- 1Institute of Clinical Medicine, China-Japan Friendship Hospital, Beijing, China
- 2Beijing University of Chinese Medicine, Beijing, China
- 3Department of Gastroenterology, People’s Hospital of Yichun, Yichun, China
- 4Department of Laboratory Medicine, China-Japan Friendship Hospital, Beijing, China
- 5Department of Rheumatology, China-Japan Friendship Hospital, Beijing, China
- 6Department of TCM Rheumatology, China-Japan Friendship Hospital, Beijing, China
- 7Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 8Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, AB, Canada
Objective: Analysis of the relationship between single nucleotide polymorphisms (SNPs) and outcomes of methotrexate (MTX) therapy for rheumatoid arthritis (RA) in China.
Materials and Methods: TYMS 28 bp VNTR (rs34743033), MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)], ATIC 347C>G (rs2372536), MTR A2756G (rs1805087), and MTRR 66A>G (rs1801394) enzyme proteins may be related to the outcomes of MTX therapy, according to our previous meta-analysis. A total of 162 patients with RA were included in our study. SNPs were evaluated using polymerase chain reaction (PCR). Disease Activity Score 28 (DAS28) was used to evaluate the clinical response, and adverse drug reactions (ADRs) were collected after physical examinations of the patients.
Results: The MTHFR 677C>T gene showed a relationship with the ADRs of MTX in the Recessive model [TT vs. (CC+CT)] (p = 0.04, OR = 2.20, 95% CI: 1.01, 4.77). In the Codominant model [CT vs. (CC+TT)], the MTHFR 677C>T gene also showed a trend of association with ADRs (p = 0.08, OR = 0.52, 95% CI: 0.25, 1.08). No significant difference was found between TYMS, MTHFR, ATIC, MTR, and MTRR gene polymorphisms and the RA response or ADRs related to MTX in our study.
Conclusion: Our results showed that the MTHFR [677C>T (rs1801133)] TT genotype is associated with ADRs to MTX in Chinese RA patients. Other SNPs, including TYMS 28bp VNTR (rs34743033), MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)], ATIC 347C>G (rs2372536), MTR A2756G (rs1805087), and MTRR 66A>G (rs1801394) gene polymorphisms, were not associated with MTX treatment outcomes. Further studies are required to validate these findings.
Introduction
Rheumatoid arthritis (RA) is characterized by synovial inflammation, cartilage damage and bone erosion (Walter et al., 2016; Fessler et al., 2017). The pathogenesis of RA, which is a systemic inflammatory disease, remains unclear. Additionally, curative therapy for RA has not been established. Currently, disease-modifying anti-rheumatic drugs (DMARDs) are usually used to delay symptom progression and achieve pain relief (Kojima et al., 2016). One DMARD, methotrexate (MTX), is widely used worldwide to treat RA because it is inexpensive, effective and safe (Swierkot et al., 2015); however, 30–40% of patients on MTX therapy fail to attain remission (Ranganathan and McLeod, 2006), and 30–40% of patients have problems with safety and tolerability (Wevers-de Boer et al., 2012). Six single nucleotide polymorphisms (SNPs) that encode proteins involved in MTX metabolism are examined in our meta-analysis (Qiu et al., 2017a,b).
Methotrexate has 1 glutamate moiety, so it is also referred to as MTX polyglutamate (MTXPG1). MTX polyglutamates (MTXPGs) can persist in the long term inside the target cell, and folylpolyglutamate synthase (FPGS) converts MTX into MTXPGs in the cells. After gamma-glutamyl hydrolase (GGH) converts MTXPGs to MTX by removing the glutamates, the drug is removed from the cells (Yamamoto et al., 2016).
Methotrexate polyglutamates inhibit thymidylate synthase (TYMS), which is a key protein involved in the de novo pyrimidine synthesis pathway. Methylenetetrahydrofolate reductase (MTHFR) is also a key enzyme in the de novo synthesis pathway (Song et al., 2014). TYMS competes with MTHFR for one of its substrates: 5,10-MTHF (Chaabane et al., 2018). MTXPGs can inhibit aminoimidazole carboxamide ribonucleotide transformylase (ATIC), causing the intracellular accumulation of aminoimidazole carboxamide ribonucleotide, which in turn inhibits adenosine-metabolizing enzymes. MTXPGs also directly inhibit methionine synthase (MTR) and methionine synthase reductase (MTRR) (Kato et al., 2012).
The aims of our study were to determine the correlation of the TYMS 28bp VNTR (rs34743033), MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)], ATIC 347C>G (rs2372536), MTR A2756G (rs1805087) and MTRR 66A>G (rs1801394) polymorphisms with the clinical response and adverse drug reactions (ADRs) to MTX treatment in Chinese RA patients.
Materials and Methods
Materials
This was a retrospective study conducted between April 2016 and April 2018 at China-Japan Friendship Hospital and People’s Hospital of Yichun of a cohort of consecutive Chinese Han patients with RA treated with MTX. A total of 162 patients from 23 provinces were included in our study. All the patients who were selected had to meet the 2010 revised classification criteria of the American College of Rheumatology and European League against Rheumatism (Aletaha et al., 2010). Each patient had been treated with MTX (mean dose = 8.92 ± 2.26 mg/week) for at least 3 months. DAS28 was used to evaluate the MTX treatment response (Versteeg et al., 2018). The DAS28 of RA patients in our study was 5.17 ± 1.5. And the mean duration of RA patients was 4 years. Patients were excluded if they had recently been pregnant or desired to become pregnant. All of this study’s protocols involving human subjects were approved by the ethics committee of People’s Hospital of Yichun (ethics ID: 2014-01). Written informed consent was obtained from each participant.
We also recorded analytical data, such as body mass index (BMI), erythrocyte sedimentation rate (ESR), C-reaction protein (CRP), rheumatoid factor (RF), cyclic citrullinated peptide (CCP), antinuclear antibody (ANA), and anti-keratin antibody (AKA) levels.
Methods
DNA Extraction
For the genetic analysis, 2 ml of peripheral blood was obtained from each patient using the standard venipuncture technique. Each sample was centrifuged, and the white blood cells were separated by pipette. The samples were stored at -80°C. DNA was extracted using the QIA amp DNA Blood Mini Kit according to the QIAGEN manufacturer’s instructions (Hilden, Florida, Germany). Quality control procedures were performed using a ND-1000 Spectrophotometer by NanoDrop Technologies (Wilmington, DE, United States) to ensure the sample and polymorphism genotyping success rates before analysis by requiring that the 260/280 was between 1.8 and 1.9 (Owen et al., 2013). We determined the precise length of the genomic DNA by gel electrophoresis using 1% agarose gel. All the purified samples were stored at -80°C until the next analyses.
Allele Genotyping
The gene polymorphisms were produced by polymerase chain reaction (PCR) and confirmed through sequencing. The forward and reverse primers were designed by Primer 5 and are described in Table 1. Six SNPs were examined in this study. All polymorphisms were genotyped using sequencing. PCR reactions were performed using 3 μl of 10x PCR reaction buffer (Osaka, Kansai, Japan), 5 μl of 100 mmol/L dNTP (Osaka, Kansai, Japan), 3 μl of each primer at a concentration of 10 μM, 14 μl of Rnase- and DNase-free water (Osaka, Kansai, Japan), and 2 μl of sample DNA at a total volume of 30 μl per single-well reaction. Assay conditions were 3 min at 94°C, 30 s at 58°C, 30 s at 94°C and 40 cycles of 72°C for 1 min. Then it was kept at 72°C for 10 min. Chromas software version 2.23 (Brisbane, QLD, Australia) was used as an absolute quantification assay to analyze the SNP assay. Each of the MTHFR677C>T (rs1801133) genes and TYMS 28bp VNTR (rs34743033) had another genetic polymorphisms, and we abandoned these data when analyzing the relationship between the gene polymorphisms and MTX treatment outcomes.
Statistical Analysis
Ethnic and gender differences in the distribution of allelic frequencies among genotypes were tested using the Hardy–Weinberg equilibrium. A p-value of <0.05 was considered to represent a significant difference in all statistical analyses. All statistical data are described as numbers and frequencies. Depending on data distribution, Student’s t-test or Mann–Whitney’s U-test was used to analyze continuous variables. Means and standard deviations (±SD) or interquartile ranges (IQRs) were used to describe the sample. Allele and genotype risk was assessed using odds ratios (ORs) and 95% confidence interval values (CIs) based on the chi-square test. When the expected count was less than 5, we used the Fisher chi-square test instead of the chi-square test. Statistical analysis was performed with Statistical Package for Social Sciences (SPSS) Version 20 (Armonk, NY, United States).
The gene polymorphisms were analyzed using five models; for example, MTRR 66A>G (rs1801394) was analyzed with the Pre-allele model (A vs. G), the Dominant model [AA vs. (AG+GG)], the Recessive model [GG vs. (AG+AA)], the Codominant model [AG vs. (AA+GG)] and the Homozygotic model (AA vs. GG).
Results
Description of the Population
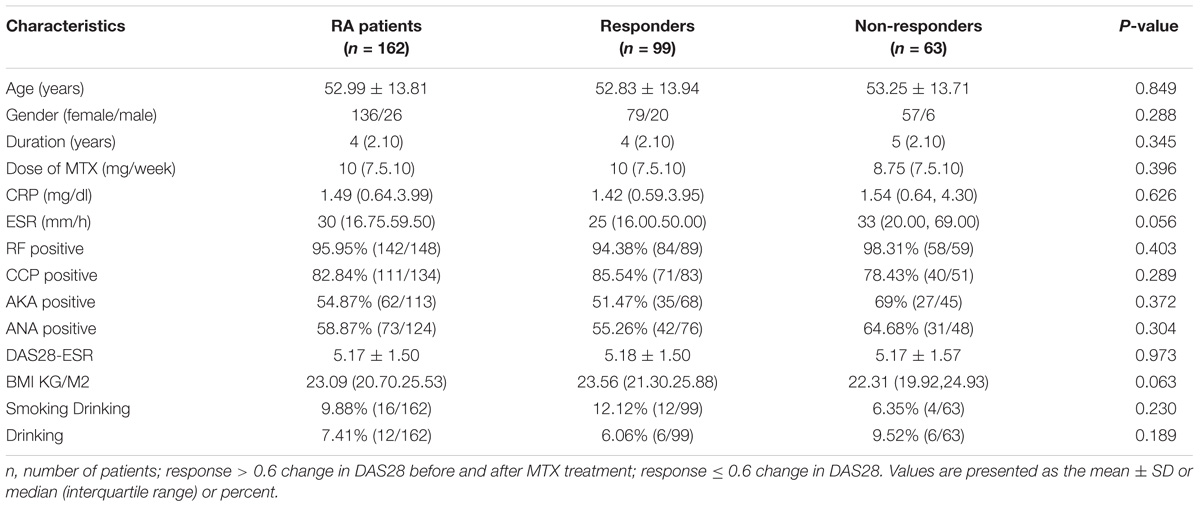
A total of 162 RA patients were observed in this study, and the characteristics of the RA group are presented in Table 2. All the patients were treated with MTX for at least 3 months. Our results showed no differences in terms of age, sex, DAS28 results, MTX dose, treatment duration, ESR, CPR, RF positive status, CCP positive status, AKA positive status, ANA positive status, individual variables of DAS28 and smoking and drinking habits between the responder and non-responder groups.
Frequencies and Polymorphisms of TYMS, MTHFR, ATIC, MTR, and MTRR
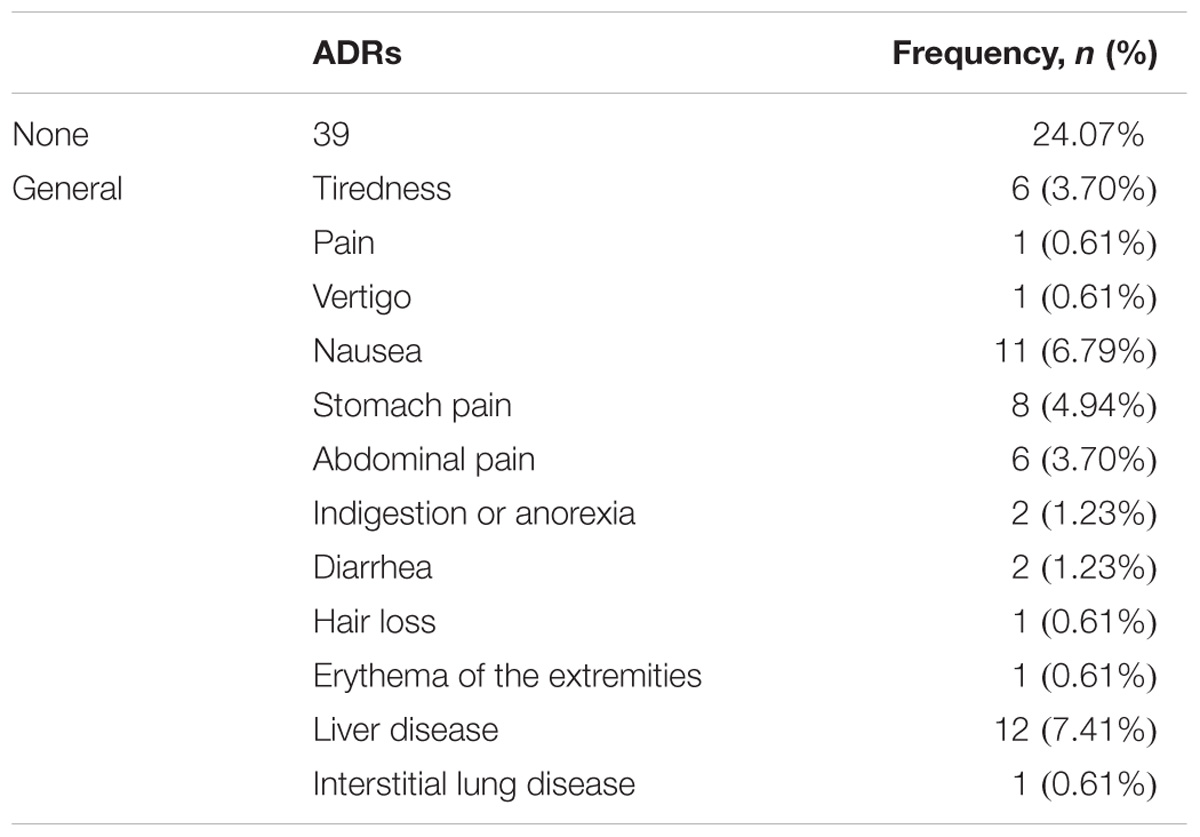
Among the 162 RA patients, 99 RA patients responded to MTX therapy, and 63 RA patients did not. During MTX therapy, 39 RA patients among 162 RA patients developed ADRs, as illustrated in Table 3. Gastrointestinal reactions (abdominal and stomach pain, diarrhea, nausea and indigestion or anorexia) were the most frequent events (29/162 patients, 17.90%); hepatic reactions were less frequent (12/162 patients, 7.41%). Interstitial lung disease, pain throughout the body, erythema of the extremities, hair loss and vertigo reactions were the least frequent ADRs, 0.61% (1/162 patients). Adverse drug events were observed after 3 months or 6 months.
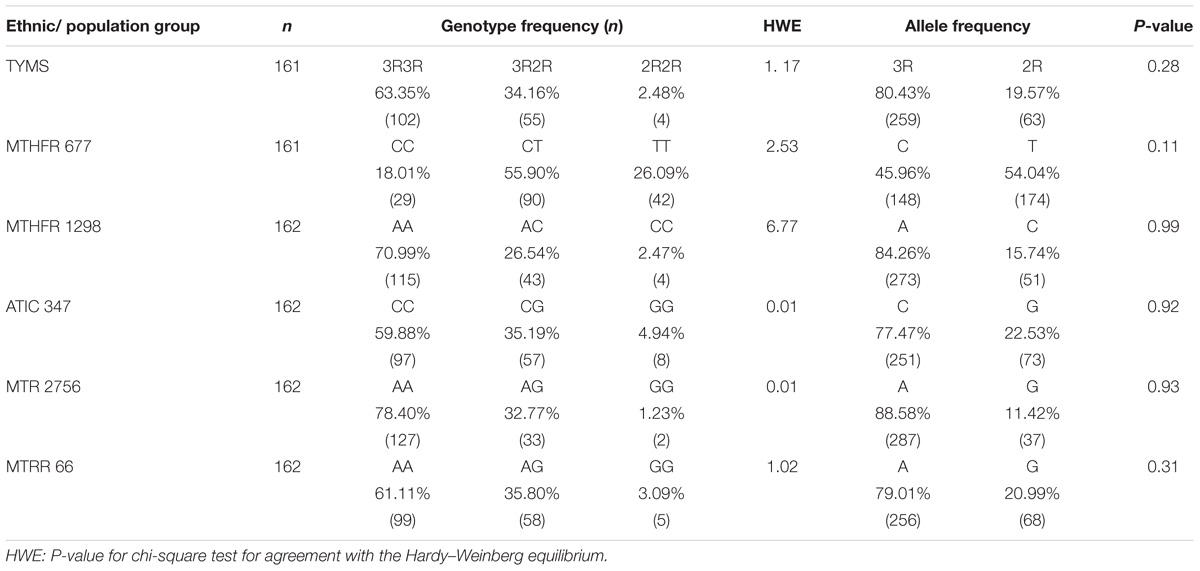
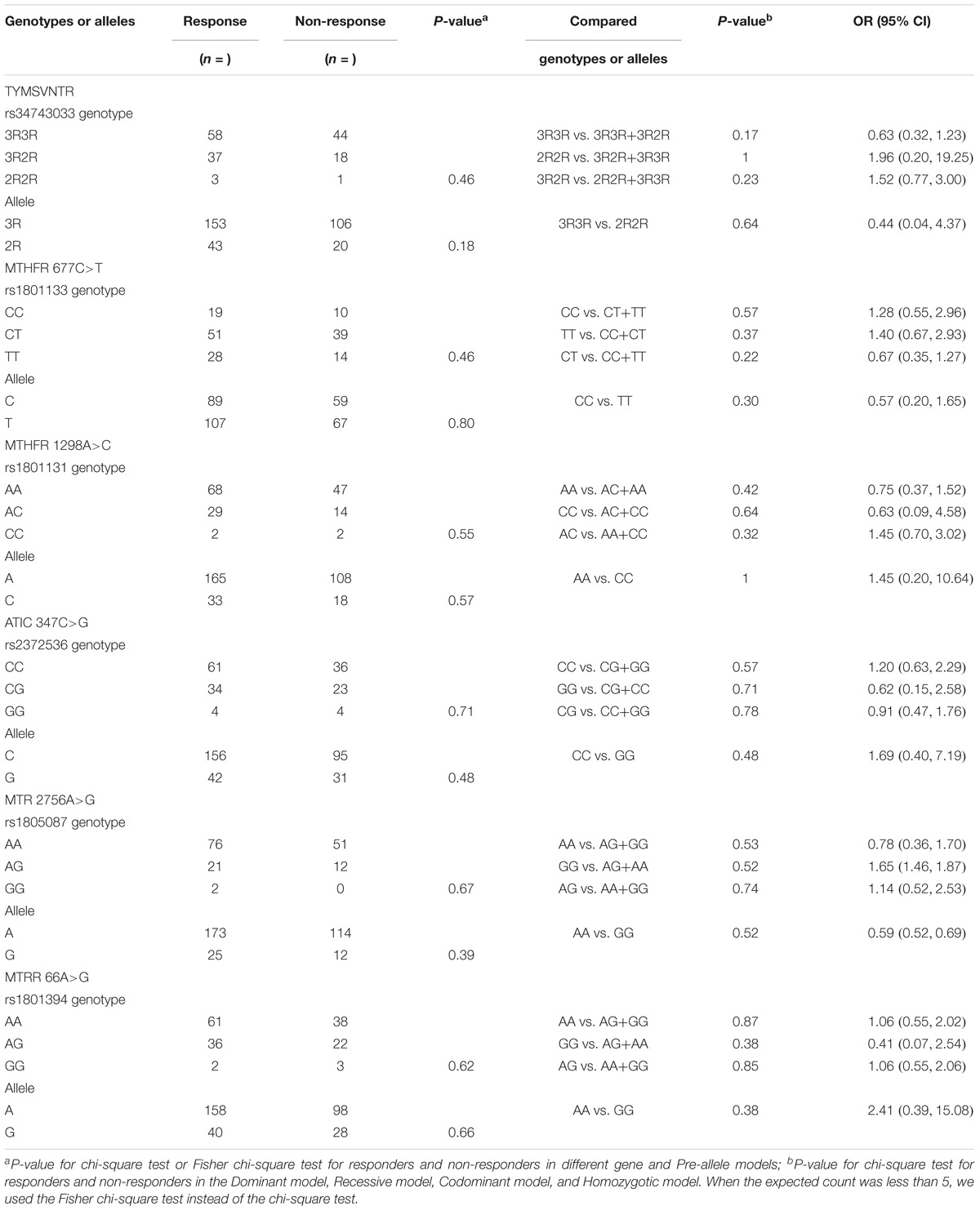
Table 4 presents a comparison of the distribution of MTX-related enzyme gene polymorphisms according to the Hardy–Weinberg equilibrium. All p-values for the chi-square test for agreement with the Hardy–Weinberg equilibrium had no statistical significance (P < 0.05). Table 5 presents the TYMS, MTHFR, ATIC, MTR, and MTRR gene polymorphism distributions of responders and non-responders. Our results showed no significant difference in the frequency distribution of genotypes, alleles, and haplotypes of TYMS 28bp VNTR (rs34743033), MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)], ATIC 347C>G (rs2372536), MTR A2756G (rs1805087) and MTRR 66A>G (rs1801394) in one case using all five statistical models (the Pre-allele model, Dominant model, Recessive model, Codominant model, and Homozygotic model).
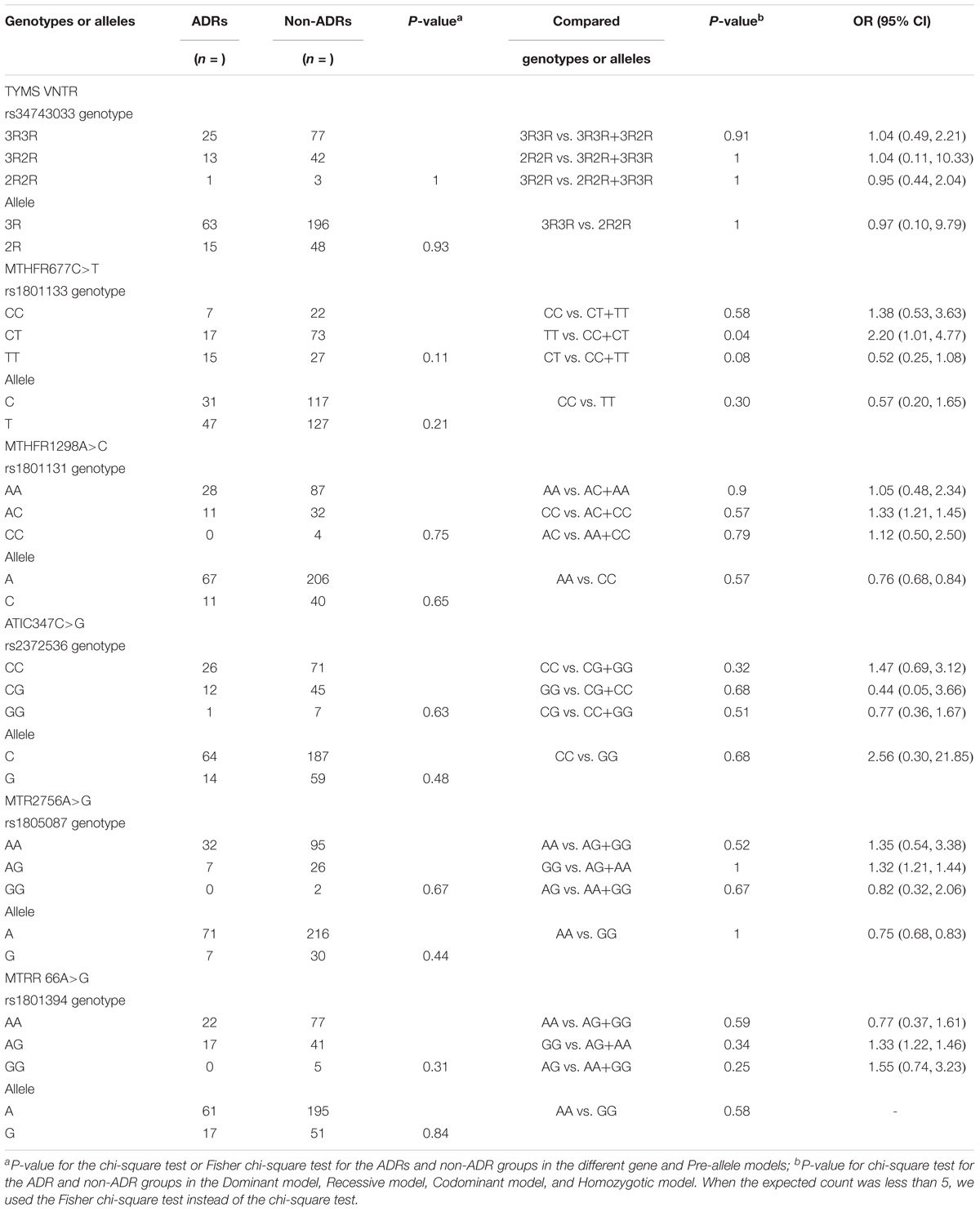
Table 6 presents the frequency distribution of TYMS 28bp VNTR (rs34743033), MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)], ATIC 347C>G (rs2372536), MTR A2756G (rs1805087) and MTRR 66A>G (rs1801394) polymorphisms in patients with and without ADRs. Only one SNP, MTHFR 677C>T (rs1801133), among the five genes was associated with an increased risk of MTX ADRs in our study. In the Recessive model [TT vs. (CC+CT)], the p-value was 0.04 (OR = 2.11, 95% CI: 1.01, 4.77). In the Codominant model [CT vs. (CC+TT)], the p-value was 0.08 (OR = 0.52, 95% CI: 0.25, 1.08), which showed a trend of association. The incidence of adverse effects in another Chinese study (Xiao et al., 2010) was 49.5% (47/95), but in our study, it was 24.07% (39/162). However, the same result was found. In the other study, the p-value of ADRs and MTHFR 677C>T (rs1801133) was 0.01, and the T allele was 0.009. Although the p-values in our study were 0.11 and 0.21, the TT genotype was 0.04. Both studies showed that MTHFR 677C>T (rs1801133) was associated with adverse effects (p < 0.05), and MTHFR 1298A>C (rs1801131) was not (p > 0.05). It is possible that the MTHFR 677C>T (rs1801133) but not the MTHFR 1298A>C (rs1801131) gene was associated with adverse effects in the Chinese population with RA.
Discussion
Analysis of TYMS, MTHFR, ATIC, MTR, and MTRR Polymorphisms Related to the Clinical Efficacy of MTX
Thymidylate synthase is located on chromosome 18q11.32 and is essential for the simultaneous conversion of deoxyuridine monophosphate (dUMP) and 5,10 methylenetetrahydrofolate (5,10-MTHF) to thymidylate monophosphate (dTMP) and dihydrofolate (DHF) (Lima et al., 2014b). Then, dTMP is phosphorylated to deoxythymidine triphosphate (dTTP), which is used for deoxyribonucleic acid (DNA) synthesis and repair (Lima et al., 2013). 5,10-methyl-THF is required for purine and pyrimidine synthesis. The promoter-enhancer region of the TYMS gene contains a double (2R) or a triple (3R) 28-base-pair (28-bp) tandem repeat polymorphism (Corre and Galibert, 2005). The enzymatic activity of TYMS increases with an increasing number of repeated 28-bp sequences because a putative Enhancer box (E-box) sequence is exhibited on the first 28-bp repeat of the 2R allele and on the first two repeats of the 3R allele (Lincz et al., 2007). In the current study, TYMS 2R/3R and 6-bp I/D polymorphisms may not be associated with non-responsiveness to or toxicity of MTX therapy in RA patients (Bae and Lee, 2018). However, Seabra Lima’s study (Lima et al., 2014a) of 233 Caucasian RA patients treated with MTX revealed that the TYMS genotype 3R3R (p = 0.005, OR = 2.34) was associated with non-response to MTX. TYMS gene polymorphisms were not related to MTX treatment outcomes in south Indian Tamil patients with RA (Muralidharan et al., 2017), and the same results were observed in our study.
The MTHFR enzyme is responsible for several crucial cellular processes, and its deficiency can have many consequences for folate status, which can influence the clinical response to MTX treatment (Jekic et al., 2013). In the case of MTHFR, a C is transited to a T in exon 4 of the SNP rs1801133, which entails the change of an alanine for a valine in position 222 of the protein; additionally, an A is transited to a C in exon 7 of the SNP rs1801131, which provokes the change of a glutamine for an alanine in position 429 of the protein (Sala-Icardo et al., 2017). Boughrara et al. (2017) showed that MTHFR [677C>T (rs1801133) and 1298A>C (rs1801131)] gene polymorphisms were not associated with the efficacy of MTX treatment in West Algerian RA patients. In south Asia, Indian RA patients with a MTHFR 1298 A allele were related to those with MTHFR 1298 CC (OR: 2.6; 95% CI: 1.1–5.8; p = 0.02) (Ghodke-Puranik et al., 2015). One meta-analysis showed that MTHFR 1298 gene polymorphisms were not associated with the outcome of MTX treatment in RA patients (Fan et al., 2017), while another meta-analysis indicated that MTHFR C677T and A1298C polymorphisms could be predictors of the risk of RA. However, data are currently insufficient to definitively confirm or refute the association between MTHFR gene polymorphisms and RA. Xiao et al. (2010) found that the MTHFR 677C>T (rs1801131) gene polymorphisms was associated with the clinical response to MTX, while MTHFR 1298A>C (rs1801133) was not. All 110 RA patients in his study were Chinese Han people from Anhui Province. In our study, neither MTHFR 677C>T (rs1801133) nor 1298A>C (rs1801131) was related to the efficacy MTX treatment for RA. The clinical baseline characteristics in Xiao’s study were similar to those of our study and included age, sex, and dose of MTX. However, in our study, the RA patients were from 23 provinces representing approximately 67.65% of China (23/34). It is possible that the inclusion of RA patients from different provinces affected the outcomes, and the inclusion of a larger samples would permit a more definite conclusion regarding the relationship between the MTHFR 677C>T gene polymorphisms and ADRs to MTX in China.
Additionally, intracellular MTXPGs are known to show greater binding affinity for ATIC compared with MTX (Baggott et al., 1986). The ATIC gene is located at chromosome 2q35, and the most common genetic polymorphism investigated is ATIC 347C>G (rs2372536) on exon 5, which results in threonine to serine substitution at position 116 of the gene (Hinks et al., 2011). AICAR will accumulate inside the cells when ATIC is inhibited, causing the release of adenosine into the extracellular space. Adenosine exerts anti-inflammatory effects by reducing neutrophil adherence and inhibiting the activity of natural killer cells, monocyte macrophages and T lymphocytes (Cronstein, 2005). Studies have reported that the ATIC rs2372536 GG genotype is associated with non-response to MTX in Caucasian patients (OR: 2.40; 95% CI: 1.30, 4.42; p = 0.005) (Kurzawski et al., 2016), but not in south Indian populations (OR: 0.98; 95% CI: 0.54–1.77; p = 0.95) (Lee and Bae, 2016; Muralidharan et al., 2016). A similar result was obtained in our present study, which showed that ATIC rs2372536 was not related MTX treatment outcomes in RA patients.
The MTR and MTRR genes play pivotal roles in both homocysteine metabolism (Leclerc et al., 1996) and folate metabolism (Muralidharan et al., 2018). The MTR gene uses the methyl group from 5-MTHF to remethylate homocysteine. The products of this reaction are methionine and tetrahydrofolate (THF) (Nikbakht et al., 2012). MTR is located at 1q 43 and is essential for maintaining adequate intracellular methionine, intracellular folate, and normal homocysteine concentrations (Leclerc et al., 1996). A polymorphism in the MTR gene A2756G (rs1805087) may decrease enzyme activity (Jacques et al., 2003). The MTRR gene is located on chromosome 5p15.3 and encodes for the enzyme MTRR, which is involved in the reductive regeneration of cob (I) alamin (Jekic et al., 2013). In the Netherlands, no associations between the MTRR and MTR gene polymorphisms and good clinical responses were found in recent-onset RA patients (Wessels et al., 2006). A study (Gonzalez-Mercado et al., 2017) of 110 Mexican RA patients who were diagnosed at least 1 year previously and had been receiving MTX for at least 3 months showed no association between MTRR gene polymorphism and MTX efficacy. Another study (Lopez-Rodriguez et al., 2018) of European Caucasian RA patients showed that MTRR fulfilled the high association standards. In Japan, Kato et al. (2012) showed that RA patients with the GG genotype at MTR A2756G (rs1805087) had a significantly higher total concentration of MTX-PGs in the red cells. This result may indicate that MTR gene polymorphisms are related to the clinical response to MTX, but there was no relationship between them either in South Indian patients (Muralidharan et al., 2018) or in our sample.
Analysis of TYMS, MTHFR, ATIC, MTR, and MTRR Polymorphisms Related to Adverse Drug Reactions to MTX
Toxicity: MTX-related adverse reactions were defined as the presentation of ADRs related to MTX at each visit. We classified the type of ADRs into System Organ Class (SOC) disorders in accordance with Common Terminology Criteria for Adverse Events (CTCAE) (Chaabane et al., 2016).
It has been reported that TYMS 28bp VNTR (Baggott et al., 1986), MTHFR 677C>T (Berkun et al., 2004; Caliz et al., 2012; Berkani et al., 2017), MTHFR 1298A>C (Berkun et al., 2004; Davis et al., 2014; Berkani et al., 2017), ATIC 347C>G (Muralidharan et al., 2016; Hakamata et al., 2018), MTR A2756G (Nikbakht et al., 2012) and MTRR 66A>G (Dervieux et al., 2006) may be associated with MTX toxicity. A study of 273 Caucasian patients with RA treated with MTX for at least 6 months showed that there were no associations between TYMS gene polymorphisms and the toxicity of MTX treatment (Swierkot et al., 2015). Another study of 185 Tunisian patients, including 35 men and 136 women with RA, showed that MTHFR A1298C, TYMS and MTR A2756G gene polymorphisms all had no association with adverse reactions to MTX (Yamamoto et al., 2016). In Spain, Salazar et al. (2014) found that two SNPs in the ATIC gene (rs16853826 and rs10197559) were associated with toxicity, but rs2372536 was not. Caliz et al. (2012) found that the MTR A2756G and MTRR A66G gene polymorphisms were not associated with increased MTX toxicity. The relationship between gene polymorphisms and ADRs to MTX were inconsistent among populations from different geographic locations.
Whether the TYMS 28bp VNTR (rs34743033), MTHFR 1298A>C (rs1801131), ATIC 347C>G (rs2372536), MTR A2756G (rs1805087), and MTRR 66A>G (rs1801394) polymorphisms are associated with ADRs to MTX, as we found, needs to be confirmed with larger samples.
Conclusion
Our results represent the first report regarding the relationship between the genetic polymorphisms of TYMS, MTHFR, ATIC, MTR, and MTRR related to the therapeutic outcome of MTX for RA in the Chinese population. Only the MTHFR 677C>T TT genotype was associated with ADRs to MTX. The strength of our study was that our patients came from all over China. However, the limitation of our study was that not all the functionally relevant genetic variants in TYMS, MTHFR, ATIC, MTR, and MTRR were included in our study, and the patient sample in our study was not large enough. Therefore, further studies with a larger sample are needed to confirm our findings.
Author Contributions
CX formulated the concept and designed the paper. SL, HY, HF, XS, LZ, JZ, JH, YX, and XL conducted the experiments. CX, HF, and HY supplied critical reagents of QIA amp DNA Blood Mini Kit and designed the primers of gene. JL contributed statistical analysis. CX and SL wrote the manuscript.
Funding
This work was supported by the International Cooperation Project of Ministry of Science and Technology (Project ID: 2014DFA31490) and the National Natural Science Foundation of China (Grant No. 81873223).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our colleagues at Beijing University of Chinese Medicine, China-Japan Friendship Hospital and the People’s Hospital of Yichun. We also thank Dr. Xiaomu Kong for advice regarding the statistical analysis. We thank the staff who helped us during the study.
References
Aletaha, D., Neogi, T., Silman, A. J., Funovits, J., Felson, D. T., Bingham, C. O. III., et al. (2010). Rheumatoid arthritis classification criteria: an american college of rheumatology/european league against rheumatism collaborative initiative. Arthritis Rheum. 62, 2569–2581. doi: 10.1002/art.27584
Bae, S. C., and Lee, Y. H. (2018). TYMS polymorphisms and responsiveness to or toxicity of methotrexate in rheumatoid arthritis. Z. Rheumatol. 77, 824–832. doi: 10.1007/s00393-018-0419-4
Baggott, J. E., Vaughn, W. H., and Hudson, B. B. (1986). Inhibition of 5-aminoimidazole-4-carboxamide ribotide transformylase, adenosine deaminase and 5′-adenylate deaminase by polyglutamates of methotrexate and oxidized folates and by 5-aminoimidazole-4-carboxamide riboside and ribotide. Biochem. J. 236, 193–200.
Berkani, L. M., Rahal, F., Allam, I., Mouaki Benani, S., Laadjouz, A., and Djidjik, R. (2017). Association of MTHFR C677T and A1298C gene polymorphisms with methotrexate efficiency and toxicity in algerian rheumatoid arthritis patients. Heliyon 3:e00467. doi: 10.1016/j.heliyon.2017.e00467
Berkun, Y., Levartovsky, D., Rubinow, A., Orbach, H., Aamar, S., Grenader, T., et al. (2004). Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann. Rheum. Dis. 63, 1227–1231. doi: 10.1136/ard.2003.016337
Boughrara, W., Benzaoui, A., Aberkane, M., Moghtit, F. Z., Dorgham, S., Lardjam-Hetraf, A. S., et al. (2017). No correlation between MTHFR c.677 C > T, MTHFR c.1298 A > C, and ABCB1 c.3435 C > T polymorphisms and methotrexate therapeutic outcome of rheumatoid arthritis in west algerian population. Inflamm. Res. 66, 505–513. doi: 10.1007/s00011-017-1034-6
Caliz, R., del Amo, J., Balsa, A., Blanco, F., Silva, L., Sanmarti, R., et al. (2012). The C677T polymorphism in the MTHFR gene is associated with the toxicity of methotrexate in a spanish rheumatoid arthritis population. Scand J. Rheumatol. 41, 10–14. doi: 10.3109/03009742.2011.617312
Chaabane, S., Marzouk, S., Akrout, R., Ben Hamad, M., Achour, Y., Rebai, A., et al. (2016). Genetic determinants of methotrexate toxicity in tunisian patients with rheumatoid arthritis: a study of polymorphisms involved in the mtx metabolic pathway. Eur. J. Drug Metab. Pharmacokinet. 41, 385–393. doi: 10.1007/s13318-015-0288-z
Chaabane, S., Messedi, M., Akrout, R., Ben Hamad, M., Turki, M., Marzouk, S., et al. (2018). Association of hyperhomocysteinemia with genetic variants in key enzymes of homocysteine metabolism and methotrexate toxicity in rheumatoid arthritis patients. Inflamm. Res. 67, 703–710. doi: 10.1007/s00011-018-1161-8
Corre, S., and Galibert, M. D. (2005). Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18, 337–348. doi: 10.1111/j.1600-0749.2005.00262.x
Cronstein, B. N. (2005). Low-dose methotrexate: a mainstay in the treatment of rheumatoid arthritis. Pharmacol. Rev. 57, 163–172. doi: 10.1124/pr.57.2.3
Davis, L. A., Polk, B., Mann, A., Wolff, R. K., Kerr, G. S., Reimold, A. M., et al. (2014). Folic acid pathway single nucleotide polymorphisms associated with methotrexate significant adverse events in United States veterans with rheumatoid arthritis. Clin. Exp. Rheumatol. 32, 324–332.
Dervieux, T., Greenstein, N., and Kremer, J. (2006). Pharmacogenomic and metabolic biomarkers in the folate pathway and their association with methotrexate effects during dosage escalation in rheumatoid arthritis. Arthritis Rheum. 54, 3095–3103. doi: 10.1002/art.22129
Fan, H., Li, Y., Zhang, L., Li, Y., and Li, W. (2017). Lack of association between MTHFR A1298C polymorphism and outcome of methotrexate treatment in rheumatoid arthritis patients: evidence from a systematic review and meta-analysis. Int. J. Rheum. Dis. 20, 526–540. doi: 10.1111/1756-185X.13100
Fessler, J., Raicht, A., Husic, R., Ficjan, A., Schwarz, C., Duftner, C., et al. (2017). Novel senescent regulatory T-Cell subset with impaired suppressive function in rheumatoid arthritis. Front. Immunol. 8:300. doi: 10.3389/fimmu.2017.00300
Ghodke-Puranik, Y., Puranik, A. S., Shintre, P., Joshi, K., Patwardhan, B., Lamba, J., et al. (2015). Folate metabolic pathway single nucleotide polymorphisms: a predictive pharmacogenetic marker of methotrexate response in indian (Asian) patients with rheumatoid arthritis. Pharmacogenomics. 16, 2019–2034. doi: 10.2217/pgs.15.145
Gonzalez-Mercado, M. G., Rivas, F., Gallegos-Arreola, M. P., Moran-Moguel, M. C., Salazar-Paramo, M., Gonzalez-Lopez, L., et al. (2017). MTRR A66G, RFC1 G80A, and MTHFR C677T and A1298C polymorphisms and disease activity in mexicans with rheumatoid arthritis treated with methotrexate. Genet. Test Mol. Biomarkers 21, 698–704. doi: 10.1089/gtmb.2017.0124
Hakamata, J., Hashiguchi, M., Kaneko, Y., Yamaoka, K., Shimizu, M., Maruyama, J., et al. (2018). Risk factors for abnormal hepatic enzyme elevation by methotrexate treatment in patients with rheumatoid arthritis: a hospital based-cohort study. Mod. Rheumatol. 28, 611–620. doi: 10.1080/14397595.2017.1414765
Hinks, A., Moncrieffe, H., Martin, P., Ursu, S., Lal, S., Kassoumeri, L., et al. (2011). Association of the 5-aminoimidazole-4-carboxamide ribonucleotide transformylase gene with response to methotrexate in juvenile idiopathic arthritis. Ann. Rheum. Dis. 70, 1395–1400. doi: 10.1136/ard.2010.146191
Jacques, P. F., Bostom, A. G., Selhub, J., Rich, S., Ellison, R. C., Eckfeldt, J. H., et al. (2003). Effects of polymorphisms of methionine synthase and methionine synthase reductase on total plasma homocysteine in the NHLBI family heart study. Atherosclerosis 166, 49–55.
Jekic, B., Lukovic, L., Bunjevacki, V., Milic, V., Novakovic, I., Damnjanovic, T., et al. (2013). Association of the TYMS 3G/3G genotype with poor response and GGH 354GG genotype with the bone marrow toxicity of the methotrexate in RA patients. Eur. J. Clin. Pharmacol. 69, 377–383. doi: 10.1007/s00228-012-1341-3
Kato, T., Hamada, A., Mori, S., and Saito, H. (2012). Genetic polymorphisms in metabolic and cellular transport pathway of methotrexate impact clinical outcome of methotrexate monotherapy in japanese patients with rheumatoid arthritis. Drug Metab. Pharmacokinet. 27, 192–199. doi: 10.2133/dmpk.DMPK-11-RG-066
Kojima, M., Nakayama, T., Kawahito, Y., Kaneko, Y., Kishimoto, M., Hirata, S., et al. (2016). The process of collecting and evaluating evidences for the development of guidelines for the management of rheumatoid arthritis, japan college of rheumatology 2014: utilization of GRADE approach. Mod. Rheumatol. 26, 175–179. doi: 10.3109/14397595.2015.1069474
Kurzawski, M., Malinowski, D., Szarmach, N., Nowak, A., Goryniak, A., Pawlik, A., et al. (2016). ATIC missense variant affects response to methotrexate treatment in rheumatoid arthritis patients. Pharmacogenomics 17, 1971–1978. doi: 10.2217/pgs-2016-0125
Leclerc, D., Campeau, E., Goyette, P., Adjalla, C. E., Christensen, B., Ross, M., et al. (1996). Human methionine synthase: cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum. Mol. Genet. 5, 1867–1874.
Lee, Y. H., and Bae, S. C. (2016). Association of the ATIC 347 C/G polymorphism with responsiveness to and toxicity of methotrexate in rheumatoid arthritis: a meta-analysis. Rheumatol. Int. 36, 1591–1599. doi: 10.1007/s00296-016-3523-2
Lima, A., Azevedo, R., Sousa, H., Seabra, V., and Medeiros, R. (2013). Current approaches for TYMS polymorphisms and their importance in molecular epidemiology and pharmacogenetics. Pharmacogenomics 14, 1337–1351. doi: 10.2217/pgs.13.118
Lima, A., Seabra, V., Bernardes, M., Azevedo, R., Sousa, H., and Medeiros, R. (2014a). Role of key TYMS polymorphisms on methotrexate therapeutic outcome in portuguese rheumatoid arthritis patients. PLoS One 9:e108165. doi: 10.1371/journal.pone.0108165
Lima, A., Seabra, V., Martins, S., Coelho, A., Araujo, A., and Medeiros, R. (2014b). Thymidylate synthase polymorphisms are associated to therapeutic outcome of advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Mol. Biol. Rep. 41, 3349–3357. doi: 10.1007/s11033-014-3197-3
Lincz, L. F., Scorgie, F. E., Garg, M. B., and Ackland, S. P. (2007). Identification of a novel single nucleotide polymorphism in the first tandem repeat sequence of the thymidylate synthase 2R allele. Int. J. Cancer 120, 1930–1934. doi: 10.1002/ijc.22568
Lopez-Rodriguez, R., Ferreiro-Iglesias, A., Lima, A., Bernardes, M., Pawlik, A., Paradowska-Gorycka, A., et al. (2018). Replication study of polymorphisms associated with response to methotrexate in patients with rheumatoid arthritis. Sci. Rep. 8:7342. doi: 10.1038/s41598-018-25634-y
Muralidharan, N., Gulati, R., Misra, D. P., and Negi, V. S. (2018). Nonassociation of homocysteine gene polymorphisms with treatment outcome in south indian tamil rheumatoid arthritis patients. Clin. Exp. Med. 18, 101–107. doi: 10.1007/s10238-017-0469-y
Muralidharan, N., Mariaselvam, C. M., Jain, V. K., Gulati, R., and Negi, V. S. (2016). ATIC 347C > G gene polymorphism may be associated with methotrexate-induced adverse events in south indian tamil rheumatoid arthritis. Pharmacogenomics 17, 241–248. doi: 10.2217/pgs.15.170
Muralidharan, N., Misra, D. P., Jain, V. K., and Negi, V. S. (2017). Effect of thymidylate synthase (TYMS) gene polymorphisms with methotrexate treatment outcome in south indian tamil patients with rheumatoid arthritis. Clin. Rheumatol. 36, 1253–1259. doi: 10.1007/s10067-017-3608-7
Nikbakht, M., MalekZadeh, K., Kumar Jha, A., Askari, M., Marwaha, R. K., Kaul, D., et al. (2012). Polymorphisms of MTHFR and MTR genes are not related to susceptibility to childhood ALL in north india. Exp. Oncol. 34,43–48.
Owen, S. A., Hider, S. L., Martin, P., Bruce, I. N., Barton, A., and Thomson, W. (2013). Genetic polymorphisms in key methotrexate pathway genes are associated with response to treatment in rheumatoid arthritis patients. Pharmacogenomics J. 13, 227–234. doi: 10.1038/tpj.2012.7
Qiu, Q., Huang, J., Lin, Y., Shu, X., Fan, H., Tu, Z., et al. (2017a). Polymorphisms and pharmacogenomics for the toxicity of methotrexate monotherapy in patients with rheumatoid arthritis: a systematic review and meta-analysis. Medicine 96:e6337. doi: 10.1097/MD.0000000000006337
Qiu, Q., Huang, J., Shu, X., Fan, H., Zhou, Y., and Xiao, C. (2017b). Polymorphisms and pharmacogenomics for the clinical efficacy of methotrexate in patients with rheumatoid arthritis: a systematic review and meta-analysis. Sci. Rep. 7:44015. doi: 10.1038/srep44015
Ranganathan, P., and McLeod, H. L. (2006). Methotrexate pharmacogenetics: the first step toward individualized therapy in rheumatoid arthritis. Arthritis Rheum. 54, 1366–1377. doi: 10.1002/art.21762
Sala-Icardo, L., Lamana, A., Ortiz, A. M., Garcia Lorenzo, E., Moreno Fresneda, P., Garcia-Vicuna, R., et al. (2017). Impact of genetic variants of ATP binding cassette B1, AICAR transformylase/IMP cyclohydrolase, folyl-polyglutamatesynthetase, and methylenetetrahydrofolatereductase on methotrexate toxicity. Reumatol. Clin. 13, 318–325. doi: 10.1016/j.reuma.2016.08.006
Salazar, J., Moya, P., Altes, A., Diaz-Torne, C., Casademont, J., Cerda-Gabaroi, D., et al. (2014). Polymorphisms in genes involved in the mechanism of action of methotrexate: are they associated with outcome in rheumatoid arthritis patients? Pharmacogenomics 15, 1079–1090. doi: 10.2217/pgs.14.67
Song, G. G., Bae, S. C., and Lee, Y. H. (2014). Association of the MTHFR C677T and A1298C polymorphisms with methotrexate toxicity in rheumatoid arthritis: a meta-analysis. Clin. Rheumatol. 33, 1715–1724. doi: 10.1007/s10067-014-2645-8
Swierkot, J. S., Karpinski, P., Pawlowska, J., Noga, L., Szechinski, J., and Wiland, P. (2015). Associations between single-nucleotide polymorphisms of RFC-1, GGH, MTHFR, TYMS, and TCII genes and the efficacy and toxicity of methotrexate treatment in patients with rheumatoid arthritis. Pol. Arch. Med. Wewn 125, 152–161.
Versteeg, G. A., Steunebrink, L. M. M., Vonkeman, H. E., Ten Klooster, P. M., van der Bijl, A. E., and van de Laar, M. (2018). Long-term disease and patient-reported outcomes of a continuous treat-to-target approach in patients with early rheumatoid arthritis in daily clinical practice. Clin. Rheumatol. 37,1189–1197. doi: 10.1007/s10067-017-3962-5
Walter, G. J., Fleskens, V., Frederiksen, K. S., Rajasekhar, M., Menon, B., Gerwien, J. G., et al. (2016). Phenotypic, functional, and gene expression profiling of peripheral CD45RA+ and CD45RO+ CD4+CD25+CD127(low) treg cells in patients with chronic rheumatoid arthritis. Arthritis Rheumatol. 68, 103–116. doi: 10.1002/art.39408
Wessels, J. A., Kooloos, W. M., De Jonge, R., De Vries-Bouwstra, J. K., Allaart, C. F., Linssen, A., et al. (2006). Relationship between genetic variants in the adenosine pathway and outcome of methotrexate treatment in patients with recent-onset rheumatoid arthritis. Arthritis Rheum. 54, 2830–2839. doi: 10.1002/art.22032
Wevers-de Boer, K., Visser, K., Heimans, L., Ronday, H. K., Molenaar, E., Groenendael, J. H., et al. (2012). Remission induction therapy with methotrexate and prednisone in patients with early rheumatoid and undifferentiated arthritis (the IMPROVED study). Ann. Rheum. Dis. 71,1472–1477. doi: 10.1136/annrheumdis-2011-200736
Xiao, H., Xu, J., Zhou, X., Stankovich, J., Pan, F., Zhang, Z., et al. (2010). Associations between the genetic polymorphisms of MTHFR and outcomes of methotrexate treatment in rheumatoid arthritis. Clin. Exp. Rheumatol. 28, 728–733.
Keywords: methotrexate, rheumatoid arthritis, single nucleotide polymorphisms, TYMS, MTHFR, ATIC, MTR, MTRR
Citation: Lv S, Fan H, Li J, Yang H, Huang J, Shu X, Zhang L, Xu Y, Li X, Zuo J and Xiao C (2018) Genetic Polymorphisms of TYMS, MTHFR, ATIC, MTR, and MTRR Are Related to the Outcome of Methotrexate Therapy for Rheumatoid Arthritis in a Chinese Population. Front. Pharmacol. 9:1390. doi: 10.3389/fphar.2018.01390
Received: 18 August 2018; Accepted: 12 November 2018;
Published: 28 November 2018.
Edited by:
Lingling Zhang, Anhui Medical University, ChinaReviewed by:
Angela Tagetti, Università degli Studi di Verona, ItalySuresh Kumar Kalangi, Indrashil University, India
Copyright © 2018 Lv, Fan, Li, Yang, Huang, Shu, Zhang, Xu, Li, Zuo and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Xiao, eGMyMDAyODEyQDEyNi5jb20=
†These authors have contributed equally to this work
 Shuang Lv
Shuang Lv HuiZhen Fan
HuiZhen Fan Jiang Li4†
Jiang Li4† Jing Huang
Jing Huang XiaoMing Shu
XiaoMing Shu Xiaoya Li
Xiaoya Li Jieyu Zuo
Jieyu Zuo Cheng Xiao
Cheng Xiao