- 1Laboratory of Genetics and Molecular Cardiology, Heart Institute (InCor), Faculdade de Medicina FMUSP, Universidade de São Paulo, São Paulo, Brazil
- 2Department of Pharmacology, Universidade Federal de São Paulo UNIFESP, São Paulo, Brazil
The ideal dose of the oral anticoagulant warfarin varies widely among patients, mainly due to genetic factors. Genetic variations that impact warfarin pharmacokinetics and the vitamin K cycle are plausible candidates for being associated with warfarin dose requirements. Therefore, the aim of this study was to assess whether polymorphisms in the ABCB1 and CYP4F2 genes were associated with stable warfarin dose requirements in Brazilian patients. This retrospective study included samples from 309 individuals. Genotyping of ABCB1 c.3435C>T and CYP4F2 c.1297G>A were performed by polymerase chain reaction followed by melting curve analysis (HRM-PCR) and TaqMan® genotyping assay, respectively. Stable doses were adjusted in a linear multiple regression model for age, gender, body mass index, self-reported race, use of amiodarone, CYP2C9 (*2 and *3), VKORC1 c.1639G>A, and ABCB1 c.3435C>T or CYP4F2 c.1297G>A. By performing a univariate analysis of variance, we found that the warfarin patients who carry ABCB1 c.3435T variant alleles (CT and TT genotypes) need fewer warfarin stable doses in comparison with the individuals that are CC wild-type: 2.5 (p = 0.003) and 4.3 (p < 0.001) mg/week less, respectively, for the overall group of patients on stable anticoagulation therapeutics (n = 309); and 5.5 (p = 0.006) and 10.2 (p < 0.001) mg/week less, respectively, for the self-declared non-white stable subgroup (n = 76). No statistically significant differences in dose requirements were observed according to CYP4F2 genotypes. In conclusion, our results suggest ABCB1 c.3435C>T variant may influence warfarin dose requirements in Brazilian patients, when associated with other genotypic, demographic and clinical factors.
Introduction
In the last 65 years, warfarin has been the most widely prescribed oral anticoagulant for treating and preventing thromboembolic events (Barnes et al., 2015). However, due to its narrow therapeutic index and high variance of pharmacokinetics and pharmacodynamics, warfarin interpatient responses are largely variable. Doses of warfarin administered out of the optimal therapeutic range commonly lead to serious adverse events, such as hemorrhagic and thromboembolic events in cases of over and under dosage, respectively. In addition to demographic and clinical factors, such as age, weight and use of concurrent medication, genetics is well established to play an important role in influencing warfarin maintenance dosage among patients (Klein et al., 2009; Johnson et al., 2011).
The key genetic variants that affect warfarin therapeutic response are presented in the VKORC1 and CYP2C9 genes, which encode the warfarin target enzyme and the primary warfarin metabolizer, respectively (Lee and Klein, 2013). However, as both these factors can explain 35–50% of the warfarin dose variability (Lee and Klein, 2013), there has been increasing interest in investigating additional clinically important genetic polymorphisms that may further explain interindividual warfarin dose variability among different populations.
Recently, studies have implicated the nonsynonymous variant presented in the CYP4F2 gene (c.1297G>A, p.Val433Met, rs2108622) in warfarin sensitivity (Caldwell et al., 2008; Cooper et al., 2008; Borgiani et al., 2009; Takeuchi et al., 2009; Cen et al., 2010; Wei et al., 2012). The CYP4F2 gene codes for the enzyme CYP4F2 (cytochrome P450 4F2), which is responsible for the primary metabolization of vitamin K in the liver. Warfarin mechanism of action consists of blocking the vitamin K redox cycle in order to diminish the concentration of the vitamin K reduced form, which is a coactivator of several coagulation factors (van Gorp and Schurgers, 2015). Thus, variations in the vitamin K bioavailability are speculated to interfere in the final response of treatment with warfarin.
Another genetic variant that has been investigated for influencing warfarin maintenance dosage is the silent polymorphism ABCB1 c.3435C>T (rs1045642) (Wadelius et al., 2004; De Oliveira Almeida et al., 2011; Kim et al., 2013). The ABCB1 gene (also called MDR1—Multidrug Resistance 1 Gene) codes for the well-known multidrug efflux pump P-glycoprotein (P-gp), responsible for restricting the absorption of many xenobiotics, as well as excreting them and their metabolites through the kidneys and liver. The referred ABCB1 polymorphism has been extensively studied and associated with the variability dosage of many drugs, including warfarin, which is one of the many substrates of P-gp (Lepper et al., 2005; Salama et al., 2006).
In this context, the aim of this study was to assess the association of ABCB1 and CYP4F2 genotypes with warfarin dose requirements in Brazilian patients.
Materials and Methods
Study Population
This retrospective study included samples from 309 on stable warfarin treatment, derived from a cohort of 832 Brazilian patients enrolled between September/2011 and March/2012 at the Heart Institute (InCor), University of São Paulo Medical School (FMUSP), São Paulo, Brazil (Santos et al., 2015). The study protocol was approved by the Institutional Ethics Committee (Register Number 0804/10), and written informed consent was obtained from all participants prior to entering the study. Patients were categorized into self-declared “racial/color” subgroups, according to the Brazilian Census criteria, as White, Intermediate (meaning Brown, “Pardo” in Portuguese) or Black. The following exclusion criteria were considered: chronic liver failure, use of other anticoagulant drugs, receiving chemotherapy and referred alcoholism. Personal demographic and clinical data were obtained through a standardized interview with a pharmacist and checked out on electronic medical records (Santos et al., 2011a; Soares et al., 2012).
The length of time between INR tests was variable, depending on how long the patient was presenting an INR within the target range and the probability of events that might affect INR. In case of target range reached, the minimum considered period between one INR measure and another was 4 weeks. The maximum was 12 weeks, in case of the patient was taking VKA therapy with consistently stable INRs. These recommendations are in accordance with anticoagulant therapy management (Holbrook et al., 2012; Witt et al., 2016). Furthermore, over the period of stable INR, there were no changes of the recommended weekly dosage. Thus, we defined stable warfarin therapeutics as three consecutive INR measures within therapeutic range (1.8–3.2), with no need to adjust warfarin dosing.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes by the standard salting-out method. Data of VKORC1 c.-1639G>A (rs9923231), CYP2C9*2 (c.430C>T, rs1799853) and CYP2C9*3 (c.1075A>C, rs1057910) genotypes were obtained from the previous study of our group (Santos et al., 2015). Genotyping of ABCB1 c.3435C>T (rs1045642) and CYP4F2 p.Val433Met (rs2108622) were performed by polymerase chain reaction followed by melting curve analysis (HRM-PCR) and TaqMan® genotyping assay (Applied Biosystems, C__16179493_40), respectively, in a Rotor-Gene Q® (Qiagen) (Santos et al., 2011b). All included patients (n = 309) presented stable warfarin maintenance doses. Positive and negative controls were added in all experiments. Furthermore, as genotyping quality control, 6% of the samples were reanalyzed and gave 100% data consistency.
Statistical Analysis
For descriptive analysis purposes of the cohort characteristics, categorical variables are presented as percentages, and continuous variables are presented as mean ± standard deviation (SD) or standard error (SE) of the mean. Chi-square and Fisher's exact tests were performed for comparative analysis of categorical variables (such as Hardy–Weinberg equilibrium calculation) according to genotypes. One-way ANOVA (analysis of variance) test was used for analysis of warfarin stable maintenance dose according to ABCB1 c.3435C>T (CC, CT or TT—categorized as 0, 1, and 2, respectively) and CYP4F2 c.1297G>A (GG, GA or AA—categorized as 0, 1, and 2, respectively) genotypes.
Mean warfarin stable doses were adjusted through a linear multiple regression model for age (years), gender (1, if male, otherwise 0), Body Mass Index (BMI) (kg/m2), self-declared race (white, brown or black—categorized as 1, 2, and 3, respectively), amiodarone use (1, if the patient administers amiodarone, otherwise 0), predicted metabolic CYP2C9 phenotypes (EM or IM + PM—categorized as 1 or 2, respectively), and VKORC1 c.-1639G>A genotypes (GG, GA or AA—categorized as 0, 1, and 2, respectively). Tukey's HSD (honest significant difference) post hoc tests were performed to identify the different groups. The level of significance was set at p ≤ 0.05. All statistical analyses were carried out using SPSS statistical package (20.0 version) and R (3.4.4 version).
Results
General Characteristics of the Study Cohort
Table 1 shows the demographic, clinical and genetic characteristics of the patients included in the study presenting stable warfarin treatment (n = 309). Of the 309 patients (mean age 64 ± 14), 154 (49.8%) were female, most of them (75.4%) declared their race/color as white, 4.6% were smokers and 10.7% used the warfarin concurrent medication amiodarone. The observed genotypic distributions for VKORC1 and frequencies of CYP2C9 predicted metabolic phenotypes are shown in Table 1. The observed required doses of warfarin ranged from 7.5 to 70 mg per week (median = 27.5, mean = 28.5 ± 11.0, in mg/week).
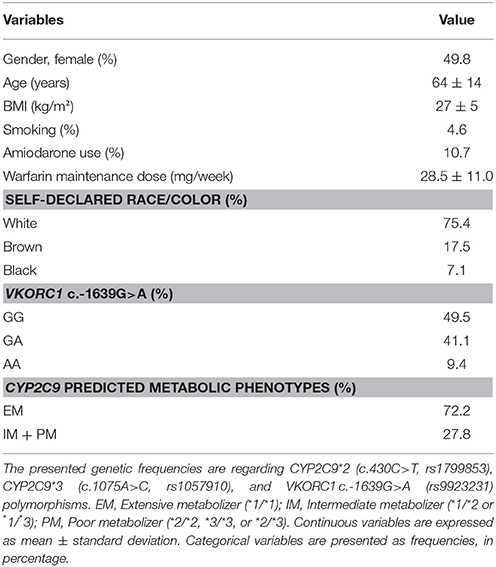
Table 1. General, clinical and genetic characteristics of the patients on stable warfarin therapeutics (n = 309).
Supplementary Tables 1, 2 show the demographic, clinical and genetic characteristics of the stable patients according with ABCB1 and CYP4F2 genotypes, respectively (Supplementary Tables 1, 2).
Frequencies of ABCB1 and CYP4F2 Polymorphisms
Table 2 summarizes the frequencies of ABCB1 and CYP4F2 genotypes of the patients. For all included stable patients (n = 309), the frequencies of the T variant allele and of the homozygous genotype for the ABCB1 c.3435C>T were 44 and 18.8%, respectively. The frequencies of the A variant allele and of the homozygous genotype for the CYP4F2 c.1297G>A were 32 and 8.7%, respectively. ABCB1 c.3435C>T and CYP4F2 c.1297G>A genotypic distributions are in accordance with Hardy-Weinberg equilibrium (Table 2).
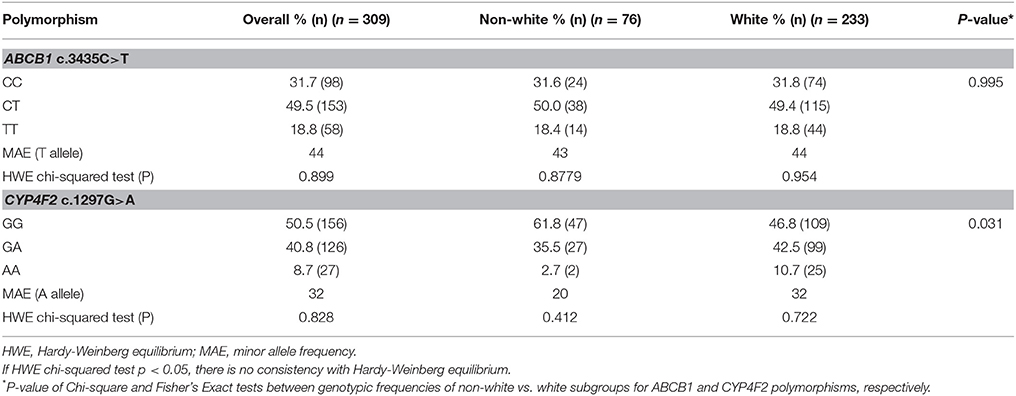
Table 2. Genotypic frequencies of ABCB1 and CYP4F2 polymorphisms of the patients with stable maintenance warfarin dose, according to self-declared racial subgroups (non-white or white).
Mean Warfarin Dose According to ABCB1 and CYP4F2 Genotypes
By performing a univariate analysis of variance (ANOVA) for the overall group of patients on stable warfarin treatment (n = 309), we found different mean warfarin doses according to ABCB1 c.3435C>T genotypes p-value (ANOVA) < 0.001; CC: 30.5 ± 0.6 (n = 98), CT: 28.0 ± 0.5 (n = 153), TT: 26.2 ± 0.6 (n = 58) (mean stable dose ± SE in mg/week). A Tukey's HSD post hoc test revealed that patients with ABCB1 3435CC genotype presented higher doses than CT or TT carriers (CC vs. TT: p < 0.001; CC vs. CT: p = 0.003; CT vs. TT: p = 0.111). The data were adjusted for age, gender, BMI, self-reported race/color, use of amiodarone, CYP2C9 (*2 and *3), and VKORC1 c.-1639G>A (Figure 1 and Table 3). The regression coefficients and respective p values are shown in Supplementary Tables 3, 4.
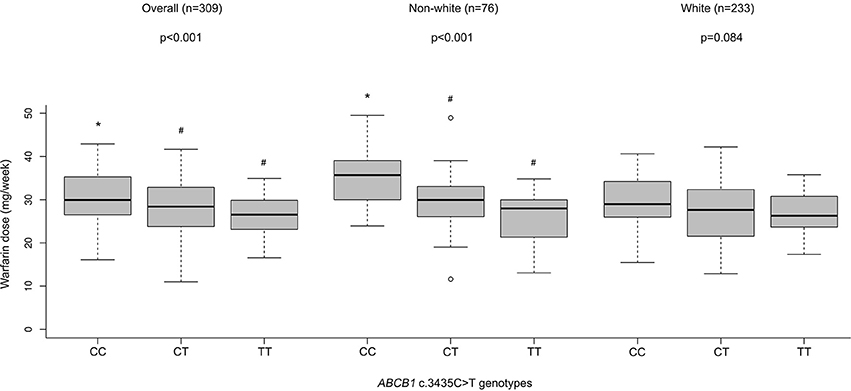
Figure 1. Warfarin dose (mg/week) according to ABCB1 genotypes in patients with stable dose. Stable maintenance dose means three consecutive INR values within the target therapeutic range (1.8–3.2). Weekly warfarin stable doses were adjusted for age, gender, BMI, self-declared race (white, brown or black), amiodarone use, predicted metabolic CYP2C9 phenotypes (EM or IM + PM), and VKORC1 c.-1639G>A genotypes. For the white (n = 233) and non-white (n = 76) patient subgroups, self-declared race was not used as an independent variable. P values of ANOVA's test are presented. Values with different superscript symbols (*, #) are significantly different according to Tukey's HSD post-hoc test. Statistically significant differences were set when p < 0.05.
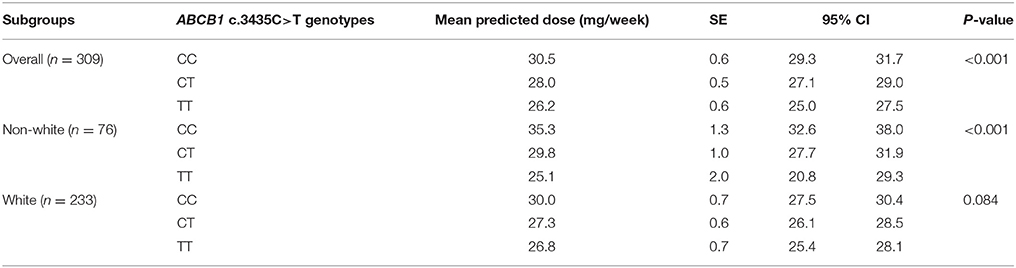
Table 3. Predicted warfarin doses by the multiple linear regression models adjusted with ABCB1 genotypes for overall, white and non-white patients' subgroups.
Additionally, we separated patients with stable dose (n = 309) into two distinct subgroups according to self-declared race/color: white (n = 233) and non-white (n = 76). For this analysis, we adjusted the dose for all covariates cited above, except self-reported race. Table 2 summarizes the frequencies of ABCB1 and CYP4F2 genotypes of the patients with stable warfarin therapy according to self-declared racial subgroups (Table 2).
For the non-white subgroup, we still found differences in the mean warfarin doses according to ABCB1 polymorphism p-value (ANOVA) < 0.001; CC = 35.3 ± 1.3 (n = 24), CT = 29.8 ± 1.0 (n = 38); TT = 25.1 ± 2.0 (n = 14) (mean stable dose ± SE in mg/week). Carriers of ABCB1 3435CC genotype presented higher doses than CT or TT, according to Tukey's HSD test (CC vs. TT: p < 0.001; CC vs. CT: p = 0.006; CT vs. TT: p = 0.063) (Figure 1 and Table 3).
Nevertheless, for the white subgroup, we have not found differences in the mean warfarin doses according to ABCB1 polymorphism p-value (ANOVA) = 0.084; CC = 30.0 ± 0.7 (n = 74), CT = 27.3 ± 0.6 (n = 115); TT = 26.8 ± 0.7 (n = 44) (mean stable dose ± SE in mg/week) (Figure 1). P values according to Tukey's HSD test were: CC vs. TT: p = 0.126; CC vs. CT: p = 0.144; CT vs. TT: p = 0.867.
Regarding the CYP4F2 p.Val433Met polymorphism, we found no significant statistical differences in the mean warfarin stable doses among the genotypic groups p-value (ANOVA) = 0.062; GG = 27.7 ± 0.4 (n = 156), GA = 29.3 ± 0.5 (n = 126), AA = 28.9 ± 0.9 (n = 27). P-values according to Tukey's HSD test were: GG vs. AA: p = 0.567; GG vs. GA: p = 0.053; GA vs. AA: p = 0.946. No significant differences were found when the white subgroup was assessed p-value (ANOVA) = 0.257; GG = 27.1 ± 0.6 (n = 109), GA = 28.3 ± 0.6 (n = 99), AA = 28.5 ± 1.1 (n = 25) (Supplementary Figure 1 and Supplementary Table 5).
For the non-white subgroup we found a slight difference in the mean warfarin doses according to CYP4F2 polymorphism: p-value (ANOVA) = 0.048; GG = 29.2 ± 1.1 (n = 47), GA = 33.1 ± 1.0 (n = 27), AA = 33.0 ± 1.9 (n = 2). P values according to Tukey's HSD test were: GG vs. AA: p = 0.705; GG vs. GA: p = 0.042; GA vs. AA: p = 1.000. However, only two patients were CYP4F2 1297AA carriers and self-declared their race/color as non-white (Supplementary Figure 1 and Supplementary Table 5).
Discussion
In the present study, we found the warfarin patients who carry ABCB1 c.3435T variant alleles (CT and TT genotypes) need, on average, fewer warfarin stable maintenance doses in comparison to the individuals that are wild-type CC carriers. Similar data were obtained in the study of Wadelius et al. They found that an ABCB1 haplotype containing only the 3435T allele as variant (called D haplotype) was over-represented [p(χ2) = 0.0242] among the low warfarin dose patients (<0.33 mg/kg, 60%) subgroup in comparison to the medium dose (0.33–0.46 mg/kg, 25%) and the high dose (>0.46 mg/kg, 15%) subgroups (p = 0.0094). On average, the patients that carried one 3435T variant allele for the D haplotype (n = 20) needed 24% lower warfarin dose and dose/body weight (BW) than the other patients (n = 181) who did not carry this haplotype (Wadelius et al., 2004).
In addition, Kim et al. found that patients with ABCB1 c.3435TT genotypes needed, descriptively, the lowest dose of warfarin compared with those with the other 2 genotypes (warfarin dose (mg/week): CC: 59.5 (n = 70); CT: 56.9 (n = 90); TT: 55.6 (n = 36). Nevertheless, no statistical differences between the genotypic subgroups were found. However, according to race, it is possible to verify that the African, Hispanic, and Asian Americans (the non-white subgroup in Kim's study), when analyzed together presented a decreasing pattern of mean warfarin doses according to the addition of the 3435T allele (Kim et al., 2013). The authors considered that the study had little statistical power due to a small number of patients in some genotypic subgroups. Likewise, other studies have also failed to find statistically significant association between ABCB1 c.3435C>T and warfarin dose requirements (Takeuchi et al., 2009; Issac et al., 2014).
Contrary to our result, a Brazilian study of De Oliveira Almeida et al. found that the ABCB1 c.3435T variant allele contributes to higher warfarin doses in thrombophilic patients (De Oliveira Almeida et al., 2011; de Oliveira Almeida et al., 2014). This finding was exclusively seen for the subgroup of patients who needed up to 70 mg of weekly warfarin dose. It is valid to point out that in the study of De Oliveira Almeida et al. the frequency of the T allele was very high (66%) in comparison to the reported ABCB1 c.3435C>T polymorphism frequency in other Brazilian studies, including the present one (around 40%) (Scheiner et al., 2010; Santos et al., 2011a). Furthermore, the patients were categorized into two subgroups: those receiving more and those receiving less than 70 mg warfarin weekly dose, which differs from the methodology of analysis of the present study.
In the literature, the studies that aimed to investigate the effect of the ABCB1 c.3435C>T polymorphism on the codified P-glycoprotein (P-gp) are quite controversial (Fromm, 2002; Sakaeda et al., 2003; Lepper et al., 2005). Some studies have reported pronounced efflux of substrate and higher mRNA expression of P-gp in the gastrointestinal tract in 3435CC carriers in comparison to 3435CT or 345TT (Greiner et al., 1999; Hoffmeyer et al., 2000), while other studies have found that these parameters are significantly lower in the wild-type individuals (Moriya et al., 2002; Nakamura et al., 2002; Morita et al., 2003). In other words, some studies have observed that the ABCB1 c.3435C>T polymorphism leads to augmented P-gp function or expression, although other studies have found that this variant causes decreased or loss of P-gp activity (Hoffmeyer et al., 2000; Hitzl et al., 2001; Kim et al., 2001; Sakaeda et al., 2003; Kimchi-Sarfaty et al., 2007). However, these studies were not performed with the same substrates and populations. So, it is likely that the effect of the ABCB1 c.3435C>T polymorphism on the P-gp activity depends on the substrate-specificity and environmental factors (Perloff et al., 2001; Lepper et al., 2005).
Moreover, the biological assumptions about how the ABCB1 c.3435C>T polymorphism affect P-gp are diverse. Despite the fact that this polymorphism is a silent mutation, as it does not lead to change of the codified isoleucine amino acid at position 1145, Kimchi-Sarfaty et al. have hypothesized that this variant may lead to a conformational P-gp change, which may affect the substrate specificity, explaining the observed affected efflux-pump function (Kimchi-Sarfaty et al., 2007). This conformational change could be due to distinct timing of cotranslational folding of the codon containing the variant. Furthermore, other so far unidentified variants that are in linkage disequilibrium with the c.3435C>T polymorphism, presented in control expression regions (e.g., promoter, enhancer) of the ABCB1 gene, may influence P-gp expression (Hoffmeyer et al., 2000; Frittitta et al., 2001). Additionally, as the ABCB1 c.3435C>T is in strong linkage disequilibrium with the non-synonymous c.2677T>G (p.Ser893Ala) and the synonymous c.1236T>C (p.Gly412=) variants, some authors hypothesize that the c.3435C>T itself does not directly influence the P-gp activity, but this common haplotype does (Salama et al., 2006; Saraeva et al., 2007).
We suggest that a possible biological explanation for our finding regarding the impact of ABCB1 c.3435C>T polymorphism in the warfarin dose requirements is that the rs1045642 variant allele leads to an altered P-gp fold, affecting the warfarin site of recognition, and therefore leads to decreased warfarin efflux activity. It would cause augmented warfarin bioavailability and therefore lower warfarin doses would be required for those patients who are CT and TT carriers.
Our study presents the following limitation: we did not assess the genetic ancestry of the patients. Instead, we used the self-declared race/color parameter, categorized according to the Brazilian Census IBGE (Brazilian Institute of Geography and Statistics) criteria. Self-declared race/color does not correlate completely with genetic ancestry in Brazilians (Pena et al., 2011), as the self-perception of color/race in Brazil is primarily accounted for by phenotypic characteristics, as well as socio, cultural and geographic factors into which the subject is inserted (Santos et al., 2009). However, this parameter is useful in the real-life setting.
The association between ABCB1 c.3435C>T and warfarin dose requirements, the main finding of this study, was observed for the overall stable group and non-white self-declared subgroup of patients, but not for those who self-declared their race/color as white. So, the above cited limitation might be, at least partially, contributing to this lack of association. The c.3435T variant allele in the ABCB1 gene is frequently rarer in brown and black than in white people (Santos et al., 2011a; Soares et al., 2012). However, in the study of Kim et al. the P-gp function was found to be similar, on average, in European Americans and African Americans, for example (Kim et al., 2001). Thus, further studies are needed to elucidate the specific impact of the ABCB1 c.3435C>T polymorphism in non-whites.
Regarding the lack of association of the CYP4F2 c.1297G>A polymorphism with warfarin dose requirements in the present study, when this variant was included as a covariate in a multiple-regression analysis along with other relevant clinical (age, gender, BMI, self-declared race, amiodarone use) and genetic factors (VKORC1 and CYP2C9 genotypes), it did not contribute significantly to the model, even when self-declared race/color subgroups were assessed. Nevertheless, we verified a descriptive bias of augmented dose in individuals that carry the polymorphic allele A, but with no statistical significance (corresponding to GA and AA genotypes): GG = 27.7 ± 0.4 (n = 156), GA = 2 9.3 ± 0.5 (n = 126), AA = 28.9 ± 0.9 (n = 27).
Through a genome wide association study (GWAS), Caldwell et al. implicated the CYP4F2 c.1297G>A polymorphism as a minor significant influencer of warfarin dose variability. The authors found that in three independent cohorts of white patients who had a stable warfarin dose (n = 1,009), the CYP4F2 c.1297G>A variant contributed to approximately 1 mg/day more in homozygous AA individuals, when compared to wild-type (Caldwell et al., 2008). Other studies reported similar results (Zhang et al., 2009; Perini et al., 2010; Botton et al., 2011).
In conclusion, our results suggest ABCB1 c.3435C>T polymorphism may marginally influence warfarin dose requirements in Brazilian patients, when associated with other genotypic, demographic and clinical factors.
Ethics Statement
This study was approved by the ethics committee and all included subjects provided written informed consent (SDC: 4519/17/019, CAPPesq: 0804/10).
Author Contributions
All authors have contributed substantially to the conception and design of this paper. LT performed the experiments, collected and analyzed the data, and wrote the paper. LM recruited the patients, collected data and critically revised the manuscript. RS recruited the patients. JK and AP provided the facilities. PS analyzed the data and critically revised the manuscript.
Funding
The authors declare that they have received grants for the support of this study from São Paulo Research Foundation (FAPESP) (grant numbers: #2013/09295-3, #2016/22507-8, and #2016/23454-5) and the Graduate Program in Medical Sciences-FMUSP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We mostly thank the participants of the study and the technical assistance provided by the staff of the Laboratory of Genetics and Molecular Cardiology, Heart Institute (InCor). We also thank São Paulo Research Foundation (FAPESP) for financially supporting the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00542/full#supplementary-material
References
Barnes, G. D., Lucas, E., Alexander, G. C., and Goldberger, Z. D. (2015). National trends in ambulatory oral anticoagulant use. Am. J. Med. 128, 1300–1305.e2. doi: 10.1016/j.amjmed.2015.05.044
Borgiani, P., Ciccacci, C., Forte, V., Sirianni, E., Novelli, L., Bramanti, P., et al. (2009). CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics 10, 261–266. doi: 10.2217/14622416.10.2.261
Botton, M. R., Bandinelli, E., Rohde, L. E., Amon, L. C., and Hutz, M. H. (2011). Influence of genetic, biological and pharmacological factors on warfarin dose in a Southern Brazilian population of European ancestry. Br. J. Clin. Pharmacol. 72, 442–450. doi: 10.1111/j.1365-2125.2011.03942.x
Caldwell, M. D., Awad, T., Johnson, J. A., Gage, B. F., Falkowski, M., Gardina, P. K., et al. (2008). CYP4F2 genetic variant alters required warfarin dose. Blood 111, 4106–4112. doi: 10.1182/blood-2007-11-122010
Cen, H. J., Zeng, W. T., Leng, X. Y., Huang, M., Chen, X., Li, J. L., et al. (2010). CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br. J. Clin. Pharmacol. 70, 234–240. doi: 10.1111/j.1365-2125.2010.03698.x
Cooper, G. M., Johnson, J. A., Langaee, T. Y., Feng, H., Stanaway, I. B., Schwarz, U. I., et al. (2008). A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112, 1022–1027. doi: 10.1182/blood-2008-01-134247
De Oliveira Almeida, V. C., De Souza Ferreira, A. C., Ribeiro, D. D., Gomes Borges, K. B., Salles Moura Fernandes, A. P., and Brunialti Godard, A. L. (2011). Association of the C3435T polymorphism of the MDR1 gene and therapeutic doses of warfarin in thrombophilic patients. J. Thromb. Haemost. 9, 2120–2122. doi: 10.1111/j.1538-7836.2011.04452.x
de Oliveira Almeida, V. C., Ribeiro, D. D., Gomes, K. B., and Godard, A. L. (2014). Polymorphisms of CYP2C9, VKORC1, MDR1, APOE and UGT1A1 genes and the therapeutic warfarin dose in Brazilian patients with thrombosis: a prospective cohort study. Mol. Diagn. Ther. 18, 675–683. doi: 10.1007/s40291-014-0121-4
Frittitta, L., Ercolino, T., Bozzali, M., Argiolas, A., Graci, S., Santagati, M. G., et al. (2001). A cluster of three single nucleotide polymorphisms in the 3'-untranslated region of human glycoprotein PC-1 gene stabilizes PC-1 mRNA and is associated with increased PC-1 protein content and insulin resistance-related abnormalities. Diabetes 50, 1952–1955. doi: 10.2337/diabetes.50.8.1952
Fromm, M. F. (2002). The influence of MDR1 polymorphisms on P-glycoprotein expression and function in humans. Adv. Drug Deliv. Rev. 54, 1295–1310. doi: 10.1016/S0169-409X(02)00064-9
Greiner, B., Eichelbaum, M., Fritz, P., Kreichgauer, H. P., von Richter, O., Zundler, J., et al. (1999). The role of intestinal P-glycoprotein in the interaction of digoxin and rifampin. J. Clin. Invest. 104, 147–153. doi: 10.1172/JCI6663
Hitzl, M., Drescher, S., van der Kuip, H., Schäffeler, E., Fischer, J., Schwab, M., et al. (2001). The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics 11, 293–298. doi: 10.1097/00008571-200106000-00003
Hoffmeyer, S., Burk, O., von Richter, O., Arnold, H. P., Brockmöller, J., Johne, A., et al. (2000). Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 97, 3473–3478. doi: 10.1073/pnas.97.7.3473
Holbrook, A., Schulman, S., Witt, D. M., Vandvik, P. O., Fish, J., Kovacs, M. J., et al. (2012). Evidence-based management of anticoagulant therapy: antithrombotic therapy and prevention of thrombosis, 9th ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 141(2 Suppl.), e152S–e184S. doi: 10.1378/chest.11-2295
Issac, M. S., El-Nahid, M. S., and Wissa, M. Y. (2014). Is there a role for MDR1, EPHX1 and protein Z gene variants in modulation of warfarin dosage? a study on a cohort of the Egyptian population. Mol. Diagn. Ther. 18, 73–83. doi: 10.1007/s40291-013-0055-2
Johnson, J. A., Cavallari, L. H., Beitelshees, A. L., Lewis, J. P., Shuldiner, A. R., and Roden, D. M. (2011). Pharmacogenomics: application to the management of cardiovascular disease. Clin. Pharmacol. Ther. 90, 519–531. doi: 10.1038/clpt.2011.179
Kim, R. B., Leake, B. F., Choo, E. F., Dresser, G. K., Kubba, S. V., Schwarz, U. I., et al. (2001). Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin. Pharmacol. Ther. 70, 189–199. doi: 10.1067/mcp.2001.117412
Kim, Y., Smith, A., and Wu, A. H. (2013). C3435T polymorphism of MDR1 gene with warfarin resistance. Clin. Chim. Acta. 425, 34–36. doi: 10.1016/j.cca.2013.07.010
Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., et al. (2007). A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 doi: 10.1126/science.1135308
Klein, T. E., Altman, R. B., Eriksson, N., Gage, B. F., Kimmel, S. E., et al. (2009). Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 360, 753–764. doi: 10.1056/NEJMoa0809329
Lee, M. T., and Klein, T. E. (2013). Pharmacogenetics of warfarin: challenges and opportunities. J. Hum. Genet. 58, 334–338. doi: 10.1038/jhg.2013.40
Lepper, E. R., Nooter, K., Verweij, J., Acharya, M. R., Figg, W. D., and Sparreboom, A. (2005). Mechanisms of resistance to anticancer drugs: the role of the polymorphic ABC transporters ABCB1 and ABCG2. Pharmacogenomics 6, 115–138. doi: 10.1517/14622416.6.2.115
Morita, Y., Sakaeda, T., Horinouchi, M., Nakamura, T., Kuroda, K., Miki, I., et al. (2003). MDR1 genotype-related duodenal absorption rate of digoxin in healthy Japanese subjects. Pharm. Res. 20, 552–556. doi: 10.1023/A:1023282312757
Moriya, Y., Nakamura, T., Horinouchi, M., Sakaeda, T., Tamura, T., Aoyama, N., et al. (2002). Effects of polymorphisms of MDR1, MRP1, and MRP2 genes on their mRNA expression levels in duodenal enterocytes of healthy Japanese subjects. Biol. Pharm. Bull. 25, 1356–1359. doi: 10.1248/bpb.25.1356
Nakamura, T., Sakaeda, T., Horinouchi, M., Tamura, T., Aoyama, N., Shirakawa, T., et al. (2002). Effect of the mutation (C3435T) at exon 26 of the MDR1 gene on expression level of MDR1 messenger ribonucleic acid in duodenal enterocytes of healthy Japanese subjects. Clin. Pharmacol. Ther. 71, 297–303. doi: 10.1067/mcp.2002.122055
Pena, S. D., Di Pietro, G., Fuchshuber-Moraes, M., Genro, J. P., Hutz, M. H., Fe Kehdy, S., et al. (2011). The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE 6:e17063. doi: 10.1371/journal.pone.0017063
Perini, J. A., Struchiner, C. J., Silva-Assunção, E., and Suarez-Kurtz, G. (2010). Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin. Pharmacol. Ther. 87, 417–420. doi: 10.1038/clpt.2009.307
Perloff, M. D., von Moltke, L. L., Störmer, E., Shader, R. I., and Greenblatt, D. J. (2001). Saint John's wort: an in vitro analysis of P-glycoprotein induction due to extended exposure. Br. J. Pharmacol. 134, 1601–1608. doi: 10.1038/sj.bjp.0704399
Sakaeda, T., Nakamura, T., and Okumura, K. (2003). Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics 4, 397–410. doi: 10.1517/phgs.4.4.397.22747
Salama, N. N., Yang, Z., Bui, T., and Ho, R. J. (2006). MDR1 haplotypes significantly minimize intracellular uptake and transcellular P-gp substrate transport in recombinant LLC-PK1 cells. J. Pharm. Sci. 95, 2293–2308. doi: 10.1002/jps.20717
Santos, P. C., Marcatto, L. R., Duarte, N. E., Gadi Soares, R. A., Cassaro Strunz, C. M., Scanavacca, M., et al. (2015). Development of a pharmacogenetic-based warfarin dosing algorithm and its performance in Brazilian patients: highlighting the importance of population-specific calibration. Pharmacogenomics 16, 865–876. doi: 10.2217/pgs.15.48
Santos, P. C., Soares, R. A., Krieger, J. E., Guerra-Shinohara, E. M., and Pereira, A. C. (2011b). Genotyping of the hemochromatosis HFE p.H63D and p.C282Y mutations by high-resolution melting with the Rotor-Gene 6000® instrument. Clin. Chem. Lab. Med. 49, 1633–1636. doi: 10.1515/CCLM.2011.654
Santos, P. C., Soares, R. A., Santos, D. B., Nascimento, R. M., Coelho, G. L., Nicolau, J. C., et al. (2011a). CYP2C19 and ABCB1 gene polymorphisms are differently distributed according to ethnicity in the Brazilian general population. BMC Med. Genet. 12:13. doi: 10.1186/1471-2350-12-13
Santos, R. V., Fry, P. H., Monteiro, S., Maio, M. C., Rodrigues, J. C., Bastos-Rodrigues, L., et al. (2009). Color, race, and genomic ancestry in Brazil: dialogues between anthropology and genetics. Curr. Anthropol. 50, 787–819. doi: 10.1086/644532
Saraeva, R. B., Paskaleva, I. D., Doncheva, E., Eap, C. B., and Ganev, V. S. (2007). Pharmacogenetics of acenocoumarol: CYP2C9, CYP2C19, CYP1A2, CYP3A4, CYP3A5 and ABCB1 gene polymorphisms and dose requirements. J. Clin. Pharm. Ther. 32, 641–649. doi: 10.1111/j.1365-2710.2007.00870.x
Scheiner, M. A., Damasceno, A. M., and Maia, R. C. (2010). ABCB1 single nucleotide polymorphisms in the Brazilian population. Mol. Biol. Rep. 37, 111–118. doi: 10.1007/s11033-009-9547-x
Soares, R. A., Santos, P. C., Machado-Coelho, G. L., do Nascimento, R. M., Mill, J. G., Krieger, J. E., et al. (2012). CYP2C9 and VKORC1 polymorphisms are differently distributed in the Brazilian population according to self-declared ethnicity or genetic ancestry. Genet. Test. Mol. Biomarkers 16, 957–963. doi: 10.1089/gtmb.2012.0019
Takeuchi, F., McGinnis, R., Bourgeois, S., Barnes, C., Eriksson, N., Soranzo, N., et al. (2009). A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 5:e1000433. doi: 10.1371/journal.pgen.1000433
van Gorp, R. H., and Schurgers, L. J. (2015). New insights into the pros and cons of the clinical use of vitamin K antagonists (VKAs) versus direct oral anticoagulants (DOACs). Nutrients 7, 9538–9557. doi: 10.3390/nu7115479
Wadelius, M., Sörlin, K., Wallerman, O., Karlsson, J., Yue, Q. Y., Magnusson, P. K., et al. (2004). Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 4, 40–48. doi: 10.1038/sj.tpj.6500220
Wei, M., Ye, F., Xie, D., Zhu, Y., Zhu, J., Tao, Y., et al. (2012). A new algorithm to predict warfarin dose from polymorphisms of CYP4F2, CYP2C9 and VKORC1 and clinical variables: derivation in Han Chinese patients with non valvular atrial fibrillation. Thromb. Haemost. 107, 1083–1091. doi: 10.1160/TH11-12-0848
Witt, D. M., Clark, N. P., Kaatz, S., Schnurr, T., and Ansell, J. E. (2016). Guidance for the practical management of warfarin therapy in the treatment of venous thromboembolism. J. Thromb. Thrombolysis 41, 187–205. doi: 10.1007/s11239-015-1319-y
Keywords: warfarin pharmacogenetics, ABCB1, MDR1, CYP4F2, warfarin stable dose
Citation: Tavares LC, Marcatto LR, Soares RAG, Krieger JE, Pereira AC and Santos PCJL (2018) Association Between ABCB1 Polymorphism and Stable Warfarin Dose Requirements in Brazilian Patients. Front. Pharmacol. 9:542. doi: 10.3389/fphar.2018.00542
Received: 15 March 2018; Accepted: 04 May 2018;
Published: 23 May 2018.
Edited by:
Rick Kittles, Irell and Manella Graduate School of Biological Sciences, City of Hope, United StatesReviewed by:
Jason Hansen Karnes, University of Arizona, United StatesAdam P. Bress, University of Utah, United States
Copyright © 2018 Tavares, Marcatto, Soares, Krieger, Pereira and Santos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulo C. J. L. Santos, cGF1bG8uY2FsZWJAdW5pZmVzcC5icg==
 Letícia C. Tavares
Letícia C. Tavares Leiliane R. Marcatto
Leiliane R. Marcatto Renata A. G. Soares1
Renata A. G. Soares1 Jose E. Krieger
Jose E. Krieger Alexandre C. Pereira
Alexandre C. Pereira Paulo C. J. L. Santos
Paulo C. J. L. Santos