
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pharmacol., 26 June 2018
Sec. Neuropharmacology
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.00683
This article is part of the Research TopicTrace Amines and the Trace Amine-Associated Receptor 1: Emerging role and clinical implicationsView all 16 articles
Since its discovery in 2001, the major focus of TAAR1 research has been on its role in monoaminergic regulation, drug-induced reward and psychiatric conditions. More recently, TAAR1 expression and functionality in immune system regulation and immune cell activation has become a topic of emerging interest. Here, we review the immunologically-relevant TAAR1 literature and incorporate open-source expression and cancer survival data meta-analyses. We provide strong evidence for TAAR1 expression in the immune system and cancers revealed through NCBI GEO datamining and discuss its regulation in a spectrum of immune cell types as well as in numerous cancers. We discuss connections and logical directions for further study of TAAR1 in immunological function, and its potential role as a mediator or modulator of immune dysregulation, immunological effects of psychostimulant drugs of abuse, and cancer progression.
The Gs-linked G-protein coupled receptor Trace Amine Associated Receptor 1 (TAAR1) is a target for a wide variety of agonists including endogenous amines and amphetamine-like drugs of abuse. Endogenous agonists include common biogenic amines as well as trace amines (TAs) (Borowsky et al., 2001; Bunzow et al., 2001). TAs are present in the mammalian nervous system at levels much lower than those of common biogenic amines and include β-phenylethylamine (β-PEA), tyramine, octopamine, tryptamine, and thyronamine (Boulton, 1976; Juorio, 1982; Burchett and Hicks, 2006). TAs resemble common biogenic amines in terms of subcellular localization, chemical structure, and metabolism (Borowsky et al., 2001; Lindemann and Hoener, 2005). TAAR1 is also a target of drugs of abuse including methamphetamine, amphetamine, and 3,4-methylenedioxymethamphetamine (MDMA) (Bunzow et al., 2001). Unlike common biogenic amines, both TAs and amphetamine-like drugs show greater selectivity for TAAR1 relative to other aminergic receptors, which we have hypothesized to underlie both signaling regulation and dysregulation related to imbalances between these signaling molecules in psychiatric conditions and in addiction (Xie and Miller, 2008). Aminergic neurotransmitter imbalance is implicated in several neuropathological conditions and therefore the major focus of TAAR1 research has been its role in monoaminergic regulation, drug-induced reward, and psychiatric conditions (Miller, 2011). Less studied is TAAR1 expression and functionality in immune cells, which is the subject of this review.
The immune system is a complex collection of organs, tissues, and cells that serve to protect the host organism from pathogens. In its simplest form it can be subdivided into the innate and adaptive immune systems based on the temporal sequence of the immune response (Janeway, 2001). While immune cell interactions are extremely complex and the lines separating branches of immunity are ambiguous at best, for the purpose of this review we will follow this basic dichotomy. Here, we review the immunologically relevant TAAR1 literature and incorporate open-source expression and cancer survival data as a mode of observational insight into physiologically-relevant topics of interest not only to the TAAR1 research community, but also to other life science investigators. We mined the NCBI Gene Expression Omnibus (GEO) profiles database (Barrett et al., 2013) which contains gene expression profiles from curated GEO Datasets (GDS) to find evidence of TAAR1 expression across various immune cell types and cancers. We then make connections and suggest logical directions for further study of TAAR1 in immunological function. We also provide complete references containing queryable NCBI GEO Profile ID and GDS accession numbers.
Microarray datasets containing normalized counts corresponding to the TAAR1 gene transcript were obtained through BioGPS, RefDIC, and NCBI GEO. Normalized counts were log2 transformed and visualized in R.
Datasets for TAAR1 expression were extracted from http://biogps.org using the search function to query for “TAAR1.”
Transcriptomic profiling data of immunologically relevant cell types was obtained from the Reference Database of Immune Cells (RefDIC, Hijikata et al., 2007). Datasets were extracted using the “Expression profile” function (available at http://refdic.rcai.riken.jp/profile.cgi) utilizing the microarray profile type and using the official gene symbol “TAAR1.” The human and mouse TAAR1 gene was selected (ID 134864 and 111174, respectively). Queries for various immune cell types were conducted using a text search within the “Change Dataset” function.
The NCBI Gene Expression Omnibus (GEO) includes the GEO Profiles database (Barrett et al., 2013) which contains gene expression profiles from curated GEO Datasets (GDS) that are searchable by gene identifier and by keywords. The GEO Profiles database was probed by cell type using the advanced search builder function to build a query containing cell types of interest. Briefly, to extract all GEO Profiles containing data for TAAR1 RNA expression in astrocytes, for example, a search was conducted using the terms “TAAR1 (AND) ASTROCYTES.” Other immune cell types were similarly queried using this search method.
GDS accession numbers, literature references, and Profile ID numbers associated with their respective GEO Profiles are included in tables to allow for direct access to the expression data discussed. Individual GEO Profiles discussed in this review may be freely accessed at https://www.ncbi.nlm.nih.gov/geoprofiles/ by entering the GEO Profile ID provided in the text search box. Similarly, entire GEO Datasets may be accessed in the same manner at https://www.ncbi.nlm.nih.gov/gds. As the microarray technology used to produce GEO Profiles commonly utilizes an algorithmic detection call to determine cutoffs for positive signals, some GEO Profiles contained samples in which the TAAR1 expression signal was below the cutoff for expression, and those samples are denoted as “Below cutoff” in tables. GEO Profiles where not all but at least one sample was positive for TAAR1 expression are denoted as “Partial” in tables, and GEO Profiles where all samples were negative for TAAR1 expression signal are denoted as “Negative.”
When available, statistical analysis within a dataset was carried out using the Analyze Dataset tool located at the bottom of the dataset of interest's page.
Antibody-based protein expression data is freely available online from the Human Protein Atlas (HPA) at www.proteinatlas.org (Uhlén et al., 2015; Thul and Lindskog, 2017). This database was generated by probing various human tissues for all protein-coding genes and is cataloged in a searchable module, allowing for tissue-specific exploration of expression patterns. For antibody-based data, the gene of interest is assigned an expression level based on staining intensity and fractional quantity of stained cells in a sample. TAAR1-specific results can be viewed directly at https://www.proteinatlas.org/ENSG00000146399-TAAR1/tissue.
A text query for the HGNC gene symbol “TAAR1” was made using the mRNA transcript search tool available from https://hive.biochemistry.gwu.edu.
Processed RNAseq data representing 41,907 RNAseq samples across 171 cancer studies was obtained from cbioportal.org using a text search for “TAAR1” in the gene query tool.
A gene query for “TAAR1” was carried out using the Genome Browser tool available at https://cancer.sanger.ac.uk/cosmic/browse/genome.
Datasets were obtained through a text search for “TAAR1” at http://syslab4.nchu.edu.tw/CRN/.
RNA-seq expression profiles for TAAR1 were obtained from the NIH National Cancer Institute Genomic Data Commons (GDC) Data Portal at https://portal.gdc.cancer.gov/. RNA-seq datasets for the TCGA cancer studies discussed can be accessed from the link above by navigating to the “Exploration” tool, then selecting “Genes,” entering “TAAR1” in the search bar, then navigating to “View Files in Repository” and finally selecting “Transcriptome Profiling.” Cancer RNA-seq expression dataset names begin with the prefix “TCGA-” followed by the abbreviation for the appropriate cancer type. Data can be downloaded as.TXT files and are searchable by text to identify data specific to TAAR1.
RNA-seq datasets can be directly accessed through the ArrayExpress website by accessing https://www.ebi.ac.uk/arrayexpress/experiments/ followed by the ArrayExpress experiment number, i.e., https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2706/.
Overall survival trends expressed as hazard ratios (HR) for 80 unique human cancer studies representing 15 cancer types were obtained from the online databases Prognoscan (Mizuno et al., 2009) and PROGgene (Goswami and Nakshatri, 2013). For each PROGgene study (n = 68) the sample population was bifurcated at the median into high- and low-TAAR1 expression groups. Study data obtained from Prognoscan (n = 12) utilized a minimum p-value approach to determine the point of bifurcation into high and low expression groups. Briefly, the HR obtained for each study can be explained as the ratio of events (deaths) in the high TAAR1 expression group to events in the low TAAR1 expression group. To perform the meta-analysis after data collection all HR values were log-transformed to normalize values around zero to enable the calculation of subgroup averages and then back transformed to produce the average HR value. Forest plots were created using the R package ggplot2.
Transcription factor analysis of the TAAR1 5' untranslated region (input sequence: Human DNA sequence from clone RP11-295F4 on chromosome 6, complete sequence, GenBank: AL513524.8) was carried out with the transcription factor (TF) binding site predictor programs LASAGNA-Search 2.0 (Lee and Huang, 2013, http://biogrid-lasagna.engr.uconn.edu/lasagna_search/) and MatInspector (Quandt et al., 1995; Cartharius et al., 2005, Bioinformatics) which is available as a free trial or part of a software suite from Genomatix available at https://www.genomatix.de/.
The perpetual circulation of blood through the body provides a constant sentry of diverse immune cells capable of monitoring homeostatic alterations. This “sentinel principle” posits that blood cells reflect changes in overall physiology and is the basis for the practice of biomarker and liquid biopsy research that is foundational to the detection of many diseases (Burczynski and Dorner, 2006; Liew et al., 2006). The blood is composed of erythrocytes, leukocytes, platelets, and plasma. The plasma represents the largest volume of the blood. It is made up largely of water which includes various proteins, clotting factors, and other metabolic constituents. Erythrocytes represent the largest cellular portion of whole blood and predominantly function as oxygen carriers in cellular respiration. The remaining 1% of the blood is made up of leukocytes, which are the immune cells of the blood, and platelets, which facilitate blood clotting in response to injury and other homeostatic disruptions. Leukocytes are the mixed cell population of white blood cells consisting of all the immune cells of the blood and include cells of both the innate and adaptive response. More than a decade ago, TAAR1 mRNA was identified in circulating human leukocytes using RT-PCR (D'Andrea et al., 2003; Nelson et al., 2007).
Here, we utilized the NCBI GEO Profiles database (Barrett et al., 2013) to perform a search of TAAR1 RNA expression in blood, which revealed detectable expression in all of the 20 datasets obtained, representing blood from humans, rhesus monkeys, and mice. Accession numbers and references for expression datasets in whole blood, platelets, and PBMCs are summarized in Table 1. Accordingly, TAAR1 RNA is present in leukocytes and TAAR1 RNA is consistently detected in whole blood.
In addition to their classic role in clotting, platelets are also thought to play a role in the initial innate immune response (Palabrica et al., 1992; Opal, 2000; Esmon, 2004; Theopold et al., 2004). Neutrophil-derived signals augment the thrombocytic response and in turn can help isolate pathogens and inhibit their entry into the system circulation (Hickey and Kubes, 2009; Massberg et al., 2010). In light of the likely possibility of cross-talk between platelets and innate immune cells it is interesting to note that there are detectable levels of TAs present in human platelets, and intra-platelet concentrations are significantly decreased upon platelet activation (D'Andrea et al., 2003). These initial data suggested that the release of TAs in response to injury or immune cell signals could have a functional role in the innate immune response. Using NCBI GEO data, we found evidence that platelets express TAAR1 RNA. TAAR1 RNA is detectable in human (Raghavachari et al., 2007, GDS3318; Piccaluga et al., 2014, GDS5405, GDS5406; Risitano et al., 2012, GDS4659) and mouse (Wright et al., 2014; Lee et al., 2016, GDS5320; Paugh et al., 2013, GDS4821) platelets, suggesting a potential for a possible feedback mechanism in platelet function (Table 1). The presence of TAAR1 RNA in platelets suggests a potential for platelet expression of TAAR1 protein and a potential for responses generated by TAAR1 agonists. It is important to note, however, that RNAs present in anucleate platelets are derived from the progenitor megakaryocyte cells from which platelets are derived, and therefore the TAAR1 RNA detected may or may not be utilized by platelets to synthesize TAAR1 protein. Platelets do, however, possess the machinery needed to translate RNA to protein and as such further study of platelet protein expression is needed to determine whether this cell type can produce functional TAAR1 protein (Weyrich et al., 2009). With this caveat in mind, TAAR1 protein expression in platelets could be a contributory mechanism of psychostimulant-induced effects on platelet-mediated immune responses, and so further investigation is necessary to examine protein expression and effects of selective TAAR1-targeted drugs on platelet function.
Peripheral blood mononuclear cells (PBMCs) are a mixed subpopulation of leukocytes lacking the more dense, granulated polymorphonuclear (PMN) cells. PBMCs include the B- and T-cells of adaptive immunity, as well as the natural killer (NK), NK T-cells, macrophage and dendritic cells of monocytic lineage of the innate immunity. Analysis of PBMC gene expression is widely utilized to identify biomarkers for disease diagnosis for an array of pathological conditions. PBMC transcriptomics are used in inflammatory conditions such as arthritis (Boyle et al., 2003; Shou et al., 2006; Wong et al., 2016), in various cancers (Burczynski et al., 2005; Showe et al., 2009; Piccolo et al., 2015), and in numerous psychiatric conditions including depression (Mendez-David et al., 2013; Fan et al., 2015), schizophrenia (Lai et al., 2011), and bipolar disorder (Herberth et al., 2011). In lieu of diseased patient populations, pharmacological cellular activation with mitogens or foreign antigens in vitro is used to model immune challenge-mediated changes in cellular function and gene expression patterns. Cellular mitogen Phytohaemagglutinin (PHA) stimulation is a well-known model of cellular activation. PHA has been shown to upregulate TAAR1 mRNA from human PBMCs relative to low TAAR1 mRNA levels at rest (Nelson et al., 2007). Analogously, PHA also induced a significant increase in rhesus monkey PBMC TAAR1 protein from low baseline levels and this upregulation augmented TAAR1 agonist-induced PKA and PKC phosphorylation (Panas et al., 2012). These data suggest that TAAR1 may be present at very low levels in normal physiological states but is upregulated in activated states, suggesting that it may be necessary in downstream responses related to cellular activation. Supporting this concept, work by Sriram et al. (2016) showed that viral antigenic exposure by HIV-1 infection upregulates TAAR1 protein in human PBMCs and that upregulation is augmented by pretreatment with the TAAR1 agonist methamphetamine (METH). Importantly, TAAR1 activation with METH increases HIV-1 viral titers and replication (Sriram et al., 2016). These data suggest that upregulation and activation of TAAR1 may be a mechanism by which anti-viral immune processes may be diminished or viral fitness is altered directly. It is intriguing to speculate that these effects may also occur in response to upregulation of TAAR1 resulting from amphetamine-like psychostimulant abuse, imbalances in TAAR1 ligand availability due to neurotransmitter level dysregulation as in psychiatric diseases, or modulations to endogenous TA levels through diet or host-microbiome interactions.
With regard to viral infections, however, our search of NCBI GEO revealed expression array studies in which human PBMC samples were devoid of TAAR1 at baseline (Table 1). For example, neither influenza virus infection nor a deficiency in the IRF7 gene, an important mediator of antiviral defenses, altered TAAR1 expression (Ciancanelli et al., 2015, GDS5626). Similarly, data derived from another study indicates that PBMCs infected with HIV alone or with tuberculosis co-infection were devoid of TAAR1 expression (Dawany et al., 2014, GDS4786). Data from another study (Cai et al., 2014, GDS5030, GDS4966) indicates that human PBMCs infected with active or latent tuberculosis lacked detectable TAAR1 expression as determined by a detection call of absent; however, one positive signal was obtained for a single uninfected sample (Cai et al., 2014, GDS4966). Furthermore, TAAR1 expression in both normal human PBMCs and those infected with Herpes simplex (Sancho-Shimizu et al., 2011, GDS4540) was absent and did not respond to the bacterial antigen lipopolysaccharide (LPS) or activation of the single-stranded DNA receptor TLR7/8 suggesting an inability of these cellular activation pathways to alter TAAR1 expression. It is important to remember, however, that the array-based technology used to determine presence of expression in these GEO datasets relies on detection calls derived from computational algorithms, and as such an “absent” call does not necessarily mean that the transcript is truly absent (Oudes et al., 2005). As such, there is a need for more TAAR1-specific expression profiling through less ambiguous methods such as RT-PCR and immunological staining.
In contrast to these virally-induced infectious diseases, our search of NCBI GEO revealed that TAAR1 RNA is detectable in PBMCs of patients with the inflammatory or immunological diseases osteoarthritis (Ramos et al., 2014, GDS5363), juvenile idiopathic arthritis (Hinze et al., 2010, GDS4267; Moncrieffe et al., 2010, GDS4272), and multiple sclerosis (Malhotra et al., 2011, GDS4147). TAAR1 RNA is also present in human PBMCs isolated from gastric, liver, and pancreatic cancers (Shi et al., 2014, GDS4882), as well as breast cancer (LaBreche et al., 2011, GDS3952). TAAR1 RNA was also present, but not differentially expressed from controls, in datasets from pulmonary hypertension (Cheadle et al., 2012, GDS5499), interleukin-10 treatment (Teles et al., 2013, GDS4551), nickel exposure (Arita et al., 2013, GDS4974), and benzene exposure (McHale et al., 2009, GDS3561), and these data are summarized in Table 1.
Taken together, our analyses found that TAAR1 expression in PBMCs varies between studies and significant modulation of expression has only been demonstrated in vitro in response to cellular activation or immunological challenge. It may be that TAAR1 functionality in the immune system may mediate alterations to immune cell maturation processes. Our analysis indicates a role for TAAR1 in PBMC-derived erythroid maturation. TAAR1 levels are significantly higher (two-tailed t-test, p = 0.05) in the earliest stage of erythropoiesis (n = 3) vs. the later 3 stages (n = 9) (Merryweather-Clarke et al., 2011, GDS3860). Activation of TAAR1 present in these early progenitor cells would predictably alter signaling cascades involved in the phenotypic development of these cells. Further investigation into the disparity of TAAR1 expression between maturation stages may lend insight into a mechanism by which erythrocytic pathologies develop.
Granulocytes, also referred to as polymorphonuclear leukocytes (PMN), are a type of innate immune cells that act as the first line of cellular defense due to their ability to be recruited to the site of infection through chemotaxis. When activated these cells migrate toward chemoattractants derived from pathogens or local macrophage (Amulic et al., 2012). Granulocytes include phagocytic neutrophils that act initially to engulf foreign invaders and eosinophils that function primarily in defense against parasitic infections. Basophils are the rarest type of granulocyte and function in the initial infection response, allergic reactions, and in the T-cell polarization necessary for the adaptive immune response (Parham, 2009). Granulocytes express gene transcripts for TAAR1 and another TAAR family member, the orphan receptor TAAR2. Both TAAR1 and TAAR2 are co-expressed in subsets of human PMN (Babusyte et al., 2013). Interestingly, expression of both TAAR1 and TAAR2 was required for the chemosensory migration of PMN toward TAs, as evidenced by non-functionality when either TAAR was knocked down (Babusyte et al., 2013). If replicable, these data suggest the possibility of TAAR1/TAAR2 signaling interactions and/or dimerization as a prerequisite for chemotaxis of PMN toward TAs. Further studies are necessary to confirm if TAAR1 and TAAR2 form functional dimers. Although studies reporting functional subsets for blood granulocytes are few, there is an emerging indication for subset-specific markers (Clemmensen et al., 2012; Pillay et al., 2012). As such, TAAR1 and TAAR2 may represent markers for a granulocyte subset capable of chemosensory migration to TAs and other TAAR1 (and potentially TAAR2) ligands. These data also raise the possibility that the potent TAAR1 agonist METH and other TAAR1-targeted compounds could act in the same manner to alter chemotactic activity of particular subsets of PMN. Potentially, drugs that selectively target TAAR1 could potentially be used to treat migratory granulocyte dysfunction. Our NCBI GEO analysis revealed array-based evidence to support TAAR1 expression in granulocytes (Lattin et al., 2008, GSE10246). Overall, expression data for mixed granulocytes, eosinophils, neutrophils, and mast cells revealed a consistent TAAR1 signal (Table 2). No basophil-specific samples were available from the GEO database, so our analysis could not determine basophilic TAAR1 expression. While the relatively low-abundance of basophils could make the detection of low-level TAAR1 difficult, their role in the initial immune response and T-cell function necessitate targeted expression analysis of this cell type. Babusyte et al. (2013) provides convincing evidence that TAAR1 is in fact present in granulocytes but does not specify cellular subsets, so cell-type specific investigation is needed. Our analysis also indicates that mast cells, which are primarily involved in the allergic reaction and histamine release, also express TAAR1 (Lattin et al., 2008, GSE10246; Ito et al., 2012, GDS4420; Geoffrey et al., 2006, GDS2742). Finally, as all the granulocytes profiled in these GEO datasets were normal cells, it would be interesting to investigate any changes in TAAR1 expression levels with cellular activation.
Monocytes represent a short-lived subset of leukocytes with phagocytic ability that matures into macrophages and dendritic cells. At the time of initial immunological challenge, these monocytic cells of the innate immune system are the first to arrive and provide a rapid response to engulf pathogens and induce inflammation. Local phagocytic macrophages possess receptors called pattern recognition receptors that can recognize conserved pathogen antigens. Phagocytic cells engulf invaders on site while also releasing cytokines and chemokines that induce inflammation and the recruitment of other innate immune cells such as neutrophils and antigen-presenting cells (APCs). APCs are a bridge between the innate and adaptive immune response, processing antigens and presenting them to helper T-cells (Th) cells that will in turn undergo clonal expansion and activate cytotoxic T-cells (Tc) and B-cells. The result of this process is pathogen-specific immunological memory consisting of clonal populations of T- and B-cells that recognize and react to the initial specific antigen expressed by the pathogen (Janeway, 2001; Mogensen, 2009; Turvey and Broide, 2010; Monie, 2017). Babusyte et al. (2013) reported that human monocytes demonstrate variation in TAAR subtype mRNA expression as 20% of monocytes screened did not express any of the TAAR genes. There was some level of monocytic expression of all TAAR genes except TAAR8, and the TAAR2 signal was particularly high in this cell type (Babusyte et al., 2013). Our analysis of transcriptomic data for TAAR1 expression in mixed monocytic cell samples obtained from NCBI GEO (n = 22) strengthen the argument for TAAR1 presence in these cells, as 72.7% of datasets with measurements for TAAR1 contained positive signals for expression above detection threshold. Table 3 includes complete references containing queryable GEO Profile ID and GDS accession numbers. Notably, the only other manuscript published to date explicitly describing TAAR expression in monocytic cell types found that primary mouse macrophage and dendritic cells are devoid of all nine TAARs and are unresponsive to LPS or mouse gamma-herpes virus. TAAR1 was similarly absent in freshly isolated bone marrow cells and maturing bone marrow-derived dendritic cells and macrophages (Nelson et al., 2007). However, in contrast to this study are the various RNA expression datasets that reveal detectable expression of TAAR1 in macrophage. A search of datasets containing a probe representing the TAAR1 gene yielded 34 results of which 24 contained macrophage samples positive for TAAR1 expression (Table 3). Nine of the remaining ten datasets had a signal for TAAR1 expression that fell below the threshold for detection. Notably, a TAAR1 signal was completely absent in only one of the 34 data sets (Table 3). Our analysis of a macrophage gene expression profile (Gleissner et al., 2010, GDS3787) revealed significant TAAR1 upregulation with in vitro exposure to CXCL4, a cytokine released by activated platelets that plays a role in T- and NK cell migration and angiostatic activity.
Natural Killer (NK) cells are a type of cytotoxic lymphocyte important in the response to viral infection, the detection of tumor formation, and in the promotion of self-tolerance (Terunuma et al., 2008). 86.7% of NK cells isolated from human buffy coat samples had some level of mRNA for TAAR1,−2,−5,−6, or −9 that was detectable (Babusyte et al., 2013). NK cells isolated from mouse spleen also had detectable levels of gene transcripts for TAAR1,−2,−3, and −5 (Nelson et al., 2007), which is in agreement with our analysis of transcriptomic profiling (Table 4) of mouse spleen and pancreas (Fehniger et al., 2007, GDS2957; Sitrin et al., 2013, GDS4948, GDS4946). Array-based expression data exists to further support TAAR1 expression in NK cells (Table 4). A unique cell type that combines phenotypic characteristics of both NK cells and T-cells, termed NKT-cells, possess enhanced killing ability and have displayed significant antitumor function in preclinical studies (Schmidt-Wolf, 1991; Kim et al., 2007). Our analysis reveal TAAR1 RNA expression is detectable in mouse NKT-cells (Verykokakis et al., 2013, GDS5602), and therefore a targeted analysis of TAAR1 presence and function in this unique cell type is needed.
T-cells are a lymphocytic subset of immune cells that are important in cell-mediated immunity. Stimulation with specific factors differentially activates T-cells to illicit the development of specific regulatory, effector, and helper functions and these phenotypes in turn shape the nature of the immune response (Barnes, 2011). Identifying physiological perturbations in T-cell function is crucial to understanding possible TAAR1 function in the immune system. There is a lack of both agreement and availability of literature exploring TAAR1 expression and function in T-cells and to date the only three manuscripts published on the topic have yielded opposing results. Observationally, the potent TAAR1 agonist METH alters T-cell function through mitochondrial injury, oxidative stress, and alterations to cytokine production (Potula et al., 2010). More specifically, work by Sriram et al. (2016) found that METH treatment increases TAAR1 mRNA and functional protein expression in human T-cells associated with a METH-induced decrease in secretion of the proinflammatory cytokine IL-2 and altered cAMP production. Importantly, these METH-induced effects were TAAR1-dependent (Sriram et al., 2016) suggesting that TAAR1 is expressed in T-cells and is capable of altering T-cell function. The same study also reported that HIV-1 positive METH users displayed enhanced expression of TAAR1 protein in T-cells of the lymph nodes compared to non-users. Elucidating the biology of HIV-1 infection in the context of concomitant chronic drug abuse is clinically important as METH-induced immunomodulation in HIV-1 infection could have a significant impact on treatment modulation and HIV-1 pathogenesis (Boddiger, 2005). Overall, current data suggest an association of TAAR1 expression, HIV-1 infection and METH and therefore more study of this possible connection is warranted. Supporting the hypothesis of lymphocytic modulation by TAAR1 is the observation that T-cells expressing TAAR1 and TAAR2 treated with TAs in vitro displayed increased secretion of IL-4, a cytokine that stimulates the proliferation of T- and B-cells (Babusyte et al., 2013). It is important to note that this effect was reportedly due to TA interaction with both TAAR1 and TAAR2, as the effect was lost with siRNA knockdown of either receptor (Babusyte et al., 2013), and therefore exploration of TAAR1/TAAR2 interactions is needed to replicate and elucidate this effect. To the contrary of the work of Sriram et al. (2016) and Babusyte et al. (2013), mouse splenic T-cells were earlier reported as devoid of TAAR1, TAAR2, TAAR3, and TAAR5 (Nelson et al., 2007). Our analysis reveals that 62% (n = 29) of the available RNA expression array datasets obtained from GEO profiles containing a TAAR1 probe were positive for measurable gene expression in T-cells in all samples, and 86% of datasets included one or more samples with measurable TAAR1 expression (Table 4). Accordingly, there is sufficient data pointing toward the presence of TAAR1 to warrant further research of its expression, signaling, and function in T-cells.
B-cells are the antibody secreting lymphocytes and have been shown to express TAAR1 and other TAAR family members. B-cells isolated from human blood express TAAR1 and TAAR2 as well as TAAR5,−6, and −9 at lower levels (Babusyte et al., 2013). Panas et al. (2012) observed expression of functional TAAR1 that exhibits METH-induced TAAR1-dependent PKA and PKC phosphorylation in immortalized rhesus monkey B-cells. As immortalized cells may exist in a state mimicking constant immune activation, these investigators sought to recapitulate the TAAR1-dependent PKA and PKC phosphorylation in primary rhesus PBMCs. Indeed, PHA-activated primary rhesus PBMCs have upregulated TAAR1 mRNA expression and display the same PKA and PKC phosphorylation when treated with METH; however TAAR2 expression was uninvestigated. Similar cellular signaling governs the TAAR1-dependent, METH-induced modulation of monoamine transporter kinetic and internalization functions, discussed elsewhere (Miller et al., 2005; Xie and Miller, 2007, 2009). Notably, mouse B-cells were shown to express mRNA for TAAR1-4 at a moderate level and low-level expression was apparent for TAAR5-9 (Nelson et al., 2007). B-cells play a critical role in allergic inflammation by synthesizing and secreting antibodies such as IgE. TAs were demonstrated to induce secretion of IgE in purified human B-cells in a TAAR1/TAAR2-dependent manner (Babusyte et al., 2013). The ability of TAAR1 and TAAR2 co-expression to trigger IgE secretion in B-cells in response to TAs represents a new mechanism by which TAs directly alter immune cell function, and also raise the possibility that selective TAAR1 compounds may act similarly.
Our analysis of complementary GEO datasets confirmed B-cell TAAR1 expression in all but one (n = 13) dataset obtained (Table 4), supporting the expression of TAAR1 in this cell type.
It is interesting to note that TAAR1 expression in B-cells may vary based on maturation stage, as we had previously speculated for PBMC-derived erythroid progenitor cells. Retrospective analysis carried out with the NCBI GEO “Analyze DataSet” tool of the 2006 study of Luckey et al., 2006, GDS1695) revealed that TAAR1 transcripts are significantly higher in plasma B-cells than in more mature memory B-cells (two-tailed t-test, p ≤ 0.05).
Neuronal expression of TAAR1 is documented in human dopaminergic brain regions including the ventral tegmental area, substantia nigra, hippocampus, amygdala, and other major regions (Borowsky et al., 2001; Espinoza et al., 2015). The interface between the immune system and the brain, commonly referred to as the neuroimmune system, is distinct in both its cellular population and physical permeability. The resident immune cells of the brain consist of astrocytes, microglia, and a unique type of macrophage that inhabits the perivascular and subarachnoid space (Engelhardt et al., 2017). The brain's lack of native T- and B-cells that make up the adaptive immune response in the periphery is due to a largely impenetrable physical barrier between the tissue of the brain and the blood circulation, known as the blood brain barrier (BBB).
Astrocytes make up the largest population of brain cells and represent a dynamic component of neurological homeostasis, cell signaling, and immunological responses (Schubert et al., 1997; Ransohoff and Brown, 2012; Bazargani and Attwell, 2016). Some of the most critical astrocytic functions are perturbed by METH, namely disruption of the blood-brain barrier (Kousik et al., 2012; Ramirez et al., 2012; Northrop and Yamamoto, 2015; Turowski and Kenny, 2015) and glutamate clearance functionality (Cisneros and Ghorpade, 2014a). The role of TAAR1 in astrocytes was addressed by Cisneros and Ghorpade (2014a,b), who showed that TAAR1 was both present and functional in primary human astrocytes and signaled through cAMP. Importantly, TAAR1 mRNA and protein was upregulated by both METH and HIV-1 exposure. METH and/or HIV-1 treatment also increased the nuclear localization of TAAR1. Even more interesting was the apparent synergistic upregulation with combinatorial METH/HIV-1 treatment, observed in both immunological staining of protein as well as by assessment of mRNA expression. TAAR1 activation functionally altered the activity of the glutamate transporter EAAT-2, signifying the ability of TAAR1 agonists to modulate extracellular glutamate and therefore potentially mediate excitatory neurotoxicity.
Our analysis (Table 5) revealed that TAAR1 RNA is present in mouse forebrain astrocytes (Lau et al., 2012, GDS3944), normal human astrocytes (Grzmil et al., 2011, GDS4467), and epidermal growth factor-treated rat astrocytes (Liu et al., 2006, GDS2146). TAAR1 is also expressed in the human astrocyte cell line U251 (Lin et al., 2015, GDS6010) and embryonic stem cell (ESC) line H9-derived astrocytes (Lafaille et al., 2012, GDS4538). Another microarray experiment by the same group assessed expression profiles in induced astrocytes deficient in the UNC-93B protein (GDS4669), which produces a phenotype unable to signal through the viral antigen recognition receptors TLR3, TLR7, and TLR9 (Casrouge et al., 2006; Tabeta et al., 2006). Interestingly, the UNC-93B deficient astrocytes expressed higher TAAR1 RNA levels than cells normally expressing the protein. Therefore, TAAR1 upregulation may occur as a compensatory mechanism when anti-viral immune processes are disturbed.
Our analysis of the dataset by (Simpson et al., 2011) (GDS4135) revealed that TAAR1 RNA expression is positively correlated with increasing Braak staging, a clinical measurement of the progression of Alzheimer's and Parkinson's disease. Conversely, our analysis of a dataset obtained from the BioGPS database in which normal mouse astrocytes were incubated with brain slices from a beta-amyloid overexpressing mouse model of Alzheimer's disease (Kurronen et al., unpublished, dataset available from http://ds.biogps.org/?dataset=E-GEOD-29317&gene=111174) revealed that TAAR1 RNA expression was reduced relative to cells incubated with normal brain slices, and this difference could not be accounted for by age or developmental stage. Affymetrix microarray technology utilizes “detection calls” as a method of determining whether a gene transcript is present and allows for correction of noisy probe sets or error (Archer and Reese, 2010). While datasets with “absent” detection calls cannot be considered positive, it is still interesting to note that TAAR1 RNA was still measurable in other astrocytic samples from six other datasets (Table 5). Complementary searches of similar Affymetrix microarray experiments in BioGPS (Wu et al., 2013) further confirmed GEO results for expression in mouse (Beckervordersandforth et al., 2010; Zamanian et al., 2012), and suggests a bias in astrocytic TAAR1 expression toward the subventricular zone and hippocampal region relative to the ventral encephalon and the olfactory bulb (Mireia and Helena, 2012, unpublished; dataset available from http://ds.biogps.org/?dataset=E-GEOD-36456&gene=111174). These data and numerous other datasets for human TAAR1 expression can be freely browsed at http://biogps.org/gene/134864.
Microglial cells are a distinct lineage of macrophage derived from the yolk sac that exists exclusively in the brain. These innate immune cells act analogously to the bone-marrow derived macrophage in the tissues and periphery to survey the microenvironment and respond to pathogens via pattern recognition receptors and toll-like receptor binding activity (Ginhoux et al., 2010; Saijo and Glass, 2011). Moreover, microglial expression of major histocompatibility complex-I (MHC-I) and MHC-II make them capable of antigen presentation to T-cells, required for adaptive immune responses (Hauser and Knapp, 2014). Our analysis of datasets accessed from NCBI GEO Profiles revealed that TAAR1 expression is detectable in the human microglial cell line HMO6 (Yoshino et al., 2011, GDS4151) and in mouse BV-2 microglia (Dirscherl et al., 2010, GDS3613). Further analysis of the array-based study by Yoshino et al. (2011, GDS4151) revealed that ethanol treatment induced a 1.4-fold increase in TAAR1 expression in HMO6 microglia relative to DMSO control, suggesting a potential interaction between the ubiquitous drug of abuse and TAAR1 modulation of functioning in the brain's major first line of immunological defense. Microglial TAAR1 signaling has yet to be directly studied, but considerable overlap exists between the immune activated state and TAAR1 expression. The potent TAAR1 agonist METH enhances HIV-1 replication in microglia and in vivo exposure to MDMA causes rat microglia to become activated (Pubill et al., 2003; Liang et al., 2008). Thomas et al. (2004) noted that the transcription factors (TF) NFκβ, cFos, and AP-1 needed for microglial activation are also upregulated by METH. Intriguingly, our analysis of the TAAR1 promoter carried out with the TF binding site predictor program LASAGNA-Search 2.0 (Lee and Huang, 2013) revealed that these same TFs are predicted to bind in the upstream untranslated region of the TAAR1 promotor, and these data are in agreement with an analogous search we conducted utilizing the similar program MatInspector (Quandt et al., 1995; Cartharius et al., 2005, Bioinformatics). In regard to astrocytes and microglia, in addition to the TAAR1 expression described in the literature, our analyses indicate that TAAR1 RNA is present in these neuroimmune cell types. Aberrant microglial activation, signaling, and function may be due to monoamine excess. Increased levels of the neurotransmitters dopamine and norepinephrine paramount to sympathetic nervous system activation may act in a positive feedback loop to mimic and perpetuate the immunologically activated state of microglia and this prolonged cellular activation could lead to cellular damage through production of reactive oxygen species and neuroinflammation. Alternatively, pathological changes in endogenous agonists for TAAR1, as seen in psychiatric disorders, could underlie alterations in microglial functions. While these proposed mechanisms are in contrast they are not mutually exclusive; it is likely that these pathways are intimately connected in a sensitive network of microenvironmental surveillance and homeostasis in response to trace and monoamine levels.
Extensive antibody-based protein expression data is freely available online from the Human Protein Atlas (HPA) at www.proteinatlas.org (Uhlén et al., 2015; Thul and Lindskog, 2017). This database was generated by probing various human tissues for all protein-coding genes and is cataloged in a searchable module, allowing for tissue-specific exploration of expression patterns. Results of a June 2017 query of the HPA for TAAR1 protein expression yielded detection of TAAR1 in bone marrow and immune organs that included the appendix, spleen, bone marrow, tonsils, and lymph nodes (Table 6). Importantly, TAAR1 protein has been detected at appreciable levels in sites of immune cell maturation and activation, namely germinal centers in both lymph nodes and tonsils. As germinal centers are the location of B-cell maturation and fine-tuning of the adaptive immune response these data suggest that TAAR1 may be important in the B-cell mediated response. Similarly, TAAR1 protein is highly expressed in both the white and red pulp of the spleen and tonsillar germinal centers. TAAR1 protein is also highly expressed in lymphoid tissue and glandular cells of the appendix as well as glandular adrenal cells. Protein expression is summarized in Table 6, and histological images and details for these data are available online at http://www.proteinatlas.org/ENSG00000146399-TAAR1/tissue/primary$+$data.
Our retrospective analysis of a microarray dataset from HIV-1 infected lymphatic tissues (Li et al., 2009, GSE16363) containing gene probes for TAAR1,−2,−3,−5,−8, and −9 obtained from NCBI GEO revealed expression of all TAARs in normal tissue and tissue in acute and asymptomatic stages as well as fully progressed AIDS. TAAR1 was expressed at the lowest level of all TAARs and TAARs−2,−3, and −5 were most abundant. To explore changes in transcript levels of TAAR1 and related TAARs over the course of infection we obtained the raw count data for all samples and calculated log2 fold-change values for each infection stage relative to control tissues. All TAARs were downregulated in the asymptomatic stage of HIV-1 infection and all TAARs except TAAR3 were upregulated in acute HIV-1 infection. It is interesting to note that for TAAR1, expression appears to increase upon initial infection, decrease once the asymptomatic phase is reached, and return to control-baseline levels with progression to AIDS. TAARs are not significantly altered in HIV-1 infection, but trend toward upregulation with acute infection and downregulation with asymptomatic infection, corresponding to active viral replication and latent infection progressions. This finding agreed with our analysis of another GEO dataset of Dengue virus-infected blood in which TAAR1 RNA expression levels were significantly lower in convalescent infection vs. active viremia (Kwissa et al., 2014, GDS5093). The apparent increase in TAAR1 RNA in periods of active viral replication suggest that its expression could be modulated by the presence of viral antigen. Similarly, the downregulation of TAAR1 seen in asymptomatic infection, which corresponds to a period of viral latency, suggests that TAAR1 could play a role in the viral life cycle.
Recent online compilation of open-source transcriptomic profiling of immunologically relevant cell types is available through the Reference Database of Immune Cells (RefDIC), http://refdic.rcai.riken.jp (Hijikata et al., 2007). Our query for the TAAR1 gene revealed detection in numerous immunologically relevant cell types in two microarray datasets for human and one for mouse immune cells, representing three different Affymetrix GeneChip platforms. The acquired dataset of the Affymetrix GeneChip Mouse Array included the raw log2 expression values for a single probe representing TAAR1 RNA levels in 19 different cell types. TAAR1 was found to be expressed at low levels in various mouse immune cells and expression varied within cell types. Highest expression levels were observed in B-cells, T-cells, dendritic cells, macrophages, and mast cells. While the majority of TAAR1 expression-level counts for all immune cell types tend to be low, the distribution of the outlier cell types with higher expression counts, namely macrophage, appear to be skewed toward the innate branch of the immune system. This is likely because of a disproportionate amount of macrophage samples, so although observationally interesting this finding is not definitive.
The online repository BioGPS (http://www.biogps.org) is another abundant source of processed and searchable microarray expression datasets (Wu et al., 2013). A retrospective analysis of three microarray datasets retrieved from BioGPS (Mabbott et al., 2013, available from http://ds.biogps.org/?dataset=BDS _00013&gene = 134864) representing 1,049 human cell samples revealed TAAR1 gene expression in various immune cells including astrocytes, peripheral blood cells, leukocytes, monocytes, macrophage, and neutrophils. TAAR1 was also detected in mixed lymphocytes and in the lymphocytic subsets T-cells, Pre-, Pro-, and normal B-cells, and NK cells. TAAR1 was also present in low-medium levels in pancreatic islets. Similarly, analysis of two microarray datasets retrieved from BioGPS representing 232 mouse cell samples revealed detectable TAAR1 RNA in myeloid progenitor cells, thymocytes, CD4+ and CD8+ T-cells, NK cells, B-cells, mast and dendritic cells, macrophage, granulocytes, microglia, lymph node, and bone marrow (Lattin et al., 2008; Wu et al., 2013, dataset available from http://ds.biogps.org/?dataset=GSE10246&gene=111174).
The potent downregulation of the tumor-promoting gene SPP-1 as a result of TAAR1 activation has been previously described in T-cells (Babusyte et al., 2013). Notably, SPP-1 upregulation is inducible by PKC activation, a signaling pathway triggered by TAAR1 agonists, and its expression is inversely related to cancer prognosis as tumors rich in the SPP-1 gene have an enhanced ability to grow and invade other tissues to ultimately metastasize (Wai and Kuo, 2007). While no data currently exists for TAAR1-specific effects on cancer progression, a literature search for TAAR1 agonists in cancer revealed a small subset of manuscripts describing amphetamine as a tumor-promoter. The potent TAAR1 agonist amphetamine has been linked to cancer pathology since at least the 1990s, when daily amphetamine injections were found to increase tumor incidence, growth and metastases in virally-induced cancers on rats (Freire-Garabal et al., 1992, 1998). Similarly, a more recent human study (Chao et al., 2008) found that recreational amphetamine use correlates with increased risk of Non-Hodgkin's lymphoma (NHL). Specifically, hazard ratios determined from the study predicted that patients with weekly or more frequent use of amphetamines were 1.75 times as likely to develop NHL vs. their control counterparts and that recent use increased that risk to 4.3 times. Patients with 3 years prior use were three times as likely to develop NHL as drug-free patients. In contrast to the cancer-promoting effects of amphetamine are those of the potent endogenous TAAR1 ligand, 3-Iodothyronamine (T1AM). T1AM is a derivative of thyroid hormone and has been shown to inhibit growth of cancerous cells in-vitro. Specifically, in-vitro incubation of MCF7 human breast adenocarcinoma cells or HepG2 heptocellular carcinoma cells with T1AM resulted in reduced proliferation in an MTT assay. Further, IC50 values of T1AM were twice as high for control human foreskin fibroblast cells (Rogowski et al., 2017).
PKA, triggered by TAAR1 activation, is thought to act upstream in activation of the transcription factor NFKB (Bhat-Nakshatri et al., 2002). Chronic inflammation in the tumor microenvironment feeds forward to activate NFKB, which in turn perpetuates the inflammatory state that allows tumors to thrive (Karin, 2009). Constitutive activation of NFKB is associated with increased cancer risk and enhanced malignancy (Hoesel and Schmid, 2013). RNAseq data from 12 published cancer studies obtained from cBioPortal (Cerami et al., 2012; Gao et al., 2013) detected RNA for TAAR1, TAAR2, TAAR5, TAAR6, TAAR8, and TAAR9 in various cancers. A TAAR1 mRNA transcript query in BioXpress (Wan et al., 2015) revealed that TAAR1 is upregulated in esophageal, lung, and stomach cancers, and downregulated in sarcoma, cervical, renal, kidney, liver, pancreas, pituitary, prostate, urinary, and uterine cancers (Figure 1). This differential expression is statistically significant in esophageal (p = 0.023) and prostate (p = 0.000043) cancers. A gene query for TAAR1 in the online Catalog of Somatic Mutations in Cancer (COSMIC, Forbes et al., 2017) using the Genome Browser tool (https://cancer.sanger.ac.uk/cosmic/browse/genome) revealed that TAAR1 is overexpressed in at least 19 cancer types and in 16% of esophageal cancers. A similar gene-specific query in the Cancer RNA-Seq Nexus (Li et al., 2016) revealed that human TAAR1 is statistically differentially expressed in breast, bladder, cervical, lung, pancreatic, stomach, renal, and thyroid cancer (Table 7). Reexamining TCGA RNAseq data with respect to the TAAR1 gene (Figure 3) revealed varying RNA expression levels across cancer types. Overall, most cancer types displayed median TAAR1 RNA expression levels of zero, which is in agreement with the well-known phenomenon of TAAR1 expression detection being challenging (Liberles and Buck, 2006). Interestingly, cancer types containing a majority of samples without detectable TAAR1 expression also contained a number of samples with vastly varied expression levels, suggesting that TAAR1 expression varies in a patient-dependent manner even within a given cancer type. Lowest non-zero levels of TAAR1 RNA expression (maximum value for log2 TAAR1 expression < 1.5, which roughly represents values three times that of non-expression) were observed in Adrenocortical carcinoma [ACC, n = 80], Lymphoid Neoplasm Diffuse Large B-cell Lymphoma [DLBC, n = 48], Glioblastoma multiforme [GBM, n = 528], Glioma [GBMLGG, n = 696], Brain Lower Grade Glioma [LGG, n = 515], Pancreatic adenocarcinoma [PAAD, n = 185], Prostate adenocarcinoma [PRAD, n = 498], Stomach and Esophageal carcinoma [STES, n = 237], Testicular Germ Cell Tumors [TGCT, n = 150], Uterine Carcinosarcoma [UCS] n = 57. In summary, TAAR1 RNA was present at the lowest levels in cancers of the brain, pancreas, prostate, adrenal gland, stomach and esophagus, sex organs, and B-cells of the blood. As previously stated a large majority of samples in each cohort datasets lacked detectable RNA as represented by a median value of zero; however, median TAAR1 RNA levels were noticeably higher in the pan-kidney cohort representing KICH + KIRC + KIRP [KIPAN, n = 1020], Kidney renal clear cell carcinoma [KIRC, n = 607], Pheochromocytoma and Paraganglioma [PCPG, n = 179], and Skin Cutaneous Melanoma [SKCM, n = 470] cohorts. Therefore, TAAR1 RNA is most highly expressed in cancers of the kidney, skin, and neuro-endocrine cancers. It is interesting to note the substantial difference in TAAR1 RNA expression levels between the anatomically closely related adrenocortical cancers and the neuroendocrine cancers pheochromocytoma/paraglioma. An RNA-seq dataset containing 675 commonly used human cancer cell lines obtained from Array Express (Klijn et al., 2015, ArrayExpress experiment E-MATB-2706), 622 of which contained data for TAAR1 RNA expression, revealed that TAAR1 RNA is most highly expressed in the pancreatic somatostatinoma cell line QGP-1, lung carcinoid tumor cell line UMC-11, and the lung adenocarcinoma cell line VMRC-LCD. This dataset is summarized in Table 8 and directly available at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2706/. Another Array Express RNA-seq dataset of long poly adenylated RNA and long non-poly adenylated RNA from ENCODE cell lines (Djebali et al., 2012, ArrayExpress experiment E-GEOD-26284) revealed that TAAR1 RNA is most highly expressed in the bone marrow neuroblastoma cell line SK-N-SH, normal human lung fibroblast cell line NHLF, human skeletal muscle cells, and the myoblast cell line HSMM. Further exploration of TAAR1 expression in cancers utilizing the COSMIC database revealed TAAR1 deletions in two cancer cell lines. a B-cell NHL subtype, mantle cell lymphoma, JEKO-1 cell line and malignant melanoma cell line Hs940.T. These data support the hypothesis that TAAR1 is modulated in cancers and therefore may serve a functional role in cancer physiology.
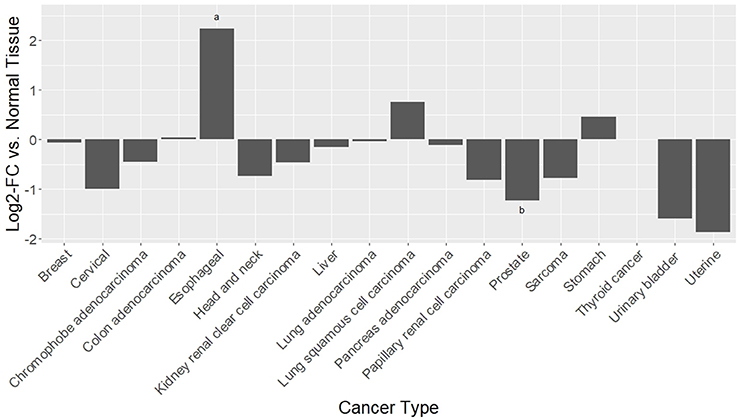
Figure 1. Differential RNAseq expression data for TAAR1 in human cancers. A TAAR1 mRNA transcript query in BioXpress (Wan et al., 2015) revealed that TAAR1 is upregulated in esophageal, lung, and stomach cancers, and downregulated in sarcoma, cervical, renal, kidney, liver, pancreas, pituitary, prostate, urinary, and uterine cancers. This differential expression is statistically significant in esophageal (*p = 0.023) and prostate (**p = 0.000043) cancers.
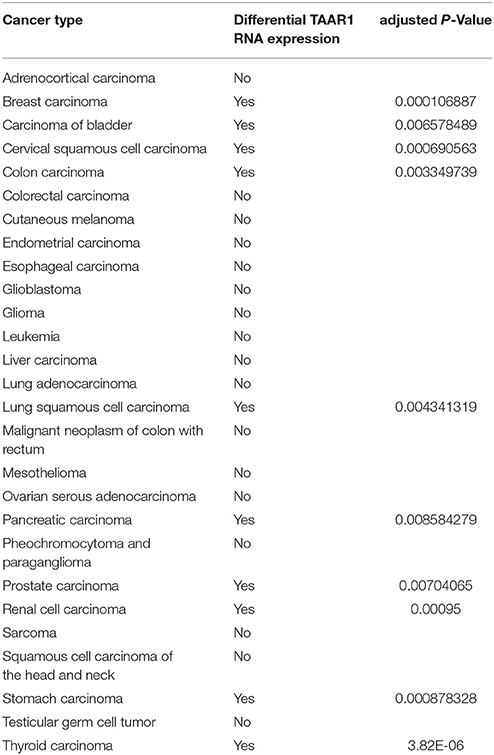
Table 7. Cancer RNA-seq Nexus datasets with differential TAAR1 RNA expression (adjusted P-value < 0.01) in differential expression analysis.
TAAR1 signaling may modulate tumor cell function to alter malignancy and tumor progression and therefore TAAR1-specific compounds could potentially have oncological therapeutic potential and represent a novel approach to modulating cancer physiology. Accordingly, our analyses of TAAR1 expression trends in overall survival cancer studies raises speculation of TAAR1 as a possible prognostic marker. Serendipitously during the writing of this review, the first manuscript to posit TAAR1 as a predictor of overall survival in cancer was published. In histologically-based experiments, Vattai et al. (2017) found TAAR1 expression to correlate with longer overall survival in early breast cancer.
Overall survival trends expressed as hazard ratios (HR) for 80 unique human cancer studies representing 15 cancer types were obtained from the online databases Prognoscan (Mizuno et al., 2009) and PROGgene (Goswami and Nakshatri, 2013). For each PROGgene study (n = 68) the sample population was bifurcated at the median into high- and low-TAAR1 expression groups. Study data obtained from Prognoscan (n = 12) utilized a minimum p-value approach to determine the point of bifurcation into high and low expression groups. Briefly, the HR obtained for each study can be explained as the ratio of events (deaths) in the high TAAR1 expression group to events in the low TAAR1 expression group (Abel et al., 1984; Mizuno et al., 2009). To perform the meta-analysis after data collection all HR values were log-transformed to normalize values around zero to enable the calculation of subgroup averages and then back transformed to produce the average HR value denoted in Figure 2. An HR value of 1 denotes that survival was not different between the high and low expression groups, whereas an HR > 1 denotes that survival was poorer in the high expression group and conversely an HR < 1 means that survival was greater in the low expression group.
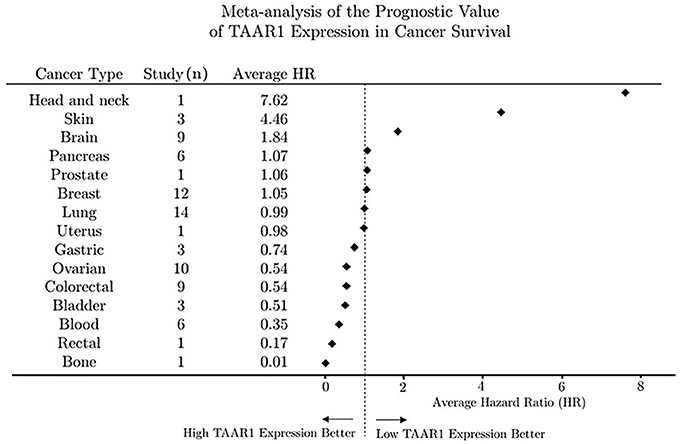
Figure 2. Meta-analysis of the prognostic value of TAAR1 expression in overall cancer survival. Forest plot of hazard ratios for survival in 80 human cancer studies representing 15 cancer types were obtained from open-source repositories (PROGgene n = 68, Prognoscan n = 12). Hazard ratios were determined for TAAR1 expression bifurcated into high and low expression and all HR values were log-transformed to normalize values around zero to enable the calculation of subgroup averages and then back transformed to produce the average HR value for each cancer type.
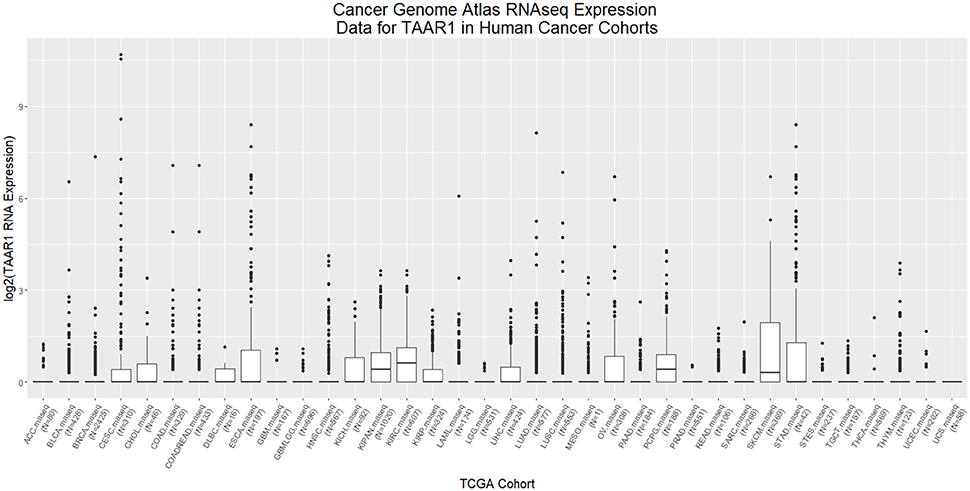
Figure 3. RNAseq analysis of TAAR1 expression in human TCGA cancer cohorts. RNAseq expression datasets representing cohorts from 36 cancer types were downloaded from The Cancer Genome Atlas (TCGA) using the R package RTCGA.rnaseq and values for the TAAR1 gene transcript were extracted, log2 transformed, and plotted with ggplot2. The minimal non-zero value for RNA expression levels was set to 1 to facilitate log2 transformation.
Our meta-analysis revealed that higher expression of TAAR1 correlates to longer median survival time in gastric, ovarian, colorectal, bladder, blood, rectal, and bone cancers. Conversely, lower TAAR1 expression correlated to longer median survival in head and neck, skin, and brain cancers (Figure 2). Breast cancer studies (n = 12) yielded an average HR of 1.05 which would suggest no differential effect of TAAR1 on cancer survival. Although these findings contradict the recent finding of Vattai et al. (2017), it is important to note that our meta-analysis did not account for progression stage, and as such a further analysis would be needed to directly compare our data with that of the early-stage breast cancer described in the 2017 manuscript. Overall survival analysis of a pancreatic cancer dataset (Grimont et al., 2015, GSE50827) stratified by cancer stage using PROGene V2 revealed that survival of stage IIB cancer that had spread from the pancreas to the lymph nodes is significantly higher (HR = 4.79, p = 0.025) vs. stage IIA cancer that lacks lymph node involvement, suggesting the possibility that TAAR1 may play a role in lymphoid cancer progression or pathways.
This review of current literature and meta-analyses of array-based evidence confirms that TAAR1 is expressed throughout various immune cell types and cancers. The ability of TAAR1 to signal in response to endogenous common biogenic amines and trace amines implicates the receptor as a mediator of aminergic regulation of immune function. Its ability to respond to exogenous amphetamine-like drugs implicates it as a modulator of at least some of the immunological actions of these drugs. Importantly, its ability to respond to dopamine implicates the receptor in the immunological action of drugs of abuse more generally, including drugs that do not directly bind to and/or activate the receptor directly, such as cocaine, but act through elevated dopamine levels. Additionally, its ability to be agonized or antagonized by newly developed synthetic drugs that are currently under development and investigation as psychiatric and addition therapeutics, respectively, mandate a greater definition of the immunological action of TAAR1-targeted drugs.
Our analyses further implicate TAAR1 as a logical target for studying the interplay between stimulant use and immune function. Modulation of neurotransmitter concentrations by METH and cocaine, for example, may act on TAAR1 either directly or indirectly to alter homeostatic immune cell signaling, in effect mimicking a prolonged state of immune activation, or by downstream changes in cellular function such as cytokine secretion, phagocytosis, and chemotaxis, for which there is emerging evidence.TAAR1-specific compounds could be efficacious therapeutics for infections and cancers, as well as valuable pharmacological tools for dissecting stimulant-induced changes and damage to immune function.
The expression of TAAR1 in various cancers and its functional significance is an intriguing yet largely unexplored area. Our analyses show that expression of TAAR1 is modulated in cancers, suggesting that TAAR1 serves a functional role in cancer physiology. We provide evidence for a differential pattern of cancer survival based on TAAR1 expression in multiple cancer types. The analyses indicate a potential prognostic value of TAAR1 detection in cancer survival that depends on the cancer type. Cancer pathologies differ widely and as such the observation that potentially protective effects of TAAR1 are type-specific supports the hypothesis that TAAR1 is utilizing cell type-specific signaling or collaborating with host cell receptor signaling in a cell type-specific manner to alter cellular function and cancer physiology.
The observational data gathered in previous decades linking amphetamine use to cancer progression needs to be revisited in light of the de-orphanization of TAAR1. As both illicit drug use and cancer incidence continue to increase globally it will be imperative to investigate TAAR1 signaling in cancer. TAAR1 signaling may play a role in the progression of many cancers, and accordingly TAAR1-specific compounds may serve as potential therapeutic additions to current clinical practices to improve survival. Pharmacologically elucidating TAAR1 signaling in cancer can lead to a better understanding of the mechanisms by which cancer eludes the immune system, and moreover, how drugs of abuse can contribute to cancer development and prognosis. Pharmacologically elucidating TAAR1 signaling, both generally with regard to TAAR1 as a protomer in receptor-receptor and/or receptor-protein complexes, or more specifically in cancer cellular phenotypes, will predictably lead to a better understanding of the mechanisms by which cancer alludes the immune system, and moreover, how drugs of abuse and other ligands for TAAR1 can contribute to cancer development, prognosis, and treatment. The current identification of TAAR1 as a marker in numerous cancers, potentially functional and therapeutically targetable, provides a logical direction for future studies on the effects of drugs targeting TAAR1. Our present analyses expand the spectrum of immune cells known to express TAAR1 and also provide a queryable roadmap for further investigation. The identification of TAAR1 across immune cell types presents an avenue for exploring both the role of TAAR1 in normal immune function as well as its potential role as a mediator or modulator of immune dysregulation. Immune susceptibilities in disease states and in particular those seen in drug users may involve aberrant TAAR1 regulation and function, and may be therapeutically targetable with drugs that interact with TAAR1.
All authors contributed to the writing of the manuscript. LF performed all meta-analyses and compiled all data presentation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by laboratory start-up funds from Northeastern University (GM).
Abel, U., Berger, J., and Wiebelt, H. (1984). CRITLEVEL: an exploratory procedure for the evaluation of quantitative prognostic factors. Methods Inform. Med. 23, 154–156. doi: 10.1055/s-0038-1635335
Amulic, B., Cazalet, C., Hayes, G. L., Metzler, K. D., and Zychlinsky, A. (2012). Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 30, 459–489. doi: 10.1146/annurev-immunol-020711-074942
An, D., Kim, K., and Lu, W. (2014). Defective Entry into Mitosis 1 (Dim1) negatively regulates osteoclastogenesis by inhibiting the expression of Nuclear Factor of Activated T-cells, Cytoplasmic, Calcineurin-dependent 1 (NFATc1). J. Biol. Chem. 289, 24366–24373. doi: 10.1074/jbc.m114.563817
Ancuta, P., Liu, K. Y., Misra, V., Wacleche, V. S., Gosselin, A., Zhou, X., et al. (2009). Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16- monocyte subsets. BMC Genomics 10:403. doi: 10.1186/1471-2164-10-403
Archer, K. J., and Reese, S. E. (2010). Detection call algorithms for high-throughput gene expression microarray data. Brief Bioinform. 2, 244–252. doi: 10.1093/bib/bbp055
Arita, A., Muñoz, A., Chervona, Y., Niu, J., Qu, Q., Zhao, N., et al. (2013). Gene expression profiles in peripheral blood mononuclear cells of Chinese nickel refinery workers with high exposures to nickel and control subjects. Cancer Epidemiol. Biomarkers Prev. 22, 261–269. doi: 10.1158/1055-9965.EPI-12-1011
Babusyte, A., Kotthoff, M., Fiedler, J., and Krautwurst, D. (2013). Biogenic amines activate blood leukocytes via trace amine-associated receptors TAAR1 and TAAR2. J. Leukoc. Biol. 93, 387–394. doi: 10.1189/jlb.0912433
Bajwa, G., DeBerardinis, R. J., Shao, B., Hall, B., Farrar, J. D., and Gill, M. A. (2016). Cutting Edge: critical role of glycolysis in human plasmacytoid dendritic cell antiviral responses. J. Immunol. 196, 2004–2009. doi: 10.4049/jimmunol.1501557
Balachander, A., Nabti, S., Sobota, R. M., Foo, S., Zolezzi, F., Lee, B. T., et al. (2015). Dendritic cell derived IL-2 inhibits survival of terminally mature cells via an autocrine signaling pathway. Eur. J. Immunol. 45, 1494–1499. doi: 10.1002/eji.201445050
Barnes, P. J. (2011). Pathophysiology of allergic inflammation. Immunol. Rev. 242, 31–50. doi: 10.1111/j.1600-065X.2011.01020.x
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, 991–995. doi: 10.1093/nar/gks1193
Bazargani, N., and Attwell, D. (2016). Astrocyte calcium signaling: the third wave. Nat. Neurosci. 19, 182–189. doi: 10.1038/nn.4201
Beckervordersandforth, R., Tripathi, P., Ninkovic, J., Bayam, E., Lepier, A., Stempfhuber, B., et al. (2010). In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7, 744–758. doi: 10.1016/j.stem.2010.11.017
Bergenfelz, C., Larsson, A. M., von Stedingk, K., Gruvberger-Saal, S., Aaltonen, K., Jansson, S., et al. (2015). Systemic monocytic-MDSCs are generated from monocytes and correlate with disease progression in breast cancer patients. PLoS ONE 10:e0127028. doi: 10.1371/journal.pone.0127028
Berglund, L. J., Avery, D. T., Ma, C. S., Moens, L., Deenick, E. K., Bustamante, J., et al. (2013). IL-21 signalling via STAT3 primes human naive B cells to respond to IL-2 to enhance their differentiation into plasmablasts. Blood 122, 3940–3950. doi: 10.1182/blood-2013-06-506865
Berisha, S. Z., Serre, D., Schauer, P., Kashyap, S. R., and Smith, J. D. (2011). Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PLoS ONE 6:e16729. doi: 10.1371/journal.pone.0016729
Bhat-Nakshatri, P., Sweeney, C. J., and Nakshatri, H. (2002). Identification of signal transduction pathways involved in constitutive NF-κB activation in breast cancer cells. Oncogene 21, 2066–2078. doi: 10.1038/sj.onc.1205243
Bielinska, A. U., Makidon, P. E., Janczak, K. W., Blanco, L. P., Swanson, B., Smith, D. M., et al. (2014). Distinct pathways of humoral and cellular immunity induced with the mucosal administration of a nanoemulsion adjuvant. J. Immun. 192, 2722–2733. doi: 10.4049/jimmunol.1301424
Bienkowska, J. R., Dalgin, G. S., Batliwalla, F., Allaire, N., Roubenoff, R., Gregersen, P. K., et al. (2009). Convergent Random Forest predictor: methodology for predicting drug response from genome-scale data applied to anti-TNF response. Genomics 94, 423–432. doi: 10.1016/j.ygeno.2009.08.008
Boddiger, D. (2005). Metamphetamine use linked to rising HIV transmission. Lancet 365, 1217–1218. doi: 10.1016/s0140-6736(05)74794-2
Bok, K., Prikhodko, V., Green, K., and Sosnovtsev, S. (2009). Apoptosis in murine norovirus-infected RAW264.7 cells is associated with downregulation of survivin. J. Virol. 83, 3647–3656. doi: 10.1128/JVI.02028-08
Boomgaarden, I., Egert, S., Rimbach, G., Wolffram, S., Müller, M. J., and Döring, F. (2010). Quercetin supplementation and its effect on human monocyte gene expression profiles in vivo. Br. J. Nutr. 104, 336–345. doi: 10.1017/S0007114510000711
Borowsky, B., Adham, N., Jones, K. A., Raddatz, R., Artymyshyn, R., Ogozalek, K. L., et al. (2001). Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 8966–8971. doi: 10.1073/pnas.151105198
Bosco, M. C., Pierobon, D., Blengio, F., Raggi, F., Vanni, C., Gattorno, M., et al. (2011). Hypoxia modulates the gene expression profile of immunoregulatory receptors in human mature dendritic cells: identification of TREM-1 as a novel hypoxic marker in vitro and in vivo. Blood 117, 2625–2639. doi: 10.1182/blood-2010-06-292136
Boulton, A. A. (1976). Identification, distribution, metabolism, and function of meta and para tyramine, phenylethylamine and tryptamine in brain. Adv. Biochem. Psychopharmacol. 15, 57–67.
Bouwens, M., Grootte Bromhaar, M., Jansen, J., Müller, M., and Afman, L. A. (2010). Postprandial dietary lipid-specific effects on human peripheral blood mononuclear cell gene expression profiles. Am. J. Clin. Nutr. 91, 208–217. doi: 10.3945/ajcn.2009.28586
Boyle, D., Rosengren, S., Bugbee, W., Kavanaugh, A., and Firestein, G. (2003). Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis Res. Ther. 5, R352–R360. doi: 10.1186/ar1004
Bunzow, J. R., Sonders, M. S., Arttamangkul, S., Harrison, L. M., Zhang, G., Quigley, D. I., et al. (2001). Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol. Pharmacol. 60, 1181–1188. doi: 10.1124/mol.60.6.1181
Burchett, S. A., and Hicks, P. T. (2006). The mysterious trace amines: protean neuromodulators of synaptic transmission in mammalian brain. Prog. Neurobiol. 79, 223–246. doi: 10.1016/j.pneurobio.2006.07.003
Burczynski, M. E., and Dorner, A. J. (2006). Transcriptional profiling of peripheral blood cells in clinical pharmacogenomic studies. Pharmacogenomics 7, 187–202. doi: 10.2217/14622416.7.2.187
Burczynski, M. E., Twine, N. C., Dukart, G., Marshall, B., Hidalgo, M., Stadler, W. M., et al. (2005). Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin. Cancer Res. 11, 1181–1189. doi: 10.1016/S0022-5347(05)00564-1
Cai, Y., Yang, Q., Tang, Y., Zhang, M., Liu, H., Zhang, G., et al. (2014). Increased complement C1q level marks active disease in human tuberculosis. PLoS ONE 9:e92340. doi: 10.1371/journal.pone.0092340
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. doi: 10.1093/bioinformatics/bti473
Casrouge, A., Zhang, S. Y., Eidenschenk, C., Jouanguy, E., Puel, A., Yang, K., et al. (2006). Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312. doi: 10.1126/science.1128346
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095
Chang, W. L. W., Coro, E. S., Rau, F. C., Xiao, Y., Erle, D. J., and Baumgarth, N. (2007). Influenza virus infection causes global respiratory tract B cell response modulation via innate immune signals. J. Immunol. 178, 1457–1467. doi: 10.4049/jimmunol.178.3.1457
Chang, X., Liu, F., Wang, X., Lin, A., Zhao, H., and Su, B. (2011). The kinases MEKK2 and MEKK3 regulate transforming growth factor-β-mediated helper T cell differentiation. Immunity 34, 201–212. doi: 10.1016/j.immuni.2011.01.017
Chang, Y. C., Chen, T. C., Lee, C. T., Yang, C. Y., Wang, H. W., Wang, C. C., et al. (2008). Epigenetic control of MHC class II expression in tumor-associated macrophages by decoy receptor 3. Blood 111, 5054–5063. doi: 10.1182/blood-2007-12-130609
Chao, C., Jacobson, L. P., Tashkin, D., Martínez-Maza, O., Roth, M. D., Margolick, J. B., et al. (2008). Recreational amphetamine use and risk of HIV-related non-Hodgkin lymphoma. Cancer Cause Control 20, 509–516. doi: 10.1007/s10552-008-9258-y
Cheadle, C., Berger, A. E., Mathai, S. C., Grigoryev, D. N., Watkins, T. N., Sugawara, Y., et al. (2012). Erythroid-specific transcriptional changes in PBMCs from pulmonary hypertension patients (pulmonary hypertension signature in PBMC). PLoS ONE 7:e34951. doi: 10.1371/journal.pone.0034951
Chen, F., Ding, X., Ding, Y., Xiang, Z., Li, X., Ghosh, D., et al. (2011). Proinflammatory caspase-2-mediated macrophage cell death induced by a rough attenuated Brucella suis strain. Infect. Immun. 79, 2460–2469. doi: 10.1128/IAI.00050-11
Chen, S. S., Claus, R., Lucas, D. M., Yu, L., Qian, J., Ruppert, A. S., et al. (2011). Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood 117, 862–871. doi: 10.1182/blood-2010-05-284638
Ciancanelli, M. J., Huang, S. X., Luthra, P., Garner, H., Itan, Y., Volpi, S., et al. (2015). Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453. doi: 10.1126/science.aaa1578
Cisneros, I. E., and Ghorpade, A. (2014a). HIV-1, Methamphetamine and Astrocyte Glutamate Regulation: Combined Excitotoxic Implications for Neuro-AIDS. Curr. HIV Res. 10, 1–15. doi: 10.2174/157016212802138832
Cisneros, I. E., and Ghorpade, A. (2014b). Methamphetamine and HIV-1-induced neurotoxicity: role of trace amine associated receptor 1 cAMP signaling in astrocytes. Neuropharmacology 85, 499–507. doi: 10.1016/j.neuropharm.2014.06.011
Cisse, B., Caton, M. L., Lehner, M., Maeda, T., Scheu, S., Locksley, R., et al. (2008). Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48. doi: 10.1016/j.cell.2008.09.016
Clemmensen, S. N., Bohr, C. T., Rørvig, S., Glenthøj, A., Mora-Jensen, H., Cramer, E. P., et al. (2012). Olfactomedin 4 defines a subset of human neutrophils. J. Leukoc. Biol. 91, 495–500. doi: 10.1189/jlb.0811417
Comer, J. E., Galindo, C. L., Zhang, F., Wenglikowski, A. M., Bush, K. L., Garner, H. R., et al. (2006). Murine macrophage transcriptional and functional responses to Bacillus anthracis edema toxin. Microb. Pathogenesis 41, 96–110. doi: 10.1016/j.micpath.2006.05.001
D'Andrea, G., Terrazzino, S., Fortin, D., Farruggio, A., Rinaldi, L., and Leon, A. (2003). HPLC electrochemical detection of trace amines in human plasma and platelets and expression of mRNA transcripts of trace amine receptors in circulating leukocytes. Neurosci. Lett. 346, 89–92. doi: 10.1016/S0304-3940(03)00573-1
Dawany, N., Showe, L. C., Kossenkov, A. V., Chang, C., Ive, P., Conradie, F., et al. (2014). Identification of a 251 gene expression signature that can accurately detect M. tuberculosis in patients with and without HIV Co-infection. PLoS ONE 9:e89925. doi: 10.1371/journal.pone.0089925
Dirscherl, K., Karlstetter, M., Ebert, S., Kraus, D., Hlawatsch, J., Walczak, Y., et al. (2010). Luteolin triggers global changes in the microglial transcriptome leading to a unique anti-inflammatory and neuroprotective phenotype. J. Neuroinflammation 7:3. doi: 10.1186/1742-2094-7-3
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Dower, K., Ellis, D. K., Saraf, K., Jelinsky, S. A., and Lin, L. L. (2008). Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J. Immunol. 180, 3520–3534. doi: 10.4049/jimmunol.180.5.3520
Dumeaux, V., Johansen, J., Børresen-Dale, A. L., and Lund, E. (2006). Gene expression profiling of whole-blood samples from women exposed to hormone replacement therapy. Mol. Cancer Ther. 5, 868–876. doi: 10.1158/1535-7163.MCT-05-0329
Dusek, J. A., Otu, H. H., Wohlhueter, A. L., Bhasin, M., Zerbini, L. F., Joseph, M. G., et al. (2008). Genomic counter-stress changes induced by the relaxation response. PLoS ONE 3:e2576. doi: 10.1371/journal.pone.0002576
Dybkaer, K., Iqbal, J., Zhou, G., Geng, H., Xiao, L., Schmitz, A., et al. (2007). Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics 8:230. doi: 10.1186/1471-2164-8-230
Edwards, J., Zhang, X., Frauwirth, K., and Mosser, D. (2006). Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 80, 1298–1307. doi: 10.1189/jlb.0406249
El Kasmi, K. C., Smith, A. M., Williams, L., Neale, G., Panopoulos, A. D., Watowich, S. S., et al. (2007). Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J. Immunol. 179, 7215–7219. doi: 10.4049/jimmunol.179.11.7215
Engelhardt, B., Vajkoczy, P., and Weller, R. O. (2017). The movers and shapers in immune privilege of the CNS. Nat. Immunol. 18, 123–131. doi: 10.1038/ni.3666
Eom, H. J., and Choi, J. (2010). p38 MAPK activation, DNA damage, cell cycle arrest and apoptosis as mechanisms of toxicity of silver nanoparticles in Jurkat T cells. Environ. Sci. Technol. 44, 8337–8342. doi: 10.1021/es1020668
Eom, H. J., Chatterjee, N., Lee, J., and Choi, J. (2014). Integrated mRNA and micro RNA profiling reveals epigenetic mechanism of differential sensitivity of Jurkat T cells to AgNPs and Ag ions. Toxicol. Lett. 229, 311–318. doi: 10.1016/j.toxlet.2014.05.019
Esmon, C. (2004). Interactions between the innate immune and blood coagulation systems. Trends Immunol. 25, 536–542. doi: 10.1016/j.it.2004.08.003
Espinoza, S., Lignani, G., Caffino, L., Maggi, S., Sukhanov, I., Leo, D., et al. (2015). TAAR1 Modulates Cortical Glutamate NMDA Receptor Function. Neuropsychopharmacology. 40, 2217–2227. doi: 10.1038/npp.2015.65
Fan, H. M., Sun, X. Y., Niu, W., Zhao, L., Zhang, Q. L., Li, W. S., et al. (2015). Altered microRNA expression in peripheral blood mononuclear cells from young patients with schizophrenia. J. Mol. Neurosci. 56, 562–571. doi: 10.1007/s12031-015-0503-z
Fang, K., Zhang, S., Glawe, J., Grisham, M. B., and Kevil, C. G. (2012). Temporal genome expression profile analysis during T-cell-mediated colitis: identification of novel targets and pathways. Inflamm. Bowel Dis. 18, 1411–1423. doi: 10.1002/ibd.22842
Favila, M. A., Geraci, N. S., Zeng, E., Harker, B., Condon, D., Cotton, R. N., et al. (2014). Human dendritic cells exhibit a pronounced type I IFN signature following Leishmania major infection that is required for IL-12 induction. J. Immunol. 192, 5863–5872. doi: 10.4049/jimmunol.1203230
Fehniger, T. A., Cai, S. F., Cao, X., Bredemeyer, A. J., Presti, R. M., French, A. R., et al. (2007). Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 26, 798–811. doi: 10.1016/j.immuni.2007.04.010
Fernandez, D. R., Telarico, T., Bonilla, E., Li, Q., Banerjee, S., Middleton, F. A., et al. (2009). Activation of mammalian target of rapamycin controls the loss of TCR in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J. Immunol. 182, 2063–2073. doi: 10.4049/jimmunol.0803600
Fleige, A., Alberti, S., Gröbe, L., Frischmann, U., Geffers, R., Müller, W., et al. (2007). Serum response factor contributes selectively to lymphocyte development. J. Biol. Chem. 282, 24320–24328. doi: 10.1074/jbc.M703119200
Fontenot, J. D., Rasmussen, J. P., Williams, L. M., Dooley, J. L., Farr, A. G., and Rudensky, A. Y. (2005). Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341. doi: 10.1016/j.immuni.2005.01.016
Forbes, S. A., Beare, D., Boutselakis, H., Bamford, S., Bindal, N., Tate, J., et al. (2017). COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783. doi: 10.1093/nar/gkw1121
Franco, M., Shastri, A. J., and Boothroyd, J. C. (2014). Infection by Toxoplasma gondii specifically induces host c-Myc and the genes this pivotal transcription factor regulates. Eukaryot. Cell 13, 483–493. doi: 10.1128/EC.00316-13
Freire-Garabal, M., Núñez, M., Balboa, J., Suárez, J., Gallego, A., and Belmonte, A. (1992). Effects of amphetamine on the development of MTV-induced mammary tumors in female mice. Life Sci. 51, PL37–40. doi: 10.1016/0024-3205(92)90416-m
Freire-Garabal, M., Nuñez-Iglesias, M. J., Rey-Méndez, M., Pereiro-Raposo, M. D., Riveiro, P., Fernández-Rial, J. C., et al. (1998). Effects of amphetamine on the development of Moloney sarcoma virus-induced tumors in mice. Oncol. Rep. 5, 381–384. doi: 10.3892/or.5.2.381
Frericks, M., Temchura, V. V., Majora, M., Stutte, S., and Esser, C. (2006). Transcriptional signatures of immune cells in aryl hydrocarbon receptor (AHR)-proficient and AHR-deficient mice. Biol. Chem. 387, 1219–1226. doi: 10.1515/BC.2006.151
Fulcher, J. A., Hashimi, S. T., Levroney, E. L., Pang, M., Gurney, K. B., Baum, L. G., et al. (2006). Galectin-1-matured human monocyte-derived dendritic cells have enhanced migration through extracellular matrix. J. Immunol. 177, 216–226. doi: 10.4049/jimmunol.177.1.216
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, PL1. doi: 10.1126/scisignal.2004088
Garaud, J. C., Schickel, J. N., Blaison, G., Knapp, A. M., Dembele, D., Ruer-Laventie, J., et al. (2011). B cell signature during inactive systemic lupus is heterogeneous: toward a biological dissection of lupus. PLoS ONE 6:e23900. doi: 10.1371/journal.pone.0023900
Geoffrey, R., Jia, S., Kwitek, A. E., Woodliff, J., Ghosh, S., Lernmark, A., et al. (2006). Evidence of a functional role for mast cells in the development of type 1 diabetes mellitus in the BioBreeding rat. J. Immunol. 177, 7275–7286. doi: 10.4049/jimmunol.177.10.7275
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Gleissner, C. A., Shaked, I., Little, K. M., and Ley, K. (2010). CXC chemokine ligand 4 induces a unique transcriptome in monocyte-derived macrophages. J. Immunol. 184, 4810–4818. doi: 10.4049/jimmunol.0901368
Goodridge, H. S., Simmons, R. M., and Underhill, D. M. (2007). Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J. Immunol. 178, 3107–3115. doi: 10.4049/jimmunol.178.5.3107
Goswami, C., and Nakshatri, H. (2013). PROGgene: gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinform. 3:22. doi: 10.1186/2043-9113-3-22
Grimont, A., Pinho, A. V., Cowley, M. J., Augereau, C., Mawson, A., Giry-Laterrière, M., et al. (2015). SOX9 regulates ERBB signalling in pancreatic cancer development. Gut 64, 1790–1799. doi: 10.1136/gutjnl-2014-307075
Grzmil, M., Morin, P. Jr, Lino, M. M., Merlo, A., Frank, S., Wang, Y., et al. (2011). MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human glioblastoma. Cancer Res. 71, 2392–2402. doi: 10.1158/0008-5472.CAN-10-3112
Hauser, K. F., and Knapp, P. E. (2014). Interactions of HIV and drugs of abuse: the importance of glia, neural progenitors, and host genetic factors. Int. Rev. Neurobiol. 118, 231–313. doi: 10.1016/B978-0-12-801284-0.00009-9
Heinemann, C., Heink, S., Petermann, F., Vasanthakumar, A., Rothhammer, V., Doorduijn, E., et al. (2014). IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4 T cells by inducing Blimp1. Nat. Commun. 5:3770. doi: 10.1038/ncomms4770
Herberth, M., Koethe, D., Levin, Y., Schwarz, E., Krzyszton, N. D., Schoeffmann, S., et al. (2011). Peripheral profiling analysis for bipolar disorder reveals markers associated with reduced cell survival. Proteomics 11, 94–105. doi: 10.1002/pmic.201000291
Hickey, M. J., and Kubes, P. (2009). Intravascular immunity: the host-pathogen encounter in blood vessels. Nat. Rev. Immunol. 9, 364–375. doi: 10.1038/nri2532
Hijikata, A., Kitamura, H., Kimura, Y., Yokoyama, R., Aiba, Y., Bao, Y., et al. (2007). Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics 23, 2934–2941. doi: 10.1093/bioinformatics/btm430
Hinze, C. H., Fall, N., Thornton, S., Mo, J. Q., Aronow, B. J., Layh-Schmitt, G., et al. (2010). Immature cell populations and an erythropoiesis gene-expression signature in systemic juvenile idiopathic arthritis: implications for pathogenesis. Arthritis Res. Ther. 12, R123. doi: 10.1186/ar3061
Hoesel, B., and Schmid, J. A. (2013). The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12:86. doi: 10.1186/1476-4598-12-86
Holmes, D., Yeh, J., Yan, D., Xu, M., and Chan, A. (2015). Dusp5 negatively regulates IL−33-mediated eosinophil survival and function. EMBO J. 34, 218–235. doi: 10.15252/embj.201489456
Hou, W., Gibbs, J. S., Lu, X., Brooke, C. B., Roy, D., Modlin, R. L., et al. (2012). Viral infection triggers rapid differentiation of human blood monocytes into dendritic cells. Blood 119, 3128–3131. doi: 10.1182/blood-2011-09-379479
Hsu, L. C., Enzler, T., Seita, J., Timmer, A. M., Lee, C. Y., Lai, T. Y., et al. (2011). IL-1β-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKβ. Nat. Immunol. 12, 144–150. doi: 10.1038/ni.1976
Ippagunta, S. K., Malireddi, R. K., Shaw, P. J., Neale, G. A., Vande Walle, L., Green, D. R., et al. (2011). The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nat. Immunol. 12, 1010–1016. doi: 10.1038/ni.2095
Irvine, K. M., Andrews, M. R., Fernandez-Rojo, M. A., Schroder, K., Burns, C. J., Su, S., et al. (2009). Colony-stimulating factor-1 (CSF-1) delivers a proatherogenic signal to human macrophages. J. Leukoc. Biol. 85, 278–288. doi: 10.1189/jlb.0808497
Ito, T., SmrŽ, D., Jung, M. Y., Bandara, G., Desai, A., SmrŽová, Š., et al. (2012). Stem cell factor programs the mast cell activation phenotype. J. Immunol. 188, 5428–5437. doi: 10.4049/jimmunol.1103366
Jabeen, R., Goswami, R., Awe, O., Kulkarni, A., Nguyen, E. T., Attenasio, A., et al. (2013). Th9 cell development requires a BATF-regulated transcriptional network. J. Clin. Invest. 123, 4641–4653. doi: 10.1172/jci69489
Janeway, C. (2001). How the immune system protects the host from infection. Microbes Infect. 3, 1167–1171. doi: 10.1016/S1286-4579(01)01477-0
Jelicic, K., Cimbro, R., Nawaz, F., Huang da, W., Zheng, X., Yang, J., et al. (2013). The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-β1 production and FcRL4 expression. Nat. Immunol. 14, 1256–1265. doi: 10.1038/ni.2746
Johnson, R. M., Kerr, M. S., and Slaven, J. E. (2012). Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J. Immunol. 188, 1896–1904. doi: 10.4049/jimmunol.1102764
Julià, A., Erra, A., Palacio, C., Tomas, C., Sans, X., Barceló, P., et al. (2009). An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS ONE 4:e7556. doi: 10.1371/journal.pone.0007556
Juorio, A. (1982). A possible role for tyramines in brain function and some mental disorders. Gen. Pharmacol. 13, 181–183. doi: 10.1016/0306-3623(82)90087-8
Kakoola, D. N., Curcio-Brint, A., Lenchik, N. I., and Gerling, I. C. (2014). Molecular pathway alterations in CD4 T-cells of nonobese diabetic (NOD) mice in the preinsulitis phase of autoimmune diabetes. Results Immunol. 4, 30–45. doi: 10.1016/j.rinim.2014.05.001
Kang, S. G., Park, J., Cho, J. Y., Ulrich, B., and Kim, C. H. (2011). Complementary roles of retinoic acid and TGF-β1 in coordinated expression of mucosal integrins by T cells. Mucosal Immunol. 4, 66–82. doi: 10.1038/mi.2010.42
Karin, M. (2009). NF-kappaB as a critical link between inflammation and cancer. CSH Perspect. Biol. 1:a000141. doi: 10.1101/cshperspect.a000141
Kazeros, A., Harvey, B. G., Carolan, B. J., Vanni, H., Krause, A., and Crystal, R. G. (2008). Overexpression of apoptotic cell removal receptor MERTK in alveolar macrophages of cigarette smokers. Am. J. Respir. Cell Mol. Biol. 39, 747–757. doi: 10.1165/rcmb.2007-0306OC
Kerkar, S. P., Chinnasamy, D., Hadi, N., Melenhorst, J., Muranski, P., Spyridonidis, A., et al. (2014). Timing and intensity of exposure to interferon-γ critically determines the function of monocyte-derived dendritic cells. Immunology 143, 96–108. doi: 10.1111/imm.12292
Khajoee, V., Saito, M., Takada, H., Nomura, A., Kusuhara, K., Yoshida, S. I., et al. (2006). Novel roles of osteopontin and CXC chemokine ligand 7 in the defence against mycobacterial infection. Clin. Exp. Immunol. 143, 260–268. doi: 10.1111/j.1365-2249.2005.02985.x
Kim, E. H., Sullivan, J. A., Plisch, E. H., Tejera, M. M., Jatzek, A., Choi, K. Y., et al. (2012). Signal integration by Akt regulates CD8 T cell effector and memory differentiation. J. Immunol. 188, 4305–4314. doi: 10.4049/jimmunol.1103568
Kim, H. M., Kang, J. S., Lim, J., Park, S. K., Lee, K., Yoon, Y. D., et al. (2007). Inhibition of human ovarian tumor growth by cytokine-induced killer cells. Arch. Pharm. Res. 30, 1464–1470. doi: 10.1007/BF02977372
Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185. doi: 10.1016/j.cmet.2006.02.002
Kissick, H. T., Dunn, L. K., Ghosh, S., Nechama, M., Kobzik, L., and Arredouani, M. S. (2014). The scavenger receptor MARCO modulates TLR-induced responses in dendritic cells. PLoS ONE 9:e104148. doi: 10.1371/journal.pone.0104148
Kitoh, A., Ono, M., Naoe, Y., Ohkura, N., Yamaguchi, T., Yaguchi, H., et al. (2009). Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31, 609–620. doi: 10.1016/j.immuni.2009.09.003
Klijn, C., Durinck, S., Stawiski, E. W., Haverty, P. M., Jiang, Z., Liu, H., et al. (2015). A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33, 306–312. doi: 10.1038/nbt.3080
Kong, S., Thiruppathi, M., Qiu, Q., Lin, Z., Dong, H., Chini, E. N., et al. (2014). DBC1 Is a suppressor of B cell activation by negatively regulating alternative NF-κB transcriptional activity. J. Immunol. 193, 5515–5524. doi: 10.4049/jimmunol.1401798
Konuma, T., Nakamura, S., Miyagi, S., Negishi, M., Chiba, T., Oguro, H., et al. (2011). Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 39, 697–709.e5. doi: 10.1016/j.exphem.2011.03.008
Kousik, S. M., Napier, T. C., and Carvey, P. M. (2012). The effects of psychostimulant drugs on blood brain barrier function and neuroinflammation. Front. Pharmacol. 3:121. doi: 10.3389/fphar.2012.00121
Koziel, J., Maciag-Gudowska, A., Mikolajczyk, T., Bzowska, M., Sturdevant, D. E., Whitney, A. R., et al. (2009). Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS ONE 4:e5210. doi: 10.1371/journal.pone.0005210
Kron, M. A., Metwali, A., Vodanovic-Jankovic, S., and Elliott, D. (2013). Nematode asparaginyl-tRNA synthetase resolves intestinal inflammation in mice with T-cell transfer colitis. Clin. Vaccine Immunol. 20, 276–281. doi: 10.1128/CVI.00594-12
Krupka, M., Seydel, K., Feintuch, C. M., Yee, K., Kim, R., Lin, C. Y., et al. (2012). Mild Plasmodium falciparum malaria following an episode of severe malaria is associated with induction of the interferon pathway in Malawian children. Infect. Immun. 80, 1150–1155. doi: 10.1128/IAI.06008-11
Kuo, C. L., Murphy, A. J., Sayers, S., Li, R., Yvan-Charvet, L., Davis, J. Z., et al. (2011). Cdkn2a is an atherosclerosis modifier locus that regulates monocyte/macrophage proliferation. Arterioscler. Thromb. Vasc. Biol. 31, 2483–2492. doi: 10.1161/ATVBAHA.111.234492
Kupfer, D. M., White, V. L., Strayer, D. L., Crouch, D. J., and Burian, D. (2013). Microarray characterization of gene expression changes in blood during acute ethanol exposure. BMC Med. Genomics 6:26. doi: 10.1186/1755-8794-6-26
Kwissa, M., Nakaya, H. I., Onlamoon, N., Wrammert, J., Villinger, F., Perng, G. C., et al. (2014). Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 16, 115–127. doi: 10.1016/j.chom.2014.06.001
Kyogoku, C., Smiljanovic, B., Grün, J. R., Biesen, R., Schulte-Wrede, U., Häupl, T., et al. (2013). Cell-specific type I IFN signatures in autoimmunity and viral infection: what makes the difference? PLoS ONE 8:e83776. doi: 10.1371/journal.pone.0083776
LaBreche, H., Nevins, J., and Huang, E. (2011). Integrating factor analysis and a transgenic mouse model to reveal a peripheral blood predictor of breast tumors. BMC Med. Genomics 4:61. doi: 10.1186/1755-8794-4-61
Lafaille, F. G., Pessach, I. M., Zhang, S. Y., Ciancanelli, M. J., Herman, M., Abhyankar, A., et al. (2012). Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491, 769–773. doi: 10.1038/nature11583
Lai, C. Y., Yu, S. L., Hsieh, M. H., Chen, C. H., Chen, H. Y., Wen, C. C., et al. (2011). MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 6:e21635. doi: 10.1371/journal.pone.0021635
Lattin, J. E., Schroder, K., Su, A. I., Walker, J. R., Zhang, J., Wiltshire, T., et al. (2008). Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 4:5. doi: 10.1186/1745-7580-4-5
Lau, C. L., Perreau, V. M., Chen, M. J., Cate, H. S., Merlo, D., Cheung, N. S., et al. (2012). Transcriptomic profiling of astrocytes treated with the Rho kinase inhibitor Fasudil reveals cytoskeletal and pro-survival responses. J. Cell Physiol. 227, 1199–1211. doi: 10.1002/jcp.22838
Lee, C., and Huang, C. H. (2013). LASAGNA-Search: an integrated web tool for transcription factor binding site search and visualization. Biotechniques 54, 141–153. doi: 10.2144/000113999
Lee, J. I., Wright, J. H., Johnson, M. M., Bauer, R. L., Sorg, K., Yuen, S., et al. (2016). Role of Smad3 in platelet-derived growth factor-C-induced liver fibrosis. Am. J. Physiol. Cell Physiol. 310, C436–C445. doi: 10.1152/ajpcell.00423.2014
Lee, J. S., Scandiuzzi, L., Ray, A., Wei, J., Hofmeyer, K. A., Abadi, Y. M., et al. (2012). B7x in the periphery abrogates pancreas-specific damage mediated by self-reactive CD8 T cells. J. Immunol. 189, 4165–4174. doi: 10.4049/jimmunol.1201241
Lee, S. M., Gardy, J. L., Cheung, C. Y., Cheung, T. K., Hui, K. P., Ip, N. Y., et al. (2009). Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS ONE 4:e8072. doi: 10.1371/journal.pone.0008072
Li, J. R., Sun, C. H., Li, W., Chao, R. F., Huang, C. C., Zhou, X. J., et al. (2016). Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 44, D944–D951. doi: 10.1093/nar/gkv1282
Li, Q., Smith, A. J., Schacker, T. W., Carlis, J. V., Duan, L., Reilly, C. S., et al. (2009). Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J. Immunol. 183, 1975–1982. doi: 10.4049/jimmunol.0803222
Liang, J., Takeuchi, H., Doi, Y., Kawanokuchi, J., Sonobe, Y., Jin, S., et al. (2008). Excitatory amino acid transporter expression by astrocytes is neuroprotective against microglial excitotoxicity. Brain Res. 1210, 11–19. doi: 10.1016/j.brainres.2008.03.012
Liberles, S. D., and Buck, L. B. (2006). A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650. doi: 10.1038/nature05066
Liew, C. C., Ma, J., Tang, H. C., Zheng, R., and Dempsey, A. A. (2006). The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J. Lab. Clin. Med. 147, 126–132. doi: 10.1016/j.lab.2005.10.005
Lin, X., Wang, R., Zhang, J., Sun, X., Zou, Z., Wang, S., et al. (2015). Insights into human astrocyte response to H5N1 infection by microarray analysis. Viruses 7, 2618–2640. doi: 10.3390/v7052618
Lindemann, L., and Hoener, M. C. (2005). A renaissance in trace amines inspired by a novel GPCR family. Trends Pharmacol. Sci. 26, 274–281. doi: 10.1016/j.tips.2005.03.007
Liu, B., Chen, H., Johns, T. G., and Neufeld, A. H. (2006). Epidermal growth factor receptor activation: an upstream signal for transition of quiescent astrocytes into reactive astrocytes after neural injury. J. Neurosci. 26, 7532–7540. doi: 10.1523/JNEUROSCI.1004-06.2006
Luckey, C. J., Bhattacharya, D., Goldrath, A. W., Weissman, I. L., Benoist, C., and Mathis, D. (2006). Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 3304–3309. doi: 10.1073/pnas.0511137103
Mabbott, N., Baillie, J., Brown, H., Freeman, T., and Hume, D. (2013). An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics 14:632. doi: 10.1186/1471-2164-14-632
Macagno, A., Molteni, M., Rinaldi, A., Bertoni, F., Lanzavecchia, A., Rossetti, C., et al. (2006). A cyanobacterial LPS antagonist prevents endotoxin shock and blocks sustained TLR4 stimulation required for cytokine expression. J. Exp. Med. 203, 1481–1492. doi: 10.1084/jem.20060136
Malhotra, S., Bustamante, M. F., Pérez-Miralles, F., Rio, J., Ruiz de Villa, M. C., Vegas, E., et al. (2011). Search for specific biomarkers of IFNβ bioactivity in patients with multiple sclerosis (IFNß bioactivity markers in multiple sclerosis). PLoS ONE 6:e23634. doi: 10.1371/journal.pone.0023634
Manel, N., Hogstad, B., Wang, Y., Levy, D. E., Unutmaz, D., and Littman, D. R. (2010). A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 470, 424–424. doi: 10.1038/nature09813
Maouche, S., Poirier, O., Godefroy, T., Olaso, R., Gut, I., Collet, J. P., et al. (2008). Performance comparison of two microarray platforms to assess differential gene expression in human monocyte and macrophage cells. BMC Genomics 9:302. doi: 10.1186/1471-2164-9-302
Massberg, S., Grahl, L., von Bruehl, M. L., Manukyan, D., Pfeiler, S., Goosmann, C., et al. (2010). Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat. Med. 16, 887–896. doi: 10.1038/nm.2184
McGovern, N., Schlitzer, A., Gunawan, M., Jardine, L., Shin, A., Poyner, E., et al. (2014). Human dermal CD14? cells are a transient population of monocyte-derived macrophages. Immunity 41, 465–477. doi: 10.1016/j.immuni.2014.08.006
McHale, C. M., Zhang, L., Lan, Q., Li, G., Hubbard, A. E., Forrest, M. S., et al. (2009). Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics 93, 343–349. doi: 10.1016/j.ygeno.2008.12.006
Mendez-David, I., El-Ali, Z., Hen, R., Falissard, B., Corruble, E., Gardier, A. M., et al. (2013). A method for biomarker measurements in peripheral blood mononuclear cells isolated from anxious and depressed mice: β-arrestin 1 protein levels in depression and treatment. Front. Pharmacol. 4:124. doi: 10.3389/fphar.2013.00124
Mense, S. M. (2006). Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol. Genomics 25, 435–449. doi: 10.1152/physiolgenomics.00315.2005
Mense, S. M., Sengupta, A., Lan, C., Zhou, M., Bentsman, G., Volsky, D. J., et al. (2006). The common insecticides cyfluthrin and chlorpyrifos alter the expression of a subset of genes with diverse functions in primary human astrocytes. Toxicol. Sci. 93, 125–135. doi: 10.1093/toxsci/kfl046
Menssen, A., Edinger, G., Grün, J. R., Haase, U., Baumgrass, R., Grützkau, A., et al. (2009). SiPaGene: a new repository for instant online retrieval, sharing and meta-analyses of GeneChip expression data. BMC Genomics 10:98. doi: 10.1186/1471-2164-10-98
Merryweather-Clarke, A. T., Atzberger, A., Soneji, S., Gray, N., Clark, K., Waugh, C., et al. (2011). Global gene expression analysis of human erythroid progenitors. Blood 117, E96–108. doi: 10.1182/blood-2010-07-290825
Mezger, M., Wozniok, I., Blockhaus, C., Kurzai, O., Hebart, H., Einsele, H., et al. (2008). Impact of mycophenolic acid on the functionality of human polymorphonuclear neutrophils and dendritic cells during interaction with Aspergillus fumigatus. Antimicrob Agents 52, 2644–2646. doi: 10.1128/aac.01618-07
Miller, G. M. (2011). The emerging role of trace amine-associated receptor 1 in the functional regulation of monoamine transporters and dopaminergic activity. J. Neurochem. 116, 164–176. doi: 10.1111/j.1471-4159.2010.07109.x
Miller, G. M., Verrico, C. D., Jassen, A., Konar, M., Yang, H., Panas, H., et al. (2005). Primate trace amine receptor 1 modulation by the dopamine transporter. J. Pharmacol. Exp. Ther. 313, 983–994. doi: 10.1124/jpet.105.084459
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2:18. doi: 10.1186/1755-8794-2-18
Mogensen, T. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/CMR.00046-08
Moncrieffe, H., Hinks, A., Ursu, S., Kassoumeri, L., Etheridge, A., Hubank, M., et al. (2010). Generation of novel pharmacogenomic candidates in response to methotrexate in juvenile idiopathic arthritis: correlation between gene expression and genotype. Pharmacogenet. Genomics 20, 665–676. doi: 10.1097/FPC.0b013e32833f2cd0
Monie, T. (2017). The canonical inflammasome: a macromolecular complex driving inflammation. Sub Cell. Biochem. 83, 43–73. doi: 10.1007/978-3-319-46503-6_2
Mosig, S., Rennert, K., Büttner, P., Krause, S., Lütjohann, D., Soufi, M., et al. (2008). Monocytes of patients with familial hypercholesterolemia show alterations in cholesterol metabolism. BMC Med. Genomics 1:60. doi: 10.1186/1755-8794-1-60
Napolitani, G., Rinaldi, A., Bertoni, F., Sallusto, F., and Lanzavecchia, A. (2005). Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6, 769–776. doi: 10.1038/ni1223
Ndolo, T., George, M., Nguyen, H., and Dandekar, S. (2006). Expression of simian immunodeficiency virus Nef protein in CD4+ T cells leads to a molecular profile of viral persistence and immune evasion. Virology 353, 374–387. doi: 10.1016/j.virol.2006.06.008
Nelson, D. A., Tolbert, M. D., Singh, S. J., and Bost, K. L. (2007). Expression of neuronal Trace Amine-associated Receptor (Taar) mRNAs in leukocytes. J. Neuroimmunol. 192, 21–30. doi: 10.1016/j.jneuroim.2007.08.006
Newell, K. A., Asare, A., Kirk, A. D., Gisler, T. D., Bourcier, K., Suthanthiran, M., et al. (2010). Immune tolerance network ST507 study group. Identification of a B cell signature associated with renal transplant tolerance in humans. J. Clin. Invest. 120, 1836–1847. doi: 10.1172/JCI39933
Nilsson, R. J., Balaj, L., Hulleman, E., van Rijn, S., Pegtel, D. M., Walraven, M., et al. (2011). Blood platelets contain tumor-derived RNA biomarkers. Blood 118, 3680–3683. doi: 10.1182/blood-2011-03-344408
Njau, F., Geffers, R., Thalmann, J., Haller, H., and Wagner, A. D. (2009). Restriction of Chlamydia pneumoniae replication in human dendritic cell by activation of indoleamine 2,3-dioxygenase. Microbes Infect. 11, 1002–1010. doi: 10.1016/j.micinf.2009.07.006
Northrop, N. A., and Yamamoto, B. K. (2015). Methamphetamine effects on blood-brain barrier structure and function. Front. Neurosci. 9:69. doi: 10.3389/fnins.2015.00069
Opal, S. (2000). Phylogenetic and functional relationships between coagulation and the innate immune response. Crit. Care Med. 28, S77–80. doi: 10.1097/00003246-200009001-00017
Oudes, A., Roach, J., Walashek, L., Eichner, L., True, L., Vessella, R., et al. (2005). Application of affymetrix array and massively parallel signature sequencing for identification of genes involved in prostate cancer progression. BMC Cancer 5:86. doi: 10.1186/1471-2407-5-86
Palabrica, T., Lobb, R., Furie, B. C., Aronovitz, M., Benjamin, C., Hsu, Y. M., et al. (1992). Leukocyte accumulation promoting fibrin deposition is mediated in vivo by P-selectin on adherent platelets. Nature 359, 848–851. doi: 10.1038/359848a0
Palau, N., Julià, A., Ferrándiz, C., Puig, L., Fonseca, E., Fernández, E., et al. (2013). Genome-wide transcriptional analysis of T cell activation reveals differential gene expression associated with psoriasis. BMC Genomics 14:825. doi: 10.1186/1471-2164-14-825
Panas, M. W., Xie, Z., Panas, H. N., Hoener, M. C., Vallender, E. J., and Miller, G. M. (2012). Trace amine associated receptor 1 signaling in activated lymphocytes. J. Neuroimmune Pharmacol. 7, 722–724. doi: 10.1007/s11481-011-9321-4
Papapanou, P. N., Sedaghatfar, M. H., Demmer, R. T., Wolf, D. L., Yang, J., Roth, G. A., et al. (2007). Periodontal therapy alters gene expression of peripheral blood monocytes. J. Clin. Periodontol. 34, 736–747. doi: 10.1111/j.1600-051X.2007.01113.x
Parnell, G. P., Tang, B. M., Nalos, M., Armstrong, N. J., Huang, S. J., Booth, D. R., et al. (2013). Identifying key regulatory genes in the whole blood of septic patients to monitor underlying immune dysfunctions. Shock 40, 166–174. doi: 10.1097/SHK.0b013e31829ee604
Parnell, G., McLean, A., Booth, D., Huang, S., Nalos, M., and Tang, B. (2011). Aberrant cell cycle and apoptotic changes characterise severe influenza A infection–a meta-analysis of genomic signatures in circulating leukocytes. PLoS ONE 6:e17186. doi: 10.1371/journal.pone.0017186
Patke, A., Mecklenbräuker, I., Erdjument-Bromage, H., Tempst, P., and Tarakhovsky, A. (2006). BAFF controls B cell metabolic fitness through a PKCβ- and Akt-dependent mechanism. J. Exp. Med. 203, 2551–2562. doi: 10.1084/jem.20060990
Paugh, B. S., Zhu, X., Qu, C., Endersby, R., Diaz, A. K., Zhang, J., et al. (2013). Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 73, 6219–6229. doi: 10.1158/0008-5472.CAN-13-1491
Petersen, B. C., Budelsky, A. L., Baptist, A. P., Schaller, M. A., and Lukacs, N. W. (2012). Interleukin-25 induces type 2 cytokine production in a steroid-resistant interleukin-17RB myeloid population that exacerbates asthmatic pathology. Nat. Med. 18, 751–758. doi: 10.1038/nm.2735
Piccaluga, P. P., Rossi, M., Agostinelli, C., Ricci, F., Gazzola, A., Righi, S., et al. (2014). Platelet-derived growth factor alpha mediates the proliferation of peripheral T-cell lymphoma cells via an autocrine regulatory pathway. Leukemia 28, 1687–1697. doi: 10.1038/leu.2014.50
Piccolo, S. R., Andrulis, I. L., Cohen, A. L., Conner, T., Moos, P. J., Spira, A. E., et al. (2015). Gene-expression patterns in peripheral blood classify familial breast cancer susceptibility. BMC Med. Genomics 8:72. doi: 10.1186/s12920-015-0145-6
Pillay, J., Kamp, V. M., van Hoffen, E., Visser, T., Tak, T., Lammers, J. W., et al. (2012). A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J. Clin. Invest. 122, 327–336. doi: 10.1172/JCI57990
Pimentel-Santos, F. M., Ligeiro, D., Matos, M., Mourão, A. F., Costa, J., Santos, H., et al. (2011). Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res. Ther. 13, R57. doi: 10.1186/ar3309
Pomié, C., Vicente, R., Vuddamalay, Y., Lundgren, B. A., van der Hoek, M., Enault, G., et al. (2011). Autoimmune regulator (AIRE)-deficient CD8 CD28low regulatory T lymphocytes fail to control experimental colitis. P. Natl. Acad. Sci. U.S.A. 108, 12437–12442. doi: 10.1073/pnas.1107136108
Potula, R., Hawkins, B. J., Cenna, J. M., Fan, S., Dykstra, H., Ramirez, S. H., et al. (2010). Methamphetamine causes mitrochondrial oxidative damage in human T lymphocytes leading to functional impairment. J. Immunol. 185, 2867–2876. doi: 10.4049/jimmunol.0903691
Pubill, D., Canudas, A. M., Pallàs, M., Camins, A., Camarasa, J., and Escubedo, E. (2003). Different glial response to methamphetamine- and methylenedioxymethamphetamine-induced neurotoxicity. Naunyn Schmiedeberg's Arch. Pharmacol. 367, 490–499. doi: 10.1007/s00210-003-0747-y
Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. (1995). MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884.
Radom-Aizik, S., Zaldivar, F., Leu, S., Galassetti, P., and Cooper, D. (2008). Effects of 30 min of aerobic exercise on gene expression in human neutrophils. J. Appl. Physiol. 104, 236–243. doi: 10.1152/japplphysiol.00872.2007
Raghavachari, N., Xu, X., Harris, A., Villagra, J., Logun, C., Barb, J., et al. (2007). Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Circulation 115, 1551–1562. doi: 10.1161/CIRCULATIONAHA.106.658641
Ramirez, S. H., Potula, R., Fan, S., Eidem, T., Papugani, A., Reichenbach, N., et al. (2012). Methamphetamine disrupts blood-brain barrier function by induction of oxidative stress in brain endothelial cells. J. Cereb. Blood Flow Metab. 29, 1933–1945. doi: 10.1038/jcbfm.2009.112
Ramos, Y. F., Bos, S. D., Lakenberg, N., Böhringer, S., den Hollander, W. J., Kloppenburg, M., et al. (2014). Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann. Rheum. Dis. 73, 1844. doi: 10.1136/annrheumdis-2013-203405
Ransohoff, R. M., and Brown, M. (2012). Review series Innate immunity in the central nervous system. J. Clin. Invest. 122, 1164–1171. doi: 10.1172/JCI58644.1164
Ricciardi, A., Elia, A. R., Cappello, P., Puppo, M., Vanni, C., Fardin, P., et al. (2008). Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol. Cancer Res. 6, 175–185. doi: 10.1158/1541-7786.mcr-07-0391
Risitano, A., Beaulieu, L., Vitseva, O., and Freedman, J. (2012). Platelets and platelet-like particles mediate intercellular RNA transfer. Blood 119, 6288–6295. doi: 10.1182/blood-2011-12-396440
Rivollier, A., He, J., Kole, A., Valatas, V., and Kelsall, B. L. (2012). Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155. doi: 10.1084/jem.20101387
Rogowski, M., Gollahon, L., Chellini, G., and Assadi-Porter, F. M. (2017). Uptake of 3-iodothyronamine hormone analogs inhibits the growth and viability of cancer cells. FEBS Open Biol. 7, 587–601. doi: 10.1002/2211-5463.12205
Rosas, M., Davies, L. C., Giles, P. J., Liao, C. T., Kharfan, B., Stone, T. C., et al. (2014). The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648. doi: 10.1126/science.1251414
Rudra, D., deRoos, P., Chaudhry, A., Niec, R. E., Arvey, A., Samstein, R. M., et al. (2012). Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13, 1010–1019. doi: 10.1038/ni.2402
Saijo, K., and Glass, C. K. (2011). Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 11, 775–787. doi: 10.1038/nri3086
Salvatore, G., Bernoud-Hubac, N., Bissay, N., Debard, C., Daira, P., Meugnier, E., et al. (2015). Human monocyte-derived dendritic cells turn into foamy dendritic cells with IL-17A. J. Lipid Res. 56, 1110–1122. doi: 10.1194/jlr.M054874
Sancho-Shimizu, V., Pérez de Diego, R., Lorenzo, L., Halwani, R., Alangari, A., Israelsson, E., et al. (2011). Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Inves. 121, 4889–4902. doi: 10.1172/JCI59259
Sanda, T., Tyner, J. W., Gutierrez, A., Ngo, V. N., Glover, J., Chang, B. H., et al. (2013). TYK2–STAT1–BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 3, 564–577. doi: 10.1158/2159-8290.cd-12-0504
Sato, S., Sanjo, H., Takeda, K., Ninomiya-Tsuji, J., Yamamoto, M., Kawai, T., et al. (2005). Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095. doi: 10.1038/ni1255
Satpathy, A. T., Briseño, C. G., Cai, X., Michael, D. G., Chou, C., Hsiung, S., et al. (2014). Runx1 and Cbfβ regulate the development of Flt3+ dendritic cell progenitors and restrict myeloproliferative disorder. Blood 123, 2968–2977. doi: 10.1182/blood-2013-11-539643
Schirmer, S. H., Bot, P. T., Fledderus, J. O., van der Laan, A. M., Volger, O. L., Laufs, U., et al. (2010). Blocking interferon {beta} stimulates vascular smooth muscle cell proliferation and arteriogenesis. J. Biol. Chem. 285, 34677–34685. doi: 10.1074/jbc.M110.164350
Schirmer, S. H., Fledderus, J. O., van der Laan, A. M., van der Pouw-Kraan, T. C., Moerland, P. D., Volger, O. L., et al. (2009). Suppression of inflammatory signaling in monocytes from patients with coronary artery disease. J. Mol. Cell Cardiol. 46, 177–185. doi: 10.1016/j.yjmcc.2008.10.029
Schmidt-Wolf, I. G. (1991). Use of a SCID mouse/human lymphoma model to evaluate cytokine-induced killer cells with potent antitumor cell activity. J. Exp. Med. 174, 139–149. doi: 10.1084/jem.174.1.139
Schubert, P., Ogata, T., Marchini, C., Ferroni, S., and Rudolphi, K. (1997). Protective mechanisms of adenosine in neurons and glial cells. Ann. N. Y. Acad. Sci. 825, 1–10.
Setoguchi, R., Taniuchi, I., and Bevan, M. J. (2009). ThPOK derepression is required for robust CD8 T cell responses to viral infection. J. Immunol. 183, 4467–4474. doi: 10.4049/jimmunol.0901428
Severa, M., Islam, S. A., Waggoner, S. N., Jiang, Z., Kim, N. D., Ryan, G., et al. (2014). The transcriptional repressor BLIMP1 curbs host defenses by suppressing expression of the chemokine CCL8. J. Immunol. 192, 2291–2304. doi: 10.4049/jimmunol.1301799
Sharma, M. K., Mansur, D. B., Reifenberger, G., Perry, A., Leonard, J. R., Aldape, K. D., et al. (2007). Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 67, 890–900. doi: 10.1158/0008-5472.can-06-0973
Sharma, R., Sharma, P. R., Kim, Y. C., Leitinger, N., Lee, J. K., Fu, S. M., et al. (2011). Correction: IL-2-controlled expression of multiple T cell trafficking genes and Th2 cytokines in the regulatory T cell-deficient scurfy mice: implication to multiorgan inflammation and control of skin and lung inflammation. J. Immunol. 186, 5012–5013. doi: 10.4049/jimmunol.1190010
Sharma-Kuinkel, B. K., Zhang, Y., Yan, Q., Ahn, S. H., and Fowler, V. G. Jr (2013). Host gene expression profiling and in vivo cytokine studies to characterize the role of linezolid and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA) murine sepsis model. PLoS ONE 8:e60463. doi: 10.1371/journal.pone.0060463
Shell, S., Hesse, C., Morris, S., and Milcarek, C. (2005). Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 280, 39950–39961. doi: 10.1074/jbc.M508848200
Shi, M., Sekar, K., Tan, C. K., Ooi, L. L., and Hui, M. K. (2014). A blood-based three-gene signature for the non-invasive detection of early human hepatocellular carcinoma. Eur. J. Cancer 50, 928–936. doi: 10.1016/j.ejca.2013.11.026
Shou, J., Bull, C. M., Li, L., Qian, H. R., Wei, T., Luo, S., et al. (2006). Identification of blood biomarkers of rheumatoid arthritis by transcript profiling of peripheral blood mononuclear cells from the rat collagen-induced arthritis model. Arthritis Res. Ther. 8, R28. doi: 10.1186/ar1883
Showe, M. K., Vachani, A., Kossenkov, A. V., Yousef, M., Nichols, C., Nikonova, E. V., et al. (2009). Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 69, 9202–9210. doi: 10.1158/0008-5472.CAN-09-1378
Simpson, J. E., Ince, P. G., Shaw, P. J., Heath, P. R., Raman, R., Garwood, C. J., et al. (2011). MRC cognitive function and ageing neuropathology study group. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer's pathology and APOE genotype. Neurobiol. Aging 32, 1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013
Sirois, M., Robitaille, L., Allary, R., Shah, M., Woelk, C. H., Estaquier, J., et al. (2011). TRAF6 and IRF7 control HIV replication in macrophages. PLoS ONE 6:e28125. doi: 10.1371/journal.pone.0028125
Sitrin, J., Ring, A., Garcia, K., Benoist, C., and Mathis, D. (2013). Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. J. Exp. Med. 210, 1153–1165. doi: 10.1084/jem.20122248
Sriram, U., Cenna, J. M., Haldar, B., Fernandes, N. C., Razmpour, R., Fan, S., et al. (2016). Methamphetamine induces trace amine-associated receptor 1 (TAAR1) expression in human T lymphocytes: role in immunomodulation. J. Leukoc. Biol. 99, 213–223. doi: 10.1189/jlb.4A0814-395RR
Stegmann, K. A., Björkström, N. K., Veber, H., Ciesek, S., Riese, P., Wiegand, J., et al. (2010). Interferon-α-induced TRAIL on natural killer cells is associated with control of hepatitis c virus infection. Gastroenterology 138, 1885.e10–1897.e10. doi: 10.1053/j.gastro.2010.01.051
Stolp, J., Chen, Y. G., Cox, S. L., Henck, V., Zhang, W., Tsaih, S. W., et al. (2012). Subcongenic analyses reveal complex interactions between distal chromosome 4 Genes controlling diabetogenic B cells and CD4 T cells in nonobese diabetic mice. J. Immunol. 189, 1406–1417. doi: 10.4049/jimmunol.1200120
Su, I. H., Dobenecker, M. W., Dickinson, E., Oser, M., Basavaraj, A., Marqueron, R., et al. (2005). Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 121, 425–436. doi: 10.1016/j.cell.2005.02.029
Szatmari, I., Pap, A., Rühl, R., Ma, J. X., Illarionov, P. A., Besra, G. S., et al. (2006). PPARγ controls CD1d expression by turning on retinoic acid synthesis in developing human dendritic cells. J. Exp. Med. 203, 2351–2362. doi: 10.1084/jem.20060141
Tabeta, K., Hoebe, K., Janssen, E. M., Du, X., Georgel, P., Crozat, K., et al. (2006). The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 7, 156–164. doi: 10.1038/ni1297
Takasaki, I., Takarada, S., Fukuchi, M., Yasuda, M., Tsuda, M., and Tabuchi, Y. (2007). Identification of genetic networks involved in the cell growth arrest and differentiation of a rat astrocyte cell line RCG-12. J. Cell. Biochem. 102, 1472–1485. doi: 10.1002/jcb.21369
Teles, R. M., Graeber, T. G., Krutzik, S. R., Montoya, D., Schenk, M., Lee, D. J., et al. (2013). Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 339, 1448–1453. doi: 10.1126/science.1233665
Terunuma, H., Deng, X., Dewan, Z., Fujimoto, S., and Yamamoto, N. (2008). Potential role of NK cells in the induction of immune responses: implications for NK cell–based immunotherapy for cancers and viral infections. Int. Rev. Immunol. 27, 93–110. doi: 10.1080/08830180801911743
Theopold, U., Schmidt, O., Söderhäll, K., and Dushay, M. S. (2004). Coagulation in arthropods: defence, wound closure and healing. Trends Immunol. 25, 289–294. doi: 10.1016/j.it.2004.03.004
Thomas, D. M., Dowgiert, J., Geddes, T. J., Francescutti-Verbeem, D., Liu, X., and Kuhn, D. M. (2004). Microglial activation is a pharmacologically specific marker for the neurotoxic amphetamines. Neurosci. Lett. 367, 349–354. doi: 10.1016/j.neulet.2004.06.065
Thul, P. J., and Lindskog, C. (2017). The human protein atlas: a spatial map of the human proteome. Science 27, 233–244. doi: 10.1002/pro.3307
Thuong, N. T., Dunstan, S. J., Chau, T. T., Thorsson, V., Simmons, C. P., Quyen, N. T., et al. (2008). Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 4:e1000229. doi: 10.1371/journal.ppat.1000229
Trandem, K., Zhao, J., Fleming, E., and Perlman, S. (2011). Highly activated cytotoxic CD8 T cells express protective IL-10 at the peak of coronavirus-induced encephalitis. J. Immunol. 186, 3642–3652. doi: 10.4049/jimmunol.1003292
Tsuge, M., Oka, T., Yamashita, N., Saito, Y., Fujii, Y., Nagaoka, Y., et al. (2014). Gene expression analysis in children with complex seizures due to influenza A(H1N1)pdm09 or rotavirus gastroenteritis. J. Neurovirol. 20, 73–84. doi: 10.1007/s13365-013-0231-5
Turowski, P., and Kenny, B. (2015). The blood-brain barrier and methamphetamine: open sesame? Front. Neurosci. 9:156. doi: 10.3389/fnins.2015.00156
Turvey, S. E., and Broide, D. H. (2010). Innate immunity. J. Allergy Clin. Immunol. 125, S24–32. doi: 10.1016/j.jaci.2009.07.016
Twiner, M. J., Ryan, J. C., Morey, J. S., Smith, K. J., Hammad, S. M., Van Dolah, F. M., et al. (2008). Transcriptional profiling and inhibition of cholesterol biosynthesis in human T lymphocyte cells by the marine toxin azaspiracid. Genomics 91, 289–300. doi: 10.1016/j.ygeno.2007.10.015
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419
van der Laan, A. M., Schirmer, S. H., de Vries, M. R., Koning, J. J., Volger, O. L., Fledderus, J. O., et al. (2012). Galectin-2 expression is dependent on the rs7291467 polymorphism and acts as an inhibitor of arteriogenesis. Eur. Heart J. 33, 1076–1084. doi: 10.1093/eurheartj/ehr220
van Leeuwen, D. M., Pedersen, M., Hendriksen, P. J., Boorsma, A., van Herwijnen, M. H., Gottschalk, R. W., et al. (2008). Genomic analysis suggests higher susceptibility of children to air pollution. Carcinogenesis 29, 977–983. doi: 10.1093/carcin/bgn065
Vanderford, T. H., Slichter, C., Rogers, K. A., Lawson, B. O., Obaede, R., Else, J., et al. (2012). Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood 119, 5750–5757. doi: 10.1182/blood-2012-02-411496
Vattai, A., Akyol, E., Kuhn, C., Hofmann, S., Heidegger, H., von Koch, F., et al. (2017). Increased trace amine-associated receptor 1 (TAAR1) expression is associated with a positive survival rate in patients with breast cancer. J. Cancer Res. Clin. 143, 1637–1647. doi: 10.1007/s00432-017-2420-8
Verway, M., Bouttier, M., Wang, T. T., Carrier, M., Calderon, M., An, B. S., et al. (2013). Vitamin D induces interleukin-1β expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 9:e1003407. doi: 10.1371/journal.ppat.1003407
Verykokakis, M., Krishnamoorthy, V., Iavarone, A., Lasorella, A., Sigvardsson, M., and Kee, B. (2013). Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J. Immunol. 191, 5973–5983. doi: 10.4049/jimmunol.1301521
Vierimaa, O., Georgitsi, M., Lehtonen, R., Vahteristo, P., Kokko, A., Raitila, A., et al. (2006). Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science 312, 1228–1230. doi: 10.1126/science.1126100
Villarroya-Beltri, C., Gutiérrez-Vázquez, C., Sánchez-Cabo, F., Pérez-Hernández, D., Vázquez, J., Martin-Cofreces, N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4:2980. doi: 10.1038/ncomms3980
Wai, P. Y., and Kuo, P. C. (2007). Osteopontin: regulation in tumor metastasis. Cancer Metast. Rev. 27, 103–118. doi: 10.1007/s10555-007-9104-9
Wan, Q., Dingerdissen, H., Fan, Y., Gulzar, N., Pan, Y., Wu, T. J., et al. (2015). BioXpress: an integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database 2015:bav019. doi: 10.1093/database/bav019
Wang, G. P., Ciuffi, A., Leipzig, J., Berry, C. C., and Bushman, F. D. (2007). HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17, 1186–1194. doi: 10.1101/gr.6286907
Waugh, E., Chen, A., Baird, M. A., Brown, C. M., and Ward, V. K. (2014). Characterization of the chemokine response of RAW264.7 cells to infection by murine norovirus. Virus Res. 181, 27–34. doi: 10.1016/j.virusres.2013.12.025
Weiss, G., Rasmussen, S., Fink, L. N., Jarmer, H., Nielsen, B. N., and Frøkiær, H. (2010). Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS ONE 5:e11065. doi: 10.1371/journal.pone.0011065
Wen, T., Mingler, M., Wahl, B., Khorki, M., Pabst, O., Zimmermann, N., et al. (2014). Carbonic anhydrase IV is expressed on IL-5-activated murine eosinophils. J. Immunol. 192, 5481–5489. doi: 10.4049/jimmunol.1302846
West, E. E., Youngblood, B., Tan, W. G., Jin, H. T., Araki, K., Alexe, G., et al. (2011). Tight regulation of memory CD8 T cells limits their effectiveness during sustained high viral load. Immunity 35, 285–298. doi: 10.1016/j.immuni.2011.05.017
Weyrich, A. S., Schwertz, H., Kraiss, L. W., and Zimmerman, G. A. (2009). Protein synthesis by platelets: historical and new perspectives. J Thromb Haemost. 7, 241–246. doi: 10.1111/j.1538-7836.2008.03211.x
Wheelwright, M., Kim, E. W., Inkeles, M. S., De Leon, A., Pellegrini, M., Krutzik, S. R., et al. (2014). All-trans retinoic acid-triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2. J. Immunol. 192, 2280–2290. doi: 10.4049/jimmunol.1301686
Wong, H. R., Cvijanovich, N., Lin, R., Allen, G. L., Thomas, N. J., Willson, D. F., et al. (2009). Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 7:34. doi: 10.1186/1741-7015-7-34
Wong, N., Kulikowski, E., Wasiak, S., Gilham, D., Calosing, C., Laura, T., et al. (2016). Apabetalone (RVX-208) decreases atherogenic, thrombotic and inflammatory mediators in vitro and in plasma of patients with cardiovascular disease (CVD). Atherosclerosis 252, e245. doi: 10.1016/j.atherosclerosis.2016.07.042
Woodruff, P. G., Koth, L. L., Yang, Y. H., Rodriguez, M. W., Favoreto, S., Dolganov, G. M., et al. (2005). A distinctive alveolar macrophage activation state induced by cigarette smoking. Am. J. Respir. Crit. Care Med. 172, 1383–1392. doi: 10.1164/rccm.200505-686OC
Woods, C. G., Fu, J., Xue, P., Hou, Y., Pluta, L. J., Yang, L., et al. (2009). Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol. Appl. Pharmacol. 238, 27–36. doi: 10.1016/j.taap.2009.04.007
Woszczek, G., Chen, L. Y., Nagineni, S., Kern, S., Barb, J., Munson, P. J., et al. (2008). Leukotriene D(4) induces gene expression in human monocytes through cysteinyl leukotriene type I receptor. J. Allergy. Clin. Immunol. 121, 215.e1–221.e1. doi: 10.1016/j.jaci.2007.09.013
Wright, J. H., Johnson, M. M., Shimizu-Albergine, M., Bauer, R. L., Hayes, B. J., Surapisitchat, J., et al. (2014). Paracrine activation of hepatic stellate cells in platelet-derived growth factor C transgenic mice: evidence for stromal induction of hepatocellular carcinoma. Int. J. Cancer 134, 778–788. doi: 10.1002/ijc.28421
Wu, C., Macleod, I., and Su, A. I. (2013). BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41, D561–D565. doi: 10.1093/nar/gks1114
Wu, K., Dong, D., Fang, H., Levillain, F., Jin, W., Mei, J., et al. (2012). An interferon-related signature in the transcriptional core response of human macrophages to Mycobacterium tuberculosis infection. PLoS ONE 7:e38367. doi: 10.1371/journal.pone.0038367
Wynn, J. L., Cvijanovich, N. Z., Allen, G. L., Thomas, N. J., Freishtat, R. J., Anas, N., et al. (2011). The influence of developmental age on the early transcriptomic response of children with septic shock. Mol. Med. 17, 1146–1156. doi: 10.2119/molmed.2011.00169
Xie, Z., and Miller, G. M. (2007). Trace amine-associated receptor 1 is a modulator of the dopamine transporter. J. Pharmacol. Exp. Ther. 321, 128–136. doi: 10.1124/jpet.106.117382.is
Xie, Z., and Miller, G. M. (2008). β-Phenylethylamine alters monoamine transporter function via trace amine-associated receptor 1: implication for modulatory roles of trace amines in brain. J. Pharmacol. Exp. Ther. 325, 617–628. doi: 10.1124/jpet.107.134247
Xie, Z., and Miller, G. M. (2009). A receptor mechanism for methamphetamine action in dopamine transporter regulation in brain. J. Pharmacol. Exp. Ther. 330, 316–325. doi: 10.1124/jpet.109.153775
Yamamoto, M., Uematsu, S., Okamoto, T., Matsuura, Y., Sato, S., Kumar, H., et al. (2007). Enhanced TLR-mediated NF-IL6–dependent gene expression by Trib1 deficiency. J. Exp. Med. 204, 2233–2239. doi: 10.1084/jem.20070183
Yang, K., Neale, G., Green, D. R., He, W., and Chi, H. (2011). The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 12, 888–897. doi: 10.1038/ni.2068
Yoshino, T., Tabunoki, H., Sugiyama, S., Ishii, K., Kim, S. U., and Satoh, J. (2011). Non-phosphorylated FTY720 induces apoptosis of human microglia by activating SREBP2. Cell. Mol. Neurobiol. 31, 1009–1020. doi: 10.1007/s10571-011-9698-x
Zahoor, M. A., Xue, G., Sato, H., Murakami, T., Takeshima, S. N., and Aida, Y. (2014). HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS ONE 9:e106418. doi: 10.1371/journal.pone.0106418
Zamanian, J. L., Xu, L., Foo, L. C., Nouri, N., Zhou, L., Giffard, R. G., et al. (2012). Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012
Zaritsky, L., Bedsaul, J., and Zoon, K. (2015). Virus multiplicity of infection affects type i interferon subtype induction profiles and interferon-stimulated genes. J. Virol. 89, 11534–11548. doi: 10.1128/JVI.01727-15
Zhang, J. (2013). Transcriptome Analysis reveals novel entry mechanisms and a central role of SRC in host defense during high multiplicity mycobacterial infection. PLoS ONE 8:e65128. doi: 10.1371/journal.pone.0065128
Zhang, Z., Martino, A., and Faulon, J. L. (2007). Identification of expression patterns of IL-2-responsive genes in the murine T cell line CTLL-2. J. Interferon Cytokine Res. 27, 991–995. doi: 10.1089/jir.2006.0169
Keywords: PBMC, platelet, granulocyte, lymphocyte, B-cell, T-cell, astrocyte, microglia
Citation: Fleischer LM, Somaiya RD and Miller GM (2018) Review and Meta-Analyses of TAAR1 Expression in the Immune System and Cancers. Front. Pharmacol. 9:683. doi: 10.3389/fphar.2018.00683
Received: 17 December 2017; Accepted: 06 June 2018;
Published: 26 June 2018.
Edited by:
Stefano Espinoza, Fondazione Istituto Italiano di Technologia, ItalyReviewed by:
Dietmar Krautwurst, Leibniz Institute for Food Systems Biology, Technical University of Munich (LSB), GermanyCopyright © 2018 Fleischer, Somaiya and Miller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory M. Miller, Z3IubWlsbGVyQG5vcnRoZWFzdGVybi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.