- 1Research Unit, Hospital Universitario N.S. de Candelaria, Universidad de La Laguna, Santa Cruz de Tenerife, Spain
- 2Genomics and Health Group, Department of Biochemistry, Microbiology, Cell Biology and Genetics, Universidad de La Laguna, Santa Cruz de Tenerife, Spain
- 3CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid, Spain
- 4Genomics Division, Instituto Tecnológico y de Energías Renovables, Santa Cruz de Tenerife, Spain
Asthma is a complex respiratory disease considered as the most common chronic condition in children. A large genetic contribution to asthma susceptibility is predicted by the clustering of asthma and allergy symptoms among relatives and the large disease heritability estimated from twin studies, ranging from 55 to 90%. Genetic basis of asthma has been extensively investigated in the past 40 years using linkage analysis and candidate-gene association studies. However, the development of dense arrays for polymorphism genotyping has enabled the transition toward genome-wide association studies (GWAS), which have led the discovery of several unanticipated asthma genes in the last 11 years. Despite this, currently known risk variants identified using many thousand samples from distinct ethnicities only explain a small proportion of asthma heritability. This review examines the main findings of the last 2 years in genomic studies of asthma using GWAS and admixture mapping studies, as well as the direction of studies fostering integrative perspectives involving omics data. Additionally, we discuss the need for assessing the whole spectrum of genetic variation in association studies of asthma susceptibility, severity, and treatment response in order to further improve our knowledge of asthma genes and predictive biomarkers. Leveraging the individual's genetic information will allow a better understanding of asthma pathogenesis and will facilitate the transition toward a more precise diagnosis and treatment.
Introduction
Asthma is a complex respiratory disease characterized by inflammation and reversible obstruction of the airways (1) that can lead to diverse symptoms such as wheeze, breathlessness, chest tightness, and cough (2). Asthma affects approximately 350 million people from all age groups worldwide (3) and causes around 350,000 deaths per year (4). Although asthma is a lifelong disease, it is considered the most common chronic condition in children and young adults (5, 6), where symptoms are usually more severe (7, 8).
A significant global burden has been attributed to asthma, which is mostly driven by direct economic costs on health care systems (9) and indirect social and economic consequences due to substantial productivity loss (10). In this regard, asthma represents one of the most important pulmonary diseases (11). However, wide differences in asthma prevalence have been estimated among countries and populations, ranging from 1.5 to 15.6% (12, 13), and also among ethnic groups within countries (14). These differences could be a result of complex interactions among environmental and genetic factors (15, 16).
Several studies support a large genetic contribution to asthma predisposition, known as heritability (17, 18), with estimates of as much as 55–74% in adults (19, 20), and almost reaching 90% in children (21). In order to elucidate the genes underlying asthma pathogenesis, several genetic approximations have been performed (22). The initial studies were linkage analyses, which are based on small panels of informative markers across the genome that were determined in multigenerational families with multiple affected individuals to allow the identification of the markers that were more frequently co-inherited with the disease (23). After a genomic region is linked to a disease, this could be followed-up using positional cloning, or the genes contained therein might serve as candidate regions for association studies in outbred population samples (23). Although the use of this approach for over 20 years allowed the identification of as few as eight asthma genes [reviewed in (24–26)], it was recognized a lack of power of this approach for detecting small effect sizes of the risk variants (27).
The use of linkage analysis decreased as it was a progressive development and use of candidate-gene association studies (23, 27). The latter were extensively used during the last two decades, mostly to refute or confirm the implication of a single biological candidate gene at a time (28), mainly by comparing the allele frequencies of a small set of single nucleotide polymorphisms (SNPs) near or within the gene of interest among asthma cases and control subjects without the disease (29).
Although candidate-gene association studies largely increased the resolution of genetic studies of asthma compared to linkage analyses, they also complicated the interpretation of overall results. The main reasons for that were that most studies have included small sample sizes and have tested a reduced number of genetic variants, which greatly decreases the power to detect significant associations. Most importantly, replication of findings in, at least, an independent study was not a standard practice. As a consequence, failure in the attempt to consistently replicate the findings in independent populations was common (23, 29). Given these criticisms, its use has progressively decreased while advances in high-throughput polymorphism genotyping platforms were occurring, leading to continuous reductions in costs and the development of key analysis methods to allow much denser genomic scans (29). These advances opened the way for genome-wide association studies (GWAS), which now allow a simultaneous exploration of hundreds of thousands of SNPs across the genome, most commonly determined in samples from unrelated cases and controls (23, 29). The main advantage of this hypothesis-free approach is the ability to detect mild effects of disease genes without any previous knowledge of the condition (23, 29). On the other side, performing GWAS could be challenging as they usually require large sample sizes and the coverage of the largest number of variants as possible to reach enough statistical power to detect significant associations with asthma (29).
Vicente et al. recently discussed the GWAS that were published from the first one in 2007 (30) until the end of 2016 (31), revealing a total of 39 common SNPs independently associated with asthma risk (22). In this review, we aimed to update the main findings of the genomic studies of asthma, treatment response and the overlap of this disease with other allergic conditions performed between 2016 and 2018. Additionally, we discuss the direction of the new generation of genetic studies of asthma to cover the unexplored variation and the forthcoming integrative omics approaches to continue disentangling the genetic predictors of asthma.
Genome-Wide Association Studies
A search on the NHGRI-EBI GWAS Catalog (32) and on PubMed records revealed that 15 GWAS of asthma and related traits had been published after the period reviewed by Vicente et al. (22), between 1st May 2016 and 19th September 2018 (Supplementary Table 1).
Asthma was defined by a physician diagnosis in most of the studies. However, some GWAS also considered other asthma definitions, such as the presence of symptoms or the prescription of any asthma medication, among others. Four GWAS focused on children (33–36) and five on adults (37–41), whereas another five attempted to identify common genetic factors between childhood and adulthood asthma (42–46). Across the 15 GWAS of asthma and related traits reviewed, the largest sample sizes were attained by those focusing on the genetic overlap of asthma and allergic diseases. The largest one included 360,838 individuals (180,129 cases and 180,709 controls) and aimed to disentangle the common genetic basis of asthma, hay fever and eczema in asthmatic children from European populations (45). The smallest comprised 949 individuals and it was focused on a highly specific phenotype, the response to asthma treatment with short-acting β2 agonists (SABA) (36) (Supplementary Table 1).
Although there is an increasing trend to include multiethnic populations in genomic studies of asthma, an underrepresentation of non-European populations is still pervasive (47, 48). In fact, the vast majority of GWAS performed between 2016 and 2018 focused on patients of European ancestry (33, 34, 39, 41–43, 45, 49), presenting a particularly poor representation of Asians and Africans-admixed populations.
In total, 451 genetic variants, including short insertions/deletions and SNPs, were reported as risk factors for asthma and related traits by the GWAS of the last 2 years. From these, 319 SNPs clustered at 167 loci that reached genome-wide significance at a threshold of p < 5 × 10−8 or p < 3 × 10−8 in the discovery or replication phases and/or after performing a meta-analysis with the results from both stages. Among these, 68 were revealed as novel asthma loci, whereas 99 had been previously associated with asthma or any allergic diseases.
In the sections below, we summarize the main findings of these GWAS, distinguishing among those that focused on asthma susceptibility; treatment response; gene-environment interactions and the overlap among asthma and allergic disorders.
Asthma Susceptibility
Eight GWAS evaluated the association with asthma (33, 35, 37, 38, 42–44, 46) (Supplementary Table 1), although only four studies revealed genome-wide significant associations (38, 43, 44, 46). These validated the association of 14 loci previously associated with asthma susceptibility (Table 1).
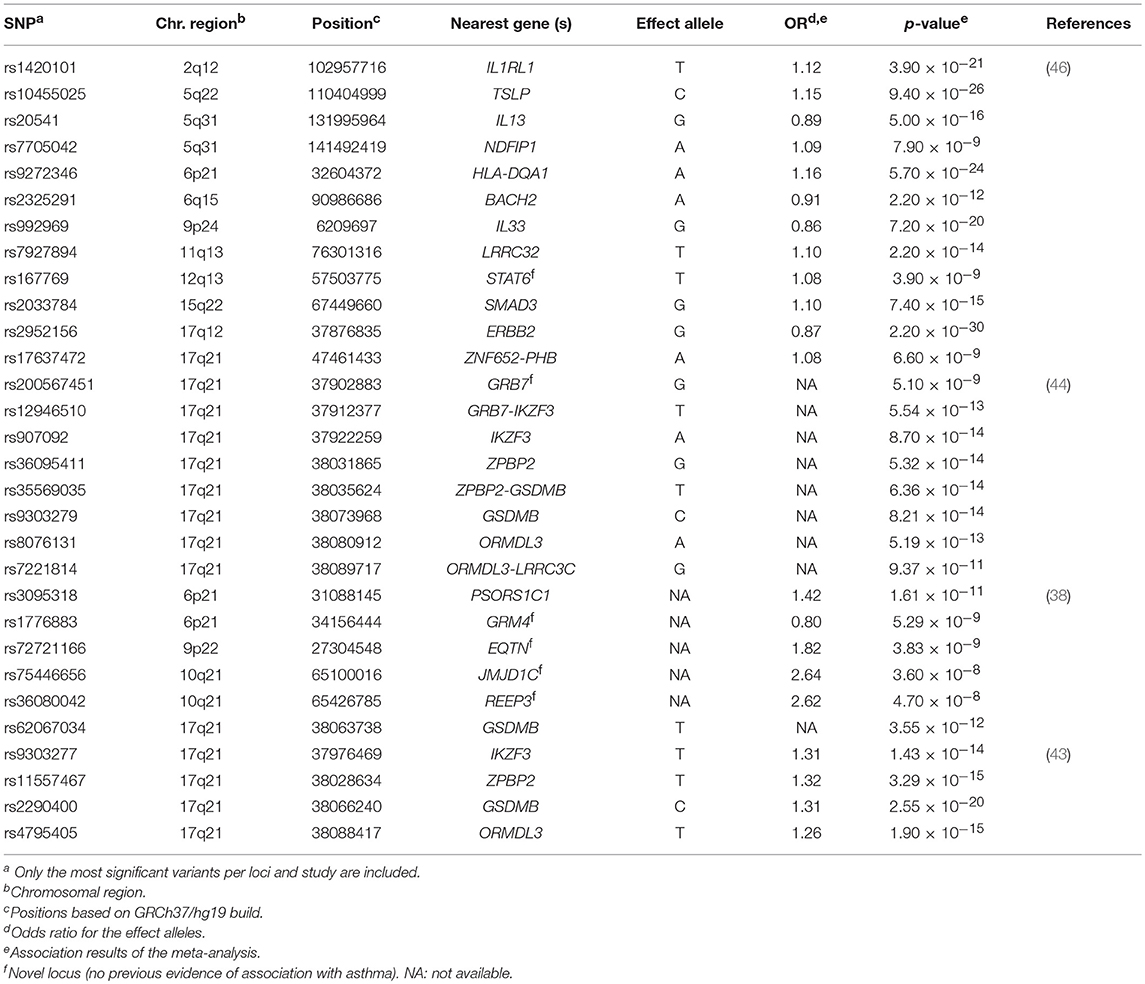
Table 1. Summary of the most significant variants identified by the genome-wide association studies of asthma susceptibility.
The well-known 17q21 asthma locus (50) has been the most replicated signal, although the main driver of this association has not been disentangled to date (43). The gene encoding the zona pellucida-binding protein 2 (ZPBP2) has been revealed as a common locus of both childhood and adulthood asthma by several studies, supported by the association of several intronic SNPs as well as variants located within the intergenic region of ZPBP2 and GSDMB (43, 44). A SNP located at the promoter region of ZPBP2 (rs11557467) showed the most significant association after performing a meta-analysis in 13,556 children and adults from several European populations (43). The risk allele was associated with asthma susceptibility (OR for the T allele = 1.32, p = 3.29 × 10−15) (43) and was also replicated in Latinos/Hispanics (44). This variant was previously evidenced to be a putative site with allele-specific nucleosome occupancy in patients with asthma (51). Similar results were found for GSDMB, with a shared signal between both European (min p = 2.55 × 10−20) (43) and Latino/Hispanic populations (min p = 8.21 × 10−14) (44). Furthermore, the association of ORMDL3 with asthma was validated in Latinos/Hispanics (min p = 1.90 × 10−15) (44), which have been also extensively associated with asthma across different populations (30, 52, 53) (Table 1). Interestingly, differences in the expression level of ZPBP2 and GSDMB have been found between European and African populations (54). In fact, early studies had revealed that SNPs associated with asthma co-regulate the expression of ORMDL3, GSDMB, and ZPBP2 in Latinos (54).
A large multiethnic GWAS performed in 23,948 asthma cases and 118,538 controls validated the association of several genes already known to be involved in asthma with functions related to immune response and other activities, such as organogenesis, cellular differentiation and transcriptional modulation, among others (46). The most significant association signal was driven by the SNP rs2952156 located at the Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2) gene, whose G allele was associated with protection for asthma (OR = 0.87, p = 2.20 × 10−30) in ethnically diverse populations (46) (Table 1).
Additionally, 6 loci not previously linked to asthma were identified in European (38), Latino/Hispanic (44) and multiethnic populations (38). In these studies, the GRM4 gene was the most frequent signal, where a higher number of variants with evidence of association with asthma susceptibility were located (min p = 5.29 × 10−9) (38). GRM4 encodes the glutamate metabotropic receptor 4, involved in synaptic neurotransmission and maintenance on normal functions of the central nervous system throughout the regulation of the adenylate cyclase cascade (55), although it has been recently linked to tumorigenesis (56). The GRM4 gene has been associated with several neurological disorders (57–59) and different types of cancer (56, 60) but, it has not been associated with any asthma-related traits and it has not been implicated in any immune-related function. However, early studies had suggested the potential implication of glutamate receptors on asthma worsening by means of triggering airways inflammation (61).
Asthma Treatment Response
The most commonly prescribed medication to treat asthma are SABA and inhaled corticosteroids (ICS) (2). Although most asthma patients treated with these medications experience a decrease in their symptoms (62), wide differences in asthma treatment response have been described among individuals and populations (63, 64). These observations suggest that genetics may play a key role in the response to asthma treatment (64, 65). Therefore, the characterization of multiple genetic markers determining therapeutic responsiveness to asthma medications could contribute in the future to identify specific pharmacogenetic profiles. This would enable clinical identification of those asthma patients that respond unsatisfactorily to these treatments or that experience adverse effects (66). Consequently, the burden of asthma could be reduced by implementing personalized asthma management and therapeutic strategies (67).
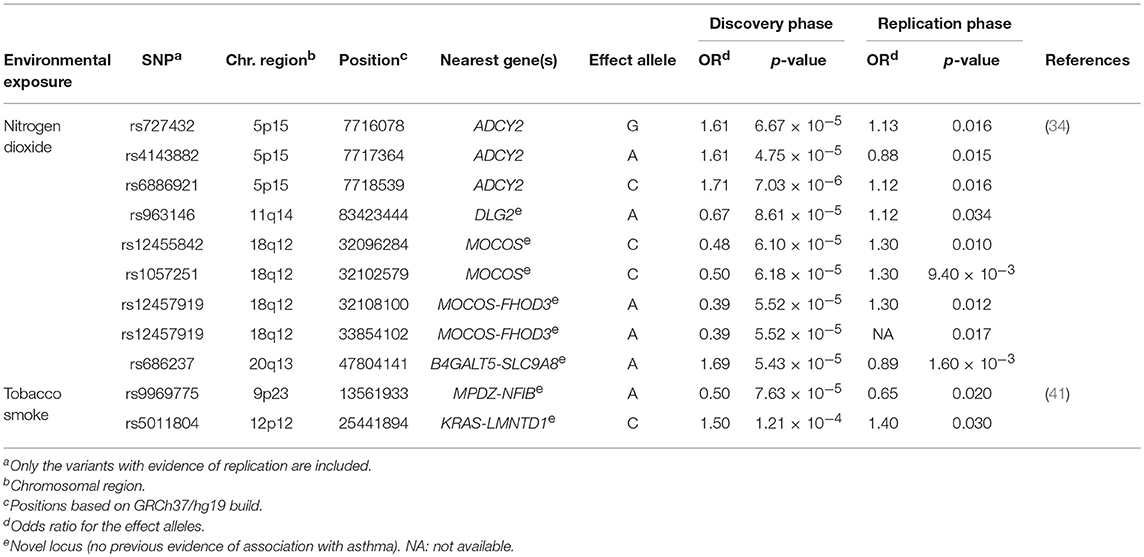
SABA are the most commonly prescribed relief asthma medication that quickly reduces bronchoconstriction throughout smooth muscle relaxation of the airways (2). Clinical response to this treatment is frequently assessed as bronchodilator response (BDR), which quantitatively measures the change in airway constriction by means of the change in forced expiratory volume in 1 s after SABA administration (68). However, high variability in BDR among individuals and populations has been described, which has been evidenced to be influenced by environmental and genetic factors (69, 70). In fact, it has been estimated that 47–92% of the total variation in BDR could be attributed to the genetic component (71, 72). Recently, a GWAS of BDR was performed in 949 children with asthma from two African American populations (36) (Supplementary Table 1). This revealed the intergenic region of SPATA31D1 and RASEF as population-specific novel loci of BDR in African American children with asthma (rs73650726, β for the A allele = 0.02, p = 7.69 × 10−9). Moreover, they found the PRKG1 to be implicated in BDR shared between African Americans and Latinos/Hispanics (min p = 3.94 × 10−8) (Table 2). This gene encodes a cyclic guanosine monophosphate-dependent protein kinase involved in several biological processes, such as the nitric-oxide signaling pathway (73, 74), which modulates vasodilation in response to β2 agonists (75). This fact together with evidence of expression of PRKG1 in pulmonary tissues suggest this could be a plausible gene of BDR in African-admixed asthmatic children (76).
Despite the large improvements in asthma therapeutic strategies in the last decades, ICS are still the most effective and commonly prescribed medication to control symptoms and prevent severe exacerbations in asthma patients (2), which consist of the most important outcome in childhood asthma (77). However, a small proportion of the genetic basis of the ICS response has been disentangled (78–80). In the period reviewed, Mosteller et al. performed the unique additional GWAS that has explored the association of genetic variants with ICS response (Supplementary Table 1). This constitutes the first GWAS of ICS response to include non-European patients. Unfortunately, they did not find any significant finding (40).
In addition to the most common types of medications used to treat asthma, there is an increasing number of emerging therapies, including biological treatments. These have been designed to act directly toward specific components of the T-lymphocyte inflammatory response involved in asthma such as, interleukin 5 (IL-5). This mediator is centrally involved in increasing immunoglobulin E levels and blood and bronchoalveolar eosinophilia in severe asthma. Therefore, the inhibition of IL-5 by using monoclonal antibodies could reduce the high levels of eosinophils (81). A few pharmacogenetic studies have recently evaluated the response to asthma therapies with anti-IL-5 monoclonal antibodies, such as mepolizumab (39) (Supplementary Table 1), which has been evidenced to reduce asthma exacerbations rates and enables asthma control (82, 83). Condreay et al. investigated the association of genetic variants with the response to asthma treatment with mepolizumab measured as number of asthma exacerbations, eosinophil count and immunoglobulin E levels in 1,192 asthma patients. Although no variants reached genome-wide significance level (p ≤ 5 × 10−8), six SNPs at 6p24 and 9p21 showed suggestive associations with mepolizumab response (Table 2) (39).
Unfortunately, despite the large efforts during the last decades, pharmacogenetic findings are still not able to predict clinical outcomes that are directly applied to asthma patients (84). As happened in the past for the asthma field, and although asthma pharmacogenetic studies have started to evolve toward GWAS approaches (64, 85), the main reason could be that most published pharmacogenomic studies continue to be performed using the candidate-gene approach (64, 80, 85).
Gene-Environment Interactions
Despite the significant contribution of genetic factors on asthma and related traits, a key role of the gene interactions with exposures to a wide variety of environmental factors has been described (15, 16, 86). Among these, early-life exposures demonstrate a high relevance in the prediction of childhood asthma development, including respiratory infections (87), gut and airway microbiome (87, 88), and tobacco smoke exposure (89). Several strategies have been used to identify gene-environment interactions during the last decades (90, 91), but their application has recently emerged in the form of genome-wide interaction studies (GWIS) (91), which are considered a powerful approach to identify novel disease loci that interact with environmental factors (41).
Two GWIS have attempted to identify gene-environment interactions involved in asthma susceptibility since 2016 (34, 41) (Supplementary Table 1). One of them explored for the first time the interaction of genetic variants with traffic-related air pollution, although previous studies used candidate gene approaches (34). Traffic air pollution measured as nitrogen dioxide levels cause deterioration of asthma symptoms by triggering exacerbations and decreasing the lung function (92, 93). This GWIS in European children revealed five loci that were suggestively associated and three of the SNPs were located at ADCY2, a known asthma locus (94). The risk alleles at ADCY2 were also associated with decreased expression of the gene in peripheral blood. Moreover, differential ADCY2 expression depending on nitrogen dioxide levels was found, suggesting that this gene could have functional implications on asthma under exposure to traffic-related air pollution (34). Similar results were found for a SNP located within the intergenic region of B4GALT5 and SLC9A8 (Table 3), which were revealed as novel plausible genes with functional implications on childhood asthma in interaction with nitrogen dioxide exposure (34).
Additionally, a GWIS of active tobacco smoking was conducted in 4,057 patients with adulthood-onset asthma of European ancestry (41) (Supplementary Table 1). It is well-known that second-hand smoke exposure to tobacco smoke increases childhood asthma risk during prenatal and postnatal stages (95–99). Although active tobacco smoke has been associated with asthma onset during adulthood (100), it is still unclear how the genetic variation could affect asthma susceptibility in interaction with tobacco smoke exposure in adults (41). The intergenic SNPs rs9969775 (OR for the A allele = 0.50, p = 7.63 × 10−5) and rs5011804 (OR for the C allele = 1.50, p = 1.21 × 10−4), which are located at the MPDZ-NFIB and KRAS-IFLTD1 loci, respectively, showed significant interactions with active tobacco smoking for late-onset asthma. These findings were validated at nominal level in an independent study (41) (Table 3). Although none of these loci showed any functions specifically related to asthma and none were previously associated with asthma or related traits, the SNP rs9969775 was postulated to be involved in the regulation of gene expression in the lung (41).
Overlap Among Asthma and Allergic Diseases
Given the firm links in the pathogenesis of asthma and other allergic diseases (20, 101), a few studies used this rationale to explore the overlapping genetic architecture among these diseases (45, 49, 102, 103), including two large-scale GWAS published between 2016 and 2018 (45, 49) (Supplementary Table 1).
Ferreira et al. carried out the largest GWAS of asthma and allergic diseases to date (45). They combined data from 360,838 children and adults from 13 different European studies, including 180,129 patients with self-reported or physician-diagnosed asthma, eczema or hay fever, and 180,709 controls (Supplementary Table 1). They reported 136 independent SNPs at 99 loci as genome-wide significant associations (p ≤ 3 × 10−8) with susceptibility to asthma or any allergic disease (Supplementary Table 2). A total of 86 variants were located at loci that were already associated with at least one of the diseases under study, whereas 50 other SNPs revealed novel loci that were shared by asthma and allergy (Table 4). A high proportion (96%) of these variants showed similar effects between asthma, eczema and hay fever. The most significant variants were located within or near loci with previous evidence of implication on asthma and/or allergic diseases such as, WNT11-LRRC32, IL18R1, TLR1, and HLA-DQA1, among others. In fact, the SNP rs7936323 of the intergenic region of WNT11 and LRRC32 genes showed the strongest evidence of association (OR for the A allele = 1.09, p = 2.20 × 10−63) (Supplementary Table 2). Altogether, the 136 SNPs identified accounted for 3.2, 3.8, and 1.2% of the total variation of asthma, hay fever and eczema, respectively. These findings partially explain the co-existence of these diseases in many patients (45) (Supplementary Table 2). Interestingly, evidence of potential functional implication on blood and pulmonary tissues was found for many of the SNPs identified. Specifically, they demonstrated that risk variants shared among asthma, hay fever and eczema are involved in the regulation of gene expression in immune response-related signaling pathways, such as in the B and T cell activation (45). These findings confirmed previous evidence (104) suggesting that these biological processes could be among the ones shared between asthma and allergic diseases (45).
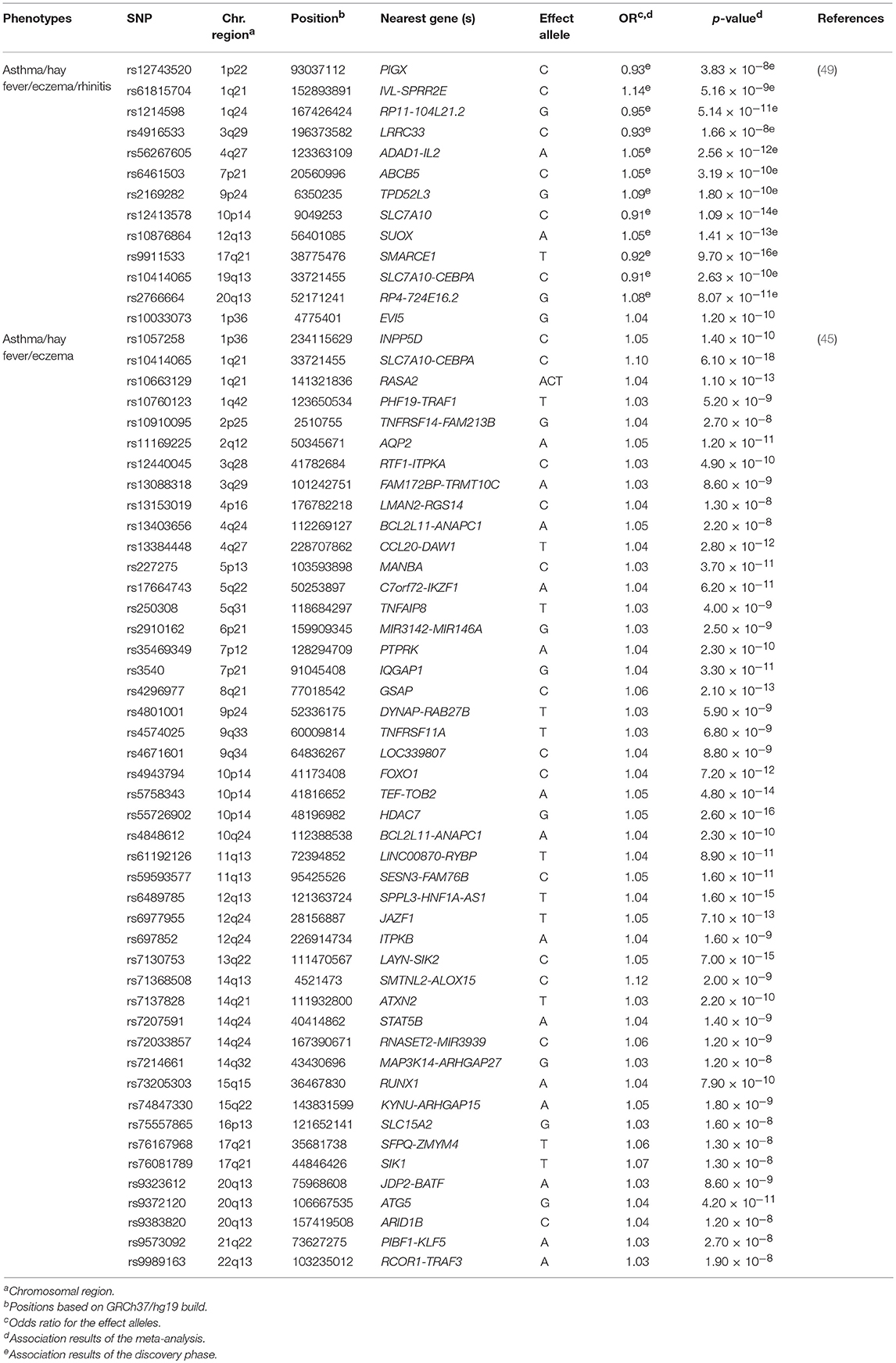
Table 4. Novel loci of asthma and allergic diseases revealed by meta-analyses published between 2016 and 2018.
Besides these findings, 29 of the genes identified by Ferreira et al. encode for proteins that are drug targets for several diseases, including allergic and auto-immune diseases. Interestingly, the protective effect of these genes was found to be correlated with the effect of drugs targeting them, attenuating allergy symptoms. These findings suggest that these could be effective targets to treat allergic diseases or asthma and thus, the proteins encoded by these should be prioritized for pre-clinical evaluation (45).
That GWAS was further complemented in a separate study with a gene-based association analysis using an algorithm that was specifically designed to identify shared risk variants among multiple phenotypes. With this approximation, which helps to increase the statistical power to detect novel risk loci, multiple variants near or within each gene are tested rather than focusing on individual SNP tests at a time (103). By relying on the information of SNPs that modify gene expression levels in different tissues and cell types, also known as expression quantitative trait loci (103, 105), they additionally revealed 19 novel risk genes for allergic diseases, which were not revealed by the previous stages of the study (45). Among these genes, nine showed functions that were closely related to well-known mechanisms involved in allergic diseases and asthma. Although further functional validation is needed, these could also represent novel drug targets (103).
Recently, Zhu et al. performed another GWAS in 110,361 Europeans in an attempt to identify genetic variants shared among asthma, hay fever, eczema and rhinitis (Supplementary Table 1). After performing a cross-trait meta-analysis, 38 loci were associated with both asthma and allergic diseases at genome-wide significance level (Supplementary Table 2). These loci were enriched in essential pathways for several tissues, such as skin, lung and whole blood, among others (49). Among these results, seven hits were novel loci that might contribute to the common genetic architecture of asthma and allergic diseases (49) (Table 4). These findings were consistent with the results reported by Ferreira et al. (45). In fact, a high proportion of the loci revealed by Zhu et al. (49) had been previously identified by Ferreira et al. (45). Interestingly, most of them had functions related to immune response, inflammation and epithelium maintenance, such as HLA-DQB1-AS1, IL1RL1, FLG-AS1, and STAT6, among others (45, 49).
Admixture Mapping
It has been evidenced that differences in asthma prevalence and severity among populations and ethnic groups could be partly explained by population-specific genetic factors. Alternative genetic scanning methods take advantage of these population specificities that are not frequently explored by means of GWAS approaches (106, 107), which could contribute to further identify asthma genes. One of such approaches is based on the exploration of the variation in genetic ancestry at chromosome-segment level (termed local ancestry) and the correlation with asthma in populations that are the result of a recent historical admixture, an approach most commonly known as admixture mapping (108, 109). Although both GWAS and admixture mapping are based on genome-wide data obtained by means of genotyping platforms, admixture mapping compares local ancestry estimates with the disease or trait (63). With this approach, a much smaller number of comparisons are involved in the scan, which highly increases the statistical power to detect associations compared to a traditional GWAS approach for a given sample size (110). Besides, this approach gives an opportunity for leveraging the specific genetic architecture of admixed populations, which have been largely underrepresented in genetic studies of asthma (111, 112). Admixed populations are characterized by high correlation over large chromosomal regions resulting from the recent admixture process, therefore these populations simplify gene mapping over longer distances (113–116). Moreover, if the trait of interest affects differentially the parental populations of the admixed, large trait differences are expected among admixed individuals, providing increased power to detect novel associations (117). Besides, the particular allelic configurations of the admixed individuals could interact with genetic risks for asthma, mitigating, or enhancing their effects in disease. In fact, causal variants are transmitted in higher proportion from the parental to the admixed populations, which leads to higher prevalence in the latter (112). As a consequence, it is expected that the proportion of the parental ancestry at those loci will vary between asthma cases and control individuals (110, 113), which would be indicative of ancestry-specific genetic risks (63). Combining admixture mapping with the traditional GWAS is a suitable strategy to identify both asthma risks that are ancestry-specific and those that are shared among different ancestry groups (63, 114). Although recently admixed populations are abundant (63), those with African admixture have been the prevalent in the asthma field (69, 109, 115).
There are two major African-admixed populations in the United States: African Americans and Latinos/Hispanics, which show different proportions of ancestry from each parental population (63). However, both show evident genetic footprints of the African admixture (116). Although very simplified, Latinos/Hispanics are usually modeled as descendants of ancient Native American, European and sub-Saharan African populations (63, 117), whereas current African Americans are modeled as descendants of an admixture event between sub-Saharan African and Europeans (63, 110, 118). Interestingly, compared to European Americans, asthma prevalence is higher in these populations, which also show a decreased response to asthma medications (14, 69, 119).
During the last decades, several loci have been associated with asthma and related traits in African Americans and Latinos/Hispanics using admixture mapping analysis as it has been reviewed by Mersha et al. (63) and Hernandez-Pacheco et al. (109). Additionally, two more admixture mapping analysis of asthma susceptibility and treatment response in African-admixed populations had been published by September 2018 (36, 120).
Spear et al. performed a genome-wide exploration in order to identify those genomic regions in which African ancestry is associated with response to asthma treatment with SABA in 949 African Americans. They found that local African ancestry at the 8p11 locus was suggestively associated with BDR in African American children with asthma, though the result did not reach significance level after considering the multiple comparisons (β = 1.49, p = 6.34 × 10−4) (36).
Additionally, Gignoux et al. revealed that the risk linked to the 18q21 locus in the admixture mapping peak in Latinos/Hispanics was driven by the Native American ancestry (OR = 1.20, p = 1.63 × 10−3), whereas the European ancestry was protective (OR = 0.86, p = 8.35 × 10−3), which was validated in an independent Hispanic/Latino population. Interestingly, this peak is located within the intergenic region of SMAD2 and ZBTB7C, none of which have been previously associated with any asthma-related trait, even in GWAS analyzing the same study populations (120–122), suggesting that admixture mapping is a powerful approach to identify novel asthma loci in admixed populations (120). The SMAD2 gene encodes a cofactor involved in regulation of the growth factor β signaling that has been extensively evidenced to play a key role in asthma (69, 123, 124).
Asthma Prediction and Translation into the Clinic
In the last 2 years, many asthma genes have been discovered and validated in independent populations, strongly supporting that these are generally involved in asthma pathogenesis. However, the genetic risk factors identified to date only represent a small proportion of total asthma heritability (22, 23, 125). Therefore, despite the uncountable advantages of the GWAS compared to previous strategies (23, 27, 126), there are a number of challenges ahead in order to better understand the genetic architecture of asthma (23, 126, 127).
One of the potential explanations of the current difficulty in explaining a larger proportion of the disease could be the reduced effect size of most genetic risks. Current reference panels and genotyping platforms are mainly designed to capture common genetic variants with are anticipated to show small effects in the disease (23, 114, 125). Therefore, the unassessed genomic variation could help to explain the missing heritability of asthma (114, 128).
Most GWAS of asthma have been limited in terms of low statistical power (23, 125, 127) due to limitations in the study design, mostly due to reduced sample sizes or the underrepresentation of genetically diverse populations, among others (20, 47, 48, 114, 129). A solution to this problem has been attempted in the last years with the emergence of large consortia gathering many asthma studies from different countries around the world (121, 122, 130, 131), which have contributed to increase the representation of patients from multiethnic populations (126, 127). However, this continuous need to increase sample sizes might have also led to heterogeneity in asthma definition by means of combination of samples with different asthma phenotypes. Consequently, this could have contributed to the reduction of the statistical power driven by the dilution of the effect size of association signals among different phenotypes (23, 125, 132–134). Thus, there is an increasing need to accurately characterize asthma patients through classification into homogenous groups (23, 134).
On the other hand, a limited number of large-scale studies have explored the role of gene-environment interactions in asthma despite robust evidence of the important contribution of environmental exposures in asthma susceptibility and severity (15, 16, 86, 135, 136). In fact, it has been evidenced underscoring the significant environmental contribution while designing a GWAS could result in reduced effect sizes (22, 137).
Last but not least, the functional implications of most asthma loci still remain unknown. Therefore, further studies are needed to increase our understanding of the impact of these on genes and cellular function, and their contribution on the molecular mechanisms underlying asthma pathophysiology (22). These have been proposed to be disentangled by means of approaches combining GWAS data with information related to biological pathways or processes (138, 139). Nonetheless, only one GWAS-based pathway enrichment analysis of asthma has been performed to date (140).
Because of all of this, our current knowledge of asthma genetics hampers our capacity to predict disease progression and treatment response, preventing its use in the clinical practice (125, 127). As a result, there is still a long way to use this knowledge and their integration with lifestyle and environment exposures (127, 141, 142) to develop precision medicine strategies for accurate prevention, diagnosis, or treatment of asthma (23).
Other Omic Studies and Integration of Multiomics
Other omics technologies, apart from genomics, are powerful tools to increase the current knowledge about asthma pathophysiology (143, 144). These are focused on data from a wide variety of biological sources: genomic modifications (epigenomics), gene transcription (transcriptomics), protein levels and chemical modifications (proteomics), endogenous and exogenous metabolites (metabolomics), and the microbiome (metagenomics), among others (23, 126, 127, 145, 146). The application of omics approximations in asthma is still incipient compared to other diseases (23, 147). Still, several studies have been performed in asthma in the last years as it was reviewed elsewhere (23, 127, 148, 149).
To our knowledge, a total of 26 asthma studies using other omics approaches have been published in the last 2 years (Supplementary Table 3). Just like recent GWAS of asthma, these studies have been equally focused on childhood and adulthood asthma since 2016. Moreover, most of them have been carried out in patients of European descent. A total of 18 studies focused on asthma susceptibility or severity (150–167); three focused on the ICS response (168–170); two explored the interactions with environmental factors (171, 172); and three inspected the overlap with other pulmonary diseases (173–175). Nonetheless, other experts have discussed the recent omics advances in asthma in this issue except for those of transcriptomics. Therefore, we focused on summarizing the main findings of studies made using this approach.
Transcriptomics provide a quantitative and qualitative characterization or RNA transcripts (176). These are mainly focused on comparing gene expression levels in cells or tissues under specific controlled conditions in order to identify differentially expressed genes that could have (alone or in combination) functional implications on the disease under study (127, 177). Rapid development of technologies has made possible the near-complete characterization of the transcriptome, first using arrays and later, by means of RNA sequencing, which has greatly promoted the genome-wide exploration of transcriptomic changes in asthma during the last years (23, 127). A number of advantages of transcriptomics studies in asthma have been extensively described (23, 126). In fact, it has been proposed to be an accurate method to characterize pathways contributing to asthma pathophysiology, and the interactions with exogenous and endogenous factors in different sample types such as, blood, sputum or lung tissues, among others (23, 146).
Transcriptomics is a powerful tool to provide or confirm a mechanistic explanation of asthma loci identified by GWAS (23). Eight transcriptomic studies of asthma have been recently performed (Supplementary Table 3). However, most of them were carried out using arrays (157, 158, 167, 170–172) and the majority focused on European populations (157, 163, 170–172) and adults (157, 163, 171, 172, 175). Only three of them explored differential gene expression in children with asthma (158, 167, 170).
The largest transcriptomic study of childhood asthma performed in the last 2 years explored array-based gene expression levels of 133 asthma patients and 11 healthy controls of Asian ancestry (167) (Supplementary Table 3). RNA was extracted from a mixed population of T cells that were isolated from peripheral blood (167). Yeh et al. classified asthma patients into three groups based on 2,048 genes differentially expressed in immune cells. These groups showed distinct inflammatory profiles, including one that clustered the patients with higher neutrophil count and the poorest treatment control, suggesting that these could correspond to those patients with the most severe asthma status. When this group was compared with asthma patients included in other groups, 163 genes were found to be upregulated. Most of these genes encoded proteins involved in glucocorticoid signaling pathway and the immune response, suggesting that this could be an accurate method to classify asthma patients based on transcriptomic data (167). In transcriptomic studies of adulthood asthma, solid or liquid airway samples are frequently used such as, sputum or lung tissues (178, 179). However, clinical procedures to obtain these samples are quite invasive and are especially impractical in children (167). For this reason, peripheral blood has been regarded as the most suitable sample for the studies in children (23, 167).
Asthma diagnosis has classically relied just on conventional clinical guidelines and biomarkers for over decades (180), which are considered very inaccurate due to the wide variety of molecular mechanisms underlying the different asthma phenotypes (181, 182). Is in this respect where integrative approaches that combine complete clinical data and the omics sources could contribute to better characterize the biological processes underlying asthma pathophysiology (182, 183), ultimately helping to define asthma subtypes and to improve the prediction of severity and treatment response.
Multiomics approaches, which incorporate information from different omics levels, have been suggested as a promising strategy to fulfill that purpose (144) as they show an increased predictive capacity (166, 184). Five multiomics studies of asthma and related traits have been performed since 2016. Most of them have combined only a few omics levels (166, 185–188) (Supplementary Table 3).
Forno et al. conducted the largest multiomics study of asthma to date (186). They proposed a novel vertical approach to combine data from different omics levels (genomics, epigenomics, transcriptomics, and proteomics) with clinical information available for 1,127 Latino/Hispanic children, including 618 asthma patients and 509 children without asthma (Supplementary Table 3). Expression of 1,645 genes was associated with cytokine levels in blood, revealing the enrichment of the cytokine signaling pathway. From the 269 genes involved in this pathway, 41 were significantly associated with more than two asthma intermediate phenotypes. As a result, this list was reduced to the IL5RA gene, which was found to be the most significant association at the following steps (186). In fact, several transcription factors previously associated with pulmonary diseases showed evidence of association with IL5RA (189–191), suggesting that these could be involved in its signaling pathway. Furthermore, low plasma levels of IL-5Rα were found in children with asthma exacerbations, whereas children with earlier age of asthma onset showed increased levels of IL-5Rα, providing firm evidence of implication of IL5RA on asthma (186).
Studies as this one suggests that vertical approaches could be a suitable strategy to perform integrative multiomics studies of asthma and even other diseases. However, further validation in independent populations and other complex diseases is needed to confirm the applicability of this method (192). In this respect, a few omics studies of asthma treatment been performed to date, opening an opportunity to identify novel markers that could be applied in the design of precision medicine approaches in asthma and novel therapeutic strategies (67, 193). Although omics approaches have promisingly broken new ground in asthma research, translation into the clinic is still very challenging due to the large amount of information that is obtained.
Unexplored Genetic Variation in Asthma
Exploring non-coding variation has been also proposed as a promising strategy to disentangle the genetic basis of complex diseases (114) such as, microRNAs (miRNAs). These are short, non-coding and single-strand RNA molecules that interact with different genomic elements and regulate gene expression at transcriptional level (148, 184, 194). Interestingly, these are involved in the regulation of the stability of immune cells and the intensity of inflammation (194). In fact, miRNAs have been proposed as potential non-invasive asthma biomarkers that could be used for asthma diagnosis (195, 196). However, although some authors have suggested the implication of miRNAs on asthma susceptibility, severity, and exacerbations (195, 197, 198), there is a lack of studies that have extensively evaluated their role in asthma (197) and further studies are needed (199).
Structural variation, including copy number variations (CNVs), has been proposed to account for part of the missing heritability of complex traits (114, 200). These involve large chromosomal segments such as, duplications or deletions with consequences on regulation of gene expression (201). It has been reported that CNVs comprise 2% of the total genetic variation (202) with effects on approximately 12% of the human genome (203). This type of variation is enriched within protein-coding genes with functions related to immune response, suggesting its implication on disorders with a significant immunological component such as, asthma (204, 205). Although structural variation has been implicated on asthma, these is an insufficient number of studies to date (206). Some have found strong evidence of association of CNVs with asthma susceptibility (206, 207). Although this type of variation might contribute to an accumulation of mutations and allergic sensitization, leading to an increase of asthma susceptibility, CNVs do not seem to be the initial trigger of asthma development (208).
A substantial proportion of the genetic risk for common diseases could also be explained by variants that are at low frequency in the population (209, 210). The rarer the variant the more likely is for the variant to be population-specific (209, 210). Besides, the pathogenic potential of variants tends to accumulate in the lower range of allele frequency. Therefore, rare variants are more likely to be more structured in populations and to have larger effects on the disease (128, 209). As a corollary, rare variants will be underrepresented in reference datasets and, therefore, remain undetected by traditional GWAS. With this scenario in mind, many rare variants with large effects may be contributing to asthma and allergic diseases (128, 209, 211). However, their study will be only available for now applying sequencing-based methods instead of genotyping arrays. Irrespective of this, endogamous populations are especially appropriate to study the role of rare and low-frequency variation in asthma (210, 212) because rare pathogenic variants are predicted to increase their frequency in these populations (213, 214). Despite this, recent studies have also demonstrated the role of rare variants on recently admixed populations (209), whose inherent characteristics also increase the possibilities to uncover the contribution of rare and low-frequency variants on asthma (215).
Predicted loss-of-function (pLoF) variants, which are likely involved in disrupting protein-coding genes, show significant scientific and clinical interest due to their utility for clinical interpretation of sequencing data. In fact, pLoF variants have been suggested to allow direct identification of causal genes (216) and provide direct mechanistic implications of association effects (217). Although this type of variation has been extensively unexplored for over decades (218), Emdin et al. have recently revealed the potential role of these variants in asthma (217). They found evidence of association of pLoF variants located at well-known IL33 and GSDMB asthma loci with lower risk of both asthma and allergic rhinitis (217). Interestingly, similar results have been found for protein-truncating variants in IL33 and GSDMB (219). These have been predicted to shorten the coding sequence by inducing loss or gain-of-function effects (220). These findings also suggested that exploration of either pLoFs or protein-truncating variants could be another powerful tool for the identification of novel therapies for asthma (217, 219).
As mentioned, the main reason for the scarce evaluation of these types of genetic variation in asthma could be attributed to the fact that research strategies on asthma genetics have focused on using SNP genotyping platforms, which are suboptimal for inferring CNVs, and do not capture rare or pLoF variants (23, 29, 114, 125), as these would be optimally detected by means of sequencing approaches. Given that simultaneous sequencing of millions of small DNA fragments is currently possible at great speed and relatively low cost thanks to large improvements in next-generation sequencing (NGS) technologies (221, 222), the interest on the impact of these types of genetic variation in asthma will continue to rise.
Estimation of Polygenic Risk Scores
Another example of the large efforts to try to accelerate the progression toward precision medicine in complex traits is an emerging approach that takes GWAS results as the start point. This consists of stratification of the whole population based on estimates of individual's genetic disease susceptibility measured as polygenic risk scores (PRSs) (223). Just like other complex diseases, the genetic architecture of asthma is polygenic, where many genes contribute to disease development (224). Hence, the overall disease risk could be considered as the result of combined effects driven by common low-risk and rare large-risk variants (225). PRSs are the result of summing up risk alleles from thousand variants revealed by the GWAS (226). Even though most common variants show small effects, combining their effects could explain a significant proportion of the disease variability or at least allow classifying patients into discrete subgroups based on different levels of probabilistic disease risk (223, 226, 227).
Although multilocus profiles of genetic risks for asthma have been constructed using small sets of variants (101), large evaluations of PRSs are lacking for asthma. Nevertheless, previous studies that focused on other complex diseases (228–236) suggest that this approach could be fruitful for asthma. For instance, Khera et al. recently estimated PRSs for five common disorders with major public health impact, including coronary artery disease (236). They found 20-fold greater coronary artery disease risk using a PRSs involving many genetic variants than previous studies based on biomarkers or the mutation panels traditionally used in the clinical practice (236).
Uncountable utilities of PRSs have been demonstrated for the study of common diseases, suggesting its plausibility in a healthcare scenario (227). In fact, it has been proposed that PRS estimation could facilitate the development of accurate preventive, diagnostic and therapeutic strategies (223, 227, 236). Moreover, given the previous evidence of genetic overlap among different diseases (35, 45, 49, 103, 227, 237), an evaluation of individual risks could be assessed simultaneously for multiple traits at a time. This would potentiate the implementation of common therapeutic strategies for different diseases (227, 236). For all these reasons, calculation of PRS has been considered as a feasible approach to translate asthma research findings into healthcare practice for early disease detection (227).
There are many technical, economic, and sociopolitical barriers that should be overcome for the use of PRSs into clinical practice. By one hand, physicians would need additional training to correctly interpret and communicate PRSs to the patients (227). On the other hand, most current PRS estimates are based on loci that were mapped using designs with an overwhelming number of European patients. Therefore, their generalizability in populations of non-European ancestries are questionable (223, 236, 238) due to the large differences in terms of effect sizes, allele frequencies and linkage disequilibrium patterns. Besides this, most PRSs have been estimated in adults. Therefore, an evaluation of their usefulness in other age groups will be needed.
For asthma, the major limitation of PRSs is related to the reduced proportion of heritability explained by the loci identified to date. Given that PRS is a quantitative measure of the individual genetic risk, the more genetic variants are incorporated into the predictive disease risk model, the better the individuals are stratified into the risk subgroups (226). In this scenario, some studies suggest that whole-genome prediction models may account for the unknown genetic risks and, therefore, be able to improve the capacity to predict disease susceptibility, outcomes and treatment response, where the contribution of rare and low-frequency variants will be particularly relevant (223, 239).
On these terms, complex diseases could be comparable to rare disorders, where rare variants with large effect sizes provide disease risk in a small proportion of the population (127, 240). Large-scale sequencing studies will be required to further assess this idea (226, 240).
Future Perspectives
Despite the large insight provided by GWAS and the admixture mapping scans during the last decades, it remains a large proportion of the missing heritability yet to be ascertained for asthma and related traits (22, 23, 125–127). The future of genetic research in asthma will be driven by NGS approaches, which are expected to significantly increase our knowledge of many other complex diseases (114, 126, 241).
The use of NGS technologies in pulmonary diseases is still emerging (242–246). More specifically, only a few asthma genetics studies have used NGS technologies (242, 243, 246) and large consortia studies are underway (247, 248). Because of its prohibitive costs for large population studies, several strategies have been proposed, such as sequencing the subjects from the extremes of the phenotype distribution (245, 246, 249) or the families where multiple individuals affected (250). The combination of NGS with conventional GWAS approaches has been suggested as another promising strategy (251).
Although the limited knowledge of genetic factors involved in asthma available to date hampers our current capacity to predict disease progression and treatment response (22, 23, 125), the use of genetic information to develop novel therapeutic targets is plausible. For instance, DeBoewe et al. recently found the association of protein-truncating variants with asthma located within widely known asthma susceptibility loci, such as IL33 or GSDMB. This reinforced the evidence suggesting the capacity of the genetic research to find potential asthma therapeutic targets. In fact, as a result of GWAS findings, several drugs targeting IL6R, IL-33, and TSLP are in development or are being evaluated in ongoing clinical trials investigating their efficacy to treat asthma and allergic diseases (22, 252, 253).
Conclusions
Our knowledge of asthma genetics has been greatly improved over the last decade because of GWAS, revealing a number of novel and firm common risk factors with small effects that overall explain a limited proportion of asthma heritability. Nonetheless, the improvements in high-throughput sequencing technologies and their anticipated cost reductions have the promise to accelerate the transition of this knowledge into the clinical practice and to progressively redirect the field toward an integrative multiomics perspective.
Author Contributions
All the authors were involved in the conception, hypotheses delineation and structuration of this article, drafted the article, and approved the final version of the manuscript.
Funding
This work was supported by the grants PI17/00610 (CF) from Instituto de Salud Carlos III (ISCIII) co-financed by the European Regional Development Funds, A way of making Europe from the European Union; AC15/00015 through Strategic Action for Health Research (AES) and European Community (EC) within the Active and Assisted Living (AAL) Programme framework (MP-Y), and the SysPharmPedia grant from the ERACoSysMed 1st Joint Transnational Call from the European Union under the Horizon 2020. NH-P was funded by a fellowship (FI16/00136) from ISCIII and co-funded by the European Social Funds from the European Union (ESF) ESF invests in your future. MP-Y was supported by the Ramón y Cajal Program (RYC-2015-17205) and by grant SAF2017-83417R by the Spanish Ministry of Economy, Industry and Competitiveness. CF was also supported by Spanish Ministry of Science, Innovation and Universities (RTC-2017-6471-1; MINECO/AEI/FEDER, UE); and by the agreement OA17/008 with Instituto Tecnológico y de Energías Renovables (ITER) to strengthen scientific and technological education, training, research, development, and innovation in Genomics, Personalized Medicine and Biotechnology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2019.00006/full#supplementary-material
Abbreviations
BDR, bronchodilator response; CNVs, copy number variations; GWAS, genome-wide association study; GWIS, genome-wide interaction study; ICS, inhaled corticosteroids; IL-5, interleukin 5; miRNA, microRNA; NGS, next-generation sequencing; OR, odds ratio; pLoF, predicted loss-of-function; PRS, polygenic risk score; SABA, short-acting β2 agonists; SNP, single nucleotide polymorphism.
References
1. Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. (2010) 42:565–9. doi: 10.1038/ng.608
2. Global Strategy for Asthma Management and Prevention. Global Initiative for Asthma (GINA) (2017). Available online at: http://ginasthma.org/ (Accessed August 20, 2018).
3. Ferkol T, Schraufnagel D. The global burden of respiratory disease. Ann Am Thorac Soc. (2014) 11:404–6. doi: 10.1513/AnnalsATS.201311-405PS
4. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet (2013) 380:2095–128. doi: 10.1016/S0140-6736(12)61728-0
5. To T, Wang C, Guan J, McLimont S, Gershon AS. What is the lifetime risk of physician-diagnosed asthma in Ontario, Canada? Am J Respir Crit Care Med. (2010) 181:337–43. doi: 10.1164/rccm.200907-1035OC
6. Facts on Asthma. World Health Organization (2011). Available online at: www.who.int/features/factfiles/asthma/en/ (Accesed September 12, 2018).
7. Nunes C, Ladeira S. Asthma, from childhood to adulthood: a prospective 20-year longitudinal study of a cohort of asthmatics. J Investig Allergol Clin Immunol. (2002) 12:242–9. doi: 10.1053/rmed.1999.076
8. Akdis CA, Agache I. Global Atlas of Asthma. Zurich: European Academy of Allergy and Clinical Immunology (2013).
9. Braman SS. The global burden of asthma. Chest (2006) 130(1 Suppl):4S–12S. doi: 10.1378/chest.130.1_suppl.4S
10. Williams SA, Wagner S, Kannan H, Bolge SC. The association between asthma control and health care utilization, work productivity loss and health-related quality of life. J Occup Environ Med. (2009) 51:780–5. doi: 10.1097/JOM.0b013e3181abb019
11. Ferrante G, La Grutta S. The Burden of Pediatric Asthma. Front Pediatr. (2018) 6:186. doi: 10.3389/fped.2018.00186
12. Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy (2004) 59:469–78. doi: 10.1111/j.1398-9995.2004.00526.x
13. Van Wonderen KE, Van Der Mark LB, Mohrs J, Bindels PJ, Van Aalderen WM, Ter Riet G. Different definitions in childhood asthma: how dependable is the dependent variable? Eur Respir J. (2010) 36:48–56. doi: 10.1183/09031936.00154409
14. Akinbami LJ, Moorman JE, Simon AE, Schoendorf KC. Trends in racial disparities for asthma outcomes among children 0 to 17 years, 2001–2010. J Allergy Clin Immunol. (2014) 134:547–53 e5. doi: 10.1016/j.jaci.2014.05.037
15. Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med. (2013) 188:309–18. doi: 10.1164/rccm.201302-0264OC
16. Oh SS, White MJ, Gignoux CR, Burchard EG. Making Precision Medicine Socially Precise. Take a Deep Breath. Am J Respir Crit Care Med. (2016) 193:348–50. doi: 10.1164/rccm.201510-2045ED
17. Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet. (2014) 15:765–76. doi: 10.1038/nrg3786
18. Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet. (2017) 49:1319–25. doi: 10.1038/ng.3931
19. Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy (2006) 36:1382–90. doi: 10.1111/j.1365-2222.2006.02512.x
20. Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. (2011) 242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x
21. Ullemar V, Magnusson PK, Lundholm C, Zettergren A, Melen E, Lichtenstein P, et al. Heritability and confirmation of genetic association studies for childhood asthma in twins. Allergy (2016) 71:230–8. doi: 10.1111/all.12783
22. Vicente CT, Revez JA, Ferreira MAR. Lessons from ten years of genome-wide association studies of asthma. Clin Transl Immunol. (2017) 6:e165. doi: 10.1038/cti.2017.54
23. Willis-Owen SAG, Cookson WOC, Moffatt MF. The genetics and genomics of asthma. Annu Rev Genomics Hum Genet. (2018) 19:223–46. doi: 10.1146/annurev-genom-083117-021651
24. Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, et al. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. (2008) 9:38. doi: 10.1186/1465-9921-9-38
25. Bouzigon E, Forabosco P, Koppelman GH, Cookson WO, Dizier MH, Duffy DL, et al. Meta-analysis of 20 genome-wide linkage studies evidenced new regions linked to asthma and atopy. Eur J Hum Genet. (2010) 18:700–6. doi: 10.1038/ejhg.2009.224
26. Moheimani F, Hsu AC, Reid AT, Williams T, Kicic A, Stick SM, et al. The genetic and epigenetic landscapes of the epithelium in asthma. Respir Res. (2016) 17:119. doi: 10.1186/s12931-016-0434-4
27. Ober C. Asthma Genetics in the Post-GWAS Era. Ann Am Thorac Soc. (2016) 13 (Suppl. 1):S85–90. doi: 10.1513/AnnalsATS.201507-459MG
28. Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic diseases: a comprehensive review of the literature. Respir Res. (2003) 4:14. doi: 10.1186/1465-9921-4-14
29. Foulkes AS. Genetic association studies, In: Gentleman R, Hornik K, Parmigiani G. editors. Applied Statistical Genetics with R. For Population-based Association Studies, (New York, NY: Springer-Verlag) (2009). p. 1–27. doi: 10.1007/978-0-387-89554-3
30. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature (2007) 448:470–3. doi: 10.1038/nature06014
31. Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. (2016) 48:709–17. doi: 10.1038/ng.3570
32. MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. (2017) 45:D896–901. doi: 10.1093/nar/gkw1133
33. Schieck M, Schouten JP, Michel S, Suttner K, Toncheva AA, Gaertner VD, et al. Doublesex and mab-3 related transcription factor 1 (DMRT1) is a sex-specific genetic determinant of childhood-onset asthma and is expressed in testis and macrophages. J Allergy Clin Immunol. (2016) 138:421–31. doi: 10.1016/j.jaci.2015.12.1305
34. Gref A, Merid SK, Gruzieva O, Ballereau S, Becker A, Bellander T, et al. Genome-Wide Interaction Analysis of Air Pollution Exposure and Childhood Asthma with Functional Follow-up. Am J Respir Crit Care Med. (2017) 195:1373–83. doi: 10.1164/rccm.201605-1026OC
35. Marques CR, Costa GN, da Silva TM, Oliveira P, Cruz AA, Alcantara-Neves NM, et al. Suggestive association between variants in IL1RAPL and asthma symptoms in Latin American children. Eur J Hum Genet. (2017) 25:439–45. doi: 10.1038/ejhg.2016.197
36. Spear ML, Hu D, Pino-Yanes M, Huntsman S, Eng C, Levin AM, et al. A genome-wide association and admixture mapping study of bronchodilator drug response in African Americans with asthma. Pharmacogenomics J. (2018). doi: 10.1038/s41397-018-0042-4 [Epub ahead of print].
37. Yatagai Y, Hirota T, Sakamoto T, Yamada H, Masuko H, Kaneko Y, et al. Variants near the HLA complex group 22 gene (HCG22) confer increased susceptibility to late-onset asthma in Japanese populations. J Allergy Clin Immunol. (2016) 138:281–3 e13. doi: 10.1016/j.jaci.2015.11.023
38. Almoguera B, Vazquez L, Mentch F, Connolly J, Pacheco JA, Sundaresan AS, et al. Identification of Four Novel Loci in Asthma in European American and African American Populations. Am J Respir Crit Care Med. (2017) 195:456–63. doi: 10.1164/rccm.201604-0861OC
39. Condreay L, Chiano M, Ortega H, Buchan N, Harris E, Bleecker ER, et al. No genetic association detected with mepolizumab efficacy in severe asthma. Respir Med. (2017) 132:178–80. doi: 10.1016/j.rmed.2017.10.019
40. Mosteller M, Hosking L, Murphy K, Shen J, Song K, Nelson M, et al. No evidence of large genetic effects on steroid response in asthma patients. J Allergy Clin Immunol. (2017) 139:797–803 e7. doi: 10.1016/j.jaci.2016.05.032
41. Vonk JM, Scholtens S, Postma DS, Moffatt MF, Jarvis D, Ramasamy A, et al. Adult onset asthma and interaction between genes and active tobacco smoking: the GABRIEL consortium. PLoS ONE (2017) 12:e0172716. doi: 10.1371/journal.pone.0172716
42. Murk W, DeWan AT. Genome-wide search identifies a gene-gene interaction between 20p13 and 2q14 in asthma. BMC Genet. (2016) 17:102. doi: 10.1186/s12863-016-0376-3
43. Nieuwenhuis MA, Siedlinski M, van den Berge M, Granell R, Li X, Niens M, et al. Combining genomewide association study and lung eQTL analysis provides evidence for novel genes associated with asthma. Allergy (2016) 71:1712–20. doi: 10.1111/all.12990
44. Yan Q, Brehm J, Pino-Yanes M, Forno E, Lin J, Oh SS, et al. A meta-analysis of genome-wide association studies of asthma in Puerto Ricans. Eur Respir J. (2017) 49:1601505. doi: 10.1183/13993003.01505-2016
45. Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. (2017) 49:1752–7. doi: 10.1038/ng.3985
46. Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. (2018) 50:42–53. doi: 10.1038/s41588-017-0014-7
47. Burchard EG, Oh SS, Foreman MG, Celedon JC. Moving toward true inclusion of racial/ethnic minorities in federally funded studies. A key step for achieving respiratory health equality in the United States. Am J Respir Crit Care Med. (2015) 191:514–21. doi: 10.1164/rccm.201410-1944PP
48. Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature (2016) 538:161–4. doi: 10.1038/538161a
49. Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. (2018) 50:857–64. doi: 10.1038/s41588-018-0121-0
50. Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax (2012) 67:762–8. doi: 10.1136/thoraxjnl-2011-201262
51. Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KC, Koka V, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. (2009) 19:118–27. doi: 10.1101/gr.084798.108
52. Ono JG, Worgall TS, Worgall S. 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr Res. (2014) 75:165–70. doi: 10.1038/pr.2013.186
53. Das S, Miller M, Broide DH. Chromosome 17q21 Genes ORMDL3 and GSDMB in Asthma and Immune Diseases. Adv Immunol. (2017) 135:1–52. doi: 10.1016/bs.ai.2017.06.001
54. Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A, et al. Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet. (2012) 131:1161–71. doi: 10.1007/s00439-012-1142-x
55. Hovelso N, Sotty F, Montezinho LP, Pinheiro PS, Herrik KF, Mork A. Therapeutic potential of metabotropic glutamate receptor modulators. Curr Neuropharmacol. (2012) 10:12–48. doi: 10.2174/157015912799362805
56. Pang Y, Zhao J, Fowdur M, Liu Y, Wu H, He M. To explore the mechanism of the GRM4 gene in osteosarcoma by RNA sequencing and bioinformatics approach. Med Sci Monit Basic Res. (2018) 24:16–25. doi: 10.12659/MSMBR.908107
57. Parihar R, Mishra R, Singh SK, Jayalakshmi S, Mehndiratta MM, Ganesh S. Association of the GRM4 gene variants with juvenile myoclonic epilepsy in an Indian population. J Genet (2014) 93:193–7. doi: 10.1007/s12041-014-0334-7
58. Muhle H, von Spiczak S, Gaus V, Kara S, Helbig I, Hampe J, et al. Role of GRM4 in idiopathic generalized epilepsies analysed by genetic association and sequence analysis. Epilepsy Res. (2010) 89:319–26. doi: 10.1016/j.eplepsyres.2010.02.004
59. Dadkhah T, Rahimi-Aliabadi S, Jamshidi J, Ghaedi H, Taghavi S, Shokraeian P, et al. A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J Affect Disord. (2017) 208:218–22. doi: 10.1016/j.jad.2016.10.008
60. Wang K, Zhao J, He M, Fowdur M, Jiang T, Luo S. Association of GRM4 gene polymorphisms with susceptibility and clinicopathological characteristics of osteosarcoma in Guangxi Chinese population. Tumour Biol. (2016) 37:1105–12. doi: 10.1007/s13277-015-3904-2
61. Said SI. Glutamate receptors and asthmatic airway disease. Trends Pharmacol Sci. (1999) 20:132–4.
62. Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. (2005) 115:233–42. doi: 10.1016/j.jaci.2004.11.014
63. Mersha TB. Mapping asthma-associated variants in admixed populations. Front Genet. (2015) 6:292. doi: 10.3389/fgene.2015.00292
64. Vijverberg SJH, Farzan N, Slob EMA, Neerincx AH, Maitland-van der Zee AH. Treatment response heterogeneity in asthma: the role of genetic variation. Exp Rev Respir Med. (2018) 12:55–65. doi: 10.1080/17476348.2018.1403318
65. Weiss ST. New approaches to personalized medicine for asthma: where are we? J Allergy Clin Immunol. (2012) 129:327–34. doi: 10.1016/j.jaci.2011.12.971
66. Ortega VE, Meyers DA, Bleecker ER. Asthma pharmacogenetics and the development of genetic profiles for personalized medicine. Pharmgenomics Pers Med. (2015) 8:9–22. doi: 10.2147/PGPM.S52846
67. Levy ML, Winter R. Asthma deaths: what now? Thorax (2015) 70:209–10. doi: 10.1136/thoraxjnl-2015-206800
68. Sears MR, Lotvall J. Past, present and future–beta2-adrenoceptor agonists in asthma management. Respir Med. (2005) 99:152–70. doi: 10.1016/j.rmed.2004.07.003
69. Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. (2004) 169:386–92. doi: 10.1164/rccm.200309-1293OC
70. Drake KA, Torgerson DG, Gignoux CR, Galanter JM, Roth LA, Huntsman S, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. (2014) 133:370–8. doi: 10.1016/j.jaci.2013.06.043
71. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (2009) 25:1754–60. doi: 10.1093/bioinformatics/btp324
72. Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. (2012) 91:839–48. doi: 10.1016/j.ajhg.2012.09.004
73. Li Z, Xi X, Gu M, Feil R, Ye RD, Eigenthaler M, et al. A stimulatory role for cGMP-dependent protein kinase in platelet activation. Cell (2003) 112:77–86. doi: 10.1016/S0092-8674(02)01254-0
74. Burgoyne JR, Madhani M, Cuello F, Charles RL, Brennan JP, Schroder E, et al. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science (2007) 317:1393–7. doi: 10.1126/science.1144318
75. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. (2012) 22:1790–7. doi: 10.1101/gr.137323.112
76. Dawes M, Chowienczyk PJ, Ritter JM. Effects of inhibition of the L-arginine/nitric oxide pathway on vasodilation caused by beta-adrenergic agonists in human forearm. Circulation (1997) 95:2293–7.
77. Jorgensen IM, Jensen VB, Bulow S, Dahm TL, Prahl P, Juel K. Asthma mortality in the Danish child population: risk factors and causes of asthma death. Pediatr Pulmonol. (2003) 36:142–7. doi: 10.1002/ppul.10305
78. Vijverberg SJ, Raaijmakers JA, Maitland-van der Zee AH. ADRB2 Arg16 and the need for collaboration in childhood asthma pharmacogenomics. Pharmacogenomics (2013) 14:1937–9. doi: 10.2217/pgs.13.195
79. Vijverberg SJ, Turner SW, Palmer CN, Tantisira KG, Maitland-van der Zee AH. Realizing personalized medicine in asthmatic children requires large-scale collaboration. Pediat Therapeut. (2015) 5:e127. doi: 10.4172/2161-0665.1000e127
80. Farzan N, Vijverberg SJ, Arets HG, Raaijmakers JA, Maitland-van der Zee AH. Pharmacogenomics of inhaled corticosteroids and leukotriene modifiers: a systematic review. Clin Exp Allergy (2017) 47:271–93. doi: 10.1111/cea.12844
81. Busse WW. Biological treatments for severe asthma: where do we stand? Curr Opin Allergy Clin Immunol. (2018) 18:509–18. doi: 10.1097/ACI.0000000000000487
82. Bel EH, Wenzel SE, Thompson PJ, Prazma CM, Keene ON, Yancey SW, et al. Oral glucocorticoid-sparing effect of mepolizumab in eosinophilic asthma. N Engl J Med. (2014) 371:1189–97. doi: 10.1056/NEJMoa1403291
83. Farne HA, Wilson A, Powell C, Bax L, Milan SJ. Anti-IL5 therapies for asthma. Cochr Database Syst Rev. (2017) 9:CD010834. doi: 10.1002/14651858.CD010834.pub3
84. Park HW, Tantisira KG, Weiss ST. Pharmacogenomics in asthma therapy: where are we and where do we go? Annu Rev Pharmacol Toxicol. (2015) 55:129–47. doi: 10.1146/annurev-pharmtox-010814-124543
85. Herrera-Luis E, Hernandez-Pacheco N, Vijverberg SJ, Flores C, Pino-Yanes M. Role of genomics in asthma exacerbations. Curr Opin Pulm Med. (2019). 25:101–12. doi: 10.1097/MCP.0000000000000533
86. Mallol J, Crane J, von Mutius E, Odhiambo J, Keil U, Stewart A. The International Study of Asthma and Allergies in Childhood (ISAAC) Phase Three: a global synthesis. Allergol Immunopathol. (2013) 41:73–85. doi: 10.1016/j.aller.2012.03.001
87. West CE, Renz H, Jenmalm MC, Kozyrskyj AL, Allen KJ, Vuillermin P, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol. (2015) 135:3–13; quiz 4. doi: 10.1016/j.jaci.2014.11.012
88. Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. (2011) 128:646–52 e1–5. doi: 10.1016/j.jaci.2011.04.060
89. Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc. (2009) 6:660–2. doi: 10.1513/pats.200907-065DP
90. Kauffmann F, Nadif R. Candidate gene-environment interactions. J Epidemiol Community Health (2010) 64:188–9. doi: 10.1136/jech.2008.086199
91. Kauffmann F, Demenais F. Gene-environment interactions in asthma and allergic diseases: challenges and perspectives. J Allergy Clin Immunol. (2012) 130:1229–40; quiz 41–2. doi: 10.1016/j.jaci.2012.10.038
92. Gehring U, Gruzieva O, Agius RM, Beelen R, Custovic A, Cyrys J, et al. Air pollution exposure and lung function in children: the ESCAPE project. Environ Health Perspect. (2013) 121:1357–64. doi: 10.1289/ehp.1306770
93. Song ZZ. Early-Life Exposure to Traffic-related Air Pollution and Lung Function in Adolescence. Am J Respir Crit Care Med. (2016) 194:385. doi: 10.1164/rccm.201603-0565LE
94. Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. (2000) 67:1154–62. doi: 10.1016/S0002-9297(07)62946-2
95. Elliot JG, Carroll NG, James AL, Robinson PJ. Airway alveolar attachment points and exposure to cigarette smoke in utero. Am J Respir Crit Care Med. (2003) 167:45–9. doi: 10.1164/rccm.2110005
96. Magnusson LL, Olesen AB, Wennborg H, Olsen J. Wheezing, asthma, hayfever, and atopic eczema in childhood following exposure to tobacco smoke in fetal life. Clin Exp Allergy (2005) 35:1550–6. doi: 10.1111/j.1365-2222.2005.02374.x
97. Alati R, Al Mamun A, O'Callaghan M, Najman JM, Williams GM. In utero and postnatal maternal smoking and asthma in adolescence. Epidemiology (2006) 17:138–44. doi: 10.1097/01.ede.0000198148.02347.33
98. Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. (2008) 177:567–73. doi: 10.1164/rccm.200710-1511PP
99. Scholtens S, Postma DS, Moffatt MF, Panasevich S, Granell R, Henderson AJ, et al. Novel childhood asthma genes interact with in utero and early-life tobacco smoke exposure. J Allergy Clin Immunol. (2014) 133:885–8. doi: 10.1016/j.jaci.2013.08.049
100. Vogelberg C, Hirsch T, Radon K, Dressel H, Windstetter D, Weinmayr G, et al. Leisure time activity and new onset of wheezing during adolescence. Eur Respir J. (2007) 30:672–6. doi: 10.1183/09031936.00152906
101. Belsky DW, Sears MR, Hancox RJ, Harrington H, Houts R, Moffitt TE, et al. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir Med. (2013) 1:453–61. doi: 10.1016/S2213-2600(13)70101-2
102. Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. (2013) 45:907–11. doi: 10.1038/ng.2686
103. Ferreira MAR, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Eleven loci with new reproducible genetic associations with allergic disease risk. J Allergy Clin Immunol. (2018). doi: 10.1016/j.jaci.2018.03.012 [Epub ahead of print].
104. Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature (2015) 518:337–43. doi: 10.1038/nature13835
105. Ferreira MA, Jansen R, Willemsen G, Penninx B, Bain LM, Vicente CT, et al. Gene-based analysis of regulatory variants identifies 4 putative novel asthma risk genes related to nucleotide synthesis and signaling. J Allergy Clin Immunol. (2017) 139:1148–57. doi: 10.1016/j.jaci.2016.07.017
106. Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. (2006) 354:1264–72. doi: 10.1056/NEJMoa054013
107. Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. (2011) 108:11983–8. doi: 10.1073/pnas.1019276108
108. Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annu Rev Genomics Hum Genet. (2010) 11:65–89. doi: 10.1146/annurev-genom-082509-141523
109. Hernandez-Pacheco N, Flores C, Oh SS, Burchard EG, Pino-Yanes M. What ancestry can tell us about the genetic origins of inter-ethnic differences in asthma expression. Curr Allergy Asthma Rep. (2016) 16:53. doi: 10.1007/s11882-016-0635-4
110. Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. (2004) 74:979–1000. doi: 10.1086/420871
111. Wells KE, Cajigal S, Peterson EL, Ahmedani BK, Kumar R, Lanfear DE, et al. Assessing differences in inhaled corticosteroid response by self-reported race-ethnicity and genetic ancestry among asthmatic subjects. J Allergy Clin Immunol. (2016) 137:1364–9 e2. doi: 10.1016/j.jaci.2015.12.1334
112. Gautam Y, Altaye M, Xie C, Mersha TB. AdmixPower: Statistical power and sample size estimation for mapping genetic loci in admixed populations. Genetics (2017) 207:873–82. doi: 10.1534/genetics.117.300312
113. Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. (1198) 63:1839–51. doi: 10.1086/302148
114. Lautenberger JA, Stephens JC, O'Brien SJ, Smith MW. Significant admixture linkage disequilibrium across 30 cM around the FY locus in African Americans. Am J Hum Genet. (2000) 66:969–78. doi: 10.1086/302820
115. Nordborg M, Tavare S. Linkage disequilibrium: what history has to tell us. Trends Genet (2002) 18:83–90. doi: 10.1016/S0168-9525(02)02557-X
116. Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet. (2002) 110:553–60. doi: 10.1007/s00439-002-0731-5
117. Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. (2005) 28:289–301. doi: 10.1002/gepi.20064
118. Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. (2005) 6:623–32. doi: 10.1038/nrg1657
119. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature (2009) 461:747–53. doi: 10.1038/nature08494
120. Gignoux CR, Torgerson DG, Pino-Yanes M, Uricchio LH, Galanter J, Roth LA, et al. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in Latinos. J Allergy Clin Immunol. (2018). doi: 10.1016/j.jaci.2016.08.057 [Epub ahead of print].
121. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. (2010) 363:1211–21. doi: 10.1056/NEJMoa0906312
122. Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. (2011) 43:887–92. doi: 10.1038/ng.888
123. Hansen G, McIntire JJ, Yeung VP, Berry G, Thorbecke GJ, Chen L, et al. CD4(+) T helper cells engineered to produce latent TGF-beta1 reverse allergen-induced airway hyperreactivity and inflammation. J Clin Invest. (2000) 105:61–70. doi: 10.1172/JCI7589
124. Sagara H, Okada T, Okumura K, Ogawa H, Ra C, Fukuda T, et al. Activation of TGF-beta/Smad2 signaling is associated with airway remodeling in asthma. J Allergy Clin Immunol. (2002) 110:249–54.
125. Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir Med. (2014) 2:405–15. doi: 10.1016/S2213-2600(14)70012-8
127. Kan M, Shumyatcher M, Himes BE. Using omics approaches to understand pulmonary diseases. Respir Res. (2017) 18:149. doi: 10.1186/s12931-017-0631-9
128. Torgerson DG, Capurso D, Mathias RA, Graves PE, Hernandez RD, Beaty TH, et al. Resequencing candidate genes implicates rare variants in asthma susceptibility. Am J Hum Genet. (2012) 90:273–81. doi: 10.1016/j.ajhg.2012.01.008
129. Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ, et al. Diversity in clinical and biomedical research: a promise yet to be fulfilled. PLoS Med. (2015) 12:e1001918. doi: 10.1371/journal.pmed.1001918
130. Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. (2010) 125:336–46 e4. doi: 10.1016/j.jaci.2009.08.031
131. Farzan N, Vijverberg SJ, Andiappan AK, Arianto L, Berce V, Blanca-Lopez N, et al. Rationale and design of the multiethnic pharmacogenomics in childhood asthma consortium. Pharmacogenomics (2017) 18:931–43. doi: 10.2217/pgs-2017-0035
132. Lotvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. (2011) 127:355–60. doi: 10.1016/j.jaci.2010.11.037
133. Ortega H, Prazma C, Suruki RY, Li H, Anderson WH. Association of CHI3L1 in African-Americans with prior history of asthma exacerbations and stress. J Asthma (2013) 50:7–13. doi: 10.3109/02770903.2012.733991
134. Ding L, Li D, Wathen M, Altaye M, Mersha TB. African ancestry is associated with cluster-based childhood asthma subphenotypes. BMC Med Genomics (2018) 11:51. doi: 10.1186/s12920-018-0367-5
135. Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. (2002) 347:911–20. doi: 10.1056/NEJMra020100
136. Beasley R. The burden of asthma with specific reference to the United States. J Allergy Clin Immunol. (2002) 109:S482–9.
137. Bonnelykke K, Ober C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J Allergy Clin Immunol. (2016) 137:667–79. doi: 10.1016/j.jaci.2016.01.006
138. Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. (2005) 23:561–6. doi: 10.1038/nbt1096
139. Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. (2010) 86:6–22. doi: 10.1016/j.ajhg.2009.11.017
140. Barreto-Luis A, Corrales A, Acosta-Herrera M, Gonzalez-Colino C, Cumplido J, Martinez-Tadeo J, et al. A pathway-based association study reveals variants from Wnt signalling genes contributing to asthma susceptibility. Clin Exp Allergy (2017) 47:618–26. doi: 10.1111/cea.12883
141. Nimmesgern E, Benediktsson I, Norstedt I. Personalized Medicine in Europe. Clin Transl Sci. (2017) 10:61–3. doi: 10.1111/cts.12446
142. Hellmann DB, Ziegelstein RC. Personomics: A new series in the green journal. Am J Med. (2017) 130:622. doi: 10.1016/j.amjmed.2017.01.029
143. Kelly RS, Virkud Y, Giorgio R, Celedon JC, Weiss ST, Lasky-Su J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:1590–5. doi: 10.1016/j.bbadis.2017.02.006
144. Pirih N, Kunej T. An updated taxonomy and a graphical summary tool for optimal classification and comprehension of omics research. OMICS (2018) 22:337–53. doi: 10.1089/omi.2017.0186
145. Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. (2012) 366:489–91. doi: 10.1056/NEJMp1114866
146. Bush A. Translating asthma: dissecting the role of metabolomics, genomics and personalized medicine. Indian J Pediatr. (2018) 85:643–50. doi: 10.1007/s12098-017-2520-0
147. Kumar M, Choudhury Y, Ghosh SK, Mondal R. Application and optimization of minimally invasive cell-free DNA techniques in oncogenomics. Tumour Biol. (2018) 40:1010428318760342. doi: 10.1177/1010428318760342
148. Galeone C, Scelfo C, Bertolini F, Caminati M, Ruggiero P, Facciolongo N, et al. Precision medicine in targeted therapies for severe asthma: is there any place for “Omics” technology? Biomed Res Int. (2018) 2018:4617565. doi: 10.1155/2018/4617565
149. Scelfo C, Galeone C, Bertolini F, Caminati M, Ruggiero P, Facciolongo N, et al. Towards precision medicine: the application of omics technologies in asthma management. F1000Res (2018) 7:423. doi: 10.12688/f1000research.14309.2
150. Checkley W, Deza MP, Klawitter J, Romero KM, Pollard SL, Wise RA, et al. Identifying biomarkers for asthma diagnosis using targeted metabolomics approaches. Respir Med. (2016) 121:59–66. doi: 10.1016/j.rmed.2016.10.011
151. Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS ONE (2016) 11:e0152724. doi: 10.1371/journal.pone.0152724
152. Reinke SN, Gallart-Ayala H, Gomez C, Checa A, Fauland A, Naz S, et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J. (2017) 49:1601740. doi: 10.1183/13993003.01740-2016
153. Cao C, Li W, Hua W, Yan F, Zhang H, Huang H, et al. Proteomic analysis of sputum reveals novel biomarkers for various presentations of asthma. J Transl Med. (2017) 15:171. doi: 10.1186/s12967-017-1264-y
154. Li N, Qiu R, Yang Z, Li J, Chung KF, Zhong N, et al. Sputum microbiota in severe asthma patients: Relationship to eosinophilic inflammation. Respir Med. (2017) 131:192–8. doi: 10.1016/j.rmed.2017.08.016
155. Arathimos R, Suderman M, Sharp GC, Burrows K, Granell R, Tilling K, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin Epigenetics (2017) 9:112. doi: 10.1186/s13148-017-0414-7
156. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:141. doi: 10.1038/s41467-017-02573-2
157. Singhania A, Wallington JC, Smith CG, Horowitz D, Staples KJ, Howarth PH, et al. Multitissue transcriptomics delineates the diversity of airway t cell functions in asthma. Am J Respir Cell Mol Biol. (2018) 58:261–70. doi: 10.1165/rcmb.2017-0162OC
158. Miller GE, Chen E, Shalowitz MU, Story RE, Leigh AKK, Ham P, et al. Divergent transcriptional profiles in pediatric asthma patients of low and high socioeconomic status. Pediatr Pulmonol. (2018) 53:710–9. doi: 10.1002/ppul.23983
159. Zhang X, Biagini Myers JM, Burleson JD, Ulm A, Bryan KS, Chen X, et al. Nasal DNA methylation is associated with childhood asthma. Epigenomics (2018) 10:629–41. doi: 10.2217/epi-2017-0127
160. Xu CJ, Soderhall C, Bustamante M, Baiz N, Gruzieva O, Gehring U, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. (2018) 6:379–88. doi: 10.1016/S2213-2600(18)30052-3
161. Kelly RS, Sordillo JE, Lasky-Su J, Dahlin A, Perng W, Rifas-Shiman SL, et al. Plasma metabolite profiles in children with current asthma. Clin Exp Allergy (2018) 48:1297–304. doi: 10.1111/cea.13183
162. Carraro S, Bozzetto S, Giordano G, El Mazloum D, Stocchero M, Pirillo P, et al. Wheezing preschool children with early-onset asthma reveal a specific metabolomic profile. Pediatr Allergy Immunol. (2018) 29:375–82. doi: 10.1111/pai.12879
163. Martinez-Nunez RT, Rupani H, Plate M, Niranjan M, Chambers RC, Howarth PH, et al. Genome-wide posttranscriptional dysregulation by MicroRNAs in human asthma as revealed by frac-seq. J Immunol. (2018) 201:251–63. doi: 10.4049/jimmunol.1701798
164. Hough KP, Wilson LS, Trevor JL, Strenkowski JG, Maina N, Kim YI, et al. Unique lipid signatures of extracellular vesicles from the airways of asthmatics. Sci Rep. (2018) 8:10340. doi: 10.1038/s41598-018-28655-9
165. Chiu CY, Lin G, Cheng ML, Chiang MH, Tsai MH, Su KW, et al. Longitudinal urinary metabolomic profiling reveals metabolites for asthma development in early childhood. Pediatr Allergy Immunol. (2018) 29:496–503. doi: 10.1111/pai.12909
166. Kelly RS, Chawes BL, Blighe K, Virkud YV, Croteau-Chonka DC, McGeachie MJ, et al. An integrative transcriptomic and metabolomic study of lung function in children with asthma. Chest (2018) 154:335–48. doi: 10.1016/j.chest.2018.05.038
167. Yeh YL, Su MW, Chiang BL, Yang YH, Tsai CH, Lee YL. Genetic profiles of transcriptomic clusters of childhood asthma determine specific severe subtype. Clin Exp Allergy (2018) 48:1164–72. doi: 10.1111/cea.13175
168. Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. (2017) 140:63–75. doi: 10.1016/j.jaci.2016.08.055
169. Turturice BA, McGee HS, Oliver B, Baraket M, Nguyen BT, Ascoli C, et al. Atopic asthmatic immune phenotypes associated with airway microbiota and airway obstruction. PLoS ONE (2017) 12:e0184566. doi: 10.1371/journal.pone.0184566
170. Qiu W, Guo F, Glass K, Yuan GC, Quackenbush J, Zhou X, et al. Differential connectivity of gene regulatory networks distinguishes corticosteroid response in asthma. J Allergy Clin Immunol. (2018) 141:1250–8. doi: 10.1016/j.jaci.2017.05.052
171. Bigler J, Boedigheimer M, Schofield JPR, Skipp PJ, Corfield J, Rowe A, et al. A severe asthma disease signature from gene expression profiling of peripheral blood from U-BIOPRED Cohorts. Am J Respir Crit Care Med. (2017) 195:1311–20. doi: 10.1164/rccm.201604-0866OC
172. Lefaudeux D, De Meulder B, Loza MJ, Peffer N, Rowe A, Baribaud F, et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol. (2017) 139:1797–807. doi: 10.1016/j.jaci.2016.08.048
173. Maniscalco M, Paris D, Melck DJ, Molino A, Carone M, Ruggeri P, et al. Differential diagnosis between newly diagnosed asthma and COPD using exhaled breath condensate metabolomics: a pilot study. Eur Respir J. (2018) 51:1701825. doi: 10.1183/13993003.01825-2017
174. Rollet-Cohen V, Bourderioux M, Lipecka J, Chhuon C, Jung VA, Mesbahi M, et al. Comparative proteomics of respiratory exosomes in cystic fibrosis, primary ciliary dyskinesia and asthma. J Proteomics (2018) 185:1–7. doi: 10.1016/j.jprot.2018.07.001
175. Mao Z, Shi Y, Cao Q, Chen Y, Sun Y, Liu Z, et al. Transcriptional regulation on the gene expression signature in combined allergic rhinitis and asthma syndrome. Epigenomics (2018) 10:119–31. doi: 10.2217/epi-2017-0072
176. Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol. (2015) 135:31–42. doi: 10.1016/j.jaci.2014.10.015
177. Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. (2011) 12:671–82. doi: 10.1038/nrg3068
178. Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. (2014) 190:1363–72. doi: 10.1164/rccm.201406-1099OC
179. Yan X, Chu JH, Gomez J, Koenigs M, Holm C, He X, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med. (2015) 191:1116–25. doi: 10.1164/rccm.201408-1440OC
180. Wenzel S. Severe asthma: from characteristics to phenotypes to endotypes. Clin Exp Allergy (2012) 42:650–8. doi: 10.1111/j.1365-2222.2011.03929.x
181. Bhakta NR, Woodruff PG. Human asthma phenotypes: from the clinic, to cytokines, and back again. Immunol Rev. (2011) 242:220–32. doi: 10.1111/j.1600-065X.2011.01032.x
182. von Mutius E, Hartert T. Update in asthma 2012. Am J Respir Crit Care Med. (2013) 188:150–6. doi: 10.1164/rccm.201303-0468UP
183. George BJ, Reif DM, Gallagher JE, Williams-DeVane CR, Heidenfelder BL, Hudgens EE, et al. Data-driven asthma endotypes defined from blood biomarker and gene expression data. PLoS ONE (2015) 10:e0117445. doi: 10.1371/journal.pone.0117445
184. Pecak M, Korosec P, Kunej T. Multiomics data triangulation for asthma candidate biomarkers and precision medicine. OMICS (2018) 22:392–409. doi: 10.1089/omi.2018.0036
185. Nicodemus-Johnson J, Myers RA, Sakabe NJ, Sobreira DR, Hogarth DK, Naureckas ET, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight (2016) 1:e90151. doi: 10.1172/jci.insight.90151
186. Forno E, Sordillo J, Brehm J, Chen W, Benos T, Yan Q, et al. Genome-wide interaction study of dust mite allergen on lung function in children with asthma. J Allergy Clin Immunol. (2017) 140:996–1003 e7. doi: 10.1016/j.jaci.2016.12.967
187. Takahashi K, Pavlidis S, Ng Kee Kwong F, Hoda U, Rossios C, Sun K, et al. Sputum proteomics and airway cell transcripts of current and ex-smokers with severe asthma in U-BIOPRED: an exploratory analysis. Eur Respir J. (2018) 51:1702173. doi: 10.1183/13993003.02173-2017
188. Lund RJ, Osmala M, Malonzo M, Lukkarinen M, Leino A, Salmi J, et al. Atopic asthma after rhinovirus-induced wheezing is associated with DNA methylation change in the SMAD3 gene promoter. Allergy (2018) 73:1735–40. doi: 10.1111/all.13473
189. Ovrevik J, Lag M, Lecureur V, Gilot D, Lagadic-Gossmann D, Refsnes M, et al. AhR and Arnt differentially regulate NF-kappaB signaling and chemokine responses in human bronchial epithelial cells. Cell Commun Signal. (2014) 12:48. doi: 10.1186/s12964-014-0048-8
190. Sisson TH, Ajayi IO, Subbotina N, Dodi AE, Rodansky ES, Chibucos LN, et al. Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am J Pathol. (2015) 185:969–86. doi: 10.1016/j.ajpath.2014.12.005
191. Lim H, Cho M, Choi G, Na H, Chung Y. Dynamic control of Th2 cell responses by STAT3 during allergic lung inflammation in mice. Int Immunopharmacol. (2015) 28:846–53. doi: 10.1016/j.intimp.2015.03.051
192. McDonald ML. Multiomics Approach to Asthma: Navigating the Network. Am J Respir Cell Mol Biol. (2017) 57:381–2. doi: 10.1165/rcmb.2017-0220ED
193. Lima JJ, Blake KV, Tantisira KG, Weiss ST. Pharmacogenetics of asthma. Curr Opin Pulm Med. (2009) 15:57–62. doi: 10.1097/MCP.0b013e32831da8be
194. Svitich OA, Sobolev VV, Gankovskaya LV, Zhigalkina PV, Zverev VV. The role of regulatory RNAs (miRNAs) in asthma. Allergol Immunopathol. (2018) 46:201–5. doi: 10.1016/j.aller.2017.09.015
195. Rijavec M, Korosec P, Zavbi M, Kern I, Malovrh MM. Let-7a is differentially expressed in bronchial biopsies of patients with severe asthma. Sci Rep. (2014) 4:6103. doi: 10.1038/srep06103
196. Milger K, Gotschke J, Krause L, Nathan P, Alessandrini F, Tufman A, et al. Identification of a plasma miRNA biomarker signature for allergic asthma: a translational approach. Allergy (2017) 72:1962–71. doi: 10.1111/all.13205
197. Fekonja S, Korosec P, Rijavec M, Jesenicnik T, Kunej T. Asthma MicroRNA regulome development using validated miRNA-target interaction visualization. OMICS (2018) 22:607–15. doi: 10.1089/omi.2018.0112
198. Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res. (2018) 19:128. doi: 10.1186/s12931-018-0828-6
199. Pua HH, Ansel KM. MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol. (2015) 36:101–8. doi: 10.1016/j.coi.2015.07.006
200. Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature (2008) 453:56–64. doi: 10.1038/nature06862
201. Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. (2011) 21:2004–13. doi: 10.1101/gr.122614.111
202. Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, et al. Mapping copy number variation by population-scale genome sequencing. Nature (2011) 470:59–65. doi: 10.1038/nature09708
203. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature (2006) 444:444–54. doi: 10.1038/nature05329
204. Lee YL, Hsiue TR, Lee YC, Lin YC, Guo YL. The association between glutathione S-transferase P1, M1 polymorphisms and asthma in Taiwanese schoolchildren. Chest (2005) 128:1156–62. doi: 10.1378/chest.128.3.1156
205. Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. (2007) 39:S22–9. doi: 10.1038/ng2054
206. Rogers AJ, Chu JH, Darvishi K, Ionita-Laza I, Lehmann H, Mills R, et al. Copy number variation prevalence in known asthma genes and their impact on asthma susceptibility. Clin Exp Allergy (2013) 43:455–62. doi: 10.1111/cea.12060
207. Oliveira P, Costa GNO, Damasceno AKA, Hartwig FP, Barbosa GCG, Figueiredo CA, et al. Genome-wide burden and association analyses implicate copy number variations in asthma risk among children and young adults from Latin America. Sci Rep. (2018) 8:14475. doi: 10.1038/s41598-018-32837-w
208. Vishweswaraiah S, Veerappa AM, Mahesh PA, Jahromi SR, Ramachandra NB. Copy number variation burden on asthma subgenome in normal cohorts identifies susceptibility markers. Allergy Asthma Immunol Res. (2015) 7:265–75. doi: 10.4168/aair.2015.7.3.265
209. Igartua C, Myers RA, Mathias RA, Pino-Yanes M, Eng C, Graves PE, et al. Ethnic-specific associations of rare and low-frequency DNA sequence variants with asthma. Nat Commun. (2015) 6:5965. doi: 10.1038/ncomms6965
210. Morin A, Madore AM, Kwan T, Ban M, Partanen J, Ronnblom L, et al. Exploring rare and low-frequency variants in the Saguenay-Lac-Saint-Jean population identified genes associated with asthma and allergy traits. Eur J Hum Genet. (2019). 27:90–101. doi: 10.1038/s41431-018-0266-4
211. Smith D, Helgason H, Sulem P, Bjornsdottir US, Lim AC, Sveinbjornsson G, et al. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLoS Genet. (2017) 13:e1006659. doi: 10.1371/journal.pgen.1006659
212. Igartua C, Mozaffari SV, Nicolae DL, Ober C. Rare non-coding variants are associated with plasma lipid traits in a founder population. Sci Rep. (2017) 7:16415. doi: 10.1038/s41598-017-16550-8
213. Henn BM, Botigue LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA. (2016) 113:E440–9. doi: 10.1073/pnas.1510805112
214. Xue Y, Mezzavilla M, Haber M, McCarthy S, Chen Y, Narasimhan V, et al. Enrichment of low-frequency functional variants revealed by whole-genome sequencing of multiple isolated European populations. Nat Commun. (2017) 8:15927. doi: 10.1038/ncomms15927
215. Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. (2008) 9:403–33. doi: 10.1146/annurev.genom.9.081307.164258
216. Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. (2013) 93:779–97. doi: 10.1016/j.ajhg.2013.10.012
217. Emdin CA, Khera AV, Chaffin M, Klarin D, Natarajan P, Aragam K, et al. Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat Commun. (2018) 9:1613. doi: 10.1038/s41467-018-03911-8
218. MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science (2012) 335:823–8. doi: 10.1126/science.1215040
219. DeBoever C, Tanigawa Y, Lindholm ME, McInnes G, Lavertu A, Ingelsson E, et al. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat Commun. (2018) 9:1612. doi: 10.1038/s41467-018-03910-9
220. Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. (2004) 36:801–8. doi: 10.1038/ng1403
221. Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. (2008) 24:133–41. doi: 10.1016/j.tig.2007.12.007
222. Rizzo JM, Buck MJ. Key principles and clinical applications of “next-generation” DNA sequencing. Cancer Prev Res. (2012) 5:887–900. doi: 10.1158/1940-6207.CAPR-11-0432
223. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Gene.t (2018) 19:581–90. doi: 10.1038/s41576-018-0018-x
224. Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. (2002) 3:779–89. doi: 10.1038/nrg910
225. Katsanis N. The continuum of causality in human genetic disorders. Genome Biol. (2016) 17:233. doi: 10.1186/s13059-016-1107-9
226. Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. (2016) 17:392–406. doi: 10.1038/nrg.2016.27
227. Schork AJ, Schork MA, Schork NJ. Genetic risks and clinical rewards. Nat Genet. (2018) 50:1210–1. doi: 10.1038/s41588-018-0213-x
228. Maas P, Barrdahl M, Joshi AD, Auer PL, Gaudet MM, Milne RL, et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol. (2016) 2:1295–302. doi: 10.1001/jamaoncol.2016.1025
229. Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. (2016) 375:2349–58. doi: 10.1056/NEJMoa1605086
230. Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation (2017) 135:2091–101. doi: 10.1161/CIRCULATIONAHA.116.024436
231. Paquette M, Chong M, Theriault S, Dufour R, Pare G, Baass A. Polygenic risk score predicts prevalence of cardiovascular disease in patients with familial hypercholesterolemia. J Clin Lipidol. (2017) 11:725–32 e5. doi: 10.1016/j.jacl.2017.03.019
232. Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med. (2017) 14:e1002258. doi: 10.1371/journal.pmed.1002258
233. Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. (2017) 109. doi: 10.1093/jnci/djw302
234. Lecarpentier J, Silvestri V, Kuchenbaecker KB, Barrowdale D, Dennis J, McGuffog L, et al. Prediction of breast and prostate cancer risks in Male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol. (2017) 35:2240–50. doi: 10.1200/JCO.2016.69.4935
235. Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, et al. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ (2018) 360:j5757. doi: 10.1136/bmj.j5757
236. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet (2018) 50:1219–24. doi: 10.1038/s41588-018-0183-z
237. Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science (2018) 360:eaap8757. doi: 10.1126/science.aap8757
238. Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. (2017) 100:635–49. doi: 10.1016/j.ajhg.2017.03.004
239. Mancuso N, Rohland N, Rand KA, Tandon A, Allen A, Quinque D, et al. The contribution of rare variation to prostate cancer heritability. Nat Genet. (2016) 48:30–5. doi: 10.1038/ng.3446
240. Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. (2013) 14:681–91. doi: 10.1038/nrg3555
241. Marx V. Next-generation sequencing: the genome jigsaw. Nature (2013) 501:263–8. doi: 10.1038/501261a
242. DeWan AT, Egan KB, Hellenbrand K, Sorrentino K, Pizzoferrato N, Walsh KM, et al. Whole-exome sequencing of a pedigree segregating asthma. BMC Med Genet. (2012) 13:95. doi: 10.1186/1471-2350-13-95
243. Campbell CD, Mohajeri K, Malig M, Hormozdiari F, Nelson B, Du G, et al. Whole-genome sequencing of individuals from a founder population identifies candidate genes for asthma. PLoS ONE (2014) 9:e104396. doi: 10.1371/journal.pone.0104396
244. Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. (2015) 47:512–7. doi: 10.1038/ng.3278
245. Qiao D, Ameli A, Prokopenko D, Chen H, Kho AT, Parker MM, et al. Whole exome sequencing analysis in severe chronic obstructive pulmonary disease. Hum Mol Genet. (2018) 27:3801–12. doi: 10.1093/hmg/ddy269
246. Mak ACY, White MJ, Eckalbar WL, Szpiech ZA, Oh SS, Pino-Yanes M, et al. Whole-Genome Sequencing of Pharmacogenetic Drug Response in Racially Diverse Children with Asthma. Am J Respir Crit Care Med. (2018) 197:1552–64. doi: 10.1164/rccm.201712-2529OC
247. Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O'Connor TD, et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun. (2016) 7:12522. doi: 10.1038/ncomms12522
248. Hersh CP, Vachani A. Whole-genome sequencing in common respiratory diseases. Ready, Set, Go! Am J Respir Crit Care Med. (2017) 196:121–2. doi: 10.1164/rccm.201703-0479ED
249. Radder JE, Zhang Y, Gregory AD, Yu S, Kelly NJ, Leader JK, et al. Extreme trait whole-genome sequencing identifies PTPRO as a novel candidate gene in emphysema with severe airflow obstruction. Am J Respir Crit Care Med. (2017) 196:159–71. doi: 10.1164/rccm.201606-1147OC
250. Hinrichs AL, Suarez BK. Incorporating linkage information into a common disease/rare variant framework. Genet Epidemiol. (2011) 35 (Suppl.1):S74–9. doi: 10.1002/gepi.20654
251. Petersen BS, Fredrich B, Hoeppner MP, Ellinghaus D, Franke A. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet. (2017) 18:14. doi: 10.1186/s12863-017-0479-5
252. Lawrence MG, Steinke JW, Borish L. Cytokine-targeting biologics for allergic diseases. Ann Allergy Asthma Immunol. (2018) 120:376–81. doi: 10.1016/j.anai.2018.01.009
Keywords: admixture mapping, asthma, genomics, genome-wide association study, multiomics, personalized medicine
Citation: Hernandez-Pacheco N, Pino-Yanes M and Flores C (2019) Genomic Predictors of Asthma Phenotypes and Treatment Response. Front. Pediatr. 7:6. doi: 10.3389/fped.2019.00006
Received: 09 November 2018; Accepted: 10 January 2019;
Published: 05 February 2019.
Edited by:
Juan Carlos Celedon, University of Pittsburgh, United StatesReviewed by:
Manuel Sanchez-Solis, Hospital Universitario Virgen de la Arrixaca, SpainJulian Ramesh Vyas, Starship Children's Health, New Zealand
Copyright © 2019 Hernandez-Pacheco, Pino-Yanes and Flores. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Pino-Yanes, bWRlbHBpbm9AdWxsLmVkdS5lcw==
Carlos Flores, Y2Zsb3Jlc0B1bGwuZWR1LmVz
 Natalia Hernandez-Pacheco
Natalia Hernandez-Pacheco Maria Pino-Yanes
Maria Pino-Yanes Carlos Flores
Carlos Flores