
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 19 February 2019
Sec. Neuropharmacology
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.00083
This article is part of the Research Topic Novel Approaches to the Neuropharmacology of Mood Disorders View all 11 articles
 Marin Veldic1*
Marin Veldic1* Ahmed T. Ahmed1
Ahmed T. Ahmed1 Caren J. Blacker1
Caren J. Blacker1 Jennifer R. Geske2
Jennifer R. Geske2 Joanna M. Biernacka1,2
Joanna M. Biernacka1,2 Kristin L. Borreggine1
Kristin L. Borreggine1 Katherine M. Moore1
Katherine M. Moore1 Miguel L. Prieto3,4
Miguel L. Prieto3,4 Jennifer L. Vande Voort1
Jennifer L. Vande Voort1 Paul E. Croarkin1
Paul E. Croarkin1 Astrid A. Hoberg1
Astrid A. Hoberg1 Simon Kung1
Simon Kung1 Renato D. Alarcon1,5
Renato D. Alarcon1,5 Nicola Keeth1
Nicola Keeth1 Balwinder Singh1
Balwinder Singh1 William V. Bobo1
William V. Bobo1 Mark A. Frye1
Mark A. Frye1Background: Pharmacogenomic testing, specifically for pharmacokinetic (PK) and pharmacodynamic (PD) genetic variation, may contribute to a better understanding of baseline genetic differences in patients seeking treatment for depression, which may further impact clinical antidepressant treatment recommendations. This study evaluated PK and PD genetic variation and the clinical use of such testing in treatment seeking patients with bipolar disorder (BP) and major depressive disorder (MDD) and history of multiple drug failures/treatment resistance.
Methods: Consecutive depressed patients evaluated at the Mayo Clinic Depression Center over a 10-year study time frame (2003–2013) were included in this retrospective analysis. Diagnoses of BP or MDD were confirmed using a semi-structured diagnostic interview. Clinical rating scales included the Hamilton Rating Scale for Depression (HRSD24), Generalized Anxiety Disorder 7-item scale (GAD-7), Patient Health Questionnaire-9 (PHQ-9), and Adverse Childhood Experiences (ACE) Questionnaire. Clinically selected patients underwent genotyping of cytochrome P450 CYP2D6/CYP2C19 and the serotonin transporter SLC6A4. PK and PD differences and whether clinicians incorporated test results in providing recommendations were compared between the two patient groups.
Results: Of the 1795 patients, 167/523 (31.9%) with BP and 446/1272 (35.1%) with MDD were genotyped. Genotyped patients had significantly higher self-report measures of depression and anxiety compared to non-genotyped patients. There were significantly more CYP2C19 poor metabolizer (PM) phenotypes in BP (9.3%) vs. MDD patients (1.7%, p = 0.003); among participants with an S-allele, the rate of CYP2C19 PM phenotype was even higher in the BP (9.8%) vs. MDD (0.6%, p = 0.003). There was a significant difference in the distribution of SLC6A4 genotypes between BP (l/l = 28.1%, s/l = 59.3%, s/s = 12.6%) and MDD (l/l = 31.4%, s/l = 46.1%, s/s = 22.7%) patients (p < 0.01).
Conclusion: There may be underlying pharmacogenomic differences in treatment seeking depressed patients that potentially have impact on serum levels of CYP2C19 metabolized antidepressants (i.e., citalopram / escitalopram) contributing to rates of efficacy vs. side effect burden with additional potential risk of antidepressant response vs. induced mania. The evidence for utilizing pharmacogenomics-guided therapy in MDD and BP is still developing with a much needed focus on drug safety, side effect burden, and treatment adherence.
In 2018, the World Health Organization (WHO) identified major depressive disorder (MDD) and bipolar disorder (BP) as leading causes of disability worldwide, negatively impacting over 360 million people (Whiteford et al., 2013; Vos et al., 2015; Ferrari et al., 2016; World Health Organization, 2018). While genetic factors are thought to contribute 59–85% to BP risk (McGuffin et al., 2003; Lichtenstein et al., 2009), and 31–42% to MDD risk (Sullivan et al., 2000) or shared genetic risk related to overlapping symptoms of bipolar and major depressive disorder (Lee et al., 2013; Doherty and Owen, 2014), there is less systematic research focused on pharmacokinetic (PK) and/or pharmacodynamic (PD) genetic variation in these two distinct patient groups. This may be of potential interest recognizing marked differences in rates of antidepressant response and antidepressant induced mania (AIM+) by diagnostic group (Frye et al., 2015).
Selective serotonin reuptake inhibitors (SSRIs) are now considered first-line treatment for MDD (Crismon et al., 1999; Anderson et al., 2008), but only an approximate 50% of patients with MDD achieve partial remission, and only 30% complete remission, with SSRI therapy (Rush et al., 2006). However, antidepressants have less evidence base in bipolar depression and may in fact contribute to mood destabilization (Frye, 2011; Sidor and Macqueen, 2011). PK and PD genetic variation (i.e., pharmacogenomics) may contribute to BP and MDD treatment-resistance (Porcelli et al., 2012).
The use of pharmacogenomics testing for mental illnesses therapy selection has increased (Drozda et al., 2014). Thus, the implementing of pharmacogenomics-guided recommendations may improve treatment outcomes for patient with treatment-resistance depression (Kung and Li, 2010; Rundell et al., 2011b). Several studies have shown improvement in antidepressant response rates associated with the use of pharmacogenomic testing in clinical settings (Hall-Flavin et al., 2012; Hall-Flavin et al., 2013; Winner et al., 2013) and a recent meta-analysis of four randomized controlled trials and two open label trials have shown the same results (Rosenblat et al., 2018). However, several other reports (Rosenblat et al., 2017; Zeier et al., 2018; Zubenko et al., 2018) have identified potential limitations of industry support and lack of blinding and control groups (Hall-Flavin et al., 2012, 2013).
The goal of this study was to assess the outcomes of PK and PD genetic variation in treatment seeking depressed patients with history of multiple drug failures/treatment resistance and assess results of genomic testing on subsequent treatment recommendations. We assessed the clinical value of pharmacogenomic testing examining the differences in psychometrics mean scores at baseline between genotyped and non-genotyped patients; and assessed the relationships between PK (CYP2D6 and CYP2C19) or PD (SLC6A4) genetic variations, and MDD/BP severity scales in pharmacogenomically-tested vs. not tested patients.
This study was approved by the Mayo Clinic Institutional Board. All participants provided written informed consent prior to enrollment, evaluation and blood draw in the Mayo Clinic Depression Center.
This was a naturalistic study. A consecutive sample of treatment-seeking adults (age 18–65) with a clinical diagnosis of MDD or BP, currently in a depressive episode, was recruited from the Mayo Clinic Depression Center between February 26, 2003 and March 27, 2013. Clinical diagnoses were confirmed by DSM-IV-TR Structured Clinical Diagnostic Interview (SCID). Inclusion criteria were based on patients who presented with long history of multiple drug failures or treatment resistance. Exclusion criteria were inability to provide written informed consent, other Axis I or II diagnoses that by clinical judgment were the main reason for seeking treatment, substance use disorder determined by clinical interview, and (+) drug screen (except nicotine and caffeine) were excluded. Data were abstracted from Electronic health record (EHR) by two reviewers (Caren J. Blacker and Kristin L. Borreggine). For data abstraction validation, 10% of the abstracted data was reviewed by Marin Veldic.
Psychometrics utilized in the consultation included the 24 item Hamilton Rating Scale for Depression (HRSD24) (Hamilton, 1960), Patient Health Questionnaire-9 (PHQ-9) (Spitzer et al., 1999), Generalized Anxiety Disorder 7 item scale (GAD-7) (Spitzer et al., 2006), and Adverse Childhood Experiences (ACE) questionnaire (Felitti et al., 1998); however, not all patients had all the scales completed at the time of consultation. Clinical demographics included age, gender, and treatment. The EHR data was reviewed to assess relevance of genotyping which was quantified as: (1) clinician providing genotype-guided recommendations (GGR), or (2) clinician providing treatment as usual (TAU), where genotyping was or was not acknowledged, but treatment was guided based on the discretion of the treating clinician.
Subjects were evaluated for the clinical treatment decision impact of genetic testing for PK [cytochrome P450 2D6 (CYP2D6) and 2C19 (CYP2C19)] and PD [serotonin transporter (SLC6A4)] genetic variation on treatment as usual in MDD or BP depressed patients. Testing was completed either with the AssureX Health GeneSight® platform or individual testing of PK or PD genes by Mayo Medical Laboratory. CYP2D6 phenotypes were defined pharmacokinetically as extensive metabolizer (EM), intermediate metabolizer (IM), poor metabolizer (PM), or ultrarapid metabolizer (URM). CYP2C19 phenotypes were defined pharmacokinetically as EM, IM, or PM. Detailed CYP2D6 and CYP2C19 allele variants are showed in Supplementary Table S1. SLC6A4, phenotypes were defined as [long/long (l/l)], [short/long (s/l)], or [short/short (s/s)]. SLC6A4 has other genetic variations that may be relevant for the analysis (Hu et al., 2006). As reviewed by Frye and colleagues, in addition to the L allele and the SLC6A4 and the SNP rs2553, known to influence the association of the 5-HTTLPR alleles with expression of the SLC6A4 gene, there is a second intron variable number of tandem repeats (VNTR) identified that would be of interest in subsequent analysis. However, these variants were not identified in earlier samples of this study (i.e., from 2003) (Frye et al., 2015). The simultaneous determination of the long and short form of SLC6A4 was performed by polymerase chain reaction (PCR) amplification of the promoter region of 5-HTT followed by Haemophilus parainfluenzae II digestion of the resulting amplicon, as described by Wendland et al. (2006). CYP2C19 and CYP2D6 genotyping was performed on genomic DNA extracted from whole blood using the xTAG Assay for P450-2C19 v2, which incorporates multiplex PCR and multiplex allele-specific primer extension (ASPE) with Luminex Molecular Diagnostics’ proprietary Universal Tag sorting system on the Luminex 100 xMAP platform. Detailed genotyping laboratory methodology is highlighted in our previous work (Mrazek et al., 2009; Mrazek et al., 2011; Frye et al., 2015).
Means and standard deviations are presented for continuous variables, and were compared by genotyping status and recommendation group using t-test or Wilcoxon Rank Sum tests. Chi-squared test and Fisher’s exact test were used to describe the differences in proportions between the genotyping status and recommendation group. The level of statistical significance was set at p < 0.05 (two-sided).
Using the SCID, 523 of the 1795 patients were diagnosed with BP and 1272 were diagnosed with MDD. 167/523 (31.9%) with BP and 446/1272 (35.1%) with MDD were genotyped. 317 subjects (18%) and 510 subjects (28%) underwent CYP2D6/CYP2C19 and SLC6A4 genotyping, respectively. Genotyped patients were less prescribed antidepressants (p = 0.009) versus other medication classes, and had significantly higher measures of self-reported anxiety (GAD-7 = 12.9 (5.6), p < 0.016) and depression (PHQ-9 = 18 (6.1), p < 0.001) in comparison to non-genotyped patients (Table 1). PK and PD genotype-guided recommendations were associated with significantly higher measures of anxiety and depression [(GAD-7 = 13.2 (5.6), p = 0.02) and (PHQ-9 = 18.1 (6.1), p = 0.005), (GAD-7 = 13.2 (5.63), p = 0.009) and (PHQ-9 = 18.0 (6.1), p < 0.001), respectively] (Table 2). There was no significant association between different genotypes and the measures of anxiety and depression.
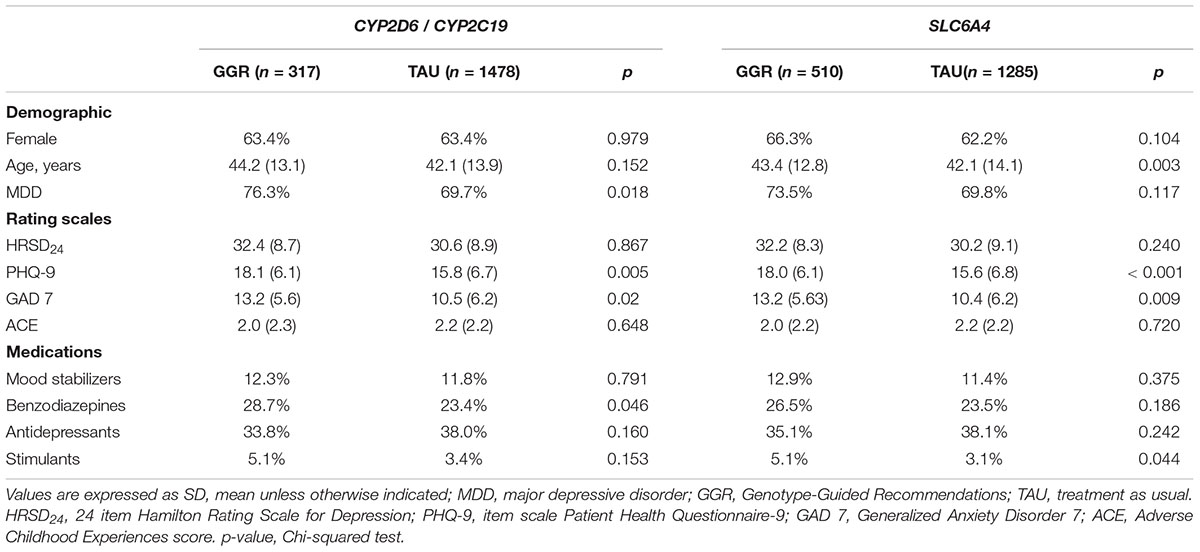
Table 2. Demographics and Clinical Characteristics of Patients with Genotype-Guided Recommendations (GGR) vs. Treatment as usual (TAU).
The pharmacogenomic profiles of CYP2C19 were: PM (3.5%), IM (27.4%), and EM (69.1%). There was a higher rate of CYP2C19 poor metabolizer phenotype in BP (9.3%) vs. MDD patients (1.7%, p = 0.003) (Table 3). Among those participants with an S-allele, the rate of CYP2C19 PM phenotype was even higher in the BP (9.8%) vs. MDD (0.6%, p = 0.003). There was no significant difference in distribution of treatment guided recommendations groups between CYP2C19 phenotypes [EM (GGR = 68.85%, TAU = 69.49%), IM (GGR = 27.05%, TAU = 28.81%), PM (GGR = 4.10%, TAU = 1.69%), (p = 0.67)] (Table 3).
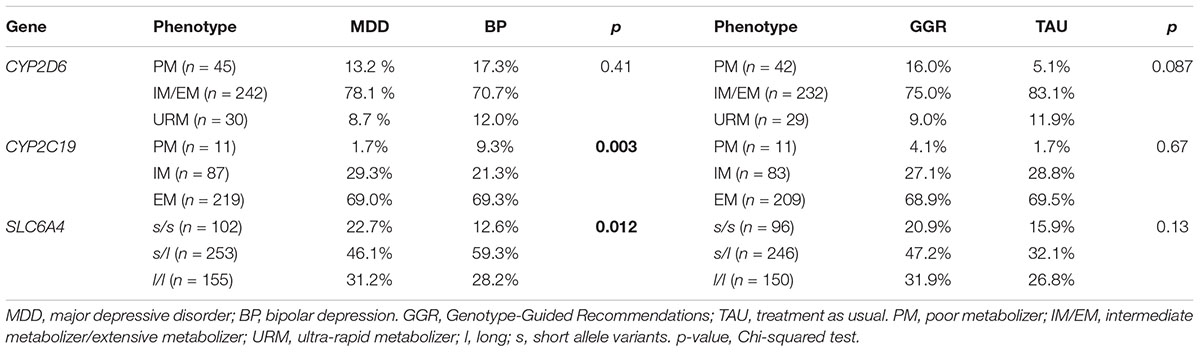
Table 3. Phenotype results by diagnose and Genotype-Guided Recommendations (GGR) vs. Treatment as usual (TAU).
The pharmacogenomic profiles of CYP2D6 were: IM/EM (76.3%), PM (14.2%), and URM (9.5%). There was no significant difference in distribution of CYP2D6 phenotypes by diagnosis (p = 0.41) (Table 2). There was no significant difference in distribution of treatment guided recommendations groups between CYP2D6 phenotypes [PM (GGR = 16%, TAU = 5.1%), EM/IM (GGR = 75%, TAU = 83.1%), URM (GGR = 9%, TAU = 11.9%), (p = 0.087)] (Table 3).
The pharmacogenomic profiles of SLC6A4 were: l/l (30.4%), s/l (49.6%), and s/s (20.0%). There was a statistically significant difference in distribution of SLC6A4 genotypes between BP (l/l = 28.2%, s/l = 59.3%, and s/s = 12.6%) and MDD (l/l = 31.2%, s/l = 46.1%, and s/s = 22.7%) patients (p = 0.012) (Table 3). Among S-allele carries, in comparison to MDD patients, there was a significantly higher rate of BP patients with PM in either CYP2D6 or CYP2C19. There was no significant difference in distribution of treatment guided recommendations groups between SLC6A4 phenotypes [l/l (GGR = 31.9%, TAU = 26.8%), s/l (GGR = 47.2%, TAU = 32.1%), and s/s (GGR = 20.9%, TAU = 15.9%) (p = 0.13)] (Table 3).
This study assessed the relationship between symptom severity, demographics, and pharmacokinetic / pharmacodynamics genetic variation among diagnostic mood disorder subgroups. There was a significant difference in CYP2C19 and SLC6A4 PK and PD phenotype distribution between BP and MDD patients with history of multiple drug failures/treatment resistance. Specifically, there were significantly higher rates of CYP2C19 PM in BP patients in comparison to MDD patients; among those participants with an S-allele, the rate of CYP2C19 PM phenotype was more than 10X higher in the BP vs. MDD.
The clinical implications of CYP2C19 and serotonin transporter genetic variation are not fully understood. It is known, however, that poor metabolizer phenotype is associated with high blood levels and increased risk of side effects. As suggested by the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline and Nassan et al. (2016), individuals on citalopram / escitalopram with CYP2C19 PM phenotype should reduce dose by 50% and /or use an alternative antidepressant (Hicks et al., 2015). There has been little investigation as to metabolizer status, blood level, and risk of antidepressant induced mania in either bipolar or unipolar depression. There is evidence to suggest that the s/s genotype is associated with increased antidepressant side-effects, including antidepressant-induced mania (Frye et al., 2015); further studies should investigate risk of Antidepressant Induced Mania (AIM+) as a function of PK-PD interaction as is being done with other antidepressant pharmacogenomic antidepressant analyses (Ahmed et al., 2018). The genotype-guided recommendations of CYP2D6, CYP2C19, and SLC6A4 were associated with significantly higher measures of anxiety and depression in comparison to treatment as usual. Like Rundell et al., 2011a, our study has found significantly higher baseline self-reported scores of depression in GGR individuals’ possible indicative of increase symptom burden and greater treatment resistance.
The results of CYP2D6 / CYP2C19 genotyping were more commonly used to make treatment recommendations in MDD than in BP. This study is limited by it cross sectional design with no longitudinal mood outcome data based on GGT vs. TAU. However, there is increasing interest and investigation identifying PK CYP2D6 and CYP2C19 genetic variants associated with clinical response to several SSRIs (Tsai et al., 2010; Mrazek et al., 2011; Gressier et al., 2015; Hicks et al., 2015). Several studies have investigated GGR vs. TAU in treatment of MDD patients. However, none have included BP patients. Studying ACE score in relationship with SLC6A4 S-allele and depression severity is also important, as there are gene and environment interactions (Caspi et al., 2003).
The decision to genotype was based on clinical factors and not pre-determined systematic criteria. Typically, patients who received genotyping might also have been self-selected and more interested in receiving it. Thus, there is inherent selection bias affecting the comparison between the two diagnostic groups. Even though the sample size was large, given the lower prevalence of CYP2C19 PM and SLC6A4 S-allele, the final number of patients with these findings were (n = 6) and, ideally, the initial sample size should be larger. This study did not have systematic follow-up to look at outcome measures of efficacy and side effects/tolerability based on these recommendations; these are important prospective studies to complete and such studies are currently underway. Our outcomes data have lacked the statistical power to accurately analyze the ancestry data; due to 89% of our population being white Caucasians, this may have affected the interpretation of our findings, this study was conducted in a clinical setting with a naturalistic study design, and is lacking standard criteria for the selection of patients for pharmacogenetic testing (Gelernter et al., 1997; Mrazek et al., 2009; Strom et al., 2012). Although, this type of design has the advantage of mimicking “real life” clinical practice, it has significant limitations when it comes to controlling for confounding. This is an issue that needs to be addressed in the future through longitudinal prospective studies with systematic genetic screening. Finally, our sample data was deficient of medication blood levels, which would clarify some of the study findings.
There may be underlying pharmacogenomic differences in treatment seeking depressed patients that potentially have impact on serum levels of CYP2C19 metabolized antidepressants (i.e., citalopram / escitalopram) contributing to rates of efficacy vs. side effect burden with additional potential risk of antidepressant response vs. induced mania. The evidence for utilizing pharmacogenomics-guided therapy in MDD and BP is still developing with a much needed focus on drug safety, side effect burden, and treatment adherence. Future work is essential; scientific and logistic barriers still exist before there can be widespread implementation of clinical genomics. Genomic science has a profound potential to individualize the drug therapy for depression.
This study was carried out in accordance with the recommendation of the Mayo Clinic Institutional Review Board with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Mayo Clinic Institutional Review Board.
ATA and JRG performed all the data analysis. CJB, KLB, and MV contributed to the acquisition of data. JMB assisted with data analysis and interpretation of findings. All co-authors provided critical revision of the manuscript for important intellectual content. MV and MAF were responsible for the study concept and design. MV an ATA drafted the manuscript. All authors critically reviewed content and approved the final version for publication.
ATA research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number T32 GM008685. MLP was supported in part by grants CONICYT PFCHA/MAGISTER BECAS CHILE/2012 – 73130844 and CONICYT FONDECYT Regular 1181365.
Mayo Clinic has a financial interest in AssureX Health and the technology referenced in this abstract.
This article was presented in part at the International College of Neuropsychopharmacology (CINP) Thematic Meeting on Treatment Resistant Depression, Prague, Czechia, July 20–22, 2017.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00083/full#supplementary-material
Ahmed, A. T., Biernacka, J. M., Jenkins, G., Rush, A. J., Shinozaki, G., Veldic, M., et al. (2018). Pharmacokinetic-pharmacodynamic interaction associated with venlafaxine-XR remission in patients with major depressive disorder with history of citalopram / escitalopram treatment failure. J. Affect Disord. 246, 62–68. doi: 10.1016/j.jad.2018.12.021
Anderson, I. M., Ferrier, I. N., Baldwin, R. C., Cowen, P. J., Howard, L., Lewis, G., et al. (2008). Evidence-based guidelines for treating depressive disorders with antidepressants: a revision of the 2000 british association for psychopharmacology guidelines. J. Psychopharmacol. 22, 343–396. doi: 10.1177/0269881107088441
Caspi, A., Sugden, K., Moffitt, T. E., Taylor, A., Craig, I. W., Harrington, H., et al. (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389. doi: 10.1126/science.1083968
Crismon, M. L., Trivedi, M., Pigott, T. A., Rush, A. J., Hirschfeld, R. M., Kahn, D. A., et al. (1999). The texas medication algorithm project: report of the texas consensus conference panel on medication treatment of major depressive disorder. J. Clin. Psychiatry 60, 142–156. doi: 10.4088/JCP.v60n0302
Doherty, J. L., and Owen, M. J. (2014). Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 6:29. doi: 10.1186/gm546
Drozda, K., Muller, D. J., and Bishop, J. R. (2014). Pharmacogenomic testing for neuropsychiatric drugs: current status of drug labeling, guidelines for using genetic information, and test options. Pharmacotherapy 34, 166–184. doi: 10.1002/phar.1398
Felitti, V. J., Anda, R. F., Nordenberg, D., Williamson, D. F., Spitz, A. M., Edwards, V., et al. (1998). Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. Am. J. Prev. Med. 14, 245–258. doi: 10.1016/S0749-3797(98)00017-8
Ferrari, A. J., Stockings, E., Khoo, J. P., Erskine, H. E., Degenhardt, L., Vos, T., et al. (2016). The prevalence and burden of bipolar disorder: findings from the global burden of disease study 2013. Bipolar Disord. 18, 440–450. doi: 10.1111/bdi.12423
Frye, M. A. (2011). Bipolar disorder — a focus on depression. New Engl. J. Med. 364, 51–59. doi: 10.1056/NEJMcp1000402
Frye, M. A., Mcelroy, S. L., Prieto, M. L., Harper, K. L., Walker, D. L., Kung, S., et al. (2015). Clinical risk factors and serotonin transporter gene variants associated with antidepressant-induced mania. J. Clin. Psychiatry 76, 174–180. doi: 10.4088/JCP.14m09127
Gelernter, J., Kranzler, H., and Cubells, J. F. (1997). Serotonin transporter protein (SLC6A4) allele and haplotype frequencies and linkage disequilibria in African- and European-American and Japanese populations and in alcohol-dependent subjects. Hum. Genet. 101, 243–246. doi: 10.1007/s004390050624
Gressier, F., Verstuyft, C., Hardy, P., Becquemont, L., and Corruble, E. (2015). Response to CYP2D6 substrate antidepressants is predicted by a CYP2D6 composite phenotype based on genotype and comedications with CYP2D6 inhibitors. J. Neural. Transm. 122, 35–42. doi: 10.1007/s00702-014-1273-4
Hall-Flavin, D. K., Winner, J. G., Allen, J. D., Carhart, J. M., Proctor, B., Snyder, K. A., et al. (2013). Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet. Genomics 23, 535–548. doi: 10.1097/FPC.0b013e3283649b9a
Hall-Flavin, D. K., Winner, J. G., Allen, J. D., Jordan, J. J., Nesheim, R. S., Snyder, K. A., et al. (2012). Using a pharmacogenomic algorithm to guide the treatment of depression. Transl. Psychiatry 2:e172. doi: 10.1038/tp.2012.99
Hamilton, M. (1960). A rating scale for depression. J. Neurol. Neurosurg. Psychiatry 23, 56–62. doi: 10.1136/jnnp.23.1.56
Hicks, J. K., Bishop, J. R., Sangkuhl, K., Muller, D. J., Ji, Y., Leckband, S. G., et al. (2015). Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134. doi: 10.1002/cpt.147
Hu, X. Z., Lipsky, R. H., Zhu, G., Akhtar, L. A., Taubman, J., Greenberg, B. D., et al. (2006). Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am. J. Hum. Genet. 78, 815–826. doi: 10.1086/503850
Kung, S., and Li, X. (2010). The clinical use of pharmacogenomic testing in treatment-resistant depression. Prim. Psychiatry 17, 46–51.
Lee, S. H., Ripke, S., Neale, B. M., Faraone, S. V., Purcell, S. M., Perlis, R. H., et al. (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45, 984–994. doi: 10.1038/ng.2711
Lichtenstein, P., Yip, B. H., Bjork, C., Pawitan, Y., Cannon, T. D., Sullivan, P. F., et al. (2009). Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239. doi: 10.1016/S0140-6736(09)60072-6
McGuffin, P., Rijsdijk, F., Andrew, M., Sham, P., Katz, R., and Cardno, A. (2003). The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry 60, 497–502. doi: 10.1001/archpsyc.60.5.497
Mrazek, D. A., Biernacka, J. M., O’kane, D. J., Black, J. L., Cunningham, J. M., Drews, M. S., et al. (2011). CYP2C19 variation and citalopram response. Pharmacogenet. Genomics 21, 1–9. doi: 10.1097/FPC.0b013e328340bc5a
Mrazek, D. A., Rush, A. J., Biernacka, J. M., O’kane, D. J., Cunningham, J. M., Wieben, E. D., et al. (2009). SLC6A4 variation and citalopram response. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150b, 341–351. doi: 10.1002/ajmg.b.30816
Nassan, M., Nicholson, W. T., Elliott, M. A., Rohrer Vitek, C. R., Black, J. L., and Frye, M. A. (2016). Pharmacokinetic pharmacogenetic prescribing guidelines for antidepressants: a template for psychiatric precision medicine. Mayo Clin. Proc. 91, 897–907. doi: 10.1016/j.mayocp.2016.02.023
Porcelli, S., Fabbri, C., and Serretti, A. (2012). Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur. Neuropsychopharmacol. 22, 239–258. doi: 10.1016/j.euroneuro.2011.10.003
Rosenblat, J. D., Lee, Y., and Mcintyre, R. S. (2017). Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? a systematic review of clinical trials and cost-effectiveness studies. J. Clin. Psychiatry 78, 720–729. doi: 10.4088/JCP.15r10583
Rosenblat, J. D., Lee, Y., and Mcintyre, R. S. (2018). The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: a meta-analysis. J. Affect. Disord. 241, 484–491. doi: 10.1016/j.jad.2018.08.056
Rundell, J. R., Harmandayan, M., and Staab, J. P. (2011a). Pharmacogenomic testing and outcome among depressed patients in a tertiary care outpatient psychiatric consultation practice. Transl. Psychiatry 1:e6. doi: 10.1038/tp.2011.7
Rundell, J. R., Staab, J. P., Shinozaki, G., Saad-Pendergrass, D., Moore, K., Mcalpine, D., et al. (2011b). Pharmacogenomic testing in a tertiary care outpatient psychosomatic medicine practice. Psychosomatics 52, 141–146. doi: 10.1016/j.psym.2010.12.023
Rush, A. J., Trivedi, M. H., Wisniewski, S. R., Nierenberg, A. A., Stewart, J. W., Warden, D., et al. (2006). Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR∗D report. Am. J. Psychiatry 163, 1905–1917. doi: 10.1176/ajp.2006.163.11.1905
Sidor, M. M., and Macqueen, G. M. (2011). Antidepressants for the acute treatment of bipolar depression: a systematic review and meta-analysis. J. Clin. Psychiatry 72, 156–167. doi: 10.4088/JCP.09r05385gre
Spitzer, R. L., Kroenke, K., and Williams, J. B. (1999). Validation and utility of a self-report version of PRIME-MD: the PHQ primary care study. Primary care evaluation of mental disorders. Patient health questionnaire. JAMA 282, 1737–1744.
Spitzer, R. L., Kroenke, K., Williams, J. W., and Löwe, B. (2006). A brief measure for assessing generalized anxiety disorder: the gad-7. Arch. Internal Med. 166, 1092–1097. doi: 10.1001/archinte.166.10.1092
Strom, C. M., Goos, D., Crossley, B., Zhang, K., Buller-Burkle, A., Jarvis, M., et al. (2012). Testing for variants in CYP2C19: population frequencies and testing experience in a clinical laboratory. Genet. Med. 14, 95–100. doi: 10.1038/gim.0b013e3182329870
Sullivan, P. F., Neale, M. C., and Kendler, K. S. (2000). Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562. doi: 10.1176/appi.ajp.157.10.1552
Tsai, M. H., Lin, K. M., Hsiao, M. C., Shen, W. W., Lu, M. L., Tang, H. S., et al. (2010). Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics 11, 537–546. doi: 10.2217/pgs.09.168
Vos, T., Barber, R. M., Bell, B., Bertozzi-Villa, A., Biryukov, S., Bolliger, I., et al. (2015). Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet 386, 743–800. doi: 10.1016/S0140-6736(15)60692-4
Wendland, J. R., Martin, B. J., Kruse, M. R., Lesch, K. P., and Murphy, D. L. (2006). Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol. Psychiatry 11, 224–226. doi: 10.1038/sj.mp.4001789
Whiteford, H. A., Degenhardt, L., Rehm, J., Baxter, A. J., Ferrari, A. J., Erskine, H. E., et al. (2013). Global burden of disease attributable to mental and substance use disorders: findings from the global burden of disease study 2010. Lancet 382, 1575–1586. doi: 10.1016/S0140-6736(13)61611-6
Winner, J. G., Carhart, J. M., Altar, C. A., Allen, J. D., and Dechairo, B. M. (2013). A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov. Med. 16, 219–227.
World Health Organization (2018). Depression and Other Common Mental Disorders. Available at: http://www.who.int/news-room/fact-sheets/detail/mental-disorders
Zeier, Z., Carpenter, L. L., Kalin, N. H., Rodriguez, C. I., Mcdonald, W. M., Widge, A. S., et al. (2018). Clinical implementation of pharmacogenetic decision support tools for antidepressant drug prescribing. Am. J. Psychiatry 175, 873–886. doi: 10.1176/appi.ajp.2018.17111282
Keywords: pharmacogenomics, cytochrome P450, CYP2C19, SLC6A4, bipolar disorder
Citation: Veldic M, Ahmed AT, Blacker CJ, Geske JR, Biernacka JM, Borreggine KL, Moore KM, Prieto ML, Vande Voort JL, Croarkin PE, Hoberg AA, Kung S, Alarcon RD, Keeth N, Singh B, Bobo WV and Frye MA (2019) Cytochrome P450 2C19 Poor Metabolizer Phenotype in Treatment Resistant Depression: Treatment and Diagnostic Implications. Front. Pharmacol. 10:83. doi: 10.3389/fphar.2019.00083
Received: 06 October 2018; Accepted: 21 January 2019;
Published: 19 February 2019.
Edited by:
Hector J. Caruncho, University of Victoria, CanadaReviewed by:
Javier Costas, Complejo Hospitalario Universitario de Santiago, SpainCopyright © 2019 Veldic, Ahmed, Blacker, Geske, Biernacka, Borreggine, Moore, Prieto, Vande Voort, Croarkin, Hoberg, Kung, Alarcon, Keeth, Singh, Bobo and Frye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marin Veldic, VmVsZGljLk1hcmluQG1heW8uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.