
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 11 June 2019
Sec. Predictive Toxicology
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.00561
A plethora of databases exist online that can assist in in silico chemical or drug safety assessment. However, a systematic review and grouping of databases, based on purpose and information content, consolidated in a single source, has been lacking. To resolve this issue, this review provides a comprehensive listing of the key in silico data resources relevant to: chemical identity and properties, drug action, toxicology (including nano-material toxicity), exposure, omics, pathways, Absorption, Distribution, Metabolism and Elimination (ADME) properties, clinical trials, pharmacovigilance, patents-related databases, biological (genes, enzymes, proteins, other macromolecules etc.) databases, protein-protein interactions (PPIs), environmental exposure related, and finally databases relating to animal alternatives in support of 3Rs policies. More than nine hundred databases were identified and reviewed against criteria relating to accessibility, data coverage, interoperability or application programming interface (API), appropriate identifiers, types of in vitro, in vivo,-clinical or other data recorded and suitability for modelling, read-across, or similarity searching. This review also specifically addresses the need for solutions for mapping and integration of databases into a common platform for better translatability of preclinical data to clinical data.
Chemical risk assessment refers to the quantification of any potential adverse effects to humans or environmental species related to exposure to chemicals, drugs, pesticides, consumer products, or any other substances. Traditionally, the assessment of chemicals, including pharmaceuticals, relied on data from animal testing; however, there are many motivations to move to a society free of such testing. In part, the new paradigm for safety assessment embraces the ethos of twenty-first century Toxicology whereby every effort is made to maximise the information that may be obtained without animal testing (Embry et al., 2014). This information may include existing knowledge on the chemical in question, or similar chemicals (the process of read-across), as well as in vitro and high throughput determination relating to mechanisms of action and effects at the cellular or organ level (Cronin et al., 2009; Kongsbak et al., 2014). Existing and experimental data are also supplemented by predictions, which may relate to toxicity, mechanisms or exposure that are collectively termed “in silico.” There is no formal definition for the process or practice of in silico chemical safety assessment; however, it needs to encompass existing knowledge and outputs from predictions of both hazard and exposure as a means of making a decision. There is also increasing interest in making this type of information gathering and assessment more translational, to gain knowledge from all sources to understand the effects on humans—patients in the case of pharmaceuticals—and how that can be translated to mechanisms and assays etc.
There are various types of data that may be considered in modern in silico chemical safety assessment. Historical, or legacy, data from toxicological testing provide one of the most important sources of information for modelling and read-across. In theory, data should be available for all endpoints that have been tested across a variety of guideline and non-standard approaches. Such data may be either available openly or be confidential business information and may encompass toxicological and physico-chemical information. These data have been the cornerstone of in silico modelling in the past and remain essential for performing safety assessment of existing chemicals. At the other end of the spectrum are upcoming resources that capture mechanistic understanding of chemicals. Such understanding has, in part at least, been facilitated by the so-called “New Approach Methodologies” (NAMs) including in vitro High-Throughput Screening (HTS) methods (bioactivity or toxicity profiling bioassays) and omics data generated by more specific genome sequencing, transcriptomics, proteomics, and metabolomics studies (Hartung et al., 2017). These, and other large data repositories, such as clinical effects and adverse drug reactions are routinely referred to as being big data. The term “big data” implies a huge volume of data collected from multiple resources and characterised by their complexity and heterogenous nature. Computational tools often manage big data or algorithms that help to capture, store, search, and analyse the data more rapidly.
Capturing a chemical's physico-chemical properties, bioactivity, and safety profiles or toxicity within databases has become a necessary part of research across many industrial sectors including pharmaceuticals, personal care products, petro-chemicals, and biocides. As a result, in silico resources have been reviewed and assessed previously by many researchers, as indicated in Table 1, which identifies 48 of these recent reviews. For example, Young (2002) reviewed web-based resources at the US National Library of Medicine (NLM) including MEDLINE®, PUBMED®, Gateway, Entrez, and TOXNET. As systems biology emerged many gene expression repositories and software were also developed (Anderle et al., 2003; Judson, 2010; Benigni et al., 2013; Fostel et al., 2014). Efforts were not limited to only gene or protein expression databases, but also included organ specific toxicity databases. The review by Fotis et al. (2018) discussed databases relating to genomics, proteomics, metabolomics, multiomics whilst the review by Papadopoulos et al. (2016) focused on such databases specifically relating to the kidney. In relation to other major organs, liver, and heart-related toxicity databases have been discussed by Luo et al. (2017) and Sato et al. (2018), respectively. These diverse types of databases have been further expanded or designed in such a way as to enable interaction with other public resources so improving accessibility for end users. Many resources have emerged that try to link or integrate the chemistry-based databases with bioactivity, pathways of toxicity, ADME, and omics data sets. The chemistry-based databases on small molecules or new compounds were discussed in detail in a number of reviews (Jonsdottir et al., 2005; Williams, 2008; Hersey et al., 2015). Some of the databases that allow for mining of the chemical information (such as 2D, 3D structures, physico-chemical properties etc.) are ChEMBL, ChEBI, PubChem, DrugBank, ZINC, etc. In drug discovery, the number of databases for target identification or prediction of activity, has grown tremendously (Oprea and Tropsha, 2006; Loging et al., 2011; Chen and Butte, 2016; Chen et al., 2016; Katsila et al., 2016; Cha et al., 2018). Other databases containing information on proteins associated with drug therapeutic effects, adverse drug reactions, and ADME properties has facilitated systematic curation and analysis of complex ligand-target data (Ji et al., 2003a). Some of the ADME, potential drug-drug interaction (DDI) information and pharmacogenomics-related databases have been cited in a number of review articles (Ekins et al., 2005; Bauer-Mehren et al., 2009; Ekins and Williams, 2010; Sim et al., 2011; Peach et al., 2012; Wishart, 2014; Ayvaz et al., 2015; Zhang et al., 2015; Przybylak et al., 2018). Fouretier et al. (2016) identified human drug safety data resources, or pharmacovigilance databases, specific to every country or subcontinent. The European Union's Innovative Medicines Initiative 2 Joint Undertaking (IMI 2) “Enhancing TRANslational SAFEty Assessment through Integrative Knowledge Management (eTRANSAFE)” project is developing an integrative data infrastructure to combine and utilise data resources, hence has stimulated the work described in this paper. The aim of eTRANSAFE is to drastically improve the feasibility and reliability of translational safety assessment during the drug development process using both publicly available resources in addition to data provided by its partners to facilitate acceptance by stakeholders, including regulatory agencies and international organisations.
Table 1. Previous review articles for identification of databases relevant to chemistry and toxicology.
In spite of many previous reviews of data sources for specific types of data (chemistry based, toxicology, omics, ADME etc.), or those predominantly focussed on a specific type of data, no review exists covering all data resources that may be required for twenty-first century toxicology and translational sciences in drug discovery. Moreover, many reviews have failed to address the importance of the full identification, mapping and integration of chemical and biological spaces. Thus, the aim of this study was to provide a comprehensive and consolidated list of in silico data resources for chemical safety assessment. This review aimed to encompass all data resources including those based on chemistry, pharmacological space, genomics, and adverse events, as well as those relevant to toxicology and human effects or clinical safety studies. The databases were assessed in groups based on their purpose and information content; data relating to each resource was recorded and summarised. It is intended that this review will provide a valuable starting point for researchers wishing to gain knowledge about a chemical substance and its exposure/effect in preclinical and clinical studies.
Initially, the different types of databases to be reviewed were established. The categories chosen for investigation were chemistry-based databases containing information on: toxicology (preclinical studies for chemicals and drugs), genes or enzymes, pathways or AOP-related, omics, protein-protein interactions, ADME, drug discovery, clinical trials, pharmacovigilance, patent-based, environmental chemical exposure, nanomaterial toxicity and animal alternatives, or 3Rs related databases. These categories were utilised to facilitate the searching and grouping/clustering of databases. An iterative process was followed to identify the multiple independent, disparate databases by searching for specific category-based databases in published review papers (Table 1), regulatory-based websites (US FDA, EPA etc.), chemical/pharmaceutical company websites and some of the specific resources on databases such as Toxnet1, Pathguide2, Fair sharing3, VLS3D4, Wikipedia5, Oxford Journal's biological databases6, and AltTox.org7 etc.
The criteria by which the databases were assessed were established and are summarised in Table 2; the criteria stipulate both essential and desirable features. Each database was screened using these criteria and only those meeting minimum requirements are included within this review. The most important criterion on which to select a database was its ease of accessibility. The databases could be considered as “open” (free to accesss or use, right to share and re-use) or “partially open” (where only partial metadata are available to access or download or not intended for commercial use). Some of the databases for which the URL links are retired were removed from the list. The availability of information on the type of Application Programming Interface (API) or the programming codes used to develop the databases was also considered as a criterion in selecting the databases. Other essential criteria include- appropriate chemical identifiers (such as SMILES, InChIs etc.), readily converted ontologies and the relevance of the endpoint(s). The desirable attributes included access to the metadata, study protocols, information on data quality assessment, ease of navigation, and other database statistics relating to the frequency of updates and number of compounds or drugs reported. Additional criteria related to the nature of the information provided or the potential use of the database and covers the type of data recorded (in vitro/ in vivo/ biomarkers/ omics/ clinical data etc.). The type of data incorporated in the database was limited not only to experimental data (in vitro, in vivo) but also included predicted data, quantitative structure-activity relationship (QSAR) models, similarity searching for chemicals or genes and read-across methods.
The list of more than 900 databases is provided in Table 3 and other detailed information of each database is compiled in an Excel spreadsheet (Supplementary Information). The information included: the name of the database; the owner of the database; any licensing requirements or restrictions on use; the URL or other information on database location; access rights (e.g., registration requirement, free access, potential to download all data etc.); endpoints covered; number and type of compounds included; granularity of data (e.g., test results, dose-response data, experimental conditions etc.) and details on data quality assurance (e.g., details of curation and assignment of quality score such as Klimisch scores) where applicable. Klimisch scores (Klimisch et al., 1997) are assigned to toxicological or physico-chemical data to assess their adequacy, relevance and reliability. The scores are as follows: 1 = reliable without restrictions; 2 = reliable with restrictions; 3 = not reliable 4 = not assignable. These scores are based on criteria such as whether the study was conducted under Good Laboratory Practice (GLP) and if key information on test substances, experimental conditions and statistical evaluation was stated.
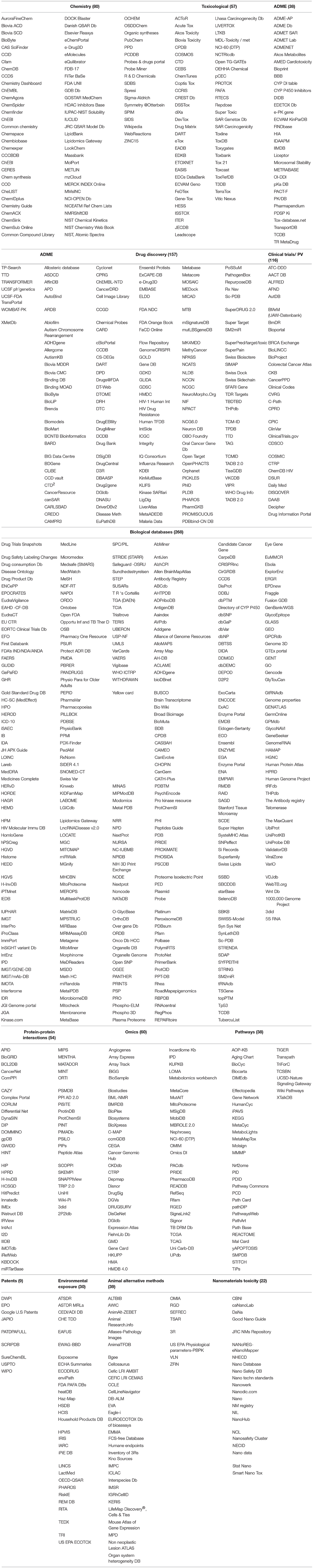
Table 3. Complete listing of all databases identified in this study grouped according to content (URL links available on-line).
Based on the criteria established herein for selecting databases, a comprehensive list of more than 900 databases was compiled (Table 3) and consolidated. The types and relative proportion of databases identified are summarised in Figure 1. The key features of the databases within each of the 13 groups identified are summarised below.
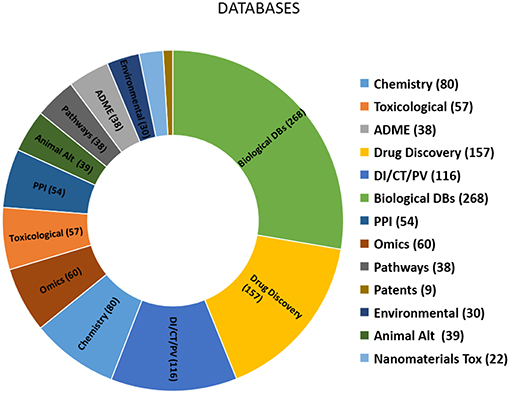
Figure 1. Chart showing the number of databases within each group. DI, Drug Information; CT, Clinical trials; PV, Pharmacovigilance; PPI, Protein-protein interactions; Animal Alt, Animal alternatives.
I. Chemistry Databases
Eighty chemistry databases were identified which relate to resources containing a large library of diverse chemicals or compounds with additional information such as name, molecular formula, structure (2D/3D), key identifiers (CAS Registry Number, IUPAC name, InChI, SMILES), physico-chemical properties, and associations with their bioactivities8. Generally, such databases enable users to perform chemical similarity searches based on different fingerprinting methods such as MACCs, Atom pairs, and Topological Torsion Fingerprints etc. However, despite potentially high numbers of compounds, many of these databases are very sparsely populated particularly with regard to high quality toxicity data. Such data resources do provide a useful and usable gateway to multiple databases such as ChEMBL, KEGG, GeneTox, Daily Med etc.
The applicability, efficiency, and diversity of any database depends on the extent of coverage of chemical space. Within the chemical databases, PubChem (Butkiewicz et al., 2017) includes the highest number of compounds (97 million compounds, 238 million substances, 264 million bioactivities, 3 million patents, and 633 data sources) in comparison to ChemSpider (Pence and Williams, 2010) (68 million chemical structures, 252 data sources), and ChEMBL (Gaulton et al., 2017) (1.8 million compounds and 1.1 million assays). However, the uniqueness of these chemistry-based databases lies in their associations with bioactivities. For example, ChEMBL provides data on bioactive molecules (drug-like properties) and links chemical, bioactivity, and genomic data. ChemSpider provides information on physical properties, biological activities (where available), interactive spectra, the name of chemical suppliers, and other miscellaneous information.
The chemistry-based databases also possess additional tools such as drawing tools, the capability to search for similar structures (both 2-D and 3-D) using similarity scorings, facilities for structure clustering, identifier exchange services, classification browsers, facilities enabling bulk download etc. For the purposes of virtual screening of compounds, building blocks or scaffolds/fragments, many databases were identified such as ZINC-15 (Sterling and Irwin, 2015), ChemSpace9, eMolecules10, Generated DataBase (GDB-DB) (Ruddigkeit et al., 2013), Biovia Screening Compounds Directory (SCD)11, Probes and Drug portal etc. (Skuta et al., 2017). Zinc-15 covers over 230 million ready-to-dock compounds in 3D-formats and 750 million purchasable compounds for screening purposes. In comparison to Zinc-15, Chemspace covers 100 million, eMolecules 1.5 million and GDB-DB 26.4 million structures for small organic molecules. Biovia SCD contains 9.7 million unique “drug-like” chemicals for HTS and lists over 21 million individual products with prices and supplier ordering information.
The Probes and Drugs portal is a partially open resource of bioactive compounds (probes, drugs, kinase inhibitors etc.) for commercial screening purposes. It contains 48 compound sets, 46,401 compounds, 34,887 standardised compounds, 18,612 scaffolds, 6,206 targets, 498,201 bioactivities, 2,455 pathways, 4,174 target/pathway classes, 483 structural alerts, and 2,791 matched structural alerts. Other DBs such as ChemDB (Chen et al., 2007) and OCHEM (Sushko et al., 2011, 2012) could be useful for providing a modeling framework to perform QSPR/QSAR studies online. ChemDB contains nearly 5 million small molecules and includes data on predicted or experimentally determined physicochemical properties (3D structure, melting temperature, and solubility). It also includes a chemical fingerprint-based method to search for similar chemicals based on atom-bond connectivity. OCHEM contains 20,56,039 records for 548 properties (physchem or ADME related) and structural alerts for endpoints such as mutagenicity, skin sensitisation, aquatic toxicity, etc.
II. Toxicological Databases
Fifty-seven databases were identified pertaining to resources providing information on the effects of drugs or xenobiotics on cells, organs or the whole body12. Data are available of many different toxicity endpoints such as: endocrine disruption, mutagenicity, carcinogenicity, skin sensitisation, teratogenicity, organ-specific toxicity etc. Data are either derived from in vitro or in vivo studies or from in silico prediction across multiple species. Of the databases identified, a noteworthy example is the US EPA's Aggregated Computational Toxicology Online Resource (AcToR) (Judson et al., 2008), covering over 500,000 chemicals. It is the warehouse for many EPA's web-based applications such as the Chemistry Dashboard (over 700,000 chemicals and includes chemical structures, experimental and predicted physicochemical, and toxicity data), Toxicity Forecaster (ToxCast) Dashboard (HTS data on over 9,000 chemicals and information on ~1,000 assay endpoints), Endocrine Disruption Screening Program in the twenty-first century Dashboard, Chemical Product Category (CPCat), and exposure databases for personal care products.
Other significant toxicological data resources include the Gene-Tox database from the National Institutes of Health (NIH) U.S. National Library of Medicine (NLM) which comprises mutagenicity data for more than 3,000 chemicals. The RepDose and FeDTex databases (Bitsch et al., 2008) are useful sources for No Observed (Adverse) Effect Level (NO(A)EL) or Lowest Observed (Adverse) Effect Level (LO(A)EL values from repeated dose studies for reproductive and developmental toxicity endpoints. RepDose consists of data on >400 chemicals investigated in 1,018 studies resulting in 6,002 specific effects. The HESS database (Sakuratani et al., 2013) contains information on 28 day repeat dose toxicity studies for 289 industrial chemicals in rats and includes additional rat metabolism datasets and information on ADME in rats and humans.
The list of databases also includes toxicogenomic related data sources such as the Comparative Toxicogenomics Database (CTD) (Davis et al., 2011), Open-TG Gates (Igarashi et al., 2015), The Data Infrastructure for Chemical Safety (diXa) (Hendrickx et al., 2015), Toxygates (Nyström-Persson et al., 2013; Natsume-Kitatani et al., 2017) etc. Open-TG-GATES stores gene expression profiles and traditional toxicological data derived from in vivo (rat) and in vitro (primary rat hepatocytes, primary human hepatocytes) exposure to 170 compounds at multiple dosages and time points. Toxygates is the new interactive version of data from Open-TG-GATES covering 24,011 samples and 170 compounds. The diXa database provides a one stop resource for toxicogenomics studies with cross-links to chemical and molecular medicine databases. diXa contains data from 34 studies involving 469 compounds and recently, all the data have been migrated to the BioStudies (EMBL-EBI) platform (Sarkans et al., 2018). BioStudies contains biological data or models and links them to external resources. At the time of writing it contains 2,552,605 files, 2,824,923 links, four projects, and 1,214,176 studies. CTD is another valuable resource which includes more than 30.5 million toxicogenomic connections relating chemicals/drugs, genes/proteins, diseases, taxa, Gene Ontology (GO) annotations, pathways, and gene interaction modules.
ChemTunes & ToxGPS consists of in vitro and in vivo toxicity endpoint specific alerting chemotypes; mechanism of action (MOA) based QSAR models, weight of evidence (WoE) outcomes, and ToxGPS datasets. Other organ specific toxicity databases inlcude: AMED cardiotoxicity (Sato et al., 2018), LiverTox (Hoofnagle et al., 2013), Liver Toxicity Knowledge Base (LTKB) (Chen et al., 2011), the National Center for Toxicological Research liver cancer database (NCTRIcdb) (Beger et al., 2004) etc. The AMED cardiotoxicity database contains data on small molecules that bind to various ion channels and potentially cause cardiotoxic risk. The data on bioactivities for hERG potassium channel were collected from ChEMBL, the NIH Chemical Genomics Center and hERGCentral. They consist of 9,259 hERG inhibitors (IC50 ≤ 10 μM) and 279,718 inactive compounds (IC50 > 10 μM). LiverTox is comprehensive resource on drug induced liver injury caused by prescription and non-prescription drugs, herbals and dietary supplements (1000's of DILI agents). LTKB has been developed by the US FDA's National Center for Toxicological Research to study drug-induced liver injury (DILI). The data are related to DILI mechanisms, drug metabolism, histopathology, therapeutic use, targets, side effects, biomarkers etc. DILIrank consists of 1,036 FDA-approved drugs that are divided into four classes, Most-DILI-concern drug (192 drugs); Less-DILI-concern drug (278 drugs), No-DILI-concern drug (312 drugs), and Ambiguous-DILI-concern drug (254 drugs). NCTRIcdb contains 999 chemicals classified as 273 liver carcinogen, 293 other carcinogen, and 304 as non-carcinogen based on studies of male and female mice and rats.
The eTOX database was developed by the European eTOX project, which was a consortium of 13 pharmaceuticals, data curators, modellers and software developers funded by the EU Innovative Medicines Initiative (IMI) Joint Undertaking for 7 years (Steger-Hartmann et al., 2009; Cases et al., 2014; Sanz et al., 2017). The database provides access to data on repeated dose toxicity and organ specific toxicity studies and contains models such as like Human outcomes module, Ontobrowser, eTox Lab, and Limtox. It covers data on 1,947 pharmaceuticals out of which 483 labelled as confidential. COSMOS (Cronin et al., 2012) was another EU funded project which aimed to develop in silico models for the prediction of human repeated dose toxicity of cosmetic ingredients to optimise safety without the use of animals by using computational models. The tools and approaches includes application of Thresholds of Toxicological Concern (TTC) of cosmetics related substances. The database includes more than 80,000 chemical records with more than 40,000 unique structures, 12,000 toxicity studies across 27 endpoints for more than 1,600 compounds.
III. ADME Databases
Thirty-eight ADME databases were identified which captured information on parameters such as area-under-the-plasma concentration-curve (AUC), maximum concentration (Cmax), Time to reach maximum concentration (Tmax), half-life (T1/2), volume of distribution (Vd), clearance (CL) etc13. These data were determined from in vitro and in vivo ADME studies involving different species (mouse, rats, dogs, monkeys) as well as humans in clinical studies. Pharmapendium (https://pharmapendium.com) is one of the most widely used commercial database (Elsevier group) in the pharmaceutical industry. The database contains information on 4,331 drugs indexed and fully searchable for more than 1.53 million PK data, 295,000 metabolising enzyme and transporters data, 1.57 million safety data, 1.69 million efficacy data and 115,000 activity data extracted from FDA/ EMA drug approval documents.
A number of databases are particularly useful for the retrieval of information on metabolites e.g., XmetDB (Spjuth et al., 2016), Metrabase (Mak et al., 2015), and Akos. XMetDB is an open resource for drugs, xenobiotics and their experimental metabolite data. It contains 162 observations from 21 scientific papers from 14 journals, covering 117 chemical structures and 95 enzymes. Akos Metabolites (Accelrys) is a restricted source containing experimental data from in vivo and in vitro studies for about 20,000 parent compounds, 100,000 transformations, and 50,000 molecules. It has indexed relevant metabolic paths for structurally related systems.
Other influential databases for PK properties include the ADME database14 which is a proprietary database from Fujitsu Kyushu Systems (Japan). This provides the latest and most comprehensive data on interactions of substances with drug metabolising enzymes and drug transporters that are specific to humans. It contains enzyme kinetic values (Km, Vmax, Ki, Kinactivation, IC50, EC50, T1/2) obtained from the literature. A recent count shows 35,776 substrates, 28,996 inhibitors, 546 activators as well as 12, 617 data for inducers of CYP450 enzymes. For other enzymes 8,608 substrates, 6,109 inhibitors, 229 activators, and 2,220 inducers were included and in the case of transporters 14,749 substrates, 6,109 inhibitors, 229 activators, and 2,220 inducers were included. The University of Washington's licensed databases such as Drug-Interaction DB (DIDB), e-PK gene, and Organ induced-DDI (OI-DDI) (Hachad et al., 2010, 2011; Yeung et al., 2015) are also very useful. DIDB has the largest manually curated collection of in vitro and in vivo data related to drug interactions in humans. Additionally, it covers pharmacokinetic profiles of drugs, QT (i.e., the time between the start of the Q wave and the end of the T wave in heart's electrical cycle) prolongation data, including results of Thorough QT (TQT) from recent New Drug Application's (NDAs) clinical trials data. e-PK gene, is based on pharmacogenomics i.e., providing in-depth analysis of the impact of genetic variants of enzymes and transporters on pharmacokinetic responses to drugs and metabolites.
OI-DDI contains information on pharmacokinetic drug exposure data for 271 compounds from publicly available renal and hepatic impairment studies presented together with the maximum change in drug exposure from drug interaction inhibition studies. Databases on transporters include TP-Search (Ozawa et al., 2004) and UCSF-Trans Portal (Morrissey et al., 2012). TP-Search enables the user to search the membrane transporters-related information by substrate/inhibitor/inducers structure or name, gene expression, functions, drug-drug interactions involving transporters (Km/Ki) etc. It coveres more than 75-membrane transporters across different tissues in mice, rats and humans. The University of California, San Francisco's UCSF-Trans Portal provides information on transporter expression, substrates, inhibitors and potential drug-drug interactions. The localisation of transporters in different organs e.g., blood-brain barrier, kidney, liver, placenta, and small intestine is available with images, notes, references, and expression data where available. UCSF Pharmacogenetics (Kroetz et al., 2009) is a restricted knowledge base providing the information on genetic variants in membrane transporters in ethnically diverse populations. The complete list of Solute Carrier Superfamily (SLC) and the ATP Binding Cassette (ABC) Superfamily are also provided within UCSF. The user can also access qPCR expression and MicroArray data. IDAAPM (Legehar et al., 2016) is a very useful and freely accessible computational resource for modelling (using KNIME workflows) to provide information on ADME properties and known adverse effects of FDA approved drugs taken from FAERS database. It contains information of about 19,226 FDA approval applications for 31,815 products, 2,505 active ingredients, 1,629 molecular structures, 2.5 million adverse effects, and 36,963 experimental drug-target bioactivity data. ADMETlab is a recent addition in this category which is useful to predict 31 ADMET endpoints prediction, systematically evaluate PK properties and druglikeness as well as performing chemical similarity searching using different fingerprint methods (Dong et al., 2018).
IV. Drug Discovery Databases
One hundred and fifty-seven databases were identified that relate directly to drug discovery including: small molecule screening, combinatorial chemistry, molecular affinity, binding, docking, enzyme interaction, activity, gene expressions, side-effects, disease, pathways, repurposing of drugs etc15. Some of the pertinent resources on drug information are DrugBank, DrugCentral, SuperDrug and the FDA's Orange Book. DrugBank (Wishart et al., 2008) is one of the most widely used databases that includes information on drugs targets, enzymes and transporters. The version available at time of writing contains 11,874 drug entries including 2,474 approved small molecule drugs, 1,177 approved biotech (protein/peptide) drugs, 129 nutraceuticals, and over 5,748 experimental drugs. Additionally, 5,131 drug target/enzyme/transporter/carrier sequences are linked to these drug entries. DrugCentral (Ursu et al., 2017) is another drug compendium covering 4,531 active ingredients, 3,807 small molecules, 279 biologics, 445 other compounds, 77,484 FDA drug labels, 34,192 prescription only drug labels, 43,292 OTC drug labels, and 97,271 pharmaceutical formulations with FDA drug labels. The web server for Drug Central also aids in finding the drug gene signature profile similarity without linking to other resources. SuperDrug (Goede et al., 2005) contains 4,605 small molecules, 3,993 Biologicals and 612 other drugs, 4,253 ATC codes, 736,562 3D conformers, 223,860 drug products, 3,006 confirmed biological targets, 1,450 predicted biological targets, and 109,698 side effects.
A small number of early drug development resources were also identified; Open Target (Koscielny et al., 2017) is useful for systematic identification and prioritisation of targets. At time of writing, it contains 21,149 targets, 2,920,121 associations, and 10,101 diseases. D3R (Gathiaka et al., 2016; Gaieb et al., 2018) is another resource containing manually curated datasets on validation and improvement of methods in computer-aided drug design. A single dataset comprises 25 or more congeneric compounds, 5–10 co-crystal structures, related affinity data and a number of known inactive compounds. Biovia MDDR database was jointly developed by BIOVIA and Clarivate Analytics. It contains 260,000 biologically relevant compounds and well-defined derivatives. E-LEA3D (Douguet, 2010) is a source for FDA's registered molecular structures −1884 approved between 1939 and 2018 with a molecular weight ≤ 2000. The 1,884 different molecular structures includes structures of enantiomers and of active metabolites. This resource is dedicated to pharmacology (molecular structures, PK, Pharmacodynamics (PD) and registration data.
Databases useful for exploring binding affinity to targets are also included in Table 3. BindingDB (Gilson et al., 2016) is an open resource of measured binding affinities, focusing on the interactions of protein considered to be drug-targets with small, drug like- molecules and it contains 1,454,892 binding data, for 7,082 protein targets, and 652,068 small molecules. AffinDB (Block et al., 2006) is a database of affinity data for structurally resolved protein-ligand complexes from the Protein Data Bank (PDB) and it contains 748 affinity data out of which 474 were covered from PDB. Brenda is a widely used resource for enzymes information including ~3 million data points from 83,000 enzymes, 137,000 literature references, and a total of 206,000 enzyme ligands providing functional and structural data (Placzek et al., 2017).
GPCR-LIgand DAtabase (GLIDA) (Okuno et al., 2006) is a G protein-coupled receptor (GPCR)-related chemical genomic database that is primarily focused on the correlation of information between GPCRs and their ligands. It contains 3,738 GPCR-related entries (links to Entrez Gene, GPCRDB, UniProt, IUPHAR, KEGG) for 649 ligand entries. For docking purposes Computed Ligand Binding Energy (CLIBE) (Chen et al., 2002) and CREDO (Schreyer and Blundell, 2013) are useful. CLIBE is a database developed by National University of Singapore and useful for the analysis of Drug Binding Competitiveness. It contains 67,184 entries for ligand binding energy, in which there are 5,978 distinctive ligands and 2,258 distinctive receptors. In contrast CREDO stores the interactions between all molecules inside macromolecular complexes from the Protein Data Bank (PDB). These molecules include proteins, nucleic acids, carbohydrates as well as small molecules. CREDO has implemented 13, different interaction types such as hydrogen bonds, halogen bonds, carbonyl interactions, and others. Other resources noted are Target Central Resource Database (TCRD), Pharos (Nguyen et al., 2017), CARLSBAD (Mathias et al., 2013), GPCR-Ligand Association Database (GLASS), and GPCR-EXP (Dai et al., 2016). TCRD and Pharos were both developed by the Illuminating the Druggable Genome (IDG) program which aimed for collecting and organising information about the most common protein targets from four families -GPCRs, kinases, ion channels, and nuclear receptors. TCRD collates many heterogenous gene/protein datasets and Pharos is a multi-modal web interface that represents the data from TCRD. Overall, the database covers 72 million associations between all mammalian genes and their attributes collected from 66 open online major resources. CARLSBAD is an aggregator of many bioactivity databases such as ChEMBL, IUPHARDb, Psychoactive Drug Screening program (PDSP) Ki, PubChem, and WOMBAT. It provides a single normalised bioactivity value (Ki, EC50 etc.) for chemical-protein target pair. It includes data for 9,32,852 activities, 8,90,323 unique structure-target pairs, 3,739 targets, and 1,301 diseases. The GLASS database is manually curated repository for experimentally-validated GPCR-ligand interactions. It contains 3,056 GPCR entries and 335,271 ligand entries. Whereas, GPCR-EXP provides information on experimental and predicted structures of GPCR. It covers data for 55 GPCRs, 282 structures across 9 species (from Protein Data Bank), and 1,076 predicted structures in human genome. Possum (Ito et al., 2012) is another standalone database for pocket similarity searching for predicted and experimentally-derived ligand binding sites covering 5,513,691 known and putative binding sites obtained from Protein Data Bank.
Allostery is a process of regulation of biological macromolecule (protein) function induced by binding of a ligand (small molecule) at an allosteric site (i.e., a distinct site other than the active site) in an efficient way to control the metabolic mechanisms or signal-transduction pathways and subsequently increasing the high receptor selectivity and lowering the target-based toxicity. This concept helped to build the Allosteric (Shen et al., 2016) database which has compiled 1,788 allosteric target entries, 77,825 allosteric modulator entries, 82,431 interactions, 1,930 allosteric sites, 56 allosteric pathways, 261 allosteric networks, and 3,350 allosteric related diseases.
For pain research, there are various databases including SuperPain (Gohlke et al., 2014) and Pain Genes database (PGDB) (Lacroix-Fralish et al., 2007). SuperPain is a database specifically relating to pain-stimulating and pain-relieving compounds, which bind or potentially bind or block to ion channels, e.g., those belonging to the family of Transient Receptor Potential (TRP) channels (TRPV1, TRPM8, TRPA1), human ether-a-go-go related gene (hERG), TREK1, P2X, Acid-sensing ion channels (ASIC) or voltage-gated sodium channels. It contains data on 8,700 ligands (experimentally identified) and 100,000 putative ligands. PGDB provides analysis of the published pain-related phenotypes of mutant mice (over 200 different mutants) and covers 430 genes associated with the pain mechanism.
Oncology is another domain where large datasets have been produced and compiled in databases such as Cancer Target Discovery and Development (CTD2) (Aksoy et al., 2017), CancerResource (Gohlke et al., 2016), canSAR (Tym et al., 2016), CancerDRD (Kumar et al., 2013), Genomics of Drug Sensitivity in Cancer (GDSC) database (Yang et al., 2013) etc. CTD2 is very useful platform for translation of high-throughput and high-content genomic and small molecule data for oncology. It contains 10,828 cancer cell-line sensitivity profiling data, 18 oncogenomic screening observations, 156 chemical-genetic interaction mappings, 66 drug-sensitivity screening results, 24 observations based on reverse phase protein arrays. Whereas, CancerResource focuses on cancer related drug-target interactions, expression, and mutation data as well as drug sensitivity data. It covers data on 48,404 compounds, 3,387 cancer-relevant protein targets, 90,744 compound-target interactions, 2,037 cell lines, 19,834 genes, 872,658 mutations, 23,016 genes (expression). CanSAR is another translational platform, which integrates genomic, protein, pharmacological, drug, and chemical data with structural biology, protein networks, and druggability data. CanSAR contains unique data on 20,316 proteins in human, 556,825 in all species, 143,698 3D structures, 400,892 chains, 12,172 cell lines and 1,962,718 unique structures, 2,148 organisms, 6,367,677 data points from 59,618 studies, and 244,099 clinical trials. Whereas, CancerDRD is a database of 148 anticancer drugs and their effectiveness against around 1,000 cancer cell lines. GDSCdb is a partially open resource containing information on drug sensitivity in cancer cells and molecular markers of drug response. It contains data on 75,000 experiments testing response to 138 anticancer drugs across almost 700 cancer cell lines.
With the advent of high—throughput in-vitro technologies to systematically investigate new indications for existing drugs has led to the development of repurposing databases for personalized medicines such as RepurposeDB (Shameer et al., 2018), repoDB (Brown and Patel, 2017), The Drug Repurposing Hub (DRH) (Corsello et al., 2017), Promiscuous (Von Eichborn et al., 2011) etc. RepurposeDB contains information on 253 drugs (74.30%) and protein drugs (25.29%) and 1,125 diseases. RepoDB another repositioning database containing information on 1,519 approved drugs, 386 terminated, 199 withdrawn, and 77 suspended drugs. DRH contains data on 10,147 compound samples, 2,247 protein targets, 6,125 unique compounds, and 663 drug indications. Promiscuous connects entities such as drugs, proteins and side effects as well as mapping the relationships between them using a network visualisation approach. To date, data are available for 10,208,308 proteins, 25,170 drugs and drug like compounds, and 23,702 drug-target interactions.
V. Drug Information, Clinical Trials, and Pharmacovigilance Databases
One hundred and sixteen up-to-date resources on drug information (medication content, packaging inserts dosing), drug safety (side-effects), clinical trials information, and pharmacovigilance are listed that are relevant for patients, researchers, pharmacists, or prescribers16. Of these DailyMed17 is the official provider of Food and Drug Administration (FDA) label information (package inserts) and has information for 98,961 drug listings as submitted to the FDA. Medicines Complete18 is another resource for authoritative information to support clinical and drug research decisions; it includes publications such as British National Formulary (BNF), BNF for children, Martindale, Stockley's Drug Interactions, Martindale's Adverse Drug reactions, Drug Administration via Enteral Feeding Tubes, AHFS Drug Information, Clarke's Analysis of Drugs and Poisons, Dale and Appelbe's Pharmacy and Medicines Law, Dietary Supplements, Drugs in Pregnancy and Lactation, Handbook on Injectable Drugs, Herbal Medicines, Injectable Drugs Guide, Kucer's the use of Antibiotics, Pediatric Injectable Drugs, Pharmaceutical Excipients, Stockley's Herbal Medicines Interactions, The Green Guide (rules and guidance for the pharmaceutical distributors) and the Orange Guide (rules and guidance for the pharmaceutical Manufacturers and distributors). It covers 600,000 plus pages of evidence-based drug information, 200 plus countries access MedicinesComplete and it has 3.7 million users. MedlinePlus19 is a general public education related resource, developed by The National Library of Medicine (NLM), a part of the US National Institutes of Health. It contains information on diseases, conditions, and wellness issues over 1,000 topics.
There are a few relevant databases related to pharmacovigilance listed in this review such as Side Effect Resource (SIDER) 4.1 (Kuhn et al., 2016), FDA Adverse Event Reporting System (FAERS) (Fang et al., 2014), Vigibase (Lindquist, 2008), and EudraVigilance (Postigo et al., 2018). SIDER 4.1 contains information (extracted from public documents and package inserts) on marketed medicines and their recorded adverse drug reactions; presently it covers data on 1,430 drugs, 5,868 side effects, 139,756 drug-side effects pairs; 39.9% of drug-side effect pairs have corresponding frequency of effect information. Another adverse event related database is FAERS (Fang et al., 2014) which contains adverse event reports, medication error reports, and product quality complaints resulting in adverse events that were submitted to FDA (from Jan 2004 and presently updated quarterly). It contains 14,160,191 total reports, 8,072,400 serious reports (excluding death) and 1,420,885 death reports. Vigibase (Lindquist, 2008) is a unique World Health Organisation (WHO) global database of individual case safety reports (ICSRs), it is linked to medical and drug classifications, including terminologies such as WHO Adverse Reaction Terminology (ART), Medical Dictionary for Regulatory Activities (MedDRA) (Morley, 2014), WHO International Statistical Classification of Diseases (ICD), the medicinal products dictionary, and WHODrug. It holds over 16 million anonymised reports of suspected adverse effects of medicines suffered by patients. EudraVigilance (Postigo et al., 2018) is the European database of Suspected Adverse Reaction reports by the European Medical Agency (EMA). MedDRA provides a single standardised international medical terminology, which can be used for regulatory communication and evaluation of data pertaining to medicinal products for human use. It supports ICH electronic communication within the ICH's Electronic Common Technical Document (eCTD) and the E2B Individual Case Safety Report. There are five levels to the MedDRA hierarchy, at the most specific level it is “Lowest Level Terms” (LLTs), >70,000 terms (e.g., feeling queasy), “Preferred Term” (e.g., nausea), “High Level term” (e.g., nausea and vomiting symptoms), “High Level group term” (GIT signs and symptoms), and “System Organ Class” (e.g., GIT disorders). ICD-1020 contains guidelines for systematic recording, coding, analysis, interpretation, and comparison of mortality-morbidity data collected in different countries. ClinicalTrials.gov (Zarin et al., 2011) is a database of privately and publicly funded clinical studies conducted throughout the world. It is a resource provided by the US NLM and the user can explore 288,732 research studies in all the 50 states and in 205 countries. Information on clinical studies (summary and protocols) are updated by the sponsor or principal investigator of the clinical study. The European Clinical Trials Database21 (EudraCT) is developed and maintained by the European Medicines Agency. The European Union Clinical Trials Register covers 33,526 clinical trials with a EudraCT protocol, of which 5,428 are clinical trials conducted with subjects 18 years old. The register also displays information on 18,700 older (12–18 years) pediatric trials.
Ontology driven databases have also emerged recently, as these can help map other databases, this listing therefore includes resources such as the Medical Subjects Headings (MeSH) (Nelson et al., 2001), National Drug File- Reference Terminology (NDF-RT) (Pathak and Chute, 2010), Unified Medical Language System (UMLS) (Humphreys et al., 1998) and The Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) (Bhattacharyya, 2016). MeSH database provides controlled vocabulary thesaurus for medical subjects and it includes medical terms (headings, subheadings, supplementary concept records, publication types) for indexing articles for PubMed. MeSH contains ~26,000 terms and is updated annually. NDF-RT is developed by the U.S. Department of Veterans Affairs, Veterans Health Administration (VHA). It organises drug list into formal representation. The categories of NDF hierarchical drug classifications are Cellular or Molecular Interactions (MoA), Chemical Ingredients, Clinical Kinetics (PK), Diseases, Manifestations or Physiologic States, Dose Forms, Pharmaceutical Preparations, Physiological Effects, Therapeutic Categories, and VA Drug Interactions. NDF-RT is updated monthly as a part of RxNorm. UMLS is a repository of biomedical vocabularies developed by US NLM. It has three components- The Metathesaurus® of inter-related medical concepts, Semantic networks (high-level categories) and the SPECIALIST Lexicon which “contains syntactic, morphological, and orthographic information for biomedical and common words in the English language”. The UMLS covers 2 milion names for some 900,000 concepts from >60 families of biomedical vocabularies, as well as 12 million relations among these concepts. SNOMED-CT is a database on structured clinical vocabulary for use in an electronic health record. It provides a standardised way to represent clinical phrases captured by the clinician and enables automatic interpretation of these. It is multinational and multilingual, and it contains 340,659 active concepts.
VI. Biological Databases
There are 268 biological databases. This category of databases refers to the resources containing information on enzymes (kinase, GPCRs, CYPs etc.) antibodies, receptors, tissue specific gene expressions/ regulations, annotated protein-peptide sequences, genetic and metabolic signaling, RNA, lipids, immune-system components etc22. Enzyme related information are provided in databases such as Enzyme Portal (Alcántara et al., 2013), GPCRdb (Pándy-Szekeres et al., 2018), International Union of Basic and Clinical Pharmacology- British Pharmacological Society (IUPHAR-BPS) Guide to Pharmacology (Hay et al., 2018), Integrated relational Enzyme database (IntEnz) (Fleischmann et al., 2004), Kinase.com, Kinweb etc. Enzyme Portal is a comprehensive database by EMBL-EBI and it contains information on enzymes, such as small-molecule chemistry, biochemical pathways, and drug compounds. It provides a summary of information from UniProt Knowledgebase, Protein Databank in Europe (PDBe), Rhea (enzyme-catalysed reactions), Reactome (biochemical pathways), IntEnz (enzyme nomenclature information), ChEBI and ChEMBL (small molecule chemistry and bioactivity), MACiE (reaction mechanism), and the Experimental Factor Ontology (EFO). GPCRdb contains data on GPCR structures and large collections of receptor mutants. The database covers data on 15,090 proteins, 418 human proteins, 3,547 species, 270 experimental structures, 184 refined structures, 144,860 ligands, 34,353 mutants, and 12,300 ligand interactions. IUPHAR/BPS Guide to Pharmacology provides data on molecular interactions between target and ligands from selected papers in pharmacology and drug discovery since 2003. It covers 2,880 total number of targets, 9,405 ligands and 1,383 approved drugs with clinical use summary.
For DNA, RNA, and gene datasets, there are number of databases available such as GenBank (Benson et al., 2013), Gene Expression Ontology (GEO) (Barrett et al., 2013), The Genotype-Tissue Expression (GTEx) portal (Stranger et al., 2017), Hugo gene Nomenclature Committee (HGNC) (Gray et al., 2016), Human Protein Atlas (HPA) (Uhlen et al., 2010), Encyclopedia of DNA element (ENCODE) (The ENCODE Project Consortium, 2011), Ensembl, TP-53 (Leroy et al., 2013), European Nucleotide Archive (ENA) (Silvester et al., 2018), European Genome-phenome Archive (EGA) (Lappalainen et al., 2015) etc. GenBank is the NIH genetic sequence database, an annotated collection of all publicly available DNA sequences. It contains publicly available nucleotide sequences for almost 260,000 formally described species. GEO is a portal for the application ontology for the domain of gene expression. Its metrics indicates 166,254 classes, 157,102 individuals, 12 properties and 50,996 maximum number of children. GTEx database is a great resource to study tissue-specific gene expression and regulation. The 11,688 samples were collected from 53 non-diseased tissue sites and 714 donors. HGNC is the worldwide authority that assigns standardised nomenclature to human genes. It approves both a short-form abbreviation known as a gene symbol and a longer and more descriptive name. HPA database consists of three parts; the Tissue Atlas provides the distribution of proteins across all major tissues and organs in the human body, the Cell Atlas provides the subcellular localisation of proteins in single cells and the Pathology Atlas shows the impact of protein levels for survival of patients with cancer. In the latest version, the Human Protein Atlas contains more than 26,000 antibodies, targeting proteins from almost 17,000 human genes.
The ENCODE consortium is supported by the National Human Genome Research Institute (NHGRI) and it has systematically mapped regions of transcription, transcription factor association, chromatin structure, and histone modification. The ENCODE portal contains over 13,000 datasets available through the portal from human, mouse, Drosophila and Caenorhabditis elegans assayed under a variety of different physiological conditions. The Ensembl database provides a bioinformatics framework to organise biology around the sequences of large genomes. Ensembl annotate genes, computes multiple alignments, predicts regulatory function, and collects disease data. TP-53 (tumor protein or cellular tumor antigen p53) is a database related to the structure of the TP53 gene, TP53 isoforms, mutation nomenclature, and the sequence of more than 5,000 tumor samples from 12 cancer types. The current release contains 80,400 tumors, 6,870 different TP53 variants. The ENA (EMBL-EBI) is a comprehensive resource on world's nucleotide sequencing information, raw sequencing data, sequence assembly information and functional annotation. It contains information on 2042.8 millions of nucleotide sequences and 5021.7 billions bases. The EGA contains data on personally identifiable genetic and phenotypic data resulting from biomedical research projects. In the entire EGA there is a total of 1,706 studies (960 cancer, 134 cardiovascular, 39 infectious, 59 inflammatory, 63 neurological, and 362 others).
VII. Protein-Protein Interaction Databases
To understand the relationships between proteins, protein-protein (PP) interaction studies are required and this has led to the creation of many valuable databases to catalog and annotate these PP interactions. Fifty-four databases are listed in Table 323. Some of the most valuable resources are Agile Protein Interactomes Data server (APID) (Alonso-Lopez et al., 2016), The Biological General Repository for Interaction Datasets (BioGRID) (Chatr-Aryamontri et al., 2015), CancerNet (Meng et al., 2015), CompPPI (Veres et al., 2015), The Database of Interacting Proteins (DIP) (Xenarios et al., 2000), Database of Macromolecular Interactions (DOMMINO) (Kuang et al., 2012), gpDB (Theodoropoulou et al., 2008), GWIDD (Kundrotas et al., 2012), Human Interactome Project (HIP) (Rual et al., 2005), Innatedb (Breuer et al., 2013), IntAct (Hermjakob et al., 2004), and Manually Annotated Targets and Drugs Online Resource (MATADOR) (Gunther et al., 2008). Possibly most relevant is APID which is a collection of protein interactomes for more than 400 organisms based in the integration of known experimentally validated protein-protein physical interactions (PPIs). It covers 375,389 interactions and 29,891 interacting proteins. BioGRID contains genetic and protein interactions curated from the primary biomedical literature for all major model organism species and humans. It contains 1,607,037 proteins and genetic interactions, 28,093 chemical associations and 726,378 post translational modifications (PMT) from major model organism species. CancerNet is a human cancer-specific miRNA-target interactions, protein-protein interactions (PPIs) and functionally synergistic miRNA pairs database. It contains interactions across 33 types of human cancers and also PPI information across 33 main normal tissues and cell types. CompPPI (Veres et al., 2015) stands for compartmentalised PPI database, which provides qualitative information on the interactions, proteins and their localizations for PPI network analysis. For human species, it covers 94,488 proteins, 266,306 localisations and 1,311,184 interactions.
The DIP database catalogues experimentally determined interactions between proteins. It combines information from a variety of sources to create a single, consistent set of protein-protein interactions. It contains 28,826 proteins, covering 834 organisms, 81,762 interactions (lists protein pairs that are known to interact with each other), results from 81,913 distinct experiments describing an interaction and 8,233 data sources. DOMMINO (Kuang et al., 2012) is based on macromolecular interactions and at time of writing it covers more than 407,000 binary interactions. The gpDB (Theodoropoulou et al., 2008) is a resource for GPCRs, G-proteins, Effectors (molecules) and their interactions. It contains 391 entries relating to G-proteins, 2,738 GPCRs entries and 1,390 effectors with data for 469 species. GWIDD (Kundrotas et al., 2012) is an integrated resource for structural studies of protein-protein interactions on a genome-wide scale covering 126,897 binary interactions, involving 43,976 proteins from 771 different organisms. HIP (Rual et al., 2005) is an open resource on human protein-protein interactome network and covers 11,999 proteins and 74,820 Interactions. Innatedb (Breuer et al., 2013) is a knowledge resource for innate immunity interactions and pathways and covers 27,172 curated interactions and 9,460 curated genes. IntAct (Hermjakob et al., 2004) is a common curation platform for 11 molecular interaction databases and containing 572,063 interactions and 107,900 interactors. MATADOR (Gunther et al., 2008) is a unique resource for protein-chemical interactions and it covers 775 drugs and their interactions with proteins.
VIII. Omics
This category is related to the resources containing datasets derived from in vitro highthroughput screening (HTS) studies covering all the datasets in metabolomics, proteomics, genomics, transcriptomics, and fluxomics. This category contains 60 databases24. A key example is ArrayExpress (Parkinson et al., 2007) which is an archive of functional genomics data stores data from high-throughput functional genomics experiments and covers 71,472 experiments and 2,311,652 bio assays. It accepts all functional genomics data generated from micoarray or next-generation sequencing (NGS) platforms. The Biochemical Genetic and Genomic (BiGG) database (King et al., 2016) is a resource based on more than 70 genome-scale metabolic networks. Genes in the BiGG models are mapped to NCBI genome annotations, and metabolites are linked to many external databases (KEGG, PubChem, and many more). BioSample (Barrett et al., 2012) contains descriptive information about almost 2 million records (a cell line, a tissue biopsy etc.) encompassing 18,000 species whereas Biostudies is a repository for descriptions of biological studies from large projects (e.g., Blueprint, Europe PubMed Central, Eurocan Platform, and diXa data warehouse) and individuals. Currently it covers 2,552,610 files, 2,824,924 links, 4 projects, and 1,214,179 studies. Cancer GenomicHub is a repository that enables data sharing across cancer genomic studies in support of precision medicine. This database is derived from 40 projects, 61 primary sites, 32,555 cases, 356,381 files, 22,147 genes, and 3,142,246 mutations datasets. The chronic kidney disease database (CKDdb) (Singh et al., 2012) contains multi-omic studies (microRNA, genomics, peptidomics, proteomics, and metabolomics) of chronic kidney disease (CKD), disease-related and diseases leading to this trait. Presently it has differential expression data from 49,395 molecule entries (redundant), of which 16,885 are unique molecules (non-redundant) from 377 manually curated studies of 230 publications. Disnor (Lo surdo et al., 2018), DrugSig (Wu et al., 2017), DisGeNet (Piñero et al., 2017), Drug Gene Interaction Database (DGIdb) (Griffith et al., 2013), Online Mendelian Inheritance in Man (OMIM) (Hamosh et al., 2005) are important resources for exploring the genes and disease domain. Disnor contains information on more than 3,700 disease-pathways and linking ~2,600 disease genes to diseases. Whereas, DrugSig contains information on drug induced gene signature for drug repositioning for more than 1,300 drugs. DisGeNet contains 561,119 gene-disease associations (GDAs), between 17,074 genes and 20,370 diseases, disorders, traits, and clinical or abnormal human phenotypes. DGIdb contains over 40,000 genes and 10,000 drugs involved in over 15,000 drug-gene interactions or belonging to one of 39 potentially druggable gene categories whereas OMIM is a widely used, an Online Catalog of Human Genes and Genetic Disorders and traits. Omics Discovery Index (Omics DI) (Perez-Riverol et al., 2017) is a great resource containing heterogeneous omics data covering 452,800 datasets, 65,200 species, 308,300 genomics, 1,600 tissues, 124,400 transcriptomics, 19 repositories, and 7,300 multiomics datasets. The Human Metabolome Database (HMDB) (Wishart et al., 2017) is a widely used database on small molecule metabolites found in the human body covering 114,110 metabolites entries (both water and lipid soluble) and linked to 5,702 protein sequences. Another useful resource on metabolites is the Metabolomics workbench (Sud et al., 2016), it contains structures and annotations of biologically relevant metabolites (>61,000 entries). MetaMapTox (Fabian et al., 2016) is a licensed resource for metabolite profiles from rat plasma and comprehensive pharmacological and toxicological data. Overall, it covers 25 specific and predictive toxicological mode of action (MoA) in eleven different target organs. RefSeq (Nasko et al., 2018) is a comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, 20,905,608 transcripts, and 100,043,962 proteins, and 73,996 organisms. MetaboLights (Kale et al., 2016) contains cross-species, cross-technique and covers metabolite structures and their reference spectra as well as their biological roles, locations and concentrations, and experimental data from metabolic experiments. PharmacoDB (Smirnov et al., 2018) is a cancer pharmacogenomic database covering 7 datasets, 41 tissues, 1,691 cell lines, 19,933 genes, 759 drugs, and 650,894 experiments. TCGA (Weinstein et al., 2013) is another resource on cancer genome data containing an array of molecular alterations underlying 206 cases of adult soft tissue sarcomas. RGED (Zhang et al., 2014) is a database of gene expression profiles in kidney disease which covers 55 RNA-sequence data, 5,299 DNA microarray, 101 cell lines, and 5,253 tissues. Connectivity Map (Lamb et al., 2006) is a very large genome-scale library of cellular signatures that catalogues transcriptional responses to chemical, genetic, and disease perturbation. It contains more than 1 million profiles resulting from perturbations of multiple cell types.
IX. Pathways-Based Databases
Pathways based toxicity databases are useful in the development of AOPs. This category contains 38 databases25. Important pathway based databases include the AOP-wiki or Wiki Pathways, Effectopedia (Vinken et al., 2017), KEGG (Kanehisa and Goto, 2000), Pathway commons (Cerami et al., 2011), Reactome (Croft et al., 2011), PathCards (Belinky et al., 2015), XTalkDB (Sam et al., 2017) etc. Effectopedia is a part of AOP Knowledge Base, which has four platforms–AOP-Wiki, Effectopedia, AOP Xplorer and Intermediate Effects DB. It contains 244 AOPs and 1,806 key events. KEGG is a vast encyclopedia of genes and genomes and classified into many modules as Pathway, Brite, Module, Orthology, Genome, Genes, Compound, Glycan, Reaction, Enzyme, Disease, Drug etc. Pathway Commons is a collection of publicly available pathway data from multiple organisms covering 4,000 pathways and 1.3M interactions. Reactome is a unique database, which includes transformations of entities such as transport from one compartment to another and interaction to form a complex, as well as the chemical transformations of classical biochemistry. Its latest version includes 2,244 human pathways, 12,047 reactions, 10,778 proteins, and 1,948 small molecules. PathCards is an integrated database of human biological pathways and their annotations. In addition, the human pathways are clustered into SuperPaths based on gene content similarity. The other DBs such as Genecards (Safran et al., 2010), MalaCards (Rappaport et al., 2014), and GeneLoc (Rosen et al., 2003) could be useful along with PathCard database. Overall, it contains 1,289 SuperPath entries, consolidated from 12 sources. XTalkDB is based on scientific literature supporting crosstalk between pairs of signaling pathways and presently contains 650 curated pathway pairs, 345 crosstalking pathway pairs, 1697-curated publications.
X. Patent Databases
Nine databases were identified relating to patents26; these are of paramount importance in drug discovery, drug formulations, production, and marketing of new molecules (Heifets and Jurisica, 2012; Papadatos et al., 2016). In addition, databases which contained information on chemical structure, synthesis, in vitro or in vivo mode of actions etc. are included. Derwent Discovery27 covers patents from 32 countries and contains 141.3 million backward citations (cited), 148.9 million forward citations (citing), 34.5 million literature citations. Whereas, the European Patent Office (EPO)28 provides free access to over 100 million patent documents. SureChEMBL (Papadatos et al., 2016) contains compounds chemistry extracted from the full text, images and attachments of patent documents from major patent authorities (WIPO, USPTO and EPO). It contains 17 million compounds extracted from 14 million patent documents.
XI. Environmental Databases
There are 30 databases with information on the effects of chemcials to the environment and non-human species29. This category of databases are related to potential, hazardous, or toxic chemicals, which are ubiquitous in nature and are present in air, water, food, soil, dust consumer goods, and are detected in the human body. The Agency for Toxic Substances and Disease Registry (ATSDR) DB (Johnson, 1995) is maintained by the U.S. Department of Health and Health Services, useful in protecting the communities from harmful health effects related to exposure to natural and man-made hazardous substances. It contains A-Z chemical lists of 275 substances and their toxicological profiles. The Hazardous Substances Data Bank (HSDB) (Fonger, 1995) is another toxicology databank from TOXNET and it covers 5,800 records on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements etc. CEDI30 and ADI DB (Sugita et al., 2006) are related to Cumulative Estimated Daily Intakes (CEDIs) and Acceptable Daily Intakes (ADIs) for a large number of food contact substances. The database contains information on over 3,000 substances.
The U.S. EPA's ECOTOX database31 provides information on adverse effects of single chemical stressors to ecologically relevant aquatic and terrestrial species. It contains data relating to 11,655 chemicals, 12,630 species obtained from 48,064 references and 919,123 individual results. The Integrated Risk Information System (IRIS) is another product from U.S. EPA and it contains basic information about the risk assessment for groups of chemicals or complex mixtures. It provides toxicity values for health effects resulting from chronic exposure to more than 500 chemicals: Reference Concentration (RfC), Reference Dose (RfD), Cancer descriptors, Oral slope factor (the slope factor used with administered doses to estimate the probability of increased cancer incidence over a lifetime) etc.
The European Centre for Ecotoxicology and Toxicology of Chemicals- Human Exposure Assessment Tools Database (ECETOC-heatDB) is a public directory of exposure data sources as well as available tools for exposure. Haz-Map® is a relational database of hazardous chemicals and occupational diseases. The database currently contains 12,086 chemical and biological agents, 240 diseases, 121 findings (signs and symptoms), hazard specific to 261 jobs, 243 job tasks, 54 industrial processes, 624 industries, and 27 non-occupational activities. LINCS (Liu et al., 2015) is a standalone library of molecular signatures describing how different types of cells respond to a variety of chemicals called “perbutagens.” At time of writing, this comprised 391 datasets covering 41,847 small molecules, 1,127 cell types derived from different organs, 978 genes, 1469 proteins/155 peptide probes, and 8 antibodies. Another noteworthy resource is the OECD QSAR Toolbox, a tool for grouping of chemicals for read-across that can be applied to data gap filling. It contains 69 profilers (e.g., DNA binding, protein binding, acute aquatic toxicity, carcinogenicity alerts, in vitro and in vivo mutagenicity alerts, keratinocyte gene expression, HESS profiler etc.) and 55 databases [CPDB, DART, Ecotox, RepDose, ToxcastDB, ZEBET, developmental toxicity database (CAESAR), EFSA Food Tox Hazard Database, EFSA Genotoxicity, REACH bioaccumulation data, GARDskin database etc.] with over 70,000 chemicals and 2,116,700 data points.
XII. Animal Alternative Databases
Thirty-nine Animal alternative databases were identified relating to data resources32, which assist researchers in complying with the 3Rs philosophy of reduction, refinement and replacement of animal use by. Examples of these databases include: Bibliography on Alternatives to Animal Testing (ALTBIB) (Liebsch et al., 2011) that provides citations from year 2000 to present year. AnimAlt-ZEBET (Grune et al., 2000) is another unique resource, which includes high quality, scientifically recognised 149 alternative methods to standard animal tests in the field of toxicology and pharmacology as well as fundamental research. Bgee provides information on gene expression patterns in 29 animal species, produced from multiple data types such as RNA-Seq, Affymetrix etc. (Bastian et al., 2019) The EURL ECVAM Database service on Alternative Methods to animal experimentation (DB-ALM)33 developed by the EU Joint Research Centre (JRC) provides evaluated information on development and applications of advanced and alternative methods to animal experimentation in biomedical sciences and toxicology, both in research and for regulatory purposes. The Tracking System for Alternative Methods toward Regulatory Acceptance (TSAR) database34 is useful in identifying alternative non-animal methods that have been proposed for regulatory safety or efficacy testing of chemicals or biological agents such as vaccines. For in vitro related DBs, Cellosaurus35 knowledge resource on cell lines is a good example covering 109,135 cell lines (81,617 human, 19,451 mouse, 1,952 rat).
The Fetal Calf Serum Free database (FCS-Free Db)36 provides a list of FCS-free media available for specific cell lines or cell types. The International Cell Line Authentication Committee (ICLAC)37 provides lists of 4,000 cell lines that are currently known to be cross-contaminated or otherwise misidentified or arising from the work of laboratories and cell line repositories worldwide. LifeMap Discovery®, Cells and Tissues Database (Edgar et al., 2013) covers data in embryonic development, stem cell differentiation, regenerative medicine, and in vivo and in vitro gene expression data curated from scientific literature and HTS data sources. Non-neoplastic Lesion ATLAS (Schmidt, 2014) is a very useful guide for standardising terminology in toxicological pathology for rodents. Another useful database is Organ System Heterogeneity DB, which provides information on the phenotypic heterogeneity of diseases, drugs and mutations in mouse genes on 26 different organ systems defined in using MedDRA ontology at the SOC (System Organ Class) level. The Interspecies database which helps to select the most appropriate animal model, which is essential for efficient extrapolation of animal data to humans or other animals. It provides information on physiological, anatomical, and biochemical parameters across species.
XIII. Nanomaterial Toxicity Databases
22 databases exist that contain information on the properties and toxicity of nanomaterials or their products38. The databases include NANoREG—eNanoMapper database (Jeliazkova et al., 2015), developed by EU FP7 eNanoMapper project and it contains toxicological data for the nanomaterials collected by 85 partners across the world. NHECD (Maimon and Browarnik, 2010) is another free database with its main objectives to study the impact of nanoparticles on health, safety, and environment. It has curated a large and developing collection of published data on environmental and health effects following exposure to nanomaterials. The EU Nano Safety Cluster is an open platform containing the Horizon 2020 projects (e.g., SmartNanoTox, NanoReg II, PATROLS etc.) addressing the safety of materials and technologies enabled by the use of nanoparticles. The Nano- Database39 is developed by DTU Environment, the Danish Ecological Council and Danish Consumer Council. It consists of assessments of the nanomaterials used in various consumer products and NanoRisk Cat categorization. There are nearly 3,036 products in this database. The Nano database40 created by Nature and Springer. The database contains data on nanomaterials, methods of production, and nano-instruments. The data have been curated from articles, patents, and other scientific sources. StatNano41 is another comprehensive database on 7,000 nanotechnology products. It also contains analytical reports on the trend of nanotechnology influence on different industries, nanostructures and nano materials.
With the advances in HTS, chemical synthesis, and biological screening (activity, potency, safety profiles), the number of commercially or publicly available databases containing this information has expanded rapidly. This review has resulted in the compilation of nearly 1,000 databases which have been systematically grouped and classified based on content and potential applications. The databases listed cover many areas including: chemical information, drug screening, toxicity (including toxicity of nanomaterials), ADME, binding, docking, clinical trials, pharmacovigilance, genes, enzymes, interactions, omics, pathways, patent information, environmental exposure, and databases providing information on alternatives to animals.
Criteria for characterising databases were considered as summarised in Table 2. The essential criteria were accessibility, relevance of endpoints, chemical identifiers, acceptable ontology, appropriate (or readily convertible) units, ease of data download in different formats and interoperability. Desirable criteria includes access to metadata (study proptocols and statistics), an assessment of the data quality, ease of use, relevant data description (e.g., classification codes) and currency of data. Ontology based databases were also listed which are useful to integrate the semantic data. For efficient database integration, flexible, and robust APIs are essential to support large datasets. One of the significant bottlenecks in database integration is identifying unique data types to ascertain the overlap between data in two or more databases.
Several factors need to be considered when using the increasing number of data resources for predictive toxicology and other purposes. A very important aspect is the accuracy and uniformity of the identity of the chemical and its chemical structure. Uniform chemical structures are often not included in databases and, on occasions, may even be incorrect. Further complications may arise as different salt forms, enantiomers, or isotopes may not be differentiated. One means to assist in the confirmation of the the identity of chemicals is to include a machine–readable representation of the chemical structure (e.g., SMILES, InChI, SDF), along with the key identifiers, such as InChIkeys. Linked to the need for correct chemical identifiers is the prerequisite for high quality chemical structures to ensure the accuracy, completeness and consistency of the information stored in databases. The process of checking the accuracy, or otherwise, of chemical structures can be undertaken using the InChIs and SMILES representations (amongst others). This helps to avoid incorrect structures by detecting duplicate chemical structures, mismatches between structures, different stereoisomer/tautomer forms, mesomeric effects, hypervalency (atom centre displays valency outside its normal value), and numerous other issues relating to chemical bonds and inorganic elements (Fourches et al., 2010). To standardise and normalise the databases, methods such as chemical structure standardisation (removal of mixtures, inorganics, organometallics, salts, solvents and fragments; normalisation of specific chemotypes/metabolites; treatment of tautomeric forms; removal of duplicates), assigning the unique IDs for the samples, de-duplication of the experimental datasets, validation of omic technologies (Sauer et al., 2017) and avoiding multiple measurements for the same parameters etc. should be applied. One critical step in the standardisation or normalisation procedure is to compile datasets with uniform unit values for a particular parameter derived from heterogonous resources. Uniform units are required to compare and analyse multiple datasets. For example, whilst many data exist for pharmacokinetics properties, there is no or little consistency even in the key parameters, often with discrepancies or differences in units/measured values. However, taking an essential propery such as intrinsic clearance (Clint) measured in vitro for enzymes as an example, the variability in units can be corrected and normalised to mL/min/g of protein.
To check the accuracy, completeness, and consistency of the databases, a number of qualitative and quantitative methods can be used. For accuracy, the quality of the data in the databases, i.e., chemical structures, can be confirmed against CAS numbers by cross-referencing with chemical name. The records for the data generated should be mentioned on the website as well as details and results of the validation along with the statistics (number of compounds covered, version number). Consistency can be checked for the data in different version updates of the database, however, this information is seldom provided. Metadata i.e., a representation of supporting data in different formats, are not systematically implemented in databases. Resolving this issue would assist in mapping datasets to each other and save time in finding a relevant study. It is also clear that many data resources contain many of the same data and sources of information. In addition, many of the data published in journals, books, or dataset compilations have been merged in single platforms (e.g., PubChem) which makes searching easier. As such, this has resulted in a great deal of overlap between existing databases and potentially the propagation of errors (i.e., an error being carried forward across data sources due to a lack of manual checking). However, overlap between the databases is not considered a major problem and it could be minimised by clearly identifying the origin of data and using primary sources where possible. Ontologies help in mapping and data integration by providing the syntax for describing classes (or concepts), properties and relationships between classes in the domain of discourse. Mapping between the schema (organisation of data as a blueprint of how the database is constructed) and domain ontologies are an important component for information integration. Whereas, metadata are data that describe the elements in a dataset (e.g., Name of the table, Type of columns, and relationships between them) and help in importing the dataset files from one resource to another or consolidate multiple databases into a single database. For many databases licensing terms and conditions are provided. Clear license information is crucial so that the end user knows what can be done with the metadata. Open licenses for databases encourage access and download of data in a machine-readable format together with their metadata. Some databases apply the license to the whole database or some third party datasests. The end users are recommended to read and understand the licensing information correctly to know whether the contents are provided for commercial or educational research purposes only. The researchers or data owners are also recommended to publish the metadata under a public domain license to ensure wide distribution and reuse. Interoperable databases, i.e., those with a user-friendly, interface, and the capacity for easy interaction and exchange with the other systems, are favoured. The choice of a particular API depends on the size of the database and the programming skills of the developer. Creating databases with greater interoperability will increase their utility and potential application in diverse areas. In addition, database mapping can be defined as the process of identifying key data sources (web pages, flat files, XML-formatted data, directly accessible DBs) by using methods such as ontologies, programming codes, graphical interfaces, and then linking those relevant databases before merging (integration) into a common platform or a single database. Mapping of different databases depends on the technical content and architecture of the datasets. For better mapping and integration easy access to the metadata are preferred. The content accessibility provided by the different databases vary: the majority of databases are open platform enabling searching of scientific data, some facilitate data downloading in different formats. Access to some databases is restricted e.g., commercially available databases.
Looking to the future and potential use of in silico resources, the integration of databases with in silico tools for predicting the properties or activities of compounds could be useful for early decision making in drug discovery and chemical risk assessment. The computational tools should not be limited to only chemistry or biology but should be able to link chemistry with activity, toxicity and the underlying mechanistic information. In other words, the tools should integrate information on dosimetry, human exposure (in silico) and in vitro toxicity screening data to provide a better chemical safety risk assessment.
An enormous variety of databases relating to in silico toxicology, prediction, and safety assessment are available; other potential uses of these databases include the identification of chemically and/or biologically similar chemicals for read-across purposes. In future development of databases key attributes to consider include data quality, accessibility, ease of downloading the data, chemical space coverage, and range of bioactiviy.
GP, MC, and JM were involved in the writing of manuscript. GP, DE, and JF conducted the literature survey and summarised and compiled the databases.
This research has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 777365 (eTRANSAFE) (https://www.etransafe.eu). This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00561/full#supplementary-material
1. ^https://toxnet.nlm.nih.gov/
3. ^https://fairsharing.org/databases/
5. ^https://en.wikipedia.org/wiki/List_of_biological_databases
6. ^https://www.oxfordjournals.org/nar/database/a/
7. ^http://alttox.org/resource-center/databases/ etc
8. ^References are included in Supplementary Data Sheet S1, section I.
10. ^https://www.emolecules.com/
12. ^References are included in Supplementary Data Sheet S1, section II.
13. ^References are included in Supplementary Data Sheet S1, section III.
14. ^http://www.fujitsu.com/jp/group/kyushu/en/solutions/industry/lifescience/admedatabase/
15. ^References are included in Supplementary Data Sheet S1, section IV.
16. ^References are included in Supplementary Data Sheet S1 section V.
17. ^https://dailymed.nlm.nih.gov/dailymed/
18. ^https://about.medicinescomplete.com/
20. ^https://icd.who.int/browse10/2016/en#/X
21. ^https://eudract.ema.europa.eu/
22. ^References are included in Supplementary Data Sheet S1, section VI.
23. ^References are included in Supplementary Data Sheet S1, section VII.
24. ^References are included in Supplementary Data Sheet S1, section VIII.
25. ^References are included in Supplementary Data Sheet S1, section IX.
26. ^References are included in Supplementary Data Sheet S1, section X.
27. ^https://clarivate.com/products/derwent-world-patents-index/
28. ^https://www.epo.org/index.html
29. ^References are included in Supplementary Data Sheet S1, section XI.
30. ^https://www.fda.gov/Food/IngredientsPackagingLabeling/PackagingFCS/CEDI/default.htm
31. ^https://cfpub.epa.gov/ecotox/
32. ^References are included in Supplementary Data Sheet S1, section XII.
33. ^https://ecvam-dbalm.jrc.ec.europa.eu/
34. ^https://tsar.jrc.ec.europa.eu/
35. ^https://web.expasy.org/cellosaurus/
38. ^References are included in Supplementary Data Sheet S1, section XIII.
Aksoy, B. A., Dancik, V., Smith, K., Mazerik, J. N., Ji, Z., Gross, B., et al. (2017). CTD2 Dashboard: a searchable web interface to connect validated results from the Cancer Target Discovery and Development Network. Database 2017. bax054. doi: 10.1093/database/bax054
Alcántara, R., Onwubiko, J., Cao, H., Matos, P. D., Cham, J. A., Jacobsen, J., et al. (2013). The EBI enzyme portal. Nucleic Acids Res. 41, D773–D780. doi: 10.1093/nar/gks1112
Alexander-Dann, B., Pruteanu, L. L., Oerton, E., Sharma, N., Berindan-Neagoe, I., Módos, D., et al. (2018). Developments in toxicogenomics: understanding and predicting compound-induced toxicity from gene expression data. Mol. Omics 14, 218–236. doi: 10.1039/C8MO00042E
Alonso-Lopez, D., Gutierrez, M. A., Lopes, K. P., Prieto, C., Santamaria, R., and De Las Rivas, J. (2016). APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res. 44, W529–W535. doi: 10.1093/nar/gkw363
Anderle, P., Duval, M., Draghici, S., Kuklin, A., Littlejohn, T. G., Medrano, J. F., et al. (2003). Gene expression databases and data mining. Biotechniques (Suppl.), 36–44. doi: 10.2144/mar03anderle
Ayvaz, S., Horn, J., Hassanzadeh, O., Zhu, Q., Stan, J., Tatonetti, N. P., et al. (2015). Toward a complete dataset of drug–drug interaction information from publicly available sources. J. Biomed. Inform. 55, 206–217. doi: 10.1016/j.jbi.2015.04.006
Barrett, T., Clark, K., Gevorgyan, R., Gorelenkov, V., Gribov, E., Karsch-Mizrachi, I., et al. (2012). BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 40, D57–D63. doi: 10.1093/nar/gkr1163
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995. doi: 10.1093/nar/gks1193
Bastian, F., Parmentier, G., Roux, J., Moretti, S., Laudet, V., and Robinson-Rechavi, M. (2019). Bgee: Integrating and Comparing Heterogeneous Transcriptome Data Among Species. Berlin; Heidelberg: Springer, 124–131.
Bauer-Mehren, A., Furlong, L. I., and Sanz, F. (2009). Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol. Syst. Biol. 5, 290–290. doi: 10.1038/msb.2009.47
Beger, R. D., Young, J. F., and Fang, H. (2004). Discriminant function analyses of liver-specific carcinogens. J. Chem. Inf. Comput. Sci. 44, 1107–1110. doi: 10.1021/ci0342829
Belinky, F., Nativ, N., Stelzer, G., Zimmerman, S., Iny Stein, T., Safran, M., et al. (2015). PathCards: multi-source consolidation of human biological pathways. Database 2015:bav006. doi: 10.1093/database/bav006
Benigni, R., Battistelli, C. L., Bossa, C., Tcheremenskaia, O., and Crettaz, P. (2013). New perspectives in toxicological information management, and the role of ISSTOX databases in assessing chemical mutagenicity and carcinogenicity. Mutagenesis 28, 401–409. doi: 10.1093/mutage/get016
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2013). GenBank. Nucleic Acids Res. 41, D36–D42. doi: 10.1093/nar/gks1195
Bianco, A. M., Marcuzzi, A., Zanin, V., Girardelli, M., Vuch, J., and Crovella, S. (2013). Database tools in genetic diseases research. Genomics 101, 75–85. doi: 10.1016/j.ygeno.2012.11.001
Bitsch, A., Escher, S., Lewin, G., Melber, C., Simetska, N., and Mangelsdorf, I. (2008). RepDose and FeDTex: Two databases focusing on systemic toxicity: first examples from analyses of repeated dose toxicity and reprotoxicity studies. Toxicol. Lett. 180, S45. doi: 10.1016/j.toxlet.2008.06.598
Block, P., Sotriffer, C. A., Dramburg, I., and Klebe, G. (2006). AffinDB: a freely accessible database of affinities for protein–ligand complexes from the PDB. Nucleic Acids Res. 34, D522–D526. doi: 10.1093/nar/gkj039
Bower, D., Cross, K. P., Escher, S., Myatt, G. J., and Quigley, D. P. (2017). CHAPTER 9 In silico toxicology: an overview of toxicity databases, prediction methodologies, and expert review. Comput. Syst. Pharmacol. Toxicol., 209–242. doi: 10.1039/9781782623731-00209
Breuer, K., Foroushani, A. K., Laird, M. R., Chen, C., Sribnaia, A., Lo, R., et al. (2013). InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 41, D1228–D1233. doi: 10.1093/nar/gks1147
Brown, A. S., and Patel, C. J. (2017). A standard database for drug repositioning. Sci. Data 4, 170029. doi: 10.1038/sdata.2017.29
Butkiewicz, M., Wang, Y., Bryant, S. H., Lowe, E. W. Jr., Weaver, D. C., and Meiler, J. (2017). High-throughput screening assay datasets from the pubchem database. Chem Inform 3:1. doi: 10.3390/molecules18010735
Cases, M., Briggs, K., Steger-Hartmann, T., Pognan, F., Marc, P., Kleinöder, T., et al. (2014). The eTOX data-sharing project to advance in silico drug-induced toxicity prediction. Int. J. Mol. Sci. 15, 21136–21154. doi: 10.3390/ijms151121136
Cerami, E. G., Gross, B. E., Demir, E., Rodchenkov, I., Babur, Ö., Anwar, N., et al. (2011). Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–D690. doi: 10.1093/nar/gkq1039
Cha, Y., Erez, T., Reynolds, I. J., Kumar, D., Ross, J., Koytiger, G., et al. (2018). Drug repurposing from the perspective of pharmaceutical companies. Br. J. Pharmacol. 175, 168–180. doi: 10.1111/bph.13798
Chatr-Aryamontri, A., Breitkreutz, B.-J., Oughtred, R., Boucher, L., Heinicke, S., Chen, D., et al. (2015). The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43, D470–D478. doi: 10.1093/nar/gku1204
Chen, B., and Butte, A. (2016). Leveraging big data to transform target selection and drug discovery. Clin. Pharmacol. Ther. 99, 285–297. doi: 10.1002/cpt.318
Chen, J. H., Linstead, E., Swamidass, S. J., Wang, D., and Baldi, P. (2007). ChemDB update—full-text search and virtual chemical space. Bioinformatics 23, 2348–2351. doi: 10.1093/bioinformatics/btm341
Chen, M., Vijay, V., Shi, Q., Liu, Z., Fang, H., and Tong, W. (2011). FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov. Today 16, 697–703. doi: 10.1016/j.drudis.2011.05.007
Chen, X., Ji, Z. L., Zhi, D. G., and Chen, Y. Z. (2002). CLiBE: a database of computed ligand binding energy for ligand–receptor complexes. Comput. Chem. 26, 661–666. doi: 10.1016/S0097-8485(02)00050-5
Chen, X., Yan, C. C., Zhang, X., Zhang, X., Dai, F., Yin, J., et al. (2016). Drug–target interaction prediction: databases, web servers and computational models. Brief. Bioinformatics 17, 696–712. doi: 10.1093/bib/bbv066
Cheng, T., Hao, M., Takeda, T., Bryant, S. H., and Wang, Y. (2017). Large-scale prediction of drug-target interaction: a data-centric review. AAPS J. 19, 1264–1275. doi: 10.1208/s12248-017-0092-6
Corsello, S. M., Bittker, J. A., Liu, Z., Gould, J., Mccarren, P., Hirschman, J. E., et al. (2017). The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 23, 405. doi: 10.1038/nm.4306
Croft, D., O'kelly, G., Wu, G., Haw, R., Gillespie, M., Matthews, L., et al. (2011). Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697. doi: 10.1093/nar/gkq1018
Cronin, M. (2005). Toxicological Information for Use in Predictive Modeling: Quality, Sources and Databases. Boca Raton, FL: Taylor and Francis.
Cronin, M. T. D. (2010). Chapter 3 Finding the Data to Develop and Evaluate (Q)SARs and Populate Categories for Toxicity Prediction. Cambridge: The Royal Society of Chemistry.
Cronin, M. T. D., Bajot, F., Enoch, S. J., Madden, J. C., Roberts, D. W., and Schwöbel, J. (2009). The in chemico-in silico interface: challenges for integrating experimental and computational chemistry to identify toxicity. Altern. Lab. Anim. 37, 513–521. doi: 10.1177/026119290903700508
Cronin, M. T. D., Madden, J. C., and Richarz, A.-N. (2012). The COSMOS Project: A Foundation for the Future of Computational Modelling of Repeat Dose Toxicity. AltTox.org.
Dai, S.-X., Li, G.-H., Gao, Y.-D., and Huang, J.-F. (2016). Pharmacophore-Map-Pick: a method to generate pharmacophore models for all human GPCRs. Mol. Inform. 35, 81–91. doi: 10.1002/minf.201500075
Davis, A. P., King, B. L., Mockus, S., Murphy, C. G., Saraceni-Richards, C., Rosenstein, M., et al. (2011). The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 39, D1067–1072. doi: 10.1093/nar/gkq813
Dong, J., Wang, N.-N., Yao, Z.-J., Zhang, L., Cheng, Y., Ouyang, D., et al. (2018). ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 10, 29–29. doi: 10.1186/s13321-018-0283-x
Douguet, D. (2010). e-LEA3D: a computational-aided drug design web server. Nucleic Acids Res. 38, W615–621. doi: 10.1093/nar/gkq322
Edgar, R., Mazor, Y., Rinon, A., Blumenthal, J., Golan, Y., Buzhor, E., et al. (2013). LifeMap Discovery™: the embryonic development, stem cells, and regenerative medicine research portal. PLoS ONE 8:e66629. doi: 10.1371/journal.pone.0066629
Ekins, S., Freundlich, J. S., Choi, I., Sarker, M., and Talcott, C. (2011). Computational databases, pathway and cheminformatics tools for tuberculosis drug discovery. Trends Microbiol. 19, 65–74. doi: 10.1016/j.tim.2010.10.005
Ekins, S., Nikolsky, Y., and Nikolskaya, T. (2005). Techniques: application of systems biology to absorption, distribution, metabolism, excretion and toxicity. Trends Pharmacol. Sci. 26, 202–209. doi: 10.1016/j.tips.2005.02.006
Ekins, S., and Williams, A. J. (2010). Precompetitive preclinical ADME/Tox data: set it free on the web to facilitate computational model building and assist drug development. Lab Chip 10, 13–22. doi: 10.1039/B917760B
Embry, M. R., Bachman, A. N., Bell, D. R., Boobis, A. R., Cohen, S. M., Dellarco, M., et al. (2014). Risk assessment in the 21st century: roadmap and matrix. Crit. Rev. Toxicol. 44, 6–16. doi: 10.3109/10408444.2014.931924
Fabian, E., Bordag, N., Herold, M., Kamp, H., Krennrich, G., Looser, R., et al. (2016). Metabolite profiles of rats in repeated dose toxicological studies after oral and inhalative exposure. Toxicol. Lett. 255, 11–23. doi: 10.1016/j.toxlet.2016.05.003
Fang, H., Su, Z., Wang, Y., Miller, A., Liu, Z., Howard, P., et al. (2014). Exploring the FDA Adverse Event Reporting System (FAERS) to generate hypotheses for disease monitoring. Clin. Pharmacol. Ther. 95, 496–498. doi: 10.1038/clpt.2014.17
Fleischmann, A., Darsow, M., Degtyarenko, K., Fleischmann, W., Boyce, S., Axelsen, K. B., et al. (2004). IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 32, D434–D437. doi: 10.1093/nar/gkh119
Fonger, G. C. (1995). Hazardous substances data bank (HSDB) as a source of environmental fate information on chemicals. Toxicology 103, 137–145. doi: 10.1016/0300-483X(95)03145-6
Fostel, J., Van Someren, E., Pronk, T., Pennings, J., Schmeits, P., Shao, J., et al. (2014). Chapter 6.2 - Toxicogenomics and Systems Toxicology Databases and Resources: Chemical Effects in Biological Systems (CEBS) and Data Integration by Applying Models on Design and Safety (DIAMONDS). San Diego, CA: Academic Press.
Fotis, C., Antoranz, A., Hatziavramidis, D., Sakellaropoulos, T., and Alexopoulos, L. G. (2018). Network-based technologies for early drug discovery. Drug Discov. Today 23, 626–635. doi: 10.1016/j.drudis.2017.12.001
Fourches, D., Muratov, E., and Tropsha, A. (2010). Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 50, 1189–1204. doi: 10.1021/ci100176x
Fouretier, A., Malriq, A., and Bertram, D. J. P. M. (2016). Open access pharmacovigilance databases: analysis of 11 databases. Pharm. Med. 30, 221–231. doi: 10.1007/s40290-016-0146-6
Gaieb, Z., Liu, S., Gathiaka, S., Chiu, M., Yang, H., Shao, C., et al. (2018). D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. J. Comput. Aided Mol. Des. 32, 1–20. doi: 10.1007/s10822-017-0088-4
Gathiaka, S., Liu, S., Chiu, M., Yang, H., Stuckey, J. A., Kang, Y. N., et al. (2016). D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J. Comput. Aided Mol. Des. 30, 651–668. doi: 10.1007/s10822-016-9946-8
Gaulton, A., Hersey, A., Nowotka, M., Bento, A. P., Chambers, J., Mendez, D., et al. (2017). The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954. doi: 10.1093/nar/gkw1074
Gilson, M. K., Liu, T., Baitaluk, M., Nicola, G., Hwang, L., and Chong, J. (2016). BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–1053. doi: 10.1093/nar/gkv1072
Goede, A., Dunkel, M., Mester, N., Frommel, C., and Preissner, R. (2005). SuperDrug: a conformational drug database. Bioinformatics 21, 1751–1753. doi: 10.1093/bioinformatics/bti295
Gohlke, B. O., Nickel, J., Otto, R., Dunkel, M., and Preissner, R. (2016). CancerResource--updated database of cancer-relevant proteins, mutations and interacting drugs. Nucleic Acids Res. 44, D932–937. doi: 10.1093/nar/gkv1283
Gohlke, B. O., Preissner, R., and Preissner, S. (2014). SuperPain--a resource on pain-relieving compounds targeting ion channels. Nucleic Acids Res. 42, D1107–1112. doi: 10.1093/nar/gkt1176
González-Medina, M., Naveja, J. J., Sánchez-Cruz, N., and Medina-Franco, J. L. (2017). Open chemoinformatic resources to explore the structure, properties and chemical space of molecules. RSC Adv. 7, 54153–54163. doi: 10.1039/C7RA11831G
Gray, K. A., Seal, R. L., Tweedie, S., Wright, M. W., and Bruford, E. A. (2016). A review of the new HGNC gene family resource. Hum. Genomics 10:6. doi: 10.1186/s40246-016-0062-6
Griffith, M., Griffith, O. L., Coffman, A. C., Weible, J. V., Mcmichael, J. F., Spies, N. C., et al. (2013). DGIdb: mining the druggable genome. Nat. Methods 10:1209. doi: 10.1038/nmeth.2689
Grune, B., Herrmann, S., Dorendahl, A., Skolik, S., Behnck-Knoblau, S., Box, R., et al. (2000). [The ZEBET database on alternative methods to animal experiments in the Internet--a concrete contribution to the protection of animals]. ALTEX 17, 127–133.
Gunther, S., Kuhn, M., Dunkel, M., Campillos, M., Senger, C., Petsalaki, E., et al. (2008). SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 36, D919–922. doi: 10.1093/nar/gkm862
Hachad, H., Overby, C. L., Argon, S., Yeung, C. K., Ragueneau-Majlessi, I., and Levy, R. H. (2011). e-PKGene: a knowledge-based research tool for analysing the impact of genetics on drug exposure. Hum. Genomics 5, 506–515. doi: 10.1186/1479-7364-5-5-506
Hachad, H., Ragueneau-Majlessi, I., and Levy, R. H. (2010). A useful tool for drug interaction evaluation: the University of Washington Metabolism and Transport Drug Interaction Database. Hum. Genomics 5, 61–72. doi: 10.1186/1479-7364-5-1-61
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A., and Mckusick, V. A. (2005). Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517. doi: 10.1093/nar/gki033
Hartung, T., Fitzgerald, R. E., Jennings, P., Mirams, G. R., Peitsch, M. C., Rostami-Hodjegan, A., et al. (2017). Systems toxicology: real world applications and opportunities. Chem. Res. Toxicol. 30, 870–882. doi: 10.1021/acs.chemrestox.7b00003
Hay, D. L., Garelja, M. L., Poyner, D. R., and Walker, C. S. (2018). Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br. J. Pharmacol. 175, 3–17. doi: 10.1111/bph.14075
Heifets, A., and Jurisica, I. (2012). SCRIPDB: a portal for easy access to syntheses, chemicals and reactions in patents. Nucleic Acids Res. 40, D428–D433. doi: 10.1093/nar/gkr919
Hendrickx, D. M., Aerts, H. J. W. L., Caiment, F., Clark, D., Ebbels, T. M. D., Evelo, C. T., et al. (2015). diXa: a data infrastructure for chemical safety assessment. Bioinformatics 31, 1505–1507. doi: 10.1093/bioinformatics/btu827
Hermjakob, H., Montecchi-Palazzi, L., Lewington, C., Mudali, S., Kerrien, S., Orchard, S., et al. (2004). IntAct: an open source molecular interaction database. Nucleic Acids Res. 32, D452–D455. doi: 10.1093/nar/gkh052
Hersey, A., Chambers, J., Bellis, L., Patrícia Bento, A., Gaulton, A., and Overington, J. P. (2015). Chemical databases: curation or integration by user-defined equivalence? Drug Discov. Today Technol. 14, 17–24. doi: 10.1016/j.ddtec.2015.01.005
Hoofnagle, J. H., Serrano, J., Knoben, J. E., and Navarro, V. J. (2013). LiverTox: a website on drug-induced liver injury. Hepatology 57, 873–874. doi: 10.1002/hep.26175
Humphreys, B. L., Lindberg, D. A., Schoolman, H. M., and Barnett, G. O. (1998). The Unified Medical Language System: an informatics research collaboration. J. Am. Med. Inform. Assoc. 5, 1–11. doi: 10.1136/jamia.1998.0050001
Igarashi, Y., Nakatsu, N., Yamashita, T., Ono, A., Ohno, Y., Urushidani, T., et al. (2015). Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 43, D921–927. doi: 10.1093/nar/gku955
Ito, J.-I., Tabei, Y., Shimizu, K., Tsuda, K., and Tomii, K. (2012). PoSSuM: a database of similar protein-ligand binding and putative pockets. Nucleic Acids Res. 40, D541–D548. doi: 10.1093/nar/gkr1130
Jeliazkova, N., Chomenidis, C., Doganis, P., Fadeel, B., Grafström, R., Hardy, B., et al. (2015). The eNanoMapper database for nanomaterial safety information. Beilstein J. Nanotechnol. 6, 1609–1634. doi: 10.3762/bjnano.6.165
Ji, Z. L., Han, L. Y., Yap, C. W., Sun, L. Z., Chen, X., and Chen, Y. Z. (2003a). Drug Adverse Reaction Target Database (DART). Drug Safety 26, 685–690. doi: 10.2165/00002018-200326100-00002
Ji, Z. L., Sun, L. Z., Chen, X., Zheng, C. J., Yao, L. X., Han, L. Y., et al. (2003b). Internet resources for proteins associated with drug therapeutic effects, adverse reactions and ADME. Drug Discov. Today 8, 526–529. doi: 10.1016/S1359-6446(03)02742-9
Johnson, B. L. (1995). ATSDR's information databases to support human health risk assessment of hazardous substances. Toxicol. Lett. 79, 11–16. doi: 10.1016/0378-4274(95)03351-K
Jonsdottir, S. O., Jorgensen, F. S., and Brunak, S. (2005). Prediction methods and databases within chemoinformatics: emphasis on drugs and drug candidates. Bioinformatics 21, 2145–2160. doi: 10.1093/bioinformatics/bti314
Judson, R. (2010). Public databases supporting computational toxicology. J. Toxicol. Environ. Health B Crit. Rev. 13, 218–231. doi: 10.1080/10937404.2010.483937
Judson, R., Richard, A., Dix, D., Houck, K., Elloumi, F., Martin, M., et al. (2008). ACToR — Aggregated computational toxicology resource. Toxicol. Appl. Pharmacol. 233, 7–13. doi: 10.1016/j.taap.2007.12.037
Kale, N. S., Haug, K., Conesa, P., Jayseelan, K., Moreno, P., Rocca-Serra, P., et al. (2016). MetaboLights: An Open-Access Database Repository for Metabolomics Data. Curr.Protoc. Bioinformatics 53, 14.13.11–14.13.18. doi: 10.1002/0471250953.bi1413s53
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Katsila, T., Spyroulias, G. A., Patrinos, G. P., and Matsoukas, M.-T. (2016). Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 14, 177–184. doi: 10.1016/j.csbj.2016.04.004
King, Z. A., Lu, J., Dräger, A., Miller, P., Federowicz, S., Lerman, J. A., et al. (2016). BiGG Models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522. doi: 10.1093/nar/gkv1049
Kiyosawa, N., Shiwaku, K., Hirode, M., Omura, K., Uehara, T., Shimizu, T., et al. (2006). Utilization of a one-dimensional score for surveying chemical-induced changes in expression levels of multiple biomarker gene sets using a large-scale toxicogenomics database. J. Toxicol. Sci. 31, 433–448. doi: 10.2131/jts.31.433
Klimisch, H. J., Andreae, M., and Tillmann, U. (1997). A systematic approach for evaluating the quality of experimental toxicological and ecotoxicological data. Regul. Toxicol. Pharmacol. 25, 1–5. doi: 10.1006/rtph.1996.1076
Kongsbak, K., Hadrup, N., Audouze, K., and Vinggaard, A. M. (2014). Applicability of computational systems biology in toxicology. Basic Clin. Pharmacol. Toxicol. 115, 45–49. doi: 10.1111/bcpt.12216
Koscielny, G., An, P., Carvalho-Silva, D., Cham, J. A., Fumis, L., Gasparyan, R., et al. (2017). Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–D994. doi: 10.1093/nar/gkw1055
Koutsoukas, A., Simms, B., Kirchmair, J., Bond, P. J., Whitmore, A. V., Zimmer, S., et al. (2011). From in silico target prediction to multi-target drug design: current databases, methods and applications. J. Proteomics 74, 2554–2574. doi: 10.1016/j.jprot.2011.05.011
Kroetz, D. L., Yee, S. W., and Giacomini, K. M. (2009). The pharmacogenomics of membrane transporters project: research at the interface of genomics and transporter pharmacology. Clin. Pharmacol. Ther. 87, 109–116. doi: 10.1038/clpt.2009.226
Kuang, X., Han, J. G., Zhao, N., Pang, B., Shyu, C.-R., and Korkin, D. (2012). DOMMINO: a database of macromolecular interactions. Nucleic Acids Res. 40, D501–D506. doi: 10.1093/nar/gkr1128
Kuhn, M., Letunic, I., Jensen, L. J., and Bork, P. (2016). The SIDER database of drugs and side effects. Nucleic Acids Res. 44, D1075–D1079. doi: 10.1093/nar/gkv1075
Kumar, R., Chaudhary, K., Gupta, S., Singh, H., Kumar, S., Gautam, A., et al. (2013). CancerDR: cancer drug resistance database. Sci. Rep. 3, 1445. doi: 10.1038/srep01445
Kundrotas, P. J., Zhu, Z., and Vakser, I. A. (2012). GWIDD: a comprehensive resource for genome-wide structural modeling of protein-protein interactions. Hum. Genomics 6:7. doi: 10.1186/1479-7364-6-7
Lacroix-Fralish, M. L., Ledoux, J. B., and Mogil, J. S. (2007). The Pain Genes Database: an interactive web browser of pain-related transgenic knockout studies. Pain 131, 3.e1–3.e4. doi: 10.1016/j.pain.2007.04.041
Lamb, J., Crawford, E. D., Peck, D., Modell, J. W., Blat, I. C., Wrobel, M. J., et al. (2006). The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. doi: 10.1126/science.1132939
Lappalainen, I., Almeida-King, J., Kumanduri, V., Senf, A., Spalding, J. D., Ur-Rehman, S., et al. (2015). The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 47, 692–695. doi: 10.1038/ng.3312
Legehar, A., Xhaard, H., and Ghemtio, L. (2016). IDAAPM: integrated database of ADMET and adverse effects of predictive modeling based on FDA approved drug data. J. Cheminform. 8, 33. doi: 10.1186/s13321-016-0141-7
Leroy, B., Fournier, J. L., Ishioka, C., Monti, P., Inga, A., Fronza, G., et al. (2013). The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res. 41, D962–969. doi: 10.1093/nar/gks1033
Liebsch, M., Grune, B., Seiler, A., Butzke, D., Oelgeschläger, M., Pirow, R., et al. (2011). Alternatives to animal testing: current status and future perspectives. Arch. Toxicol. 85, 841–858. doi: 10.1007/s00204-011-0718-x
Lindquist, M. (2008). VigiBase, the WHO Global ICSR database system: basic facts. Drug Inf. J. 42, 409–419. doi: 10.1177/009286150804200501
Liu, C., Su, J., Yang, F., Wei, K., Ma, J., and Zhou, X. (2015). Compound signature detection on LINCS L1000 big data. Mol. Biosyst. 11, 714–722. doi: 10.1039/c4mb00677a
Lo surdo, P., Calderone, A., Iannuccelli, M., Licata, L., Peluso, D., Castagnoli, L., et al. (2018). DISNOR: a disease network open resource. Nucleic Acids Res. 46, D527–D534. doi: 10.1093/nar/gkx876
Loging, W., Rodriguez-Esteban, R., Hill, J., Freeman, T., and Miglietta, J. (2011). Cheminformatic/bioinformatic analysis of large corporate databases: Application to drug repurposing. Drug Discov. Today Ther. Strateg. 8, 109–116. doi: 10.1016/j.ddstr.2011.06.004
Luo, G., Shen, Y., Yang, L., Lu, A., and Xiang, Z. (2017). A review of drug-induced liver injury databases. Arch. Toxicol. 91, 3039–3049. doi: 10.1007/s00204-017-2024-8
Madden, J. C. (2013). Chapter 5 Sources of Chemical Information, Toxicity Data and Assessment of Their Quality. Cambridge: The Royal Society of Chemistry.
Madden, J. C., Pawar, G., Cronin, M. T. D., Webb, S., Tan, Y.-M., and Paini, A. (2019). In silico resources to assist in the development and evaluation of physiologically-based kinetic models. Comput. Toxicol. 11, 33–49. doi: 10.1016/j.comtox.2019.03.001
Maimon, O., and Browarnik, A. (2010). NHECD - Nano Health and Environmental Commented Database. Boston, MA: Springer.
Mak, L., Marcus, D., Howlett, A., Yarova, G., Duchateau, G., Klaffke, W., et al. (2015). Metrabase: a cheminformatics and bioinformatics database for small molecule transporter data analysis and (Q)SAR modeling. J. Cheminform. 7, 31. doi: 10.1186/s13321-015-0083-5
Mathias, S. L., Hines-Kay, J., Yang, J. J., Zahoransky-Kohalmi, G., Bologa, C. G., Ursu, O., et al. (2013). The CARLSBAD database: a confederated database of chemical bioactivities. Database 2013:bat044. doi: 10.1093/database/bat044
Meng, X., Wang, J., Yuan, C., Li, X., Zhou, Y., Hofestädt, R., et al. (2015). CancerNet: a database for decoding multilevel molecular interactions across diverse cancer types. Oncogenesis 4, e177. doi: 10.1038/oncsis.2015.40
Morley, G. (2014). Adverse event reporting: a brief overview of MedDRA. Medical Writing 23, 113–116. doi: 10.1179/2047480614Z.000000000208
Morrissey, K. M., Wen, C. C., Johns, S. J., Zhang, L., Huang, S. M., and Giacomini, K. M. (2012). The UCSF-FDA transportal: a public drug transporter database. Clin. Pharmacol. Ther. 92, 545–546. doi: 10.1038/clpt.2012.44
Nasko, D. J., Koren, S., Phillippy, A. M., and Treangen, T. J. (2018). RefSeq database growth influences the accuracy of k-mer-based species identification. Genome Biology. 19:165. doi: 10.1101/304972
Natsume-Kitatani, Y., Nyström-Persson, J., Igarashi, Y., Satoh, D., and Mizuguchi, K. (2017). Integrated toxicogenomics analysis with Toxygates for inferring molecular mechanisms. Genomics Comput. Biol. 3, e37. doi: 10.18547/gcb.2017.vol3.iss1.e37
Nelson, S. J., Johnston, W. D., and Humphreys, B. L. (2001). Relationships in Medical Subject Headings (MeSH). Dordrecht: Springer.
Nguyen, D.-T., Mathias, S., Bologa, C., Brunak, S., Fernandez, N., Gaulton, A., et al. (2017). Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 45, D995–D1002. doi: 10.1093/nar/gkw1072
Nicola, G., Liu, T., and Gilson, M. K. (2012). Public domain databases for medicinal chemistry. J. Med. Chem. 55, 6987–7002. doi: 10.1021/jm300501t
Nyström-Persson, J., Igarashi, Y., Ito, M., Morita, M., Nakatsu, N., Yamada, H., et al. (2013). Toxygates: interactive toxicity analysis on a hybrid microarray and linked data platform. Bioinformatics 29, 3080–3086. doi: 10.1093/bioinformatics/btt531
Okuno, Y., Yang, J., Taneishi, K., Yabuuchi, H., and Tsujimoto, G. (2006). GLIDA: GPCR-ligand database for chemical genomic drug discovery. Nucleic Acids Res. 34, D673–D677. doi: 10.1093/nar/gkj028
Opassi, G., Ges,ù, A., and Massarotti, A. (2018). The hitchhiker's guide to the chemical-biological galaxy. Drug Discov. Today 23, 565–574. doi: 10.1016/j.drudis.2018.01.007
Oprea, T. I., and Tropsha, A. (2006). Target, chemical and bioactivity databases – integration is key. Drug Discov. Today Technol. 3, 357–365. doi: 10.1016/j.ddtec.2006.12.003
Ozawa, N., Shimizu, T., Morita, R., Yokono, Y., Ochiai, T., Munesada, K., et al. (2004). Transporter database, TP-Search: a web-accessible comprehensive database for research in pharmacokinetics of drugs. Pharm. Res. 21, 2133–2134. doi: 10.1023/B:PHAM.0000048207.11160.d0
Pándy-Szekeres, G., Munk, C., Tsonkov, T. M., Mordalski, S., Harpsøe, K., Hauser, A. S., et al. (2018). GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446. doi: 10.1093/nar/gkx1109
Papadatos, G., Davies, M., Dedman, N., Chambers, J., Gaulton, A., Siddle, J., et al. (2016). SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–D1228. doi: 10.1093/nar/gkv1253
Papadopoulos, T., Krochmal, M., Cisek, K., Fernandes, M., Husi, H., Stevens, R., et al. (2016). Omics databases on kidney disease: where they can be found and how to benefit from them. Clin. Kidney J. 9, 343–352. doi: 10.1093/ckj/sfv155
Parkinson, H., Kapushesky, M., Shojatalab, M., Abeygunawardena, N., Coulson, R., Farne, A., et al. (2007). ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 35, D747–D750. doi: 10.1093/nar/gkl995
Pathak, J., and Chute, C. G. (2010). Analyzing categorical information in two publicly available drug terminologies: RxNorm and NDF-RT. J. Am. Med. Inform. Assoc. 17, 432–439. doi: 10.1136/jamia.2009.001289
Peach, M. L., Zakharov, A. V., Liu, R., Pugliese, A., Tawa, G., Wallqvist, A., et al. (2012). Computational tools and resources for metabolism-related property predictions. 1. Overview of publicly available (free and commercial) databases and software. Future Med. Chem. 4, 1907–1932. doi: 10.4155/fmc.12.150
Pence, H. E., and Williams, A. (2010). ChemSpider: an online chemical information resource. J. Chem. Educ. 87, 1123–1124. doi: 10.1021/ed100697w
Perez-Riverol, Y., Bai, M., Da Veiga Leprevost, F., Squizzato, S., Park, Y. M., Haug, K., et al. (2017). Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 35, 406. doi: 10.1038/nbt.3790
Piñero, J., Bravo, À., Queralt-Rosinach, N., Gutiérrez-Sacristán, A., Deu-Pons, J., Centeno, E., et al. (2017). DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839. doi: 10.1093/nar/gkw943
Placzek, S., Schomburg, I., Chang, A., Jeske, L., Ulbrich, M., Tillack, J., et al. (2017). BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 45, D380–D388. doi: 10.1093/nar/gkw952
Polen, H. H., Zapantis, A., Clauson, K. A., Jebrock, J., and Paris, M. (2008). Ability of online drug databases to assist in clinical decision-making with infectious disease therapies. BMC Infect. Dis. 8, 153. doi: 10.1186/1471-2334-8-153
Postigo, R., Brosch, S., Slattery, J., Van Haren, A., Dogné, J.-M., Kurz, X., et al. (2018). eudravigilance medicines safety database: publicly accessible data for research and public health protection. Drug Safety 41, 665–675. doi: 10.1007/s40264-018-0647-1
Przybylak, K. R., Madden, J. C., Covey-Crump, E., Gibson, L., Barber, C., Patel, M., et al. (2018). Characterisation of data resources for in silico modelling: benchmark datasets for ADME properties. Expert Opin. Drug Metab. Toxicol. 14, 169–181. doi: 10.1080/17425255.2017.1316449
Rana, B. K., Bourne, P. E., and Insel, P. A. (2012). Receptor Databases and Computational Websites for Ligand Binding. Totowa, NJ: Humana Press.
Rappaport, N., Twik, M., Nativ, N., Stelzer, G., Bahir, I., Stein, T. I., et al. (2014). MalaCards: a comprehensive automatically-mined database of human diseases. Curr. Protoc. Bioinformatics 47, 1.24.21–21.24.19. doi: 10.1002/0471250953.bi0124s47
Rigden, D. J., Fernández-Suárez, X. M., and Galperin, M. Y. (2016). The 2016 database issue of Nucleic Acids Research and an updated molecular biology database collection. Nucleic Acids Res. 44, D1–D6. doi: 10.1093/nar/gkv1356
Rosen, N., Chalifa-Caspi, V., Shmueli, O., Adato, A., Lapidot, M., Stampnitzky, J., et al. (2003). GeneLoc: exon-based integration of human genome maps. Bioinformatics 19, i222–i224. doi: 10.1093/bioinformatics/btg1030
Rual, J. F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., et al. (2005). Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178. doi: 10.1038/nature04209
Ruddigkeit, L., Blum, L. C., and Reymond, J.-L. (2013). Visualization and virtual screening of the chemical universe database GDB-17. J. Chem. Inf. Model. 53, 56–65. doi: 10.1021/ci300535x
Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., et al. (2010). GeneCards Version 3: the human gene integrator. Database 2010:baq020. doi: 10.1093/database/baq020
Sakuratani, Y., Zhang, H. Q., Nishikawa, S., Yamazaki, K., Yamada, T., Yamada, J., et al. (2013). Hazard Evaluation Support System (HESS) for predicting repeated dose toxicity using toxicological categories. SAR QSAR Environ. Res. 24, 351–363. doi: 10.1080/1062936X.2013.773375
Sam, S. A., Teel, J., Tegge, A. N., Bharadwaj, A., and Murali, T. M. (2017). XTalkDB: a database of signaling pathway crosstalk. Nucleic Acids Res. 45, D432–D439. doi: 10.1093/nar/gkw1037
Sanz, F., Pognan, F., Steger-Hartmann, T., Díaz, C., Etox, Cases, M., et al. (2017). Legacy data sharing to improve drug safety assessment: the eTOX project. Nature Reviews Drug Discovery 16, 811. doi: 10.1038/nrd.2017.177
Sarkans, U., Gostev, M., Athar, A., Behrangi, E., Melnichuk, O., Ali, A., et al. (2018). The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 46, D1266–D1270. doi: 10.1093/nar/gkx965
Sato, T., Yuki, H., Ogura, K., and Honma, T. (2018). Construction of an integrated database for hERG blocking small molecules. PLoS ONE 13:e0199348. doi: 10.1371/journal.pone.0199348
Sauer, U. G., Deferme, L., Gribaldo, L., Hackermüller, J., Tralau, T., Van Ravenzwaay, B., et al. (2017). The challenge of the application of 'omics technologies in chemicals risk assessment: background and outlook. Regul. Toxicol. Pharmacol. 91, S14–S26. doi: 10.1016/j.yrtph.2017.09.020
Schmidt, C. W. (2014). NTP nonneoplastic lesion atlas: a new tool for toxicologic pathology. Environ. Health Perspect. 122, A76–A79. doi: 10.1289/ehp.122-A76
Schreyer, A. M., and Blundell, T. L. (2013). CREDO: a structural interactomics database for drug discovery. Database 2013:bat049. doi: 10.1093/database/bat049
Shameer, K., Glicksberg, B. S., Hodos, R., Johnson, K. W., Badgeley, M. A., Readhead, B., et al. (2018). Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Brief. Bioinformatics 19, 656–678. doi: 10.1093/bib/bbw136
Shen, Q., Wang, G., Li, S., Liu, X., Lu, S., Chen, Z., et al. (2016). ASD v3.0: unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 44, D527–D535. doi: 10.1093/nar/gkv902
Silvester, N., Alako, B., Amid, C., Cerdeño-Tarrága, A., Clarke, L., Cleland, I., et al. (2018). The European Nucleotide Archive in 2017. Nucleic Acids Res. 46, D36–D40. doi: 10.1093/nar/gkx1125
Sim, S. C., Altman, R. B., and Ingelman-Sundberg, M. (2011). Databases in the area of pharmacogenetics. Hum. Mutat. 32, 526–531. doi: 10.1002/humu.21454
Singh, S. K., Malik, A., Firoz, A., and Jha, V. (2012). CDKD: a clinical database of kidney diseases. BMC Nephrol. 13:23. doi: 10.1186/1471-2369-13-23
Skuta, C., Popr, M., Muller, T., Jindrich, J., Kahle, M., Sedlak, D., et al. (2017). Probes &Drugs portal: an interactive, open data resource for chemical biology. Nat. Methods 14, 759–760. doi: 10.1038/nmeth.4365
Smalter Hall, A., Shan, Y., Lushington, G., and Visvanathan, M. (2013). An overview of computational life science databases & exchange formats of relevance to chemical biology research. Comb. Chem. High Throughput Screen. 16, 189–198. doi: 10.2174/1386207311316030004
Smirnov, P., Kofia, V., Maru, A., Freeman, M., Ho, C., El-Hachem, N., et al. (2018). PharmacoDB: an integrative database for mining in vitro anticancer drug screening studies. Nucleic Acids Res. 46, D994–D1002. doi: 10.1093/nar/gkx911
Spjuth, O., Rydberg, P., Willighagen, E. L., Evelo, C. T., and Jeliazkova, N. (2016). XMetDB: an open access database for xenobiotic metabolism. J. Cheminform. 8, 47. doi: 10.1186/s13321-016-0161-3
Steger-Hartmann, T., Pognan, F., Sanz, F., and Diaz, C. A. (2009). In silico prediction of in vivo toxicities (eTox)—The Innovative Medicines Initiative Approach. Toxicol. Lett. 189, S258. doi: 10.1016/j.toxlet.2009.06.374
Sterling, T., and Irwin, J. J. (2015). ZINC 15 – Ligand Discovery for Everyone. J. Chem. Inf. Model. 55, 2324–2337. doi: 10.1021/acs.jcim.5b00559
Stranger, B. E., Brigham, L. E., Hasz, R., Hunter, M., Johns, C., et al. (2017). Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat. Genet. 49, 1664. doi: 10.1038/ng.3969
Sud, M., Fahy, E., Cotter, D., Azam, K., Vadivelu, I., Burant, C., et al. (2016). Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470. doi: 10.1093/nar/gkv1042
Sugita, T., Sasaki, S., Tanaka, K., Toda, M., Uneyama, C., Yamamoto, M., et al. (2006). [Development of the databases for ADI (acceptable daily intake) and relevant information on food additives, pesticides and veterinary drugs]. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho. Hokoku, 69–73.
Sushko, I., Novotarskyi, S., Korner, R., Pandey, A. K., Rupp, M., Teetz, W., et al. (2011). Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J. Comput. Aided Mol. Des. 25, 533–554. doi: 10.1007/s10822-011-9440-2
Sushko, I., Salmina, E., Potemkin, V. A., Poda, G., and Tetko, I. V. (2012). ToxAlerts: a Web server of structural alerts for toxic chemicals and compounds with potential adverse reactions. J. Chem. Inf. Model. 52, 2310–2316. doi: 10.1021/ci300245q
The ENCODE Project Consortium (2011). A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9:e1001046. doi: 10.1371/journal.pbio.1001046
Theodoropoulou, M. C., Bagos, P. G., Spyropoulos, I. C., and Hamodrakas, S. J. (2008). gpDB: a database of GPCRs, G-proteins, effectors and their interactions. Bioinformatics 24, 1471–1472. doi: 10.1093/bioinformatics/btn206
Toropov, A. A., Toropova, A. P., Raska, I., Leszczynska, D., and Leszczynski, J. (2014). Comprehension of drug toxicity: software and databases. Comput. Biol. Med. 45, 20–25. doi: 10.1016/j.compbiomed.2013.11.013
Tym, J. E., Mitsopoulos, C., Coker, E. A., Razaz, P., Schierz, A. C., Antolin, A. A., et al. (2016). canSAR: an updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 44, D938–D943. doi: 10.1093/nar/gkv1030
Uhlen, M., Oksvold, P., Fagerberg, L., Lundberg, E., Jonasson, K., Forsberg, M., et al. (2010). Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28:1248. doi: 10.1038/nbt1210-1248
Ursu, O., Holmes, J., Knockel, J., Bologa, C. G., Yang, J. J., Mathias, S. L., et al. (2017). DrugCentral: online drug compendium. Nucleic Acids Res. 45, D932–D939. doi: 10.1093/nar/gkw993
Veres, D. V., Gyurkó, D. M., Thaler, B., Szalay, K. Z., Fazekas, D., Korcsmáros, T., et al. (2015). ComPPI: a cellular compartment-specific database for protein–protein interaction network analysis. Nucleic Acids Res. 43, D485–D493. doi: 10.1093/nar/gku1007
Vinken, M., Knapen, D., Vergauwen, L., Hengstler, J. G., Angrish, M., and Whelan, M. (2017). Adverse outcome pathways: a concise introduction for toxicologists. Arch. Toxicol. 91, 3697–3707. doi: 10.1007/s00204-017-2020-z
Von Eichborn, J., Murgueitio, M. S., Dunkel, M., Koerner, S., Bourne, P. E., and Preissner, R. (2011). PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 39, D1060–1066. doi: 10.1093/nar/gkq1037
Weinstein, J. N., Collisson, E. A., Mills, G. B., Shaw, K. R., Ozenberger, B. A., Ellrott, K., et al. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120. doi: 10.1038/ng.2764
Williams, A. J. (2008). Public chemical compound databases. Curr. Opin. Drug Discov. Dev. 11, 393–404.
Wishart, D. S. (2014). Online Databases and Web Servers for Drug Metabolism Research. New Jersey, NJ: Wiley-VCH.
Wishart, D. S., Feunang, Y. D., Marcu, A., Guo, A. C., Liang, K., Vázquez-Fresno, R., et al. (2017). HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617. doi: 10.1093/nar/gkx1089
Wishart, D. S., Knox, C., Guo, A. C., Cheng, D., Shrivastava, S., Tzur, D., et al. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–906. doi: 10.1093/nar/gkm958
Wooden, B., Goossens, N., Hoshida, Y., and Friedman, S. L. (2017). Using big data to discover diagnostics and therapeutics for gastrointestinal and liver diseases. Gastroenterology 152, 53–67.e53. doi: 10.1053/j.gastro.2016.09.065
Wu, H., Huang, J., Zhong, Y., and Huang, Q. (2017). DrugSig: a resource for computational drug repositioning utilizing gene expression signatures. PLoS ONE 12:e0177743. doi: 10.1371/journal.pone.0177743
Xenarios, I., Rice, D. W., Salwinski, L., Baron, M. K., Marcotte, E. M., and Eisenberg, D. (2000). DIP: the database of interacting proteins. Nucleic Acids Res. 28, 289–291. doi: 10.1093/nar/28.1.289
Yang, W., Soares, J., Greninger, P., Edelman, E. J., Lightfoot, H., Forbes, S., et al. (2013). Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961. doi: 10.1093/nar/gks1111
Yeung, C. K., Yoshida, K., Kusama, M., Zhang, H., Ragueneau-Majlessi, I., Argon, S., et al. (2015). Organ impairment-drug-drug interaction database: a tool for evaluating the impact of renal or hepatic impairment and pharmacologic inhibition on the systemic exposure of drugs. CPT Pharmacometr. Syst. Pharmacol. 4, 489–494. doi: 10.1002/psp4.55
Young, R. R. (2002). Genetic toxicology: web resources. Toxicology 173, 103–121. doi: 10.1016/S0300-483X(02)00026-4
Zarin, D. A., Tse, T., Williams, R. J., Califf, R. M., and Ide, N. C. (2011). The ClinicalTrials.gov Results Database — Update and Key Issues. N. Engl. J. Med. 364, 852–860. doi: 10.1056/NEJMsa1012065
Zhang, G., Zhang, Y., Ling, Y., and Jia, J. (2015). Web Resources for Pharmacogenomics. Genomics Proteomics Bioinformatics 13, 51–54. doi: 10.1016/j.gpb.2015.01.002
Zhang, Q., Yang, B., Chen, X., Xu, J., Mei, C., and Mao, Z. (2014). Renal Gene Expression Database (RGED): a relational database of gene expression profiles in kidney disease. Database 2014:bau092. doi: 10.1093/database/bau092
Keywords: databases, in silico, chemicals, drugs, safety assessment
Citation: Pawar G, Madden JC, Ebbrell D, Firman JW and Cronin MTD (2019) In Silico Toxicology Data Resources to Support Read-Across and (Q)SAR. Front. Pharmacol. 10:561. doi: 10.3389/fphar.2019.00561
Received: 21 January 2019; Accepted: 03 May 2019;
Published: 11 June 2019.
Edited by:
Olavi R. Pelkonen, University of Oulu, FinlandReviewed by:
Christoph Helma, In Silico Toxicology, SwitzerlandCopyright © 2019 Pawar, Madden, Ebbrell, Firman and Cronin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark T. D. Cronin, bS50LmNyb25pbkBsam11LmFjLnVr
†Present Address: Gopal Pawar, Pharmacy and Therapeutics Section, School of Clinical and Experimental Medicine, University of Birmingham, Birmingham, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.