- 1BK21 PLUS Program for Creative Veterinary Science Research, Research Institute for Veterinary Science and College of Veterinary Medicine, Seoul National University, Seoul, South Korea
- 2Department of Medical Genetics, College of Medicine, Hallym University, Chuncheon, South Korea
- 3Department of Biomedical Laboratory Science, College of Health Science, Yonsei University, Wonju, South Korea
Introduction
The Helicobacter bacterial genus comprises of spiral-shaped gram-negative bacteria with flagella that colonize the gastro-intestinal (GI) tract of humans and various mammals (Solnick and Schauer, 2001). In particular, Helicobacter pylori was classified as a group 1 carcinogen by the International Agency for Research on Cancer (IARC) in 1994, and has been shown to occur with a high prevalence in humans, although this varies between geographical regions, ethnic groups, and various populations (Kusters et al., 2006; Goh et al., 2011). To date, more than 37 Helicobacter species have been identified in addition to H. pylori (Péré-Védrenne et al., 2017). Furthermore, non-H. pylori Helicobacters (NHPH) have been shown to infect both humans and animals, and NHPH infections are associated with intestinal carcinoma, and mucinous adenocarcinoma (Swennes et al., 2016). Despite the demonstrated association between NHPH and disease, most studies to date have investigated H. pylori in humans; thus, it is necessary to characterize NHPH and elucidate its role in the GI tract of wild rodents which are potential Helicobacter carriers (Taylor et al., 2007; Mladenova-Hristova et al., 2017).
Helicobacter apodemus, a spiral curved rod bacterium with a single flagella, was first identified in the GI tract of the Korean striped field mouse (Apodemus agrarius) in Korea, and shown to exhibit urease, oxidase, and catalase activity (Jeon et al., 2015). Since then, rodents colonized with H. apodemus have been found all over the world, including within the Xinjiang-Uygur Autonomous Region of China, Cambridge in the United States, and animal facilities in Sweden (Goto et al., 2004; Johansson et al., 2006; Miller et al., 2014). A previous study suggested that H. apodemus has the potential to cause rectal prolapse and colorectal cancer in rodents (Miller et al., 2014; Zhang et al., 2017), while another suggested that it may act as a rodent pathobiont, normally activating regulatory T-cells to maintain immune tolerance, but activating effector T-cells to contribute to inflammation and disease pathogenesis (Chai et al., 2017). Rodent H. apodemus colonization has been shown to be significantly decreased after treatment with azithromycin (compared to other antibiotics such as amoxicillin, or cefaclor), and similarly, after administration of Lactobacillus casei Zhang, and vitamin K2 (Khan et al., 2016; Zhang et al., 2017).
Nevertheless, continued research is essential to elucidate the molecular mechanisms by which H. apodemus alternately causes GI tract inflammation and GI tolerance in rodents, depending upon host health. The current study was therefore conducted to identify the genomic characteristics and specificity of H. apodemus, and to reveal its potential role in the rodent GI tract. Specifically, the genome of H. apodemus str. SCJK1 isolated from Apodemus agrarius was completely sequenced, and subjected to a comparative genomic analysis with 17 genome sequences of other Helicobacter species. It is hoped that the data in this study will serve as the basis for further studies of H. apodemus-related bacterium, and furthermore, enable future in-depth biomedical research regarding the immunological and pathological role of H. apodemus in the rodent GI tract.
Materials and Methods
H. apodemus Isolation and DNA Extraction
In May 2015, fresh fecal samples from wild A. agrarius were collected, and transported to the laboratory at 4°C. The fecal samples were homogenized in PBS, spread onto modified Charcoal-Cefoperazone-Deoxycholate agar (mCCDA) with a selective supplement (Oxoid), and micro-aerobically incubated at 42°C for 6 days. After incubation, suspected colonies were transferred to blood agar, and micro-aerobically incubated at 42°C for 2 days. Genomic DNA was extracted from each colony confirmed to be H. apodemus (via Polymerase Chain Reaction (PCR) (Miller et al., 2014), and 16S rRNA sequencing analyses) using MGTM Genomic DNA Purification kit (Macrogen, Korea). The quality of the extracted genomic DNA was evaluated using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Whole-Genome Sequencing, Genome Assembly, and Gene Annotation
The whole-genome sequencing of H. apodemus str. SCJK1 was carried out using a PacBio RS α sequencer (Pacific Biosciences, Menlo Park, CA, USA). A 20 kb DNA library was prepared using a SMRTbellTM template Prep Kit, and sequenced using a P6-C4 chemistry reagent kit (Pacific Biosciences, Menlo Park, CA, USA). The obtained sub-reads were assembled de novo using Hierarchical Genome Assembly Process v. 3.0 and SMRT Analysis v. 2.3 (default options) software (Pacific Biosciences, Menlo Park, CA, USA) (Chin et al., 2013). The reads were polished using Quiver v. 1.0 software (Pacific Biosciences, Menlo Park, CA, USA) to ensure a higher level of accuracy and lower error rate (Chin et al., 2013). Genes were annotated according to the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP, https://www.ncbi.nlm.nih.gov/genome/annotation_prok/), and “Clusters of Orthologous Group (COG)” categories were assigned using the NCBI COGs database (2014 version, https://www.ncbi.nlm.nih.gov/COG/). A summary of the generated sequencing data is included in Supplementary Table 1. Sequences for Helicobacter-related virulence genes presented in the Virulence Factor Database (VFDB, www.mgc.ac.cn/VFs/) were used to predict the H. apodemus str. SCJK1 virulence factor.
Comparative Genomic Analysis of H. apodemus With Other Helicobacter Species
A total of 17 genome sequences of other Helicobacter spp. were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/), and used to conduct a comparative genomic analysis, including JRPC_s (GCA_ 000765745.1), H. himalayensis (GCA_ 001602095.1), H. mustelae (GCA_ 000091985.1), H. cinaedi (GCA_ 000349975.1), H. bilis (GCA_ 001999985.1), H. cetorum (GCA_ 000259275.1), H. acinonychis (GCA_000009305.1), H. felis (GCA_000200595.1), H. hepaticus (GCA_000007905.1), H. pullorum (GCA_001298055.1), H. typhlonius (GCA_001460635.1), H. rodentium (GCA_ 000687535.1), H. Canadensis (GCA_000162575.1), JRPB_s (GCA_000765695.1), H. trogontum (GCA_000765905.1), and two H. pylori (GCA_001653455.1 and GCA_000013245.1) (Supplementary Table 2). The orthologous Average Nucleotide Identity (OrthoANI) algorithm was used to measure the genetic relatedness between H. apodemus str. SCJK1 and the other Helicobacter spp., and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) dendrogram was constructed based on the OrthoANI value (Lee et al., 2016). Pan-genome Orthologous Groups (POGs) were determined using the BIOiPLUG Comparative Genomics Database (https://www.bioiplug.com/), and a heat map and UPGMA dendrogram were constructed based on these data (i.e., the presence/absence of a POG).
Quality Assurance
To ensure a pure culture, a single H. apodemus colony was transferred three times. Furthermore, contamination was excluded by comparing three 16S rRNA gene fragments found in the H. apodemus str. SCJK1 genome using EzBioCloud DB software (https://www.bioiplug.com/).
Ethics Approval
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Yonsei University Wonju Campus (YWC-151203-1).
Results and Discussion
Characterization of H. apodemus Genome Features
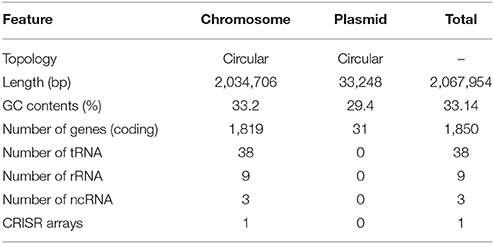
The complete genome of H. apodemus str. SCJK1 was shown to be composed of two circular contigs (one chromosome 2,034,706 bp in length, and one plasmid 33,248 bp in length) (Figure 1A), and to have an average GC content of 33.14% (chromosome, 33.2%; plasmid, 29.4%). Both size and GC content were found to be similar to those of the other analyzed Helicobacter spp., which were 1,886,022 ± 318,568 bp, and 37.3 ± 3.2%, respectively (mean ± standard deviation; Supplementary Table 2). Furthermore, the H. apodemus str. SCJK1 genome was identified to harbor a total of 1,850 coding, and 103 pseudo genes, as well as 38 transfer (tRNA), 9 ribosomal (rRNA; including three each of 5s, 16s, and 23s rRNA), and 3 non-coding (ncRNA) RNA sequences (Table 1). The various genes identified on the H. apodemus chromosome were assigned to COG categories, and thus predominantly classified as J (translation, ribosomal structure, and biogenesis, 8.27%), M (cell-wall/membrane/envelope biogenesis, 7.66%), R (general function prediction only, 7.00%), E (amino-acid transport and metabolism, 6.85%), C (energy production and conversion, 5.93%), and L (replication, recombination, and repair, 5.27%)-type genes. Moreover, the H. apodemus plasmid was annotated as predominantly carrying U (intracellular trafficking, secretion, and vesicular transport, 31.25%), L (18.75%), and X (mobilome: prophages, transposons, 12.5%)-type genes.
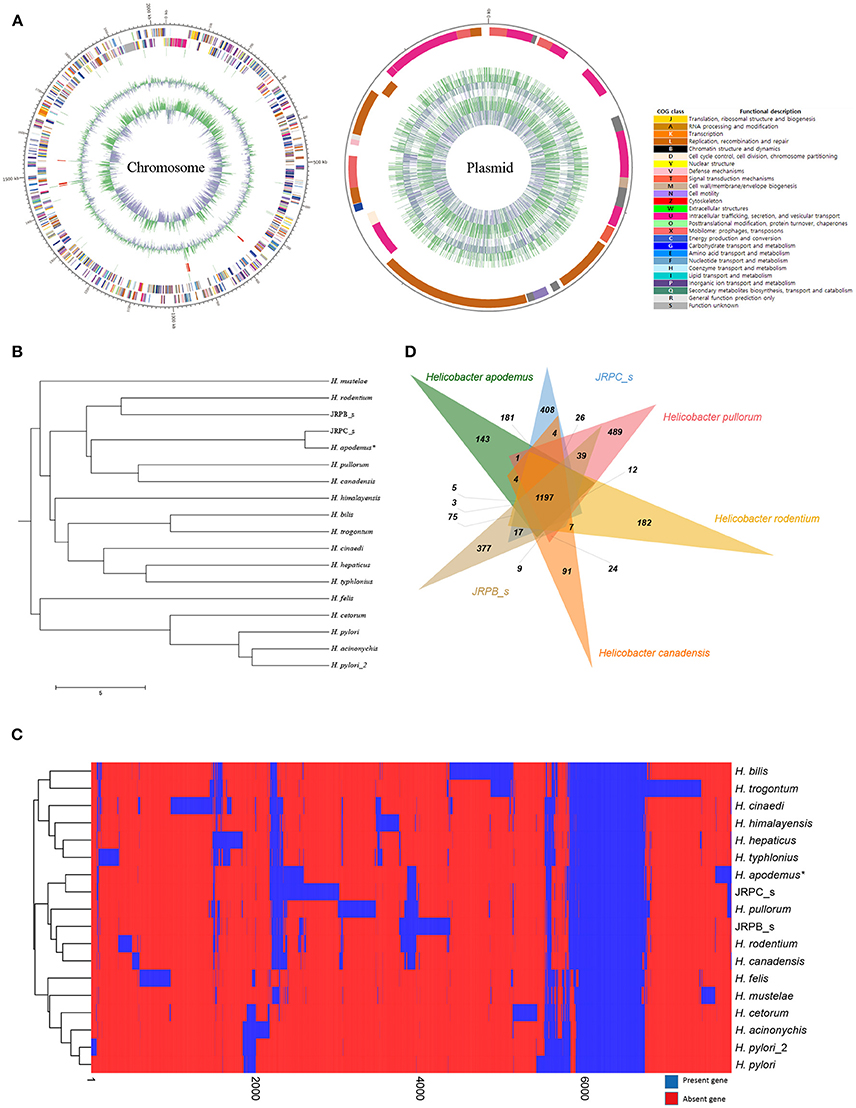
Figure 1. Complete genome sequence and comparative genomic analysis of Helicobacter apodemus str. SCJK1 with respect to 17 other Helicobacter spp. (A) Circular map of the H. apodemus str. SCJK1 genome, and Clusters of Orthologous Group (COG) categories within the genome, assigned using the NCBI COGs database. (B) Unweighted Pair Group Method with Arithmetic Mean (UPGMA) dendrogram of the analyzed Helicobacter spp. based on the orthologous Average Nucleotide Identity (Ortho ANI) value. (C) Heat map and UPGMA dendrogram of the analyzed Helicobacter spp. based on the presence or absence of Pan-genome Orthologous Groups (POG). (D) Venn diagram of six-clustered Helicobacter spp. based on POG analysis. *H. apodemus str. SCJK1 genome used in the current study.
Virulence Genes
In Helicobacter spp., a total of 91 virulence genes were identified, and classified into seven categories, and 17 subcategories. Specifically, these comprised of acid resistance (urease), adherence (adherence-associated lipoprotein (alp) A, alpB, blood group antigen binding adhesins, H. pylori adhesin (hpa) A, hopZ, horB, peb1, and sialic acid binding adhesins), immune evasion (lipopolysaccharide Lewis antigens), immune-modulator (neutrophil-activating protein and outer inflammatory protein), motility (flagella), secretion system (cag pathogenicity island (cag-PAI)-type IV secretion system), and toxin (cytolethal distending toxin, vacuolating cytotoxin) genes (Supplementary Table 3). H. apodemus str. SCJK1 was found to harbor 47 of these 91 virulence genes, including all of the urease-related “acid resistance” genes, 30 of the 38 flagella-related “motility” genes, and three of the “toxin” genes. Of the 10 possible “adherence” genes, H. apodemus str. SCJK1 carried only peb1 (PEB1-related gene). In addition, the strain harbored only one (futA) of the three “immune evasion” genes, and one (napA) of the two “immune modulator” genes. The H. apodemus str. SCJK1 plasmid was shown to include the CagX, V, E, and C (virB 9, 8, 4, and 2, respectively) “secretion system” genes, known to be associated with the cag-PAI type IV secretion system.
Three Helicobacter strains, H. acinonychis str. Sheeba, H. hepaticus ATCC 51449, and H. pylori HPAG1, were used to infer virulence factors in the H. apodemus str. SCJK1 genome (Supplementary Table 3). Of the identified virulence genes, peb1 (“adherence” category), and cdtA and B (“toxin” category, and “cytolethal distending toxin” subcategory) were shown to be present in the H. apodemus and H. hepaticus, but not in the H. pylori genome (Tomb et al., 1997). Furthermore, peb1 has been previously reported to be expressed on the surface of all Campylobacter jejuni and C. coli bacteria, and to thereby mediate intestinal colonization, indicating that it is a prominent target for the immune system (Pei and Blaser, 1993). Consistent with these observations, peb 1 of H. hepaticus was expected to be involved in colonization of the intestine, and according to the results of this study, it may also be involved in intestinal colonization of H. apodemus (Suerbaum et al., 2003). In addition, the cytolethal distending toxin gene, consisting of cdtA, cdtB, and cdtC, has been shown to be expressed by GI pathogens including Campylobacter spp., Escherichia coli, and Helicobacter spp., and associated with irreversible G2/M cell-cycle arrest, which results in the gradual expansion of the nucleoli, and corresponding loss of the cytoplasm (Young et al., 2000; Taylor et al., 2003). Accordingly, H. hepaticus, which is known to carry the cdt gene, has been shown to be associated with chronic GI tract inflammation, and the onset of irritable bowel disease (IBD) in rodents (Whary and Fox, 2004; Young et al., 2004; Ge et al., 2007). The present study showed that H. apodemus also harbors the cdtA and cdtB genes; thus, further study should be conducted to investigate whether H. apodemus exerts similar impact on the rodent GI tract. In addition, both H. pylori and H. apodemus carry genes known to be associated with the cag-PAI type IV secretion system, responsible for horizontal gene transfer between bacterial cells (Rohde et al., 2003). H. pylori has been previously reported to mediate the pathogenesis of gastric adenocarcinoma and mucosa-associated lymphatic tissue (MALT) lymphoma, by injecting cagA (which is a bacterial gene that promotes cell proliferation and differentiation) into gastric epithelial cells using the cag-PAI type IV secretion system (Odenbreit et al., 2000; Cascales and Christie, 2003). However, the H. apodemus genome did not include cag A; thus, the role of the cag-PAI type IV secretion system in the H. apodemus genome should be studied further.
Comparative Genomic Analysis
The genome of H. apodemus str. SCJK1 was compared with those of 17 other Helicobacter spp. In the constructed UPGMA dendrogram (based on orthoANI values), H. apodemus was shown to be not only closely related, but of the same species (orthoANI value 97.35) as JRPC_s, which was isolated from rats in the United States in 2003 (Lee et al., 2016). These results indicate that H. apodemus exists not only in A. agrarius, but also in other rodents, and also, demonstrate the low genetic difference between the isolates collected from the two different host types. Furthermore, H. apodemus was clustered with H. pullorum (isolated from fresh chicken meat in Portugal, 2012), H. canadensis (no isolation information), H. rodentium (isolated from a C57bl/6x129 mouse, 1995), and JRPB_s (isolated from a mouse in the United States, 2011), according to the calculated orthoANI values of 73.46, 73.52, 72.92, and 72.65, respectively (Figure 1B). The calculated relatedness between these species was consistent with the findings of previous studies, in which primers used in an H. rodentium-specific PCR assay cross-reacted with both H. apodemus and H. rodentium (Shen et al., 1997; Miller et al., 2014). However, the results of the present study were not consistent with the data presented in phylogenetic tree (based on 16S rRNA sequence comparisons) in which H. apodemus was clustered with H. mesocricetorum, H. ganmani, and H. rodentium, but not with H. pullorum or H. canadensis (Jeon et al., 2015).
In the heat map and UPGMA dendrogram (based on POG analysis), H. apodemus str. SCJK1 was clustered with JRPC_s, H. pullorum, H. canadensis, H. rodentium, and JRPB_s, consistent with orthoANI value-based results (Figure 1C). The consistency in results from orthoANI values-based and POG-based analysis indicated that H. apodemus was genetically similar to the above Helicobacter species (H. pullorum, H. canadensis, H. rodentium, and JRPB_s). In addition, the six strains shared 1,197 POGs, of which 143 were identified only in the strains analyzed in the present study (Figure 1D). H. apodemus strains (i.e., those isolated in the current study, and JRPC_s) shared 1,647 POGs, which were expected to have specific characteristics of H. apodemus.
Future Directions
Studies using whole-genome sequencing technology have made important advances in rodent GI microbial research; however, the present study is the first to analyze the complete genome sequence of H. apodemus from wild A. agrarius, which acts as a pathobiont in wild rodents. It is hoped that the results of this analysis, together with those of the conducted comparative genomic analysis, will serve as the basis for further biomedical studies investigating the immunological and inflammatory effects of H. apodemus on the rodent GI tract.
Data Access
The genome sequence of Helicobacter apodemus str. SCJK1 was deposited in the GeneBank under accession number CP021886-CP021887.
Author Contributions
SC conceived and designed the study. J-UA and WK analyzed the genome sequencing data. JGS, JKS, and B-YJ performed sampling, and prepared the manuscript. JK was a major contributor, both in experiments and writing the manuscript. All authors have read and approved the final manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF-2018R1A2B6002396) and Korea Mouse Phenotyping Project (2016M3A9D5A01952417) of the Ministry of Science, ICT and Future Planning.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00838/full#supplementary-material
References
Cascales, E., and Christie, P. J. (2003). The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149. doi: 10.1038/nrmicro753
Chai, J. N., Peng, Y., Rengarajan, S., Solomon, B. D., Ai, T. L., Shen, Z., et al. (2017). Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2:eaal5068. doi: 10.1126/sciimmunol.aal5068
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10:563. doi: 10.1038/nmeth.2474
Ge, Z., Rogers, A. B., Feng, Y., Lee, A., Xu, S., Taylor, N. S., et al. (2007). Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell. Microbiol. 9, 2070–2080. doi: 10.1111/j.1462-5822.2007.00939.x
Goh, K. L., Chan, W. K., Shiota, S., and Yamaoka, Y. (2011). Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter 1, 1–9. doi: 10.1111/j.1523-5378.2011.00874.x
Goto, K., Jiang, W., Zheng, Q., Oku, Y., Kamiya, H., Itoh, T., et al. (2004). Epidemiology of Helicobacter infection in wild rodents in the Xinjiang-Uygur autonomous region of China. Curr. Microbiol. 49, 221–223. doi: 10.1007/s00284-004-4287-6
Jeon, W. J., Dong, H.-J., Shin, J. H., Kim, I. Y., Ho, H., Oh, S. H., et al. (2015). Helicobacter apodemus sp. nov., a new Helicobacter species identified from the gastrointestinal tract of striped field mice in Korea. J. Veter. Sci. 16, 475–481. doi: 10.4142/jvs.2015.16.4.475
Johansson, S. K., Feinstein, R. E., Johansson, K.-E., and Lindberg, A. V. (2006). Occurrence of Helicobacter species other than H. hepaticus in laboratory mice and rats in Sweden. Comp. Med. 56, 110–113.
Khan, I., Azhar, E. I., Abbas, A. T., Kumosani, T., Barbour, E. K., Raoult, D., et al. (2016). Metagenomic analysis of antibiotic-induced changes in gut microbiota in a pregnant rat model. Front. Pharmacol. 7:104. doi: 10.3389/fphar.2016.00104
Kusters, J., van Vliet, A., and Kuipers, E. (2006). Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19, 449–490. doi: 10.1128/CMR.00054-05
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Miller, C. L., Muthupalani, S., Shen, Z., and Fox, J. G. (2014). Isolation of Helicobacter spp. from mice with rectal prolapses. Comp. Med. 64, 171–178.
Mladenova-Hristova, I., Grekova, O., and Patel, A. (2017). Zoonotic potential of Helicobacter spp. J. Microbiol. Immunol. Infect. 50, 265–269. doi: 10.1016/j.jmii.2016.11.003
Odenbreit, S., Püls, J., Sedlmaier, B., Gerland, E., Fischer, W., and Haas, R. (2000). Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287, 1497–1500. doi: 10.1126/science.287.5457.1497
Pei, Z., and Blaser, M. J. (1993). PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J. Biol. Chem. 268, 18717–18725.
Péré-Védrenne, C., Flahou, B., Loke, M. F., Ménard, A., and Vadivelu, J. (2017). Other Helicobacters, gastric and gut microbiota. Helicobacter 22:e12407. doi: 10.1111/hel.12407
Rohde, M., Püls, J., Buhrdorf, R., Fischer, W., and Haas, R. (2003). A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49, 219–234. doi: 10.1046/j.1365-2958.2003.03549.x
Shen, Z., Fox, J., Dewhirst, F., Paster, B., Foltz, C., Yan, L., et al. (1997). Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int. J. Syst. Evol. Microbiol. 47, 627–634. doi: 10.1099/00207713-47-3-627
Solnick, J. V., and Schauer, D. B. (2001). Emergence of diverse helicobacterspecies in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14, 59–97. doi: 10.1128/CMR.14.1.59-97.2001
Suerbaum, S., Josenhans, C., Sterzenbach, T., Drescher, B., Brandt, P., Bell, M., et al. (2003). The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. U.S.A. 100, 7901–7906. doi: 10.1073/pnas.1332093100
Swennes, A. G., Parry, N. M., Feng, Y., Sawyer, E., Lohr, B., Twedt, D. C., et al. (2016). Enterohepatic Helicobacter spp. in cats with non-haematopoietic intestinal carcinoma: a survey of 55 cases. J. Med. Microbiol. 65, 814–820. doi: 10.1099/jmm.0.000274
Taylor, N. S., Ge, Z., Shen, Z., Dewhirst, F. E., and Fox, J. G. (2003). Cytolethal distending toxin: a potential virulence factor for Helicobacter cinaedi. J. Infect. Dis. 188, 1892–1897. doi: 10.1086/379837
Taylor, N. S., Xu, S., Nambiar, P., Dewhirst, F. E., and Fox, J. G. (2007). Enterohepatic Helicobacter species are prevalent in mice from commercial and academic institutions in Asia, Europe, and North America. J. Clin. Microbiol. 45, 2166–2172. doi: 10.1128/JCM.00137-07
Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., et al. (1997). Corrections: the complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 389:412. doi: 10.1038/38792
Whary, M. T., and Fox, J. G. (2004). Natural and experimental Helicobacter infections. Comp. Med. 54, 128–158.
Young, V. B., Knox, K. A., Pratt, J. S., Cortez, J. S., Mansfield, L. S., Rogers, A. B., et al. (2004). In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect. Immun. 72, 2521–2527. doi: 10.1128/IAI.72.5.2521-2527.2004
Young, V. B., Knox, K. A., and Schauer, D. B. (2000). Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect. Immun. 68, 184–191. doi: 10.1128/IAI.68.1.184-191.2000
Keywords: Helicobacter apodemus, korean striped field mouse, wild rodents, whole genome sequencing, comparative genomic analysis
Citation: Kim J, Kim W, An J-U, Suh JG, Seong JK, Jeon B-Y and Cho S (2018) Complete Genome Sequencing and Comparative Genomic Analysis of Helicobacter Apodemus Isolated From the Wild Korean Striped Field Mouse (Apodemus agrarius) for Potential Pathogenicity. Front. Pharmacol. 9:838. doi: 10.3389/fphar.2018.00838
Received: 18 May 2018; Accepted: 11 July 2018;
Published: 31 July 2018.
Edited by:
Annalisa Bruno, Università degli Studi G. d'Annunzio Chieti e Pescara, ItalyReviewed by:
Satish Ramalingam, SRM University, IndiaSuresh Kumar Kalangi, Indrashil University, India
Copyright © 2018 Kim, Kim, An, Suh, Seong, Jeon and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seongbeom Cho, Y2hvc2VAc251LmFjLmty
 Junhyung Kim
Junhyung Kim Woohyun Kim
Woohyun Kim Jae-Uk An
Jae-Uk An Jun Gyo Suh
Jun Gyo Suh Je Kyung Seong1
Je Kyung Seong1 Seongbeom Cho
Seongbeom Cho