- 1Division of Systems Medicine and Gene Therapy, Saitama Medical University, Saitama, Japan
- 2Department of Urology, Graduate School of Medicine, Chiba University, Chiba, Japan
- 3Department of Systems Aging Science and Medicine, Tokyo Metropolitan Institute of Gerontology, Tokyo, Japan
Breast and prostate cancers are the most prevalent cancers in females and males, respectively. These cancers exhibit sex hormone dependence and thus, hormonal therapies are used to treat these cancers. However, acquired resistance to hormone therapies is a major clinical problem. In addition, certain portions of these cancers initially exhibit hormone-independence due to the absence of sex hormone receptors. Therefore, precise and profound understanding of the cancer pathophysiology is required to develop novel clinical strategies against breast and prostate cancers. Metabolic reprogramming is currently recognized as one of the hallmarks of cancer, as exemplified by the alteration of glucose metabolism, oxidative phosphorylation, and lipid metabolism. Dysregulation of metabolic enzymes and their regulators such as kinases, transcription factors, and other signaling molecules contributes to metabolic alteration in cancer. Moreover, accumulating lines of evidence reveal that long non-coding RNAs (lncRNAs) regulate cancer development and progression by modulating metabolism. Understanding the mechanism and function of lncRNAs associated with cancer-specific metabolic alteration will therefore provide new knowledge for cancer diagnosis and treatment. This review provides an overview of recent studies regarding the role of lncRNAs in metabolism in breast and prostate cancers, with a focus on both sex hormone-dependent and -independent pathways.
Breast and Prostate Cancers
In developed countries, breast and prostate cancers are the most common malignancies in women and men, respectively (1). In both breast and prostate cancers, growth and survival are mainly controlled by sex steroid hormones, which include estrogens and androgens, respectively (2). These unique properties have been targeted in treating breast and prostate cancers with hormone therapy (3). However, during long-term hormone therapy, acquired resistance develops in many patients (4), and there are few effective treatments for hormone-refractory cancers. In addition, from the tumor diagnosis, breast cancers negative for estrogen receptor α (ERα) such as triple-negative breast cancer (TNBC) exhibit no response to hormone therapy (5). In the case of prostate cancer, most cancers are initially androgen-dependent and respond to hormone therapies; however, they eventually acquire endocrine therapy resistance and become castration-resistant prostate cancer (CRPC) (6). Thus, understanding both mechanisms of hormone dependence and hormone resistance is important for the development of diagnosis and treatment options of breast and prostate cancers.
Long Non-Coding RNA (lncRNA)
Recent cancer research has focused on the roles and importance of lncRNAs in cancer development. Although up to 70% of the human genome is actively transcribed, only 2% of transcripts are translated into proteins (7). Transcripts that do not encode proteins are called non-coding RNAs, among them those with a length >200 nucleotides are categorized as lncRNAs (8). LncRNAs exert their function by regulating a variety of intracellular processes, for example, (1) recruiting epigenetic modifiers and transcription factors to the enhancer/promoter of the lncRNA target gene, (2) forming RNA-RNA binding with its target mRNA for mRNA degradation, (3) acting as a decoy, enabling avoidance of the associated protein from transcriptional regulatory regions, and (4) acting as a molecular sponge or as a competitive endogenous RNA (ceRNA) for microRNA (4, 9–11). In terms of the contribution of lncRNAs to cancer metabolism, characterization of lncRNAs associated with glucose metabolism has proceeded (12, 13) probably because the Warburg effect with aerobic glycolysis has been considered as a central event for cancer metabolism. Recently, lncRNA functions in other metabolic pathways including mitochondrial oxidative phosphorylation (OXPHOS) and lipid biosynthesis have been reported. Moreover, it has been shown that lncRNAs also modulate hormone sensitivity and resistance in breast and prostate cancers (4, 10, 14). These findings suggest that the metabolism-associated lncRNAs would be new therapeutic targets for hormone-refractory breast and prostate cancers. Further intensive analyses will reveal the precise characteristics of these lncRNAs including their expression profiles, binding factors, and three-dimensional structures, using advanced technologies such as high-throughput sequencing, mass spectrometry, and bioinformatics. Based on the molecular aspects of lncRNAs, their expressions and functions could be modulated by nucleic acid therapy using antisense oligonucleotide and RNA interference technique, as well as by small-molecule drugs that bind to lncRNAs. Moreover, effector proteins of lncRNAs will be also potential targets for modulating the lncRNA-associated signaling (4, 7). In addition, lncRNA delivery and lncRNA-inducing drugs should be important to modify the functions of these lncRNAs. While efforts are underway to improve the molecular technology and skills for analyzing lncRNAs, a comprehensive understanding for their roles in cancer metabolism remains to be elusive and further investigation will be required to apply these molecules for cancer therapy. In the present review, we provide an overview of the recent findings regarding lncRNA involvement in cancer metabolism, with a focus on breast and prostate cancers (Figure 1).
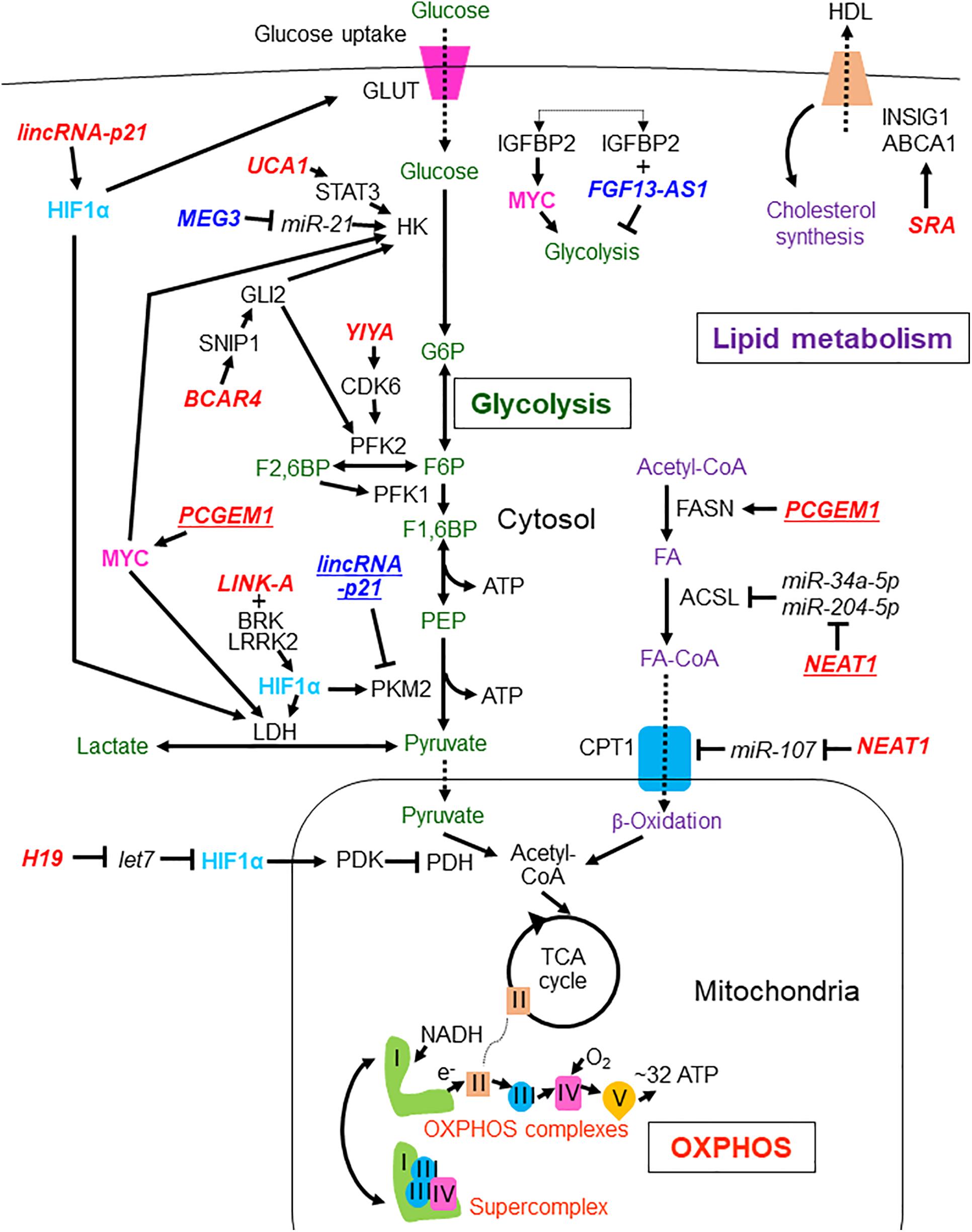
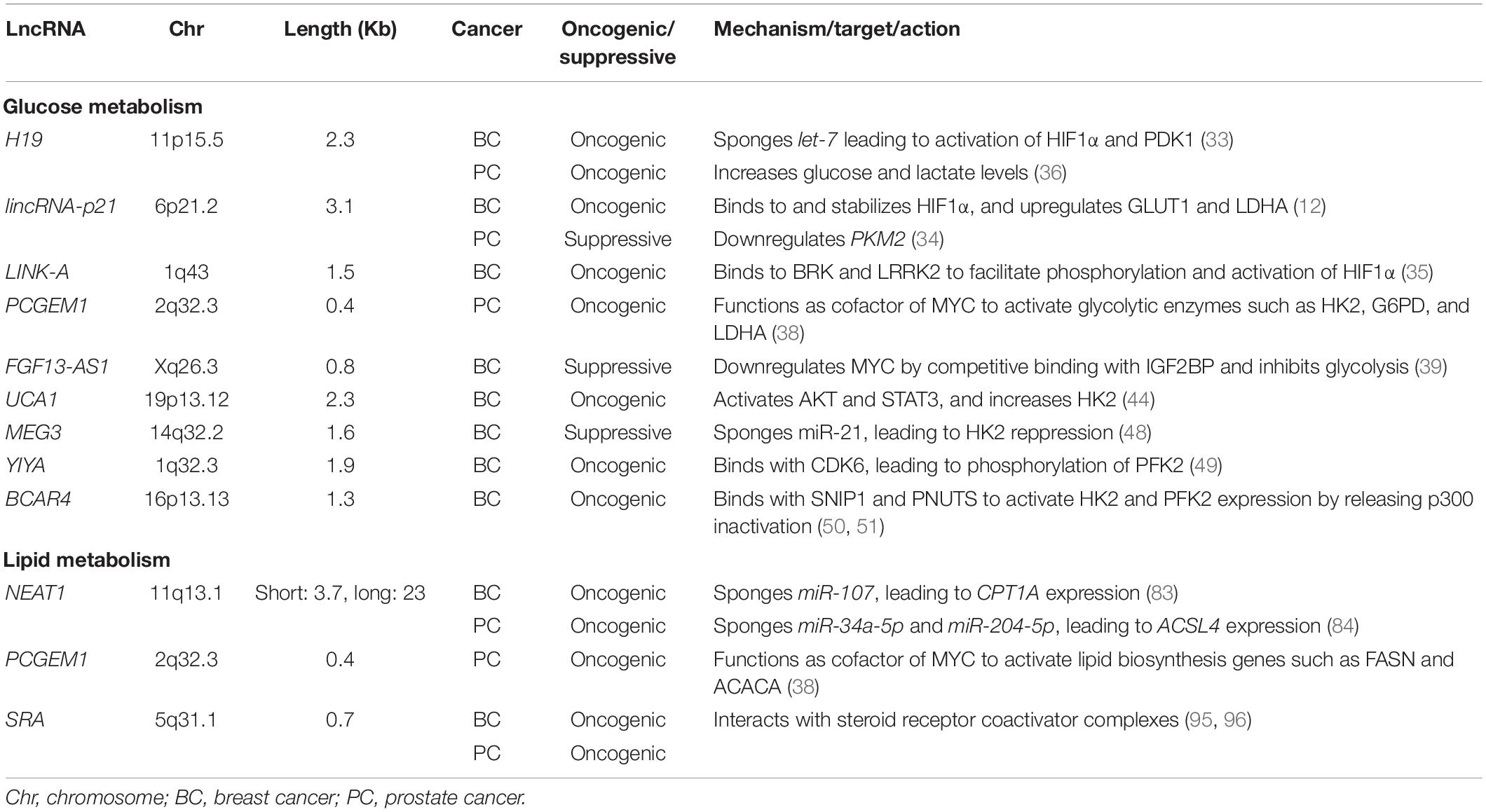
Figure 1. LncRNAs involved in the regulation of glycolysis, OXPHOS, and lipid metabolism in breast and prostate cancers. LncRNAs reported in breast and prostate cancers are shown without and with underlines, respectively. Tumor suppressive lncRNAs are indicated by dark blue characters while oncogenic lncRNAs are shown with red characters. The metabolites and processes of glycolysis, oxidative phosphorylation (OXPHOS), and lipid metabolism are shown with green, orange, and purple characters, respectively. Transcription factors HIF1α and MYC are presented with light blue and pink characters, respectively. lincRNA-p21, long intergenic non-coding RNA-p21;UCA1, urothelial carcinoma associated 1; MEG3, maternally expressed 3; FGF13-AS1, FGF13 antisense RNA 1; BCAR4, breast cancer anti-estrogen resistance 4; LINK-A, long intergenic non-coding RNA for kinase activation; SRA, steroid receptor RNA activator; PCGEM1, prostate cancer gene expression marker 1; NEAT1, nuclear enriched abundant transcript 1; GLUT, glucose transporter; HK, hexokinase; G6P, glucose-6-phosphate; PFK2, phosphofructokinase-2/fructose bisphosphatase-2; F2,6BP, fructose 2,6-bisphosphate; PFK1, phosphofructokinase-1; F1,6BP, fructose 1,6-bisphosphate; PEP, phosphoenolpyruvate; PKM2, pyruvate kinase isozymes M2; LDH, lactate dehydrogenase; HDL, high-density lipoprotein; INSIG1, insulin-induced gene 1; ABCA1, ATP binding cassette subfamily A member 1; FASN, fatty acid synthase; ACSL, Acyl-CoA synthetase long chain family; FA, fatty acid; CPT1, carnitine palmitoyltransferase 1; PDH, pyruvate dehydrogenase; TCA, tricarboxylic acid; OXPHOS, oxidative phosphorylation; STAT3, signal transducer and activator of transcription 3; HIF1α, hypoxia-inducible factor 1A; GLI2, glioma-associated oncogene homolog 2; SNIP1, Smad nuclear-interacting protein 1; CDK6, cyclin-dependent kinase 6; MYC, MYC proto-oncogene; BRK, breast tumor kinase; LRRK2, leucine rich-repeat kinase 2; PDK, pyruvate dehydrogenase kinase; Plus (+) means binding of lncRNA to protein.
Glucose Metabolism and lncRNA
Metabolic reprogramming is one of the hallmarks of cancer. Tumor cells preferentially use glycolysis for ATP production even in the presence of oxygen (15). However, the ATP production efficiency of glycolysis itself is not high compared with that of OXPHOS. Glycolysis-dependent cancer cells will be prone to undergo metabolic reprogramming as they eventually need to compensate for lower energy production efficiency by glycolysis due to high turnover of cell proliferation. As one strategy for the improvement of energy production efficiency, cancer cells often facilitate glucose uptake and usage by upregulating glucose transporters and glycolytic enzymes (16). For instance, the glucose transporter (GLUT) family, which is responsible for cellular glucose uptake, is upregulated in multiple cancer types (17). Hexokinases (HK), which convert glucose to glucose-6-phosphate (G6P) at the first step of glycolysis, are particularly activated in cancer cells wherein they function as a driver of glycolysis (18). Phosphofructokinase-2/fructose-bisphosphatase-2 (PFK2), which is responsible for the synthesis of fructose-2,6-bisphosphate, is also upregulated in various cancers (19). Pyruvate kinase catalyzes the final step of glycolysis to produce pyruvate (20), and pyruvate kinase M2 (PKM2) among the 4 pyruvate kinase isomers is assumed to play a potential role in tumorigenesis of several cancers including breast (21) and prostate cancer (22). Pyruvate dehydrogenase kinase (PDK) phosphorylates and inactivates the pyruvate dehydrogenase complex, which synthesizes acetyl coenzyme A as a critical substrate of tricarboxylic acid (TCA) cycle from pyruvate. Inhibition of pyruvate dehydrogenase complex activity by PDK-mediated phosphorylation is involved in cancer pathophysiology (23–25). Lactate dehydrogenase A (LDHA) is an enzyme that converts pyruvate to lactate, and its overexpression is also observed in many types of cancers (26). Oncogenes such as MYC proto-oncogene (MYC) and hypoxia-inducible factor 1A (HIF1α) have been also shown as master regulators for glucose metabolic alteration in cancer cells (27, 28). For example, the expression of glycolytic enzymes such as HK2, PFK, and LDHA is cooperatively regulated by MYC and HIF1α (29–31). In the following sections, we will discuss several major lncRNAs involved in the metabolic pathways in breast and prostate cancers.
H19
H19 is an lncRNA that regulates glucose metabolism in both breast and prostate cancers. In breast cancer, H19 acts as a ceRNA that sequesters microRNA let-7, leading to the upregulation of the let-7 target gene HIF1A that encodes HIF1α protein. This resulting HIF1α upregulation leads to an increase in PDK1 expression (32). PDK1 is the most dominant subtype of the PDK family that is involved in breast cancer progression and metastasis (32–34). Interestingly, hormone therapy drugs such as tamoxifen or fulvestrant elevate H19 expression in ERα-positive breast cancer cells, which contributes to the acquired resistance to these drugs (35). In addition, H19 depletion decreases the growth and the glucose and lactate levels in prostate cancer DU145 and PC-3 cells whereas the mechanism remains to be clarified (36).
Long Intergenic Non-coding RNA-p21 (lincRNA-p21)
LincRNA-p21 is inducible by hypoxia or HIF1α function in breast cancer cells (4), and binds to HIF1α and its E3 ubiquitin ligase von Hippel-Lindau (VHL), thereby disrupting the VHL-HIF1α interaction. This dissociation attenuates VHL-mediated HIF1α ubiquitination and results in HIF1α stabilization as a positive feedback loop for HIF1α-dependent pathways. It is revealed that lincRNA-p21 promotes hypoxia-induced lactate production and glucose uptake in breast cancer cells through the upregulation of GLUT1 and LDHA (4). In contrast, lincRNA-p21 is often downregulated in prostate cancer, and lincRNA-p21 silencing activates PKM2 and glycolysis in LNCaP and DU145 prostate cancer cells (37). The differential effects of lincRNA-p21 on glycolysis may be attributed to a cancer type-specific metabolic pathway regulation between breast and prostate cancers.
Long Intergenic Non-coding RNA for Kinase Activation (LINK-A)
LINK-A has been identified as a cytoplasmic lncRNA that is preferentially expressed in TNBC (38). LINK-A binds with both breast tumor kinase (BRK) and leucine rich-repeat kinase 2 (LRRK2), and this process is stimulated by the signaling from heparin-binding epidermal growth factor (HB-EGF)-triggered epidermal growth factor receptor (EGFR) and transmembrane glycoprotein non-metastatic melanoma protein B (GPNMB) complex formation. BRK phosphorylates the Tyr 565 of HIF1α, which interferes with the Pro 564 hydroxylation-mediated HIF1α degradation, whereas LRRK2 phosphorylates HIF1α Ser 797 which enhances HIF1α transcriptional activity. These HIF1α phosphorylation events facilitate glycolysis reprogramming, i.e., glucose uptake and lactate production, in TNBC (38).
Prostate Cancer Gene Expression Marker 1 (PCGEM1)
PCGEM1 was originally identified as an androgen-induced prostate-specific lncRNA. Its overexpression is highly associated with prostate tumors, and it promotes cancer cell proliferation (39, 40). In addition to hormonal regulation, PCGEM1 functions as a coactivator of MYC and is estimated to induce expression of glycolytic enzymes such as HK2, glucose-6-phosphate dehydrogenase (G6PD), and LDHA (41). PCGEM1 overexpression increases glucose uptake and lactate production along with the pentose phosphate shunt to provide a sufficient supply of nucleotides and lipids in prostate cancer.
FGF13 Antisense RNA 1 (FGF13-AS1)
FGF13-AS1 binds with an RNA-binding protein, insulin-like growth factor 2 mRNA binding protein (IGF2BP), which interacts with the MYC mRNA (42). Binding of FGF13-AS1 with IGF2BP disrupts the interaction between IGF2BP and MYC mRNAs, leading to a decrease in the half-life of MYC mRNA. FGF13-AS1 overexpression decreases glucose consumption and lactate production in TNBC subtype MDA-MB-231 cells, while FGF13-AS1 depletion increases them in luminal subtype MCF-7 cells. These results indicate that FGF13-AS1 suppresses the glucose metabolism in these breast cancer subtypes. FGF13-AS1 also suppresses the spheroid formation and stemness properties of these breast cancer subtypes (42).
Urothelial Carcinoma Associated 1 (UCA1)
UCA1 participates in glucose metabolism in several cancer types (43–46). Particularly, in tumor suppressor Merlin-deficient breast cancer cells, upregulated UCA1 stimulates glucose consumption and lactate production through the activation of serine/threonine kinase AKT and signal transducer and activator of transcription 3 (STAT3) with a simultaneous increase in HK2 expression (47). Moreover, UCA1 has been implicated in hormone therapy resistance in breast cancer cells as it sponges miR-18a, leading to HIF1α activation or inhibition of mTOR signaling (48, 49), suggesting its multiple functions in breast cancer metabolism.
Maternally Expressed 3 (MEG3)
As shown in other type of cancer (50), the lncRNA MEG3 is downregulated in breast cancer tissues (51). MEG3 acts as a molecular sponge of miR-21, resulting in the repression of HK2 protein levels and the glycolytic pathway (i.e., glucose consumption and lactate production) in both MCF-7 and MDA-MB-231 cells (51).
YIYA
YIYA is an lncRNA expressed in approximately 40% of clinical breast cancer tumors and is associated with poor recurrence-free survival of the patients (52). This report also indicates that YIYA binds with cyclin-dependent kinase 6 (CDK6) to enhance CDK6-dependent phosphorylation of PFK2. In breast cancer cells, YIYA stimulates cell proliferation with an increase in the glycolytic pathway (i.e., elevation of glucose consumption and lactate production) (52).
Breast Cancer Anti-Estrogen Resistance 4 (BCAR4)
BCAR4 has been identified as a TNBC-upregulated lncRNA that is essential for breast cancer metastasis (53). This report indicates that BCAR4 associates with Smad nuclear-interacting protein 1 (SNIP1) and serine/threonine-protein phosphatase 1 regulatory subunit 10 (PPP1R10/PNUTS) to promote the transcriptional signaling by non-canonical hedgehog signal mediator glioma-associated oncogene homolog 2 (GLI2) by discharging the inhibitory role of SNIP1 on p300 histone acetyltransferase activity. More recently, it was demonstrated that BCAR4 is upregulated by the Hippo pathway downstream effector, Yes-associated protein (YAP), and modulates Hedgehog signaling to activate transcription of HK2 and PFK2 in TNBC cells (54). Notably, BCAR4 induces glucose uptake and lactate production in these cells. Furthermore, high expression of both BCAR4 and YAP is associated with poor survival of patients with breast cancer, suggesting a critical role for the YAP-BCAR4-glycolysis axis in this disease.
OXPHOS With lncRNA
As aerobic glycolysis has been assumed as a major energy resource for cancer metabolism, mitochondrial OXPHOS has been rather considered as a second-grade metabolic pathway in cancers. Nevertheless, recent lines of evidence indicate that OXPHOS is also crucial for energy metabolism in cancers (55). The OXPHOS system comprises five enzyme complexes embedded in the inner mitochondrial membrane: complex I (NADH dehydrogenase), complex II (succinate ubiquinone oxidoreductase), complex III (ubiquinol cytochrome c oxidoreductase), complex IV (cytochrome c oxidoreductase), and complex V (ATP synthase). While the contributions of glycolysis and OXPHOS to ATP biosynthesis vary among cell types as shown by a meta-analysis, the contribution ratios of OXPHOS were almost equally ∼80% in both normal cells and cancer cells (56). Moreover, it has been revealed that OXPHOS rather remains the most significant source of energy production in tumors, suggesting that mitochondrial respiration is not generally impaired in cancer cells (57).
Several studies indicate that OXPHOS expression and function are both upregulated in breast and prostate cancers. Upregulation of OXPHOS gene expression and enzyme activity is detected in patients with breast cancer (58). In tumor suppressor RB1-deficient TNBC, OXPHOS is highly upregulated to promote cancer cell proliferation (59, 60). Moreover, cytochrome c oxidase subunit 7a-related polypeptide (COX7RP), which was originally identified as an estrogen-responsive gene, promotes mitochondrial respiration and cell proliferation in breast cancer cells by accelerating the formation of mitochondrial supercomplexes, which contributes to efficient ATP synthesis by assembling complexes I, III, and IV (61).
It has been recently shown that a compound-induced mitochondrial fragmentation leading to the disruption of OXPHOS and ATP synthesis inhibits the proliferation of prostate cancer PC3 cells (62). An integrated analysis of RNA-sequencing datasets and proteome data indicates that OXPHOS-related genes are upregulated in prostate cancer and negatively associated with STAT3 expression (63). Another integrated analysis elucidates a preservative role of MYC on OXPHOS function in prostate cancer tumorigenesis (64). In this report, MYC knockdown in PC3 cells reduced the levels of oxidative phosphorylation and TCA cycle metabolites along with decreased oxygen consumption rates (OCR), leading to diminished ATP production. Docetaxel-resistant PC3 cells show a metabolic shift from glycolysis toward mitochondrial respiration (65). Interestingly, RNA-seq analysis of prostate tissues shows remarkable enrichment of OXPHOS-related genes in CRPC (66). Moreover, a study analyzing substrate-specific OXPHOS capacities using primary human prostate tissues revealed that the malignant tissues exhibits a significant metabolic shift toward higher succinate oxidation by complex II, particularly in high-grade prostate tumors (67).
OXPHOS upregulation, however, is rather limited to particular cancer subtypes (68–70). In addition, OXPHOS activity can be coordinated by many factors including mitochondrial DNA content, expression levels of respiration complexes subunits and regulatory genes, and the cellular environment. Up to date, few studies have been performed for the analysis of OXPHOS activity in the association with lncRNAs. Further analysis will clarify the role of lncRNA on OXPHOS in different cancer types, including breast and prostate cancers.
Integrated analysis of the mitochondrial proteome and the gene expression dataset showed that the lncRNA small nucleolar RNA host gene 3 (SNHG3) is related to ovarian cancer survival as well as the expression of energy metabolism-related genes such as complex III subunit ubiquinol-cytochrome c reductase hinge protein (UQCRH) (71). In silico analysis speculates that SNHG3 can function by sponging miR-186-5p. In gastric cancer tissues, expression levels of lncRNA MIF-AS1 (lncMIF-AS1) and complex IV subunit NADH dehydrogenase1 alpha subcomplex 4 (NDUFA4) are higher compared with those in non-cancerous tissues (72). Mechanistically, lncMIF-AS1 sponges miR-212-5p to activate NDUFA4 expression. lncMIF-AS1 overexpression promotes proliferation and decreases apoptosis of gastric cancer cells through the activation of oxygen consumption and ATP production. The lncRNA cytoplasmic endogenous regulator of oxidative phosphorylation 1 (Cerox1) sponges miR-488-3p and elevates the expression of several subunits of complex I in mouse neuroblastoma cells (73).
Lipid Metabolism With lncRNA
Altered regulation in the synthesis and utilization of lipids is also a hallmark of cancers as it meets the high energy demand required for cancer proliferation and survival. In contrast to normal tissues and cells, malignant tumors prefer to synthesize lipids through the de novo pathway (74, 75). Increased expression or abnormal activity of key lipogenic enzymes such as fatty acid synthase (FASN) and acetyl coenzyme A (acetyl-CoA) carboxylase is often attributed to the high growth rate and lipogenic phenotype of tumor cells (76–80). Moreover, carnitine palmitoyl transferase (CPT) 1 and 2, which are rate-limiting enzymes involved in mitochondrial fatty acid transportation, play crucial roles in increasing fatty acid oxidation required for the cellular fuel demands of breast cancer cells (81). CPT1A/CPT2 were highly expressed in recurrent human breast cancers and are associated with poor prognosis. Moreover, increased fatty acid oxidation has also been suggested as a source of energy (82), along with increased steroidogenesis by upregulation of Acyl-CoA synthetase long-chain family member (ACSL) 3 (ACSL3) in prostate cancer (83). ACSL4, as well as ACSL3, which is an enzyme that converts fatty acids to acyl-CoA, is also involved in the loss of androgen sensitivity and acquisition of castration resistance, leading to cancer growth and invasion in prostate cancer (83–85). We mention several lncRNAs involved in breast and prostate cancers in the following sections.
Nuclear Enriched Abundant Transcript 1 (NEAT1)
In breast and prostate cancers, NEAT1 stimulates the TCA cycle by promoting the use of free fatty acids as fuel. NEAT1 upregulates CPT1A expression by inhibiting miR-107 to promote the progression of breast cancer cells (86). CPT1A synthesizes acylcarnitines, which are transported from the cytosol into the mitochondria, and their acyl groups are metabolized through the TCA cycle. NEAT1 also affects ACSL4 expression by competitively sponging both miR-34a-5p and miR-204-5p in prostate cancer (87). The alteration of ACSL4 expression is essential in the development and progression of breast and prostate cancers: ER expression is inversely correlated with ACSL4 expression in breast cancer (84), and ACSL4 is also involved in the loss of steroid hormone sensitivity and the acquisition of castration resistance in prostate cancer (85, 88). Recent findings indicate that NEAT1 affects mitochondrial dynamics and functions by regulating the sequestration of mRNAs encoding mitochondrial proteins in nuclear bodies called paraspeckles (89, 90).
PCGEM1
In addition to regulating glucose metabolism as described above, PCGEM1 widely regulates metabolic gene expression, including lipid metabolism in prostate cancer cells (41). The expression levels of several enzymes involved in lipid biosynthesis, such as those of FASN and acetyl-CoA carboxylase alpha (ACACA), are decreased by PCGEM1 knockdown. PCGEM1-overexpressing prostate cancer cells showed an increase in the cellular level of citrate, indicating enhanced fatty acid synthesis.
Steroid Receptor RNA Activator (SRA)
SRA was originally identified as a lncRNA that coactivates steroid hormone receptor transcriptional activity by associating with steroid receptor coactivator-containing complexes (91). Furthermore, SRA has also been revealed to interact with other transcription factors (92, 93). SRA overexpression has been found in various tumors including prostate cancer (91, 94). Both androgen receptor-dependent and -independent mechanisms are involved in SRA-mediated prostate cancer progression (95, 96). SRA expression is also significantly upregulated in breast cancer tissues (97). In MCF-7 breast cancer cells, SRA silencing decreased mRNA levels of insulin-induced gene 1 protein (INSIG1) and cholesterol transporter ATP-binding cassette transporter ABCA1, suggesting its role in lipids/cholesterol homeostasis (98, 99).
Conclusion
In this review, we summarized the functions and mechanisms of cancer metabolism-related lncRNAs especially in breast and prostate cancers (Table 1). We consider that these lncRNA functions may play critical roles in cancer pathophysiology from the viewpoint of their contribution to metabolic reprogramming. Moreover, the functions of cancer metabolism-related lncRNAs will be modulated by hormone status in breast and prostate cancers. Elucidating the mechanisms underlying metabolic alterations regulated by lncRNAs will lead to the development of new diagnostic and therapeutic options for both breast and prostate cancers.
Author Contributions
SK, TT, KI, KH-I, and SI: conception, providing the data and design. SK and TT: manuscript writing. KI, KH-I, and SI: conception and final approval of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Support Project of Strategic Research Center in Private Universities from the MEXT; grants from the Japan Society for the Promotion of Science (15K15353 and 20K21667 to SI, 17H04205 and 20H03734 to KH-I, and 20K21636 to KI), Practical Research for Innovative Cancer Control (JP18ck0106194 to KI) and Project for Cancer Research and Therapeutic Evolution (P-CREATE, JP18cm0106144 to SI) from the Japan Agency for Medical Research and Development (AMED), and Takeda Science Foundation to SI and KI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
2. Folkerd EJ, Dowsett M. Influence of sex hormones on cancer progression. J Clin Oncol. (2010) 28:4038–44. doi: 10.1200/jco.2009.27.4290
3. Ulm M, Ramesh AV, McNamara KM, Ponnusamy S, Sasano H, Narayanan R. Therapeutic advances in hormone-dependent cancers: focus on prostate, breast and ovarian cancers. Endocr Connect. (2019) 8:R10–26. doi: 10.1530/ec-18-0425
4. Takeiwa T, Ikeda K, Mitobe Y, Horie-Inoue K, Inoue S. Long noncoding RNAs involved in the endocrine therapy resistance of breast cancer. Cancers (Basel). (2020) 12:1424. doi: 10.3390/cancers12061424
5. Kansara S, Pandey V, Lobie PE, Sethi G, Garg M, Pandey AK. Mechanistic involvement of long non-coding RNAs in oncotherapeutics resistance in triple-negative breast cancer. Cells. (2020) 9:1511. doi: 10.3390/cells9061511
6. Chaturvedi AP, Dehm SM. Androgen receptor dependence. Adv Exp Med Biol. (2019) 1210:333–50. doi: 10.1007/978-3-030-32656-2_15
7. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. (2012) 9:703–19. doi: 10.4161/rna.20481
8. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. (2015) 21:1253–61. doi: 10.1038/nm.3981
10. Mitobe Y, Takayama KI, Horie-Inoue K, Inoue S. Prostate cancer-associated lncRNAs. Cancer Lett. (2018) 418:159–66. doi: 10.1016/j.canlet.2018.01.012
11. Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. (2017) 108:2107–14. doi: 10.1111/cas.13352
12. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. (2014) 53:88–100. doi: 10.1016/j.molcel.2013.11.004
13. Takayama K, Inoue S. The emerging role of noncoding RNA in prostate cancer progression and its implication on diagnosis and treatment. Brief Funct Genomics. (2016) 15:257–65. doi: 10.1093/bfgp/elv057
14. Mitobe Y, Ikeda K, Suzuki T, Takagi K, Kawabata H, Horie-Inoue K, et al. ESR1-stabilizing long noncoding RNA TMPO-AS1 promotes hormone-refractory breast cancer progression. Mol Cell Biol. (2019) 39:e00261-19. doi: 10.1128/MCB.00261-19
15. Zheng J. Energy metabolism of cancer: glycolysis versus oxidative phosphorylation. Oncol Lett. (2012) 4:1151–7. doi: 10.3892/ol.2012.928
16. Phan LM, Yeung SC, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. (2014) 11:1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001
17. Ancey PB, Contat C, Meylan E. Glucose transporters in cancer – from tumor cells to the tumor microenvironment. FEBS J. (2018) 285:2926–43. doi: 10.1111/febs.14577
18. Xu S, Herschman HRA. Tumor agnostic therapeutic strategy for hexokinase 1-null/hexokinase 2-positive cancers. Cancer Res. (2019) 79:5907–14. doi: 10.1158/0008-5472.CAN-19-1789
19. Yi M, Ban Y, Tan Y, Xiong W, Li G, Xiang B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: a pair of valves for fine-tuning of glucose metabolism in human cancer. Mol Metab. (2019) 20:1–13. doi: 10.1016/j.molmet.2018.11.013
20. Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. (2012) 23:560–6. doi: 10.1016/j.tem.2012.06.010
21. Tahtouh R, Wardi L, Sarkis R, Hachem R, Raad I, El Zein N, et al. Glucose restriction reverses the Warburg effect and modulates PKM2 and mTOR expression in breast cancer cell lines. Cell Mol Biol (Noisy-le-grand). (2019) 65:26–33. doi: 10.14715/cmb/2019.65.7.6
22. Talesa VN, Ferri I, Bellezza G, Love HD, Sidoni A, Antognelli C. Glyoxalase 2 is involved in human prostate cancer progression as part of a mechanism driven by PTEN/PI3K/AKT/mTOR signaling with involvement of PKM2 and ERα. Prostate. (2017) 77:196–210. doi: 10.1002/pros.23261
23. Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J Natl Cancer Inst. (2017) 109:djx071. doi: 10.1093/jnci/djx071
24. Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. (2014) 71:2577–604. doi: 10.1007/s00018-013-1539-2
25. Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab (Lond). (2014) 11:10. doi: 10.1186/1743-7075-11-10
26. Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. (2013) 65:904–10. doi: 10.1002/iub.1216
27. Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. (1997) 94:6658–63. doi: 10.1073/pnas.94.13.6658
28. Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. (2000) 275:21797–800. doi: 10.1074/jbc.C000023200
29. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. (2008) 7:11–20. doi: 10.1016/j.cmet.2007.10.002
30. Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. (2008) 8:705–13. doi: 10.1038/nrc2468
31. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
32. Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. (2018) 37:1062–74. doi: 10.1038/onc.2017.368
33. Du J, Yang M, Chen S, Li D, Chang Z, Dong Z. PDK1 promotes tumor growth and metastasis in a spontaneous breast cancer model. Oncogene. (2016) 35:3314–23. doi: 10.1038/onc.2015.393
34. Dupuy F, Tabariès S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. (2015) 22:577–89. doi: 10.1016/j.cmet.2015.08.007
35. Basak P, Chatterjee S, Bhat V, Su A, Jin H, Lee-Wing V, et al. Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER+ breast cancer cells. Cell Physiol Biochem. (2018) 51:1518–32. doi: 10.1159/000495643
36. Sun SC, Zhao H, Liu R, Wang BL, Liu YQ, Zhao Y, et al. Expression of long non-coding RNA H19 in prostate cancer and its effect on the proliferation and glycometabolism of human prostate cancer cells. Zhonghua Nan Ke Xue. (2017) 23:120–4.
37. Wang X, Xu Y, Wang X, Jiang C, Han S, Dong K, et al. LincRNA-p21 suppresses development of human prostate cancer through inhibition of PKM2. Cell Prolif. (2017) 50:e12395. doi: 10.1111/cpr.12395
38. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. doi: 10.1038/ncb3295
39. Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. (2000) 97:12216–21. doi: 10.1073/pnas.97.22.12216
40. Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. (2004) 23:605–11. doi: 10.1038/sj.onc.1207069
41. Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. (2014) 111:18697–702. doi: 10.1073/pnas.1415669112
42. Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick M, et al. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. (2019) 450:63–75. doi: 10.1016/j.canlet.2019.02.008
43. Li J, Wang M, Chen X. Long non-coding RNA UCA1 modulates cell proliferation and apoptosis by regulating miR-296-3p/Myc axis in acute myeloid leukemia. Cell Cycle. (2020) 19:1454–65. doi: 10.1080/15384101.2020.1750814
44. Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. (2016) 7:13616. doi: 10.1038/ncomms13616
45. Fan L, Huang C, Li J, Gao T, Lin Z, Yao T. Long non-coding RNA urothelial cancer associated 1 regulates radioresistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int J Mol Med. (2018) 42:2247–59. doi: 10.3892/ijmm.2018.3778
46. Hu ML, Wang XY, Chen WM. TGF-β1 upregulates the expression of lncRNA UCA1 and its downstream HXK2 to promote the growth of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. (2018) 22:4846–54. doi: 10.26355/eurrev_201808_15620
47. Mota MSV, Jackson WP, Bailey SK, Vayalil P, Landar A, Rostas JW III, et al. Deficiency of tumor suppressor Merlin facilitates metabolic adaptation by co-operative engagement of SMAD-Hippo signaling in breast cancer. Carcinogenesis. (2018) 39:1165–75. doi: 10.1093/carcin/bgy078
48. Li X, Wu Y, Liu A, Tang X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumour Biol. (2016) 37:14733–43. doi: 10.1007/s13277-016-5348-8
49. Wu C, Luo J. Long non-coding RNA (lncRNA) urothelial carcinoma-associated 1 (UCA1) enhances tamoxifen resistance in breast cancer cells via inhibiting mTOR signaling pathway. Med Sci Monit. (2016) 22:3860–7. doi: 10.12659/msm.900689
50. Ji L, Li X. Long noncoding RNA MEG3 is a tumor suppressor in choriocarcinoma by upregulation of microRNA-211. J Cell Physiol. (2019) 234:22911–20. doi: 10.1002/jcp.28853
51. Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys. (2019) 661:22–30. doi: 10.1016/j.abb.2018.10.021
52. Xing Z, Zhang Y, Liang K, Yan L, Xiang Y, Li C, et al. Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. (2018) 78:4524–32. doi: 10.1158/0008-5472.Can-17-0385
53. Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. (2014) 159:1110–25. doi: 10.1016/j.cell.2014.10.013
54. Zheng X, Han H, Liu GP, Ma YX, Pan RL, Sang LJ, et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. (2017) 36:3325–35. doi: 10.15252/embj.201797609
55. Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. (2018) 24:2482–90. doi: 10.1158/1078-0432.CCR-17-3070
56. Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. (2004) 313:459–65. doi: 10.1016/j.bbrc.2003.11.136
57. Vaupel P, Mayer A. Availability, not respiratory capacity governs oxygen consumption of solid tumors. Int J Biochem Cell Biol. (2012) 44:1477–81. doi: 10.1016/j.biocel.2012.05.019
58. Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, et al. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. (2011) 10:4047–64. doi: 10.4161/cc.10.23.18151
59. Zacksenhaus E, Shrestha M, Liu JC, Vorobieva I, Chung PED, Ju Y, et al. Mitochondrial OXPHOS Induced by RB1 deficiency in breast cancer: implications for anabolic metabolism, stemness, and metastasis. Trends Cancer. (2017) 3:768–79. doi: 10.1016/j.trecan.2017.09.002
60. Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, Ju Y, et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest. (2016) 126:3739–57. doi: 10.1172/JCI81568
61. Ikeda K, Horie-Inoue K, Suzuki T, Hobo R, Nakasato N, Takeda S, et al. Mitochondrial supercomplex assembly promotes breast and endometrial tumorigenesis by metabolic alterations and enhanced hypoxia tolerance. Nat Commun. (2019) 10:4108. doi: 10.1038/s41467-019-12124-6
62. Elshaarawy RFM, Alfaifi MY, Shati AA, Alshehri MA, Elbehairi SEI, Hafez HS. Role of Pd(II)-Chitooligosaccharides-Gboxin analog in oxidative phosphorylation inhibition and energy depletion: targeting mitochondrial dynamics. Chem Biol Drug Des. (2020): doi: 10.1111/cbdd.13703 [Epub ahead of print]
63. Oberhuber M, Pecoraro M, Rusz M, Oberhuber G, Wieselberg M, Haslinger P, et al. STAT3-dependent analysis reveals PDK4 as independent predictor of recurrence in prostate cancer. Mol Syst Biol. (2020) 16:e9247. doi: 10.15252/msb.20199247
64. Agarwal E, Altman BJ, Seo JH, Ghosh JC, Kossenkov AV, Tang HY, et al. Myc-mediated transcriptional regulation of the mitochondrial chaperone TRAP1 controls primary and metastatic tumor growth. J Biol Chem. (2019) 294:10407–14. doi: 10.1074/jbc.AC119.008656
65. Ippolito L, Marini A, Cavallini L, Morandi A, Pietrovito L, Pintus G, et al. Metabolic shift toward oxidative phosphorylation in docetaxel resistant prostate cancer cells. Oncotarget. (2016) 7:61890–904. doi: 10.18632/oncotarget.11301
66. Takayama KI, Fujimura T, Suzuki Y, Inoue S. Identification of long non-coding RNAs in advanced prostate cancer 2 associated with androgen receptor splicing factors. Commun Biol. (2020) 3:393. doi: 10.1038/s42003-020-01120-y
67. Schöpf B, Weissensteiner H, Schäfer G, Fazzini F, Charoentong P, Naschberger A, et al. OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. Nat Commun. (2020) 11:1487. doi: 10.1038/s41467-020-15237-5
68. Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. (2012) 22:547–60. doi: 10.1016/j.ccr.2012.08.014
69. Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. (2013) 12:329–41. doi: 10.1016/j.stem.2012.12.013
70. Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. (2014) 514:628–32. doi: 10.1038/nature13611
71. Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. (2018) 150:343–54. doi: 10.1016/j.ygyno.2018.06.013
72. Li L, Li Y, Huang Y, Ouyang Y, Zhu Y, Wang Y, et al. Long non-coding RNA MIF-AS1 promotes gastric cancer cell proliferation and reduces apoptosis to upregulate NDUFA4. Cancer Sci. (2018) 109:3714–25. doi: 10.1111/cas.13801
73. Sirey TM, Roberts K, Haerty W, Bedoya-Reina O, Rogatti-Granados S, Tan JY, et al. The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. eLife. (2019) 8:e45051. doi: 10.7554/eLife.45051
74. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. (2007) 7:763–77. doi: 10.1038/nrc2222
75. Chavarro JE, Kenfield SA, Stampfer MJ, Loda M, Campos H, Sesso HD, et al. Blood levels of saturated and monounsaturated fatty acids as markers of de novo lipogenesis and risk of prostate cancer. Am J Epidemiol. (2013) 178:1246–55. doi: 10.1093/aje/kwt136
76. Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin Cancer Res. (1997) 3:2115–20.
77. Swinnen JV, Vanderhoydonc F, Elgamal AA, Eelen M, Vercaeren I, Joniau S, et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int J Cancer. (2000) 88:176–9. doi: 10.1002/1097-0215(20001015)88:23.0.co;2-3
78. Turyn J, Schlichtholz B, Dettlaff-Pokora A, Presler M, Goyke E, Matuszewski M, et al. Increased activity of glycerol 3-phosphate dehydrogenase and other lipogenic enzymes in human bladder cancer. Horm Metab Res. (2003) 35:565–9. doi: 10.1055/s-2003-43500
79. Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. (2000) 16:202–8. doi: 10.1016/s0899-9007(99)00266-x
80. Ligibel JA, Alfano CM, Courneya KS, Demark-Wahnefried W, Burger RA, Chlebowski RT, et al. American Society of Clinical Oncology position statement on obesity and cancer. J Clin Oncol. (2014) 32:3568–74. doi: 10.1200/jco.2014.58.4680
81. Han S, Wei R, Zhang X, Jiang N, Fan M, Huang JH, et al. CPT1A/2-mediated FAO enhancement-A metabolic target in radioresistant breast cancer. Front Oncol. (2019) 9:1201. doi: 10.3389/fonc.2019.01201
82. Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. (2006) 9:230–4. doi: 10.1038/sj.pcan.4500879
83. Migita T, Takayama KI, Urano T, Obinata D, Ikeda K, Soga T, et al. ACSL3 promotes intratumoral steroidogenesis in prostate cancer cells. Cancer Sci. (2017) 108:2011–21. doi: 10.1111/cas.13339
84. Monaco ME, Creighton CJ, Lee P, Zou X, Topham MK, Stafforini DM. Expression of long-chain fatty Acyl-CoA synthetase 4 in breast and prostate cancers is associated with sex steroid hormone receptor negativity. Transl Oncol. (2010) 3:91–8. doi: 10.1593/tlo.09202
85. Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. (2008) 99:314–20. doi: 10.1038/sj.bjc.6604472
86. Xiong Y, Liu Z, Li Z, Wang S, Shen N, Xin Y, et al. Long noncoding RNA nuclear paraspeckle assembly transcript 1 interacts with microRNA107 to modulate breast cancer growth and metastasis by targeting carnitine palmitoyltransferase1. Int J Oncol. (2019) 55:1125–36. doi: 10.3892/ijo.2019.4869
87. Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia Y, et al. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. (2020) 65:109422. doi: 10.1016/j.cellsig.2019.109422
88. Wu X, Deng F, Li Y, Daniels G, Du X, Ren Q, et al. ACSL4 promotes prostate cancer growth, invasion and hormonal resistance. Oncotarget. (2015) 6:44849–63. doi: 10.18632/oncotarget.6438
89. Alvarez-Paggi D, Hannibal L, Castro MA, Oviedo-Rouco S, Demicheli V, Tórtora V, et al. Multifunctional Cytochrome c: learning new tricks from an old dog. Chem Rev. (2017) 117:13382–460. doi: 10.1021/acs.chemrev.7b00257
90. Wang Y, Hu SB, Wang MR, Yao RW, Wu D, Yang L, et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol. (2018) 20:1145–58. doi: 10.1038/s41556-018-0204-2
91. Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. (1999) 97:17–27. doi: 10.1016/s0092-8674(00)80711-4
92. Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. (2004) 15:549–58. doi: 10.1016/j.molcel.2004.06.044
93. Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. (2006) 11:547–60. doi: 10.1016/j.devcel.2006.08.003
94. Lanz RB, Razani B, Goldberg AD, O’Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc Natl Acad Sci USA. (2002) 99:16081–6. doi: 10.1073/pnas.192571399
95. Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. (2003) 369(Pt 1):163–71. doi: 10.1042/BJ20020743
96. Agoulnik IU, Weigel NL. Coactivator selective regulation of androgen receptor activity. Steroids. (2009) 74:669–74. doi: 10.1016/j.steroids.2009.02.007
97. Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, et al. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. (2000) 60:6266–71.
98. Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai MJ, et al. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. (2010) 24:1090–105. doi: 10.1210/me.2009-0427
Keywords: long non-coding RNA, cancer metabolism, glucose, oxidative phosphorylation, lipid, breast cancer, prostate cancer
Citation: Kamada S, Takeiwa T, Ikeda K, Horie-Inoue K and Inoue S (2020) Long Non-coding RNAs Involved in Metabolic Alterations in Breast and Prostate Cancers. Front. Oncol. 10:593200. doi: 10.3389/fonc.2020.593200
Received: 10 August 2020; Accepted: 11 September 2020;
Published: 06 October 2020.
Edited by:
Guoliang Huang, Guangdong Medical University, ChinaReviewed by:
Ming Yang, Tianjin Academy of Agricultural Sciences, ChinaCinzia Antognelli, University of Perugia, Italy
Copyright © 2020 Kamada, Takeiwa, Ikeda, Horie-Inoue and Inoue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Inoue, c2lub3VlQHRtaWcub3IuanA=; c2lub3VlMDdAZ21haWwuY29t
 Shuhei Kamada1,2
Shuhei Kamada1,2 Toshihiko Takeiwa
Toshihiko Takeiwa Kazuhiro Ikeda
Kazuhiro Ikeda Satoshi Inoue
Satoshi Inoue