay, A and C) were determined. The Figure presents mean values ± SD of two independent experiments with six biological replicates each.
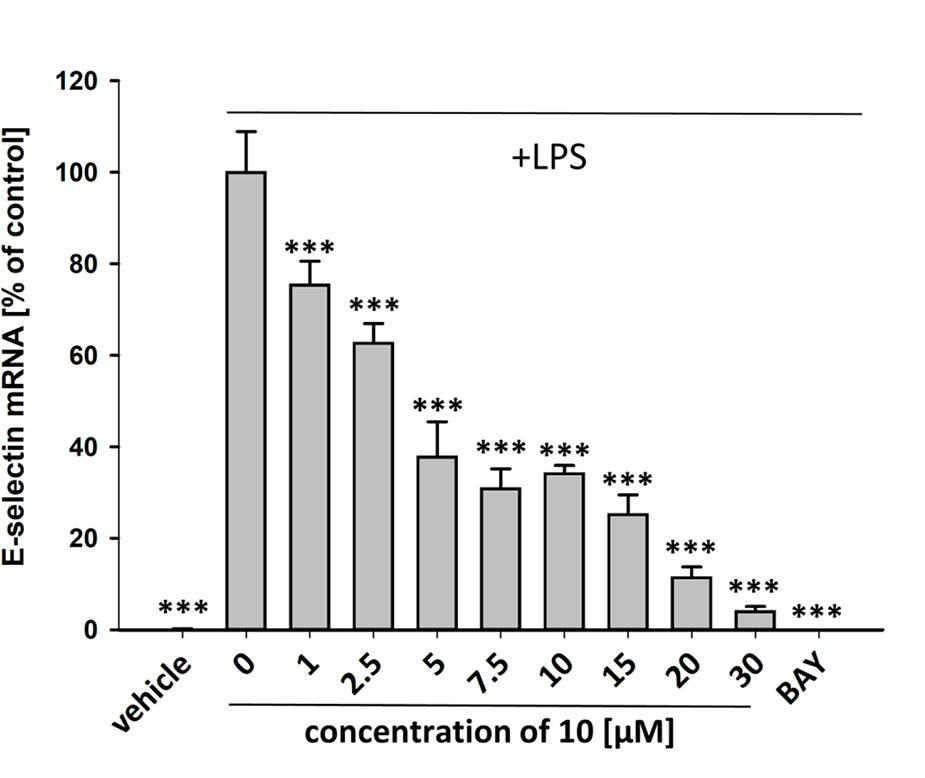
Figure 8 Inhibition of E-selectin mRNA expression in HUVECtert cells stimulated with LPS. Cells were pretreated with test compounds for 30 min followed by stimulation with LPS (20 ng/ml) for 4 hours. Basal values refer to vehicle-stimulated cells. Bay-11–7082 (5 µM) served as positive control. Results are normalized to β2-microglobulin. Data are presented as mean ± SE (n=4). P-values vs LPS-stimulated group are shown as ***<0.001.
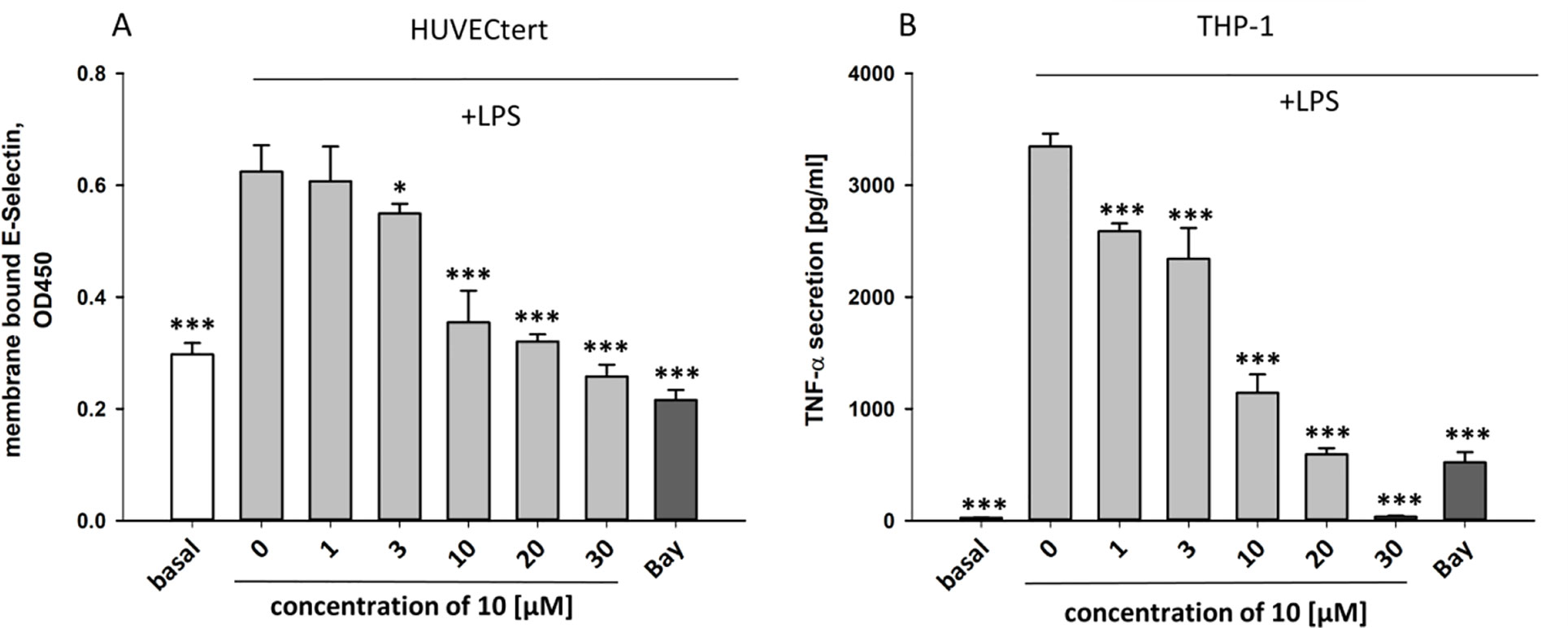
Figure 9 Inhibition of (A) protein expression of membrane bound E-selectin in HUVECtert cells and (B) TNF-α protein secretion in THP-1cells stimulated with LPS. Cells were pretreated with different concentrations of β-damascenone (10) for 30 min followed by stimulation with LPS (30 ng/ml) for 4 hours. Basal values refer to unstimulated cells. Bay-11–7082 (5 µM) served as positive control. Blank values were subtracted from readouts. Data are presented as mean ± SE (n=6). P-values vs LPS-stimulated group are shown as *< 0.5, **<0.01 ***<0.001.
The experiments described in the previous paragraph were performed using cells stimulated by bacterial LPS. Further experiments analyzed whether 10 can also inhibit effects induced by pro-inflammatory cytokines that bind to different receptors than LPS. To this end, endothelial cells were stimulated with TNF-α, IL-1β, or LPS in the absence or presence of 10. All stimuli upregulated E-selectin mRNA in HUVECtert, and the induction was significantly inhibited by 10 (Figure 10). Similarly, 10 inhibited the induction of TNF-α mRNA in THP-1 cells treated by all three pro-inflammatory stimuli (Figure 10). Since these stimuli activate different receptors with different downstream signaling steps, we hypothesized that 10 inhibits the transduction of inflammatory signals at a post-receptor level. The signaling pathway, in which all three upstream pathways converge, is the NF-κB signaling pathway (Sun, 2017).
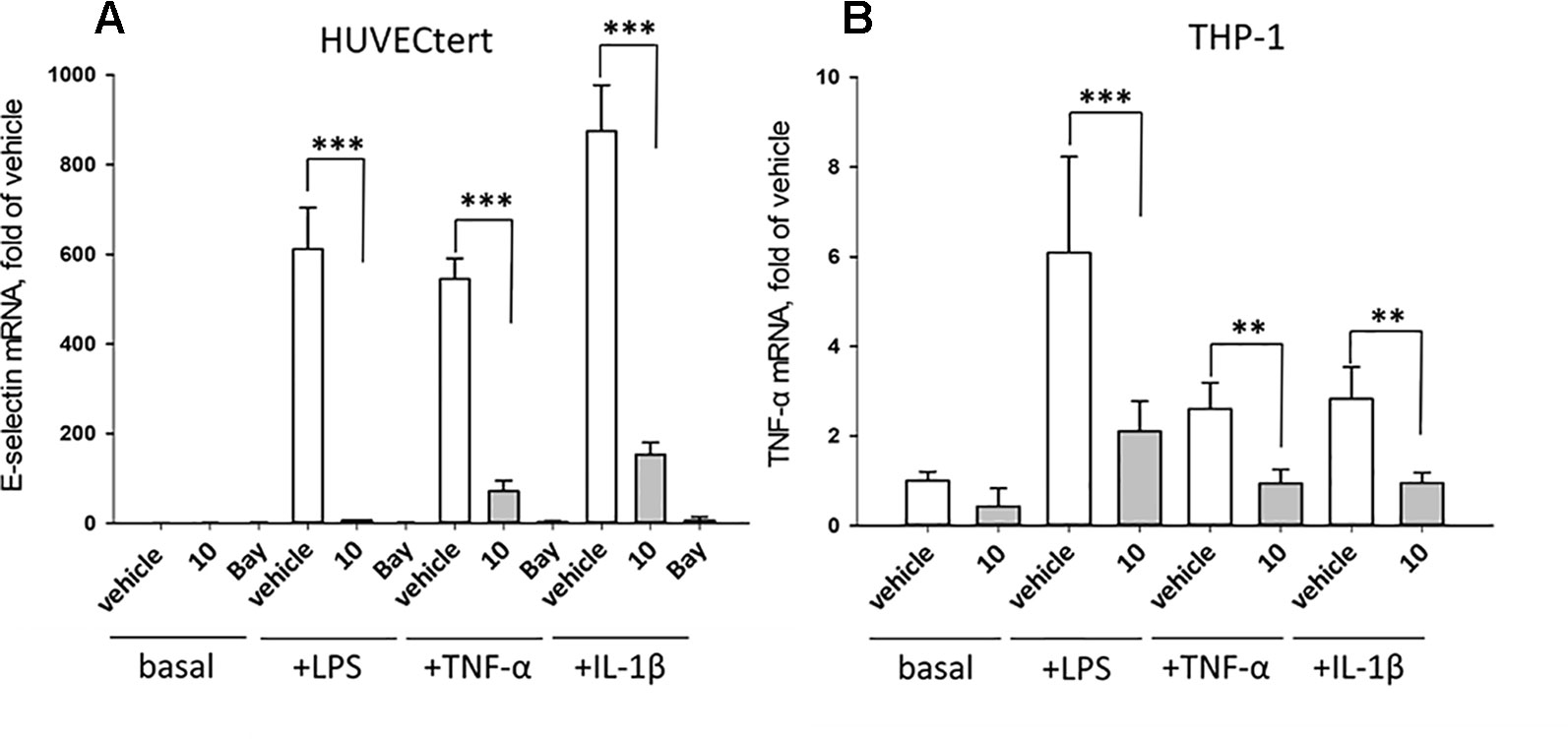
Figure 10 Inhibition of (A) E-selectin mRNA expression in HUVECtert and (B) TNF-α mRNA expression in THP-1 cells stimulated with different agonists of NF-κB pathway. Cells were pretreated with β-damascenone (10) for 30 min followed by stimulation with LPS (30 ng/ml), TNF-α (0.3 ng/ml) or IL-1β (1 ng/ml) for 4 hours. Basal values refer to vehicle-stimulated cells. Isolation of total RNA, cDNA synthesis and real-time PCR were performed as described in materials and methods section. Results are normalized to β2-microglobulin. Data are presented as mean ± SE (n=4). P-Values are shown as **<0.01 ***<0.001
In order to directly test whether the anti-inflammatory activity of 10 may result from inhibiting the NF-κB signaling pathway, HEK 293 cells stably transfected with a NF-κB-driven luciferase gene were used. 10 inhibited the NF-κB-driven reporter gene transactivation concentration-dependently (Figure 11A). The IC50 value obtained was 21.3 µM. The effect was not due to cytotoxicity of 10 as documented by the Cell Tracker Green fluorescence (Figure 11C). 10 is an active electrophile, and in contrast to other megastigmane aglycones, 10 possesses two active sites. This characteristic feature makes 10 a reactive Michael acceptor able to react with thiol groups found in many signal-transducing proteins (Gerhauser et al., 2009). We, therefore, tested whether the inhibitory effect of 10 can be inhibited by adding glutathione acting as decoy for reactive Michael acceptors (Heilmann et al., 2001). Figure 11B shows that 5 mM glutathione largely blocked the NF-κB inhibitory activity of 10 as well as that of the positive control parthenolide, which is also a reactive Michael acceptor (Kwok et al., 2001).
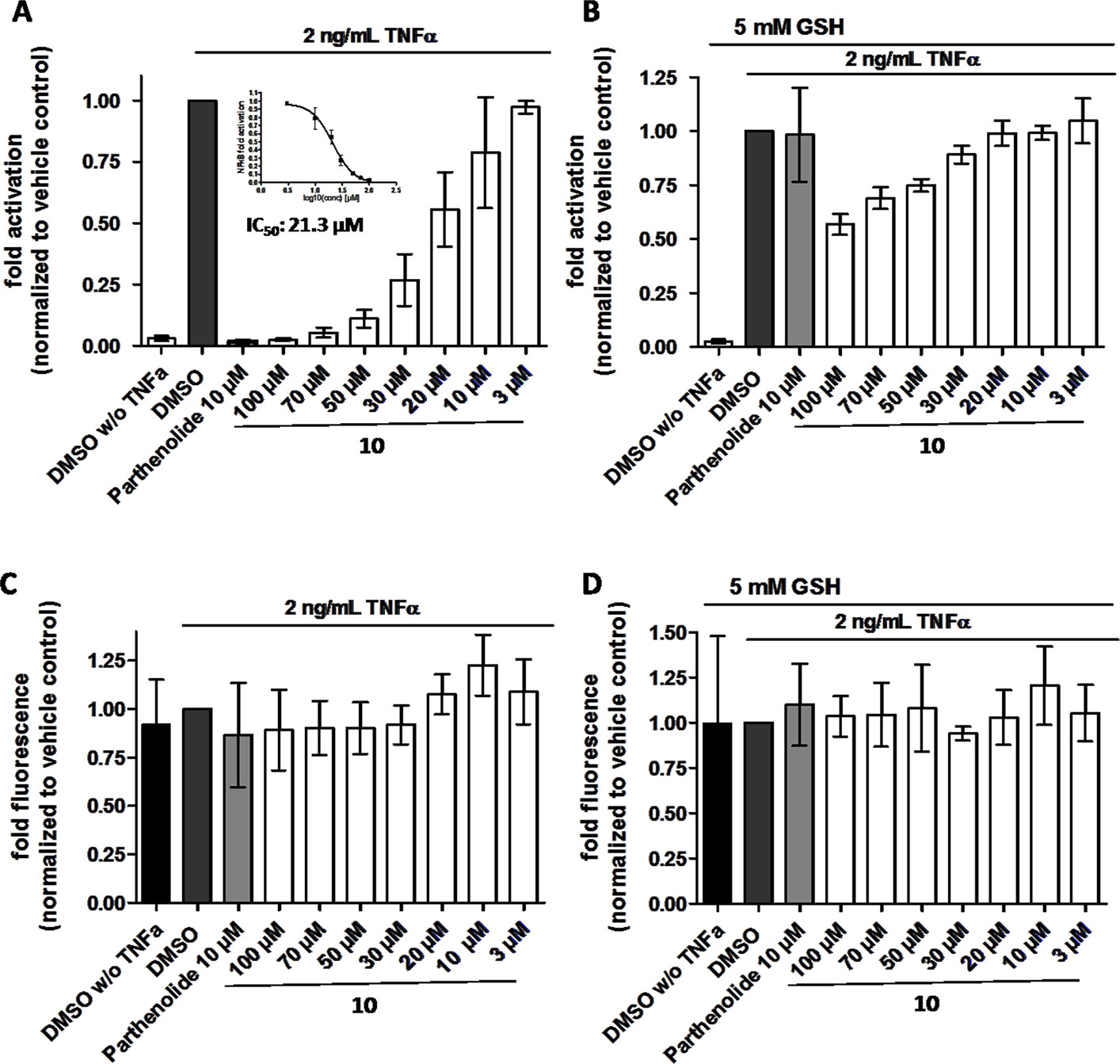
Figure 11 β-Damascenone (10) inhibits TNF-α (2 ng/ml)-induced NF-κB-driven luciferase reporter gene transactivation concentration dependently; the inhibition is reversed in the presence of 5 mM glutathione (GSH). HEK293/NF-κB-luc cells were loaded with cell tracker green (CTG), a probe for vital staining. After 24 hours cells were pretreated for 1 hour with the indicated compounds, and in (B) in addition with 5 mM GSH (as indicated), and activated with TNF-α (2 ng/ml) for 4 hours. Then, (A and B) luciferase activity and (C and D) CTG fluorescence were measured. Luciferase activity is shown normalized to CTG fluorescence. Parthenolide (10 µM) served as positive control. Data are presented as mean ± SD (n = 4) and are normalized to the vehicle control DMSO.
Electrophiles have been shown to covalently modify IKKβ, thus leading to its inactivation (Kapahi et al., 2000; Kwok et al., 2001). This kinase phosphorylates the NF-κB inhibitor IκB and targets it for proteasomal degradation, finally leading to NF-κB activation. Thus, inactivation of IKKβ by electrophiles results in inhibition of NF-κB independently of the upstream mechanisms that induced activation of IKKβ.
In summary, the major finding of this investigation is that β-damascenone is a major active compound of Epipremnum pinnatum, and that it inhibits NF-κB signaling pathway in vitro in human cellular systems that had been activated with different inflammatory stimuli, and that this is most likely mediated by the electrophilic property of the compound. Our results are in agreement with the data of Gerhauser et al., 2009), who showed that 10 was able to inhibit iNOS expression in LPS-stimulated murine macrophages and to activate the transcription factor Nrf2. Also, the activation of Nrf2 can be promoted by an electrophilic insult that covalently modifies Keap1, which finally leads to Nrf2 accumulation, nuclear translocation and transcriptional activation of respective target genes (Matzinger et al., 2018). Thus, 10 as an electrophilic compound can simultaneously activate Nrf2 and inhibit NF-κB, finally resulting in an antioxidant defense and suppression of pro-inflammatory target genes. Such a pharmacodynamic profile may be especially beneficial for treatment of acute inflammatory conditions, e.g., ischemia-reperfusion injury, which is usually accompanied by severe oxidative stress. Therefore, our results show that megastigmane derivatives, in particular β-damascenone and its precursors, may be responsible for the anti-inflammatory effects of Epipremnum pinnatum via inhibition of the NF-κB pathway. It is most likely that its α,β-unsaturated carbonyl moiety is responsible for this effect. Since α,β-unsaturated carbonyl moieties interact with many signal-transduction proteins, it needs to be clarified whether β-damascenone expresses its effects only by targeting proteins within the NF-κB and Nrf2 signaling cascade, or whether it also affects other signaling pathways.
Author Contributions
Bio-activity guided isolation was performed by S-PP. Structure elucidation including LC-MS and GC-MS was performed by S-PP and OK. S-PP performed the COX-2 mRNA assay. Analysis of NF-κB dependent genes, TNF-α and E-selectin ELISA was performed by TP. NK and TP did cell viability assays. SH, SL, and JR performed the Luciferase reporter gene assays. Graphs and statistical analysis were made by contributors who performed the experiments. S-PP wrote the original draft of the manuscript. RB, VB, VD, and TP designed and conceived the study, contributed to writing and revisions of the manuscript. All authors contributed to writing and revisions of the manuscript.
Funding
The work was partially supported by a grant from the Austrian Science Fund (P 27682-B30 to VB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to acknowledge C. Berg, Graz Botanical Gardens, and H. Böchzelt, Joanneum Research, for providing and identifying Epipremnum leaf samples. Furthermore, we want to thank K.B. Jensen, NAWI Graz Central Lab "Environmental, Plant & Microbial Metabolomics", K. Hellauer, Department of Pharmaceutical Chemistry, University of Graz, and Barbara Braunböck-Müller, Department of Pharmacognosy, University of Vienna, for their support in performing experiments, and Lothar Streeck GmbH & Co. Flavors & Fragrances for providing megastigmatrienone/tabanone.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01351/full#supplementary-material
References
Arulpriya, P., Lalitha, P. (2011). The potential use of Rhaphidophora aurea at various aspects: a review (1958-2011). J. Pharm. Res. 4 (11), 4006–4010.
Boyce, P. C. (1998). The genus Epipremnum Schott (Araceae-Monsteroideae-Monstereae) in West and Central Malesia. Blumea 43, 183–213.
Chan, L. W., Cheah, E. L. C., Saw, C. L. L., Weng, W., Heng, P. W. S. (2008). Antimicrobial and antioxidant activities of Cortex Magnoliae officinalis and some other medicinal plants commonly used in South-East Asia. Chin. Med. 3, 15. doi: 10.1186/1749-8546-3-15
Das, S. K., Sengupta, P., Mustapha, M. S., Kifayatudullah, M., Goussudin, M. (2015). Phytochemical investigation and antioxidant screening of crude leaves extract from Epipremnum aureum. Int. J. Pharmacognosy Phytochem. Res. 7 (4), 684–689.
Fakhrudin, N., Waltenberger, B., Cabaravdic, M., Atanasov, A. G., Malainer, C., Schachner, D. (2014). Identification of plumericin as a potent new inhibitor of the NF-kappaB pathway with anti-inflammatory activity in vitro and in vivo. Br. J. Pharmacol. 171, 1676–1686. doi: 10.1111/bph.12558
Gerhauser, C., Klimo, K., Hummer, W., Holzer, J., Petermann, A., Garreta-Rufas, A. (2009). Identification of 3-hydroxy-beta-damascone and related carotenoid-derived aroma compounds as novel potent inducers of Nrf2-mediated phase 2 response with concomitant anti-inflammatory activity. Mol. Nutr. Food Res. 53, 1237–1244. doi: 10.1002/mnfr.200800492
Goh, S. H. (1998). Bioactive principles from folkloric anti-neoplastic plants and from bioprospecting the malaysian forest. Pure Appl. Chem. 70 (11), 2133.
Gribble, G. W. (1991). “Natural products containing a cyclohexane, cyclohexene, or cyclohexadiene subunit,” in In Second Supplements to the 2nd Edition of Rodd's Chemistry of Carbon Compounds (Amsterdam: Elsevier), 375–445. doi: 10.1016/B978-044453347-0.50066-5
Hamzah, F., binti Mohd Yusop, F. H., Abdul Manaf, S. F., Yahya, A. (2015). Epipremnum pinnatum activity on chick pancreatic lipase. Adv. Mater. Res. 1113, 182–186. doi: 10.4028/www.scientific.net/AMR.1113.182
Hayden, M. S., Ghosh, S. (2012). NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234. doi: 10.1101/gad.183434.111
Heilmann, J., Wasescha, M. R., Schmidt, T. J. (2001). The influence of glutathione and cysteine levels on the cytotoxicity of helenanolide type sesquiterpene lactones against KB cells. Bioorg. Med. Chem. 9, 2189–2194. doi: 10.1016/S0968-0896(01)00131-6
Herderich, M., Feser, W., Schreier, P. (1992). Vomifoliol 9-O-β-d-glucopyranosyl-4-O-β-d-xylopyranosyl-6-O-β-d-glucopyranoside: a trisaccharide glycoside from apple fruit. Phytochem. 31, 895–897. doi: 10.1016/0031-9422(92)80035-D
Hoesel, B., Schmid, J. A. (2013). The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12, 86. doi: 10.1186/1476-4598-12-86
Hou, X., Rivers, J., Leon, P., McQuinn, R. P., Pogson, B. J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends In Plant Sci. 21, 792–803. doi: 10.1016/j.tplants.2016.06.001
Kapahi, P., Takahashi, T., Natoli, G., Adams, S. R., Chen, Y., Tsien, R. Y. (2000). Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Ikappa B kinase. J. Biol. Chem. 275, 36062–36066. doi: 10.1074/jbc.M007204200
Kwok, B. H., Koh, B., Ndubuisi, M. I., Elofsson, M., Crews, C. M. (2001). The anti-inflammatory natural product parthenolide from the medicinal herb Feverfew directly binds to and inhibits IkappaB kinase. Chem. Biol. 8, 759–766. doi: 10.1016/S1074-5521(01)00049-7
Lajter, I., Pan, S.-P., Nikles, S., Ortmann, S., Vasas, A., Csupor-Loffler, B. (2015). Inhibition of COX-2 and NF-kappaB1 gene expression, NO Production, 5-LOX, and COX-1 and COX-2 enzymes by extracts and constituents of Onopordum acanthium. Plant Med. 81, 1270–1276. doi: 10.1055/s-0035-1546242
Lan, T. L., Sulaiman, S. F., Najimudin, N. (2007). Anticancer medicinal plant, Epipremnum pinnatum (L.) Engl. chlorofom extract elicited both apoptotic and non-apoptotic call deaths in T-47D mammary carcinoma cells. KMITL Sci. Tech. J. 7. 24–43.
Linnet, A., Latha, P. G., Gincy, M. M., Anuja, G., Suja, S. R., Shyamal, S. (2010). Anti-inflammatory, analgesic and anti-lipid peroxidative effects of Raphidophora pertusa (Roxb.) Schott. and Epipremnum pinnatum (Linn.) Engl. aerial parts. Indian J. Natural Prod. Resour. 1, 5–10.
Liu, T., Zhang, L., Joo, D., Sun, S.-C. (2017). NF-κB signaling in inflammation. Signal Transduction Targeted Ther. 2, 17023. doi: 10.1038/sigtrans.2017.23
Liu, Y., Wang, X., Erwann, R., Mu, Y. eds. (2011) Removal of benzene by pot plants in test rooms. in: 2011 International Conference on Remote Sensing, Environment and Transportation Engineering. doi: 10.1109/RSETE.2011.5966174
Matzinger, M., Fischhuber, K., Heiss, E. H. (2018). Activation of Nrf2 signaling by natural products-can it alleviate diabetes? Biotechnol. Adv. 36, 1738–1767. doi: 10.1016/j.biotechadv.2017.12.015
May, M. J., Ghosh, S. (1998). Signal transduction through NF-κB. Immunol. Today 19, 80–88. doi: 10.1016/S0167-5699(97)01197-3
McClatchey, W. (1996). The ethnopharmacopoeia of Rotuma. J. Ethnopharmacol. 50, 147–156. doi: 10.1016/0378-8741(95)01343-1
Meshram, A., Srivastava, N. (2015). Epipremnum aureum (Jade pothos): a multipurpose plant with its medicinal and pharmacological properties. J. Crit. Rev. 2, 21–25.
Morikawa, T., Zhang, Y., Nakamura, S., Matsuda, H., Muraoka, O., Yoshikawa, M. (2007). Bioactive constituents from chinese natural medicines. Xxii. Absolute structures of new megastigmane glycosides, sedumosides E1, E2, E3, F1, F2, and G, from Sedum sarmentosum (Crassulaceae). Chem. Pharm. Bull. 55, 435–441. doi: 10.1248/cpb.55.435
Osamu, M., Masayuki, Y., Toshio, M., Kiyofumi, N. (2009). Lipid metabolism-improving agents containing Sedum sarmetosum extracts, megastigmanes, or flavonoids, and pharmaceuticals or foods containing them. JP 2009209045. Kinki University, Japan; Eishin Shoji K. K.
Oskolkova, O. V., Godschachner, V., Bochkov, V. N. (2017). Off-target anti-inflammatory activity of the p2x7 receptor antagonist AZ11645373. Inflammation 40 (2), 530–536. doi: 10.1007/s10753-016-0499-8
Otsuka, H., Hirata, E., Shinzato, T., Takeda, Y. (2003). Stereochemistry of megastigmane glucosides from Glochidion zeylanicum and Alangium premnifolium. Phytochem. 62, 763–768. doi: 10.1016/S0031-9422(02)00614-3
Otsuka, H., Yao, M., Kamada, K., Takeda, Y. (1995). Alangionosides G-M: glycosides of megastigmane derivatives from the leaves of Alangium premnifolium. Chem. Pharmaceut. Bull. 43, 754–759. doi: 10.1248/cpb.43.754
Pahl, H. L. (1999). Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853. doi: 10.1038/sj.onc.1203239
Ren, X. (2010). Traditional Chinese medicine ointment (external application) for treating uratoma. CN 101897836.
Shen, Y., Terazawa, M. (2001). Dihydroroseoside, a new cyclohexanone glucoside, from the leaves of shirakamba (Betula platyphylla Sukatchev var. japonica Hara). J. Wood Sci. 47, 145–148. doi: 10.1007/BF00780564
Siew, Y.-Y., Zareisedehizadeh, S., Seetoh, W.-G., Neo, S.-Y., Tan, C.-H., Koh, H.-L. (2014). Ethnobotanical survey of usage of fresh medicinal plants in Singapore. J. Ethnopharmacol. 155, 1450–1466. doi: 10.1016/j.jep.2014.07.024
Sonawane, C. S., Jagdale, D. M., Patil, S. D., Patil, L. J., Kadam, V. J. (2011). Phytochemical screening and in vitro antimicrobial activity studies of Epipremnum aureum Linn. leaves extracts. Der Pharmacia Sin. 2, 267–272.
Sun, S.-C. (2017). The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558. doi: 10.1038/nri.2017.52
Tada, Y., Matsuzaki, T., Tanaka, Y. (2010). Isolation and characterization of formaldehyde-responsive genes from golden pothos (Epipremnum aureum). Plant Biotechnol. 27, 325–331. doi: 10.5511/plantbiotechnology.27.325
Tan, M. L., Muhammad, T. S. T., Najimudin, N., Sulaiman, S. F. (2005). Growth arrest and non-apoptotic programmed cell death associated with the up-regulation of c-myc mRNA expression in T-47D breast tumor cells following exposure to Epipremnum pinnatum (L.) Engl. hexane extract. J. Ethnopharmacol. 96, 375–383. doi: 10.1016/j.jep.2004.07.005
Tommasi, N., Piacente, S., Simone, F. de, Pizza, C. (1996). Constituents of Cydonia vulgaris: isolation and structure elucidation of four new flavonol glycosides and nine new α-ionol-derived glycosides. J. Agric. Food Chem. 44, 1676–1681. doi: 10.1021/jf950547a
Umehara, K., Hattori, I., Miyase, T., Ueno, A., Hara, S., Kageyama, C. (1988). Studies on the constituents of leaves of Citrus unshiu MARCOV. Chem. Pharm. Bull. 36, 5004–5008. doi: 10.1248/cpb.36.5004
WHO (2009). Medicinal plants in Papua New Guinea: information on 126 commonly used medicinal plants in Papua New Guinea. Manila, Philippines: World Health Organization Western Pacific Region.
Wong, K. T., Tan, B. K. H., Sim, K. Y., Goh, S. H. (1996). A Cytotoxic Melanin Precursor, 5,6-Dihydroxyindole, from the Folkloric Anti-Cancer Plant Rhaphidophora korthalsii. Natural Prod. Lett. 9, 137–140. doi: 10.1080/10575639608044937
Wu, Z., Raven, P. H., Hong, D. Y. (2010). Flora of China: Acoraceae through Cyperaceae. St. Louis: Science Press, Beijing, and Missouri Botanical Garden Press.
Xiang, J., Zhang, Y. (2014). Chinese medicinal composition for treating qi-stagnation and blood stasis type femoral head necrosis. CN 104274640.
Yu, L.-L., Hu, W.-C., Ding, G., Li, R.-T., Wei, J.-H., Zou, Z.-M. (2011). Gusanlungionosides A-D, potential tyrosinase inhibitors from Arcangelisia gusanlung. J. Natural Prod. 74, 1009–1014. doi: 10.1021/np100900k
Zhang, Z., Zhang, W., Ji, Y.-P., Zhao, Y., Wang, C.-G., Hu, J.-F. (2010). Gynostemosides A-E, megastigmane glycosides from Gynostemma pentaphyllum. Phytochem. 71, 693–700. doi: 10.1016/j.phytochem.2009.12.017
Zong, C. (2014). Chinese medicinal preparation for treating infectious monocytosis and preparation methods thereof. CN 104225408.
Keywords: β-damascenone, megastigmane, Epipremnum pinnatum, COX-2, IL-8, NF-κB, gene expression
Citation: Pan S-P, Pirker T, Kunert O, Kretschmer N, Hummelbrunner S, Latkolik SL, Rappai J, Dirsch VM, Bochkov V and Bauer R (2019) C13 Megastigmane Derivatives From Epipremnum pinnatum: β-Damascenone Inhibits the Expression of Pro-Inflammatory Cytokines and Leukocyte Adhesion Molecules as Well as NF-κB Signaling. Front. Pharmacol. 10:1351. doi: 10.3389/fphar.2019.01351
Received: 08 July 2018; Accepted: 25 October 2019;
Published: 28 November 2019.
Edited by:
Anna Karolina Kiss, Medical University of Warsaw, PolandReviewed by:
Javier Echeverria, Universidad de Santiago de Chile, ChileMichael Heinrich, UCL School of Pharmacy, United Kingdom
Adam Matkowski, Wroclaw Medical University, Poland
Copyright © 2019 Pan, Pirker, Kunert, Kretschmer, Hummelbrunner, Latkolik, Rappai, Dirsch, Bochkov and Bauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rudolf Bauer, cnVkb2xmLmJhdWVyQHVuaS1ncmF6LmF0
 San-Po Pan
San-Po Pan Teresa Pirker
Teresa Pirker Olaf Kunert
Olaf Kunert Nadine Kretschmer
Nadine Kretschmer Scarlet Hummelbrunner3
Scarlet Hummelbrunner3 Verena M. Dirsch
Verena M. Dirsch Valery Bochkov
Valery Bochkov Rudolf Bauer
Rudolf Bauer