- 1Biomedicine Research Center, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 2Key Laboratory of Protein Modification and Degradation, Guangzhou Medical University, Guangzhou, China
Non-coding RNAs (ncRNAs) are defined as RNA molecules that do not encode proteins, but recent evidence has proven that peptides/proteins encoded by ncRNAs do indeed exist and usually contain less than 100 amino acids. These peptides/proteins play an important role in regulating tumor energy metabolism, epithelial to mesenchymal transition of cancer cells, the stability of the c-Myc oncoprotein, and the ubiquitination and degradation of proliferating cell nuclear antigen (PCNA). These peptides/proteins represent promising drug targets for fighting against tumor growth or biomarkers for predicting the prognosis of cancer patients. In this review, we summarize the characteristics of peptides/proteins that have recently been identified as putative ncRNA translation products and their outlook for small molecule peptide drugs, drug targets, and biomarkers.
Introduction
Non-coding RNAs (ncRNAs) comprise a class of RNA nucleic acid molecules that are transcribed from DNA but do not encode proteins (Guttman et al., 2013). However, ncRNAs are involved in many diseases (Cho, 2011; Esteller, 2011; Hu et al., 2015; Zhou et al., 2016; Nan et al., 2017; Luo et al., 2018) and utilize a variety of mechanisms to participate in gene regulation, including DNA methylation, chromatin modifications, transcriptional regulation, mRNA splicing, translation control, RNA stability, and so on (Muro et al., 2011; Pelechano and Steinmetz, 2013; Xing et al., 2014; Huang and Shan, 2015).
For a long time, there has been widespread recognition that ncRNA are unable to encode proteins (Guttman et al., 2013). However, with the development of deep ribosome profiling sequencing (Ribo-Seq) technology, mass spectrometry and algorithms including fragment length organization similarity score (FLOSS; Ingolia et al., 2014), ORF-RATER (Fields et al., 2015), and Ribo taper (Calviello et al., 2016), a subset of ncRNA have been identified to be able to encode peptides (<100 amino acids) or proteins, such as HOXB-AS3 (Huang et al., 2017), SPAR (Matsumoto et al., 2017), FBXW7-185aa (Yang et al., 2018), SHPRH-146aa (Zhang et al., 2018), miPEP-200a, and miPEP-200b (Fang et al., 2017; Figure 1). HOXB-AS3 and SPAR are encoded by long non-coding RNA (lncRNA). HOXB-AS3 plays an important role in regulating tumor energy metabolism, and SPAR interacts with the lysosomal v-ATPase to negatively regulate mTORC1 activation. FBXW7-185aa and SHPRH-146aa are encoded by circular RNA (circRNA), regulating the stability of the c-Myc oncoprotein and mediating ubiquitination and degradation of proliferating cell nuclear antigen (PCNA). miPEP-200a and miPEP-200b are encoded by primary miRNA (pri-miRNA). miPEP-200a and miPEP-200b affect epithelial to mesenchymal transition of cancer cells. We describe a bank of biologically functional peptides/proteins encoded by lncRNA, circRNA, and pri-miRNA. The biological activity of ncRNA encoded peptides/proteins may provide possibilities for developing new therapies for current refractory and other diseases.
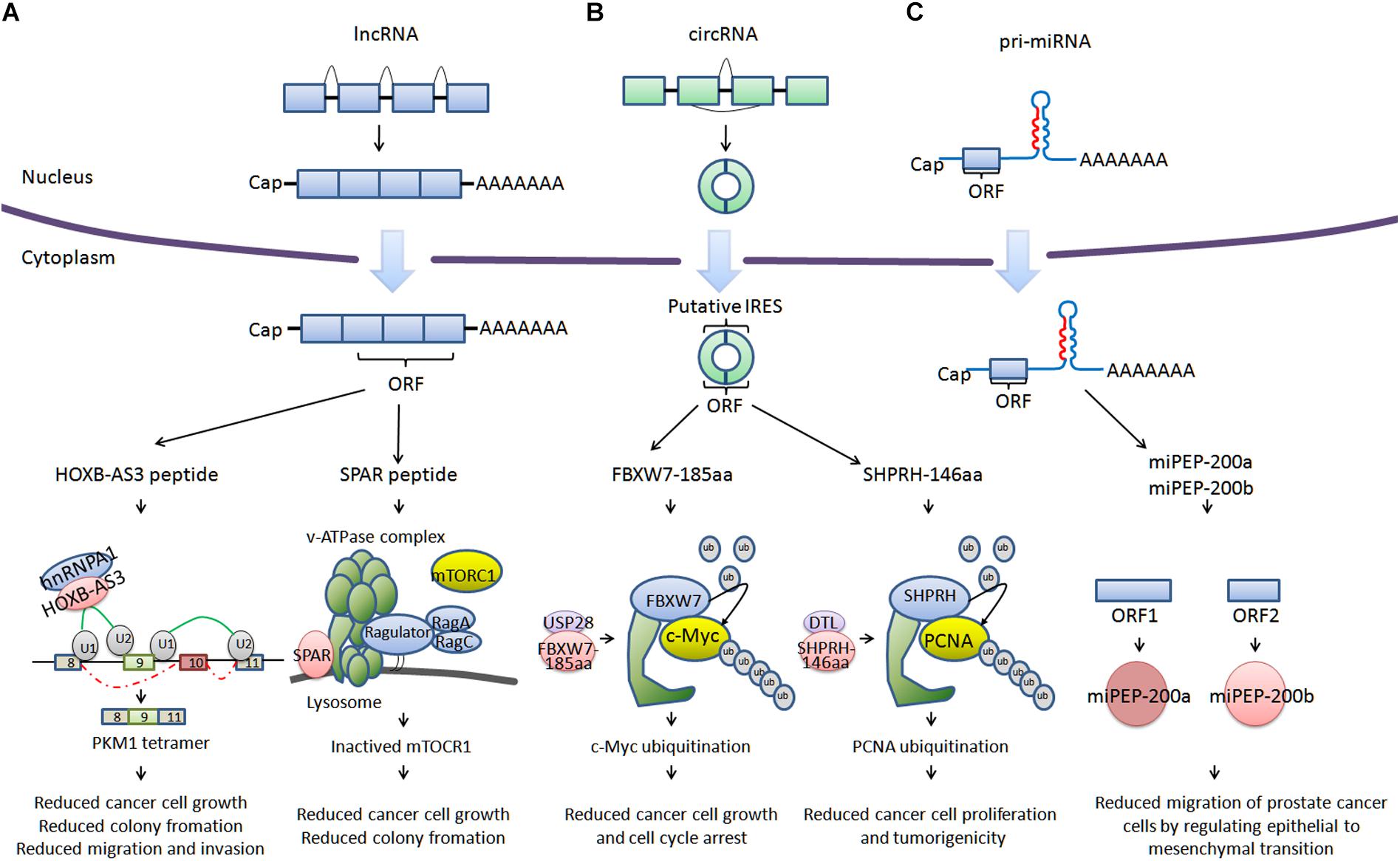
FIGURE 1. Cancer-related peptides/proteins encoded by ncRNAs. (A) Cancer-related peptides encoded by lncRNA. HOXB-AS3 is encoded by the lncRNA HOXB-AS3. HOXB-AS3 regulates tumor energy metabolism by antagonizing hnRNP A1, mediating PKM splicing. SPAR is encoded by LINC00961. SPAR interacts with the lysosomal v-ATPase to negatively regulate mTORC1 activation. (B) Cancer-related proteins encoded by circRNA. FBXW7-185aa is encoded by circ-FBXW7. FBXW7-185aa regulates the stability of the c-Myc oncoprotein. SHPRH-146aa is encoded by circ-SHPRH. SHPRH-146aa mediates the ubiquitination and degradation of PCNA. (C) Cancer-related proteins encoded by pri-miRNA. miPEP-200a and miPEP-200b are encoded by pri-miRNA (miR-200a and miR-200b). miPEP-200a and miPEP-200b affect the epithelial to mesenchymal transition of prostate cancer cells.
Peptides/proteins encoded by ncRNAs represent promising drug targets for fighting against tumor growth or biomarkers for predicting the prognosis of cancer patients. In this review, we summarize the characteristics of peptides/proteins encoded by ncRNA and their outlook for small molecule peptide drugs, drug targets, and biomarkers.
Peptides/Small Proteins Encoded by NCRNA as Promising Candidates in Antitumor Drugs
Advantages and Development Status of Anti-tumor Peptide/Small Protein Drugs
It is generally known that some peptide/protein drugs (such as antibacterial peptides) have been widely used in the field of animal medicine and have achieved good results (Brogden et al., 2003). In addition, the use of peptides/small protein drugs has a long successful history in the treatment of human disease, for instance, insulin for the treatment of diabetes (Kaur et al., 2018). Some anti-tumor peptides/small proteins such as mifamurtide (Brosa et al., 2014), interferon-γ (INF-γ; Berek, 2000), and interleukin-2 (IL-2; Liang et al., 2012), have been used in clinics and achieved a certain curative efficacy. Although chemotherapy is the major treatment method for cancer, it has fierce side effects, including the insufficient uptake of drugs in tumor cells, non-specific cytotoxicity of chemotherapeutic drugs and the rapid emergence of drug resistance, which largely limit clinical application (Gandhi et al., 2014; He et al., 2016). Compared with traditional chemotherapy agents, peptide and protein drugs possess unique advantages, including high specificity and activity, low immunogenicity and less cytotoxicity to normal tissues (Leader et al., 2008). Therefore, peptide and protein therapeutics have emerged as crucial approaches in recent clinical cancer treatment. The anti-cancer mechanisms of peptides and proteins are mainly divided into two types: one is usually related to direct induction of apoptosis in tumor cells via specific pathways, and the other is related to direct inhibition of tumor cell growth through targeting tumor angiogenesis or stimulating an immune response (Vaishya et al., 2015; Redington et al., 2017). Some protein drugs, such as tumor necrosis factor alpha (TNF-α), INF-γ, and IL-2, have been widely used in clinical cancer treatment (Berek, 2000; Calzascia et al., 2007; Liang et al., 2012). Nevertheless, despite TNF-α, INF-γ, and IL-2 possessing anti-tumor potency, the systemic clinical applications of high doses of these drugs are severely limited due to their systemic toxicities, for instance, inflammatory response, and underlying cardiotoxicity (Ashkenazi et al., 1999; Berek, 2000; Liang et al., 2012). Therefore, it is necessary and urgent to continue looking for higher efficiency and lower toxicity anti-tumor drugs.
Application Prospect of Peptides/Small Proteins Encoded by lncRNAs in the Treatment of Tumors
Only 2% of the human genome is able to code for proteins or peptides, and the rest is actively transcribed into ncRNAs (Fang et al., 2017). Surprisingly, a limited number of “ncRNAs” have recently shown the ability to encode proteins or peptides. HOXB-AS3 was annotated as lncRNA in the NCBI and lncRNA databases. Our research team has recently discovered that the lncRNA HOXB-AS3 could actually encode a conserved 53-aa peptide, called HOXB-AS3 peptide (Huang et al., 2017). In vitro studies showed that HOXB-AS3, rather than its lncRNA, inhibited proliferation, migration, invasion, and colony formation of colon cancer cells (CRC) via antagonizing hnRNP A1 protein (Huang et al., 2017). More importantly, the HOXB-AS3 peptide clearly impaired the in vivo growth of CRC xenografts and decreased the number and size of lung metastatic nodules (Huang et al., 2017). Therefore, HOXB-AS3 has great potential for the treatment of colon cancer via antagonizing hnRNP A1. Moreover, a conserved lncRNA LINC00961, encodes a novel peptide, called “small regulatory peptide of amino acid response” (SPAR). SPAR locates at the late endosome/lysosome and interacts with the lysosomal v-ATPase to negatively regulate mTORC1 activation (Matsumoto et al., 2017).
Application Prospect of Peptides/Small Proteins Encoded by circRNAs in the Treatment of Tumors
Very few circRNAs have been shown to encode small proteins with antitumor activity based on small-ORFs. Zhang et al. (2018) reported that a novel antitumor protein, SHPRH-146aa, was encoded by the circular RNA SHPRH and was driven by internal ribosome entry site (IRES) elements. SHPRH-146aa was able to protect full-length SHPRH from degradation by the ubiquitin-proteasome. Stabilized SHPRH inhibited glioblastoma cell proliferation and tumorigenicity via sequentially ubiquitinating PCNA as an E3 ligase (Zhang et al., 2018). In addition, Yang et al. (2018) also found that a novel 21-kDa protein, named FBXW7-185aa, was encoded by the circular RNA FBXW7. The FBXW7-185aa protein can reduce the half-life of the c-Myc oncoprotein, which is known to regulate the transcription of numerous genes and pathways, via antagonizing ubiquitin carboxyl-terminal hydrolase 28 (USP28) induced c-Myc stabilization in glioma cells (Yang et al., 2018). Therefore, the FBXW7-185aa protein can inhibit the proliferation and cell cycle acceleration of malignant glioma cells by downregulating the protein expression of c-Myc (Yang et al., 2018).
Application Prospect of Peptides/Small Proteins Encoded by pri-miRNAs in the Treatment of Tumors
Impressively, Fang et al. (2017) found that two proteins (miPEP-200a and miPEP-200b) were encoded by miR-200a and miR-200b, respectively. The experiment results indicated that miPEP-200a and miPEP-200b suppressed the migration of prostate cancer cells through regulating the epithelial to mesenchymal transition of tumor cells.
Application of Peptide/Small Protein Drugs Encoded by ncRNA
The ideal drug not only has specificity and pharmacological activity, but can also reach the target site to play a role. Whether these cancer-suppressive peptides/small proteins (SPAR, HOXB-AS3, FBXW7-185aa, SHPRH-146aa, miPEP-200a, and miPEP-200b) encoded by ncRNAs, are secreted into human serum remains unknown. Whether they can enter tumor cells through transporters on the cell membrane and play an inhibitory role remains unknown. Due to the potential developmental value and clinical applications, these problems will become a hot topic of research. In the past few decades, nanoscale carriers have become one of the key strategies for enhancing cancer treatment, which is attributed to the markedly enhanced tumor accumulation of drugs, prolonged blood circulation of the drug-loaded nanoparticles, and improved intracellular delivery efficiency (Elsabahy and Wooley, 2012; He et al., 2016). Therefore, if these peptides/small proteins have a very short half-life in vivo and are difficult to pass the biofilm barrier, they could be wrapped in nanomaterials to avoid being quickly metabolized and be delivered to tumor cells through nanoparticles to play an anti-cancer role. Moreover, these peptides/small proteins could interact with chemical drugs to treat tumors through nanoscale codelivery systems. Recombinant human adenovirus-p53 injection has become a new method for the clinical treatment of cancer. After the coding sequence of these peptides/small proteins is recombined with adenovirus, it could be injected into patients to treat tumors.
Peptides/Small Proteins Encoded by NCRNA as Promising Cancer Drug Targets
Targeted Therapy
Targeted therapy refers to a type of treatment that uses drugs or other substances to attack specific targeted molecules (e.g., certain enzymes, proteins, DNA, RNA or other molecules), thereby maximizing efficacy and minimizing toxicity (Sawyers, 2008). This is also called “molecularly targeted therapy” and “precision medicine.” The purpose of targeted therapy in cancer is to restrict the growth and survival of cancer cells without injury to normal cells. In view of tumor heterogeneity, several types of targeted therapies, including monoclonal antibodies (e.g., Rituximab and Infliximab) (Maifrede et al., 2018), angiogenesis inhibitors (e.g., Pemetrexed and FGFR inhibitors) (Rojas et al., 2016), hormone therapies (e.g., Palbociclib) (Lynce et al., 2018), immune therapies (e.g., Durvalumab) (Raja et al., 2018), and rapamycin target inhibitor (e.g., Everolimus) (Kornblum et al., 2018), have been approved for cancer treatment. Additionally, tumor suppressor have also been studied as targets for targeted therapies in cancer. An example is the most important tumor suppressor protein, p53, which is associated with carcinogenesis by missense mutation (Brosh and Rotter, 2009). It was reported that 96.7% of high-grade serous ovarian carcinoma (HGSOC) cases contain pathogenic TP53 mutations (Ahmed et al., 2010). More recently, Soragni et al. (2016) designed a peptide inhibitor of p53 amyloid aggregation, ReACp53, which penetrated cells and restored the p53 function of tumor suppression in HGSOCs.
Application of ncRNA Encoded Peptides/Proteins as Promising Therapy Targets
ncRNA encoded peptides/proteins (HOXB-AS3, FBXW7-185aa, SHPRH-146aa, miPEP-200a, and miPEP-200b) have been proved to suppress tumorigenesis, which has enriched the research of ncRNAs in cancer development. Whether or not these tumor suppressor ncRNAs present mutations, like TP53 does, in tumors is still unknown. Strategies for rescuing or strengthening the function of tumor suppressor peptides/proteins, including vaccination with synthesized peptides or viral vector vaccines that encode relevant peptides sequences for cancer therapies, are now in development (Efremova et al., 2017; Radvanyi, 2018). Meanwhile, in the human genome, increasing evidence suggests that numerous ncRNAs are functional and play pivotal roles in many aspects of biology (Esteller, 2011). Maybe there are some hidden oncopeptides/oncoproteins encoded by ncRNAs that need to be identified; these hidden oncopeptides/oncoproteins may be exploited as novel targets for targeted therapies in cancer.
Peptides/Proteins Encoded by NCRNA as Promising Cancer Biomarkers
Biomarkers in Clinical Detection
With the advancement of treatment modalities, the survival rate and quality of life for early-stage cancer patients have been improved. However, due to the lack of specific symptoms and signs of early-stage cancer, and limited by cancer heterogeneity, only a portion of cancer patients are diagnosed early. Most cancer patients experience tumor recurrence and metastasis at regional or distant sites, which seriously shortens their survival time and greatly diminished quality of life. It is obvious that cancer bring devastating effects on patients and their families, which is a tremendous burden on society. Bray estimated there would be 22.2 million new cancer cases in 184 countries by 2030 (Bray et al., 2012). Cancer has been a severe challenge to society, and the early detection of cancer is highly important for clinical work.
It has been widely recognized that cancer is a very heterogeneous disease, and implementing precision medicine for patients is widely accepted. The molecular subtype of cancer provides us a new perspective: for different molecular subtypes, we can develop different, appropriate therapies and monitor the disease progression in the most suitable way. For instance, patients with triple-negative breast cancer (TNBC) have an increased risk of death compared with women with other types of breast cancer (Mayer et al., 2014). Gene expression analyses recently showed that TNBC has six distinct subtypes, each displaying a unique biology and showing distinct responses to standard treatment (Lehmann et al., 2011; Masuda et al., 2013). It is urgent to develop more biology-specific cancer biomarkers so that patients can receive the most appropriate therapy. Developing more suitable biomarkers for different molecular typing of tumors will be an inevitable biological challenge for cancer treatment. Therefore, biology-specific cancer biomarkers urgently need to be discovered and developed.
Peptides/Proteins Encoded by ncRNA as Promising Cancer Biomarkers
HOXB-AS3 Encoded by lncRNA Is a Potential Prognostic Biomarker
Our research team reported that the HOXB-AS3, encoded by the lncRNA HOXB-AS3, has been proven to play an important role in the development of cancer metabolism reprogramming. In mice, the in vivo growth of CRC xenografts with HOXB-AS3 ORF and 5′ UTR-ORF stably transfected cells was clearly impaired, including inhibition of tumor growth and lung metastasis. The HOXB-AS3 levels were decreased in primary CRC tissues compared with their paired non-tumoral tissues. Comparing the correlations between the level of HOXB-AS3 and clinic pathological features in 90 CRC cases, we found that patients with the clinical stage between IIB to IV have significantly lower levels of HOXB-AS3. The mean overall survival time for CRC patients with high levels of HOXB-AS3 was 1.6 times that of CRC patients with low levels of HOXB-AS3. Kaplan-Meier survival analyses revealed that patients with lower HOXB-AS3 levels were at increased risk of CRC-related death. Therefore, the low level of the HOXB-AS3 was correlated with a poor prognosis in CRC patients (Huang et al., 2017).
The Potential of Peptides/Proteins Encoded by circRNAs as Cancer Biomarkers
Nu Zhang’s research team reported that circ-FBXW7 can encode a 21-kDa protein, called FBXW7-185aa, which is assumed as a prognostic marker for glioblastoma. The mice injected with glioblastoma multiforme cells that stably overexpressed circ-FBXW7 had longer survival times and exhibited a much lower tumorigenicity than mice injected with the corresponding control cells. In 38 pairs of glioblastoma samples compared with their paired tumor-adjacent tissues, the expression of FBXW7-185aa was obviously decreased. The protein expression was investigated to demonstrate that glioblastoma patients with higher FBXW7-185aa expression had an increased total survival time (Yang et al., 2018).
Another study focused on SHPRH-146aa, which is encoded by circ-SHPRH, and showed that the SHPRH-146aa expression was significantly downregulated in 60 glioblastoma samples compared with their adjacent normal tissues. The subcutaneous injection with SHPRH-146aa stably overexpressing cells resulted in substantially lower tumorigenicity compared with the injection of the corresponding control cells in mice. Cancer patients with higher SHPRH-146aa expression had a longer survival time than those with lower SHPRH-146aa expression in glioblastoma patient samples. The result suggested SHPRH-146aa is a prognostic marker for glioblastoma in clinics (Zhang et al., 2018).
The Potential of Peptides/Proteins Encoded by pri-miRNAs as Cancer Biomarkers
The primary microRNA, miR-200a and miR-200b, that encode proteins (miPEP-200a and miPEP-200b) can inhibit the migration of prostate cancer cells, and it was observed that low-level expression of miPEP-200a and miPEP-200b were associated with poor survival outcome in cancer patients. This evidence indicated that miPEP-200a and miPEP-200b have the potential to be used as diagnostic and prognostic markers (Fang et al., 2017).
The Expectation of Peptides/Proteins Encoded by ncRNA Used as the Cancer Biomarkers
Accumulating evidence has shown that peptides/proteins encoded by ncRNA represent a promising source for prognostic biomarkers. Although the differential expression and prognostic correlation of the five peptides/proteins HOXB-AS3, SHPRH-146aa, FBXW7-185aa, miPEP-200a and miPEP-200b in cancer have been confirmed by the IHC analysis of paraffin sections of tumor tissues and western blot analysis of protein samples, we do not see a clear report that the peptides/proteins encoded by ncRNA are detected in body fluid (including blood, serum, urine, chest fluid, etc.). If these biology-specific peptides/proteins can be found in body fluid, the widespread progress of using them as cancer biomarkers in clinical applications will be greatly promoted. The tremendous research space of peptides/proteins encoded by ncRNAs used as cancer biomarkers will encourage researchers to continuously devote great effort in this field so that they can meet the requirements of being reliable, cost-effective, and capable of precise detection and monitoring.
Conclusion
ncRNA-encoded peptides/proteins open up a new field. The peptides/proteins mentioned in this review affect multiple steps in the development of tumorigenesis. These peptides/proteins regulate tumor energy metabolism, the stability of oncoproteins and the epithelial to mesenchymal transition of cancer cells. They are the new stars of the future drug targets for fighting against tumor growth or may be used as biomarkers for predicting the prognosis of cancer patients.
However, the ncRNA-encoded peptides/proteins that have been discovered so far are only the beginning, and those that are undiscovered will provide a wealth of opportunities for small molecule peptide drugs, drug targets, and biomarkers. Currently, whether these ncRNA-encoded peptides/proteins can be used in clinical practice requires a large sample of clinical studies. Follow-up treatment effects of ncRNA-encoded peptides/proteins and detecting these as biomarkers in more samples will be needed.
Author Contributions
G-RY conceived the general idea. SZ, JW, YH, and NM wrote the first draft. G-RY revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81772998 and 81672393), the R&D Plan of Guangzhou (201704020115), the Yangcheng Scholars Program from the Ministry of Education of Guangzhou (1201561583), Innovative Research Team of Ministry of Education of Guangzhou (1201610015), the R&D Plan of Guangdong (2017A020215096), and the National Funds of Developing Local Colleges and Universities (B16056001).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmed, A. A., Etemadmoghadam, D., Temple, J., Lynch, A. G., Riad, M., Sharma, R., et al. (2010). Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 221, 49–56. doi: 10.1002/path.2696
Ashkenazi, A., Pai, R. C., Fong, S., Leung, S., Lawrence, D. A., Marsters, S. A., et al. (1999). Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 104, 155–162. doi: 10.1172/JCI6926
Berek, J. S. (2000). Interferon plus chemotherapy for primary treatment of ovarian cancer. Lancet 356, 6–7. doi: 10.1016/S0140-6736(00)02422-3
Bray, F., Jemal, A., Grey, N., Ferlay, J., and Forman, D. (2012). Global cancer transitions according to the human development index (2008-2030): a population-based study. Lancet Oncol. 13, 790–801. doi: 10.1016/S1470-2045(12)70211-5
Brogden, K. A., Ackermann, M., McCray, P. J., and Tack, B. F. (2003). Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 22, 465–478. doi: 10.1016/S0924-8579(03)00180-8
Brosa, M., Garcia, D. M., Mora, J., Villacampa, A., Pozo, T., Adán, C., et al. (2014). Economic considerations on the use of mifamurtide in the treatment of osteosarcoma in Spain. Value Health 17, A526–A527. doi: 10.1016/j.jval.2014.08.1662
Brosh, R., and Rotter, V. (2009). When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 9, 701–713. doi: 10.1038/nrc2693
Calviello, L., Mukherjee, N., Wyler, E., Zauber, H., Hirsekorn, A., Selbach, M., et al. (2016). Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170. doi: 10.1038/nmeth.3688
Calzascia, T., Pellegrini, M., Hall, H., Sabbagh, L., Ono, N., Elford, A. R., et al. (2007). TNF-α is critical for antitumor but not antiviral T cell immunity in mice. J. Clin. Invest. 117, 3833–3845. doi: 10.1172/JCI32567
Cho, W. C. (2011). Grand challenges and opportunities in deciphering the role of non-coding RNAs in human diseases. Front. Genet. 2:1. doi: 10.3389/fgene.2011.00001
Efremova, M., Finotello, F., Rieder, D., and Trajanoski, Z. (2017). Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Front. Immunol. 8:1679. doi: 10.3389/fimmu.2017.01679
Elsabahy, M., and Wooley, K. L. (2012). Design of polymeric nanoparticles for biomedical delivery applications. Chem. Soc. Rev. 41, 2545–2561. doi: 10.1039/c2cs15327k
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi: 10.1038/nrg3074
Fang, J., Morsalin, S., Rao, V. N., and Reddy, E. S. P. (2017). Decoding of non-coding DNA and non-coding RNA: pri-micro RNA-encoded novel peptides regulate migration of cancer cells. J. Pharm. Sci. Pharm. 3, 23–27. doi: 10.1166/jpsp.2017.1070
Fields, A. P., Rodriguez, E. H., Jovanovic, M., Stern-Ginossar, N., Haas, B. J., Mertins, P., et al. (2015). A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell 60, 816–827. doi: 10.1016/j.molcel.2015.11.013
Gandhi, N. S., Tekade, R. K., and Chougule, M. B. (2014). Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J. Control. Release 194, 238–256. doi: 10.1016/j.jconrel.2014.09.001
Guttman, M., Russell, P., Ingolia, N. T., Weissman, J. S., and Lander, E. S. (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. doi: 10.1016/j.cell.2013.06.009
He, C., Tang, Z., Tian, H., and Chen, X. (2016). Co-delivery of chemotherapeutics and proteins for synergistic therapy. Adv. Drug Deliv. Rev. 98, 64–76. doi: 10.1016/j.addr.2015.10.021
Hu, G., Yang, T., Zheng, J., Dai, J., Nan, A., Lai, Y., et al. (2015). Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol. Carcinog. 54(Suppl. 1), E192–E204. doi: 10.1002/mc.22314
Huang, C., and Shan, G. (2015). What happens at or after transcription: insights into circRNA biogenesis and function. Transcription 6, 61–64. doi: 10.1080/21541264.2015.1071301
Huang, J. Z., Chen, M., Chen, D., Gao, X. C., Zhu, S., Huang, H., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 68, 171–184. doi: 10.1016/j.molcel.2017.09.015
Ingolia, N. T., Brar, G. A., Sternginossar, N., Harris, M. S., Talhouarne, G. J. S., Jackson, S. E., et al. (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379. doi: 10.1016/j.celrep.2014.07.045
Kaur, R., Kaur, M., and Singh, J. (2018). Endothelial dysfunction and platelet hyperactivity in type 2 diabetes mellitus: molecular insights and therapeutic strategies. Cardiovasc. Diabetol. 17:121. doi: 10.1186/s12933-018-0763-3
Kornblum, N., Zhao, F., Manola, J., Klein, P., Ramaswamy, B., Brufsky, A., et al. (2018). Randomized phase II trial of fulvestrant plus everolimus or placebo in postmenopausal women with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer resistant to aromatase inhibitor therapy: results of PrE0102. J. Clin. Oncol. 36, 1556–1563. doi: 10.1200/JCO.2017.76.9331
Leader, B., Baca, Q. J., and Golan, D. E. (2008). Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 7, 21–39. doi: 10.1038/nrd2399
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y., et al. (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767. doi: 10.1172/JCI45014
Liang, X., De Vera, M. E., Buchser, W. J., Romo, D. V. C. A., Loughran, P., Beer Stolz, D., et al. (2012). Inhibiting systemic autophagy during interleukin 2 immunotherapy promotes long-term tumor regression. Cancer Res. 72, 2791–2801. doi: 10.1158/0008-5472.CAN-12-0320
Luo, Y., Liang, M., Yao, W., Liu, J., Niu, Q., Chen, J., et al. (2018). Functional role of lncRNA LOC101927497 in N-methyl-N′-nitro-N-nitrosoguanidine-induced malignantly transformed human gastric epithelial cells. Life Sci. 193, 93–103. doi: 10.1016/j.lfs.2017.12.007
Lynce, F., Shajahan-Haq, A. N., and Swain, S. M. (2018). CDK4/6 inhibitors in breast cancer therapy: current practice and future opportunities. Pharmacol. Ther. 191, 65–73. doi: 10.1016/j.pharmthera.2018.06.008
Maifrede, S., Nieborowskaskorska, M., Sullivan, K., Dasgupta, Y., and Podszywalowbartnicka, P. (2018). Tyrosine kinase inhibitor-induced defects in DNA repair sensitize FLT3(ITD)-positive leukemia cells to PARP1 inhibitors. Blood 132, 67–77. doi: 10.1182/blood-2018-02-834895
Masuda, H., Baggerly, K. A., Wang, Y., Zhang, Y., Gonzalez-Angulo, A. M., Meric-Bernstam, F., et al. (2013). Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 19, 5533–5540. doi: 10.1158/1078-0432.CCR-13-0799
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mayer, I. A., Abramson, V. G., Lehmann, B. D., and Pietenpol, J. A. (2014). New strategies for triple-negative breast cancer–deciphering the heterogeneity. Clin. Cancer Res. 20, 782–790. doi: 10.1158/1078-0432.CCR-13-0583
Muro, E. M., Mah, N., and Andradenavarro, M. A. (2011). Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie 93, 1916–1921. doi: 10.1016/j.biochi.2011.07.024
Nan, A., Chen, L., Zhang, N., Liu, Z., Yang, T., Wang, Z., et al. (2017). A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch. Toxicol. 91, 1671–1684. doi: 10.1007/s00204-016-1837-1
Pelechano, V., and Steinmetz, L. M. (2013). Gene regulation by antisense transcription. Nat. Rev. Genet. 14:880. doi: 10.1038/nrg3594
Radvanyi, L. G. (2018). Targeting the cancer mutanome of breast cancer. Nat. Med. 24, 703–704. doi: 10.1038/s41591-018-0065-z
Raja, R., Kuziora, M., Brohawn, P., Higgs, B. W., Gupta, A., Dennis, P. A., et al. (2018). Early reduction in ctDNA predicts survival in lung and bladder cancer patients treated with durvalumab. Clin. Cancer Res. doi: 10.1158/1078-0432.CCR-18-0386 [Epub ahead of print].
Redington, J. M., Breydo, L., and Uversky, V. N. (2017). When good goes awry: the aggregation of protein therapeutics. Protein Pept. Lett. 24, 340–347. doi: 10.2174/0929866524666170209153421
Rojas, L., Cardona, A. F., Arrieta, O., Wills, B., and Corrales-Rodriguez, L. (2016). PD2.04 (also presented as P1.42): PEM/CBP/BEV followed by maintenance PEM/BEV in hispanic patients with NSCLC: outcomes according to TS, ERCC1 and VEGF. J. Thorac. Oncol. 11, S177–S178. doi: 10.1016/j.jtho.2016.08.018
Soragni, A., Janzen, D. M., Johnson, L. M., Lindgren, A. G., Thaiquynh, N. A., Tiourin, E., et al. (2016). A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103. doi: 10.1016/j.ccell.2015.12.002
Vaishya, R., Khurana, V., Patel, S., and Mitra, A. K. (2015). Long-term delivery of protein therapeutics. Expert Opin. Drug Del. 12, 415–440. doi: 10.1517/17425247.2015.961420
Xing, Z., Lin, A., Li, C., Liang, K., Wang, S., Liu, Y., et al. (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125. doi: 10.1016/j.cell.2014.10.013
Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C., Xiao, F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Ins. 110, 304–315. doi: 10.1093/jnci/djx166
Zhang, M., Huang, N., Yang, X., Luo, J., Yan, S., Xiao, F., et al. (2018). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. doi: 10.1038/s41388-017-0019-9
Keywords: long non-coding RNA (lncRNA), circular RNA (circRNA), primary miRNA (pri-miRNA), small ORF, small peptide, drug target, biomarker
Citation: Zhu S, Wang J, He Y, Meng N and Yan G-R (2018) Peptides/Proteins Encoded by Non-coding RNA: A Novel Resource Bank for Drug Targets and Biomarkers. Front. Pharmacol. 9:1295. doi: 10.3389/fphar.2018.01295
Received: 19 September 2018; Accepted: 22 October 2018;
Published: 13 November 2018.
Edited by:
Zhi Shi, Jinan University, ChinaReviewed by:
Shengtao Zhou, West China Second University Hospital of Sichuan University, ChinaWeiyi Fang, Southern Medical University, China
Copyright © 2018 Zhu, Wang, He, Meng and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang-Rong Yan, jxygr007@yahoo.com; jxygr007@126.com
†These authors have contributed equally to this work
 Song Zhu
Song Zhu Jizhong Wang1,2†
Jizhong Wang1,2† Nan Meng
Nan Meng Guang-Rong Yan
Guang-Rong Yan