
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 14 May 2019
Sec. Experimental Pharmacology and Drug Discovery
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.00490
This article is part of the Research Topic Sigma Receptors View all 16 articles
 Anna Tesei1*†
Anna Tesei1*† Michela Cortesi1†
Michela Cortesi1† Sara Pignatta1
Sara Pignatta1 Chiara Arienti1
Chiara Arienti1 Giulio Massimo Dondio2
Giulio Massimo Dondio2 Chiara Bigogno2
Chiara Bigogno2 Alessio Malacrida3
Alessio Malacrida3 Mariarosaria Miloso3
Mariarosaria Miloso3 Cristina Meregalli3
Cristina Meregalli3 Alessia Chiorazzi3
Alessia Chiorazzi3 Valentina Carozzi3
Valentina Carozzi3 Guido Cavaletti3
Guido Cavaletti3 Marta Rui4
Marta Rui4 Annamaria Marra4
Annamaria Marra4 Daniela Rossi4
Daniela Rossi4 Simona Collina4*
Simona Collina4*Introduction: Pancreatic cancer (PC) is one of the most lethal tumor worldwide, with no prognosis improvement over the past 20-years. The silent progressive nature of this neoplasia hampers the early diagnosis, and the surgical resection of the tumor, thus chemotherapy remains the only available therapeutic option. Sigma receptors (SRs) are a class of receptors proposed as new cancer therapeutic targets due to their over-expression in tumor cells and their involvement in cancer biology. The main localization of these receptors strongly suggests their potential role in ER unfolded protein response (ER-UPR), a condition frequently occurring in several pathological settings, including cancer. Our group has recently identified RC-106, a novel pan-SR modulator with good in vitro antiproliferative activities toward a panel of different cancer cell lines. In the present study, we investigated the in vitro properties and pharmacological profile of RC-106 in PC cell lines with the aim to identify a potential lead candidate for the treatment of this tumor.
Methods: Pancreatic cancer cell lines Panc-1, Capan-1, and Capan-2 have been used in all experiments. S1R and TMEM97/S2R expression in PC cell lines was quantified by Real-Time qRT-PCR and Western Blot experiments. MTS assay was used to assess the antiproliferative effect of RC-106. The apoptotic properties of RC-106 was evaluated by TUNEL and caspase activation assays. GRP78/BiP, ATF4, and CHOP was quantified to evaluate ER-UPR. Proteasome activity was investigated by a specific fluorescent-based assay. Scratch wound healing assay was used to asses RC-106 effect on cell migration. In addition, we delineated the in vivo pharmacokinetic profile and pancreas distribution of RC-106 in male CD-1 mice.
Results: Panc-1, Capan-1, and Capan-2 express both SRs. RC-106 exerts an antiproliferative and pro-apoptotic effect in all examined cell lines. Cells exposure to RC-106 induces the increase of the expression of ER-UPR related proteins, and the inhibition of proteasome activity. Moreover, RC-106 is able to decrease PC cell lines motility. The in vivo results show that RC-106 is more concentrated in pancreas than plasma.
Conclusion: Overall, our data evidenced that the pan-SR modulator RC-106 is an optimal candidate for in vivo studies in animal models of PC.
Pancreatic cancer remains one of the most lethal tumor types for both men and women and, it represents the 11th most common cancer worldwide (Ilic and Ilic, 2016). WCRF reported that in 2018 there were 460,000 new cases, which mainly affected developed countries (Weledji et al., 2016). For this type of tumor, beneficial pharmaceutical approaches result challenging to develop, since the etiology as well as the triggering factors associated with PC remain undefined (Kim and Ahuja, 2015). Relying on the negative prognosis – the average 5-year survival rate is 6% or less (Siegel et al., 2014) – and on the lack of a concrete cure, PC urgently requires effective therapeutic strategies.
Over the past few decades, SRs, have been widely associated with aging- and mitochondria-associated disorders, such as Parkinson’s and Alzheimer’s disease and cancer (Martin et al., 1976; Su, 1982; Vaupel, 1983; Quirion et al., 1987; Maurice and Lockhart, 1997; Skuza, 2003; Peviani et al., 2014; Collina et al., 2017a,b). Moreover, although no endogenous SRs ligands have ever been found, and the specific role played by this orphan receptor family in cell biology has yet to be clarified, SRs are considered as potential therapeutic targets for neurodegenerative diseases and cancer. Accumulating evidence strongly suggests a pivotal role of these proteins in ER-UPR pathways, whose activation is frequently detected in many solid tumors (Shuda et al., 2003; Corazzari et al., 2017). In particular, the triggering of the UPR machinery in cancer is the result of neoplastic cells spreading in unfavorable environments characterized by hypoxia, low pH, high levels of ROS and inadequate glucose and amino acid supply, conditions that could compromise the correct ER protein folding. Under such stress conditions, SRs are activated to allow the cells survival, as broadly demonstrated by the direct involvement of S1R in UPR pathways (Hayashi, 2015; Penke et al., 2017). The decrease of Ca++ ion level in ER, the accumulation of misfolded or aggregated protein within the ER, the rise of ROS level due to stress conditions promote the exit of S1R from a dormant state and its activation as chaperon protein. Accordingly, the correct Ca++ signaling from the ER to the mitochondria, the transmission of the ER stress signal to the nucleus and the consequent increase of antistress and antioxidant proteins production are guaranteed (Hayashi and Su, 2007; Mori et al., 2013; Wang et al., 2015).
Only recently S2R has been cloned and its identity as TMEM97 has been postulated (Alon et al., 2017). TMEM97 is a transmembrane protein involved in cholesterol homeostasis, and its dysregulation has been associated to ER stress and to activation of the UPR, thus causing cellular lipid accumulation (Colgan et al., 2011). Notably, UPR is classically related to the maintenance of cellular homeostasis in secretory cells (i.e., pancreatic and immune cells), where the high demand for protein synthesis and secretion leads to proteostasis and cellular stress (Hetz, 2012; Moore and Hollien, 2012). Indeed, pancreatic cells have high hormone and enzyme secretory functions and possess highly developed ER. The role of ER stress in PC pathobiology and inflammation has been increasingly recognized as an important factor in tumorigenesis and chemoresistance (Yadav et al., 2014). Nonetheless, PC is extremely rich in stroma, is hypoxic and deficient in metabolites (Vasseur et al., 2010). A similar behavior can be found when cells grow under chronic metabolic stress conditions, favoring the activation of adaptive mechanisms, such as UPR and autophagy (Kondo et al., 2005; Moenner et al., 2007) the latter frequently associated to SR overexpression (Zeng et al., 2012; Mir et al., 2013). Altogether, these findings pointed out SRs as potential targets useful for inhibiting UPR machinery in PC.
Our research team is active in the SR modulation and recently we identified compound RC-106 endowed with pan-SR modulatory activity (S1R antagonist and S2R agonist profile) and in vitro antiproliferative properties toward a panel of cancer cell lines (i.e., Capan-2, MDA-MB 231, PC3, and U87) (Rui et al., 2016; Rossi et al., 2017). These encouraging results led us to further investigate its potential in PC treatment. After preparing RC-106 in a suitable amount to support the whole study, we deepened its antitumor properties and evaluated its capability to interfere with ER stress conditions. Lastly preliminary PK and biodistribution studies have been performed, to verify if RC-106 is able to reach the target tissue.
Reagents and solvents for synthesis were obtained from Sigma-Aldrich (Italy). Solvents were purified according to the guidelines in Purification of Laboratory Chemicals. Melting points were measured on SMP3 Stuart Scientific apparatus and are uncorrected. For FT-IR analysis a Spectrum One PerkinElmer spectrophotometer equipped with a MIRacleTM ATR device was used. The IR spectra were scanned over wavenumber range of 4000–650 cm-1 with a resolution of 4 cm-1. Analytical thin-layer chromatography (TLC) was carried out on silica gel precoated glass backed plates (Fluka Kieselgel 60 F254, Merck); visualized by UV radiation, acidic ammonium molybdate (IV), or potassium permanganate. FC was performed with Silica Gel 60 (particle size 230e400 mesh, purchased from Merck). Proton NMR spectra were recorded on Bruker Avance 400 spectrometer operating at 400 MHz. 13C NMR spectra were recorded on 500 MHz spectrometer, operating at 125 MHz, with complete proton decoupling. UPLC-UV-ESI/MS analyses were carried out on a Acuity UPLC Waters LCQ FLEET system using an ESI source operating in positive ion mode, controlled by ACQUIDITY PDA and 4 MICRO (Waters). Analyses were run on a ACQUITY BEH C18 (50 mm × 2.1 mm, 1.7 mm) column, at room temperature, with gradient elution (solvent A: water containing 0.1% of formic acid; solvent B: methanol containing 0.1% of formic acid; gradient: 10% B in A to 100% B in 3 min, followed by isocratic elution 100% B for 1.5 min, return to the initial conditions in 0.2 min) at a flow rate of 0.5 mL min-1. Detailed synthetic procedure and characterization of intermediates and RC-106 are reported in the Supplementary Material.
Pancreatic adenocarcinoma Panc-1, Capan-1, and Capan-2, cell lines were purchased by the ATCC. All cell lines were grown in culture medium composed of DMEM/Ham’s F12 (1:1; Euroclone) supplemented with fetal calf serum (10%; Euroclone), glutamine (2 mM; Euroclone), and insulin (10 μg/mL; Sigma-Aldrich, St. Louis, MO, United States). All experiments were performed on cells in the exponential growth phase and checked periodically for mycoplasma contamination by MycoAlertTM Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
Spheroids were obtained as previously described (Zanoni et al., 2016). Briefly, a rotatory cell culture system RCCS (Synthecon Inc., Houston, TX, United States) was used. The rotary systems were placed inside a humidified 37°C, 5% CO2 incubator and all procedures were performed in sterile conditions. Single cell suspensions of about 1 × 106 cells/ml of Panc-1 were placed in the 50 mL rotating chamber at an initial speed of 12 rpm. Speed was increased as cells formed aggregates to avoid sedimentation. The culture medium was changed every 4 days and tumor spheroids with an equivalent diameter ranging from about 500–1300 μm were obtained in around 15 days. After formation, spheroids were transferred into a 96-well low-attachment culture plates (Corning Inc., Corning, NY, United States; one spheroid/well), containing 100 μL of fresh culture medium per well.
Cytotoxicity was assayed using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Milan, Italy). Cells were seeded onto a 96-well plate at a density of 3 × 103 cells per well. Cell lines were exposed to increasing concentrations of the drug, ranging from 0.1 to 100 μM. The effect of the drug was evaluated after 24, 48, and 72 h of continued exposure. Two independent experiments were performed in octuplicate. The OD of treated and untreated cells was determined at a wavelength of 490 nm using a fluorescence plate reader.
Dose response curves were created by Excel software. IC50 values were determined graphically from the plot.
Cell viability of Panc-1 spheroids was measured using a 3D cell viability assay (Promega, Milan, Italy). Briefly, homogeneous spheroids were removed from the 96-well low-attachment culture plate and placed separately in single wells of a 96-well opaque culture plate (BD Falcon). CellTiter-Glo® 3D reagent was added to each well and the luminescence signal was read after 30 min with the GloMax® bioluminescent reader (Promega).
The analysis of morphological parameters were performed as previously described (Piccinini et al., 2017). Briefly, an inverted Olympus IX51 microscope (Olympus Corporation, Tokyo, Japan), equipped with a Nikon Digital Sight DS-Vi1 camera (CCD vision sensor, square pixels of 4.4 μm side length, 1600 × 1200 pixel resolution, 8-bit gray level; Nikon Instruments, Spa. Florence, Italy) was used to take images and for morphological analyses. The open-source ReViSM software tools was used to achieve morphological 3D, such as volume and sphericity, and to select morphologically homogeneous spheroids. For the experiments, Panc-1 spheroids characterized by spherical shape and by a diameter size ranging from 500 to 600 μM were selected.
Total cellular RNA was extracted using TRIzol reagent (Life technologies) in accordance with manufacturer’s instruction and quantified using the Nanodrop MD-1000 spectrophotometer system. Reverse transcription reactions were performed in 20 μL of nuclease free water containing 400 ng of total RNA using iScript cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, CA). Real-Time PCR was run using 7500 Fast Real-Time PCR system (Applied Biosystems) and TaqMan assays to detect the expression of SIGMAR1, TMEM97, GRP78/BiP, ATF4, and CHOP genes.
Reactions were carried out in triplicate at a final volume of 20 μL containing 40 ng of cDNA template, TaqMan universal PCR Master Mix (2X), and selected TaqMan assays (20X). Samples were maintained at 50°C for 2 min, then at 95°C for 10 min followed by 40 amplification cycles at 95°C for 15 s, and at 60°C for 30 s.
The amount of mRNA was normalized to the endogenous genes GAPDH and HPRT-1.
TUNEL assay was performed as previously described (Tesei et al., 2007). Briefly, cells were fixed in 1% formaldehyde in PBS on ice for 15 min, suspended in 70% ice cold ethanol and stored overnight at 20°C. Cells were then washed twice in PBS and re-suspended in PBS containing 0.1% Triton X-100 for 5 min at 48°C. Thereafter, samples were incubated in 50 μL of solution containing TdT and FITC conjugated dUTP deoxynucleotides 1:1 (Roche Diagnostic GmbH, Mannheim, Germany) in a humidified atmosphere for 90 min at 37°C in the dark, washed in PBS, counterstained with propidium iodide (2.5 μg/mL, MP Biomedicals, Verona, Italy) and RNAse (10 kU/mL, Sigma–Aldrich) for 30 min at 48°C in the dark and analyzed by flow cytometry. Flow cytometric analysis was performed using a FACS Canto flow cytometer (Becton Dickinson, San Diego, CA, United States). Data acquisition and analysis were performed using FACSDiva software (Becton Dickinson). Samples were run in triplicate and 10,000 events were collected for each replicate.
Western Blot were performed as previously described (Arienti et al., 2016). Briefly, Cell proteins were extracted with M-PER (Thermo Fisher Scientific) supplemented with Halt Protease Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific).
Mini-PROTEANTGXTM precast gels (4–20% and any kD; BIO-RAD) were run using Mini-PROTEAN Tetra electrophoresis cells and then electroblotted by Trans-Blot TurboTM Mini PVDF Transfer Packs (BIORAD). The unoccupied membrane sites were blocked with T-TBS 1X (Tween 0.1%) and 5% non-fat dry milk to prevent non-specific binding of antibodies and probed with specific primary antibodies overnight at 4°C. This was followed by incubation with the respective secondary antibodies. The antibody-antigen complexes were detected with Immun-StarTM WesternCTM kit (BIO-RAD).
The following primary antibodies were used: anti-sigma receptor (S18): sc-22948 (Santa Cruz Biotechnology inc.), anti-TMEM97, anti-caspase-3, and anti-caspase-9. Anti-vinculin (sc-5573) from Santa Cruz Biotechnology and anti-actin from Sigma Aldrich Inc., were used as housekeeping. Quantity One Software was used for analysis.
Cells were seeded in 6-well plates at density of 250 × 103 cells/well. Cells were treated with increasing concentration of RC-106 and after 24 h total protein extracts were obtained: cells were washed 2 times with PBS and lysed with 100 μL of lysis buffer (Hepes 5 mM pH 7.5, NaCl 150 mM, Glycerol 10%, Triton X100 1%, MgCl2 1.5 mM, EGTA 5 mM). Protein concentration of samples was quantified using Bradford method. Proteasome activity was quantified as described below. Proteasome solution was composed by 40 μg of proteins, 10 μL of 10X proteasome buffer (Hepes pH 7.5 250 mM, EDTA pH 8.0 5 mM, NP-40 0.5%, SDS 0.01%) and 10 μL of 10 mM proteasome substrate (N-Succinyl-Leu-Leu-Val-Tyr-7-Amido-4-Methylcoumarin, 7.6 mg/mL; Sigma-Aldrich, United States). 100 μL of proteasome solution was loaded in wells of a black 96-well plate. The plate was then incubated at 37°C for 2 h and the fluorescence was measured in a microplate reader (excitation 380 nm, emission 460 nm; BMG-Labtech, Germany).
Cells were seeded in a 6-well plate and were incubated at 37°C until confluence of 90–100% was reached. Culture medium was then replaced by serum free medium. After 24 h, a scratch was made on cell monolayer using a plastic tip and wells were washed 2 times with PBS to remove detached cells and debris. Culture medium, with or without RC-106, was added to each well. Micrographs of the scratches were taken at 0 h, immediately after the scratch, and at 24, 48, and 72 h. Cell migration area was quantified using IMAGEJ software.
The experiments were performed in agreement with the Italian Law D. L.vo 4 marzo 2014, n. 26. The treatments involved male CD-1 mice and a unique number on the tail identified each animal. Mice were housed, in groups of four, in cages suitable for the species. After 5 days of adaptation to the local housing conditions, animals were housed in a single, exclusive, air-conditioned room to provide a minimum of 15 air changes/hour. The environmental controls were set to maintain the temperature at around 22°C and the relative humidity within the range 50 to 60%, along with an approximate 12:12 h light/dark cycle automatically controlled. Food (Mucedola Standard GLP diet) and water were available ad libitum throughout the entire duration of the study. All animals were weighted on the day of the treatment.
Mice (n = 4/time point) received an intraperitoneal administration (i.p., 10 mL/kg) of RC-106 at 10 mg/kg. CD-1 male mice were exsanguinated under anesthesia (isoflurane) from the aorta at the following time points: 5, 10, 30, 120, 240, and 480 min. Blood samples were collected in tubes containing heparin, gently mixed and immediately placed on ice. Afterward, they have been centrifuged (3500 × g, at 4°C for 15 min), the obtained plasma has been collected and transferred to individually labeled tubes and frozen at -20°C until the analysis. Plasma samples were used for quantification of RC-106. Pancreas was taken by surgical resection after 20 min from the last treatment, washed in saline, dried on paper, weighted and frozen at -20°C. The organ was homogenized using a Velp OV5 homogenizer with 20 mM ammonium formiate buffer in a ratio of 1 g of tissue per 10 mL of buffer.
20 mg/mL stock solution (s.s.) of RC-106 was prepared by dissolving the compound in DMSO. 1 mL of 5% Tween80 in H2O was slowly added to 500 μL of s.s. under stirring. Then 8.5 mL of water was gently spiked to obtain the 1 mg/mL formulation of RC-106.
Standard curves of RC-106 were prepared for plasma and pancreas homogenate, and analyzed together with each QC and unknown sample set. For the PK and pancreas distribution sample analysis, plasma and pancreas homogenate samples (50 μL) were spiked in 200 μL of IS in MeOH (0.1 μg/mL of RC-33), followed by 2 min vortex mixing. Samples were centrifuged and transferred in UFLC vials. 5 μL aliquots of the collected samples were injected into the LC-MS/MS system. Standard calibration graphs were constructed by linear least-squares regression analysis on the analyte/IS area ratio plotted against sample concentration. Calibration ranges were from 5 to 1000 ng/mL for plasma, and from 5 to 500 ng/mL for pancreas homogenate. Accuracy values were determined in triplicates at three different concentrations (high, medium, and low) in the range of linearity of the calibration curves.
Analyses were acquired on a Shimadzu AC20 UFLC system interfaced with an API 3200 Triple Quadrupole detector (AB Sciex). Data acquisition and control were performed using the AnalystTM 6.1 (Applied Biosystems) Software. A Phenomenex Gemini-NX C18 (50 mm × 2 mm, 5 μm) column was selected to carry out the analytical evaluations. A gradient method was set up (Supplementary Table S1) and it provided the employment of water and methanol, both containing 0.1% of formic acid, at a flow rate of 0.3 mL/min. The LC eluate was directly introduced into the MS interface using the ESI in the positive ion mode. The MRM transitions m/z 181.2 were tracked (Supplementary Table S2).
We studied an easy to handle synthetic route suitable to dispose of RC-106 in a g-scale amount. The synthetic route is outlined in Scheme 1. Briefly, a Heck reaction between 4-bromobiphenyl and (E)-ethyl crotonate, using Palladium acetate microencapsulated in polyurea matrix (Pd EnCat®) as catalyst allowed to obtain the α,β-unsaturated ester (E)-1 which was easily reduced to give allyl alcohol (E)-2, and then converted into RC-106 according to Frøyen and Juvvik (1995). The use of Pd EnCat® simplified the work-up procedure and more important avoided the heavy metal contamination of the product, which could compromise the in vitro and in vivo studies.

Scheme 1. Synthesis of RC-106. Reagents and conditions: (a) (E)-ethyl crotonate, Pd EnCat® 40, TEAC, NaOAc, DMF anhydrous, N2 atm., 105°C; (b) LiAlH4 (1M in THF), Et2O anhydrous, N2 atm., 0°C; (c) Ph3P, NBS, N2, -15/18°C; (d) 4-benzylpiperidine, Et3N, N2 atm., from -15/-18°C to r.t.
We explored the expression of S1R and TMEM97/S2R genes in a panel of cell lines representative of pancreatic adenocarcinoma. The expression of S1R and TMEM97/S2R was evaluated in cells derived both from primary tumor and metastatic site (i.e., liver), characterized by different doubling time and different mutational status of p53, KRAS, P16/CDKN2A, and SMAD 4 (Table 1), the major driver-genes involved in the pathogenesis of PC (Sipos et al., 2003).
The expression level of SRs was determined by Real-Time qRT-PCR. We used cervix adenocarcinoma HeLa as reference sample, because of its high expression of both S1R and TMEM97/S2R (Bartz et al., 2009; Ebrahimi-Fakhari et al., 2015; Miki et al., 2015). All analyzed cell lines express SRs and no correlation between the tumor site and the expression level of both targets, as well as respect to the mutational status of p53 and KRAS was evidenced. In particular, S1R was expressed at similar levels in the PC cell lines. Conversely, differences about the expression of TMEM97/S2R have been evidenced in the three cell lines investigated, with the highest expression in Capan-1 (4-fold respect to the control line) and the lowest in Capan-2 cells (Figure 1). Basing on these results, we took into account the three cell lines to perform the biological evaluation.
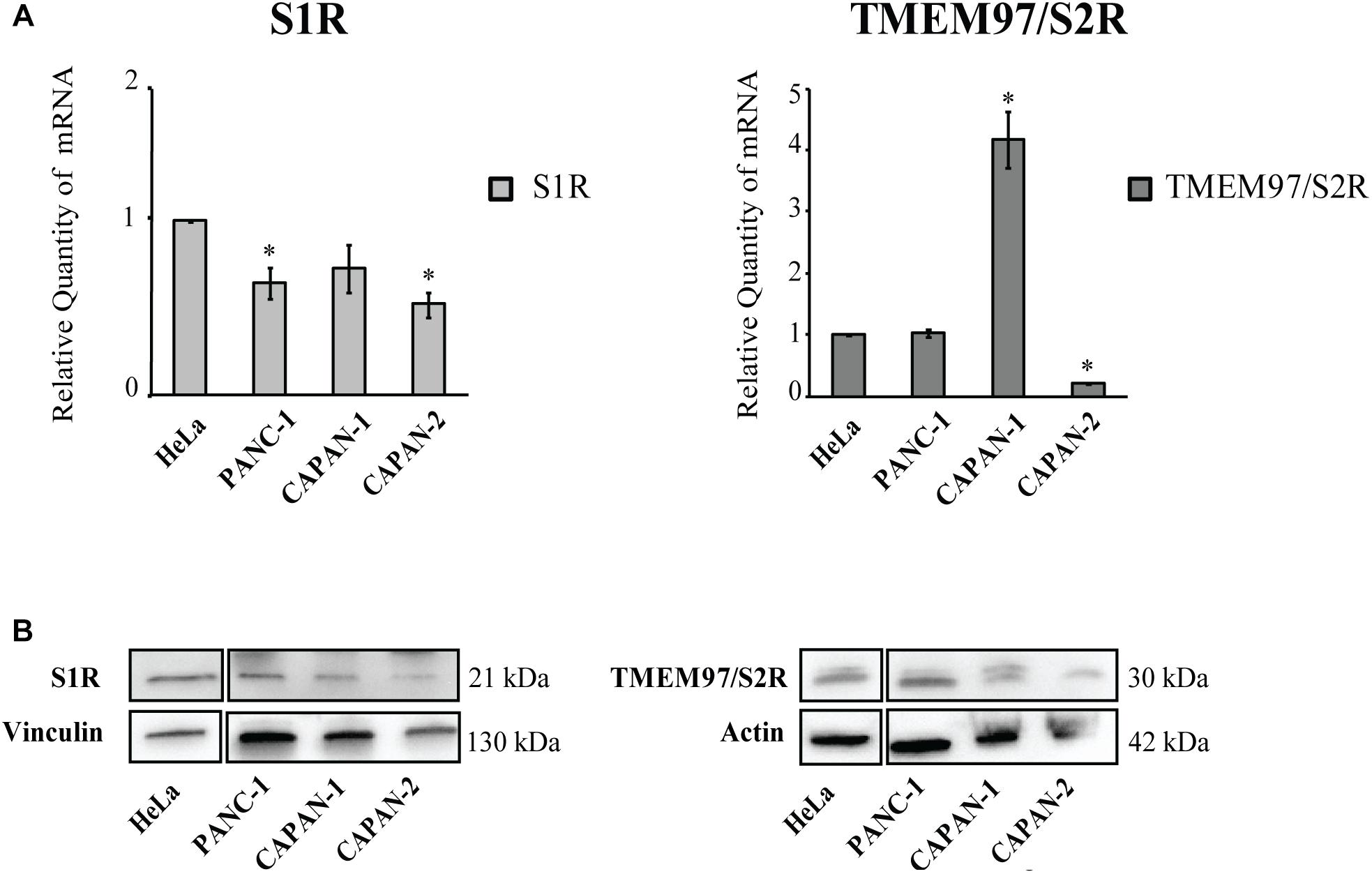
Figure 1. Relative Quantification (RQ) of the target genes Sigma 1 and TMEM97/S2R. (A) Analysis were performed with Real-Time PCR. RNA expression was normalized to GAPDH and HPRT-1. The RNA gene expression was relative to HeLa cell line (RQ = 1). Values are the mean ± SD of three independent experiments. ∗P < 0.05 vs. CTR. (B) Western Blot analysis of TMEM97 and S1R in PC cell lines. HeLa were used as reference sample. Images are representative of two independent experiments.
We evaluated the in vitro cytotoxic activity of RC-106 by MTS assay. Cells were treated for 24, 48, and 72 h with increasing concentrations, ranging from 0.1 to 100 μM. RC-106 was effective in all cell lines tested independently from the exposure time (IC50 values ranging from 33 to 57 μM, Figure 2A). Encouraged by these results, we investigated the capability of RC-106 to penetrate three dimensional structures mimicking tumor micronodules of about 500–600 μm in diameter. Panc-1 cells grown as 3D spheroids were treated with increasing concentrations of RC-106 (12.5–50 μM for 48 h, Figure 2B). The results obtained with Panc-1 spheroids with a diameter up to 600 μm (IC50 = 39.55 μM, Figures 2B,C) are in line with those observed in 2D culture.
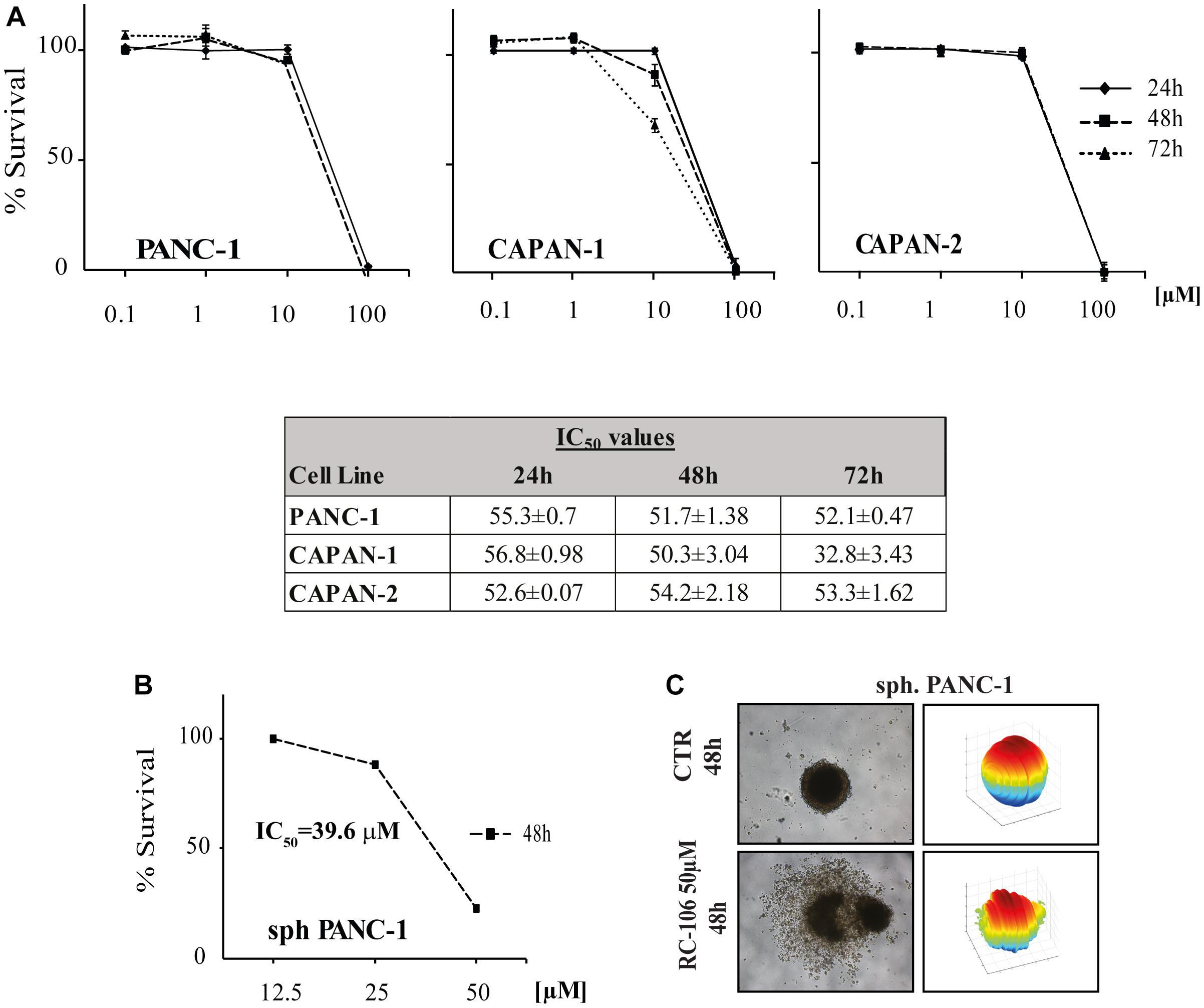
Figure 2. Cell viability of 2D and 3D cell lines. (A) In vitro cytotoxic activity of RC-106 was evaluated in three PC cell lines. Cells were exposed to increasing concentration of the molecule for 24, 48, and 72 h. MTS assay was used to determine cell viability. Values are the mean ± SD of three independent experiments. (B) Homogeneous-size and shape pancreatic adenocarcinoma spheroids were treated with RC-106 for 48 h at concentration ranging from 12.5 to 50 μM. Cell viability was measured using CellTiter-Glo 3D assay. (C) 3D spheroids shape reconstructed on representative brightfield images of Panc-1 spheroids treated with 50 μM of RC-106 for 48 h. The corresponding 3D-shape of Panc-1 spheroids were obtained using ReViSM software tools.
The apoptotic properties of RC-106 was evaluated by TUNEL assay. The exposure time (48 h) and the drug concentration (50 μM) have been chosen according to the data resulting from cell viability assay. TUNEL assay showed a significant induction of apoptosis in treated samples compared to the untreated controls, with a percentage of apoptotic cells ranging from 53.25% ± 4.7 (Panc-1) to 78.55% ± 5.6 (Capan-1) (Figure 3A). Hence, we investigated the activation of caspase cascade by Western Blot analysis, treating cells with RC-106 at different exposure times. We found that both caspases 3 and 9 were cleaved, in all cell lines after the treatment, indicating the activation of the intrinsic apoptotic pathway. To sum up, RC-106 was able to activate both caspases in all the considered cell lines, but after different exposure times and concentrations. In detail, in Panc-1 and Capan-2 cell lines this event occurred after an exposure of 48 h to RC-106 at 25 μM concentration, whereas in Capan-1 cell line after 12 h at 50 μM concentration (Figure 3B).
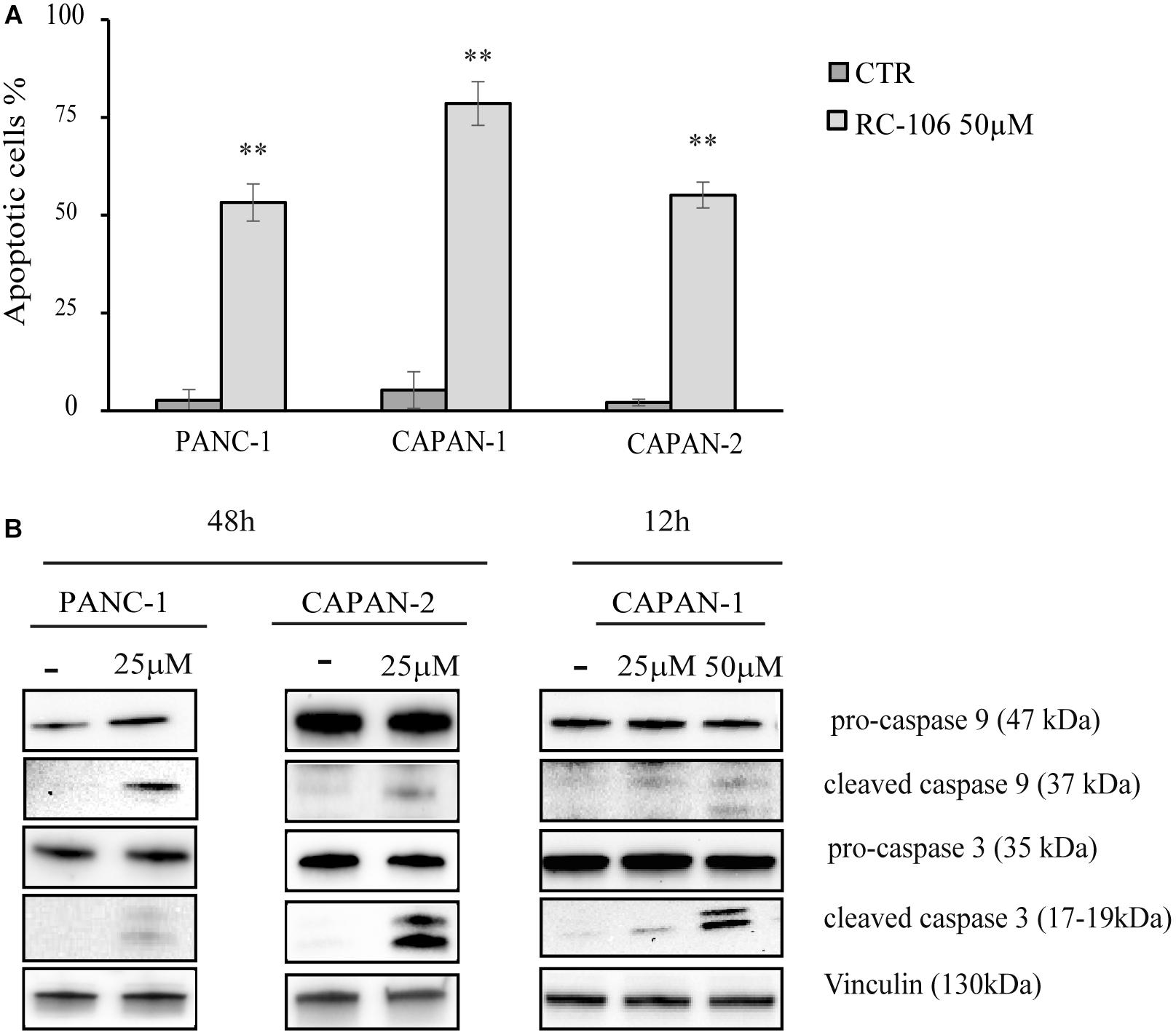
Figure 3. Apoptosis analysis. (A) TUNEL assay performed on Panc-1, Capan-1, and Capan-2 cell lines. Cells were treated with RC-106 50 μM for 48 h. Values are the mean ± SD of three independent experiments. ∗∗P < 0.01 vs. CTR. (B) Western Blot analysis of caspase 3 and 9 activation after 48 h treatment with RC-106 25 μM (Panc-1 and Capan-2) and after 12 h treatment with RC-106 50 μM (Capan-1). Images are representative of two independent experiments.
The expression of the ER stress master proteins GRP78/BiP, ATF4, and CHOP, commonly used for the detection of UPR activation (Samali et al., 2010), was analyzed by Real-Time qRT-PCR. In general, the mRNA expression of all the investigated ER markers highly increased after the exposure to 50 μM of RC-106. In the two cell lines derived from primitive pancreatic tumor, Panc-1 and Capan-2, the trend is similar. In particular, GRP78/BiP and ATF4 mRNA levels increased after 24 h of treatment, while CHOP mRNA levels considerably increased after 12 h, then slightly declined after 24 h (Figure 4A). The highest increase in expression of CHOP was individuated in Capan-2 (70 fold higher than untreated cells). A different behavior was observed for the metastatic cell line Capan-1, where a faster switch-off of all ER markers was evidenced already starting from 12 h after the beginning of treatment.
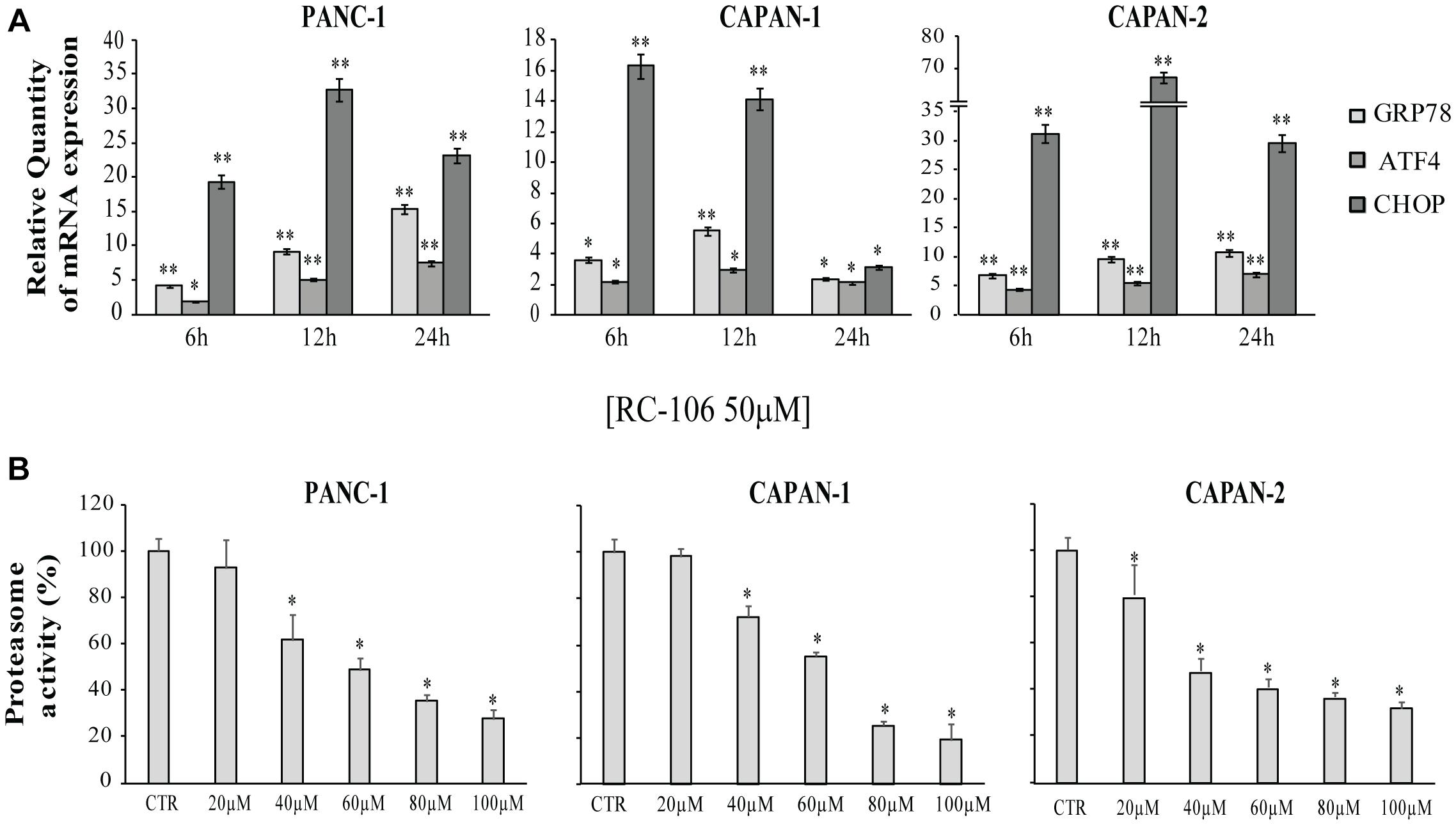
Figure 4. Relative Quantification (RQ) of the ER stress and UPR marker genes. (A) GRP78, ATF4, and CHOP mRNA expression levels were measured after a treatment with RC-106 50 μM for 6,12, and 24 h. Analysis were performed with Real-Time PCR. RNA expression was normalized to GAPDH and HPRT-1. In each time point tested the RNA gene expression was relative to the corresponding untreated control (RQ = 1). Values are the mean ± SD of three independent experiments. (∗P < 0.05 vs. CTR; ∗∗P < 0.01 vs. CTR). (B) Graphs represent the proteasome activity of PANC-1, Capan-1, and Capan-2, treated with increasing concentration of RC-106 (20–100 μM) for 24 h. Data are expressed as the average percentage ± SD of at least three independent experiments and are compared to untreated controls (CTR 100%; ∗P < 0.05 vs. CTR).
All the cell lines were treated with increasing concentrations of RC-106 (20–100 μM) to evaluate in vitro RC-106 proteasome effect. After 24 h of treatment, RC-106 was able to reduce proteasome activity in a dose dependent manner in all the PC investigated (Figure 4B). Capan-2 resulted the most sensible cell line as showed by the lowest concentration used to inhibit proteasome activity (20 μM). Instead the greatest proteasome inhibition is observed in Capan-1 cells but at highest concentration used (100 μM).
Scratch wound healing assay was performed to assess the effect of RC-106 on cell migration. After the scratch, cells were treated with increasing concentration of RC-106 (20–60 μM) and cell migration was evaluated after 24, 48, and 72 h. Capan-1 untreated cells migrated normally to refill the scratch present on cell monolayer. Cell migration was significantly reduced after 48 h of treatment with RC-106 (c = 20 μM). Conversely, RC-106 at concentrations of 40 and 60 μM reduced cell migration already after 24 h of treatment, whereas at major times these concentrations resulted too toxic, promoting cellular death. Capan-2 untreated cells migrated normally and continued to fill the empty space of the scratch for all considered times. RC-106 20 and 40 μM significatively reduceded Capan-2 cells migration ability after 48 and 72 h of treatment. RC-106 60 μM is too toxic and, as for Capan-1 cells, it was not possibile to quantify cell migration inhibition at 48 and 72 h. Panc-1 untreated cells migrated only for the first 24 h, then they slow down and stop migration. RC-106 reduced Panc-1 cell migration in a dose dependent manner, but only in cells treated with 60 μM, migration was significatively reduced for all considered time points (Figure 5).
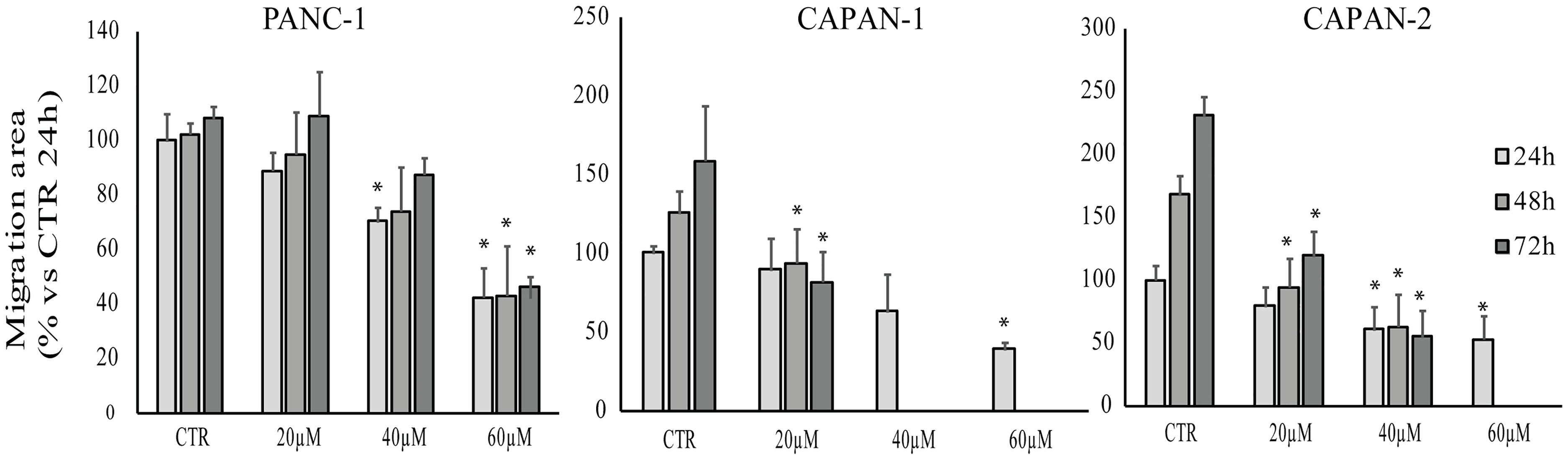
Figure 5. Effect of RC-106 on Capan-1, Capan-2, and Panc-1 cell migration. Migration area of cells was quantified after 24, 48, and 72 h of treatment with increasing concentrations of RC-106 (20–60 μM). Data are expressed as the average percentage ± SD of at least three independent experiments and are compared to controls (CTR, 100%; ∗P < 0.05 vs. respective CTR).
We investigated the in vivo PK profile and pancreas distribution of RC-106 in male CD-1 mice. Basing on our experience, we developed a rapid and sensitive UFLC-MS/MS method for detecting and quantifying RC-106 in biological matrices (Rossi et al., 2013; Marra et al., 2016a,b). Briefly, chromatographic elutions were achieved on a reverse phase column and eluting under a gradient conditions (Supplementary Table S1). LC eluates were directly introduced into the MS interface using the ESI source and detected in positive ion mode (Supplementary Table S2). According to the structure of RC-106, parent ion m/z 356.5 and product ion m/z 181.2 – MRM transitions – were monitored during the analyses. Quantification of RC-106 in plasma or pancreas homogenate were performed by generating 7 concentrations-calibration curves (5–1000 ng/mL for plasma, and 5–500 ng/mL for pancreas homogenate), employing RC-33 as IS, 0.1 μg/mL in MeOH. Accordingly, concentrations of RC-106 at each time point were extrapolated from the corresponding calibration curve. The developed method resulted suitable to separate RC-106 from endogenous interferences. Afterward, CD-1 male mice received intraperitoneal administration at a concentration of 10 mg/kg. Plasma PK parameters are listed in Supplementary Table S3. RC-106 showed a maximal concentration (Cmax) in plasma of 973.3 ng/mL (Tmax of 5 min) with an area under the curve (AUC0-t) of 67986.7 ng/mL∗min (Figure 6 and Supplementary Table S4). Interestingly, RC-106 reached high concentrations also in pancreas with AUC0-t of 1729315.7 ng/mL∗min, thus showing AUC0-t pancreas/AUC0-t plasma of about 25 times (Figure 6 and Supplementary Table S4).
Chemotherapy is the only therapeutic strategy effective in counteracting PC. Nevertheless, the pharmaceutical panorama counts very few effective molecules, since the etiology of this tumor is still elusive and specific therapeutic targets have not been identified yet. Recently, our research team highlighted that the PC cell lines express both S1R and S2R/TMEM97. Therefore, molecules acting via SRs pathway may play a positive role in counteracting PC. In the present work, we deepened the in vitro properties of the pan-SR modulator RC-106 and evaluated its PK profile to define its potential as lead compound.
Pancreatic cancer cell lines Panc-1, Capan-1, and Capan-2, harboring a mutational status representative of clinical tumors and expressing both SRs, have been selected to delineate the in vitro RC-106 profile, and used in all the experiments. The citoxicity tests clearly showed that RC-106 exerts a strong antiproliferative and pro-apoptotic action in all considered cell lines, with IC50 values in the micromolar range. To straighten these data, we exploit the 3D cell culture spheroids, an in vitro model mimicking in vivo features, thus providing better read-outs for drug screening (Carragher et al., 2018). The analysis of 3D morphological parameters of Panc-1 cells, the only able to grow as 3D structure, showed a complete disaggregation of spheroid organization and cytoarchitecture, thus confirming both the strong cytotoxic activity of RC-106 and its good penetration capability.
The cytotoxic activity of RC-106 seems to be mostly attributable to the induction of the intrinsic apoptotic pathways. Herein, we focused on the failure of the adaptive response to restore protein-folding homeostasis. In fact, when UPR is inadequate to restore ER proteostasis, the pathway alternates its signaling toward a terminal UPR, leading to cellular death. To study the role of SRs in ER stress, we measured the expression of the key factors GRP78/BiP, ATF4, and CHOP. In detail, GRP78/BiP is one of the best characterized ER chaperones (Lee, 2005), whereas ATF4 and CHOP are both markers for the shift of the UPR signaling into the alternate signaling program called the “terminal UPR” (Oyadomari and Mori, 2004; Maly and Papa, 2014; Hetz and Papa, 2018).
The cellular exposure to RC-106 induces a relevant increase of the considered key regulators of ER stress, being GRP78/BiP, ATF4, and CHOP overexpressed. To sum up, results of our experiments demonstrated that the antitumor activity of RC-106 is related to the triggering of the “terminal UPR,” confirming the key role of SRs as ER Stress gatekeepers (Tesei et al., 2018). It is worth noting that some compounds able to activate the terminal “UPR” have already reached the clinic for the treatment of several neoplasia, including PC (Hetz et al., 2013; Wang et al., 2018). Among them, bortezomib an inhibitor of proteasome enzyme complex (Chen et al., 2011) deserve to be mentioned, even if its therapeutic use is hampered by its toxic side effects (Field-Smith et al., 2006; Chen et al., 2011; Kharel et al., 2018). Since previous works reported that the silencing or the presence of loss-of-function mutations of S1R lead to an imbalance of protein degradation (Fukunaga et al., 2015; Dreser et al., 2017; Kim, 2017), we extent the evaluation to proteasome inhibition activity. RC-106 resulted able to inhibit the proteasome activity in all the examined cell lines in a dose dependent manner. As a last step of cell biology investigation, we performed the scratch wound healing assay suitable for estimating the local spreading of cancer cells in the tissues/organs. The results showed that RC-106 is able to decrease PC cell motility in a dose dependent manner, suggesting its therapeutic efficacy also in advanced disease.
Taken together the aforementioned results suggest RC-106 as a valuable candidate for the treatment of PC. Considering that tissue distribution in target organ is at the core of drug discovery and development process, having a direct impact on pharmacology, we conclude our study performing PK and pancreas distribution evaluations. The results show that RC-106 is 25 times more concentrated in pancreas than plasma, reaching a concentration similar or even higher (Cmax about 70 μM) than those required to be effective in all the in vitro experiments considered in this work.
Pancreatic cancer treatment is one of the most relevant challenges that the scientific community will have to face in the 21st century. Although novel approaches for PC have been recently proposed, chemotherapy still remains the only effective option to mitigate and counteract the devastating outcome. We herein propose RC-106, a pan-SR modulator with S1R antagonist and S2R agonist profile discovered by our research team, as a valuable compound for in vivo investigation. Obtained results clearly demonstrated that it is effective against PC, via apoptotic pathways, driven by both SR modulation and proteasome complex inhibition. We also deepen the mechanism of action, studying the role played by SR as ER gatekeepers. The so-obtained results demonstrated that RC-106 is able to modulate UPR in response to ER stress, enhancing the expression of GRP78/BiP, ATF4, and CHOP. Furthermore, RC-106 affected not only the viability of PC lines, but also their metastatic potential. Not last in importance, our lead compound it is able to reach the target tissue.
In conclusion, basing on pharmacological and PK profile we suggest the pan-SR modulator RC-106, as an optimal candidate for proof of concept in vivo studies in animal models of PC.
Autorizzazione ministeriale 433/2016-PR: the procedures were authorized by the national authority (Istituto Superiore di Sanità, authorization number 433/2016-PR) and adhered to all the applicable institutional and governmental guidelines for the treatment of laboratory animals (Italian D.L.vo n. 26/2014).
AT and SC: conceptualization. AT, MM, GD, DR, and SC: experimental design and methodology. MC, SP, CA, AlM, CM, AC, VC, MR, and AnM: investigation. MC, AlM, MR, and AnM: writing-original draft preparation. AT, MC, GD, MM, MR, and SC: writing-review and editing. AT, GC, and SC: supervision. AT and SC: project administration. All authors contributed to manuscript revision, and read and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00490/full#supplementary-material
ATCC, American Type Culture Collection; ATF4, activating transcription factor 4; CHOP, C/EBP homologous protein; CTR, control; DMSO, dimethyl sulfoxide; ER, endoplasmic reticulum; ESI, electrospray ionization; FC, flash chromatography; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GRP78, 78-kDa glucose regulated protein; HPRT-1, hypoxanthine phosphoribosyltransferase 1; IS, internal standard; LC, liquid chromatography; MAM, mitochondria associated ER membrane; M-PER, Mammalian Protein Extraction Reagent; MRM, multiple reaction monitoring; MS, mass spectrometry; NMR, nuclear magnetic resonance; OD, optical density; PBS, phosphate buffered saline; PC, Pancreatic cancer; PK, pharmacokinetic; QC, quality control; RCCS, rotary cell culture system; ROS, reactive oxygen species; S1R, sigma 1 receptor; S2R, sigma 2 receptor; SD, standard deviation; SRs, sigma receptors; TdT, terminal deoxynucleotidyl transferase; TLC, thin layer chromatography; TMEM97, transmembrane protein 97; UFLC, ultra-fast liquid chromatography; UPLC, ultra performance liquid chromatography; UPR, unfolded protein response; UV, ultraviolet; WCRF, World Cancer Research Fund.
Alon, A., Schmidt, H. R., Wood, M. D., Sahn, J. J., Martin, S. F., and Kruse, A. C. (2017). Identification of the gene that codes for the σ2 receptor. Proc. Natl. Acad. Sci. U.S.A. 114, 7160–7165. doi: 10.1073/pnas.1705154114
Arienti, C., Zanoni, M., Pignatta, S., Del Rio, A., Carloni, S., Tebaldi, M., et al. (2016). Preclinical evidence of multiple mechanisms underlying trastuzumab resistance in gastric cancer. Oncotarget 7, 18424–18439. doi: 10.18632/oncotarget.7575
Bartz, F., Kern, L., Erz, D., Zhu, M., Gilbert, D., Meinhof, T., et al. (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10, 63–75. doi: 10.1016/j.cmet.2009.05.009
Carragher, N., Piccinini, F., Tesei, A., Trask, O. J., Bickle, M., and Horvath, P. (2018). Concerns, challenges and promises of high-content analysis of 3D cellular models. Nat. Rev. Drug Discov. 17:606. doi: 10.1038/nrd.2018.99
Chen, D., Frezza, M., Schmitt, S., Kanwar, J., and Dou, Q. P. (2011). Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239–253.
Colgan, S. M., Al-Hashimi, A. A., and Austin, R. C. (2011). Endoplasmic reticulum stress and lipid dysregulation. Expert Rev. Mol. Med. 13:e4. doi: 10.1017/S1462399410001742
Collina, S., Bignardi, E., Rui, M., Rossi, D., Gaggeri, R., Zamagni, A., et al. (2017a). Are sigma modulators an effective opportunity for cancer treatment? A patent overview (1996-2016). Expert Opin. Ther. Pat. 27, 565–578. doi: 10.1080/13543776.2017.1276569
Collina, S., Rui, M., Stotani, S., Bignardi, E., Rossi, D., Curti, D., et al. (2017b). Are sigma receptor modulators a weapon against multiple sclerosis disease? Future Med. Chem. 9, 2029–2051. doi: 10.4155/fmc-2017-0122
Corazzari, M., Gagliardi, M., Fimia, G. M., and Piacentini, M. (2017). Endoplasmic reticulum stress, unfolded protein response, and cancer cell fate. Front. Oncol. 7:78. doi: 10.3389/fonc.2017.00078
Dreser, A., Vollrath, J. T., Sechi, A., Johann, S., Roos, A., Yamoah, A., et al. (2017). The ALS-linked E102Q mutation in Sigma receptor-1 leads to ER stress-mediated defects in protein homeostasis and dysregulation of RNA-binding proteins. Cell Death Differ. 24, 1655–1671. doi: 10.1038/cdd.2017.88
Ebrahimi-Fakhari, D., Wahlster, L., Bartz, F., Werenbeck-Ueding, J., Praggastis, M., Zhang, J., et al. (2015). Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum. Mol. Genet. 25, 3588–3599. doi: 10.1093/hmg/ddw204
Field-Smith, A., Morgan, G. J., and Davies, F. E. (2006). Bortezomib (Velcadetrade mark) in the treatment of multiple myeloma. Ther. Clin. Risk Manag. 2, 271–279. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18360602 (accessed January 11, 2019).
Frøyen, P., and Juvvik, P. (1995). One-pot synthesis of secondary or tertiary amines from alcohols and amines via alkoxyphosphonium salts. Tetrahedron Lett. 36, 9555–9558.
Fukunaga, K., Shinoda, Y., and Tagashira, H. (2015). The role of SIGMAR1 gene mutation and mitochondrial dysfunction in amyotrophic lateral sclerosis. J. Pharmacol. Sci. 127, 36–41. doi: 10.1016/j.jphs.2014.12.012
Hayashi, T. (2015). Sigma-1 receptor: the novel intracellular target of neuropsychotherapeutic drugs. J. Pharmacol. Sci. 127, 2–5. doi: 10.1016/j.jphs.2014.07.001
Hayashi, T., and Su, T. P. (2007). Sigma-1 receptor chaperones at the ER- mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610. doi: 10.1016/j.cell.2007.08.036
Hetz, C. (2012). The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102. doi: 10.1038/nrm3270
Hetz, C., Chevet, E., and Harding, H. P. (2013). Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12, 703–719. doi: 10.1038/nrd3976
Hetz, C., and Papa, F. R. (2018). The unfolded protein response and cell fate control. Mol. Cell 69, 169–181. doi: 10.1016/j.molcel.2017.06.017
Ilic, M., and Ilic, I. (2016). Epidemiology of pancreatic cancer. World J. Gastroenterol. 22, 9694–9705. doi: 10.3748/wjg.v22.i44.9694
Kharel, P., Uprety, D., Chandra, A. B., Hu, Y., Belur, A. A., and Dhakal, A. (2018). Bortezomib-induced pulmonary toxicity: a case report and review of literature. Case Rep. Med. 2018, 1–5. doi: 10.1155/2018/2913124
Kim, F. J. (2017). Introduction to sigma proteins: evolution of the concept of sigma receptors. Handb. Exp. Pharmacol. 244, 1–11. doi: 10.1007/164_2017_41
Kim, V. M., and Ahuja, N. (2015). Early detection of pancreatic cancer. Chin. J. Cancer Res. 27, 321–331. doi: 10.3978/j.issn.1000-9604.2015.07.03
Kondo, Y., Kanzawa, T., Sawaya, R., and Kondo, S. (2005). The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 5, 726–734. doi: 10.1038/nrc1692
Lee, A. S. (2005). The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35, 373–381. doi: 10.1016/j.ymeth.2004.10.010
Maly, D. J., and Papa, F. R. (2014). Druggable sensors of the unfolded protein response. Nat. Chem. Biol. 10, 892–901. doi: 10.1038/nchembio.1664
Marra, A., Rossi, D., Maggi, L., Corana, F., Mannucci, B., Peviani, M., et al. (2016a). Development of easy-to-use reverse-phase liquid chromatographic methods for determining PRE-084. RC-33 and RC-34 in biological matrices. The first step for in vivo analysis of sigma1 receptor agonists. Biomed. Chromatogr. 30, 645–651. doi: 10.1002/bmc.3609
Marra, A., Rossi, D., Pignataro, L., Bigogno, C., Canta, A., Oggioni, N., et al. (2016b). Toward the identification of neuroprotective agents: g-scale synthesis, pharmacokinetic evaluation and CNS distribution of (R)-RC-33, a promising SIGMA1 receptor agonist. Future Med. Chem. 8, 287–295. doi: 10.4155/fmc.15.191
Martin, W. R., Eades, C. G., Thompson, J. A., Huppler, R. E., and Gilbert, P. E. (1976). The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J. Pharmacol. Exp. Ther. 197, 517–532.
Maurice, T., and Lockhart, B. P. (1997). Neuroprotective and anti-amnesic potentials of sigma (σ) receptor ligands. Prog. Neuropsychopharmacol. Biol. Psychiatry 21, 69–102. doi: 10.1016/S0278-5846(96)00160-1
Miki, Y., Tanji, K., Mori, F., and Wakabayashi, K. (2015). Sigma-1 receptor is involved in degradation of intranuclear inclusions in a cellular model of huntington’s disease. Neurobiol. Dis. 74, 25–31. doi: 10.1016/j.nbd.2014.11.005
Mir, S. U. R., Schwarze, S. R., Jin, L., Zhang, J., Friend, W., Miriyala, S., et al. (2013). Progesterone receptor membrane component 1/Sigma-2 receptor associates with MAP1LC3B and promotes autophagy. Autophagy 9, 1566–1578. doi: 10.4161/auto.25889
Moenner, M., Pluquet, O., Bouchecareilh, M., and Chevet, E. (2007). Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 67, 10631–10634. doi: 10.1158/0008-5472.CAN-07-1705
Moore, K. A., and Hollien, J. (2012). The unfolded protein response in secretory cell function. Annu. Rev. Genet. 46, 165–183. doi: 10.1146/annurev-genet-110711-155644
Mori, T., Hayashi, T., Hayashi, E., and Su, T. P. (2013). Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8:e76941. doi: 10.1371/journal.pone.0076941
Oyadomari, S., and Mori, M. (2004). Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381–389. doi: 10.1038/sj.cdd.4401373
Penke, B., Fulop, L., Szucs, M., and Frecska, E. (2017). The role of sigma-1 receptor, an intracellular chaperone in neurodegenerative diseases. Curr. Neuropharmacol. 16, 97–116. doi: 10.2174/1570159X15666170529104323
Peviani, M., Salvaneschi, E., Bontempi, L., Petese, A., Manzo, A., Rossi, D., et al. (2014). Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation. Neurobiol. Dis. 62, 218–232. doi: 10.1016/j.nbd.2013.10.010
Piccinini, F., Tesei, A., Zanoni, M., and Bevilacqua, A. (2017). ReViMS: software tool for estimating the volumes of 3-D multicellular spheroids imaged using a light sheet fluorescence microscope. Biotechniques 63, 227–229. doi: 10.2144/000114609
Quirion, R., Chicheportiche, R., Contreras, P. C., Johnson, K. M., Lodge, D., William Tam, S., et al. (1987). Classification and nomenclature of phencyclidine and sigma receptor sites. Trends Neurosci. 10, 444–446. doi: 10.1016/0166-2236(87)90094-4
Rossi, D., Pedrali, A., Gaggeri, R., Marra, A., Pignataro, L., Laurini, E., et al. (2013). Chemical, pharmacological, and in vitro metabolic stability studies on enantiomerically pure RC-33 compounds: promising neuroprotective agents acting as σ1 receptor agonists. ChemMedChem 8, 1514–1527. doi: 10.1002/cmdc.201300218
Rossi, D., Rui, M., Di Giacomo, M., Schepmann, D., Wünsch, B., Monteleone, S., et al. (2017). Gaining in pan-affinity towards sigma 1 and sigma 2 receptors. SAR studies on arylalkylamines. Bioorg. Med. Chem. 25, 11–19. doi: 10.1016/j.bmc.2016.10.005
Rui, M., Rossi, D., Marra, A., Paolillo, M., Schinelli, S., Curti, D., et al. (2016). Synthesis and biological evaluation of new aryl-alkyl(alkenyl)-4-benzylpiperidines, novel sigma receptor (SR) modulators, as potential anticancer-agents. Eur. J. Med. Chem. 124, 649–665. doi: 10.1016/j.ejmech.2016.08.067
Samali, A., Fitzgerald, U., Deegan, S., and Gupta, S. (2010). Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell Biol. 2010:830307. doi: 10.1155/2010/830307
Shuda, M., Kondoh, N., Imazeki, N., Tanaka, K., Okada, T., Mori, K., et al. (2003). Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J. Hepatol. 38, 605–614. doi: 10.1016/S0
Siegel, R., Ma, J., Zou, Z., and Jemal, A. (2014). Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29. doi: 10.3322/caac.21208
Sipos, B., Möser, S., Kalthoff, H., Török, V., Löhr, M., and Klöppel, G. (2003). A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch. 442, 444–452. doi: 10.1007/s00428-003-0784-4
Skuza, G. (2003). Potential antidepressant activity of sigma ligands. Pol. J. Pharmacol. 55, 923–934.
Su, T. P. (1982). Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J. Pharmacol. Exp. Ther. 223, 284–290.
Tesei, A., Cortesi, M., Zamagni, A., Arienti, C., Pignatta, S., Zanoni, M., et al. (2018). Sigma receptors as endoplasmic reticulum stress “gatekeepers” and their modulators as emerging new weapons in the fight against cancer. Front. Pharmacol. 9:711. doi: 10.3389/fphar.2018.00711
Tesei, A., Rosetti, M., Ulivi, P., Fabbri, F., Medri, L., Vannini, I., et al. (2007). Study of molecular mechanisms of pro-apoptotic activity of NCX 4040, a novel nitric oxide-releasing aspirin, in colon cancer cell lines. J. Transl. Med. 5:52. doi: 10.1186/1479-5876-5-52
Vasseur, S., Tomasini, R., Tournaire, R., and Iovanna, J. L. (2010). Hypoxia induced tumor metabolic switch contributes to pancreatic cancer aggressiveness. Cancers 2, 2138–2152. doi: 10.3390/cancers2042138
Vaupel, D. B. (1983). Naltrexone fails to antagonize the sigma effects of PCP and SKF 10,047 in the dog. Eur. J. Pharmacol. 92, 269–274.
Wang, J., Shanmugam, A., Markand, S., Zorrilla, E., Ganapathy, V., and Smith, S. B. (2015). Sigma 1 receptor regulates the oxidative stress response in primary retinal Müller glial cells via NRF2 signaling and system x, the Na-independent glutamate-cystine exchanger. Free Radic. Biol. Med. 86, 25–36. doi: 10.1016/j.freeradbiomed.2015.04.009
Wang, M., Law, M. E., Castellano, R. K., and Law, B. K. (2018). The unfolded protein response as a target for anticancer therapeutics. Crit. Rev. Oncol. Hematol. 127, 66–79. doi: 10.1016/j.critrevonc.2018.05.003
Weledji, E. P., Enoworock, G., Mokake, M., and Sinju, M. (2016). How grim is pancreatic cancer? Oncol. Rev. 10:294. doi: 10.4081/oncol.2016.294
Yadav, R. K., Chae, S.-W., Kim, H.-R., and Chae, H. J. (2014). Endoplasmic reticulum stress and cancer. J. Cancer Prev. 19, 75–88. doi: 10.15430/JCP.2014.19.2.75
Zanoni, M., Piccinini, F., Arienti, C., Zamagni, A., Santi, S., Polico, R., et al. (2016). 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 6:19103. doi: 10.1038/srep19103
Keywords: Pancreatic cancer, pan-sigma receptor modulators, endoplasmic reticulum stress, unfolded protein response, proteasome inhibition
Citation: Tesei A, Cortesi M, Pignatta S, Arienti C, Dondio GM, Bigogno C, Malacrida A, Miloso M, Meregalli C, Chiorazzi A, Carozzi V, Cavaletti G, Rui M, Marra A, Rossi D and Collina S (2019) Anti-tumor Efficacy Assessment of the Sigma Receptor Pan Modulator RC-106. A Promising Therapeutic Tool for Pancreatic Cancer. Front. Pharmacol. 10:490. doi: 10.3389/fphar.2019.00490
Received: 22 January 2019; Accepted: 17 April 2019;
Published: 14 May 2019.
Edited by:
Tangui Maurice, INSERM U1198 Mécanismes Moléculaires dans les Démences Neurodégénératives, FranceReviewed by:
Franck Borgese, INSERM U1091 Institut de Biologie de Valrose, FranceCopyright © 2019 Tesei, Cortesi, Pignatta, Arienti, Dondio, Bigogno, Malacrida, Miloso, Meregalli, Chiorazzi, Carozzi, Cavaletti, Rui, Marra, Rossi and Collina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Tesei, YW5uYS50ZXNlaUBpcnN0LmVtci5pdA== Simona Collina, c2ltb25hLmNvbGxpbmFAdW5pcHYuaXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.