
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 17 January 2019
Sec. Predictive Toxicology
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.01548
This article is part of the Research Topic The Molecular Mechanism of the Toxicity and Metabolism of Mycotoxins View all 8 articles
 Lingling Wang1,2†
Lingling Wang1,2† Jun Wu1,2†
Jun Wu1,2† Zhiwen Liu1
Zhiwen Liu1 Yutao Shi1
Yutao Shi1 Jinqiu Liu1
Jinqiu Liu1 Xiaofan Xu1
Xiaofan Xu1 Shuxian Hao3
Shuxian Hao3 Peiqiang Mu1,2
Peiqiang Mu1,2 Fengru Deng1,2
Fengru Deng1,2 Yiqun Deng1,2*
Yiqun Deng1,2*Aflatoxin B1 (AFB1) is one of the most hazardous mycotoxins contamination in food and feed products, which leads to hepatocellular carcinoma in humans and animals. In the present study, we isolated and characterized an AFB1 degrading bacteria CG1061 from chicken cecum, exhibited an 93.7% AFB1 degradation rate by HPLC. 16S rRNA gene sequence analysis and a multiplex PCR experiment demonstrated that CG1061 was a non-pathogenic Escherichia coli. The culture supernatant of E. coli CG1061 showed an 61.8% degradation rate, whereas the degradation rates produced by the intracellular extracts was only 17.6%, indicating that the active component was constitutively secreted into the extracellular space. The degradation rate decreased from 61.8 to 37.5% when the culture supernatant was treated with 1 mg/mL proteinase K, and remained 51.3% when that treated with 100°C for 20 min. We postulated that AFB1 degradation was mediated by heat-resistant proteins. The content of AFB1 decreased rapidly when it was incubated with the culture supernatant during the first 24 h. The optimal incubation pH and temperature were pH 8.5 and 55°C respectively. According to the UPLC Q-TOF MS analysis, AFB1 was bio-transformed to the product C16H14O5 and other metabolites. Based on the results of in vitro experiments on chicken hepatocellular carcinoma (LMH) cells and in vivo experiments on mice, we confirmed that CG1061-degraded AFB1 are less toxic than the standard AFB1. E. coli CG1061 isolated from healthy chicken cerum is more likely to colonize the animal gut, which might be an excellent candidate for the detoxification of AFB1 in food and feed industry.
Microbiotas presenting in human and animal intestinal tracts with a high population density, and great diversity, play a crucial role in the growth and health of the hosts. In poultry, 1 g wet weight of cecal content may contain 1011 bacteria, which contribute to enhanced nutrient absorption, xenobiotic compounds transformation, and improvements in immune function(Worrell et al., 1989; Kollarczik et al., 1994; Hooper et al., 2001). Based on accumulating evidence, gut microbes efficiently transform mycotoxins into less toxic compounds. Bacteria have been isolated from animal intestines and confirmed to detoxify deoxynivalenol and zearalenone (Guan et al., 2009; Yu et al., 2010; Gao et al., 2018). Aflatoxins (AFs) are a group of toxic and carcinogenic fungal metabolites produced mainly by the fungi Aspergillus flavus and Aspergillus parasiticus. AFs contamination in cereal grains is a serious problem worldwide, particularly in sub-tropical and tropical areas (Guan et al., 2011). Of the AFs, aflatoxin B1 (AFB1) is the most hazardous member and has been classified as a Group I naturally occurring carcinogen by the International Agency for Research on Cancer [IARC] (1993). In this study, we intend to isolate AFB1 degrading bacteria from chicken cecum for the development of microbial or enzyme products for applications in the food and feed industries.
Several strategies have been reported to detoxify AFs, including physical, chemical, and biological methods. Physical methods include absorption, heating and irradiation (Aziz and Moussa, 2002; Arzandeh and Jinap, 2011; Di Gregorio et al., 2014). Chemical methods include ammonization, solvent extraction and oxidation (Lee and Cucullu, 1978; Norred and Morrissey, 1983; Akbas and Ozdemir, 2006; Zhu et al., 2016). These physical and chemical detoxification methods have many limitations, such as nutritional losses, expensive equipment and time consuming. In comparison, biological methods was found to be most efficient and showed the greatest specificity and environmental soundness. Nocardia corynebacterioides was the first reported AFB1-detoxifying microorganism (Ciegler et al., 1966). Bacterial and fungal AFB1 degradation strains have been continuously to be reported in the subsequent decades. The AFB1 degrading fungi Pleurotus ostreatus, Armilleriella tabescens, and Trametes versicolor have been proved to possess a detoxification function (Liu et al., 1998; Motomura et al., 2003; Alberts et al., 2009; Das et al., 2014). Some bacteria isolated from soil, feces, nuts and other environments effectively degrade AFB1, such as Rhodococcus erythropolis, Mycobacterium fluoranthenivorans, Stenotrophomonas maltophilia, Enterobacteriaceae sp., Myxococcus fulvus, Bacillus subtilis, and Pseudomonas putida (Alberts et al., 2006; Guan et al., 2008; El-Deeb et al., 2013; Samuel et al., 2014; Juri et al., 2015). Although many microorganisms have been reported as degraders of AFB1, there was little information about the structure of the degraded products, few studies confirmed the toxicity of the degradation products by cytotoxicity assessment on hepatocellular or animal toxicity test. The currently known microbial degradation metabolites of AFB1 and their levels of toxicity were summarized by Verheecke et al. (2016).
The objectives of this study were: (1) to isolate new AFB1 degrading bacteria from chicken cecum; (2) to investigate the optimal degradation conditions; and (3) to analysis the degraded AFB1 products and confirm the toxicity of the degraded AFB1 products.
The protocol of the study was approved by Laboratory Animal Ethics committee of South China Agricultural University in China (Permit No. 00181878). All the animal experiments in this research were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals of Guangdong province, China. The date of approval by the ethics committee was September 9, 2017.
Eight healthy broiler chickens (about 12 months old) were purchased for cecal collection in markets near South China Agricultural University, Guangzhou, China. The chickens were treated with cervical dislocation for euthanasia. Approximately 5 g of the cecal contents were mixed with 20 mL of sterile Luria-Bertani (LB) medium (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, in 1 L of distilled water, pH 7.0). The mixture remained static for 10 min and the supernatant were preserved in glycerol at a final concentration of 15% v/v at –80°C until further analysis. AFB1 was purchased from Sigma-Aldrich (St. Louis, MO, United States). A stock solution of 250 μg/mL AFB1 was dissolved in DMSO and stored at -20°C.
Eight cecal samples were incubated in LB medium containing 2.5 μg/mL AFB1 at 37°C for 72 h shaken at 180 rpm in the dark. AFB1 was added to sterile LB medium at a final concentration of 2.5 μg/mL and incubated under the same conditions as the control. Cultures were centrifuge at 1.2 × 104rpm for 5 min. The amount of AFB1 in the supernatant was detected using a previously published method with slight modifications as mentioned subsequently (Guan et al., 2008). AFB1 was extracted three times with chloroform (1:2, v/v). The chloroform extracts were evaporated, and the residue was dissolved in methanol, filtered (Merck Millipore, Germany, 0.22 μm) and then analyzed by high-performance-liquid chromatography (HPLC) using an Agilent 1260 Infinity liquid chromatography system (Agilent Technologies, Germany) equipped with a G1321B FLD detector and ZORBAX SB-C18 column. The mobile phase was composed of a mixture of solvent A (water/acetonitrile/formic acid, 92/5/0.8, v/v/v) and solvent B (methanol/acetonitrile/formic acid, 92/5/0.8, v/v/v), and separation was performed at a flow rate of 1.0 mL/min. The time of analysis was 35 min. The volume of injection was 10 μl. The retention time of AFB1 was 27.6 min. The experiments were conducted with three repeats. The AFB1 degradation rate was calculated using the following formula:
(1-AFB1 peak area of treatment/AFB1 peak area of control) × 100%
All the AFB1 degradation rate mentioned in this article were calculated using this formula.
Ten microliters of sample with the highest AFB1 degradation were serially diluted with sterilized distilled water, spread on an LB plate, and incubated at 37°C for 24 h. Fifty colonies on the plate were picked and inoculated to sterile LB medium at a final concentration of 2.5 μg/mL AFB1, and incubated at 37°C for 72 h with 180 rpm in the dark. Sterile LB medium was incubated at same condition as the control. AFB1 degradation rate was determined by HPLC which performed as mentioned above. AFB1 degrading isolates were selected and purified for further study. Genomic DNA was extracted using agenomic DNA Miniprep Kit (Axygen, China). The 16S rRNA gene was amplified by PCR using universal primers (27F, 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R, 5′-GGTTACCTTGTTACGACTT-3′). The sequences were BLAST on the National Center for Biotechnology Information website and the phylogenetic tree was constructed using MEGA 7.0.
In an attempt to identify the virulence gene of our isolate E. coli CG1061, a multiplex PCR was developed to detection 10 virulence genes (ipaH, aatA, eltA, vtx2, eae, aggR, vtx1, aaiC, estA-porcine, and estA-human) with a Diarrhoeagenic E. coli PCR Kit (Statens Serum Institut, Denmark) which were widely used for detection the virulence of E. coli. The details description of these virulence genes has been reported (Persson et al., 2007). Amplification conditions were 94°C for 6 min, followed by 35 cycles of 94°C for 50 s, 57°C for 40 s and 72°C for 50 s, and finally 72°C for 3 min. Amplicons were analyzed by electrophoresis on 1.5% (w/v) agarose gels under standard conditions.
Escherichia coli CG1061 was cultivated in LB liquid medium for 48 h at 37°C with shaking at 180 rpm. After centrifugation at 1.2 × 104rpm for 5 min at 4°C, supernatant and pellets of cultures were collected respectively.
The pellets of cultures were washed three times with PBS, resuspended and lysed on ice using an ultrasonic cell disintegrator (JY92-2D, Xinzhi Instruments, Ningbo, China). The disintegrated cell suspension was centrifuged at 8.0 × 103 rpm for 10 min at 4°C. The supernatant was filtered and incubated with 2.5 μg/mL AFB1 at 37°C for 24 h, with shaking at 180 rpm in the dark. PBS containing 2.5 μg/mL AFB1 was used as the control.
The supernatant of cultures were filtered (Merck Millipore, Germany, 0.22 μm) and prepared for next experiments. For the heat treatment, the culture supernatant was dipped into boiling water bath for 20 min. For the proteinase K treatments, culture supernatant was treated with 1 mg/mL proteinase K, incubated at 58°C for 3 h. Then all samples were incubated with 2.5 μg/mL AFB1 at 37°C for 24 h, with shaking at 180 rpm in the dark. Sterile LB medium containing 2.5 μg/mL AFB1 was used as the control. The experiments were conducted with three repeats.
Culture supernatant was incubated for 0, 12, 24, 36, 48, 60, and 72 h at 37°C and a pH of 7.0. Sterile LB medium was incubated in same length of time as their control respectively. The assays were performed in the presence of at 2.5 μg/mL AFB1, with shaking at 180 rpm in the dark. The experiments were conducted with three repeats.
The effects of pH on AFB1 degradation were studied by adjusting the pH of the culture supernatant to a pH of 4.3, 4.9, 6.5, 7.4, 8.5, and 8.9 with Na2HPO4 and KH2PO4 buffer solutions and then incubating the samples at 37°C for 24 h. Sterile LB medium was incubated in same pH value as their control respectively. All assays were performed at 2.5 μg/mL AFB1 in the dark without shaking. The experiments were conducted with three repeats.
The culture supernatant was incubated at 4, 25, 37, 55, and 70°C in a pH 7.0 solution for 24 h to confirm the effects of temperature on AFB1 degradation. Sterile LB medium was incubated in same temperature condition as their control respectively. All assays were performed at 2.5 μg/mL AFB1 in the dark without shaking. The experiments were conducted with three repeats.
Escherichia coli CG1061 was cultured in LB medium containing AFB1 at a final concentration of 12.5 μg/mL for 72 h at 37°C, with shaking at 180 rpm in the dark. The AFB1 degradation products were extracted three times with chloroform (1:2, v/v). The chloroform extracts were evaporated, the residue was dissolved in methanol. After filtered (Merck Millipore, Germany, 0.22 μm), the AFB1 degradation products were separated using an Agilent 1290 series ultra-high-performance liquid chromatography (UPLC) system. An Agilent Eclipse Plus C18 column was used for separation (50 × 2.1 mm, 1.8 μm). The inject volume was 2 μl. The mobile phase consisted of acetonitrile (A) and water with 0.2% formic acid (B) and 10 mM ammonium formate at a flow rate of 0.3 mL/min. Mass spectrometry analyses were performed using a quadrupole tandem time-of-flight (Q-TOF) G6450B mass spectrometer system in the positive ion mode. Instrument Parameters: gas temperature 300°C, gas flow 8 l/min, Ion spray voltage 4000 V. Mass spectra were acquired in a full-scan analysis in the range of m/z 50–1000.
The toxicity of CG1061-degraded AFB1 and standard AFB1 was examined in chicken hepatocellular carcinoma LMH cells using the MTT (3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide) method. LMH cells obtained from the ATCC (Catalog No. CRL-2117). E. coli CG1061 was cultured in LB medium containing AFB1 at a final concentration of 12.5 μg/mL for 72 h at 37°C, with shaking at 180 r/min in the dark. The supernatant of cultures were filtered and prepared as CG1061-degraded AFB1 samples. Sterile LB medium containing 12.5 μg/mL AFB1 was incubated under the same conditions to prepare standard AFB1 samples. Both samples were diluted with William’s E medium containing 10% (w/v) fatal bovine serum (FBS) at final concentrations of 3.75, 2.00, 1.00, 0.50, and 0.25 μg/mL. Sterile LB medium was diluted with William’s E medium containing 10% (w/v) FBS as the control. LMH cells were incubated with diluted samples in 96-well plates at 37°C for 48 h in a 5% CO2 incubator (Thermo, Forma 370), followed by the addition of MTT and an additional incubation for 3 h. Then the formed crystals were dissolved in DMSO for 30 min. The absorption at 490 nm was recorded using a microplate reader (Bio-Rad, Model-680). The experiments were conducted with five repeats. The percentage of cell viability was calculated using the following formula:
(OD490 of sample-OD490 of Blank)/(OD490 of control - OD490 of Blank) × 100%
Fifteen 1-week-old male nude mice were purchased from Guangdong Medical Experimental Center (China) and were randomly allocated to three groups (n = 5). Group I, the control only treated with saline; Group II, treated with 100 μg/kg standard AFB1 of body weight (BW); Group III, treated with 100 μg/kg CG1061-degraded AFB1 of BW. The chloroform extracts and the residue were dissolved in methanol, then diluted in saline and administrated to mice though intraperitoneal injection every alternative day for 5 weeks. After that, all animals were anesthetized by exposure to 10% chloral hydrate. The liver tissue was separated and homogenized in PBS buffer, then centrifuged at 2.5 × 103 rpm for 10 min at 4°C. The supernatant was collected for measurement of alkaline phosphate (ALP) activities, superoxide dismutase (SOD) activities and reduced glutathione (GSH), which are considered as common biochemical markers (Wei et al., 1993; Vipin et al., 2017). These indices were determined by a microplate reader (Bio-Rad, Model-680) using assay kits provided by Jianchen (Nanjing, China). All the animals were maintained at a temperature of 25°C with a 12 h light/dark cycle and were provided with a standard diet and purified drinking water.
All experiments were conducted with at least three repeats and the data are presented as the means ± standard deviation (SD). One-Way analysis of variance (ANOVA) was performed for the data of degradation and toxicity experiments using the software package SPSS version 16.0, and minimum significant differences were calculated using Duncan’s multiple range test.
The results showed that three out of all eight cecal samples were detected for more than 80% degradation rate. Sample No.8 showed the highest AFB1 degradation rate for 93.9%. Fifty colonies picked up from Sample No.8 were detected of AFB1 degradation activity by HPLC. The results demonstrated that twelve out of all 50 colonies were detected for more than 80% degradation rate, The left strains had less or no degradation activity at all. Among the isolates, the degradation rate of CG1061 was highest for 93.7% at a final concentration of 2.5 μg/mL AFB1 when the cell counts was 109 cfu/mL. It is a gram-negative bacterium, presenting white colonies of approximately 2 mm in diameter on LB plates. The sequence of its 16S rRNA gene (accession number, MG786579 in GenBank) had a 97% similarity with that of Escherichia coli. Phylogenetic tree constructed from analysis of 16S rRNA gene sequences showed the relationships of CG1061 and some strains within Escherichia, Salmonellae, and Shigellae (Figure 1). The phylogenetic position of strain CG1061 was nearest the type strain of Escherichia coli ATCC 11775. E. coli is the predominant facultative anaerobe in human and animal colonic flora, and covers some pathogenic strains contributing to diarrheal disease (Nataro and Kaper, 1998). In the multiplex PCR (Figure 2), E. coli CG1061 produced only the 16S rDNA control band, but not the reported virulence genes. So, CG1061 is a non-pathogenic E. coli.

Figure 1. Phylogenetic tree constructed from 16S rRNA gene sequences showing the position of the strain E. coli CG1061 using MEGA 7.0. The neighbor-joining method was used and bootstrapped with 1000 replications of each sequence.
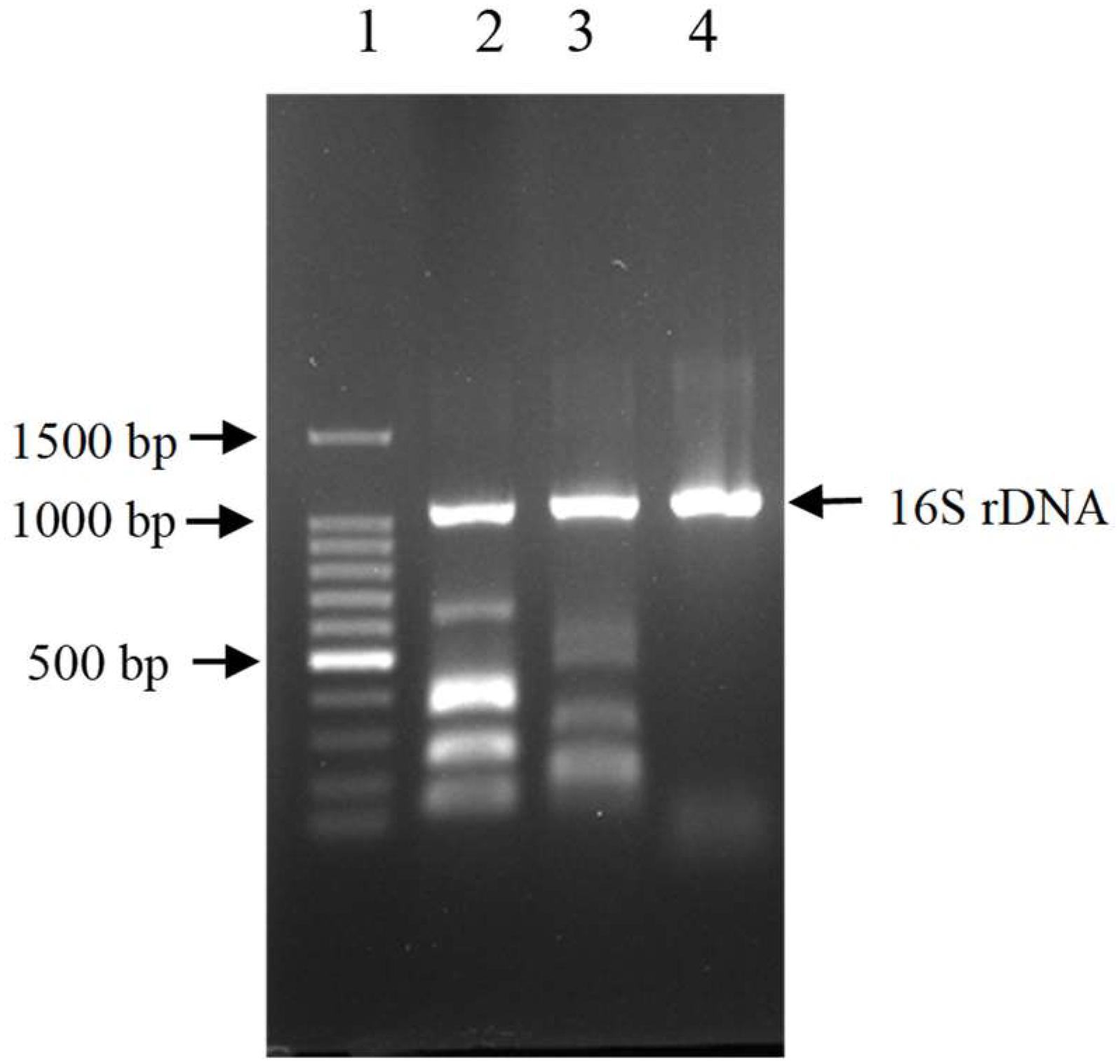
Figure 2. Electrophoretic patterns of simultaneous multi-PCR showing the absence of virulence gene in strain E. coli CG1061. Lane 1: 100 bp DNA marker, 1500, 1000, 900, 800, 700, 600, 500, 400, 300, 200, 100 bp (from top to bottom); lane 2 (control 1): 16S rDNA, ipaH, vtx2, eae, vtx1 and estA-porcine (from top to bottom); lane 3 (control 2): 16S rDNA, aatA, eltA, aggR, aaiC, and estA- human (from top to bottom); lane 4: strain CG1061.
The culture supernatant of the 48 h culture of E. coli CG1061 showed an 61.8% degradation rate for AFB1, whereas only 17.6% (Table 1) degradation rate was detected for the intracellular extracts, demonstrating that the active component was constitutively secreted into the extracellular space. When the culture supernatant was treated with proteinase K, a significant decrease in degradation rate from 61.8 to 37.5% occurred. After heating of 100°C for 20 min, the degradation rate was 51.3%, indicating that the active components maybe heat-resistant proteins.
Culture supernatant of E. coli CG1061 was incubated with AFB1 for 72 h, and AFB1 degradation rates were determined every 12 h by HPLC (Figure 3A). In the first 12 h, AFB1 degradation rates increased rapidly from 0 to 63.5%, then reach to 79.0% at 24 h, 86.3% at 48 h, and 91.9% at 72 h. The results demonstrated that culture supernatant of E. coli CG1061 could significantly reduce AFB1 content in first 24 h (reduced nearly 80%). The impact of the incubation pH on the AFB1 degradation by the E. coli CG1061 culture supernatant was investigated and is shown in Figure 3B. The degradation rates of alkaline conditions were higher than that of acid conditions. The highest degradation rate was 44.8% at pH 8.5. Figure 3C shows the effect of the incubation temperature on AFB1 degradation. Over a broad temperature range from 37 to 70°C, the degradation rates were more than 50%. The highest degradation activity appeared at 55°C (degradation rate, 63.5%). The culture supernatant did not lose its degradation activity at 70°C, indicating that the active components involved were thermostable.
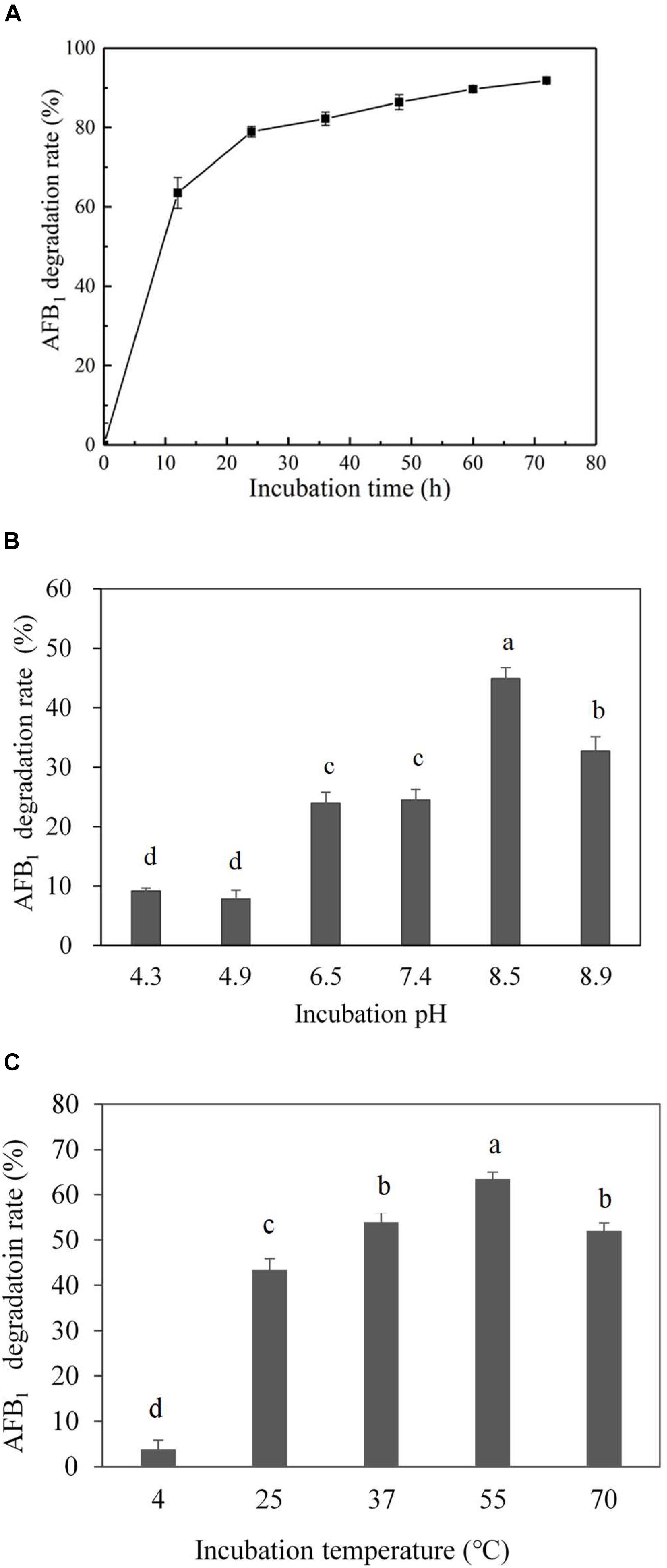
Figure 3. Effect of incubation time, pH and temperature on degradation rate by the culture supernatant of E. coli CG1061. (A) The dynamics of AFB1 degradation by the culture supernatant of E. coli CG1061. The experiments were performed in the presence of 2.5 μg/mL AFB1 at 37°C, pH 7.0. (B) Effects of pH on the degradation of E. coli CG1061 culture supernatant. The experiments were performed in the presence of 2.5 μg/mL AFB1 at 37°C for 24 h. (C) Effects of incubation temperature on the degradation of E. coli CG1061 culture supernatant. The experiments were performed in the presence of 2.5 μg/mL AFB1 at pH 7.0 for 24 h. Different letters among groups means significant different by Duncan’s multiple range test (P < 0.05).
The degraded products were analyzed by UPLC Q-TOF MS to identify the possible degradation products. The products of AFB1 appeared at retention times of 4.965 min and showed the m/z ion peaks at 287.09 for the protonated adduct [M+H] +. It was further analyzed by MS-MS to determine the exact masses of the fragmentation ions (Figure 4). It corresponded to molecular formula C16H14O5. Except this, other possible products of low abundance were also identified as C8H4O3, C26H25N3O12S, C17H12O7, and C17H15O5 by the metabolite analysis software MET ID.
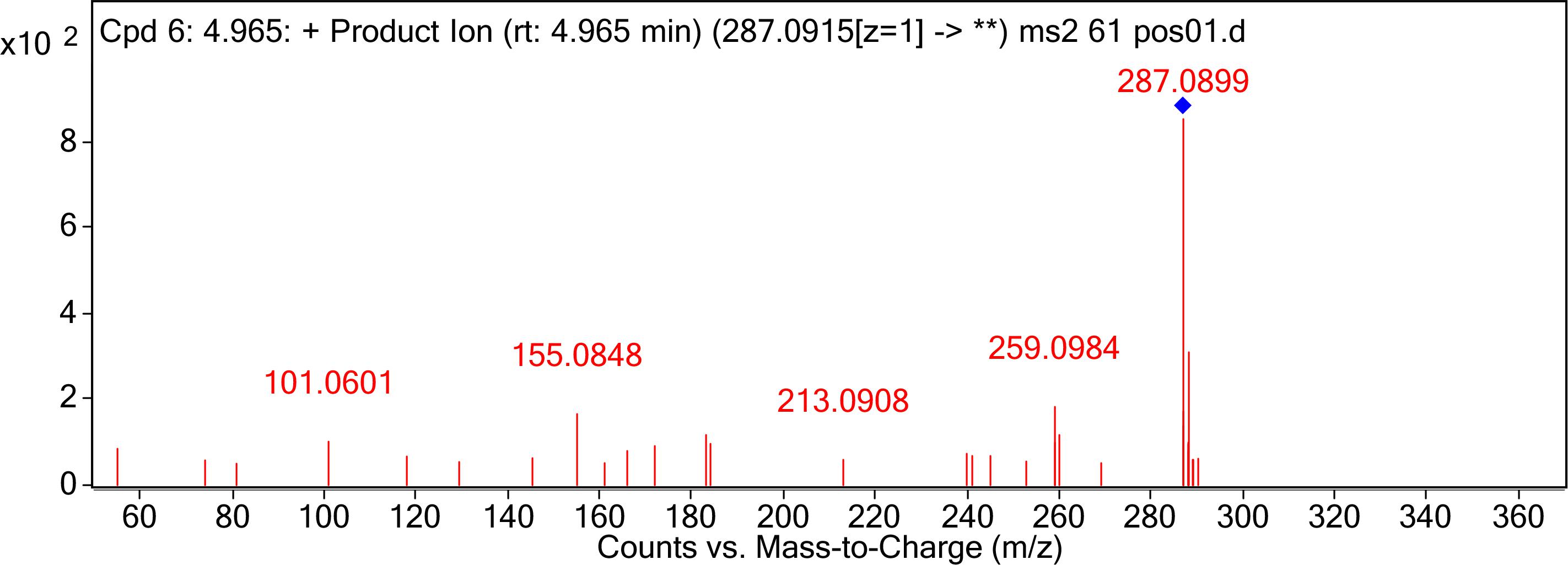
Figure 4. MS/MS spectra analysis of degraded AFB1 products. MS/MS spectra of degradation product with 287.09 m/z. Mass spectrometry analyses were performed using a (Q-TOF) mass spectrometer system in the positive ion mode. Mass spectra were acquired in a full-scan analysis in the range of m/z 50–1000.
In the vitro experiment, CG1061-degraded AFB1 and standard AFB1 were diluted to final concentrations of 3.75, 2.00, 1.00, 0.50, and 0.25 μg/mL. The viability of LMH cells treated with standard AFB1 was from 79.2% (0.25 μg/mL) to 20.1% (3.75 μg/mL), and the cell viability treated with CG1061-degraded AFB1 was from 93.4% (0.25 μg/mL) to 70.5% (3.75 μg/mL) (Figure 5). The viability of cells treated with CG1061-degraded AFB1 was obviously higher than that of standard AFB1 at the same concentration, implying that the cytotoxicity of CG1061-degraded AFB1 was much lower than that of standard AFB1 on LMH cells.
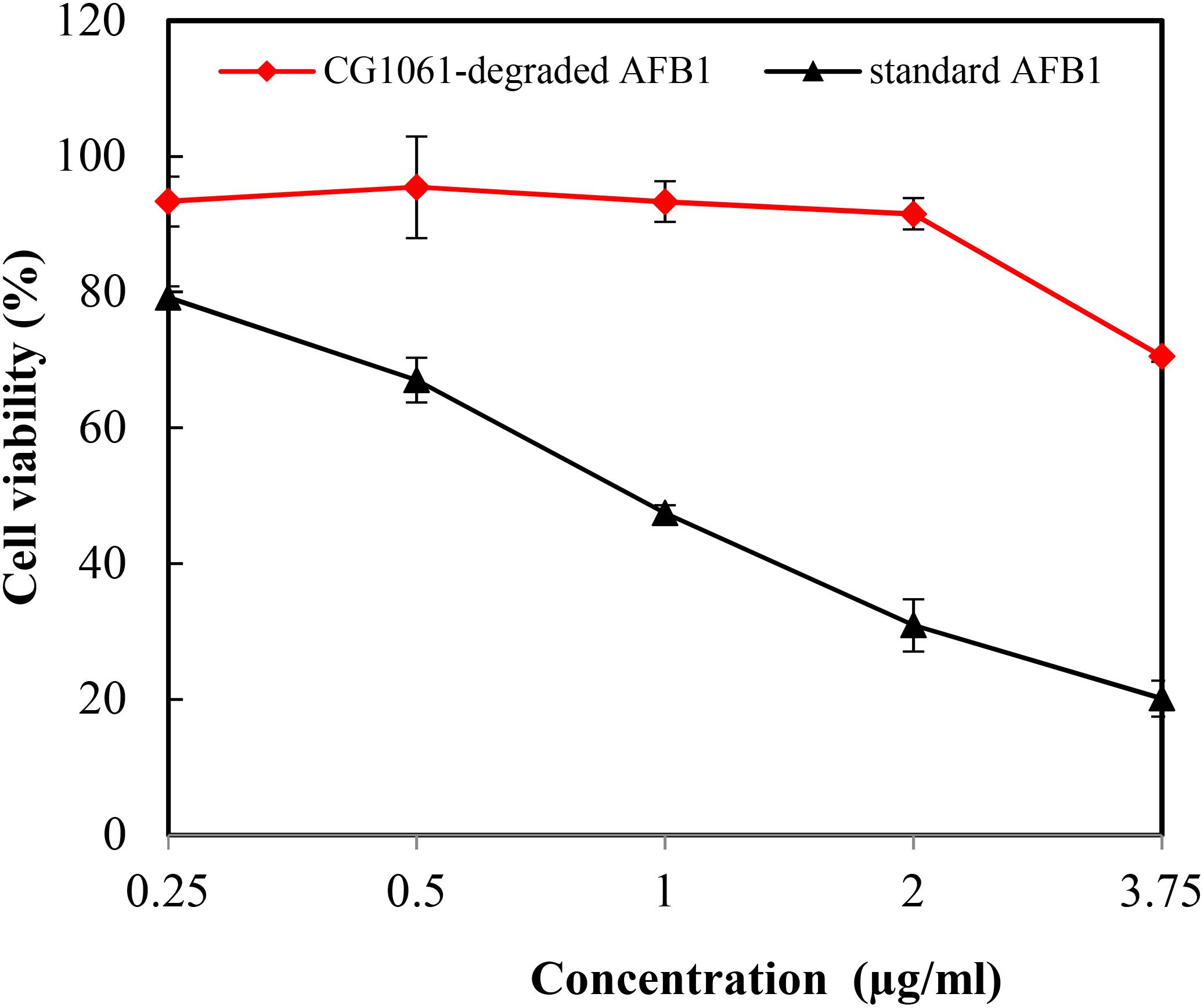
Figure 5. Viability of the LMH cells treated with standard AFB1 and with CG1061-degraded AFB1. The experiments were performed by the MTT method. LMH cells were incubated with samples in 96-well plates at 37°C for 48 h in a 5% CO2 incubator.
In vivo experiment, after animals were administrated with standard AFB1 or CG1061-degraded AFB1, the activity of hepatic ALP, SOD, GSH were measured respectively. As shown in Table 2, the activity of hepatic ALP treated with standard AFB1 was significant increased (P < 0.05) to 148% compared with the control. ALP was an important disease biomarker (Wei et al., 1993), and its significant increase indicated that the administration of AFB1 (100 μg/kg of BW) caused liver damage. The activity of ALP did not exhibit any significant differences between that treated with CG1061-degraded AFB1 and the control. For the level of SOD and GSH, there is no significant change among all the groups. Based on the results of in vitro and in vivo experiments, we concluded that AFB1 has been transformed into lower toxicity products by E. coli CG1061.
The present study we successfully isolated an AFB1 detoxifying bacteria, Escherichia coli CG1061 from chicken cecum. E. coli CG1061 could eliminate AFs effectively, which reduced AFB1 by 93.7% after aerobically cultured for 72 h. The further studies indicated that the active component maybe kinds of heat-resistant compounds, and it is secreted into the extracellular space. In many cases, the supernatant of culture is identified as the degrading matrix and the active component was proved to be heat-resistant. Wang et al. (2017) reported AFB1 degradation by the cell-free supernatant of TADC7 microbial consortium was stable up to 90°C. Xia et al. (2017) reported the AFB1-degrading activity of isolate Bacillus subtilis JSW-1 was predominantly attributed to the cell-free supernatant and this activity was found to be heat stable but sensitive to proteinase K treatment. However, the enzymes responsible for AFB1 degradation was not purified and identified, the mechanisms involved are still unknown.
In our research, culture supernatant of CG1061 degraded AFB1 for nearly 80% in first 24 h. The results were similar to those reported for Bacillus subtilis UTBSP1 (Farzaneh et al., 2012) and Bacillus licheniformis CFR1 (Rao et al., 2017). Alberts observed 33.2% of residual AFB1 after an incubation in the presence of Rhodococcus erythropolis extracellular extracts for 72 h at 30°C (Alberts et al., 2006). Mycobacterium DSM44556 degraded more than 90% of AFB1 within 4 h at 30°C (Teniola et al., 2005). Wang et al., 2017 reported the degradation rate of a Fusarium sp. WCQ3361 cell-free culture in 1 min at 30°C was 70.2%. There were great differences in degradation efficiency from strains to strains. Next, we explored the optimal degradation conditions, the highest degradation activities of culture supernatant of E. coli CG1061 appeared at pH 8.5 and 55°C. Xu et al. (2017) purified AF-degrading enzyme from Bacillus shackletonii L7, which exhibited the highest activity at pH 8.0 and 70°C. The degradation rate of a Fusarium sp. supernatant at 90°C was greater than 73% (Wang et al., 2017), and its optimal reaction pH value was 7.0. Motomura et al., 2003 reported optimum activities of an extracellular enzyme from fungi Pleurotus ostreatus at pH values ranging from 4.0 to 5.0 and at the temperature of 25°C. These reports showed the optimal degradation conditions varied with difference microorganisms, suggesting that there may be multiple AFB1 metabolic types in microorganisms. Some strains showed high degradation activity at high temperature (more than 70°C), the mechanism of AFB1 degradation requires further study.
The enzymes responsible for degradation may have different target sites on AFB1 molecule. They were reported for catalyzing the bisfuran ring or coumarin group of AFB1. The different action sites lead to different AFB1 degradation products. In this study the metabolite C16H14O5 degradated by E. coli CG1061 was determined by UPLC Q-TOF MS. Among of reported degradation products of AFB1, the molecular formula of AFD1 is C16H14O5. AFD1 was chemically-transformed from AFB1 during the ammonization process (Grove et al., 1984). AFD1 is produced by the opening of the lactone ring of AFB1. The lactone ring is the actual group of AFB1 responsible for its toxic effects (Vanhoutte et al., 2016). Samuel et al. (2014) and Eshelli et al. (2015) reported the bio-transformation of AFB1 to AFD1 by Pseudomonas putida and Rhodococcus erythropolis respectively. In this research, whether the product C16H14O5 was AFD1 or its isomer remain to be further studied. Liu et al. (1998) reported that Armillariella tabescense multienzyme exhibits detoxifying activity by opening the difuran ring of AFB1. Besides these, aflatoxicol, AFD2 and AFB2a have been reported as AFB1 degradation products (Nakazato et al., 1990; Samuel et al., 2014; Chen et al., 2015). In some cases, no degradation products were detected (Alberts et al., 2006; Farzaneh et al., 2012).
AFB1 degradation is not always link to less toxic product. So, it is crucial to confirm their toxicity using reliable methods. Approaches of toxicity research of the AFB1 metabolites mainly included Ames assay (mutagenicity assessment), SOS-chromotest (mutagenicity assessment), MTT test (cytotoxicity assessment) at present (Alberts et al., 2009; Samuel et al., 2014; Risa et al., 2018). AFB1 exposure always leads to liver damage. To determine the toxicity of AFB1 degradation products of E. coli CG1061, we performed the MTT test on chicken hepatocellular carcinoma cells and the animal toxicity test conducted with mice in this study, which demonstrated the degradation metabolites less toxic than parent AFB1. In the animal test, the activity of hepatic ALP was significantly increased (P < 0.05) when treated with standard AFB1, but for the level of SOD and GSH there was no significant change under same treatment. The results indicated that the activity of hepatic ALP was more sensitive to AFB1 than SOD and GSH which were connected with oxidative stress and the antioxidant activity. Kojima and Sakurada (1976) reported ALP activity increased more significantly than that of the other enzymes in the liver of metabolic abnormalities mice.
In summary, non-pathogenic E. coli 1061 was reported for effectively removing AFB1, its degradation active component was constitutively produced and thermo-stability. The optimal incubation time, pH and temperature were 24 h, pH 8.5 and 55°C respectively. AFB1 was bio-transformed to new compounds, which were proved less toxicity by toxicity assessment. From the finding of this investigation, Inoculants of E. coli 1061 or enzymes prepared from culture supernatant can be used for detoxification of AFB1-contaminated food and feed raw materials.
LW designed the experiments and wrote the manuscript. JW analyzed the data and revised the draft. ZL performed the MTT test and animal experiments. JL co-performed the animal experiments. YS detected AFB1 degrading rates. XX performed the pathogenicity of strains. SH isolated strains from chicken cecum. PM analyzed the data. FD co-designed the experiments. YD defined the research theme and supervised the conduct of experiments.
The research was supported by grants from the Natural Science Foundation of China (31600102), the Natural Science Foundation of Guangdong Province (2015A030312005), Science and Technology Program of Guangzhou City (201804020067), Science and Technology Key Program of Guangzhou City (201607010178), and Science and Technology Planning Project of Guangdong Province (2016A020210025).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Akbas, M. Y., and Ozdemir, M. (2006). Effect of different ozone treatments on aflatoxin degradation and physicochemical properties of pistachios. J. Sci. Food Agricult. 86, 2099–2104. doi: 10.1002/jsfa.2579
Alberts, J. F., Engelbrecht, Y., Steyn, P. S., Holzapfel, W., and van Zyl, W. (2006). Biological degradation of aflatoxin B-1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 109, 121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019
Alberts, J. F., Gelderblom, W. C. A., Botha, A., and van Zyl, W. H. (2009). Degradation of aflatoxin B-1 by fungal laccase enzymes. Int. J. Food Microbiol. 135, 47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022
Arzandeh, S., and Jinap, S. (2011). Effect of initial aflatoxin concentration, heating time and roasting temperature on aflatoxin reduction in contaminated peanuts and process optimisation using response surface modelling. Int. J. Food Sci. Technol. 46, 485–491. doi: 10.1111/j.1365-2621.2010.02514.x
Aziz, N. H., and Moussa, L. A. A. (2002). Influence of gamma-radiation on mycotoxin producing moulds and mycotoxins in fruits. Food Control 13, 281–288. doi: 10.1016/s0956-7135(02)00028-2
Chen, Y. J., Kong, Q., Chi, C., Shan, S. H., and Guan, B. (2015). Biotransformation of aflatoxin B-1 and aflatoxin G(1) in peanut meal by anaerobic solid fermentation of Streptococcus thermophilus and Lactobacillus delbrueckii subsp bulgaricus. Int. J. Food Microbiol. 211, 1–5. doi: 10.1016/j.ijfoodmicro.2015.06.021
Ciegler, A., Lillehoj, E. B., Peterson, R. E., and Hall, H. H. (1966). Microbial detoxification of aflatoxin. Appl. Microbiol. 14, 934–939.
Das, A., Bhattacharya, S., Palaniswamy, M., and Angayarkanni, J. (2014). Biodegradation of aflatoxin B1 in contaminated rice straw by Pleurotus ostreatus MTCC 142 and Pleurotus ostreatus GHBBF10 in the presence of metal salts and surfactants. World J. Microbiol. Biotechnol. 30, 2315–2324. doi: 10.1007/s11274-014-1657-5
Di Gregorio, M. C., de Neeff, D. V., Jager, A. V., Corassin, C. H., Carao, A. C. D., de Albuquerque, R., et al. (2014). Mineral adsorbents for prevention of mycotoxins in animal feeds. Toxin Rev. 33, 125–135. doi: 10.3109/15569543.2014.905604
El-Deeb, B., Altalhi, A., Khiralla, G., Hassan, S., and Gherbawy, Y. (2013). Isolation and characterization of endophytic bacilli bacterium from maize grains able to detoxify aflatoxin B1. Food Biotechnol. 27, 199–212. doi: 10.1080/08905436.2013.811083
Eshelli, M., Harvey, L., Edrada-Ebel, R., and McNeil, B. (2015). Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0. Toxins 7, 439–456. doi: 10.3390/toxins7020439
Farzaneh, M., Shi, Z. Q., Ghassempour, A., Sedaghat, N., Ahmadzadeh, M., Mirabolfathy, M., et al. (2012). Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 23, 100–106. doi: 10.1016/j.foodcont.2011.06.018
Gao, X. J., Mu, P. Q., Wen, J. K., Sun, Y., Chen, Q. M., and Deng, Y. Q. (2018). Detoxification of trichothecene mycotoxins by a novel bacterium, Eggerthella sp DII-9. Food Chem. Toxicol. 112, 310–319. doi: 10.1016/j.fct.2017.12.066
Grove, M. D., Plattner, R. D., and Peterson, R. E. (1984). Detection of aflatoxin D1 in ammoniated corn by mass spectrometry-mass spectrometry. Appl. Environ. Microbiol. 48, 887–889.
Guan, S., He, J. W., Young, J. C., Zhu, H. H., Li, X. Z., Ji, C., et al. (2009). Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 290, 290–295. doi: 10.1016/j.aquaculture.2009.02.037
Guan, S., Ji, C., Zhou, T., Li, J. X., Ma, Q. G., and Niu, T. G. (2008). Aflatoxin B-1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 9, 1489–1503. doi: 10.3390/ijms9081489
Guan, S., Zhou, T., Yin, Y., Xie, M., Ruan, Z., and Young, J. C. (2011). Microbial strategies to control aflatoxins in food and feed. World Mycotoxin J. 4, 413–424. doi: 10.3920/wmj2011.1290
Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G., and Gordon, J. I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884. doi: 10.1126/science.291.5505.881
International Agency for Research on Cancer [IARC] (1993). Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Lyon: IARC.
Juri, M. G. F., Dalcero, A. M., and Magnoli, C. E. (2015). In vitro aflatoxin B-1 binding capacity by two Enterococcus faecium strains isolated from healthy dog faeces. J. Appl. Microbiol. 118, 574–582. doi: 10.1111/jam.12726
Kojima, Y., and Sakurada, T. (1976). Increase in alkaline phosphatase activity in the liver of mice bearing ehrlich ascites tumor. Cancer Res. 36, 23–27.
Kollarczik, B., Gareis, M., and Hanelt, M. (1994). In vitro transformation of the Fusarium mycotoxins deoxynivalenol and zearalenone by the normal gut microflora of pigs. Nat. Toxins 2, 105–110. doi: 10.1002/nt.2620020303
Lee, L. S., and Cucullu, A. F. (1978). Conversion of aflatoxin B1 to aflatoxin D1 in ammoniated peanut and cottonseed meals. J. Agric. Food Chem. 26, 881–884. doi: 10.1021/jf60218a036
Liu, D.-L., Yao, D.-S., Liang, R., Ma, L., Cheng, W.-Q., and Gu, L.-Q. (1998). Detoxification of aflatoxin B1 by enzymes isolated from Armillariella tabescens. Food Chem. Toxicol. 36, 563–574. doi: 10.1016/s0278-6915(98)00017-9
Motomura, M., Toyomasu, T., Mizuno, K., and Shinozawa, T. (2003). Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 158, 237–242. doi: 10.1078/0944-5013-00199
Nakazato, M., Morozumi, S., Saito, K., Fujinuma, K., Nishima, T., and Kasai, N. (1990). Interconversion of aflatoxin B1 and aflatoxicol by several fungi. Appl. Environ. Microbiol. 56, 1465–1470.
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. doi: 10.1128/CMR.11.1.142
Norred, W. P., and Morrissey, R. E. (1983). Effects of long-term feeding of ammoniated, aflatoxin-contaminated corn to Fischer 344 rats. Toxicol. Appl. Pharmacol. 70, 96–104. doi: 10.1016/0041-008x(83)90182-5
Persson, S., Olsen, K. E. P., Scheutz, F., Krogfelt, K. A., and Gerner-Smidt, P. (2007). A method for fast and simple detection of major diarrhoeagenic Escherichia coli in the routine diagnostic laboratory. Clin. Microbiol. Infect. 13, 516–524. doi: 10.1111/j.1469-0691.2007.01692.x
Rao, K. R., Vipin, A. V., Hariprasad, P., Appaiah, K. A. A., and Venkateswaran, G. (2017). Biological detoxification of Aflatoxin B-1 by Bacillus licheniformis CFR1. Food Control 71, 234–241. doi: 10.1016/j.foodcont.2016.06.040
Risa, A., Krifaton, C., Kukolya, J., Kriszt, B., Cserhati, M., and Tancsics, A. (2018). Aflatoxin B1 and zearalenone-detoxifying profile of rhodococcus type strains. Curr. Microbiol. 75, 907–917. doi: 10.1007/s00284-018-1465-5
Samuel, M. S., Sivaramakrishna, A., and Mehta, A. (2014). Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegradation 86, 202–209. doi: 10.1016/j.ibiod.2013.08.026
Teniola, O. D., Addo, P. A., Brost, I. M., Farber, P., Jany, K. D., Alberts, J. F., et al. (2005). Degradation of aflatoxin B-1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp nov DSM44556(T). Int. J. Food Microbiol. 105, 111–117. doi: 10.1016/j.ijfoodmicro.2005.05.004
Vanhoutte, I., Audenaert, K., and De Gelder, L. (2016). Biodegradation of mycotoxins: tales from known and unexplored worlds. Front. Microbiol. 7:20. doi: 10.3389/fmicb.2016.00561
Verheecke, C., Liboz, T., and Mathieu, F. (2016). Microbial degradation of aflatoxin B1: current status and future advances. Int. J. Food Microbiol. 237, 1–9. doi: 10.1016/j.ijfoodmicro.2016.07.028
Vipin, A. V., Rao, K. R., Kurrey, N. K., Appaiah, K. A. A., and Venkateswaran, G. (2017). Protective effects of phenolics rich extract of ginger against Aflatoxin B-1-induced oxidative stress and hepatotoxicity. Biomed. Pharmacother. 91, 415–424. doi: 10.1016/j.biopha.2017.04.107
Wang, C. Q., Li, Z. Y., Wang, H., Qiu, H. Y., Zhang, M. H., Li, S., et al. (2017). Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agric. 97, 1342–1348. doi: 10.1002/jsfa.7872
Wei, J. S., Chung, N. C., Wei, L. L., Tzeng, W. F., Liu, T. Z., and Wang, J. Y. (1993). High-molecular-mass alkaline phosphatase as a tumor marker for colorectal cancer: comparison of two test methods. Clin. Chem. 39, 540–543.
Worrell, N. R., Mallett, A. K., Cook, W. M., Baldwin, N. C., and Shepherd, M. J. (1989). The role of gut micro-organisms in the metabolism of deoxynivalenol administered to rats. Xenobiotica 19, 25–32. doi: 10.3109/00498258909034673
Xia, X., Zhang, Y., Li, M. Y., Garba, B., Zhang, Q., Wang, Y., et al. (2017). Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Cont. 75, 92–98. doi: 10.1016/j.foodcont.2016.12.036
Xu, L., Eisa Ahmed, M. F., Sangare, L., Zhao, Y., Selvaraj, J. N., Xing, F., et al. (2017). Novel aflatoxin-degrading enzyme from Bacillus shackletonii L7. Toxins 9:36. doi: 10.3390/toxins9010036
Yu, H., Zhou, T., Gong, J. H., Young, C., Su, X. J., Li, X. Z., et al. (2010). Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 10:9. doi: 10.1186/1471-2180-10-182
Keywords: aflatoxin B1, Escherichia coli, degradation, detoxification, chicken cecum
Citation: Wang L, Wu J, Liu Z, Shi Y, Liu J, Xu X, Hao S, Mu P, Deng F and Deng Y (2019) Aflatoxin B1 Degradation and Detoxification by Escherichia coli CG1061 Isolated From Chicken Cecum. Front. Pharmacol. 9:1548. doi: 10.3389/fphar.2018.01548
Received: 30 September 2018; Accepted: 18 December 2018;
Published: 17 January 2019.
Edited by:
Hani El-Nezami, The University of Hong Kong, Hong KongReviewed by:
Carol Verheecke-Vaessen, Cranfield University, United KingdomCopyright © 2019 Wang, Wu, Liu, Shi, Liu, Xu, Hao, Mu, Deng and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiqun Deng, eXFkZW5nQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.