- Department of Neurology, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang, China
Oxidized-low density lipoprotein (ox-LDL) can induce injury of endothelial cells, causing atherosclerosis, which is an important initial event in several cardiovascular diseases. Long non-coding RNAs (lncRNAs) have emerged as regulators of diverse biological processes, but their specific biological functions and biochemical mechanisms in ox-LDL-induced endothelial cell injury have not been well investigated. Here, we describe the initial functional analysis of a poorly characterized human lncRNA ZEB1 antisense 1 (ZEB1-AS1). We found that ox-LDL treatment could induce a decreased cell viability and an increased cell apoptosis in endothelial cells, and knockdown of ZEB1-AS1 significantly reversed this effect. Mechanistically, ox-LDL treatment could sequester p53 from binding to ZEB1-AS1 promoter region, causing transcriptional activation and upregulation of ZEB1-AS1. Moreover, enhanced ZEB1-AS1 could upregulate Nucleotide-Binding Oligomerization Domain 2 (NOD2) expression through recruiting leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) to stabilize NOD2 mRNA. Experimental data showed that knockdown of NOD2 or LRPPRC dramatically abrogated the functional role of ZEB1-AS1 in ox-LDL-induced endothelial cell injury. In summary, we demonstrated that lncRNA ZEB1-AS1 regulates the ox-LDL-induced endothelial cell injury via an LRPPRC-dependent mRNA stabilization mechanism. Therefore, ZEB1-AS1 may serve as a multi-potency target to overcome endothelial cell injury, atherosclerosis and other cardiovascular diseases.
Introduction
The dysfunction of endothelial cells (ECs), which is closely related with endothelial damage, play key roles in the initiation and progression of atherosclerosis (AS) (Blann, 1999). AS is the primary factor underlying cardiovascular and cerebrovascular diseases with high morbidity and mortality rates (Kanemitsu et al., 2011). Evidence has indicated that oxidized low-density lipoprotein (ox-LDL), are predominantly engulfed by macrophages, resulting in increased pro-inflammatory cytokine expression and further endothelial cell injury, thus contributing to progression of AS and other cardiovascular diseases (Chen et al., 2002). Although ox-LDL is a critical regulator during ECs injury, the precise regulatory mechanism is still not well known.
Long non-coding RNAs (lncRNAs) are a class of poor conserved endogenous RNAs longer than 200 nucleotides that do not encode proteins but regulate gene expression (Loewen et al., 2014). On a functional level, lncRNAs are involved in complex biological processes through diverse mechanisms. These comprise, among others, gene regulation by titration of transcription factors, alternative splicing, sponging of microRNAs, and recruitment of chromatin modifying enzymes (Kornienko et al., 2013; Fatica and Bozzoni, 2014; Nakagawa and Kageyama, 2014; Ni et al., 2017). Accumulating evidence indicates that lncRNAs play diverse roles in the AS and ECs dysfunction, such as MALAT1 (Gast et al., 2018), ANRIL (Holdt and Teupser, 2018), and H19 (Pan, 2017). Previously, we identified that lncRNA XIST (X-inactive specific transcript) regulates ox-LDL-mediated ECs injury through modulation of miR-320/NOD2 axis (Xu et al., 2018). However, functional potentials of other lncRNAs are waiting to be revealed.
The lncRNA ZEB1-antisense 1 (ZEB1-AS1, transcript ID: ENST00000607166.1) derives from the promoter region of ZEB1, a transcriptional factor that plays important roles in physiology and tumorigenesis (Li et al., 2016). As a famous epithelial-mesenchymal transition (EMT) promoter, ZEB1 plays important roles in ECs transition and cell adhesion (Gu et al., 2015). For example, ZEB1 could transcriptionally promote the expression of intercellular adhesion molecule-1 (ICAM-1), inducing the monocyte-macrophage adhesion and atherosclerotic lesion formation (Abou-Jaoude et al., 2018). However, whether lncRNA ZEB1-AS1 is involved in the ox-LDL-mediated ECs injury and AS, and if so, what is the underlying mechanism is still unknown.
In our previous study, we revealed the essential role of NOD2, which plays important roles in innate immunity by recognizing bacterial lipopolysaccharides (Caruso et al., 2014), in ox-LDL-induced ECs. Here, we sought to identify the functional role of ZEB1-AS1 in ox-LDL-mediated injury of human umbilical vein endothelial cells (HUVECs) and investigated its function by regulating NOD2 and interacting with leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC), which could promote RNA stability (Siira et al., 2017; Chen et al., 2019). We revealed that knockdown of ZEB1-AS1 alleviated the ox-LDL-induced ECs apoptosis. Mechanistically, ZEB1-AS1 guided LRPPRC to promote the stability of NOD2 mRNA, contributing to increase nucleotide-binding oligomerization domain 2 (NOD2) protein expression. Therefore, targeting ZEB1-AS1 could be a potential therapeutic strategy leading to lessen ECs injury related disease, such as AS.
Materials and Methods
Cell Culture and Reagents
HUVECs were purchased from American Tissue Culture Collection (ATCC, Manassas, VA, United States), and were cultured in Dulbecco’s modified Eagle (DMEM, Gibco, Carlsbad, CA, United States) medium with 10% fetal bovine serum (FBS) (Gibco BRL, Grand Island, NY, United States) at 37°C in 5% CO2. ox-LDL was purchased from Sigma-Aldrich (St. Louis, MO, United States) and used at the concentration of 100 μg/ml as previously described (Che et al., 2017; Xu et al., 2018).
Vector Construction and Cell Transduction
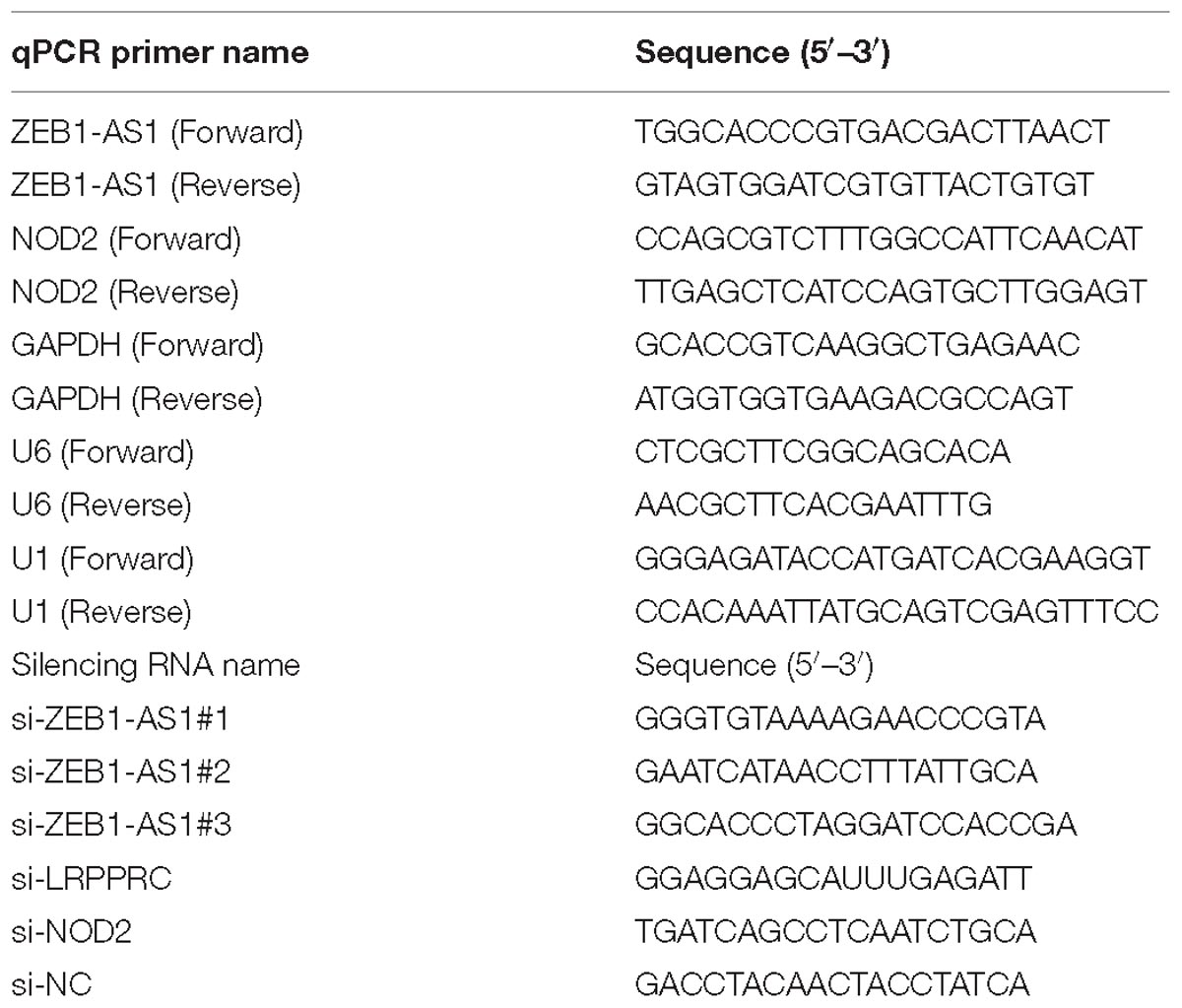
Short interfering RNA (siRNA) oligonucleotides targeting ZEB1-AS1, LRPPRC, NOD2 and negative control siRNAs were designed and purchased from GenePharma (Shanghai, China). The siRNA sequences are listed in Table 1. SiRNA transfections were performed using 75 nM siRNA and Lipofectamine RNAimax (Life Technologies, Waltham, MA, United States). The overexpression vector with full-length of ZEB1-AS1 was cloned into plasmids by GenePharma. The sequences for siRNAs were shown in Table 1.
Reverse Transcription (RT) and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from HUVECs by using the Qiagen RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Dnase I was used to eliminate the potential DNA pollutions. and RT and qPCR kits were used to evaluate the expression of target RNAs. RT (20 μl) reactions were performed using the PrimeScript®RT reagent kit (Takara, Dalian, China) and incubated for 30 min at 37°C followed by 5 s at 85°C. The quality of RT- samples were controlled by testing the density and purity. The 260/280 values should be between 1.8 and 2.0, and the density should be more than 200 μg/ml. For qPCR, 2 μl of diluted RT product was mixed with 23 μl reaction buffer provided by Takara (Takara Inc., Dalian, China) to a final volume of 25 μl. All reactions were carried out using an Eppendorf Mastercycler EP Gradient S (Eppendorf, Germany) under the following conditions: 95°C for 30 s followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The transcript expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for the normalization of detected RNAs using the comparative 2−ΔΔCq method (Wagner, 2013). The primer sequences for qPCR are presented in Table 1.
Rapid Amplification of cDNA Ends (RACE)
Both 5′ RACE and 3′ RACE were performed. In 3′ RACE, cDNAs were generated using an Oligo-dT primer that complemented the natural polyA tail of mRNAs. PCR was then used to amplify cDNA product from the 3′ end of ZEB1-AS1 with a sense specific primer and Oligo-dT primer. In 5′ RACE, an antisense gene specific primer was used to produce cDNA from the 5′ end of ZEB1-AS1. Next, a string of identical nucleotides (dATP) were added to the 3′ end of the cDNA. PCR was then carried out to amplify cDNA from the 5′ end using an antisense specific primer and Oligo-dT primer. We used a priming strategy in which both the 5′ and 3′ RACE reactions were primed using the same primer sequence, albeit reverse complemented, to ensure amplification of a contiguous long transcript.
Cell Viability Assay
The cell viability was assayed using the MTT Kit (Dojindo, Rockville, MD, United States). In brief, cells were seeded into a 96-well plate and then treated with ox-LDL for 24 h at the concentration of 100 μg/ml. After, cells were treated with the MTT reagent and further cultured for 2 h. The optical density at 450 nm was measured with a spectrophotometer (Thermo Electron Corporation, Waltham, MA, United States).
Flow Cytometry Apoptosis Assay
Cells were collected after ox-LDL treatment or transient transfection for 48 h, then washed with PBS and trypsin containing 0.025% trypsin-EDTA to get the single-cell suspensions. They were then fixed in ice-cold 70% ethanol and with Annexin-V FITC and propidium iodide (PI) solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Apoptosis was detected by on a BD FACSCalibur instrument.
TUNEL Assay
For TUNEL assay, cells cultured in chamber slide were fixed with 4% paraformaldehyde for 30 min. Then cells were stained with TUNEL kit on ice according to the manufacturer’s instructions (Vazyme, TUNEL Bright-Red Apoptosis Detection Kit, A113). TUNEL-positive cells were counted under fluorescence microscopy (DMI4000B, Leica, Mannheim, Germany).
Cytosolic/Nuclear Fraction and RNA Florescent in situ Hybridization (RNA FISH)
The cellular fraction was isolated to locate the sublocation of ZEB1-AS1. Briefly, 107 cells were harvested, resuspended in 1 ml of ice-cold RNase-free PBS, 1 ml of buffer C1 (1.28 M Sucrose, 40 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 4% Triton X-100) and 3 ml of RNase-free water, and incubated for 15 min on ice. The cells were then centrifuged for 15 min at 3,000 × g, and the supernatant containing the cytoplasmic constituents and the nuclear pellet were retained for RNA extraction. GAPDH was set as the positive control for cytoplasm, and U1 served as the positive controls for the nucleus.
For RNA FISH, HUVECs were seeded and fixed with 4% paraformaldehyde, treated with 0.5% Triton in PBS, followed by pre-hybridization. They were then hybridized with the biotin-labeled ZEB1-AS1 probe (5 μM) overnight at 42°C. The anti-biotin monoclonal antibody and Alexa Fluor®647-conjugated secondary antibody (Invitrogen, Shanghai, China) were used for detecting ZEB1-AS1. Cells were washed with PBS and then placed on cover slips with prolong gold antifade reagent containing DAPI (Cell Signaling Technology, Cambridge, MA, United States). Stained cells were observed under an Olympus microscope supporting a Hamamatsu 1394 ORCA-ERA video camera and the images were stored using Slidebook Digital Microscopy Software (Intelligent Imaging Innovations). The ZEB1-AS1 probes were synthesized by Sangon (Shanghai, China).
RNA Pulldown
The RNA pulldown assay was performed using a Magnetic RNA-Protein Pull-down Kit (Thermo Scientific) according to manufacturer’s instructions. HUVECs (2 × 107) were cross-linked for each hybridization reaction. The cell lysates were hybridized with a mixture of biotinylated DNA probes for 4 h at 37°C. The binding proteins were separated by electrophoresis.
RIP Assay and ChIP Assay
The RNA immunoprecipitation (RIP) was performed using the EZ-Magna RIP kit (Millipore, Burlington, MA, United States) according to the manufacturer’s instructions. Briefly, 107 cells were lysed with RIP lysis buffer using one freeze-thaw cycle. Cell extracts were co-immunoprecipitated with anti-LRPPRC (ab205022, Abcam, Cambridge, MA, United States) antibody and the retrieved RNA was subjected to qRT-PCR analysis. IgG antibody (EMD Millipore, cat. no. 12–371) was used as a negative control. For qRT-PCR analysis, GAPDH was used as a non-specific control.
An EZ-Magna ChIP kit (Millipore) was used for the ChIP assay according to the manufacturer’s protocol. Briefly, cells were treated with formaldehyde and incubated for 10 min to generate DNA-protein cross-links. Cell lysates were then sonicated to generate chromatin fragments of 200–300 bp and immunoprecipitated with p53 antibody (Abcam, cat. no. ab131442) or IgG antibody (EMD Millipore, cat. no. 12–371). RNA was recovered and analyzed by qPCR.
Immunofluorescence Assay
HUVECs were fixed in 4% formaldehyde for 15 min and then washed with PBS. The fixed cells were treated with pepsin and dehydrated through ethanol, and further permeabilized in Triton X100 (Sigma-Aldrich) for 20 min. Goat serum was used for blocked, and then cells were incubated with anti-NOD2 antibody (Abcam, ab36836, 1:500, Cambridge, MA, United States) for overnight at 4°C, and then incubated with the appropriate rhodamine-conjugated secondary antibody for 1 h at room temperature. The cells were then washed and incubated with DAPI (Invitrogen) for nuclear staining. The cells were visualized for immunofluorescence with a fluorescence microscopy (DMI4000B, Leica).
Western Blots and Antibodies
RIPA buffer (Sigma-Aldrich) was used to lyse the cells to obtain total protein lysates. Protein concentration was measured using the BCA method (Sigma-Aldrich). The quantified protein (25 μg) was transferred onto polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich) following SDS-PAGE gel electrophoresis. Then, the membrane was blocked with 5% non-fat dry milk in tri-buffered saline plus Tween (TBS-T) buffer for 2 h at room temperature and incubated with anti-LRPPRC antibody (1:1000, ab205022, Abcam), anti-NOD2 antibody (1:1000, ab36836, Abcam) or anti-GAPDH antibody (Invitrogen, cat. no. PA1-987) at 4°C overnight, followed by Horseradish peroxidase-conjugated (HRP) secondary antibody (1:5000, Abcam, cat. no. ab7090) at room temperature for 1 h.
Statistical Analysis
All in vitro experiments were performed using triple parallel samples and repeated triple times. Kolmogorov–Smirnov test was applied for data analysis with the distribution of each group samples. Data were presented as median (interquartile range). Mann–Whitney U-test was used for the comparison of datasets containing two groups. The Kruskal–Wallis test (post hoc Mann–Whitney U-test with Bonferroni’s) was used for analyzing statistical difference among multiple groups. A two-sided P < 0.05 was considered as statistical significance. Statistical analysis was performed using Prism 5 (GraphPad Software Inc., San Diego, CA, United States).
Results
Knockdown of lncRNA ZEB1-AS1 Alleviates ox-LDL-Induced Cell Injury in HUVECs
To investigate the functional role of ZEB1-AS1 in ox-LDL-induced HUVECs injury, we treated the HUVECs with 100 μg/ml ox-LDL. As shown in Figure 1A, ox-LDL treatment significantly suppressed cell viability of HUVECs at a time-dependent manner. Then, we characterized lncRNA ZEB1-AS1. 5- and 3-RACE assay was employed to determine the transcriptional start and termination sites of ZEB1-AS1. The full length of ZEB1-AS1 was shown to be of 2503 nt containing 1 exon, and located at Chr10: 31316528–31319030. We silenced ZEB1-AS1 in HUVECs by using specific silencing oligonucleotides (Figure 1B), and si-ZEB1-AS1#2 was used for further experiments. Knockdown of ZEB1-AS1 showed little effect on cell viability under normal condition and significantly increased cell viability when HUVECs was treated with ox-LDL (Figure 1C). Moreover, ox-LDL treatment (24 h) increased the release of LDH, however, this effect was dramatically reversed by knockdown of ZEB1-AS1 (Figure 1D). Flow cytometry analysis revealed that ox-LDL treatment increased HUVECs apoptosis, and ZEB1-AS1 silencing reversed this effect (Figure 1E). Similarly, TUNEL assay showed that knockdown of ZEB1-AS1 suppressed the ox-LDL-induced cell death (Figure 1F). Together, our results indicate that knockdown of ZEB1-AS1 could effectively abrogate the ox-LDL-induced HUVECs injury.
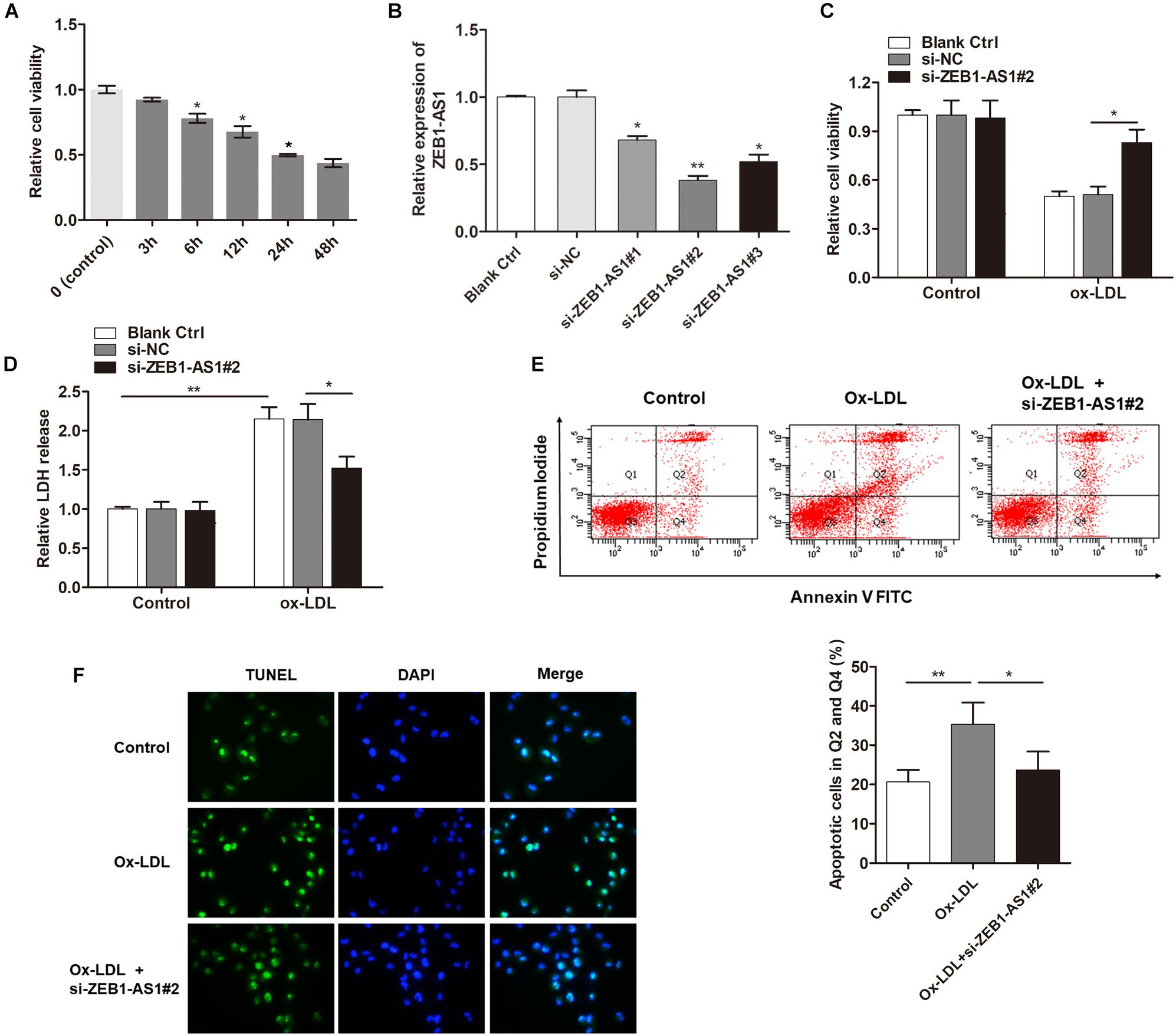
Figure 1. Silence of lncRNA ZEB1-AS1 abrogates oxidized-low density lipoprotein (ox-LDL)-induced cell injury in HUVECs. (A) HUVECs were treated with 100 μg/ml ox-LDL for respective time durations. MTT assay showed that ox-LDL treatment decreased cell viability, ∗P < 0.05 (Mann–Whitney U-test) was used for analyzing statistical difference among multiple groups compared to previous group. (B) qRT-PCR showed that ZEB1-AS1 was silenced by transfection of specific ZEB1-AS1 siRNAs (si-ZEB1-AS1#1, si-ZEB1-AS1#2, and si-ZEB1-AS1#3), ∗P < 0.05, ∗∗P < 0.01 (Kruskal–Wallis test) compared to si-NC and blank control cell group (without transfection). (C,D) HUVECs (treated with ox-LDL or not) were transfected with si-NC or si-ZEB1-AS1#2. Cell viability (C) and LDH release (D) were detected were measured by MTT assay and ELISA, respectively, ∗P < 0.05 (Mann–Whitney U-test), ∗∗P < 0.01 (Kruskal–Wallis test). (E) HUVECs apoptosis was detected by using flow cytometry after treatment with ox-LDL and/or knockdown of ZEB1-AS1 for 24 h. The data showed that ZEB1-AS1 knockdown reversed the apoptosis induced by ox-LDL treatment. ∗P < 0.05, ∗∗P < 0.01 (Mann–Whitney U-test). (F) Nuclear apoptosis was evaluated by using TUNEL assay in HUVECs treated with ox-LDL and/or si-ZEB1-AS1#2. ZEB1-AS1 knockdown reversed the apoptosis induced by ox-LDL treatment.
ox-LDL Upregulates lncRNA ZEB1-AS1 Expression via Modulating p53 Modification at ZEB1-AS1 Promoter Region
To investigate the underlying mechanism by which ZEB1-AS1 mediates ox-LDL-induced cell injury, we determined the regulation of ZEB1-AS1 expression by ox-LDL. When HUVECs were treated with 100 μg/ml ox-LDL, we found that ZEB1-AS1 expression was increased in a time-dependent manner (Figure 2A). To verify how ox-LDL regulates ZEB1-AS1 expression, we analyzed the promoter region of ZEB1-AS1 by using the online transcription factor prediction software, JASPAR. Two potential binding sites of p53 were verified at the promoter region of ZEB1-AS1 (Figure 2B). Then, p53 was overexpressed in HUVECs by transfection of specific p53 overexpression vector (Figure 2C). Overexpression of p53 significantly inhibited ZEB1-AS1 expression (Figure 2D), suggesting that p53 suppresses the transcription of ZEB1-AS1 gene. To prove the direct interaction between p53 and ZEB1-AS1, we performed ChIP experiment by using specific primers. As shown in Figure 2E, the antibody directed against p53 immunoprecipitated the DNA fragments containing the potential binding sites of p53 at ZEB1-AS1 promoter region. Moreover, treatment with ox-LDL significantly decreased the enrichment levels of p53 at ZEB1-AS1 promoter region (Figure 2F). Therefore, our results demonstrated that ox-LDL decreased the direct binding of p53 to the ZEB1-AS1 promoter region, which relieved the p53-induced suppressive effect on ZEB1-AS1 gene transcription.
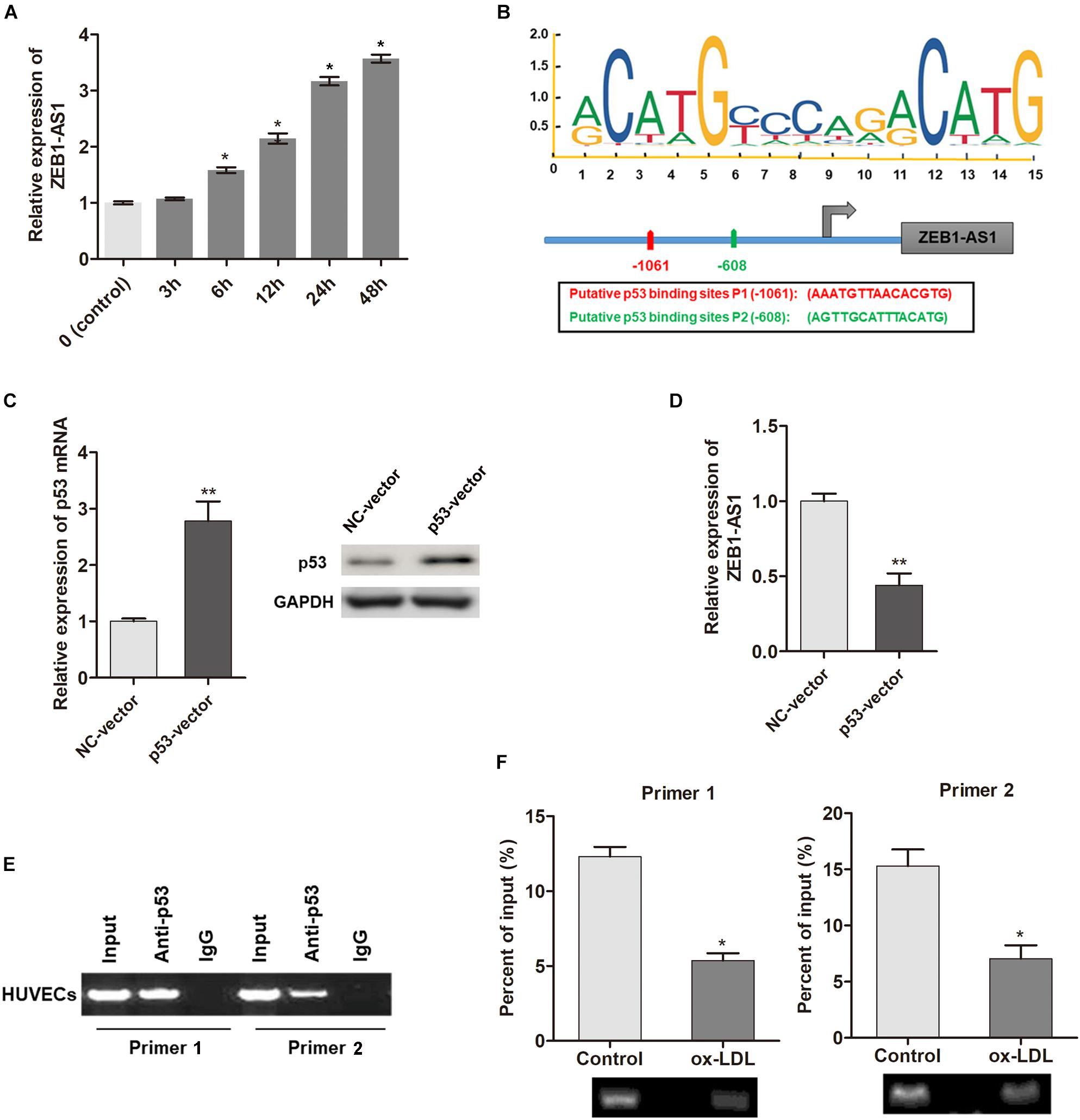
Figure 2. ox-LDL decreased p53 binding at ZEB1-AS1 promoter region, thereby activating the transcription of ZEB1-AS1. (A) The relative expression of ZEB1-AS1 was detected by qRT-PCR in HUVECs under normal control or ox-LDL condition, and ox-LDL treatment increased ZEB1-AS1 expression in a time-dependent manner. ∗P < 0.05 (Mann–Whitney U-test) compared to previous group. (B) Two binding sites of p53 (–1149, –534) at ZEB1-AS1 promoter were predicted by using online JASPAR software. (C) p53 was overexpressed in HUVECs by transfection of specific overexpression vector, ∗∗P < 0.01 (Mann–Whitney U-test). (D) Relative expression of ZEB1-AS1 was measured via qRT-PCR in cells overexpressed p53, ∗∗P < 0.01 (Mann–Whitney U-test). (E) ChIP assay was performed by using p53 antibody and two primers to detect the immunoprecipitations. (F) The enrichment of p53 at ZEB1-AS1 promoter region was evaluated via ChIP assay in cells treated with ox-LDL or not. The enrichment was significantly suppressed by ox-LDL treatment, ∗P < 0.05 (Mann–Whitney U-test).
ZEB1-AS1 Regulates ox-LDL-Induced ECs Injury via the Upregulation of NOD2 Level
Our previous study showed that NOD2 played a critical role in ECs injury, thus we assume ZEB1-AS1 may contribute to the ox-LDL-induced ECs injury through targeting NOD2. Immunofluorescence showed that ox-LDL treatment significantly increased the expression of NOD2 (Figure 3A). Western blot analysis further confirmed the upregulated NOD2 protein upon ox-LDL treatment (Figure 3B). Gain- or loss- functional assays showed that knockdown of ZEB1-AS1 significantly decreased NOD2 level at both transcript and protein levels (Figure 3C). Then, we tested the functional role of NOD2 in ox-LDL/ZEB1-AS1-mediated ECs injury by overexpression of ZEB1-AS1 and knockdown of NOD2 (Figures 3D,E). As shown in Figure 3F, enhanced expression of ZEB1-AS1 decreased HUVECs viability under ox-LDL treatment, however, this effect was significantly attenuated by NOD2 knockdown. Similarly, knockdown of NOD2 reversed the ZEB1-AS1-induced HUVECs apoptosis according to flow cytometry analysis (Figure 3G). These data showed that ZEB1-AS1 regulates ECs injury via the upregulation of NOD2.
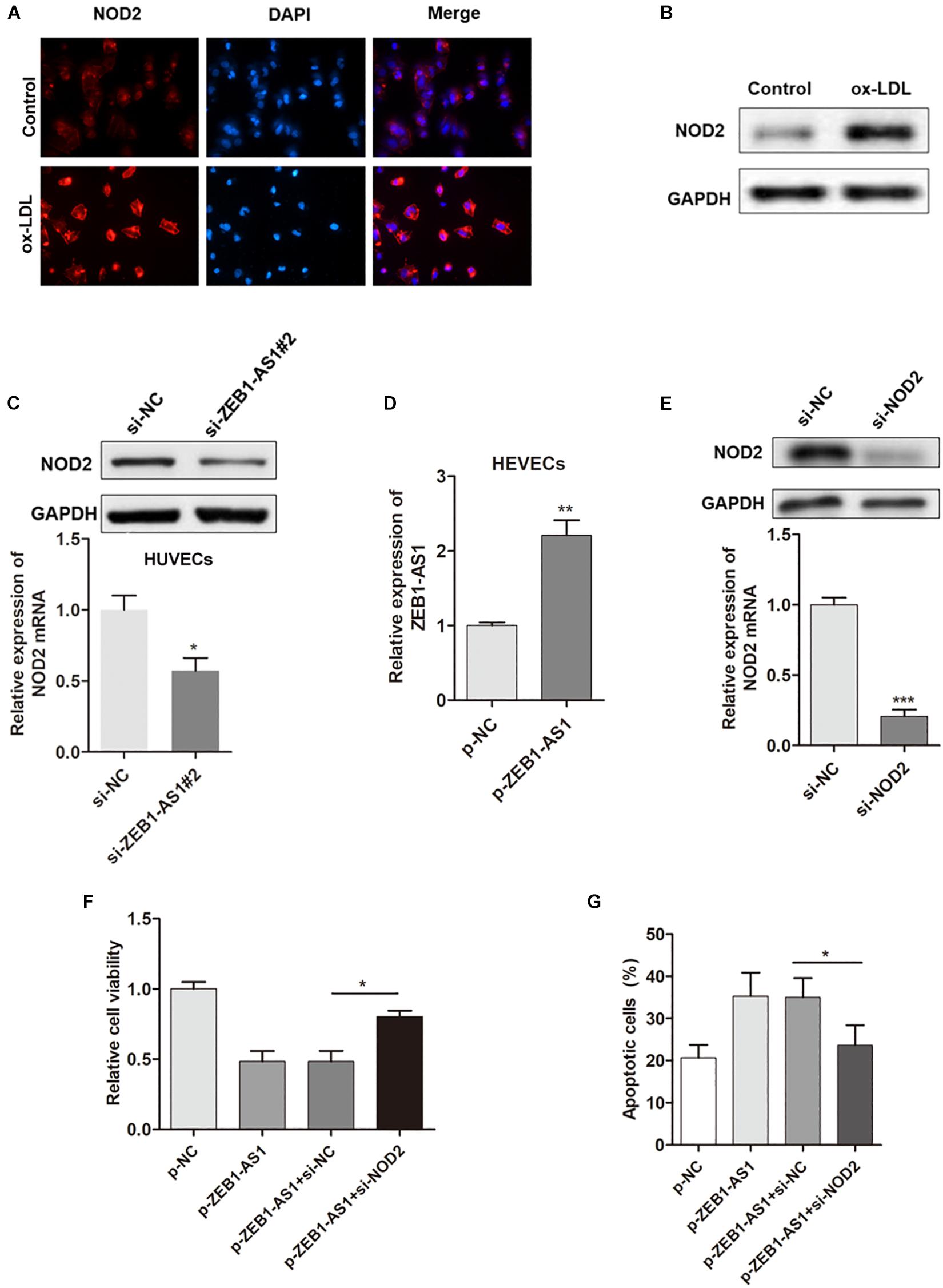
Figure 3. NOD2 was verified as a functional target of ZEB1-AS1 in ox-LDL-induced ECs injury. (A) The expression of NOD2 protein in HUVECs under normal or ox-LDL condition was assessed by using immunofluorescence. ox-LDL significantly increased NOD2 expression, ∗P < 0.05 (Mann–Whitney U-test). (B) Western blot analysis showed that ox-LDL treatment decreased NOD2 protein level. (C) The expression of NOD2 mRNA (left panel) and protein (right panel) was downregulated by ZEB1-AS1 knockdown, ∗P < 0.05 (Mann–Whitney U-test). (D) ZEB1-AS1 was overexpressed in HUVECs by transfection of ZEB1-AS1 overexpression plasmid, ∗∗P < 0.01 (Mann–Whitney U-test). (E) NOD2 was silenced in HUVECs at both transcript (left panel) and protein (right panel) levels by transfection of specific siRNA, ∗∗∗P < 0.001 (Mann–Whitney U-test). (F,G) Cell viability (F) and apoptosis (G) were evaluated by MTT assay and flow cytometry assay, respectively. Knockdown of NOD2 attenuated the functional effect induced by ZEB1-AS1 overexpression, ∗P < 0.05 (Mann–Whitney U-test).
ZEB1-AS1 Directly Interacts With LRPPRC to Play Key Roles in ECs Injury
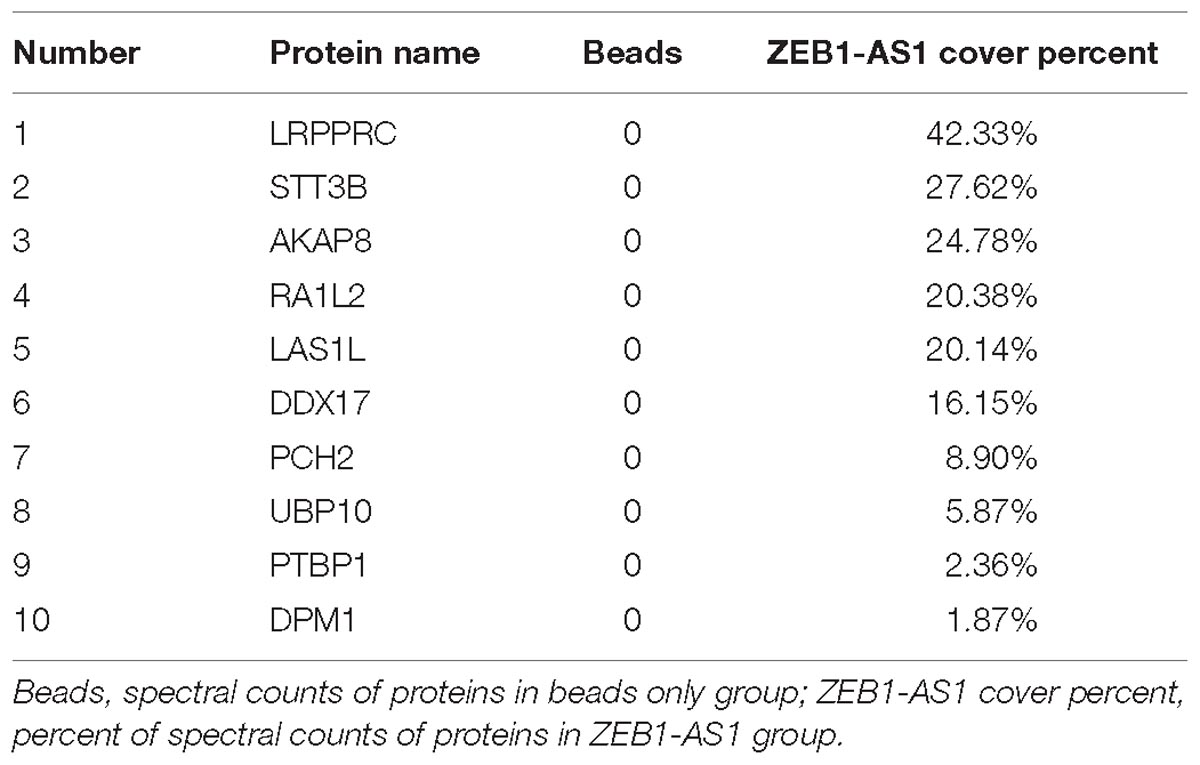
The subcellular localization of a lncRNA is associated closely with its biological mechanism. Cellular fractionation assays and RNA florescent in situ hybridization (RNA-FISH) showed that ZEB1-AS1 was distributed mainly in the cytoplasm in ECs cells (Figures 4A,B). lncRNAs located in the cytoplasm are usually associated with post-transcriptional regulation (Chen et al., 2017; Lu et al., 2018). To investigate the underlying mechanism by which ZEB1-AS1 regulates NOD2, we determined the second structure of ZEB1-AS1. Based on minimum free energy (MFE) and partition function1, we predicted that nucleotides 421–630 nt in the ZEB1-AS1 transcript, formed a stem-loop structure (Figure 4C), which is critical for the physical interaction with proteins. To identify ZEB1-AS1-interacting proteins in HUVECs, we performed RNA pulldown assays using in vitro transcribed biotinylated ZEB1-AS1 followed by mass spectrometry. A list of proteins were identified (Table 2). One overtly differential band (∼150 kDa) appeared after silver staining and was identified as leucine rich pentatricopeptide repeat containing (LRPPRC) by mass spectrometry (Figures 4D,E). Consistent with ZEB1-AS1 localization, LRPPRC was also distributed mainly in the cytoplasm in HUVECs (Figure 4F). We further confirmed the special interaction between ZEB1-AS1 and LRPPRC using RNA pull down and RNA immunoprecipitation (RIP) (Figures 4G,H). Our results suggest that ZEB1-AS1 directly interacts with LRPPRC in HUVECs.
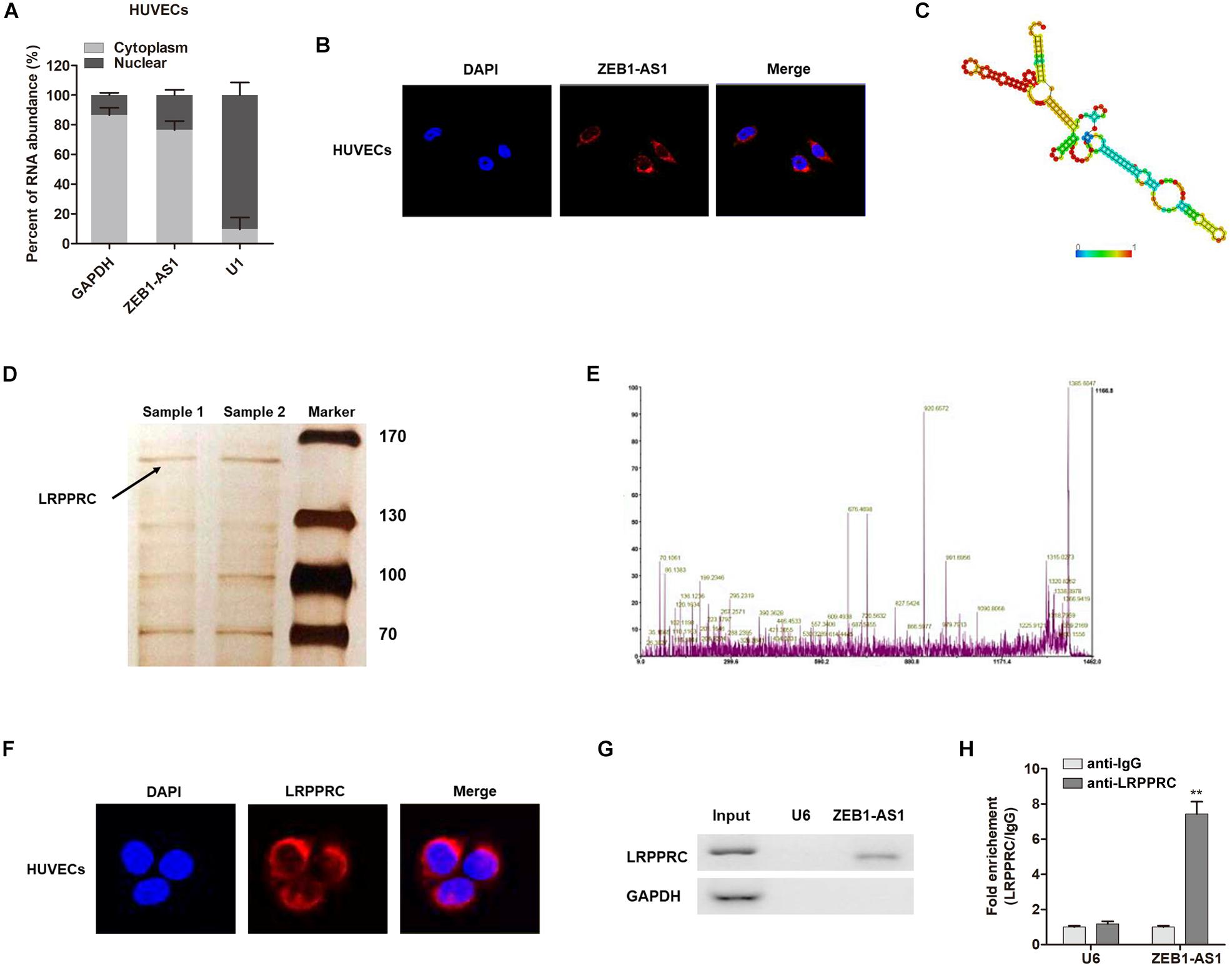
Figure 4. ZEB1-AS1 directly interacts with LRPPRC in ECs. (A). Nuclear fraction experiment and qRT-PCR detected the abundance of ZEB1-AS1 in the nucleus and cytoplasm. GAPDH was set as the positive control for cytoplasm, and U1 served as the positive controls for the nucleus. (B) The subcellular distribution of ZEB1-AS1 was visualized by RNA Fluorescent in situ hybridization (FISH) in HUVECs. ZEB1-AS1 was mainly distributed in cytoplasm of HUVECs. (C). Prediction of 421–630 nt ZEB1-AS1 structure was based on minimum free energy (MFE) and partition function (http://rna.tbi.univie.ac.at/). (D) RNA pulldown assay was performed using ZEB1-AS1 incubated with cytoplasm extracts of HUVECs, followed by silver staining. An arrow indicates LRPPRC. (E) Verification of LRPPRC protein by using mass spectrometry. (F) The subcellular distribution of LRPPRC (cytoplasm) was visualized by immunofluorescence in HUVECs. (G) The interaction between ZEB1-AS1 and LRPPRC was confirmed by RNA pulldown and western blotting. GAPDH served as negative controls. (H) RIP was performed using anti-LRPPRC and control IgG antibodies, followed by qRT-PCR to examine the enrichment of ZEB1-AS1 and U6. U6 served as negative controls. ∗∗P < 0.01 compared respective IgG group (Mann–Whitney U-test).
ZEB1-AS1 Enhances mRNA Stability of NOD2 via Recruiting LRPPRC
It is reported that LRPPRC play key roles in promoting mRNA stability (Siira et al., 2017; Chen et al., 2019). Hence, we assume that ZEB1-AS1 may guide LRPPRC to enhance the stabilization of NOD2 mRNA, thereby contributes to the ox-LDL-induced ECs injury. To prove this hypothesis, we silenced LRPPRC in HUVECs (Figure 5A) and found that LRPPRC knockdown eliminated the ZEB1-AS1-mediated upregulation of NOD2 expression in both mRNA and protein levels (Figure 5B). To determine whether ZEB1-AS1 directly interacts with NOD2 mRNA, we performed a biotinylated oligonucleotide pulldown assay. We found that endogenous NOD2 mRNA was co-precipitated with ZEB1-AS1 in HUVECs (Figure 5C). In addition, RIP assay revealed that NOD2 mRNA was enriched in LRPPRC precipitates, indicating that ZEB1-AS1 and LRPPRC could directly interact with NOD2 (Figure 5D). Furthermore, we treated HUVECs with actinomycin D (ActD), which allowed us to measure the decay of pre-existing mRNA. The data showed that knockdown of ZEB1-AS1 or LRPPRC resulted in a decreased half-life of NOD2 mRNA in HUVECs (Figure 5E), whereas overexpression of ZEB1-AS1 increased its half-life (Figure 5F). Moreover, knockdown of LRPPRC eliminated the increased half-life induced by overexpression of ZEB1-AS1 (Figure 5F). Taken together, our data indicated that ZEB1-AS1 guides LRPPRC to stabilize NOD2 mRNAs, contributing to its increased expression in HUVECs.
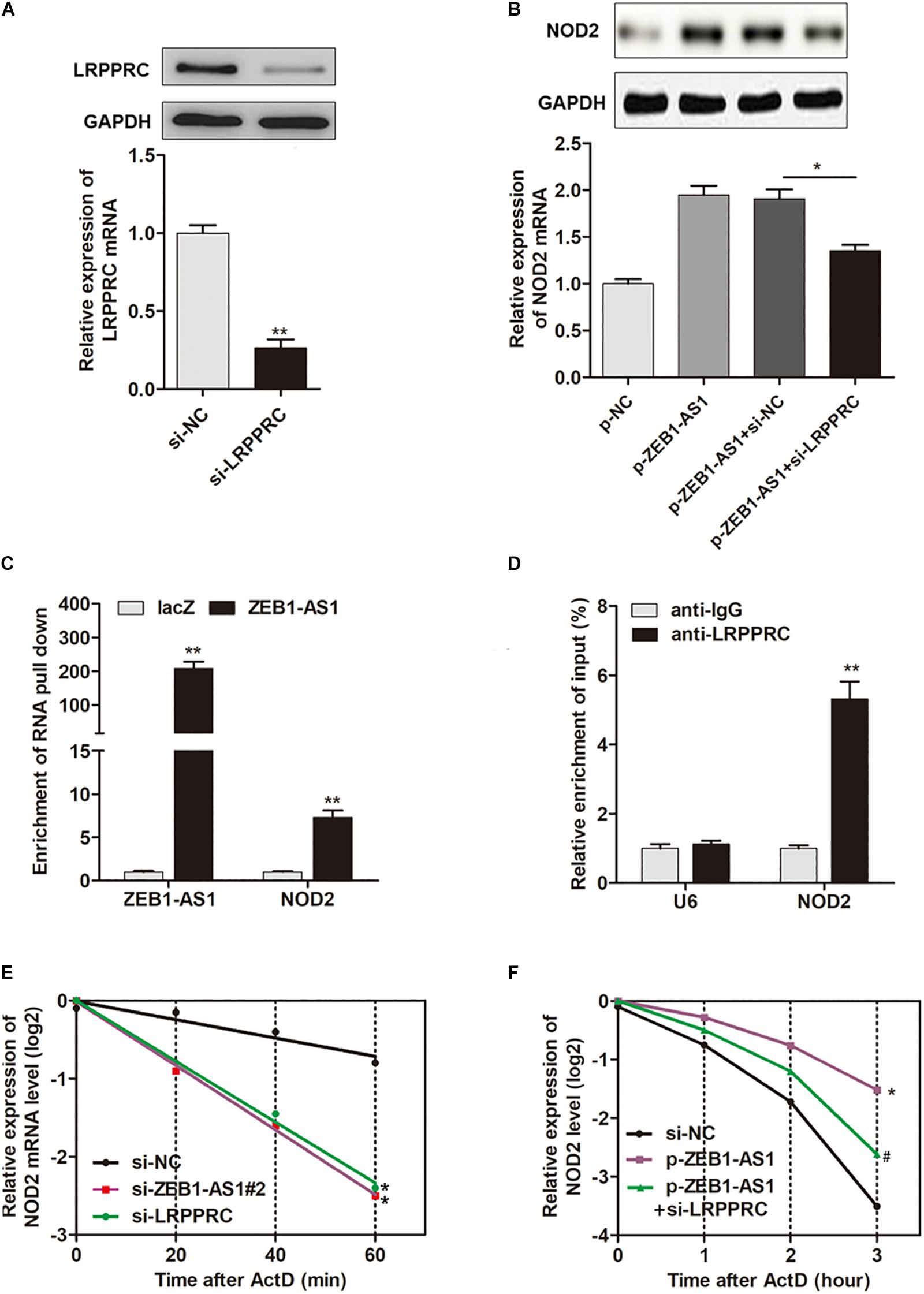
Figure 5. ZEB1-AS1 regulates mRNA stability of NOD2 via recruiting LRPPRC. (A) LRPPRC was silenced by specific siRNAs at both transcript (down panel) and protein (upper panel) levels, ∗∗P < 0.01 (Mann–Whitney U-test). (B) NOD2 expression was detected in HUVECs overexpressed with ZEB1-AS1 and/or silenced with LRPPRC, ∗P < 0.05 (Mann–Whitney U-test). (C) HUVECs lysates were incubated with in vitro synthesized, biotin-labeled control LacZ DNA probes or DNA probes against ZEB1-AS1 for the biotinylated oligonucleotide pulldown assay. The precipitates from the pulldown were analyzed by qRT-PCR to detect the interacting mRNAs. ∗∗P < 0.01 (Mann–Whitney U-test) compared to respective LacZ probes. (D) RIP was performed using anti-LRPPRC and control IgG antibodies, followed by qRT-PCR to examine the enrichment of NOD2 and U6. U6 served as negative controls. ∗∗P < 0.01 (Mann–Whitney U-test) compared to respective IgG controls. (E) HUVECs expressing si-ZEB1-AS1#2 or si-LRPPRC were treated with actinomycin D (5 μg/mL) for the indicated periods of time. ∗P < 0.05 (Mann–Whitney U-test) compared to si-NC group (F). HUVECs expressing si-NC, or p-ZEB1-AS1, or p-ZEB1-AS1 + si-LRPPRC were treated with actinomycin D (5 μg/mL) for the indicated periods of time. Total RNA was purified and then analyzed using qRT-PCR to examine the mRNA half-life of NOD2. ∗P < 0.05 (Mann–Whitney U-test) compared to si-NC group, and #P < 0.05 (Mann–Whitney U-test) compared to p-ZEB1-AS1 group.
ZEB1-AS1 Modulates ox-LDL-Induced ECs Injury by Stabilization of NOD2 mRNA
After having validated the direct interaction between ZEB1-AS1, LRPPRC, and NOD2, we sought to verify whether ZEB1- AS1 mediates ox-LDL-induced HUVECs injury by stabilization of NOD2. As expected, knockdown of LRPPRC increased cell viability and decreased cell apoptosis (Figures 6A,B). Moreover, silence of LRPPRC abrogated the effect on HUVECs injury induced by ZEB1-AS1 overexpression (Figures 6C,D). These results clearly revealed that ZEB1-AS1 modulates ox-LDL-induced ECs injury by stabilization of NOD2 mRNA.
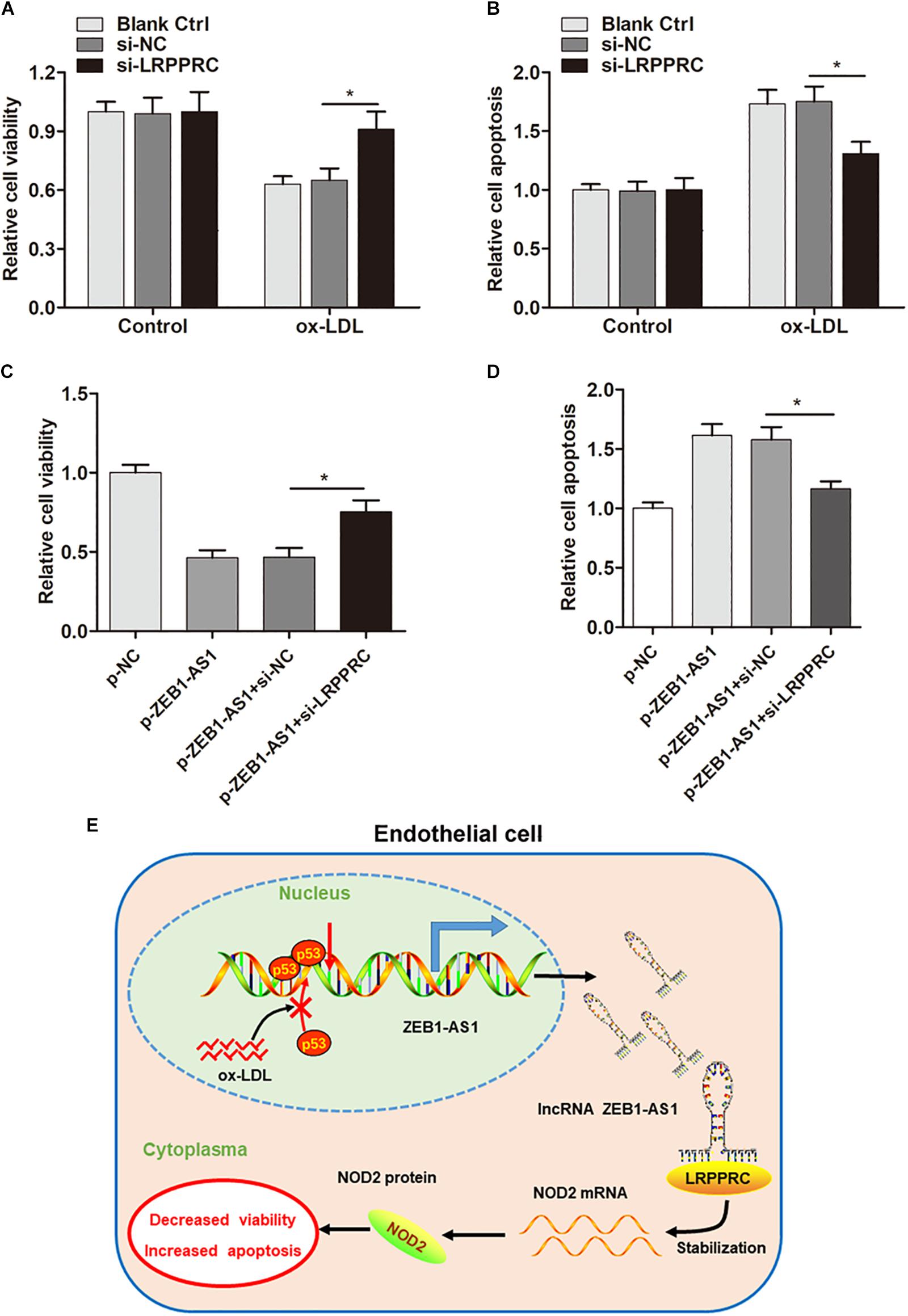
Figure 6. ZEB1-AS1 modulates ox-LDL-induced ECs injury by stabilization of NOD2 mRNA. (A,B) HUVECs (treated with ox-LDL or not) were transfected with si-NC or si-LRPPRC. Cell viability (A) and LDH release (B) were detected were measured by MTT assay and flow cytometry, respectively, ∗P < 0.05 (Mann–Whitney U-test). (C,D) Cell viability (C) and apoptosis (D) were evaluated by MTT assay and flow cytometry assay, respectively, ∗P < 0.05 (Mann–Whitney U-test). (E) A scheme of the proposed mechanisms. ox-LDL treatment increases ZEB1-AS1 expression by sequestering p53 from binding to the promoter region of ZEB1-AS1 gene, which activates the transcription and upregulates ZEB1-AS1 level in HUVECs. Overexpression of ZEB1-AS1 upregulates NOD2 level by guiding LRPPRC to stabilize its mRNA, resulting in decreased viability and increased apoptosis of ECs.
Overall, our study demonstrated that ox-LDL could upregulate ZEB1-AS1 expression level in ECs, which thereby promoted ECs injury by post-transcriptionally stabilization of NOD2 mRNA in an LRPPRC-dependent manner (Figure 6E), suggesting that ZEB1-AS1 may be a promising therapeutic target for patients with AS or ECs injury–related diseases.
Discussion
Numerous studies in recent years have helped to gain a better understanding of the molecular mechanisms during ECs injury and AS progression (Sumiyoshi and Asada, 1988; Pech-Amsellem et al., 1996; Li et al., 2017). However, the specific regulatory model is still largely unknown, and it is of much importance to discover new molecular signatures which may be useful for AS prevention and therapy. Our previous study revealed the functional role of lncRNA XIST in ox-LDL-induced cell injury (Xu et al., 2018). However, the roles of lncRNAs in ox-LDL-induced ECs injury is still largely unknown. In this study, we found that another lncRNA, ZEB1-AS1, mediated ox-LDL-induced ECs injury. Mechanistically, ox-LDL upregulated ZEB1-AS1 level via decreasing p53 modification at the promoter region of ZEB1-AS1 gene. ZEB1-AS1 could enhance the expression of NOD2 by post-transcriptionally stabilization of NOD2 mRNA dependent on LRPPRC.
lncRNAs are reported to regulate biological functions via diverse mechanism: guider; decoy; scaffold effect on DNA, RNA or protein; and post-transcriptional effects (Ulitsky and Bartel, 2013). The expression of ZEB1-AS1 has been reported and was identified as an oncogene in several cancer types, such as gastric cancer (Xu et al., 2017), prostate cancer (Su et al., 2017), colorectal cancer (Xiong et al., 2018), and glioma (Lv et al., 2016). In addition, one study by Wang et al. (2018) demonstrated that lncRNA ZEB1-AS1 could promote renal fibrosis in diabetic nephropathy. However, the functional role of ZEB1-AS1 in AS or other ECs injury-related disease is not reported. Li et al. (2017) reported the length of ZEB1-AS1 was 2,449 bp, lacking more in-depth data (Caruso et al., 2014). The discrepancies above might be explained by different cell lines used in different studies or different splicing variants of ZEB1-AS1. To the best of our knowledge, this is the first study to systematically evaluate the role of ZEB1-AS1 in ECs injury and the underlying regulatory mechanism. We found that ZEB1-AS1 is critical for ox-LDL-induced cell injury and may serve as a therapeutic target.
We investigated the underlying mechanism by which ox-LDL regulates ZEB1-AS1 expression during ECs injury. Recent studies revealed that activation or inhibition of transcription via the various modifications at gene promoters is the major factor that controls gene expression in a temporal and spatial manner, resulting in the establishment and maintenance of biological changes (Piunti and Pasini, 2011). Since ZEB1-AS1 is reported to be suppressed by p53 (Wang et al., 2018), we investigated whether ox-LDL influence the expression of ZEB1-AS1 by regulation of p53 binding. Luckily, we identified that p53 was associated with ZEB1-AS1, resulting a suppression of ZEB1-AS1 transcription, which is consistent with the previous report. More importantly, treatment with ox-LDL could dramatically decrease the binding level of p53, which finally released the activation of ZEB1-AS1.
The subcellular localization of lncRNA is associated closely with its biological mechanism. Our results showed that ZEB1-AS1 was distributed mainly in the cytoplasm in ECs, indicating it may regulate ECs injury at the post-transcriptional level. Previously, we reported that NOD2 is critical for ECs cell apoptosis and AS progression (Liu et al., 2013; Yuan et al., 2013; Kim, 2015). NOD2, a major pattern recognition receptor integrating ER stress and inflammation, is essential for antimicrobial innate immunity and tissue homeostasis (Liu et al., 2013). Interactions between pattern recognition receptors shape innate immune responses to particular classes of pathogens, such as ECs injury and AS (Oh et al., 2005), which may account for the important function of NOD2 in ECs injury. Our study further verified the regulation of NOD2 by ZEB1-AS1 to affect the initiation and progression of ECs injury. More importantly, ZEB1-AS1 directly bind to LRPPRC to influence the mRNA stability of NOD2. LRPPRC, an RNA binding protein, regulates mRNA stability and polyadenylation mainly in mitochondria, but also in the cytoplasm and nucleus (Tsuchiya et al., 2004; Chujo et al., 2012; Ruzzenente et al., 2012; Volpon et al., 2017). We found that both ZEB1-AS1 and LRPPRC were distributed mainly in the cytoplasm in HUVECs. Knockdown ZEB1-AS1 or LRPPRC resulted in a decrease in the half-life of NOD2 in HUVECs. Overall, we identified a novel mechanism by which ZEB1-AS1 guides LRPPRC to stabilize NOD2 mRNA, which increases NOD2 protein level and enhances ox-LDL-induced ECs injury.
Recently, small molecules designed to target the folded structure of pathologic non-coding RNA have shown significant anti-disease effects in vivo by selectively modulating non-coding RNAs in lesion cells, such as cancer (Velagapudi et al., 2016). Additionally, siRNA or locked nucleic acids can serve as therapeutic agents that specifically target lncRNAs in vivo (Disney and Angelbello, 2016). In the future, inhibition of ECs injury and AS progression via small molecules that specifically target ZEB1-AS1 might represent potential therapeutic methods.
Conclusion
We report the discovery that ZEB1-AS1 functionally participates in ox-LDL-induced ECs injury via LRPPRC-mediated stabilization of NOD2. Uncovering the precise role of ZEB1-AS1/LRPPRC/NOD2 pathway in the progression of ox-LDL-induced ECs death and AS will not only increase our knowledge of ox-LDL-induced AS, but also enable the development of novel therapeutic strategies to overcome oxidation product-induced diseases.
Author Contributions
XX and CL designed and mainly did the study. CM, ZD, and YD helped and did the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abou-Jaoude, A., Badique, L., Mlih, M., Awan, S., Guo, S., Lemle, A., et al. (2018). Loss of the adaptor protein shca in endothelial cells protects against monocyte macrophage adhesion, LDL-oxydation, and atherosclerotic lesion formation. Sci. Rep. 8:4501. doi: 10.1038/s41598-018-22819-3
Blann, A. D. (1999). Endothelial cell damage and the development or progression of atherosclerosis. Clin. Sci. 97, 119–121. doi: 10.1042/cs0970119
Caruso, R., Warner, N., Inohara, N., and Nunez, G. (2014). NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908. doi: 10.1016/j.immuni.2014.12.010
Che, J., Liang, B., Zhang, Y., Wang, Y., Tang, J., and Shi, G. (2017). Kaempferol alleviates ox-LDL-induced apoptosis by up-regulation of autophagy via inhibiting PI3K/Akt/mTOR pathway in human endothelial cells. Cardiovasc. Pathol. 31, 57–62. doi: 10.1016/j.carpath.2017.08.001
Chen, D. L., Lu, Y. X., Zhang, J. X., Wei, X. L., Wang, F., Zeng, Z. L., et al. (2017). Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics 7, 4836–4849. doi: 10.7150/thno.20942
Chen, M., Masaki, T., and Sawamura, T. (2002). LOX-1, the receptor for oxidized low-density lipoprotein identified from endothelial cells: implications in endothelial dysfunction and atherosclerosis. Pharmacol. Ther. 95, 89–100. doi: 10.1016/s0163-7258(02)00236-x
Chen, Z., Chen, X., Xie, R., Huang, M., Dong, W., Han, J., et al. (2019). DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing il-11-stat3 signaling and ccnd1 expression. Mol. Ther. 27, 326–341. doi: 10.1016/j.ymthe.2018.12.015
Chujo, T., Ohira, T., Sakaguchi, Y., Goshima, N., Nomura, N., Nagao, A., et al. (2012). LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 40, 8033–8047. doi: 10.1093/nar/gks506
Disney, M. D., and Angelbello, A. J. (2016). Rational design of small molecules targeting oncogenic noncoding rnas from sequence. Acc. Chem. Res. 49, 2698–2704. doi: 10.1021/acs.accounts.6b00326
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Gast, M., Rauch, B. H., Nakagawa, S., Haghikia, A., Jasina, A., Haas, J., et al. (2018). Immune system-mediated atherosclerosis caused by deficiency of long noncoding RNA MALAT1 in ApoE-/- mice. Cardiovasc. Res. 115, 302–314. doi: 10.1093/cvr/cvy202
Gu, K., Li, M. M., Shen, J., Liu, F., Cao, J. Y., Jin, S., et al. (2015). Interleukin-17-induced EMT promotes lung cancer cell migration and invasion via NF-kappaB/ZEB1 signal pathway. Am. J. Cancer Res. 5, 1169–1179.
Holdt, L. M., and Teupser, D. (2018). Long noncoding RNA ANRIL: lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front. Cardiovasc. Med. 5:145. doi: 10.3389/fcvm.2018.00145
Kanemitsu, S., Tanabe, S., Ohue, K., Miyagawa, H., Miyake, Y., and Okabe, M. (2011). Improve morbidity and mortality in coronary artery bypass graft surgery for severe atherosclerosis. Ann. Vasc. Dis. 4, 93–98. doi: 10.3400/avd.oa.10.01044
Kim, H. J. (2015). Role of nucleotide-binding and oligomerization domain 2 protein (NOD2) in the development of atherosclerosis. Korean J. Physiol. Pharmacol. 19, 479–484. doi: 10.4196/kjpp.2015.19.6.479
Kornienko, A. E., Guenzl, P. M., Barlow, D. P., and Pauler, F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. doi: 10.1186/1741-7007-11-59
Li, A., Peng, W., Xia, X., Li, R., Wang, Y., and Wei, D. (2017). Endothelial-to-mesenchymal transition: a potential mechanism for atherosclerosis plaque progression and destabilization. DNA Cell Biol. 36, 883–891. doi: 10.1089/dna.2017.3779
Li, T., Xie, J., Shen, C., Cheng, D., Shi, Y., Wu, Z., et al. (2016). Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 35, 1575–1584. doi: 10.1038/onc.2015.223
Liu, H. Q., Zhang, X. Y., Edfeldt, K., Nijhuis, M. O., Idborg, H., Back, M., et al. (2013). NOD2-mediated innate immune signaling regulates the eicosanoids in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 33, 2193–2201. doi: 10.1161/ATVBAHA.113.301715
Loewen, G., Jayawickramarajah, J., Zhuo, Y., and Shan, B. (2014). Functions of lncRNA hotair in lung cancer. J. Hematol. Oncol. 7:90. doi: 10.1186/s13045-014-0090-4
Lu, Q. C., Rui, Z. H., Guo, Z. L., Xie, W., Shan, S., and Ren, T. (2018). LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J. Cell. Mol. Med. 22, 1527–1537. doi: 10.1111/jcmm.13420
Lv, Q. L., Hu, L., Chen, S. H., Sun, B., Fu, M. L., Qin, C. Z., et al. (2016). A long noncoding rna zeb1-as1 promotes tumorigenesis and predicts poor prognosis in glioma. Int. J. Mol. Sci. 17:E1431. doi: 10.3390/ijms17091431
Nakagawa, S., and Kageyama, Y. (2014). Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochim. Biophys. Acta 1839, 215–222. doi: 10.1016/j.bbagrm.2013.10.009
Ni, W., Zhang, Y., Zhan, Z., Ye, F., Liang, Y., Huang, J., et al. (2017). A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J. Hematol. Oncol. 10:91. doi: 10.1186/s13045-017-0449-4
Oh, H. M., Lee, H. J., Seo, G. S., Choi, E. Y., Kweon, S. H., Chun, C. H., et al. (2005). Induction and localization of NOD2 protein in human endothelial cells. Cell Immunol. 237, 37–44. doi: 10.1016/j.cellimm.2005.09.006
Pan, J. X. (2017). LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 322–328.
Pech-Amsellem, M. A., Myara, I., Storogenko, M., Demuth, K., Proust, A., and Moatti, N. (1996). Enhanced modifications of low-density lipoproteins (LDL) by endothelial cells from smokers: a possible mechanism of smoking-related atherosclerosis. Cardiovasc. Res. 31, 975–983. doi: 10.1016/0008-6363(96)00059-4
Piunti, A., and Pasini, D. (2011). Epigenetic factors in cancer development: polycomb group proteins. Future Oncol. 7, 57–75. doi: 10.2217/fon.10.157
Ruzzenente, B., Metodiev, M. D., Wredenberg, A., Bratic, A., Park, C. B., Camara, Y., et al. (2012). LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456. doi: 10.1038/emboj.2011.392
Siira, S. J., Spahr, H., Shearwood, A. J., Ruzzenente, B., Larsson, N. G., Rackham, O., et al. (2017). LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 8:1532. doi: 10.1038/s41467-017-01221-z
Su, W., Xu, M., Chen, X., Chen, N., Gong, J., Nie, L., et al. (2017). Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer 16:142. doi: 10.1186/s12943-017-0711-y
Sumiyoshi, A., and Asada, Y. (1988). Endothelial injury as an initiating factor: mechanism and significance of endothelial cell injury in the etiology of atherosclerosis. Nihon Rinsho 46, 515–519.
Tsuchiya, N., Fukuda, H., Nakashima, K., Nagao, M., Sugimura, T., and Nakagama, H. (2004). LRP130, a single-stranded DNA/RNA-binding protein, localizes at the outer nuclear and endoplasmic reticulum membrane, and interacts with mRNA in vivo. Biochem. Biophys. Res. Commun. 317, 736–743. doi: 10.1016/j.bbrc.2004.03.103
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
Velagapudi, S. P., Cameron, M. D., Haga, C. L., Rosenberg, L. H., Lafitte, M., Duckett, D. R., et al. (2016). Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 113, 5898–5903. doi: 10.1073/pnas.1523975113
Volpon, L., Culjkovic-Kraljacic, B., Sohn, H. S., Blanchet-Cohen, A., Osborne, M. J., and Borden, K. L. B. (2017). A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 23, 927–937. doi: 10.1261/rna.060137.116
Wagner, E. M. (2013). Monitoring gene expression: quantitative real-time rt-PCR. Methods Mol. Biol. 1027, 19–45. doi: 10.1007/978-1-60327-369-5_2
Wang, J., Pang, J., Li, H., Long, J., Fang, F., Chen, J., et al. (2018). lncRNA ZEB1-AS1 Was Suppressed by p53 for renal fibrosis in diabetic nephropathy. Mol. Ther. Nucleic Acids 12, 741–750. doi: 10.1016/j.omtn.2018.07.012
Xiong, W. C., Han, N., Wu, N., Zhao, K. L., Han, C., Wang, H. X., et al. (2018). Interplay between long noncoding RNA ZEB1-AS1 and miR-101/ZEB1 axis regulates proliferation and migration of colorectal cancer cells. Am. J. Transl. Res. 10, 605–617.
Xu, T. P., Wang, Y. F., Xiong, W. L., Ma, P., Wang, W. Y., Chen, W. M., et al. (2017). E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 8:e2837. doi: 10.1038/cddis.2017.205
Xu, X., Ma, C., Liu, C., Duan, Z., and Zhang, L. (2018). Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem. Biophys. Res. Commun. 503, 586–592. doi: 10.1016/j.bbrc.2018.06.042
Keywords: endothelial dysfunction, oxidative low-density lipoprotein, ZEB1-AS1, nucleotide-binding oligomerization domain 2, leucine rich pentatricopeptide repeat containing
Citation: Xu X, Ma C, Duan Z, Du Y and Liu C (2019) lncRNA ZEB1-AS1 Mediates Oxidative Low-Density Lipoprotein-Mediated Endothelial Cells Injury by Post-transcriptional Stabilization of NOD2. Front. Pharmacol. 10:397. doi: 10.3389/fphar.2019.00397
Received: 11 January 2019; Accepted: 29 March 2019;
Published: 16 April 2019.
Edited by:
Francesco Squadrito, University of Messina, ItalyReviewed by:
Shizuka Uchida, University of Louisville, United StatesMatthew A. Nystoriak, University of Louisville, United States
Simona Greco, Policlinico San Donato (IRCCS), Italy
Copyright © 2019 Xu, Ma, Duan, Du and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Liu, xuxiaohui0a@163.com
 Xiaohui Xu
Xiaohui Xu Chao Liu
Chao Liu