
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 15 September 2020
Sec. Thoracic Oncology
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.561598
This article is part of the Research TopicInfluence of Potential Diagnostic Biomarkers in Lung CancerView all 26 articles
 Muyun Peng1,2
Muyun Peng1,2 Qi Huang1,2
Qi Huang1,2 Wei Yin1,2
Wei Yin1,2 Sichuang Tan1,2
Sichuang Tan1,2 Chen Chen1,2
Chen Chen1,2 Wenliang Liu1,2
Wenliang Liu1,2 Jingqun Tang1,2
Jingqun Tang1,2 Xiang Wang1,2
Xiang Wang1,2 Bingyu Zhang1,2
Bingyu Zhang1,2 Min Zou1,2
Min Zou1,2 Jina Li1,2
Jina Li1,2 Wenhui Su3,4
Wenhui Su3,4 Lientu Wang5
Lientu Wang5 Lihan Chin5*
Lihan Chin5* Fenglei Yu1,2*
Fenglei Yu1,2*Background: Routine clinical surveillance involves serial radiographic imaging following radical surgery in localized non-small cell lung cancer (NSCLC). However, such surveillance can detect only macroscopic disease recurrence and is frequently inconclusive. We investigated if detection of ctDNA before and after resection of NSCLC identifies the patients with risk of relapse, and furthermore, informs about response to management.
Methods: We recruited a total of 77 NSCLC patients. A high-throughput 127 target-gene capture technology and a high-sensitivity circulating single-molecule amplification and resequencing technology (cSMART) assay were used to detect the somatic mutations in the tumor tissues as well as the plasma of NSCLC patients before and after surgery to monitor for minimal residual disease (MRD). Kaplan-Meier and Cox regression analysis were performed to evaluate the relapse-free survival (RFS) and overall survival (OS) of patients with predictor variables.
Results: Patients with a higher stage (III/IV) and preoperative ctDNA-positive status demonstrated a significant 2.8-3.4-fold risk and 3.8-4.0-fold risk for recurrence and death, respectively. Preoperative ctDNA-positive patients associated with a lower RFS (HR = 3.812, p = 0.0005) and OS (HR = 5.004, p = 0.0009). Postoperative ctDNA-positive patients also associated with a lower RFS (HR = 3.076, p = 0.0015) and OS (HR = 3.195, p = 0.0053). Disease recurrence occurred among 63.3% (19/30) of postoperative ctDNA-positive patients. Most of these patients 89.5% (17/19) had detectable ctDNA within 2 weeks after surgery and was identified in advance of radiographic findings by a median of 12.6 months.
Conclusion: Advanced stage and preoperative ctDNA-positive are strong predictors of RFS and OS in localized NSCLC patients undergoing complete resection. Postoperative detection of ctDNA increases chance to detect early relapse, thus can fulfill an important role in stratifying patients for immediate further treatment with adjuvant and neoadjuvant therapy.
Lung cancer is a leading cause of cancer-related mortality, with an estimated nearly 2.1 million new cancer cases diagnosed leading to an estimated 1.8 million deaths worldwide in 2018 (1). In the United States, it is estimated that approximately 230,000 new cases of lung cancer will be diagnosed, and about 140,000 people will die from the disease in 2019 (2). For early-stage non-small cell lung cancer (NSCLC) patients, the best chance for cure is a complete (R0) resection by anatomic lung resection with mediastinal lymph node dissection. The 5-year overall survival (OS) of this approach is 61.5% and 5-year recurrence-free survival (RFS) is 59.0% (3). Following a radical surgery, serial radiographic imaging is routine. Because such surveillance detects only macroscopic recurrence, it is frequently inconclusive due to postoperative normal tissue changes. Given the population health burden of lung cancer, there is an imperative to develop a sensitive and specific biomarker that can detect the molecular residual disease (MRD) before macroscopic recurrence.
Plasma circulating tumor DNA (ctDNA) are DNA fragments in the blood that contain tumor-specific somatic alterations. It can be collected at minimal discomfort to the patient. The detection of ctDNA is a promising strategy for the prognosis and surveillance of solid tumors (4–6). It can predict the recurrence of non-metastatic breast, colon and pancreas cancers (7–9). Likewise, it can be detected in the postoperative blood sample in 94% of lung patients experiencing recurrence, and the results precede radiographic findings by a median of 5.2 months in 72% of patients (10). Being able to identify microscopic remnants of tumor cells and metastases can drastically change treatment algorithms, especially for localized lung cancer. In this study, we set out to determine whether preoperative or postoperative ctDNA positive status can predict survival outcomes in patients with localized lung cancer.
Furthermore, the detection of a positive EGFR mutation in the plasma of NSCLC patient during third cycle of treatment by erlotinib or chemotherapy has been associated with a reduced progression-free survival (PFS) and OS (11). We also addressed the hypothesis that post-surgical ctDNA detection could guide personalized interventions of adjuvant and neoadjuvant therapy.
A total of 81 patients diagnosed with solitary lung nodules intended for surgery at the Thoracic Surgery Department of The Second Xiangya Hospital were collected into our study between February 2014 and December 2015. All patients gave their written informed consent for specimen collection, provision of clinical information, and biomarker analysis before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Second Xiangya Hospital, Central South University, Changsha (Project identification code: 2014S006).
Eligible patients were age >18 years old with solitary lung nodules who agreed to the curative-intent treatment in this study. Preoperatively, the patients received chest computer tomography (CT) scans. Their blood samples were collected 1–7 days before surgery. Postoperatively, the patients received surveillance CT scans or positron emission tomography (PET)-CT scans at a series of scheduled time-points (2 weeks, 3, 6, 12, 18, and 24 months). Blood samples were also collected from patients during these follow-up visits. Patients with stage II or higher, if their physical condition permits, were further treated by adjuvant chemotherapy or target therapy (with or without radiotherapy) 1 month after surgery. If a recurrence or metastasis was suspected, a biopsy was performed to confirm the diagnosis. If a biopsy was not possible, then surgery, chemotherapy, radiotherapy or targeted therapy would be administered according to the specific situation.
Tumor specimens were collected by surgery. Macro-dissection was performed to enrich the tumor tissue percentage to around 80% before DNA extraction. Histologic evaluation of stained FFPE tumor sections was used to confirm the diagnosis of NSCLC. For the clinical staging of disease, the criteria from the TNM staging system of the International Association for the Study of Lung Cancer (version 8) was used.
Genomic DNA and total RNA Isolation from FFPE specimen blocks or scrapings from cytological slides were performed using the AmoyDx FFPE DNA and RNA Kits with nucleic acid purification spin columns (Amoy Diagnostics, Xiamen, China). By spectroscopy analysis, all purified DNA and RNA samples were verified to be of high quality for mutation analysis. Matching 10-mL blood samples, collected in Streck tubes (Streck, La Vista, NE, United States), was taken by venipuncture within 48 h of tumor specimen collection. DNA for the cSMART assay was prepared from 2 mL of purified plasma using a commercially available kit (QIAamp DNA Blood MiniKit, Qiagen, Hilden, Germany). The concentration of the purified DNA was measured by the Qubit® dsDNA HS Assay Kit (Life Technologies, Grand Island, NY, United States).
A novel cSMART assay (12, 13) was used for the detection and quantitation of hotspot oncogenic ctDNA mutations targeting 127 recurrent mutations in lung cancer (Supplementary Table S1). In brief, 50 ng of FFPE DNA was fragmented in NEB Next dsDNA fragmentase buffer (New England Biolabs, MA, United States) to an average size of 300 bp. DNA libraries were generated as previously described (14) except that a degenerate 4 bp barcode sequence was incorporated into the sequencing adaptor for uniquely identifying and counting single allelic molecules. Single DNA molecules were circularized and targeted with back-to-back primers located within 20 to 48 bp from the mutation loci to ensure maximum sensitivity and specificity for mutation detection. The ctDNA status was classified as detectable (ctDNA-positive) or undetectable (ctDNA-negative) according to mutation ratio. A mutation ratio equal to 0 is defined as ctDNA-negative and greater than 0 is defined as ctDNA-positive.
Data were summarized using descriptive statistics. The ctDNA variables in the different groups were compared using the Kruskal–Wallis non-parametric test. Fisher’s exact test was used to test for the association between ctDNA detection and histological subtypes. The primary outcome measure was RFS as evaluated by standard RECIST criteria. RFS was defined as the length of time after surgery that the patient survives without any signs or symptoms of lung cancer. OS was defined as the length of time from the date of diagnosis until death. Curves for RFS and OS were constructed using the Kaplan–Meier method and compared using the log-rank test. Multivariable Cox proportional hazards regression analyses for RFS and OS were performed using the Wald test to assess the predictive ability of preoperative and postoperative ctDNA. Analyses were performed using statistical software Graphpad Prism (version 7.0) and SPSS (version 22). Power calculation for Cox proportional hazards regression was estimated using the R package “powerSurvEpi1.” We used the powerEpi function that takes into account the correlation between two covariates, which we considered to be stage and preoperative ctDNA status. For RFS and OS, taking a minimum postulated hazard ratio (HR) of 2.7 and 3.7, power estimates were 69 and 77%, respectively, at a type I error rate of 0.05.
After accounting for missing data, 77 patients underwent surgery for localized lung cancer were included in the final analysis (Figure 1). We collected a total of 77 tissue samples. Accompanying this was the collection of 77 preoperative blood samples that belonged to 77 patients and 199 postoperative blood samples drawn at different times from 71 patients (Supplementary Table S2).
Table 1 and Supplementary Table S3 summarizes the clinical characteristics of patients. Their median age was 60.3 years old, have a male to female ratio of 2.7 and most were never smokers (49, 63.6%). Those diagnosed with adenocarcinoma, squamous carcinoma, and others, the numbers were 40 (51.9%), 30 (39.0%), and 7 (9.1%) patients, respectively. Those with tumors located in the right-upper, right-middle, right-lower, left-upper, and left -lower lobes, the numbers were 26 (33.8%), 2 (2.6%), 18 (23.3%), 20 (26.0%), and 11 (14.3%) patients, respectively. Those diagnosed with disease stages I, II, III, and IV, the numbers were 41 (53.2%), 18 (23.4%), 16 (20.8%), and 2 (2.6%) patients, respectively. The two cases of stage IV patients were accidentally found to have pleural implant metastasis during operation.
All the 77 patients harbored at least one mutation in their tumor tissue, with an average of 1.2 gene mutations per patient (Supplementary Table S4). The most frequent mutations observed in tissues located in the TP53 (60%), EGFR (21%), KEAP1 (9%), NAV3 (8%), CDKN2A (8%), and PIK3CA (8%) genes (Supplementary Figure S1).
Preoperative ctDNA-positive status was detected in 46 of 77 patients (59.7%). They constituted 43.9% (18/41), 72.2% (13/18), 81.3% (13/16), and 100% (2/2) of patients with stage I, II, III, and IV, respectively (Figure 2A). They further associated with males, never smokers, lung squamous carcinoma and visceral pleural invasion (p < 0.01) (Figure 2C). There was no association between preoperative ctDNA-positive status and age or BMI. Among 46 patients with positive preoperative ctDNA, the average number of gene mutation is 1.46, with one gene mutation detected in 29 cases, 2 in 13 cases, and 3 in 4 cases (Supplementary Table S2).
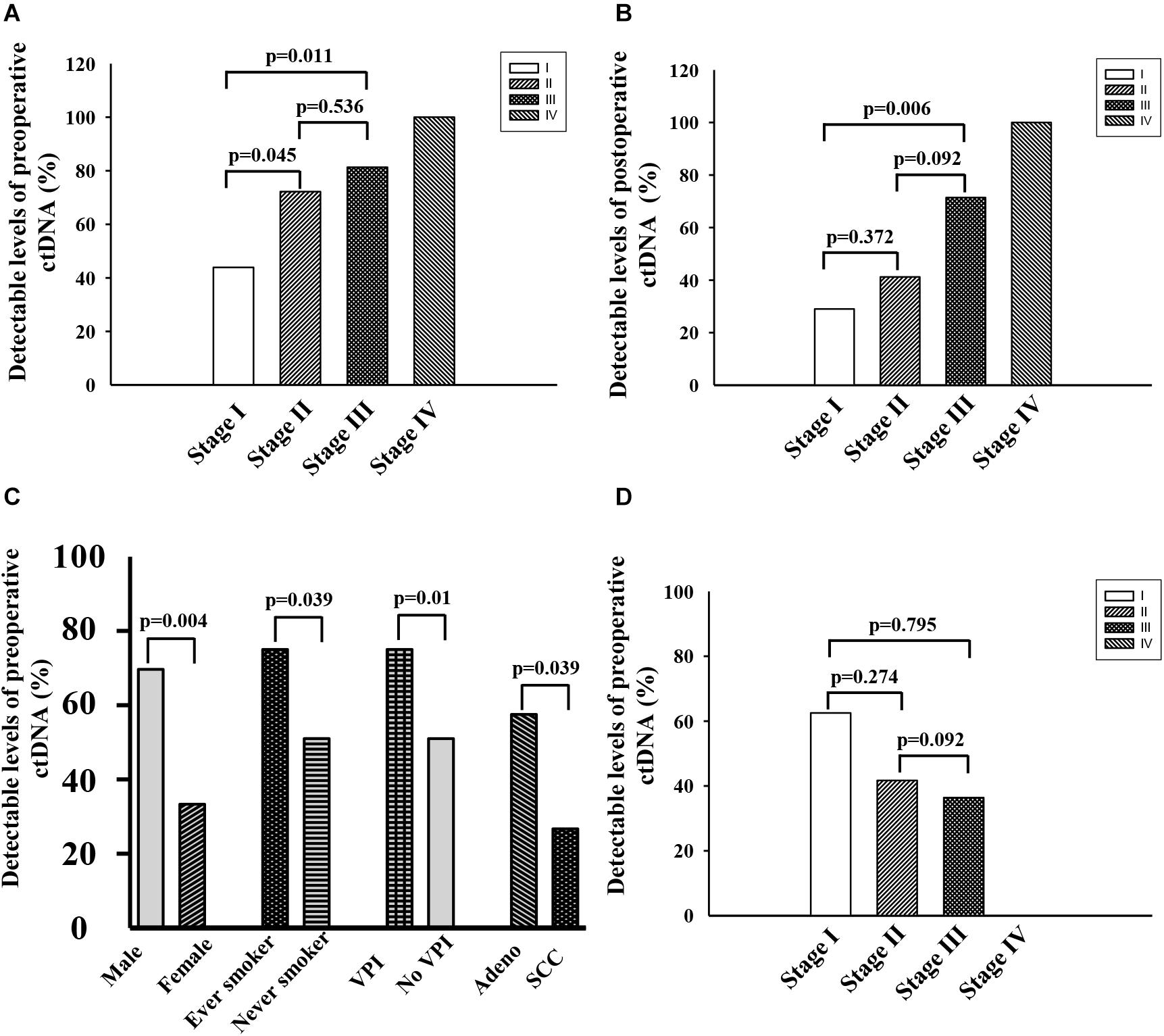
Figure 2. (A) Fraction of patients with detectable preoperative ctDNA in NSCLC at cancer stage I∼IV. (B) Fraction of patients with detectable postoperative ctDNA in NSCLC at cancer stage I∼IV. (C) Characteristics associated with positive preoperative ctDNA. (D) Negative conversion ratio of preoperative ctDNA at cancer stage I∼III.
Postoperative ctDNA-positive status was detected in 30 of 71 patients (42.25%). They formed 29.0% (11/38), 41.2% (7/17), 71.4% (10/14), and 100.0% (2/2) of patients with stage I, II, III, and IV, respectively (Figure 2B). Among the 41 patients with detectable preoperative ctDNA, 22 of them continued to have detectable postoperative ctDNA. The negative conversion ratios were 62.50% (10/16), 41.67% (5/12), 36.36% (4/11), and 0.0% (0/2) of patients with stage I, II, III, and IV, respectively (Figure 2D).
During the median follow-up period of 46 months in this study, 35 (45%) patients had a cancer recurrence, and 25 (32.47%) patients died from cancer recurrence/metastasis. In stage I–III patients, those with preoperative ctDNA-positive status had a lower RFS and OS than those who did not (P < 0.001; Figures 3A,B and Supplementary Tables S2, S5). During the follow-up, patients with postoperative ctDNA-positive status were detected in at least a one-time point for 28 (39%) patients. Among these, 17 of 28 (61%) patients experienced a recurrence, and 13 of them eventually died due to recurrence/metastasis. In stage I–III patients, those with postoperative ctDNA-negative status had a higher RFS and OS than ctDNA-positive patients (P < 0.05; Figures 3C,D and Supplementary Tables S2, S5).
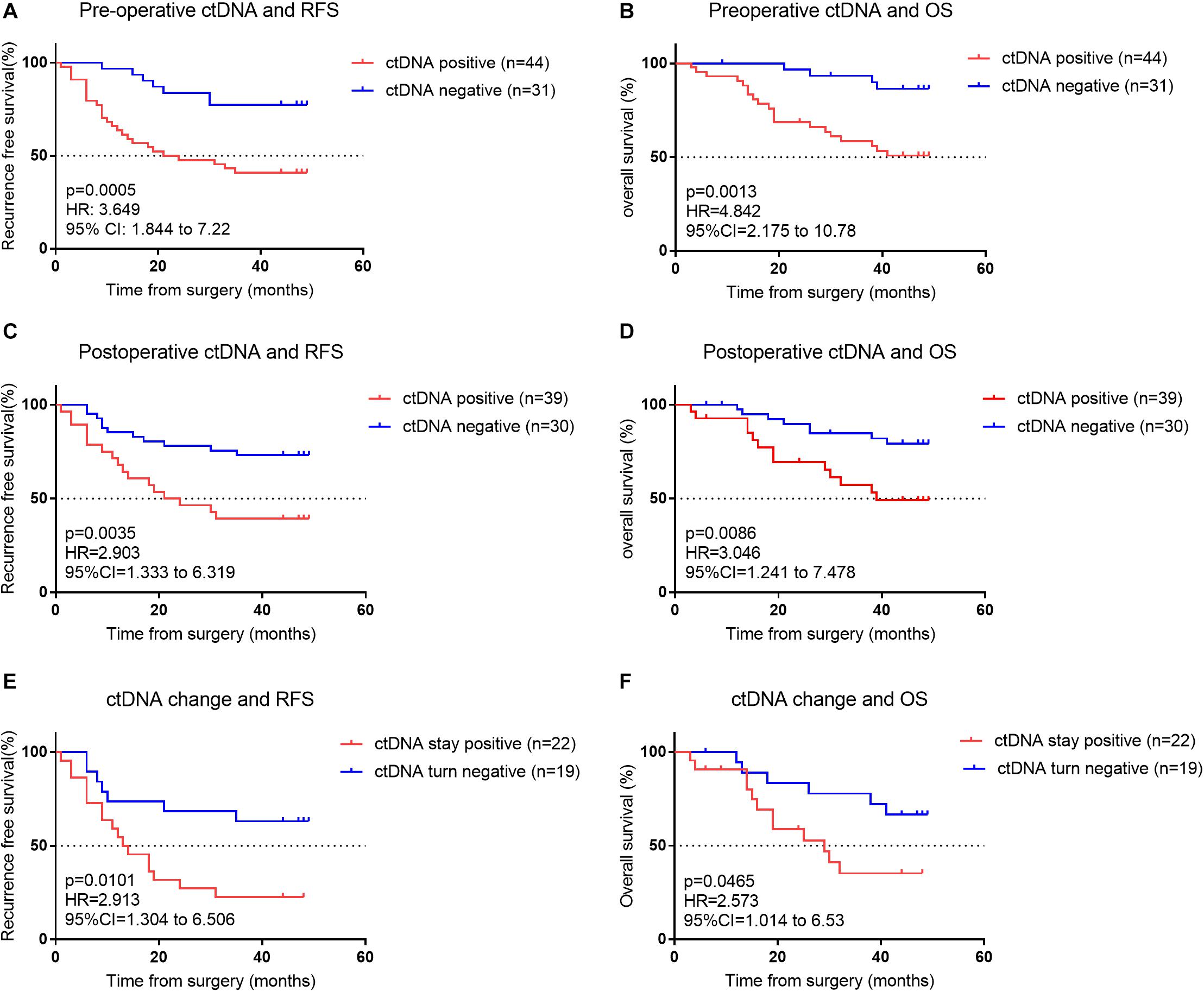
Figure 3. (A) Kaplan–Meier estimates of recurrence-free survival (RFS) for all assessable patients undergoing curative intent surgery for early NSCLC cancer, stratified by pre-operative ctDNA status: detectable (positive) versus undetectable (negative). (B) Kaplan–Meier estimate for overall survival (OS) for matched patients, stratified by pre-operative ctDNA status. (C) Kaplan–Meier estimates for RFS, stratified by postoperative ctDNA status. (D) Kaplan–Meier estimates for OS, stratified by post-operative ctDNA status. (E,F) Kaplan–Meier estimates for RFS (E) and OS (F) for patients’ whose ctDNA status changes from detectable (positive) pre-operatively to undetectable (negative) post-operatively compared with patients whose ctDNA status remains undetectable (negative) both pre- and post-operatively.
We investigated patients whose ctDNA turned negative after surgery, indicating their lesion was completely removed. Among the 41 patients who were preoperative ctDNA-positive, 22 continued to have detectable postoperative ctDNA. Patients with undetectable postoperative ctDNA (ctDNA turned negative) had a significantly better RFS and OS than detectable postoperative ctDNA (ctDNA stayed positive) (p < 0.05; Figures 3E,F and Supplementary Tables S2, S5). Postoperative ctDNA preceded the radiographic findings or clinical symptoms by a median of 12.6 months in 19 (63.3%) of patients. In these 19 patients, 17 were postoperative ctDNA-positive within 2 weeks after surgery, implying ctDNA was not fully cleared after surgery and they likely had an MRD. We also found that among the 31 patients who were preoperative ctDNA-negative, 8 have newly detectable postoperative ctDNA.
Independent univariate analysis showed that stage, preoperative and postoperative ctDNA statuses were significant clinicopathological factors for RFS and OS (Table 2). After multivariate analysis, only stage and preoperative ctDNA status remained significant for RFS and OS (p < 0.05). The results indicated that having a higher stage (III/IV) compared to a lower stage (I/II) conferred a 2.8- or 3.8-fold risk of recurrence/metastasis or death, respectively. Also, having a preoperative ctDNA-positive status, compared to ctDNA-negative status, conferred a 3.4- or 4.0-fold risk of recurrence/metastasis or death, respectively.
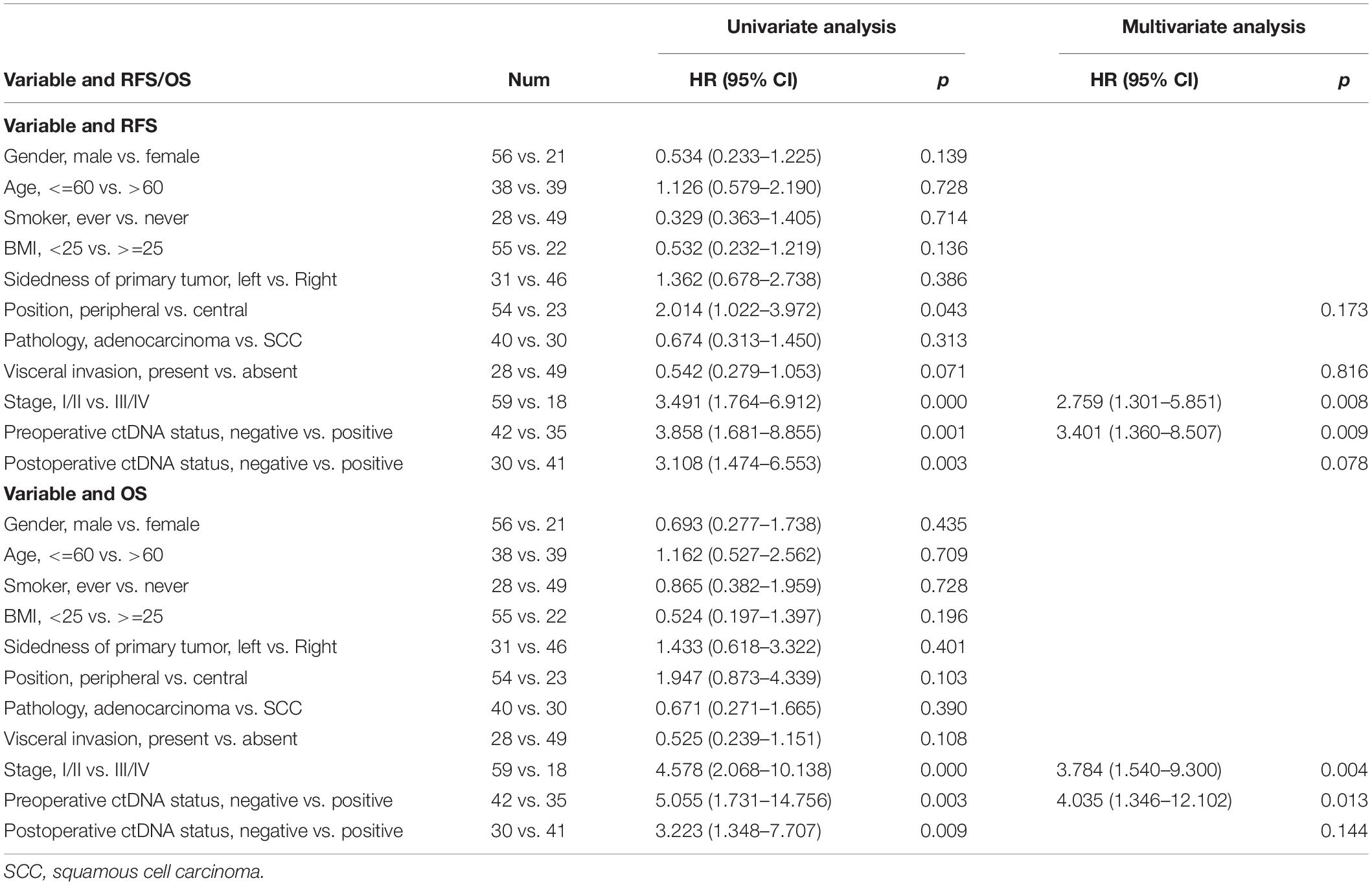
Table 2. Recurrence-free survival (RFS) and overall survival (OS) analysis by clinicopathologic variables and pre- and post-operative ctDNA status.
Our findings indicated that among other factors, having a lower disease stage and a preoperative ctDNA-negative status were two strong predictors of a better outcome in RFS and OS. The Kaplan-Meier for RFS and OS stratified by ctDNA detection status also showed that before and after surgery, the ctDNA-negative NSCLC patients had superior RFS and OS compared with ctDNA-positive patients.
We found the ctDNA detection rate dropped from 59.7 to 42.25% after the curative resection of the primary tumor, with most reductions in stage I and II patients. A study of 41 patients tracking ctDNA mutation frequency of 6 tumor driver genes (EGFR, KRAS, TP53, BRAF, PIK3CA, and ERBB2) within 10 days before and after surgical resection, also reported a ctDNA mutation frequency decreased from a median of 8.88 to 0.28%, with most reductions seen in patients with stage I disease (15). In another study of 76 NSCLC patients who underwent curative-intent surgery, the ctDNA mutation frequency decreased from 7.94% ± 4.78% before surgery to 0.28% ± 0.32% after surgery (p < 0.001) (16). These results imply that earlier stage lung cancer patients are less likely to have residual disease after resection.
Early-detection strategies have the potential to reduce cancer morbidity and mortality (17). Our previous study have shown that ctDNA can be used in the early diagnosis of lung cancer (18). In this cohort, even for stage I patients, 48.8% of patients have detectable levels of ctDNA in their plasma. In stage III disease, more than four-fifths of patients have detectable ctDNA. Another study has shown ctDNA is detectable in 47% of patients with stage I cancers of any type, and 55, 69, and 82% of patients with stage II, III, and IV cancers, respectively (19).
At present, most solid tumors are treatable by surgery, and even when occult metastasis has occurred, adjuvant therapy or additional surgery can contribute to cure in certain patients (20). Studies are demonstrating the necessity and efficacy of adjuvant and neoadjuvant therapy in early-stage NSCLC patients (21, 22). However, adjuvant chemotherapy in early-stage NSCLC provide an absolute survival benefit of only 4–5% compared to observation or best supportive care (21). Because the current staging model does not accurately identify the molecular residual disease (MRD) or micro-metastases, it could misguide patient selection. Validation of predictive biomarkers is urgently required to facilitate patient selection and risk stratification. Postoperative ctDNA monitoring can help with the identification of patients who could receive the most benefit from neoadjuvant and adjuvant therapy.
Postoperative ctDNA detection of patients correlates with better monitoring of relapse (9). Abbosh et al., report that ctDNA-positive status associated with the relapse of disease after intent-to-cure surgery in NSCLC patients. In that study, 13 of 14 patients experienced relapse had measurable ctDNA before demonstrating clinically evident disease and detection of ctDNA preceded the radiographic diagnosis by a median interval of 70 days. In another study that assessed the MRD in patients with lung cancer using CAPP-Seq, ctDNA was detected after curative-intent therapies among 20 of 37 patients, all of whom had disease recurrence and ctDNA associated with disease relapse earlier than CT imaging by a median lead time of 5.2 months in 72% of lung cancer patients (10). On the contrary, a very few, 1 out of 10 patients, exhibited persistent or recurrent ctDNA levels who did not relapse during a follow-up period of median 775 days (23). The time interval between the postoperative increase in ctDNA levels and the clinical diagnosis of cancer recurrence opens up a window of opportunity for intervention.
Our study focused on the effect of preoperative and postoperative ctDNA status on survival in resectable lung cancer. This study has a relatively long follow-up time, and the conclusion is persuasive. Nevertheless, this article has certain disadvantages. In our cohort, 25 patients received adjuvant therapy, of whom 21 received adjuvant chemotherapy. Because the designed time point of blood collection is not completely coincident with adjuvant chemotherapy, 8 patients lack the comparison before and after chemotherapy, and 7 patients have negative ctDNA before and after adjuvant treatment. Therefore, our data cannot evaluate the impact of adjuvant chemotherapy on ctDNA. However, previous studies have confirmed the possible role of circulating DNA in judging the efficacy of chemotherapy (24, 25). A well-designed clinical trial is needed to verify the role of ctDNA in evaluating the efficacy of neoadjuvant therapy for lung adenocarcinoma.
In summary, when used in conjunction with AJCC staging, ctDNA provides a relatively precise risk stratification. The serial monitoring of ctDNA in a larger cohort of patients receiving neoadjuvant or adjuvant treatment in the future would provide a more robust indication regarding patient selection.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Xiangya Hospital, Central South University, Changsha. The patients/participants provided their written informed consent to participate in this study.
MP, FY, and LW contributed to the conception and design of the study. MP, QH, WY, ST, CC, WL, JT, XW, BZ, MZ, JL, and FY organized the database. MP, LC, and WS performed the statistical analysis. MP and LC wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was supported by the Science Program of Health Department of Hunan Province (C2016067), Hunan Provincial Natural Science Foundation (2018JJ3764), and Key Research and Development Program of Hunan Province (2019SK2253). They co-funded the testing and follow-up costs of the study.
LC and LW were employed by the company Berry Oncology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.561598/full#supplementary-material
FIGURE S1 | The most frequently detected mutations in tumor tissue and ctDNA.
TABLE S1 | 127 gene panel.
TABLE S2 | Plasma ctDNA detection indices at each time point for all patients.
TABLE S3 | Characteristics of all patients.
TABLE S4 | Tissue capture results of all 77 patients.
TABLE S5 | Recurrence free survival and overall survival for all patients.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
3. Hong TH, Kim J, Shin S, Kim HK, Choi YS, Zo JI, et al. Clinical outcomes of microscopic residual disease after bronchial sleeve resection for non-small cell lung cancer. J Thoracic Cardiovas Surgery. (2020). S0022–5223:30518-3. doi: 10.1016/j.jtcvs.2020.02.079
4. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. (2013) 497:108–12. doi: 10.1038/nature12065
5. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New Engl J Med. (2013) 368:1199–209. doi: 10.1056/NEJMoa1213261
6. De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. (2014) 25:1729–35. doi: 10.1093/annonc/mdu239
7. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Trans Med. (2015) 7:302ra133. doi: 10.1126/scitranslmed.aab0021
8. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Trans Med. (2016) 8:346ra92. doi: 10.1126/scitranslmed.aaf6219
9. Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. (2019) 30:1580–90. doi: 10.1093/annonc/mdz227
10. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discovery. (2017) 7:1394–403. doi: 10.1158/2159-8290.CD-17-0716
11. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. (2015) 21:3196–203. doi: 10.1158/1078-0432.CCR-14-2594
12. Lv W, Wei X, Guo R, Liu Q, Zheng Y, Chang J, et al. Noninvasive prenatal testing for Wilson disease by use of circulating single-molecule amplification and resequencing technology (cSMART). Clin Chem. (2015) 61:172–81. doi: 10.1373/clinchem.2014.229328
13. Chen K, Zhao H, Shi Y, Yang F, Wang LT, Kang G, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res. (2019) 25:7058–67. doi: 10.1158/1078-0432.CCR-19-1213
14. Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn. (2013) 33:409–15. doi: 10.1002/pd.4033
15. Guo N, Lou F, Ma Y, Li J, Yang B, Chen W, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep. (2016) 6:33519. doi: 10.1038/srep33519
16. Chen K, Zhang J, Guan T, Yang F, Lou F, Chen W, et al. Comparison of plasma to tissue DNA mutations in surgical patients with non-small cell lung cancer. J Thoracic Cardiovasc Surgery. (2017) 154:1123–31.e2. doi: 10.1016/j.jtcvs.2017.04.073
17. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. (2013) 339:1546–58. doi: 10.1126/science.1235122
18. Peng M, Xie Y, Li X, Qian Y, Tu X, Yao X, et al. Resectable lung lesions malignancy assessment and cancer detection by ultra-deep sequencing of targeted gene mutations in plasma cell-free DNA. J Med Genet. (2019) 56:647–53. doi: 10.1136/jmedgenet-2018-105825
19. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. (2014) 6:224ra24. doi: 10.1126/scitranslmed.3007094
20. Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. (2007) 369:1742–57. doi: 10.1016/S0140-6736(07)60781-8
21. Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. (2008) 26:3552–9. doi: 10.1200/JCO.2007.13.9030
22. Sandler JE, D’Aiello A, Halmos B. Changes in store for early-stage non-small cell lung cancer. J Thoracic Dis. (2019) 11:2117–25. doi: 10.21037/jtd.2019.05.34
23. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. (2017) 545:446–51. doi: 10.1038/nature22364
24. Han X, Han Y, Tan Q, Huang Y, Yang J, Yang S, et al. Tracking longitudinal genetic changes of circulating tumor DNA (ctDNA) in advanced Lung adenocarcinoma treated with chemotherapy. J Transl Med. (2019) 17:339. doi: 10.1186/s12967-019-2087-9
Keywords: non-small cell lung cancer, circulating tumor DNA, circulating single molecule amplification and re-sequencing technology, prognostic biomarker, minimal residual disease
Citation: Peng M, Huang Q, Yin W, Tan S, Chen C, Liu W, Tang J, Wang X, Zhang B, Zou M, Li J, Su W, Wang L, Chin L and Yu F (2020) Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 10:561598. doi: 10.3389/fonc.2020.561598
Received: 13 May 2020; Accepted: 20 August 2020;
Published: 15 September 2020.
Edited by:
Jarushka Naidoo, Johns Hopkins Medicine, United StatesReviewed by:
Rachel E. Sanborn, Providence Portland Medical Center, United StatesCopyright © 2020 Peng, Huang, Yin, Tan, Chen, Liu, Tang, Wang, Zhang, Zou, Li, Su, Wang, Chin and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihan Chin, cWlubGloYW42NDhAYmVycnlvbmNvbG9neS5jb20=; Fenglei Yu, eXVmZW5nbGVpQGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.