- 1McKusick-Zhang Center for Genetic Medicine, Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, School of Basic Medicine Peking Union Medical College, Beijing, China
- 2The Research Center for Medical Genomics, China Medical University, Shenyang, China
- 3Developmental Medicine Department, King Abdullah International Medical Research Center (KAIMRC), King Saud Bin Abdulaziz University for Health Sciences, Ministry of National Guard-Health Affairs (MNGHA), Riyadh, Saudi Arabia
- 4Medical Genomics Research Department, King Abdullah International Medical Research Center (KAIMRC), King Saud Bin Abdulaziz University for Health Science, Ministry of National Guard-Health Affairs (MNGHA), Riyadh, Saudi Arabia
- 5Department of Biochemistry, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan
Split hand/split foot malformation (SHFM) or ectrodactyly is characterized by a deep median cleft of the hand or foot, hypoplasia or aplasia of the metacarpals, metatarsals, and phalanges. It is a clinically and genetically heterogeneous group of limb malformations. This study aimed to identify the pathogenic variant in a consanguineous Pakistani family with autosomal recessive SHFM. Peripheral blood samples were obtained, DNA was extracted, WNT10B coding and noncoding regions were PCR amplified and Sanger sequencing was performed using workflow suggested by Thermo Fisher Scientific. A novel homozygous nonsense variant (c.1098C>A; p.Cys366*) was identified in the WNT10B gene in the index patients, which probably explains SHFM type 6 in this family in comparison with similar data from the literature.
Introduction
Split-hand/split-foot malformation (SHFM; OMIM:225300) or ectrodactyly is a rare limb developmental disorder. It is characterized by a deep median cleft of the hand or foot corresponding to the central rays of the autopod (1, 2). Phalangeal aplasia and hypoplasia of metacarpals and metatarsals are also signature features of SHFM. It has an incidence rate of ~1 in 90,000 live births (3, 4). Both individuals and families with SHFM can have a highly variable presentation ranging from minor clefting at the central ray to severe, lobster claw-like deformity (5, 6). It can exist as an isolated entity or as a complex syndrome. Few patients have been reported exhibiting craniofacial defects, ectodermal dysplasia or intellectual disability (5–7). Typically, SHFM is inherited as an autosomal dominant trait with incomplete penetrance; however, several cases have been reported with an autosomal recessive or X-linked inheritance pattern (7–9). To date, eight types of non-syndromic SHFM were described and mapped to different chromosomes. Four autosomal dominant types have been mapped to chromosomes 2q31 (SHFM-5; MIM 606708), 3q27 (SHFM-4; MIM 605289), 7q21 (SHFM-1; MIM 183600), 10q24 (SHFM-3; MIM 246560), and the X-linked form to Xq26.3 (SHFM-2; MIM 313350) (10–15). The autosomal recessive SHFM-6 (MIM 225300) was mapped to chromosome 12q13.12 harboring the wingless-type MMTV integration site family member 10 (WNT10B, MIM 601906) (8, 9). Mutations in several genes have been associated with SHFM including TP63, DLX5, DLX6, ZAK, FGFR1, EPS15L1, and WNT10B; however, the clinical features are often indistinguishable (16–18). In the zebrafish, the expression of TP63, DLX5, DLX6, FGFR1, and WNT10B was shown in the fin's apical ectodermal ridge (AER) cells (16). A similar pattern was also observed in the mouse limbs (19).
In this study, we described a consanguineous family of Pakistani origin showing SHFM with recessive inheritance. Sanger sequencing of WNT10B was used to possibly detect a novel homozygous nonsense mutation that co-segregating with SHFM phenotype within the studied family. We provided a brief phenotype comparison of this family to other cases described in the literature.
Case Presentation
Methods
Ethics and Consent Approval
The study design and protocol were approved by the Institutional Review Board (IRB) at Quaid-i-Azam University Islamabad Pakistan, the Ethical Review Committee (ERC) of Peking Union Medical College (Beijing, China), and China Medical University (Shenyang, China). For minors, written informed consent was signed by their parents.
Patients and DNA Sample Collection
The two index patients (IV:1, IV:2) of Pakistani origin were children from a consanguineous marriage (Figure 1A). Blood samples were collected from individuals III:1, III:2, IV:1, IV:2, and IV:3. The QIAquick DNA extraction kit (QIAamp, Qiagen, Valencia, CA, USA) was used for genomic DNA extraction. The DNA quantity and quality were assessed using nanodrop-2000 spectrophotometer (Thermo Scientific, Schaumburg, IL, USA).
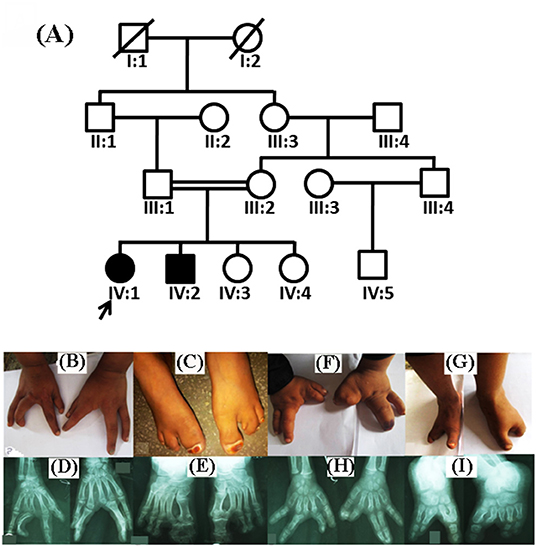
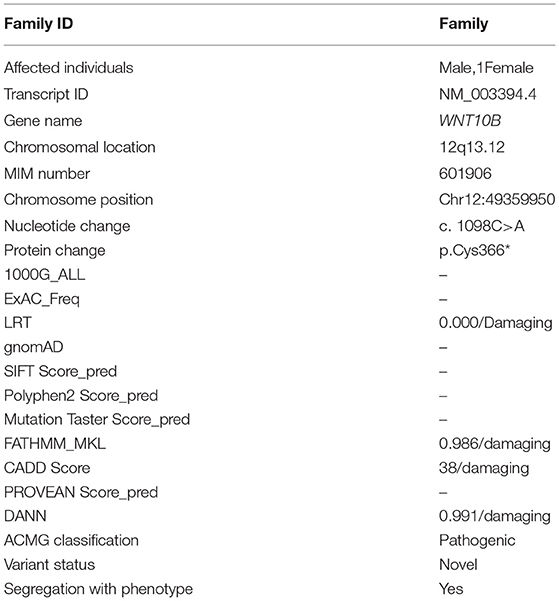
Figure 1. Pedigree analysis and clinical features of split hand/foot malformation (SHFM) observed in the family. (A) A consanguineous pedigree showing two affected members (IV:1 and IV:2) in the fourth generation. Affected individuals in the pedigree are shown with shaded symbols and unaffected with open symbols. (B,C) 14 years old affected individual (IV:1), showing fusion of proximal phalanx with a median cleft in both hands, and clinodactyly in the left hand. Feet showing complex pre axial syndactyly. (D,E) Radiographic examination of the affected individual (IV:1) revealed median cleft in both hands, aplasia, with complex syndactyly, while feet showed complex preaxail syndactyly of the toes and 1st and 2nd metatarsals. (F,G) Affected individual (IV:2) having aplasia in both hands and feet, missing digits and complex syndactyly, feet exhibiting typical median cleft phenotypes. (H,I) Radiographs of the same affected individual revealed missing digits in both hands and feet, complex preaxial and postaxial syndactyly.
Sanger Sequencing of WNT10B
Genomic sequence of WNT10B gene, including exons, introns, 5′ untranslated region (UTR) and 3′ UTR, was retrieved from the University of California Santa Cruz genome database browser (UCSC; http://genome.ucsc.edu/). Oligonucleotide primer pairs of the five exons in WNT10B were designed with Gene Runner software (version 5.0.69 Beta; Hastings Software, Inc., Hastings, NY) (Table 1). For all family members, all coding and noncoding regions of the WNT10B was PCR-amplified in 20 μL reaction volume with 10 pMol of each primer pair and sequenced by Sanger's method after purification. Standard sequencing protocol was followed using BigDye® Terminator v3.1 and Cycle Sequencing Kit (Applied Biosystems, USA). BioEdit tool was applied to analyze and detect mutation in sequenced data using NCBI GeneBank accession number [NG_023347.1] as a reference for alignment. In addition, 200 ethnically-matched control samples were sequenced to assess the allele frequency of the novel variant.
Results
Clinical and Radiological Examinations
Both patients (IV:1 and IV:2) had a physical examination and X-rays performed by an orthopedic surgeon. Patients during the time of recruitment for genetic analysis were 14 (IV:1) and 12 (IV:2) years of age. Both patients of the family exhibited SHFM phenotype with involvement of hands and feet segregated in an autosomal recessive manner. Parents of the affected patients were normal and healthy. The patient IV:1 had complex syndactyly, clinodactyly, dysplastic hands and feet (Figures 1B–E). In contrast, the patient IV:2 had syndactyly in the left hand and both feet, aplasia of the radial ray of hand and hallux valgus deformity in the big toe. The distal phalanx of the middle finger was also missing. Photos of the hands and feet are shown in Figures 1F–I. No other dysmorphic features were observed. Both patients had an average intellect and were attending regular school classes with satisfactory performance.
Radiological Examinations
During the radiological evaluation, the patient IV:1 showed an absence of the first metacarpal and multiple phalanges in both hands. Both feet had deep midline cleft and syndactyly (Figures 1B–E). Radiographic examination of Patient IV:2 showed the absence of multiple metatarsals and phalanges in both feet (Figures 1F–I). Detailed clinical information of both patients were compared to previously reported SHFM cases as seen in Table 2.

Table 2. Clinical features and WNT10B variations detected in the present family and those families reported previously.
Mutation Confirmation
Analysis of Sanger sequencing shown in Figure 2A identified a homozygous nonsense variant (c.1098C>A; p.Cys366*) in exon 5 of the WNT10B gene in both affected individuals (IV:1 and IV:2) of the family. Their parents were heterozygous for the same variant (Figure 2B). This variant leads to premature termination of the 389 amino acid protein in the main WNT domain (Figure 2C). Other vertebrate species indicated in Figure 1D share similar variant and an overall gene sequence.
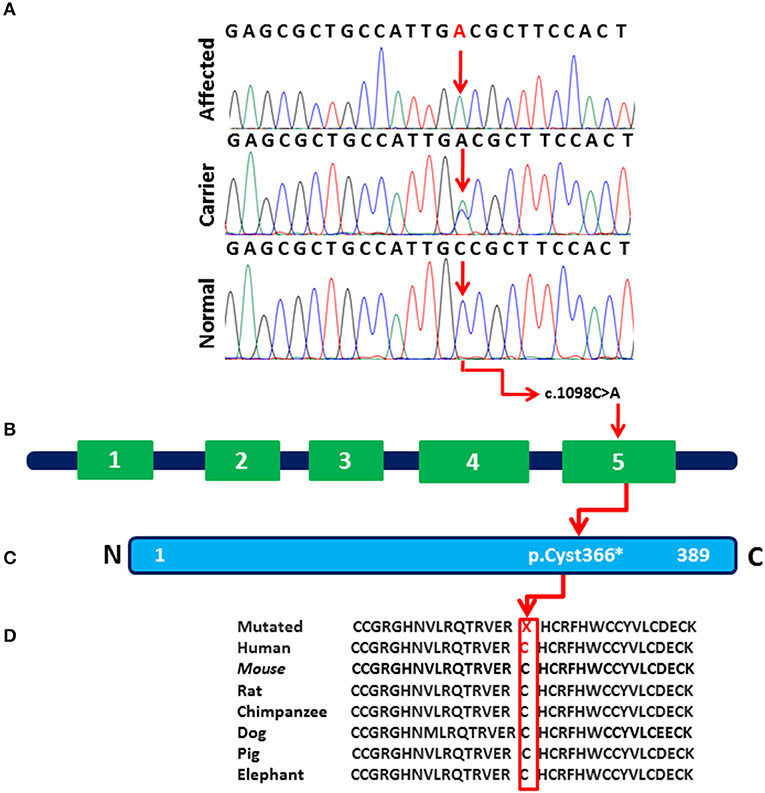
Figure 2. (A) Sequence chromatogram of the WNT10B gene showing a homozygous nonsense mutation (c.1098C>A; p.Cys366*) in affected individuals (IV:1 and IV:2), a heterozygous variant in (III:1, III:2, IV:3) and control reference in 200 ethnically-matched normal individuals. (B) The exon/intron structure of the WNT10B gene along with reported mutation causing SHFM phenotypes. Variant (c.1098C>A) marked with black color represents our index patients. (C) Variant c.1098C>A (p.Cys366*) detected in the present study affect the last region (amino acids 367–389) of the WNT10B domain. (D) The amino acids mutated in the affected individuals are conserved in different species.
In-silico Analysis
The novelty and pathogenicity of the identified variant were predicted using different online in silico analysis tools including Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org/), Protein Variation Effect Analyzer (PROVEAN, http://provean.jcvi.org), Mutation Taster (http://www.mutationtaster.org/), Varsome (https://varsome.com/), and Combined Annotation Dependent Depletion (CADD, https://cadd.gs.washington.edu/). Finally, for the interpretation of variants, American College of Medical Genetics and Genomics (ACMG) 2015 guidelines were used (20) (Table 3). The variant (c.1098C>A; p.Cys366*) was not observed in gnomAD (https://gnomad.broadinstitute.org/), 1000 Genomes (http://www.1000genomes.org/), dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), and in the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org) and was not present in 200 ethnically-matched control individuals.
Discussion
In this study, we identified a novel homozygous nonsense variant c.1098C>A in WNT10B in a consanguineous family having SHFM. The nonsense variant (c.1098C>A; p.Cys366*) results in a Cysteine amino acid substitution to stop codon at a position 366 which results in a shorter protein formation (21). A number of families has been reported previously from Pakistan having different mutations in the WNT10B leading to SHFM phenotypes (22–24). Khan et al. (9), Aziz et al. (24), and Ullah et al. (25), studied several Pakistani consanguineous SHFM families using linkage analysis followed by direct sequencing and identified pathogenic variants in the WNT10B gene (9, 24, 25). The Clinical features of affected members in these families exhibited SHFM phenotype, which is inherited in the form of autosomal recessive pattern. The SHFM features observed in our patients show similarities to those reported previously (8, 9, 24, 26). Detailed clinical comparisons of our patients with that reported earlier are presented in Table 2.
During organogenesis, Wnt signaling plays a significant role in proximal-distal outgrowth as well as dorso-ventral patterning of limb formations (27). Wnt signaling is essential in cartilage, bone, muscle and joint development (28). Other WNT genes such as WNT3, WNT4, WNT6, WNT7A, WNT7B, WNT9B, WNT10A, and WNT16 as well as WNT10B show higher expression throughout the limb bud ectoderm in all phases of mouse limb formation with the exception of the apical ectodermal ridge where Wnt10b expression is only seen at embryonic day 11.5 (E11.5) (29, 30).
A few individuals were described carrying WNT10B variants exhibiting developmental tooth alterations, low bone mass or obesity but causality was not established (25, 26, 31, 32). Such phenotypes were not observed in our patients. Until now, 20 different types of (missense, nonsense, splice site, and frameshift) mutation have been identified in the WNT10B gene causing different human disorders including obesity, dental anomalies, and SHFM (Table 4) (HGMD: http://www.hgmd.org). Among the 20 HGMD reported mutations, only 12 mutations showed associations with SHFM anomalies. In line with previous reports, the WNT10B mutation (p.Cys366*) identified in this study is predicted to affect the development of the hands and feet resulting in SHFM type 6. Our findings support the vital function of WNT10B in the human skeleton development.
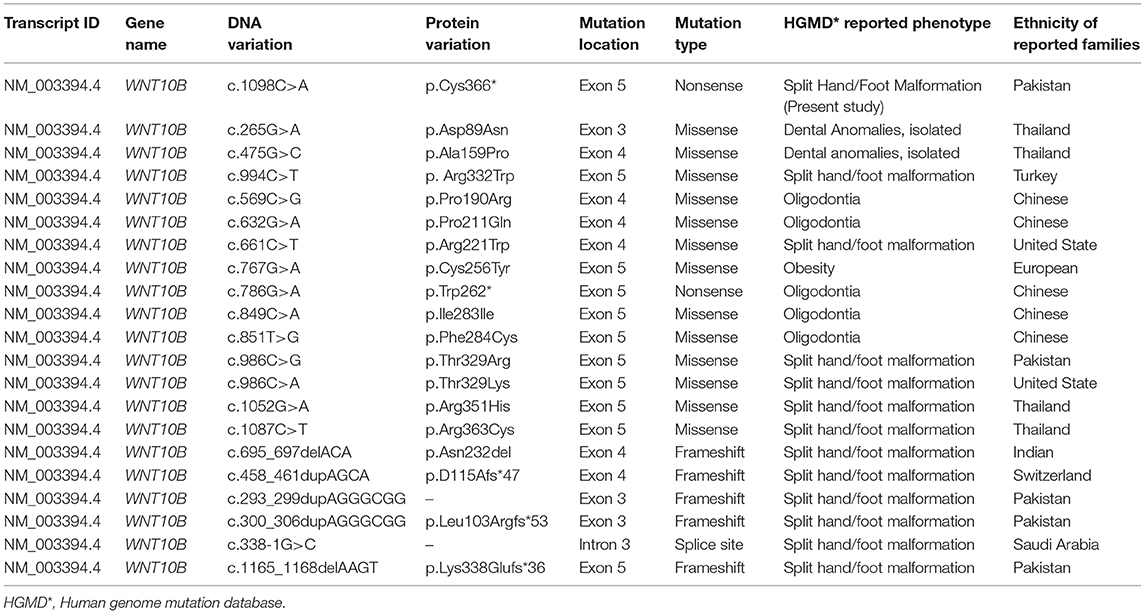
Table 4. Mutations identified in WNT10B gene and associated HGMD disorders identified in different ethnic groups.
Conclusion
We have reported a novel sequence variant (c.1098C>A; p.Cys366*) in the WNT10B gene in a consanguineous Pakistani family presenting with SHFM type 6. This study further extended the spectrum of mutations in the WNT10B gene which might be helpful in proper molecular diagnosis and genetic counseling.
Web Resources
1000 Genomes—http://www.1000genomes.org/;
Exome Variant Server—http://evs.gs.washington.edu/EVS/;
GnomAD—https://gnomad.broadinstitute.org;
dbSNP—http://www.ncbi.nlm.nih.gov/SNP/;
OMIM—http://www.omim.org/;
HGMD—http://www.biobase-international.com/products/hgmd
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The study design and protocol were approved by the Institutional Review Board (IRB) at Quaid-i-Azam University Islamabad Pakistan, the Ethical Review Committee (ERC) of Peking Union Medical College (Beijing, China), and China Medical University (Shenyang, China). For minors, written informed consent was signed by their parents. Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
AK and RW participated in the design of the study, performed PCR, gene sequencing, and manuscript writing. AK, MU, MAA, and SH studied family, collected blood samples, and extracted DNA. MAn, WA, and MAl participated in manuscript preparation and XZ collected funds and supervised the study progress. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2016YFC0905100), the CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant Nos. 2017-I2M-B&R-05 and 2016-I2M-1-002), the National Natural Science Foundation of China (NSFC) (Grant No. 81230015), the Beijing Municipal Science and Technology Commission (Grant No. Z151100003915078), and the Central Research Institutes of Basic Research and Public Service Special Operations (Grant No. 2018PT32024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Dr. Peter Georgics, the University of Michigan for helpful comments on the manuscript. We thank all the individuals for their participation in this study.
References
1. Bernardini L, Palka C, Ceccarini C, Capalbo A, Bottillo I, Mingarelli R. Complex rearrangement of chromosomes 7q21.13-q22.1 confirms the ectrodactyly-deafness locus and suggests new candidate genes. Am J Med Genet A. (2008) 146A:238–44. doi: 10.1002/ajmg.a.32093
2. Klar AJ. Split hand/foot malformation genetics supports the chromosome 7 copy segregation mechanism for human limb development. Philos Trans R Soc Lond B Biol Sci. (2016) 371:20150415. doi: 10.1098/rstb.2015.0415
3. Gane BD, Natarajan P. Split-hand/feet malformation: a rare syndrome. J Fam Med Prim Care. (2016) 5:168–9. doi: 10.4103/2249-4863.184656
4. Yu P, Yang W, Han D, Wang X, Guo S, Li J, et al. Mutations in WNT10B are identified in individuals with oligodontia. Am J Hum Genet. (2016) 99:195–201. doi: 10.1016/j.ajhg.2016.05.012
5. Duijf PH, van Bokhoven H, Brunner HG. Pathogenesis of split-hand/split-foot malformation. Hum Mol Genet. (2003) 12:R51–60. doi: 10.1093/hmg/ddg090
6. Elliott AM, Evans JA. The association of split hand foot malformation (SHFM) and congenital heart defects. Birth Defects Res A Clin Mol Teratol. (2008) 82:425–34. doi: 10.1002/bdra.20452
7. Umair M, Ullah A, Abbas S, Ahmad F, Basit S, Ahmad W. First direct evidence of involvement of a homozygous loss-of-function variant in the EPS15L1 gene underlying split-hand/split-foot malformation. Clin Genet. (2018) 93:699–702. doi: 10.1111/cge.13152
8. Ugur SA, Tolun A. Homozygous WNT10b mutation and complex inheritance in split-hand/foot malformation. Hum Mol Genet. (2008) 17:2644–53. doi: 10.1093/hmg/ddn164
9. Khan S, Basit S, Zimri FK, Ali N, Ali G, Ansar M, et al. A novel homozygous missense mutation in WNT10B in familial split-hand/foot malformation. Clin Genet. (2012) 82:48–55. doi: 10.1111/j.1399-0004.2011.01698.x
10. Boles RG, Pober BR, Gibson LH, Willis CR, McGrath J, Roberts DJ, et al. Deletion of chromosome 2q24-q31 causes characteristic digital anomalies: case report and review. Am J Med Genet. (1995) 55:155–60. doi: 10.1002/ajmg.1320550204
11. Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. (2000) 67:59–66. doi: 10.1086/302972
12. Al Ghamdi A, Al-Qattan MM, Hadadi A, Alabdulrahman A, Almuzzaini B, Alatwi N, et al. A classification system for split-hand/ foot malformation (SHFM): a proposal based on 3 pedigrees with WNT10B mutations. Eur J Med Genet. (2019) 14:103738. doi: 10.1016/j.ejmg.2019.103738
13. Nunes ME, Schutt G, Kapur RP, Luthardt F, Kukolich M, Byers P, et al. A second autosomal split hand/split foot locus maps to chromosome 10q24-q25. Hum Mol Genet. (1995) 4:2165–70. doi: 10.1093/hmg/4.11.2165
14. Raas-Rothschild A, Manouvrier S, Gonzales M, Farriaux JP, Lyonnet S, Munnich A. Refined mapping of a gene for split hand-split foot malformation (SHFM3) on chromosome 10q25. J Med Genet. (1996) 33:996–1001. doi: 10.1136/jmg.33.12.996
15. Faiyaz-Ul-Haque M, Zaidi SH, King LM, Haque S, Patel M, Ahmad M, et al. Fine mapping of the X-linked split-hand/split-foot malformation (SHFM2) locus to a 5.1-Mb region on Xq26.3 and analysis of candidate genes. Clin Genet. (2005) 67:93–7. doi: 10.1111/j.1399-0004.2004.00369.x
16. Merlo GR, Paleari L, Mantero S, Genova F, Beverdam A, Palmisano GL, et al. Mouse model of split hand/foot malformation type I. Genesis. (2002) 33:97–101. doi: 10.1002/gene.10098
17. Shamseldin HE, Faden MA, Alashram W, Alkuraya FS. Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J Med Genet. (2012) 49:16–20. doi: 10.1136/jmedgenet-2011-100556
18. Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. (2002) 2:607–16. doi: 10.1016/S1534-5807(02)00166-1
19. Lo Iacono N, Mantero S, Chiarelli A, Garcia E, Mills AA, Morasso MI, et al. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. (2008) 135:1377–88. doi: 10.1242/dev.011759
20. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. (2015) 17:405–24. doi: 10.1038/gim.2015.30
21. Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. (2005) 118(Pt 9):1773–6. doi: 10.1242/jcs.01701
22. Umair M, Ahamd F, Bilal M, Asiri A, Younus M, Khan A. A comprehensive review of genetic skeletal disorders reported from Pakistan: a brief commentary. Meta Gene. (2019) 20:100559. doi: 10.1016/j.mgene.2019.100559
23. Umair M, Hayat A. Nonsyndromic split-hand/foot malformation: recent classification. Mol Syndromol. (2019) 10:243–54. doi: 10.1159/000502784
24. Aziz A, Irfanullah, Khan S, Zimri FK, Muhammad N, Rashid S, et al. Novel homozygous mutations in the WNT10B gene underlying autosomal recessive split hand/foot malformation in three consanguineous families. Gene. (2014) 534:265–71. doi: 10.1016/j.gene.2013.10.047
25. Ullah A, Gul A, Umair M, Irfanullah, Ahmad F, Aziz A, et al. Homozygous sequence variants in the WNT10B gene underlie split hand/foot malformation. Genet Mol Biol. (2018) 41:1–8. doi: 10.1590/1678-4685-gmb-2016-0162
26. Kantaputra PN, Kapoor S, Verma P, Intachai W, Ketudat Cairns JR. Split hand-foot malformation and a novel WNT10B mutation. Eur J Med Genet. (2018) 61:372–5. doi: 10.1016/j.ejmg.2018.02.001
27. Salva JE, Merrill AE. Signaling networks in joint development. Dev Dyn. (2017) 246:262–74. doi: 10.1002/dvdy.24472
28. Stevens JR, Miranda-Carboni GA, Singer MA, Brugger SM, Lyons KM, Lane TF. Wnt10b deficiency results in age-dependent loss of bone mass and progressive reduction of mesenchymal progenitor cells. J Bone Miner Res. (2010) 25:2138–47. doi: 10.1002/jbmr.118
29. Witte F, Dokas J, Neuendorf F, Mundlos S, Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Exp Patterns. (2009) 9:215–23. doi: 10.1016/j.gep.2008.12.009
30. Christodoulides C, Scarda A, Granzotto M, Milan G, Dalla Nora E, Keogh J, et al. WNT10B mutations in human obesity. Diabetologia. (2006) 49:678–84. doi: 10.1007/s00125-006-0144-4
31. Zhou H, Mak W, Zheng Y, Dunstan CR, Seibel MJ. Osteoblasts directly control lineage commitment of mesenchymal progenitor cells through Wnt signaling. J Biol Chem. (2008) 283:1936–45. doi: 10.1074/jbc.M702687200
Keywords: Pakistani family, SHFM, autosomal recessive mode, gene variant, non-sense mutation
Citation: Khan A, Wang R, Han S, Umair M, Alshabeeb MA, Ansar M, Ahmad W, Alaamery M and Zhang X (2020) A Novel Homozygous Nonsense Mutation p.Cys366* in the WNT10B Gene Underlying Split-Hand/Split Foot Malformation in a Consanguineous Pakistani Family. Front. Pediatr. 7:526. doi: 10.3389/fped.2019.00526
Received: 11 June 2019; Accepted: 04 December 2019;
Published: 09 January 2020.
Edited by:
Merlin G. Butler, University of Kansas Medical Center, United StatesReviewed by:
Muzammil Ahmad Khan, Gomal University, PakistanFarooq Ahmad, Woman University Swabi, Pakistan
Copyright © 2020 Khan, Wang, Han, Umair, Alshabeeb, Ansar, Ahmad, Alaamery and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Zhang, eHVlemhhbmcmI3gwMDA0MDtwdW1jLmVkdS5jbg==
†These authors have contributed equally to this work
 Amjad Khan
Amjad Khan Rongrong Wang1†
Rongrong Wang1† Shirui Han
Shirui Han Muhammad Umair
Muhammad Umair Mohammad A. Alshabeeb
Mohammad A. Alshabeeb Xue Zhang
Xue Zhang