- 1Institute of Molecular Biology and Pathology, National Research Council, Rome, Italy
- 2Department of Biology and Biotechnology, Sapienza University of Rome, Rome, Italy
Central Nervous System tumors are the leading cause of cancer-related death in children, and medulloblastoma has the highest incidence rate. The current therapies achieve a 5-year survival rate of 50–80%, but often inflict severe secondary effects demanding the urgent development of novel, effective, and less toxic therapeutic strategies. Historically identified on a histopathological basis, medulloblastoma was later classified into four major subgroups—namely WNT, SHH, Group 3, and Group 4—each characterized by distinct transcriptional profiles, copy-number aberrations, somatic mutations, and clinical outcomes. Additional complexity was recently provided by integrating gene- and non-gene-based data, which indicates that each subclass can be further subdivided into specific subtypes. These deeper classifications, while getting over the typical tumor heterogeneity, indicate that different forms of medulloblastoma hold different molecular drivers that can be successfully exploited for a greater diagnostic accuracy and for the development of novel, targeted treatments. Long noncoding RNAs are transcripts that lack coding potential and play relevant roles as regulators of gene expression in mammalian differentiation and developmental processes. Their cell type- and tissue-specificity, higher than mRNAs, make them more informative about cell- type identity than protein-coding genes. Remarkably, about 40% of long noncoding RNAs are expressed in the brain and their aberrant expression has been linked to neuro-oncological disorders. However, while their involvement in gliomas and neuroblastomas has been extensively studied, their role in medulloblastoma is still poorly explored. Here, we present an overview of current knowledge regarding the function played by long noncoding RNAs in medulloblastoma biology.
Introduction
Medulloblastoma (MB), with an estimated 5000–8000 cases/year worldwide (1, 2), is an aggressive tumor arising in the cerebellum. It mainly affects children and is a major cause of mortality in pediatric oncology (3). While the previous classification of MB by the World Health Organization (WHO) was largely based on histological features (4), the new classification in 2016 exploited molecular parameters to catalog the large variety of tumors of the Central Nervous System (CNS) (5). Rational molecular-based classification was supported by the advancement of sequencing technologies allowing extensive genomic/transcriptomic studies. This classification benefits from the integration between histological and molecular parameters and led to no longer considering MB as a unique pathology. Several subclasses of MB have been unveiled, each displaying dysregulated genes—the driver genes—altered by single nucleotide mutations, somatic copy-number aberrations, or by defects in transcriptional or post-transcriptional gene regulation.
In this review, we highlight the link between MB tumors and the emerging class of regulatory long noncoding RNAs (lncRNAs), and their potential as promising cancer biomarkers and novel therapeutic agents.
Medulloblastoma
Recent genomic and transcriptomic analyses on a large cohort of primary tumors assigned MBs to four molecularly distinct subgroups (6, 7). They include the extensively characterized WNT and SHH subgroups, and the Group 3 (G3) and Group 4 (G4), whose pathogenesis and signaling pathways are poorly defined.
WNT
Approximately 10% of all MB patients belong to this subgroup, characterized by the most favorable prognosis with 95% of survival (7, 8). WNT tumors, which exhibit classic histology, are recognizable by a WNT gene expression signature. Nuclear accumulation of β-catenin is considered a biomarker for WNT signaling pathway activation. This subgroup often carries heterozygous TP53 mutations, as well as mutations in the DEAD-box helicase gene DDX3X and in chromatin modifiers genes, such as SMARCA4 and CREBBP, indicating the implication of altered epigenome in the development of this disease. Integration of gene expression and DNA methylation profiles indicated that WNT subgroup comprises at least two subtypes, WNTα, mainly enriched for children and characterized by monosomy 6, and WNTβ, including mainly adults without monosomy 6 (9).
SHH
SHH MBs represent approximately 30% of all MB cases, characterized by an intermediate prognosis, with survival rates ranging from 60 to 80% (7). SHH tumors, mainly exhibiting desmoplastic histology, display an aberrant activation of the SHH signaling, due to mutations of negative regulators of SHH pathway, such as PTCH1 and SUFU, and copy number aberrations of SHH target genes, such as MYCN and GLI2 (7). TP53 mutations are found in about 30% of childhood SHH MBs and are associated with extremely poor outcomes. Recent analyses suggest that SHH subgroup consists of four distinct subtypes. It includes SHHα, enriched for MYCN and GLI2 amplifications, with the worst prognosis; SHHβ, harboring PTEN gene deletions and frequently metastatic; SHHγ displaying scarce copy number aberrations and SHHδ, that is enriched for TERT gene promoter mutations and has a favorable prognosis (9).
Group 3
G3 is the most aggressive subgroup accounting for about 25% of all MBs, about half of them being metastatic at diagnosis (10). These tumors display a MYC signature, being characterized by amplification of the MYC proto-oncogene and exhibiting aberrant MYC expression in almost all cases (7). G3 shows intra-tumoral heterogeneity, including three further subtypes: G3α and G3β with a more favorable prognosis compared to G3γ, which frequently harbors increased MYC copy number (9).
Group 4
G4, the most common subtype, accounts for 35% of all MBs. These tumors are often metastatic at diagnosis and have intermediate prognosis. It is the most enigmatic subgroup, characterized by a neuronal gene expression signature, resembling that of glutamatergic neurons (7). Common alterations pertain to inactivating mutations in KDM6A gene, duplication of SNCAIP gene, and amplification of MYCN and CDK6 proto-oncogenes. G4 has been re-classified into G4α, characterized by MYCN and CDK6 amplifications, G4β, strongly enriched for SNCAIP duplications and putative PRDM6 overexpression, and G4γ enriched for focal CDK6 amplification (9).
Long Noncoding RNAs
RNA is considered as the most “rediscovered” biological macromolecule (11, 12) since, starting from the informational role assigned to mRNAs in 1961 (13, 14), novel unexpected functions have been attributed to RNA in the last three decades. In the 1980s, its capacity to catalyze biochemical reactions was associated with its ability to fold into complex tridimensional structures (15). In the early 1990s, regulatory functions were attributed to two long RNAs lacking protein-coding capacity, H19 (16, 17), and XIST (18, 19). Since then, a huge number of noncoding RNAs, both short and long in size, was discovered in parallel with the finding that more than half of the transcriptome encodes non-proteinogenic transcripts. Among them, the lncRNAs number in the tens of thousands and include also circular RNAs, covalently closed RNA circles derived from back-splicing of linear transcripts (20). LncRNAs are >200 nucleotides and represent very versatile molecules for their unique ability to specifically recognize both nucleic acids and protein partners via base-pairing and modular tridimensional structures, respectively. They are flexibly involved in important biological processes, such as development, cell differentiation and growth, thanks to their main functions of gene expression regulators and the genome structure architects.
Mechanisms of Action
LncRNAs may be engaged in fine-scale modulation of gene expression as well as in large-scale control of developmental programs. They may act through a variety of mechanisms, depending on their cellular localization. Some of them are exclusively localized in the nucleus, others in the cytoplasm, others change their localization during development or differentiation, and still others show both localizations. In the latter case, a single lncRNA might have multiple molecular functions.
Nuclear lncRNAs
Nuclear lncRNAs (Figure 1A) can be found in the nucleoplasm or associated with chromatin (21). Typically, these latter are supposed to control protein-coding gene expression at the epigenetic level by recruiting chromatin modifiers to specific genomic loci. This is achieved through their scaffolding activity, by which they interact simultaneously with distinct protein complexes, and through their capability to act as “molecular guides,” that ensure the specificity of target recognition (21). This function can be carried out in cis or in trans. The cis-acting RNAs are typically low-abundant, and regulated genes are located in the proximity of their transcription site; trans-acting RNAs are more abundant and can modulate the expression of genes at independent loci (21). Notably, perturbations of the epigenetic regulation were recognized as causative of malignancies (22), and some cancer-related lncRNAs, such as XIST (23), HOTAIR (24–26), NBAT (27), and LINC-PINT (28), were reported to direct epigenetic modifications (29).
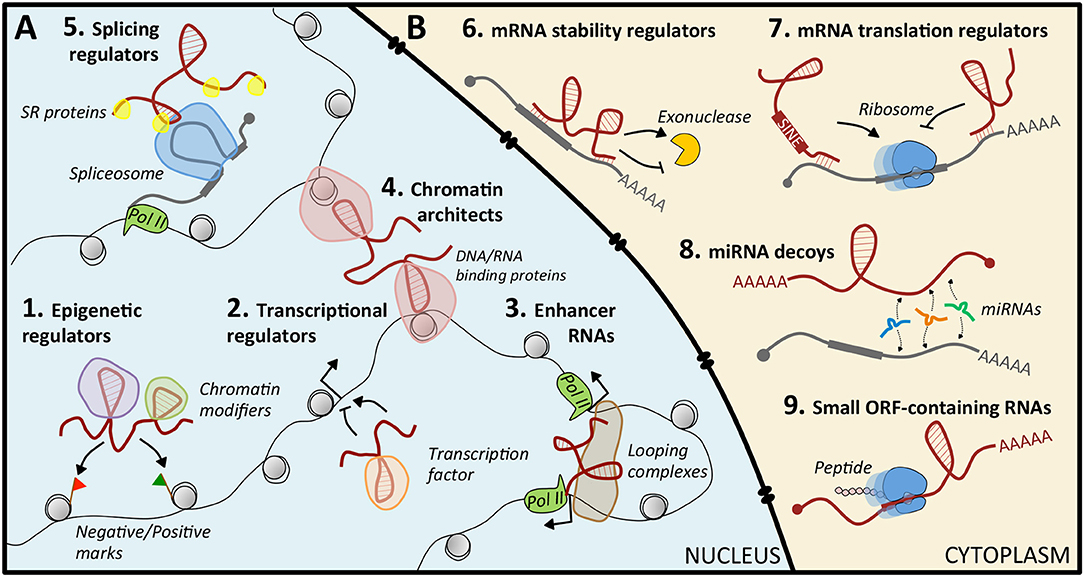
Figure 1. Mechanisms of action of lncRNAs. (A) In the nucleus, lncRNAs can regulate gene expression by guiding epigenetic (1) or transcription (2) factors to specific loci, by acting as enhancers (3), by structuring the three-dimensional conformation of chromatin (4), or by recruiting splicing factors (5). (B) In the cytoplasm, they act by modulating mRNA stability (6) or translation (7), by sequestering miRNAs (8), or by coding small peptides (9).
Nuclear lncRNAs can also act as regulators of transcriptional programs, by recruiting transcription activators or repressors to specific loci (30, 31), as enhancer RNAs that exert enhancer-like functions (32, 33), as chromosome architects and nuclear organizers that contribute to the formation of specific sub-nuclear structures (21, 34, 35), or as regulators of alternative splicing (36).
Cytoplasmic lncRNAs
Cytoplasmic lncRNAs (Figure 1B) regulate gene expression at the post-transcriptional level, often exploiting their sequence complementarity with transcripts deriving from the same genomic locus or from independent loci. Upon specific target recognition, they are able to modulate mRNA stability, both positively as BACE1-AS (37) and TINCR (38), and negatively as ½-sbsRNAs (39), or translation, as lincRNA p21 (40). Another role is that of decoys for microRNAs (miRNAs): in this case, the lncRNA functions as a competing endogenous RNA (ceRNA) that sequesters miRNAs from their mRNA targets, causing translational de-repression. This activity is based on regulatory crosstalk between multiple transcripts (41, 42). Notably, lncRNA-mediated ceRNA networks in cancer are continuously emerging (43, 44). However, only for a very limited number, such as Gas5 (45), linc-RoR (46, 47), NORAD (48), and linc-NeD125 (49), this function has been characterized: their aberrant enrichment or local increased concentration in pathological conditions can culminate in tumorigenesis. Finally, some lncRNAs may contain short open reading frames producing small, functional peptides (50).
The role of lncRNAs in CNS
The CNS of mammals is a very sophisticated system in which neuronal and glial cells structurally and functionally interact to guarantee the proper brain activity. Numerous evidence correlates the evolutionary increase in human brain complexity with the expanding number of lncRNAs (51, 52). Accordingly, 40% of human annotated lncRNAs are expressed in the brain, where they display neuro-anatomical and/or cell-type specific expression, and about 30% of lncRNAs appears to be primate-specific (31, 53). Notably, compared to lncRNAs from other tissues, the brain-specific lncRNAs are: (i) the most evolutionarily conserved species, (ii) predicted to retain conserved secondary structures, and (iii) preferentially adjacent to protein-coding genes involved in neuronal differentiation and function (54). Overall, these findings indicate that brain-specific lncRNAs likely possess conserved functions and are crucially implicated in higher-order cognitive abilities as well as in establishing neural cell-type diversity and function. This hypothesis is sustained by their spatiotemporal expression, which is exquisitely regulated during NS development (55) and in response to neuronal activity (56). So far, a growing body of literature shows that lncRNAs influence every step of neurodevelopment, from early stages of differentiation to synaptogenesis (57–59). In vitro studies revealed some lncRNAs, such as RMST (60), TUNA (61), DALI (62), and PAUPAR (63), that control complex gene expression programs underlying the neurogenic commitment of pluripotent embryonic stem cells. This is mainly achieved through their action of “guide” RNAs that convey transcriptional and/or epigenetic factors on the promoters of neuronal genes. In vivo analyses identified other species such as GOMAFU (56, 64, 65), EVF2 (66), PNKY (67), and linc-BRN1B (68) that, through the recruitment of epigenetic, transcriptional, or splicing factors, govern the balance between self-renewal and neuronal differentiation. LncRNAs also contribute to synaptogenesis and neuronal plasticity, which underlies learning, memory, and cognition, by regulating crucial proteins that control neurite elaboration (69), translation in synapses (70, 71), and ion channel subunits (72).
lncRNAs in Neuro-oncological Disorders
Based on their crucial role in NS development and function, lncRNA qualitative and/or quantitative alterations may profoundly impact on different neurological pathologies, including neurodevelopmental, neurodegenerative, neuro-immunological, and neuro-oncological disorders (73, 74). In the latter settings, lncRNAs have drawn extensive attention as molecules that may drive tumorigenesis. In addition, they can serve as predictors of cancer sub-types as well as potential therapeutic targets.
It is widely understood that mutations, epigenetic alterations or somatic copy number aberrations in the noncoding portion of the genome underlie cancer pathology (75). Accordingly, recent studies indicated that lncRNAs are highly deregulated in cancer, where they participate as tumor-suppressors or oncogenes in tumor initiation and progression. Notably, most lncRNAs displaying aberrant expression are cancer-type unique (76). However, despite the identification of a large number of lncRNAs in neurological cancers, only for a few of them mechanisms of action have been experimentally clarified.
Extensive studies have been carried out in gliomas, the most prevalent types of primary intracranial carcinoma (77). Several lncRNAs associated with glioma stemness (78–80), proliferation, and migration (81–84) have been identified, and most of them function as miRNA decoys (81).
Neuroblastoma (NB) is a pediatric tumor of the sympathetic NS, accounting for more than 7% of childhood malignancies (85). The molecular link between deregulated lncRNA expression and NB tumorigenic features is emerging (86), and several deregulated lncRNAs during NB pathogenesis have been uncovered (87–92).
Our knowledge of lncRNA function in MB physiopathology is still fragmentary. Genome-wide association studies may help to understand how genetic polymorphisms in lncRNA loci contribute to MB predisposition (93). Furthermore, in spite of the numerous high-throughput expression studies carried out so far, lncRNAs have been largely disregarded. However, re-annotation of array-based data and integration of cancer phenotype associations allowed prioritizing disease-related lncRNAs in tumors, including MB (94), demonstrating the potential of data re-analyses. In another study, a de-novo genome-wide inspection of MB subgroup-specific chromosomal alterations identified the first G3 MB gene fusions (6). They involve the 5′-end of PVT1, a lncRNA hosting the putative MB oncogene miR-1024 (95, 96). In the PVT1-MYC fusion, the induction of miR-1024 and the associated malignant phenotype may be explained through an oncogenic positive feedback-loop, established by MYC on its response elements on PVT1 promoter (6).
Other studies focused on the role played in MB by previously identified noncoding oncogenes. Among them, UCA1 (97) and CRNDE (98, 99) are upregulated in MB samples. UCA1 knockdown in MB cells results in the arrest of cell cycle progression, suppression of cell migration, and proliferation (100). Similarly, in vitro downregulation of CRNDE blocked cell cycle, inhibited proliferation and aggregation, while increasing apoptosis. Tumor growth was also reduced in MB mouse models silenced for CRNDE (101). Inversely, the lncRNA HOTAIR (102) is downregulated in MB samples, whereas its target genes HOXD8 and HOXD10 are upregulated (103). The misbalance of these crucial developmental genes may partially account for the embryonic origin and the pediatric onset of MB. However, their mechanisms of action are presently unknown.
Mechanistic insights into the role of lncRNAs in MB biology have been carried out only for a very few species, as discussed below.
Mechanisms of Action of lncRNAs in Mb
The colon cancer upregulated transcript CCAT1 (104) is a prototype of oncogenic lncRNA, associated with several carcinomas, where it promotes cell proliferation, invasion, migration, and chemoresistence (105–107). In MB, its expression is upregulated in 20 unstratified tumor samples and also in at least four MB cell lines (108). CCAT1 knockdown in MB cells causes the decrease of cell proliferation rate, (depending on CCNA and CDK2 gene repression), cell migration, and invasion. Its in vivo depletion reduces the volume of subcutaneous tumors of xenotransplanted mice (108). CCAT1 has been proposed to play its oncogenic role by altering the phosphorylated, active status of components of the tumorigenic MAPK pathway. In combination with previous reports indicating CCAT1 as a miRNA sponge (109–111), this study suggests that CCAT1 may control tumorigenesis through multiple activities.
Another lncRNA implicated in MB is ANRIL (112), which plays a pivotal role in multiple cancers as an epigenetic regulator of its neighbor tumor-suppressors CDKN2A/B (113, 114). ANRIL expression is upregulated in MB cells, where its knockdown lowers cell viability and migration while increasing apoptosis, by deranging the expression of several apoptotic factors (115). ANRIL has been shown to act as a decoy for miR-323, a miRNA identified in neurons (116) and characterized as a glioma tumor-suppressor (117, 118). Consistently, miR-323 silencing counteracted the abovementioned ANRIL-dependent cell phenotypes. This regulative axis impinges on BRI3, a miR-323 target gene (119) encoding for a brain-expressed transmembrane factor (120). BRI3 activates MAPK, AKT and WNT signaling cascades, already associated with MB progression (121–123), through a double mechanism: BRI3 upregulation enhances the phosphorylation of p38, MAPK, ERK, and AKT kinases and stimulates the accumulation of Wnt3a, Wnt5a, and β-catenin. The dysregulation of such pathways may partially explain the apoptotic phenotypes observed upon imbalance of ANRIL/miR-323/BRI3 module (115).
More recently, the lncRNA LOXL1-AS1, the antisense transcript to the LOXL1 genomic locus, whose variants are strongly associated with the exfoliation syndrome (124), was found to be overexpressed in MB tissues. In vitro and in vivo experiments revealed that it controls cell viability, proliferation, cell cycle, and metastasis by activating the PI3K-AKT pathway (125).
Recently, the ceRNA mechanism has emerged as a crucial pathogenic pathway in MB. Linc-NeD125 was the first ceRNA identified in MB and, generally, in tumors of the CNS (49). It was identified in NB cells as the precursor of miR-125-b1 (126), a neuronal-enriched miRNA (127) involved in neural cell differentiation (128), function (129) and NB and MB cell proliferation, and apoptosis (130, 131). Notably, linc-Ned125 is significantly and specifically upregulated in primary G4 MBs, compared to the other subgroups. In this context, it functions as a miRNA decoy. Linc-NeD125 interacts with miR-19a-3p, miR-19b-3p, and miR-106a-5p that pleiotropically control the expression of four G4 MB driver genes, namely KDM6A, MYCN, CDK6, and SNCAIP (7) (Figure 2). Through this mechanism, linc-NeD125 causes the driver gene translational de-repression, contributing to G4 MB tumorigenesis and/or to the maintenance of cancer cell identity. This study highlighted linc-NeD125 as a novel potential G4 driver gene, as well as a specific biomarker and a potential therapeutic target. Accordingly, its knockdown in G4-derived cells caused a significant reduction of cell proliferation, migration, and invasion (49).
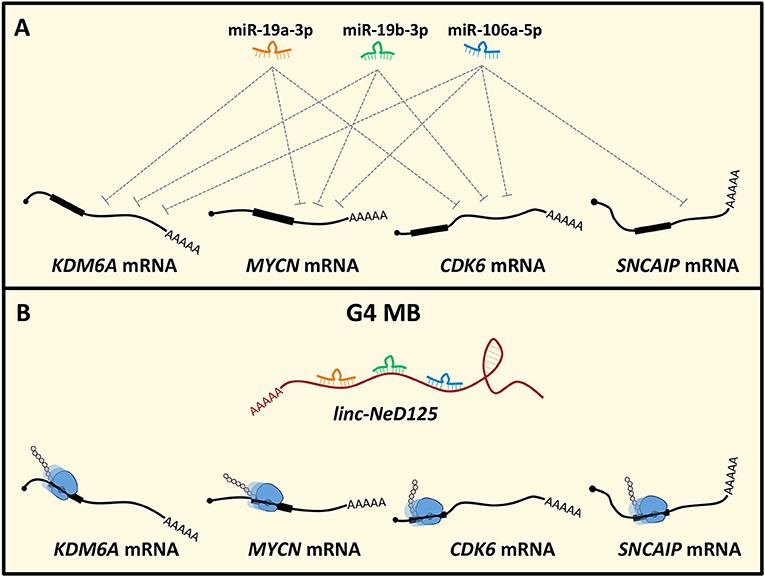
Figure 2. Linc-NeD125-based network in MB. (A) MiR-19a-3p, miR-19b-3p, and miR-106a-5p pleiotropically control KDM6A, MYCN, CDK6, SNCAIP gene expression. (B) In G4 MB, linc-NeD125 is upregulated and, functioning as a decoy for the three miRNAs, causes translational derepression of the G4 MB driver genes KDM6A, MYCN, CDK6, SNCAIP.
The second example of ceRNA in MB is the lncRNA Nkx2-2as, that behaves as a tumor-suppressor in SHH MB subgroup. It is highly down-regulated in MB cells derived from a SHH mouse model and it suppresses the malignant phenotype of MB cells, functioning as a sponge for miR-103/107 and miR-548 m. This activity causes the depression of the tumor-suppressor genes BTG2/Tis21/PC3 and LATS1/2, promoting tumor growth in vitro and in vivo (132).
Future Directions
The main challenges in fighting cancer are the identification of specific biomarkers, for timely diagnosis and prognosis, and novel tumor-driver genes, which can be therapeutically targeted for suppressing tumor growth. The former function would help the choice of pre-operative treatments and facilitate the tumor follow-up examinations. Unfortunately, very few biomarkers are known for pediatric tumors (133) and in MB <20 protein-coding genes have been characterized as promising candidates. However, most of these biomarkers were identified from single studies and from heterogeneous tumor types, lacking tumor-specificity (133). The recent categorization of MB into at least four subtypes, with distinct features, led the scientists to consider them as distinct pathologies with likely different responses to therapy. This new perspective triggered the search for novel MB-subgroup specific biomarkers and therapeutic targets. For both issues lncRNAs are very challenging (75). Since many of them are uniquely expressed in specific cancer types, they may function as powerful cancer biomarkers (134). In addition, for their ability to fold into complex tridimensional structures that increase their stability, they can be easily detected into body fluids as urine, blood, and cerebrospinal fluids, making the tumor diagnosis less invasive (75). Notably, lncRNAs are also considered new relevant targets for cancer therapy as highly tissue-specific drivers of cancer phenotypes. Finally, in this search for lncRNAs as novel molecules that distinguish clinically relevant cancer subtypes and predict tumor behavior, the circular RNAs are proving to be effective cancer biomarkers for their abundance, stability, and specificity (135).
Author Contributions
PL and EC reviewed the literature and wrote the manuscript. JR prepared the figures and contributed to the text discussion.
Funding
This work was partially supported by a grant from the Epigen-Epigenomics Flagship Project to EC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with the authors PL and EC.
Acknowledgments
We apologize to colleagues whose work was not discussed or cited in this review due to the space limitation.
References
1. Ivanov DP, Coyle B, Walker DA, Grabowska AM. In vitro models of medulloblastoma: choosing the right tool for the job. J Biotechnol. (2016) 236:10–25. doi: 10.1016/j.jbiotec.2016.07.028
2. Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon (2013)
3. Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. (2012) 123:465–72. doi: 10.1007/s00401-011-0922-z
4. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. Lyon: IARC (2007).
5. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. (2016) 131:803–20. doi: 10.1007/s00401-016-1545-1
6. Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. (2012) 488:49–56. doi: 10.1038/nature11327
7. Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. (2012) 12:818–34. doi: 10.1038/nrc3410
8. Wang J, Garancher A, Ramaswamy V, Wechsler-Reya RJ. Medulloblastoma: from molecular subgroups to molecular targeted therapies. Annu Rev Neurosci. (2018) 41:207–32. doi: 10.1146/annurev-neuro-070815-013838
9. Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. (2017) 31:737–54.e6. doi: 10.1016/j.ccell.2017.05.005
10. Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. (2012) 123:473–84. doi: 10.1007/s00401-012-0958-8
12. Ballarino M, Morlando M, Fatica A, Bozzoni I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Invest. (2016) 126:2021–30. doi: 10.1172/JCI84419
13. Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. (1961) 190:576–81. doi: 10.1038/190576a0
14. Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. (1961) 3:318–56. doi: 10.1016/S0022-2836(61)80072-7
15. Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. (1982) 31:147–57. doi: 10.1016/0092-8674(82)90414-7
16. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. (1990) 10:28–36. doi: 10.1128/MCB.10.1.28
17. Gabory A, Jammes H, Dandolo L. The H19 locus: role of an im- printed non-coding RNA in growth and development. Bioessays. (2010) 32:473–80. doi: 10.1002/bies.200900170
18. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. (1991) 349:38–44. doi: 10.1038/349038a0
19. Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. (2011) 130:223–36. doi: 10.1007/s00439-011-1008-7
20. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
21. Engreit JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. (2016) 17:756–70. doi: 10.1038/nrm.2016.126
22. Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. (2013) 153:38–55. doi: 10.1016/j.cell.2013.03.008
23. Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. (2013) 152:1308–23. doi: 10.1016/j.cell.2013.02.016
24. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
25. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. (2011) 71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021
26. Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long noncoding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin. (2014) 46:1–5. doi: 10.1093/abbs/gmt117
27. Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. (2015) 6:32410–25. doi: 10.18632/oncotarget.5609
28. Marin-Bejar O, Mas AM, González J, Martinez D, Athie A, Morales X, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. (2017) 18:202. doi: 10.1186/s13059-017-1331-y
29. Morlando M, Fatica A. Alteration of epigenetic regulation by long noncoding RNAs in cancer. Int J Mol Sci. (2018) 19:pii: E570. doi: 10.3390/ijms19020570
30. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. (2009) 10:155–9. doi: 10.1038/nrg2521
31. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
32. Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. (2013) 51:606–17. doi: 10.1016/j.molcel.2013.07.022
33. Mousavi K, Zare H, Koulnis M, Sartorelli V. The emerging roles of eRNAs in transcriptional regulatory networks. RNA Biol. (2014) 11:106–10. doi: 10.4161/rna.27950
34. Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. (2014) 21:198–206. doi: 10.1038/nsmb.2764
35. Rinn J, Guttman M. RNA and dynamic nuclear organization. Long noncoding RNAs may function as organizing factors that shape the cell nucleus. Science. (2014) 12: 345:1240–1. doi: 10.1126/science.1252966
36. Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. (2018) 46:2169–84. doi: 10.1093/nar/gky095
37. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med. (2008) 14:723–30. doi: 10.1038/nm1784
38. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. (2013) 493:231–5. doi: 10.1038/nature11661
39. Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. (2011) 470:284–8. doi: 10.1038/nature09701
40. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. (2012) 47:648–55. doi: 10.1016/j.molcel.2012.06.027
41. Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. (2015) 16:113–26. doi: 10.1038/nrg3853
42. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
43. Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. (2016) 7:13479–90. doi: 10.18632/oncotarget.7266
44. Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. (2018) 19:E1310. doi: 10.3390/ijms19051310
45. Li W, Zhao W, Lu Z, Zhang W, Yang X. Long non-coding RNA GAS5 promotes proliferation, migration and invasion by regulation of miR-301a in esophageal cancer. Oncol Res. (2018) 26:1285–94. doi: 10.3727/096504018X15166193231711
46. Fu Z, Li G, Li Z, Wang Y, Zhao Y, Zheng S, et al. Endogenous miRNA Sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov. (2017) 3:17004. doi: 10.1038/cddiscovery.2017.4
47. Wang L, Yu X, Zhang Z, Pang L, Xu J, Jiang J, et al. Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J Exp Clin Cancer Res. (2017) 36:182. doi: 10.1186/s13046-017-0658-2
48. Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. (2017) 16:169. doi: 10.1186/s12943-017-0738-0
49. Laneve P, Po A, Favia A, Legnini I, Alfano V, Rea J, et al. The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget. (2017) 8:31003–15. doi: 10.18632/oncotarget.16049
50. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. (2015) 160:595–606. doi: 10.1016/j.cell.2015.01.009
51. Clark BS, Blackshaw S. Understanding the role of lncRNAs in nervous system development. Adv Exp Med Biol. (2017) 1008:253–82. doi: 10.1007/978-981-10-5203-3_9
52. Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. (2011) 17:337–46. doi: 10.1016/j.molmed.2011.02.002
53. Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. (2008) 105:716–21. doi: 10.1073/pnas.0706729105
54. Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. (2009) 5:e1000617. doi: 10.1371/journal.pgen.1000617
55. Luo H, Sun S, Li P, Bu D, Cao H, Zhao Y. Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS ONE. (2013) 8:e70835. doi: 10.1371/journal.pone.0070835
56. Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu, et al. The long non- coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. (2014) 19:486–94. doi: 10.1038/mp.2013.45
57. Knauss J, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. (2013) 235:200–14. doi: 10.1016/j.neuroscience
58. Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. (2015) 88:861–77. doi: 10.1016/j.neuron.2015.09.045
59. Quan Z, Zheng D, Qing H. Regulatory roles of long non-coding RNAs in the central nervous system and associated neurodegenerative diseases. Front Cell Neurosci. (2017) 11:175. doi: 10.3389/fncel.2017.00175
60. Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncod- ing RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. (2013) 51:349–59. doi: 10.1016/j.molcel.2013.07.017
61. Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. (2014) 53:1005–19. doi: 10.1016/j.molcel.2014.01.021
62. Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. (2014) 3:e04530. doi: 10.7554/eLife.04530
63. Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. (2014) 33:296–311. doi: 10.1002/embj.201386225
64. Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. (2007) 120 (Pt 15):2498–506. doi: 10.1242/jcs.009357
65. Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. (2011) 16:479–90. doi: 10.1111/j.1365-2443.2011.01502.x
66. Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. (2009) 12:1020–7. doi: 10.1038/nn.2371
67. Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz CC, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. (2015) 16:439–47. doi: 10.1016/j.stem.2015.02.007
68. Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. (2013) 2:e01749. doi: 10.7554/eLife.01749
69. Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upre- gulation. Nat Biotechnol. (2012) 30:453–9. doi: 10.1038/nbt.2158
70. Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. (2002) 22:10232–41. doi: 10.1523/JNEUROSCI.22-23-10232.2002
71. Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. (2003) 112:317–27. doi: 10.1016/S0092-8674(03)00079-5
72. Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. (2013) 16:1024–31. doi: 10.1038/nn.3438
73. Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. (2010) 1338:20–35. doi: 10.1016/j.brainres.2010.03.110
74. Chen Y, Zhou J. LncRNAs: macromolecules with big roles in neurobiology and neurological diseases. Metab Brain Dis. (2017) 32:281–91. doi: 10.1007/s11011-017-9965-8
75. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–463. doi: 10.1016/j.ccell.2016.03.010
76. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. (2015) 28:529–40. doi: 10.1016/j.ccell.2015.09.006
77. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. (2018) 17:61. doi: 10.1186/s12943-018-0812-2
78. Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, et al. Targeting the notch-regulated non-coding RNA TUG1for glioma treatment. Nat Commun. (2016) 7:13616. doi: 10.1038/ncomms13616
79. Zhao X, Liu Y, Zheng J, Liu X, Chen J, Liu L, et al. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim Biophys Acta. (2017) 1864:1605–17. doi: 10.1016/j.bbamcr.2017.06.020
80. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. (2015) 359:75–86. doi: 10.1016/j.canlet.2014
81. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long non-coding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta- catenin pathway by scaffolding EZH2. Clin Cancer Res. (2018) 24:684–95. doi: 10.1158/1078-0432.CCR-17-0605
82. Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. (2015) 6:537–46. doi: 10.18632/oncotarget.2681
83. Deguchi S, Katsushima K, Hatanaka A, Shinjo K, Ohka F, Wakabayashi T, et al. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene. (2017) 36:4629–40. doi: 10.1038/onc.2017.88
84. Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H, et al. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. (2016) 24:1806–22. doi: 10.1038/mt.2016.103
85. Lanzkowsky P. Chapter 24: Neuroblastoma. In: Lanzkowsky P, Lipton J, Fish J, editors. Manual of Pediatric Hematology and Oncology., 6th ed. Academic Press (2016) p. 1054–8.
86. Pandey GK, Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. (2015) 6:18265–75. doi: 10.18632/oncotarget.4251
87. Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. (2014) 26:722–37. doi: 10.1016/j.ccell.2014.09.014
88. Buechner J, Einvik C. N-myc and noncoding RNAs in neuroblastoma. Mol Cancer Res. (2012) 10:1243–53. doi: 10.1158/1541-7786.MCR-12-0244
89. Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. (2010) 24:4033–46. doi: 10.1096/fj.10-157032
90. Liu PY, Erriquez D, Marshall GM, Tee AE, Polly P, Wong M, et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J Natl Cancer Inst. (2014) 106:dju113. doi: 10.1093/jnci/dju113
91. Atmadibrata B, Liu PY, Sokolowski N, Zhang L, Wong M, Tee AE, et al. The novel long noncoding RNA linc00467 promotes cell survival but is down-regulated by N-Myc. PLoS ONE. (2014) 9:e88112. doi: 10.1371/journal.pone.0088112
92. Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, et al. Transcribed-ultra conserved region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer. (2009) 9:441. doi: 10.1186/1471-2407-9-441
93. Chen YD, Zhang N, Qiu XG, Yuan J, Yang M. LncRNA CDKN2BAS rs2157719 genetic variant contributes to medulloblastoma predisposition. J Gene Med. (2018) 20:e300. doi: 10.1002/jgm.3000
94. Xu C, Qi R, Ping Y, Li J, Zhao H, Wang L, et al. Systemically identifying and prioritizing risk lncRNAs through integration of pan-cancer phenotype associations. Oncotarget. (2017) 8:12041–51. doi: 10.18632/oncotarget.14510
95. Shtivelman E, Bishop JM. The PVT gene frequently amplifies with MYC in tumor cells. Mol Cell Biol. (1989) 9:1148–54. doi: 10.1128/MCB.9.3.1148
96. Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. (2007) 213:511–8. doi: 10.1002/jcp.21133
97. Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. (2016) 20:2819–24.
98. Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynhian AT, et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. (2011) 2:829–40. doi: 10.1177/1947601911431081
99. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. (2011) 25:1915–27. doi: 10.1101/gad.17446611
100. Zhengyuan X, Hu X, Qiang W, Nanxiang L, Junbin C, Wangming Z. Silencing of urothelial carcinoma associated 1 inhibits the proliferation and migration of medulloblastoma cells. Med Sci Monit. (2017) 23:4454–61. doi: 10.12659/MSM.904675
101. Song H, Han LM, Gao Q, Sun Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur Rev Med Pharmacol Sci. (2016) 20:2588–97. doi: 10.26355/eurrev_201712_13925
102. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
103. Chakravadhanula M, Ozols VV, Hampton CN, Zhou L, Catchpoole D, Bhardwaj RD. Expression of the HOX genes and HOTAIR in atypical teratoid rhabdoid tumors and other pediatric brain tumors. Cancer Genet. (2014) 207:425–8. doi: 10.1016/j.cancergen.2014.05.014
104. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. (2012) 130:1598–606. doi: 10.1002/ijc.26170
105. Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. (2016) 7:62474–89. doi: 10.18632/oncotarget.11518
106. Ma X, Sheng S, Wu J, Jiang Y, Gao X, Cen X, et al. LncRNAs as an intermediate in HPV16 promoting myeloid-derived suppressor cell recruitment of head and neck squamous cell carcinoma. Oncotarget. (2017) 8:42061–75. doi: 10.18632/oncotarget.14939
107. Zhang E, Han L, Yin D, He X, Hong L, Si X, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. (2017) 45:3086–101. doi: 10.1093/nar/gkw1247
108. Gao R, Zhang R, Zhang C, Zhao L, Zhang Y. Long noncoding RNA CCAT1 promotes cell proliferation and metastasis in human medulloblastoma via MAPK pathway. Tumori J. (2018) 104:43–50. doi: 10.5301/tj.5000662
109. Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. (2016) 39:330–6. doi: 10.14348/molcells.2016.2308
110. Wang ZH, Guo XQ, Zhang QS, Zhang JL, Duan YL, Li GF, et al. Long non-coding RNA CCAT1 promotes glioma cell proliferation via inhibiting microRNA-410. Biochem Biophys Res Commun. (2016) 480:715–20. doi: 10.1016/j.bbrc.2016.10.047
111. Zhou B, Wang Y, Jiang J, Jiang H, Song J, Han T, et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. (2016) 68:201–10. doi: 10.1002/iub.1474
112. Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. (2007) 67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004
113. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. (2011) 30:1956–62. doi: 10.1038/onc.2010.568
114. Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. (2010) 38:662–74. doi: 10.1016/j.molcel.2010.03.021
115. Zhang H, Wang X, Chen X. Potential role of long non-coding RNA ANRIL in pediatric medulloblastoma through promotion on proliferation and migration by targeting miR-323. J Cell Biochem. (2017) 118:4735–44. doi: 10.1002/jcb.26141
116. Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. (2004) 101:360–5. doi: 10.1073/pnas.2333854100
117. Lavon I, Zrihan D, Granit A, Einstein O, Fainstein N, Cohen MA, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. (2010) 12:422–33. doi: 10.1093/neuonc/nop061
118. Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. (2013) 11:10. doi: 10.1186/1479-5876-11-10
119. Yang L, Xiong Y, Hu XF, Du YH. MicroRNA-323 regulates ischemia/ reperfusion injury-induced neuronal cell death by targeting BRI3. Int J Clin Exp Pathol. (2015) 8:10725–33.
120. Vidal R, Calero M, Révész T, Plant G, Ghiso J, Frangione B. Sequence, genomic structure and tissue expression of Human BRI3, a member of the BRI gene family. Gene. (2001) 266:95–102. doi: 10.1016/S0378-1119(01)00374-2
121. Guldal CG, Ahmad A, Korshunov A, Squatrito M, Awan A, Mainwaring LA, et al. An essential role for p38 MAPK in cerebellar granule neuron precursor proliferation. Acta Neuropathol. (2012) 123:573–86. doi: 10.1007/s00401-012-0946-z
122. Włodarski P, Grajkowska W, Łojek M, Rainko K, Józwiak J. Activation of Akt and Erk pathways in medulloblastoma. Folia Neuropathol. (2006) 44:214–20.
123. Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. (1992) 68:33–51. doi: 10.1016/0092-8674(92)90204-P
124. Hauser MA, Aboobakar IF, Liu Y, Miura S, Whigham BT, Challa P, et al. Genetic variants and cellular stressors associated with exfoliation syndrome modulate promoter activity of a lncRNA within the LOXL1 locus. Hum Mol Genet. (2015) 24:6552–63. doi: 10.1093/hmg/ddv347
125. Gao R, Zhang R, Zhang C, Liang W, Tang W. LncRNA LOXL1-AS1 promotes the proliferation and metastasis of medulloblastoma by activating the PI3K/AKT pathway. Anal Cell Pathol. (2018) 2018:9275685. doi: 10.1155/2018/9275685
126. Bevilacqua V, Gioia U, Di Carlo V, Tortorelli AF, Colombo T, Bozzoni I, et al. Identification of linc-NeD125, a novel long non coding RNA that hosts miR-125b-1 and negatively controls proliferation of human neuroblastoma cells. RNA Biol. (2015) 12:1323–37. doi: 10.1080/15476286.2015.1096488
127. Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. (2005) 21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x
128. Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. MiR-125 potentiates early neural specification of human embryonic stem cells. Development. (2012) 139:1247–57. doi: 10.1242/dev.073627
129. Akerblom M, Petri R, Sachdeva R, Klussendorf T, Mattsson B, Gentner B, et al. MicroRNA-125 distinguishes developmentally generated and adult-born olfactory bulb interneurons. Development. (2014) 141:1580–8. doi: 10.1242/dev.101659
130. Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, et al. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci USA. (2007) 104:7957–62. doi: 10.1073/pnas.0700071104
131. Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer. (2009) 124:568–77. doi: 10.1002/ijc.23948
132. Zhang Y, Wang T, Wang S, Xiong Y, Zhang R, Zhang X, et al. Nkx2-2as suppression contributes to the pathogenesis of Sonic hedgehog medulloblastoma. Cancer Res. (2018) 78:962–73. doi: 10.1158/0008-5472.CAN-17-1631
133. Russell MD, Young AM, Karri SK. Biomarkers of pediatric brain tumors. Front Pediatr. (2013) 1:7. doi: 10.3389/fped.2013.00007
134. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
Keywords: nervous system, pediatric tumor, medulloblastoma, long noncoding RNAs, oncogenes, tumor suppressors, diagnostic biomarkers, therapeutic targets
Citation: Laneve P, Rea J and Caffarelli E (2019) Long Noncoding RNAs: Emerging Players in Medulloblastoma. Front. Pediatr. 7:67. doi: 10.3389/fped.2019.00067
Received: 08 June 2018; Accepted: 18 February 2019;
Published: 14 March 2019.
Edited by:
Andrea Carai, Bambino Gesù Children Hospital (IRCCS), ItalyReviewed by:
Anshu Malhotra, Emory University, United StatesFrancesca Del Bufalo, Bambino Gesù Children Hospital (IRCCS), Italy
Copyright © 2019 Laneve, Rea and Caffarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Caffarelli, ZWxpc2EuY2FmZmFyZWxsaUB1bmlyb21hMS5pdA==
 Pietro Laneve
Pietro Laneve Jessica Rea
Jessica Rea Elisa Caffarelli
Elisa Caffarelli