- 1Jiangsu Collaborative Innovation Center of Regional Modern Agriculture & Environmental Protection/Jiangsu Key Laboratory for Eco-Agricultural Biotechnology around Hongze Lake, School of Life Sciences, Huaiyin Normal University, Huaian, China
- 2Department of Urology, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, Huaian, China
20(S)-ginsenoside Rh2 (Rh2), a well-known protopanaxadiol-type ginsenoside from Panax ginseng has especially gained attention for its anticancer activities on various types of human cancer cells. However, the molecular mechanism through which Rh2 promotes apoptosis in hepatocellular carcinoma (HePG2) cells is not known at the transcriptome level. Rh2 can specifically inhibit the proliferation of HePG2 in a dose- and time-dependent manner. Moreover, Rh2 can significantly increase the apoptosis which was related with an increase in protein expression levels of caspase-3, caspase-6, and poly (ADP-ribose) polymerase. Comparison of RNA-seq transcriptome profiles from control group and Rh2-treated group yielded a list of 2116 genes whose expression was significantly affected, which includes 971 up-regulated genes and 1145 down-regulated genes. The differentially expressed genes in p53 signaling pathway and DNA replication may have closely relationships to the cells apoptosis caused by Rh2 treatment. The results of qPCR validation showed that dynamic changes in mRNA, such as CDKN1A, CCND2, PMAIP1, GTSE1, and TP73.
Introduction
Ginseng (Panax ginseng C.A.Mey) has been known as the "King of Herbs" since ancient times in eastern Asia (Hossen et al., 2017). It is a perennial herb that grows in cool habitats and belongs to the Araliaceae family (Park and Cho, 2009). The root of ginseng been widely used in traditional medicine in eastern Asian countries to promote health (Yu et al., 2015). Ginsenosides are the main essential bioactive ingredients that have various pharmacological activities, including anti-inflammatory, anti-allergic, anti-fatigue, anti-stress, and anti-cancer activities (Oh et al., 2014). More than 100 ginsenosides from Panax species have been identified and categorized into four types: protopanaxadiol, protopanaxatriol, oleanolic acid, and ocotillol-type (Nag et al., 2012). Ginsenoside Rh2 is a metabolite produced by Rg3, Rb1, Rb2, and Rc by human intestinal bacteria after absorption (Chi et al., 2005). Ginsenoside Rh2 is classified into 20(S) and 20(R) forms according to the different arrangement of the hydroxyl group at the C-20 position, and 20(S)-Rh2 was found to possess strong cytoxic activities to several cancer cells (Liu et al., 2010; Kim et al., 2017). 20(S)-Rh2 has a number of pharmacological properties, such as anti-inflammatory, anticancer, cardioprotection, and neuroprotective effects (Zhang et al., 2012). Rh2 has especially gained attention for its broad-spectrum anti-proliferative effect on a variety of human cancer cells (Oh et al., 1999; Kim et al., 2004; Qi et al., 2011; Guo et al., 2014; Shi et al., 2014; Qian et al., 2016; Yang et al., 2016; Wang et al., 2018). In addition, additional in vivo work showed that Rh2 also can significantly suppress the growth of uterine leiomyoma in SD rats (Zhu et al., 2015). The tumor growth inhibitory activity of Rh2 was also found in H22 tumor-bearing mice (Chen et al., 2017) and nude mice bearing human ovarian cancer cells (Nakata et al., 1998).
The molecular pathways affected by Rh2 in cancer cells are mainly through apoptosis-related pathways. for example, Rh2 induces apoptosis in colorectal cancer cells through activation of p53 (Li et al., 2011). in human leukemia Reh cells, Rh2 could induce apoptosis through the mitochondrial pathway (Xia et al., 2014). Rh2 also suppresses proliferation of hepatocellular carcinoma (HCC) cells by targeting EZH2 to regulate CDKN2A-2B gene cluster transcription (Li et al., 2017). Although apoptosis and other related pathways and genes have been identified in Rh2-treated cancer cells, the molecular mechanism through which Rh2 promotes cancer cell apoptosis is not known at the transcriptome level. To the best of our knowledge, this study is the first transcriptome analysis of the anti-proliferative effects of Rh2 on hepg2 cells. The results will aid the identification of functional genes and provide a more comprehensive understanding of the role of Rh2 in promoting apoptosis in human hematoma cells.
Materials and Methods
Materials
20(S)-Rh2 was obtained from Chengdu Must Bio-Technology Co., Ltd. (Chengdu, Sichuan, China) and the purity (> 98%) was determined by high-performance liquid chromatography. 3-4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Sigma-Aldrich (St. Louis, MO, USA). The fetal bovine serum was obtained from Corning (Medford, MA, USA). Penicillin–streptomycin solution (10,000 unit/10,000 µg/ml) was purchased from Invitrogen–Gibco (Carlsbad, CA, USA). Minimum essential medium was obtained from Sigma-Aldrich (Poole, UK). Protease and phosphatase inhibitors cocktail tablets were bought from Roche (Mannheim, Germany). Phosphate buffered saline (PBS) tables was purchased from Amresco (Solon, OH, USA). The antibodies for caspase-3, -6, and poly (ADP-ribose) polymerase (PARP) were obtained from Cell Signaling Technology (Beverly, MA, USA). Trizol reagent was purchased from Ambion (Austin, TX, USA). Reverse transcription kit was bought from Invitrogen (Carlsbad, CA, USA). Luna universal qPCR master mix was obtained from New England BioLabs (Ipswich, MA, USA). Annexin-PI kit was obtained from BD Biosciences (San Jose, CA, USA). All cell culture suppliers were purchased from Coster (Cambridge, MA, USA).
Cell Line and Cell Culture
The HepG2 cell line was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Wuhan, China) and cultured in minimum essential medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 95% air–5% CO2 in an humidified CO2 incubator (Heracell 150i, Thermo Fisher Scientific, Waltham, MA, USA).
Cell Viability
The cytotoxicity of Rh2 on HepG2 cells was determined using the MTT assay as described previously (Bai et al., 2019). Briefly, cells were seeded (1 × 104 cells/well) in 96-well plate for overnight. Then the cells were incubated with an increasing concentration of Rh2 (0–60 µM) for 24 and 48 h, respectively. The supernatant was removed via aspiration and 100 µl of MTT solution (0.5 mg/ml) was added and cells were continuously cultured until 100 µl of MTT stop solution (10% SDS, 0.01 M hydrochloric acid) was added to each well to solubilize the formazan. The absorbance was measured at 550 nm using a microplate reader (Tecan Infinite M200 Pro, Männedorf, Switzerland).
Cell Morphology Analysis
To examine the effect of Rh2 on the morphology of HepG2 cells, cells (5 × 104 cells/well) were seeded on sterilized cover glasses in 6-well plate for overnight. Then the cells were incubated with Rh2 (20 or 50 µM) for 24 h and the cell morphology was observed with Olympic (Tokyo, Japan). The changes of microstructure were surveyed under scanning electron microscope (SEM). Treated cells were washed with PBS three times and fixed with 2.5% glutaraldehyde solution for 1 h at room temperature. Subsequently, cells were rinsed again with PBS three times, and then dehydrated using graded ethanol (30%, 50%, 70%, 90%, 100%). Cells were fixed onto a copper stub and coated with gold using a sputter coater under vacuum, and then the surface morphology of cells was characterized by SEM (FEI Quanta 450 FEG, Hillsboro, USA).
Annexin V-Fitc Apoptosis Assay
The cells for apoptosis were examined using Annexin V-FITC apoptosis detection kit. Cells (4 × 105 cells/well) were seeded in 6-well plate for overnight. Then the cells were incubated with Rh2 (20 or 50 µM) for 24 h. After treatment, the cells were collected by trypsinization with 0.25% EDTA and centrifuged at 1500 rpm for 3 min to remove the medium. Then the precipitation was washed with pre-cold PBS and resuspended in 100 µl 1 × binding buffer containing 5 µl FITC-Annexin V and 5 µl propidium iodide. After incubation in the dark for 15 min, the fluorescence intensity was evaluated under an Accuri C6 Plus flow cytometer (Becton, Dickinson and company, CA, USA).
RNA Extraction, Library Construction, and Sequencing
HepG2 cells were seeded in 40 mm dishes at a density of 1 × 106 cells/well. Following treatment with 50 µM Rh2 for 24 h, the cells were washed twice with cold PBS and collected by centrifuged for 3 min. Total RNA of each sample was extracted with a Trizol reagent kit (Invitrogen). RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and checked by agarose gel electrophoresis. After enrichment of mRNA using Oligo(dT) beads, it was fragmented into short fragments in fragmentation buffer and reverse transcripted into cDNA with random primers. Then, DNA polymerase I, RNase H, dNTP, and buffer were used to synthesize the second-strand cDNA. The synthesized product were purified with a PCR purification kit (Qiagen, Venlo, The Netherlands), end repaired, poly(A) added, and ligated to Illumina sequencing adapters. After that, the ligated products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced with Illumina HiSeq2500 (Gene Denovo Biotechnology Co. Guangzhou, China).
RNA-Seq Data Analysis
In order to ensure the quality of the data for the following analysis, the raw data of six samples were firstly processed through in-house perl scripts. In this step, clean data were obtained by removing reads containing adapter, reads containing ploy-N, and low quality reads from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality.
Differentially Expressed Genes
RNAs differential expression between control and Rh2 treated groups was analyzed using DESeq2 software (Love et al., 2014). We defined the genes with false discovery rate (FDR) < 0.05 and absolute fold change (FC) ≥ 2 as differentially expressed genes (DEGs).
Gene Ontology and Pathway Enrichment Analysis
Gene Ontology (GO) is a widely used ontology in the bioinformatics field, which offers a semantic vocabulary standard to define and describe the functions of genes and proteins, and can be updated and applied in any organism (Myhre et al., 2006). Hierarchical system was used in GO for gene classification and putting identical genes at the same level. The vocabulary of genes and proteins involved in GO covers three aspects of biology: molecular function (MF), cellular component (CC), and biological process (BP). Terms is the basic units of GO, which were also known as GO classes. Each GO term has a human readable name and a GO ID, and belongs to one of the three sub-ontologies.
Genes often interact with other genes when performing their functions in BP. We often classify genes involved in same biological functions into pathways, which are helpful for us to further understand the biological functions of genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a pathway-related database resource for understanding high-level functions and utilities of the biological systems (Lin and Feng, 2012). KEGG pathway enrichment analysis in DEGs comparing with the whole genome background can identify DEGs into significantly enriched metabolic pathways or signaling pathways.
In the present study, we use the online DAVID functional annotation clustering tool (https://david.ncifcrf.gov) by choosing the default option to determine the significantly represented GO categories and KEGG enriched pathways for DEGs of HepG2 cells response to Rh2 treatment. The calculated p value was corrected with FDR ≤0.05 as a threshold under the FDR correction. At the same time, GO terms and KEGG Pathways of DEGs meeting this condition were defined as significantly enriched GO terms and KEGG pathways.
Real Time Quantitative RT-PCR
HepG2 cells were seeded in 40 mm dishes at a density of 1 × 106 cells/well. Following treatment with 50 µM Rh2 for 24 h, the RNA extraction was carried out as described in the section of RNA extraction, library construction, and sequencing. A 20 µl PCR reaction system was consist with 2 µl cDNA, 10 µl SYBR mixture, 1 µl forward primer, 1 µl reverse primer, and 6 µl deionized water. After mixing, the PCR reaction was performed using CFX-96™ Real-Time instrument (Bio-Rad, CA, USA). The GAPDH gene was used as a house keeping gene to normalize the expression level of the test genes, and the relative gene expression level was analyzed using the 2−ΔΔCT method. All of the samples were analyzed in triplicate. Primers were synthesized by Sangon Biotech (Shanghai, China) and were listed in Table 1.
Western Blot Analysis
HepG2 cells were seeded in 40 mm dishes at a density of 1 × 106 cells/well. Following treatment with 20 or 50 µM Rh2 for 24 h, the cells were washed twice with cold PBS and centrifuged for 3 min, and the supernatant was removed. The cells were lysed in 200 µl of lysis buffer and protein content was measured with Bradford reagent using bovine serum albumin as a standard. Equal amounts of protein were resolved on 10%–15% SDS-polyacrylamide gels for 2 h at 120 V and then transferred to PVDF membranes. The membranes were blocked for 2 h using 5% milk powder in Tris-buffered saline containing Tween 20 (1 × TTBS). After incubation, the membranes were washed with 1 × TTBS and incubated overnight with the primary antibody (1:1000). The membranes were washed and incubated for 1 h with horseradish peroxidase-conjugated secondary anti-IgG (1:500). The protein bands were visualized using the eECL Western Blot Kit (CWBIO, China) according to the manufacturer's instructions and photographed using the Tanon 5200 Multi imaging system (Tanon,Shanghai, China).
Statistical Analysis
The results have been represented as the means standard deviation (SD). The significance was analyzed using SPSS 20.0 package (SPSS Inc., Chicago, IL, USA). Differences among samples were compared using a Duncan's multiple range test and p values less than 5% were considered to be statistically different.
Results and Discussion
Rh2 Inhibits HepG2 Cell Proliferation
HCC is the most malignant primary human liver cancer and has a poor outcome after combined surgical treatment, radiotherapy, and chemotherapy (Lin, 2014). Although our understanding of the mechanism of HCC carcinogenesis has improved, the prognosis of HCC remains poor. Hence, there is an urgent need to develop effective therapies for HCC (Shang et al., 2016). There is growing interest in developing more effective therapeutic agents from natural resources.
To confirm the effects of Rh2 on HepG2 human hepatocarcinoma cells, we performed an MTT assay (Figure 1) and found that Rh2 inhibited HepG2 cell growth in a concentration-dependent manner with an IC50 of 51.97 and 45.46 µM at 24 and 48 h, respectively, which is consistent with the literature (Chen et al., 2015; Shi et al., 2016). To confirm the cytotoxic effects of Rh2 on HepG2, cell morphology was visualized under an inverted microscope and by SEM. As shown in Figure 2A, the untreated cells were spread out and flattened, whereas the Rh2-treated cells were round, shrunken, and showed altered adherence. Moreover, cell numbers were reduced. As shown in Figure 2B, control cells had a normal cell architecture with a distinct cytoskeleton, whereas the Rh2-treated cells had atypical morphological changes, including cell shrinkage and incomplete membranes.
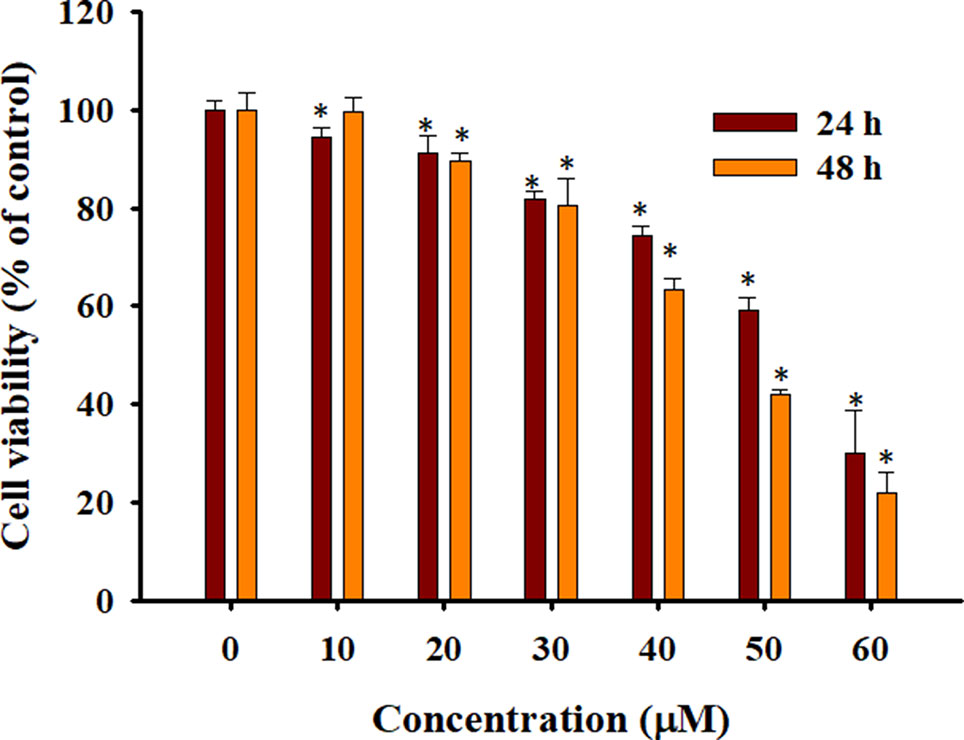
Figure 1 Cytotoxicity of 20(S)-ginsenoside Rh2 (Rh2) on HepG2 cells. Cells were treated with various concentrations of Rh2 for 24 or 48 h and cell viability was evaluated using the 3-4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The values are presented as the mean ± SD (n = 3). Within one column,*p < 0.05 vs. control group.
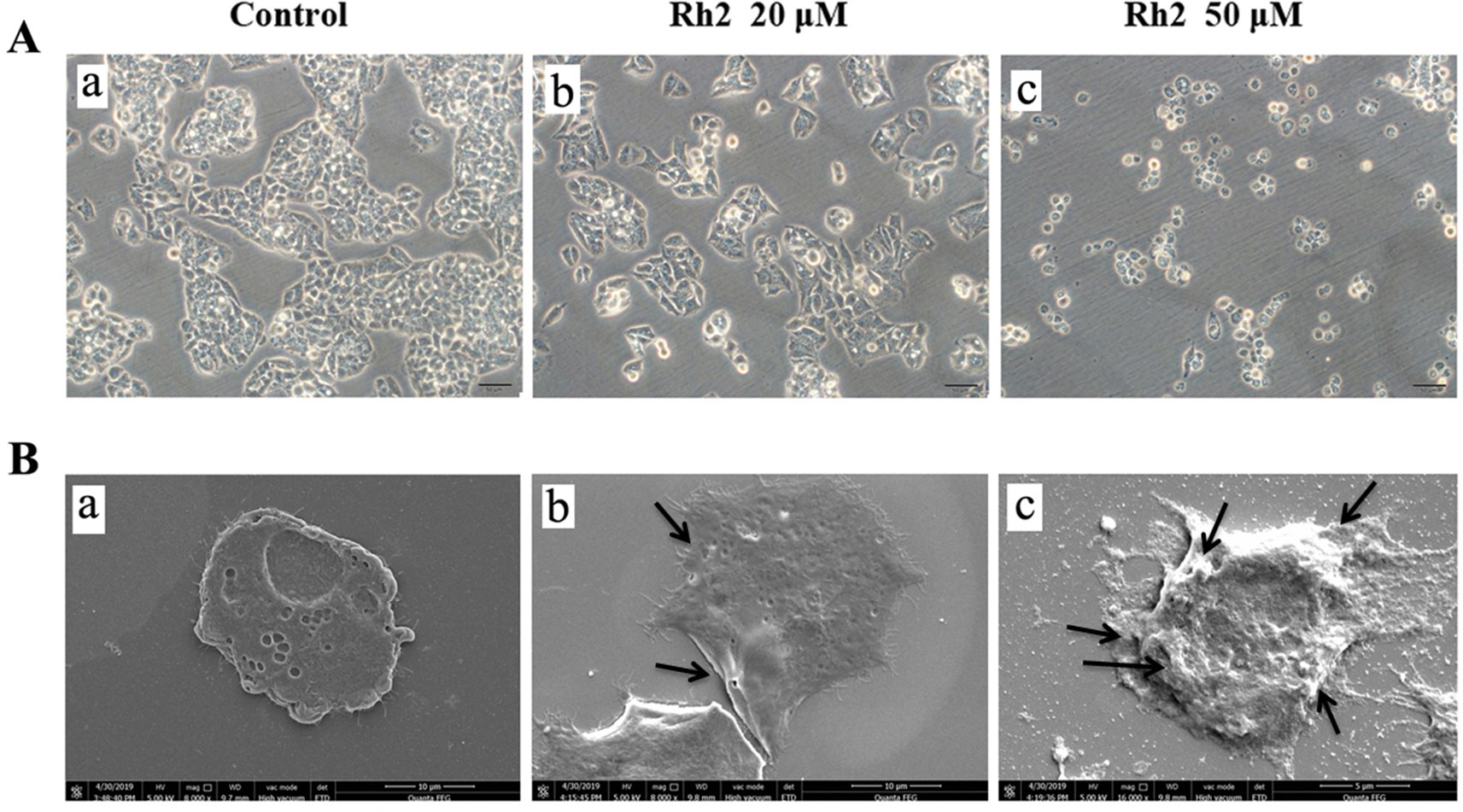
Figure 2 Photomicrographs of HepG2 cells after incubation with Rh2 under a light microscope (A) and SEM (B): (a) control, (b) 20 µM Rh2 treatment, and (c) 50 µM Rh2 treatment. Arrows indicated the damage cell membrane.
Rh2 Induces Apoptosis in HepG2 Cells
Apoptosis, also known as programmed cell death, is an important regulatory mechanism by which the body maintains cell structure and normal tissue development (Bennetts and Pierce, 2010). Inducing tumor cell apoptosis is an important way to inhibit tumor growth. Rh2 inhibits HCC cells in many ways, including inhibiting cell proliferation, inducing cell differentiation, promoting cell apoptosis, inhibiting cell invasion and metastasis, reducing drug resistance, and improving immunity (Li et al., 2017). Although the anti-HCC cell activity of Rh2 has been extensively reviewed, only a few studies have investigated the molecular responses in HCC HepG2 cells (Li et al., 2011; Cheong et al., 2015; Chen et al., 2016; Shi et al., 2016). In this study, we investigated the anti-cancer effects of Rh2 on HepG2 cells and the molecular mechanism by which Rh2 promotes HepG2 cell apoptosis at the transcriptome level.
To confirm the increased apoptotic effects of Rh2 on HepG2 cells, the cell lines were treated with 20 and 50 µM Rh2 for 24 h. After treatment, the cells were examined for apoptosis via flow cytometry using Annexin V-FITC/PI double staining (Figure 3A). The percentage apoptosis increased from 0.057 in the control group to 46.5% and 66.07% (Figure 3B) at 20 and 50 µM, respectively. These results indicate that the anti-proliferative effects of Rh2 on HepG2 cells are mostly cytostatic, rather than cytotoxic. Caspases are a family of cysteine proteases and the primary drivers of apoptotic cell death, and play a critical role in the transduction of apoptosis signals (Vaculova and Zhivotovsky, 2008). Among the major caspases, caspases 3 and 6 are executioners of tumor growth and progression. PARP also plays important roles in apoptosis, the DNA damage response, and cell cycle regulation (Smulson et al., 2000). Interestingly, apoptosis is a genetically regulated cell suicide program mediated by activation of effector caspases 3 and 6. The effects of Rh2 on the expressions of apoptosis-related proteins were determined by assessing the expression levels of caspases 3 and 6 and PARP protein in HepG2 cells via Western blotting. As shown in Figure 4, treatment with 50 µM Rh2 dramatically increased caspase 3 and 6 expression in HepG2 cells but had less of an effect on cleaved-PARP protein. These results indicate that Rh2 promotes the apoptosis of hepatoma cells by upregulating the expression of caspase family proteins.
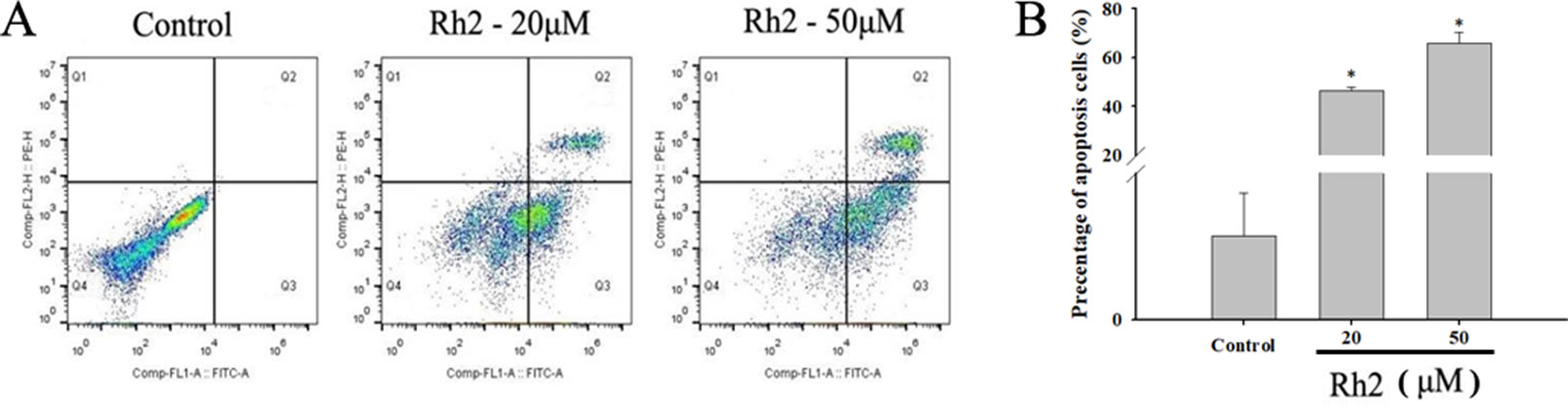
Figure 3 Effects of Rh2 on cell apoptosis in HepG2 cells. (A) Cells were treated with various concentrations of Rh2 for 24 h and the apoptosis rate was determined using Annexin V-FITC/PI staining. (B) Histogram showing the percentage of apoptotic cells. The values are presented as the mean ± SD (n = 3). *p < 0.05 vs. control group.
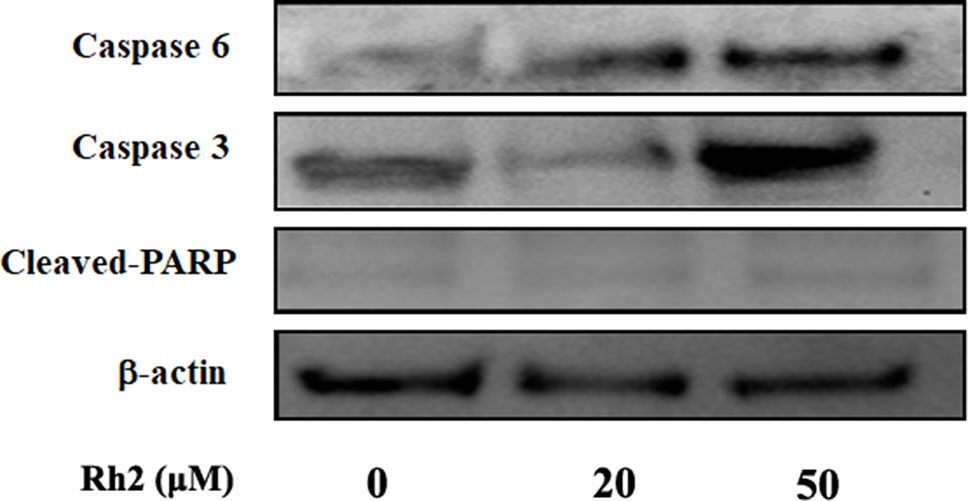
Figure 4 The effects of Rh2 on apoptosis-related proteins expression in HepG2 cells. Cells were treated with various concentrations of Rh2 for 24 h. After preparing the total protein, caspases 3 and 6, and cleaved-PARP were measured via Western blotting.
Data Generation and Comparison With RNA-Seq Analyses
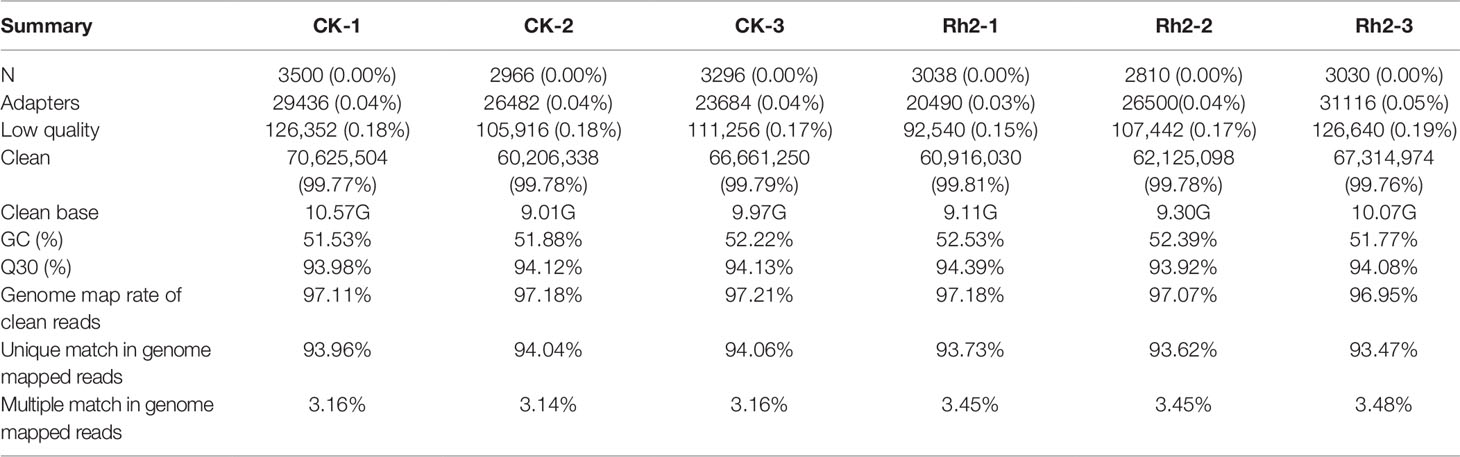
To identify what genes were regulated via Rh2 treatment, and further elucidate the molecular mechanism by which Rh2 inhibits HepG2 cell proliferation, transcriptome analyses were carried out on Rh2-treated and control HepG2 cell samples. Illumina HiSeq2500 was used for sequencing. As shown in Table 2, there were 70,625,504 (99.77%), 60,206,338 (99.78%), and 66,661,250 (99.79%) clean reads in control groups CK-1 to -3 and 60,916,030 (99.81%), 62,125,098 (99.78%), and 67,314,974 (99.76%) in Rh2-treated groups 1–3, respectively. The Q30 proportion of all six samples exceeded 93%, indicating that the quality of the clean reads obtained was high, and the results met the requirement for the next step in bioinformatics analyses. The rates of clean reads for all six samples exceeded 97%, indicating that the utilization rate of sequencing data was high.
DEG Response to Rh2 Treatment
To identify the DEG response to Rh2 treatment, the differential expression multiple between different samples was determined from the level of gene expression. Using FC ≥ 2 and FDR < 0.05 as criteria, 2116 DEGs were identified between the control and Rh2-treated groups. Next, the DEG clusters were analyzed in a heat map (Figure 5A). DEGs identified in biological replicates clustered together, indicating good reproducibility of the treatments. The DEG statistics indicated 971 upregulated genes and 1145 downregulated genes (Figure 5B).
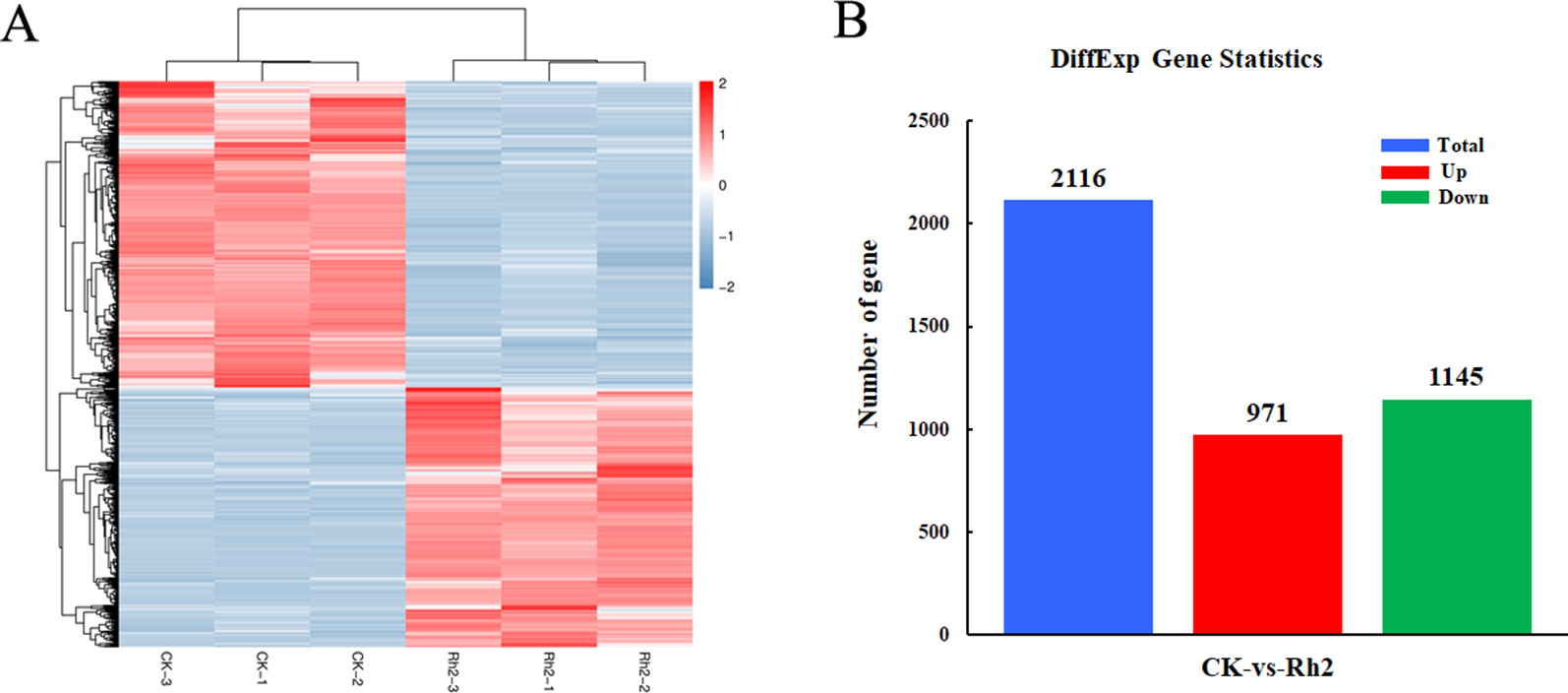
Figure 5 Differentially expressed genes between control and Rh2-treated cells using a heat map (A) and the distribution pattern (B).
GO Enrichment Analyses of DEGs
The DEGs in the CK vs. Rh2 groups were analyzed for GO functional enrichment. Each group of DEGs was annotated into three classifications: MF, CC, and BP. The top 25 functional enriched classes of each group are shown in Figure 6. In MF, the DEGs were mainly enriched for protein dimerization activity, electron carrier activity, heme binding, and heparin binding (Figure 6A). In CC, the DEGs were mainly enriched in the extracellular region part, microsome, endoplasmic reticulum, and vesicular fraction (Figure 6B). In BP, the DEGs were mainly enriched in oxidation reduction, response to wounding, wound healing, and response to cAMP (Figure 6C).
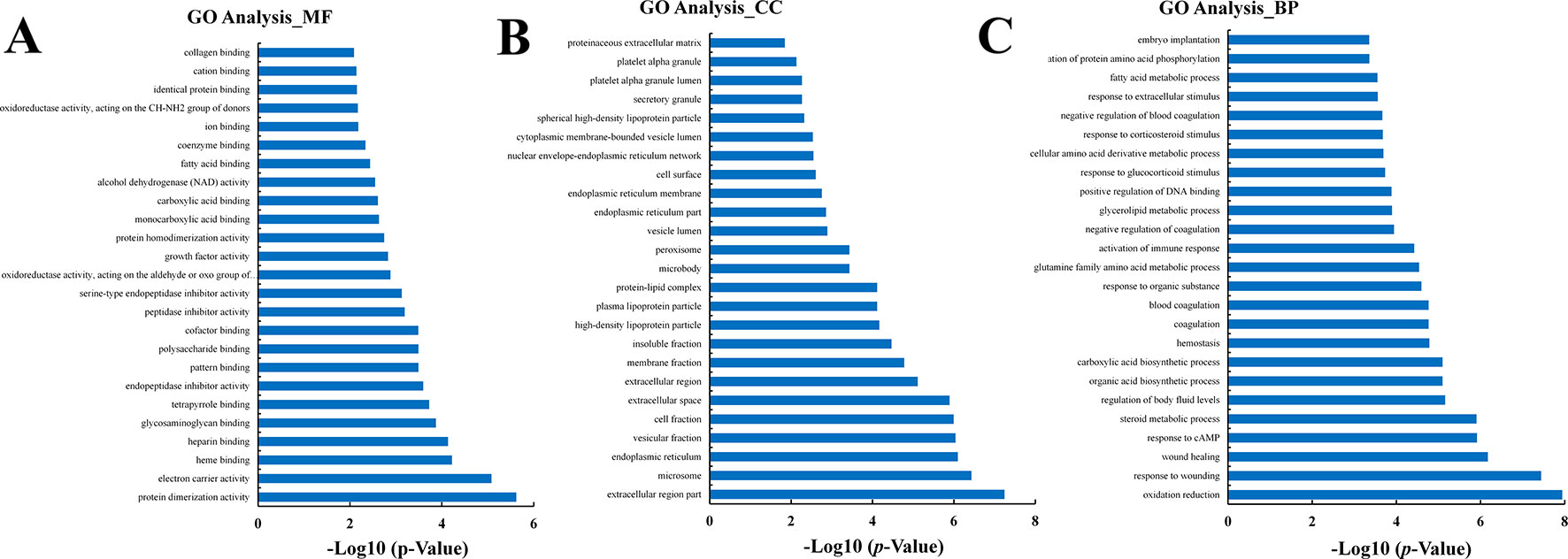
Figure 6 The classification of Gene Ontology (GO) functional enrichment analyses with the differentially expressed genes (DEGs) for CK vs. Rh2. The top 25 functional enriched classes of DEGs annotated by molecular function (MF) (A), cell components (B), and biological processes (BP) (C) sub-ontology of GO enrichment analyses.
KEGG Pathway Enrichment Analyses of DEGs
KEGG pathway analyses of the DEGs in CK vs. Rh2 were performed online at OmicShare. Pathways and all of the DEGs clustered into six categories: organismal systems, metabolism, human diseases, environmental information processing, cellular process, and genetic information processing (Figure 7A). The environmental information processing category includes signal transduction, signaling molecules and interaction, and membrane transport. The cellular processing category includes transport and catabolism, cell growth and death, cellular community-eukaryotes, and cell motility. A scatterplot was used to show the top signaling pathways in the KEGG enrichment analyses. As shown in Figure 7B, the apoptosis-related signaling pathways, including the p53 signaling pathway and DNA replication, were significantly enriched. The DEGs in these two pathway categories may have close relationships to the cell apoptosis caused by Rh2 treatment, highlighting the modulation of Rh2 in tumorigenesis
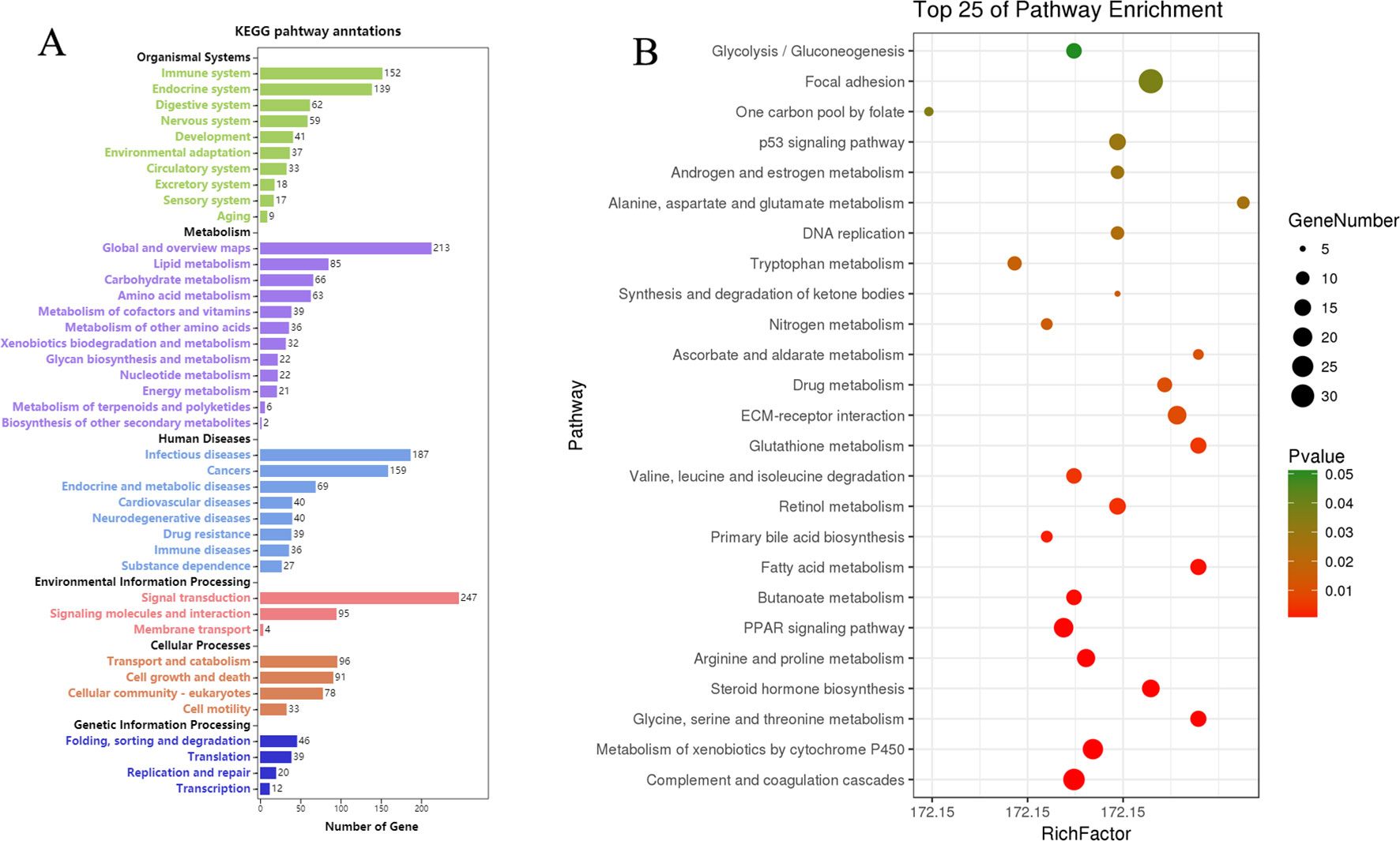
Figure 7 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses of DEGs for CK vs. Rh2. (A) KEGG pathway annotation and classification. (B) Scatterplot of the top 25 pathways in KEGG enrichment.
DEGs in Apoptosis and Cell Cycle-Related Pathways
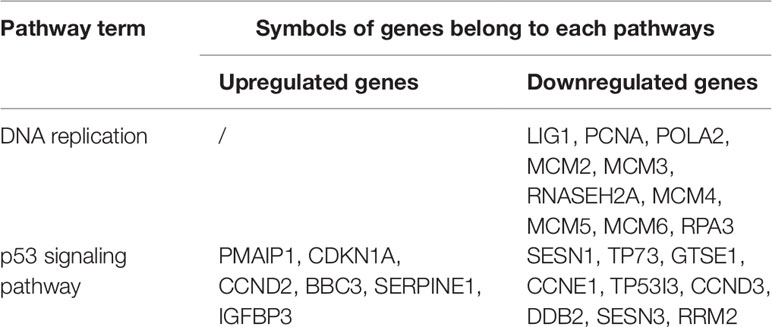
Transcription factor p53 is regulated by its phosphorylation (Zhang et al., 2002). p53 is the upstream regulator of caspase family proteins in oxidative stress and DNA damage induced by various stress conditions (Vakifahmetoglu et al., 2006). The function of p53 is to induce mitochondria to release cytochrome C, which directly improves caspase 9 activity (Schuler et al., 2000). The activation of caspase 3 is promoted by the activation of caspase 9 (Kaushal et al., 2004). The p53-mediated apoptotic signaling pathway is important for cancer treatment with multiple chemotherapies (Zhan et al., 2014). Recent studies have shown that various natural products can induce apoptosis, which is closely associated with the p53 pathway (Laux et al., 2004; Yeruva et al., 2005; Volate et al., 2010). Table 3 lists the DEGs in KEGG pathways related to cell apoptosis and the cell cycle. Fifteen DEGs were annotated in the p53 signaling pathway: six were upregulated and nine were downregulated.
Differential Expression of Representative DEGs
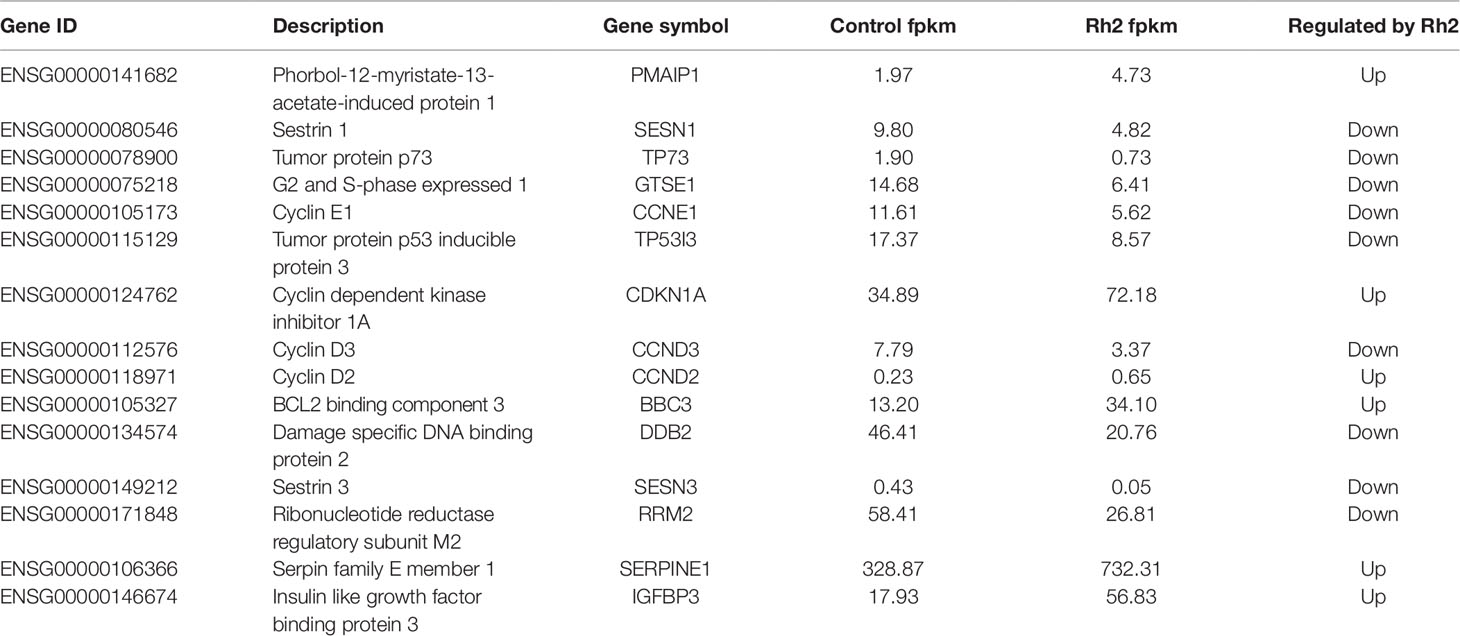
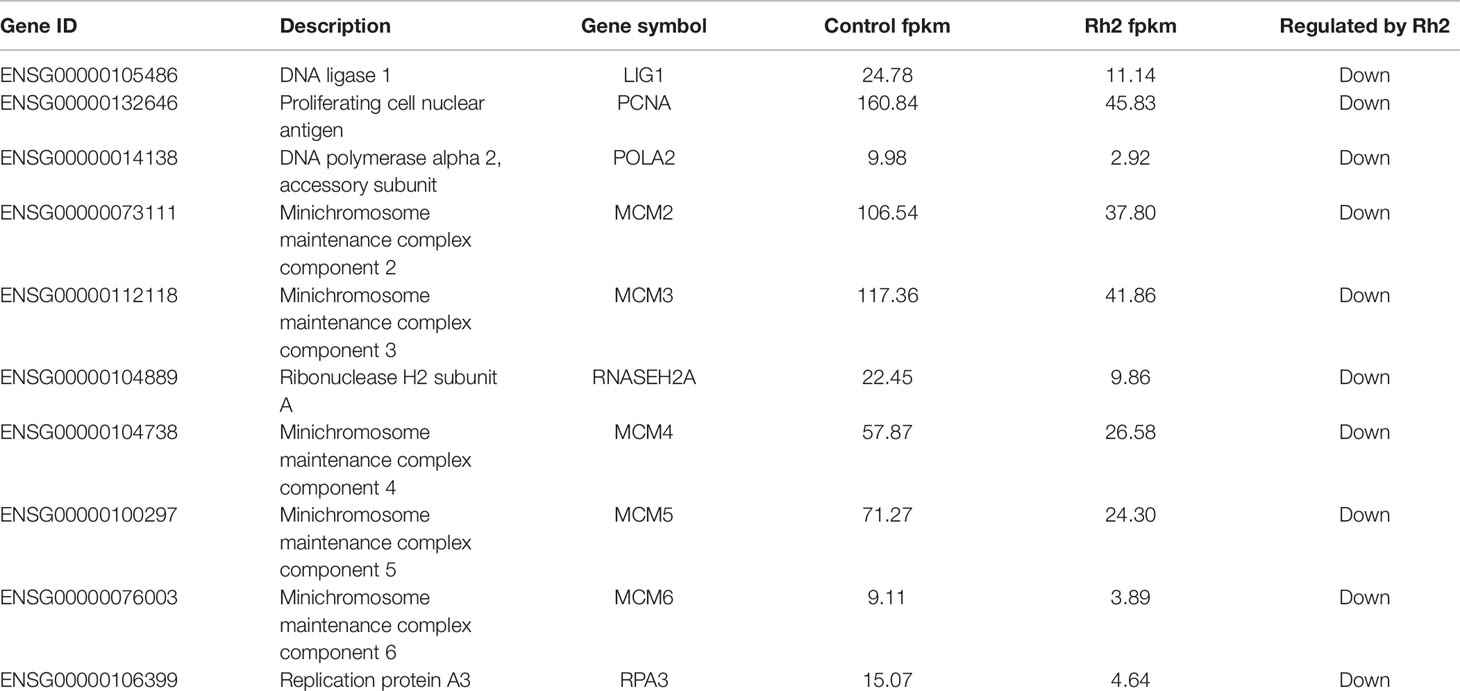
Based on the enrichment results of GO and KEGG, 15 DEGs belonging to the p53 signaling pathway were selected as representative DEGs for a heat map and are listed in Table 4. The color represents the relative expression of the genes. Red represents upregulation and green represents downregulation. As shown in Figure 8 and Table 4, after Rh2 exposure, six genes (PMAIP1, CDKN1A, CCND2, BBC3, SERPINE1, and IGFBP3) were significantly upregulated, while nine (SESN1, TP73, GTSE1, CCNE1, TP53I3, CCND3, DDB2, SESN3, and RRM2) were significantly downregulated. In addition, the 10 DEGs annotated in the DNA replication pathway were all downregulated (Tables 3 and 5).
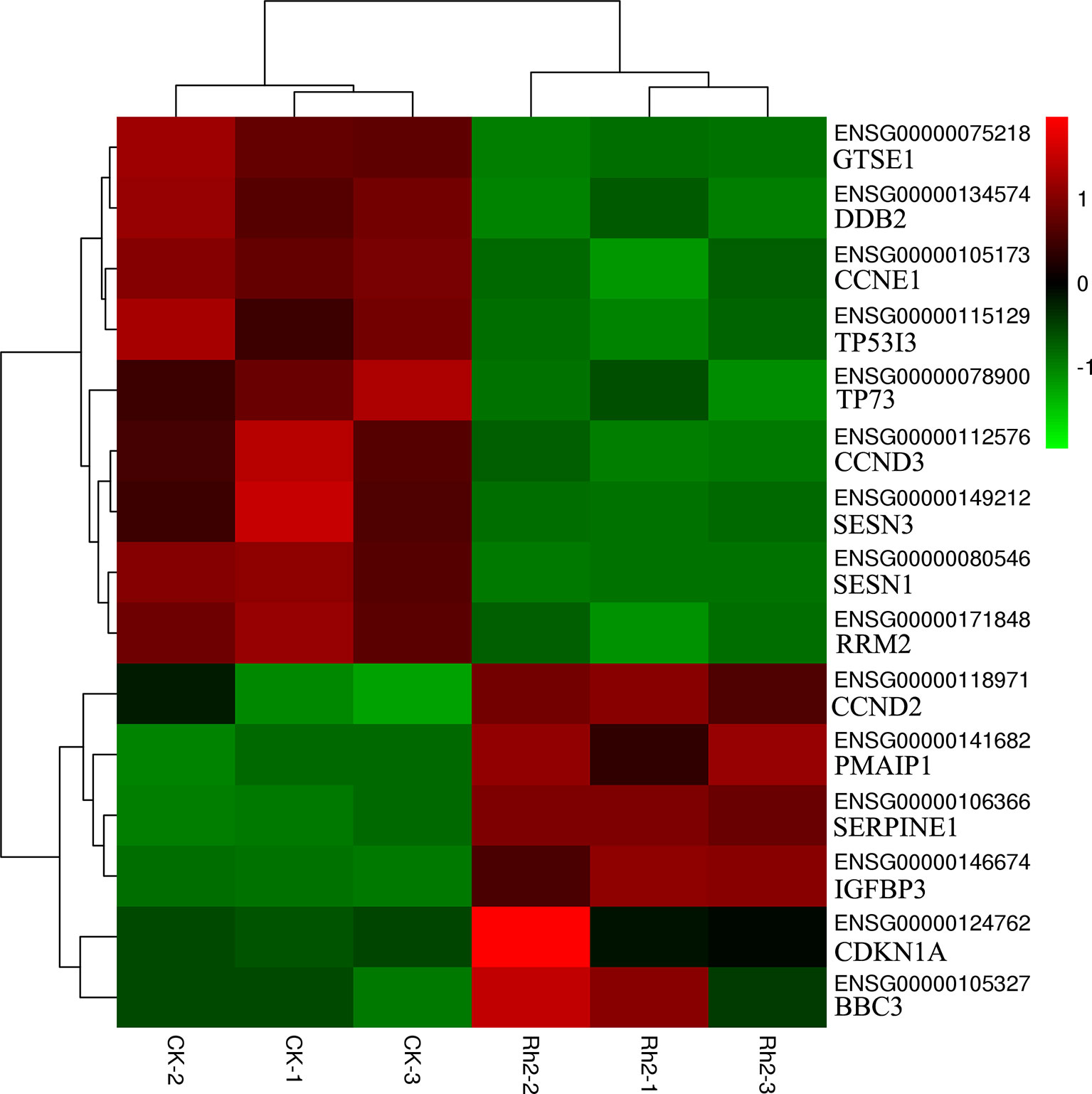
Figure 8 Heat map of representative DEGs in the apoptosis-related p53 signaling pathway by cluster analyses.
RT-qPCR Verification of DEGs
RT-qPCR analyses were used to verify the accuracy of the transcriptome sequencing data. Eleven DEGs in the p53 signaling pathway were selected for qPCR analyses, including PMAIP1, SESN1, TP73, GTSE1, CCNE1, TP53I3, CDKN1A, CCND3, CCND2, BBC3, and DDB2. As shown in Figure 9, the DEGs PMAIP1, CDKN1A, CCND2, and BBC3 were significantly upregulated, while SESN1, TP73, GTSE1, CCNE1, TP53I3, and DDB2 were significantly downregulated on qPCR analyses. The expression of CCND3 was slightly downregulated. This differs slightly from the selection criteria (FC ≥ 2, FDR< 0.05) for DEGs. In general, the relative expression of the selected DEGs showed similar tendencies to the RNA-Seq results (Tables 3 and 4), indicating that the sequencing results were reliable.
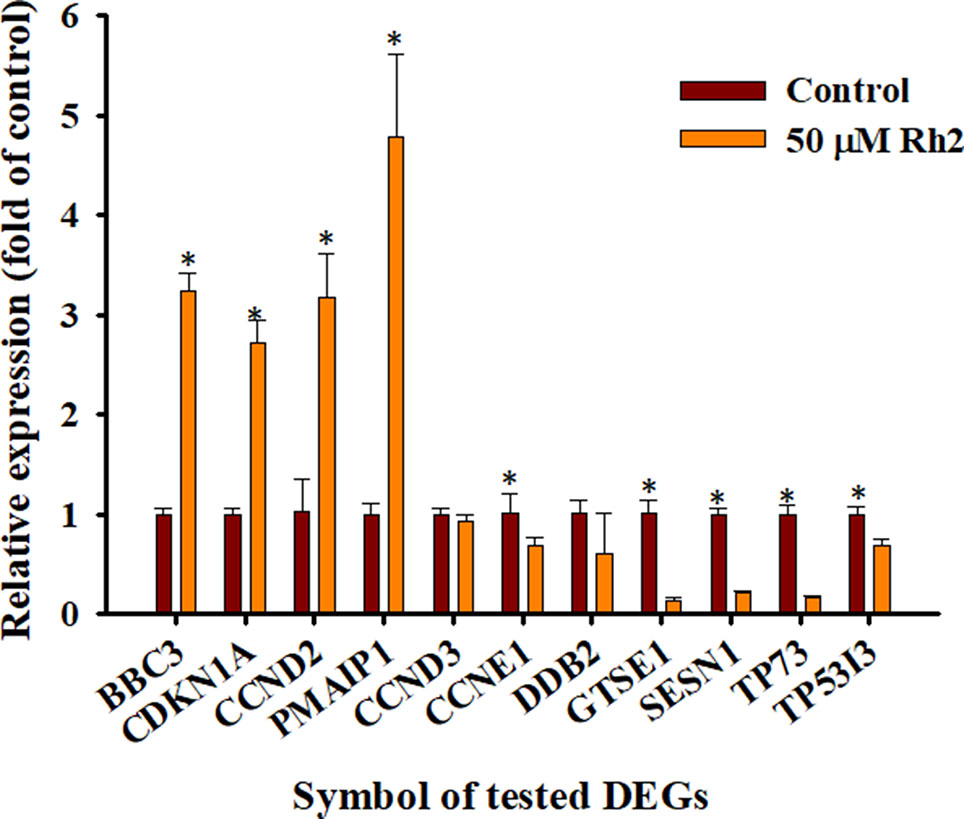
Figure 9 RT-qPCR verification of the selected DEGs. The mRNA expression levels of the test DEGs in the CK vs. Rh2 groups were quantified using RT-qPCR and the housekeeping gene GAPDH was used to normalize the relative expression level. Within the same gene, *p < 0.05 vs. control group.
Conclusions
We demonstrated that Rh2 can inhibit proliferation and induce apoptosis in HepG2 cells. Investigations of the possible mechanism of the anti-proliferation effect revealed that Rh2 induced marked apoptosis that was related to changes in mRNAs in the p53 signaling pathway. Additional studies are currently underway to investigate its overall in vivo anticancer effect and potential mechanisms.
Data Availability Statement
The data generated for this study can be found in the Sequence Read Archive, accession number PRJNA548682.
Author Contributions
WH contributed to design of the study. JiZ, WL, QY, YC and JinZ performed all the experiments. JiZ and WH wrote the manuscript. JiaZ and GF contributed to manuscript revision. All authors read and approved the submitted version.
Funding
This study was financially supported by National Natural Science Foundation of China (31600281), Huai'an 2017 Annual Promotion Project for Science and Technology International Cooperation (HAC201702), Natural Science Foundation of Jiangsu Province (BK20171269), 333 Project of Jiangsu Province (BRA2017241), and Natural Science Foundation of the Jiangsu Higher Education Institutions of China (18KJB320001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yunyao Jiang (School of Pharmaceutical Sciences, Tsinghua University) for helpful discussions on transcriptome results.
References
Bai, Y., Jiang, Y., Liu, T., Li, F., Zhang, J., Luo, Y., et al. (2019). Xinjiang herbal tea exerts immunomodulatory activity via TLR2/4-mediated MAPK signaling pathways in RAW264. 7 cells and prevents cyclophosphamide-induced immunosuppression in mice. J. Ethnopharmacol. 228, 179–187. doi: 10.1016/j.jep.2018.09.032
Bennetts, P. S., Pierce, J. D. (2010). Apoptosis: understanding programmed cell death for the CRNA. AANA J. 78 (3), 237–245.
Chen, F., Deng, Z. Y., Zhang, B., Xiong, Z. X., Zheng, S. L., Tan, C. L., et al. (2015). Esterification of ginsenoside Rh2 enhanced its cellular uptake and antitumor activity in human HepG2 cells. J. Agric. Food. Chem. 64 (1), 253–261. doi: 10.1021/acs.jafc.5b05450
Chen, F., Sun, Y., Zheng, S. L., Qin, Y., McClements, D. J., Hu, J. N., et al. (2017). Antitumor and immunomodulatory effects of ginsenoside Rh2 and its octyl ester derivative in H22 tumor-bearing mice. J. Funct. Foods. 32, 382–390. doi: 10.1016/j.jff.2017.03.013
Chen, F., Zheng, S. L., Hu, J. N., Sun, Y., He, Y. M., Peng, H., et al. (2016). Octyl ester of ginsenoside Rh2 induces apoptosis and G1 cell cycle arrest in human HepG2 cells by activating the extrinsic apoptotic pathway and modulating the Akt/p38 MAPK signaling pathway. J. Agric. Food. Chem. 64 (40), 7520–7529. doi: 10.1021/acs.jafc.6b03519
Cheong, J. H., Kim, H., Hong, M. J., Yang, M. H., Kim, J. W., Yoo, H., et al. (2015). Stereoisomer-specific anticancer activities of ginsenoside Rg3 and Rh2 in HepG2 cells: disparity in cytotoxicity and autophagy-inducing effects due to 20 (S)-epimers. Biol. Pharm. Bull. 38 (1), 102–108. doi: 10.1248/bpb.b14-00603
Chi, H., Kim, D. H., Ji, G. E. (2005). Transformation of ginsenosides Rb2 and Rc from Panax ginseng by food microorganisms. Biol. Pharm. Bull. 28 (11), 2102–2105. doi: 10.1248/bpb.28.2102
Guo, X. X., Li, Y., Sun, C., Jiang, D., Lin, Y. J., Jin, F. X., et al. (2014). p53-dependent Fas expression is critical for Ginsenoside Rh2 triggered caspase-8 activation in HeLa cells. Protein Cell. 5 (3), 224–234. doi: 10.1007/s13238-014-0027-2
Hossen, M. J., Hong, Y. D., Baek, K.-S., Yoo, S., Hong, Y. H., Kim, J. H., et al. (2017). In vitro antioxidative and anti-inflammatory effects of the compound K-rich fraction BIOGF1K, prepared from Panax ginseng. J. Ginseng Res. 41 (1), 43–51. doi: 10.1016/j.jgr.2015.12.009
Kang, S., Im, K., Kim, G., Min, H. (2017). Antiviral activity of 20(R)-ginsenoside Rh2 against murine gammaherpesvirus. J. Ginseng Res. 41 (4), 496–502. doi: 10.1016/j.jgr.2016.08.010
Kaushal, G. P., Liu, L., Kaushal, V., Hong, X., Melnyk, O., Seth, R., et al. (2004). Regulation of caspase-3 and-9 activation in oxidant stress to RTE by forkhead transcription factors, Bcl-2 proteins, and MAP kinases. Am. J. Physiol.-Renal Physiol. 287 (6), F1258–F1268. doi: 10.1152/ajprenal.00391.2003
Kim, J. H., Yi, Y. S., Kim, M. Y., Cho, J. Y. (2017). Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J.Ginseng Res. 41 (4), 435–443. doi: 10.1016/j.jgr.2016.08.004
Kim, H. S., Lee, E. H., Ko, S. R., Choi, K. J., Park, J. H., Im, D. S. (2004). Effects of ginsenosides Rg 3 and Rh 2 on the proliferation of prostate cancer cells. Arch. Pharmacal Res. 27 (4), 429. doi: 10.1007/BF02980085
Laux, M. T., Aregullin, M., Berry, J. P., Flanders, J. A., Rodriguez, E. (2004). Identification of a p53-dependent pathway in the induction of apoptosis of human breast cancer cells by the natural product, resveratrol. J. Altern. Complement Med. 10 (2), 235–239. doi: 10.1089/107555304323062211
Li, B., Zhao, J., Wang, C. Z., Searle, J., He, T. C., Yuan, C. S., et al. (2011). Ginsenoside Rh2 induces apoptosis and paraptosis-like cell death in colorectal cancer cells through activation of p53. Cancer Panax ginseng Lett. 301 (2), 185–192. doi: 10.1016/j.canlet.2010.11.015
Li, Q., Li, B., Dong, C., Wang, Y. (2017). 20 (S)-Ginsenoside Rh2 suppresses proliferation and migration of hepatocellular carcinoma cells by targeting EZH2 to regulate CDKN2A-2B gene cluster transcription. Eur. J. Pharmacol. 815, 173–180. doi: 10.1016/j.ejphar.2017.09.023
Lin, C. M., Feng, W. (2012). Microarray and synchronization of neuronal differentiation with pathway changes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) databank in nerve growth factor-treated PC12 cells. Curr. Neurovasc. Res. 9 (3), 222–229. doi: 10.2174/156720212801618983
Lin, R. P. G. (2014). Clinical study of hepatectomy combined with Jianpi Huayu therapy for hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 15 (14), 5951–5957. doi: 10.7314/APJCP.2014.15.14.5951
Liu, J., Shimizu, K., Yu, H., Zhang, C., Jin, F., Kondo, R. (2010). Stereospecificity of hydroxyl group at C-20 in antiproliferative action of ginsenoside Rh2 on prostate cancer cells. Fitoterapia. 87 (7), 902–905. doi: 10.1016/j.fitote.2010.05.020
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Myhre, S., Tveit, H., Mollestad, T., Lægreid, A. (2006). Additional gene ontology structure for improved biological reasoning. Bioinformatics 22 (16), 2020–2027. doi: 10.1093/bioinformatics/btl334
Nag, S. A., Qin, J., Wang, W., Wang, M. H., Wang, H., Zhang, R. (2012). Ginsenosides as anticancer agents: in vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front. Pharmacol. 3, 25. doi: 10.3389/fphar.2012.00025
Nakata, H., Kikuchi, Y., Tode, T., Hirata, J., Kita, T., Ishii, K., et al. (1998). Inhibitory effects of ginsenoside Rh2 on tumor growth in nude mice bearing human ovarian cancer cells. Jpn. J. Cancer Res. 89 (7), 733–740. doi: 10.1111/j.1349-7006.1998.tb03278.x
Oh, M., Choi, Y., Choi, S., Chung, H., Kim, K., Kim, S. I., et al. (1999). Anti-proliferating effects of ginsenoside Rh2 on MCF-7 human breast cancer cells. Int. J. Oncol. 14 (5), 869–944. doi: 10.3892/ijo.14.5.869
Oh, S. J., Lee, S., Choi, W. Y., Lim, C. J. (2014). Skin anti-photoaging properties of ginsenoside Rh2 epimers in UV-B-irradiated human keratinocyte cells. Z. Naturforsch. C: Biosci. 39 (4), 673–682. doi: 10.1007/s12038-014-9460-x
Park, J., Cho, J. (2009). Anti-inflammatory effects of ginsenosides from Panax ginseng and their structural analogs. Afr. J. Biotechnol. 8 (16), 3682–3690. doi: 10.5897/AJB2009.000-9371
Qi, X. D., Hou, J. C., Yu, H. T., Zhang, C. J. (2011). 20 (S)-Ginsenoside-Rh2 and 20 (R)-Ginsenoside-Rh2 activate IkappaB phosphorylation expression in human lung adenocarcinoma A549 cells. Adv. Mat Res. 268, 1205–1210. doi: 10.4028/www.scientific.net/AMR.268-270.1205
Qian, J., Li, J., Jia, J. G., Jin, X., Yu, D. J., Guo, C. X., et al. (2016). Ginsenoside-Rh2 inhibits proliferation and induces apoptosis of human gastric cancer SGC-7901 side population cells. Asian Pac J. Cancer Prev. 17 (4), 1817–1821. doi: 10.7314/APJCP.2016.17.4.1817
Schuler, M., Bossy-Wetzel, E., Goldstein, J. C., Fitzgerald, P., Green, D. R. (2000). p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. Int. J. Biol. Chem. 275 (10), 7337–7342. doi: 10.1074/jbc.275.10.7337
Shang, N., Arteaga, M., Zaidi, A., Cotler, S. J., Breslin, P., Ding, X., et al. (2016). FAK kinase activity is required for the progression of c-MET/β-catenin-driven hepataocellular carcinoma. Gene Expr. 17 (1), 79–88. doi: 10.3727/105221616X691604
Shi, Q., Li, J., Feng, Z., Zhao, L., Luo, L., You, Z., et al. (2014). Effect of ginsenoside Rh2 on the migratory ability of HepG2 liver carcinoma cells: recruiting histone deacetylase and inhibiting activator protein 1 transcription factors. Mol. Med. Rep. 10 (4), 1779–1785. doi: 10.3892/mmr.2014.2392
Shi, Q., Shi, X., Zuo, G., Xiong, W., Li, H., Guo, P., et al. (2016). Anticancer effect of 20 (S)-ginsenoside Rh2 on HepG2 liver carcinoma cells: Activating GSK-3β and degrading β-catenin. Oncol. Rep. 36 (4), 2059–2070. doi: 10.3892/or.2016.5033
Smulson, M., Simbulan-Rosenthal, C., Boulares, A., Yakovlev, A., Stoica, B., Iyer, S., et al. (2000). Roles of poly (ADP-ribosyl) ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv. Enzyme Regul. 1 (40), 183–215. doi: 10.1016/S0065-2571(99)00024-2
Vaculova, A., Zhivotovsky, B. (2008). Chapter eight caspases: determination of their activities in apoptotic cells. Methods Enzymol. 442, 157–181. doi: 10.1016/S0076-6879(08)01408-0
Vakifahmetoglu, H., Olsson, M., Orrenius, S., Zhivotovsky, B. (2006). Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25 (41), 5683. doi: 10.1038/sj.onc.1209569
Volate, S. R., Kawasaki, B. T., Hurt, E. M., Milner, J. A., Kim, Y. S., White, J., et al. (2010). Gossypol induces apoptosis by activating p53 in prostate cancer cells and prostate tumor-initiating cells. Mol. Cancer Ther. 9 (2), 461–470. doi: 10.1158/1535-7163.MCT-09-0507
Wang, Y., Xu, H., Lu, Z., Yu, X., Lv, C., Tian, Y., et al. (2018). Pseudo-Ginsenoside Rh2 induces A549 cells apoptosis via the Ras/Raf/ERK/p53 pathway. Exp. Ther. Med. 15 (6), 4916–4924. doi: 10.3892/etm.2018.6067
Xia, T., Wang, J. C., Xu, W., Xu, L. H., Lao, C. H., Ye, Q. X., et al. (2014). 20 (S)-Ginsenoside Rh2 induces apoptosis in human leukaemia Reh cells through mitochondrial signaling pathways. Biol. Pharm. Bull. 37 (2), 248–254. doi: 10.1248/bpb.b13-00667
Yang, J., Yuan, D., Xing, T., Su, H., Zhang, S., Wen, J., et al. (2016). Ginsenoside Rh2 inhibiting HCT116 colon cancer cell proliferation through blocking PDZ-binding kinase/T-LAK cell-originated protein kinase. J. Ginseng Res. 40 (4), 400–408. doi: 10.1016/j.jgr.2016.03.007
Yeruva, L. V., Elegbede, J. A., Toy, B. J., Wang, R. C. (2005). The apoptosis-inducing activity of natural plant products involved regulation of events downstream of the p53 pathway. Proc. Am Assoc Cancer Res. 65 (9 Supplement), 1389.
Yu, G. J., Choi, I. W., Kim, G. Y., Kim, B. W., Park, C., Hong, S. H., et al. (2015). Anti-inflammatory potential of saponins derived from cultured wild ginseng roots in lipopolysaccharide-stimulated RAW 264.7 macrophages. Int. J. Med. Mol. Adv. Sci. 35 (6), 1690–1698. doi: 10.3892/ijmm.2015.2165
Zhan, Y. S., Tan, S. W., Mao, W., Jiang, J., Liu, H. L., Wu, B. (2014). Chemotherapy mediates intestinal injury via p53/p53 upregulated modulator of apoptosis (PUMA) signaling pathway. J. Dig. Dis. 15 (8), 425–434. doi: 10.1111/1751-2980.12157
Zhang, J., Krishnamurthy, P. K., Johnson, G. V. (2002). Cdk5 phosphorylates p53 and regulates its activity. J. Neurochem. 81 (2), 307–313. doi: 10.1046/j.1471-4159.2002.00824.x
Zhang, J., Zhou, F., Wu, X., Zhang, X., Chen, Y., Zha, B. S., et al. (2012). Cellular pharmacokinetic mechanisms of adriamycin resistance and its modulation by 20 (S)-ginsenoside Rh2 in MCF-7/Adr cells. Br. J. Pharmacol. 165 (1), 120–134. doi: 10.1111/j.1476-5381.2011.01505.x
Keywords: 20(S)-ginsenoside Rh2, hepatocellular carcinoma, anti-proliferation, p53 signaling pathway, RNA-seq
Citation: Zhang J, Li W, Yuan Q, Zhou J, Zhang J, Cao Y, Fu G and Hu W (2019) Transcriptome Analyses of the Anti-Proliferative Effects of 20(S)-Ginsenoside Rh2 on HepG2 Cells. Front. Pharmacol. 10:1331. doi: 10.3389/fphar.2019.01331
Received: 09 June 2019; Accepted: 18 October 2019;
Published: 07 November 2019.
Edited by:
Jae Youl Cho, Sungkyunkwan University, South KoreaReviewed by:
Mi-Yeon Kim, Soongsil University, South KoreaKi Sung Kang, Gachon University, South Korea
Xihong Zhao, Wuhan Institute of Technology, China
Copyright © 2019 Zhang, Li, Yuan, Zhou, Zhang, Cao, Fu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weicheng Hu, aHVfd2VpY2hlbmdAMTYzLmNvbQ==; Guangbo Fu, ZmdiMjAwQHZpcC4xNjMuY29t
†These authors have contributed equally to this work
 Ji Zhang1†
Ji Zhang1† Weicheng Hu
Weicheng Hu