
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 15 October 2020
Sec. Cancer Molecular Targets and Therapeutics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.554331
 Longshan Zhang1
Longshan Zhang1 Yao Fan2
Yao Fan2 Xiaoqing Wang1
Xiaoqing Wang1 Mi Yang1
Mi Yang1 XiXi Wu1
XiXi Wu1 Weiqiang Huang1
Weiqiang Huang1 Jin Lan3
Jin Lan3 Liwei Liao1
Liwei Liao1 Wenqi Huang1
Wenqi Huang1 Lu Yuan1
Lu Yuan1 Hua Pan1
Hua Pan1 Yuting Wu1
Yuting Wu1 Longhua Chen1*
Longhua Chen1* Jian Guan1*
Jian Guan1*Carbohydrate sulfotransferase 4 (CHST4) plays an important role in lymphocyte homing and is abnormally expressed in several cancer types; however, its precise function in tumor development and progression is unknown. Here we confirm that CHST4 is aberrantly expressed in various tumor subtypes. In particular, we found that CHST4 expression was downregulated in hepatitis B virus-related hepatocellular carcinoma (HBV-HCC) tumors compared to paired normal tissue. We also showed that CHST4 overexpression inhibited the proliferation and metastasis of HCC cells in vitro. Clinically, CHST4 was identified as an independent prognostic factor for HBV-HCC patients. We further illuminated the anti-tumor role and mechanism of CHST4 in HBV-HCC by constructing a FENDRR–miR-10b-5p–CHST4 competing endogenous RNA network. We found that downregulation of CHST4 expression may promote HBV expression and regulate ribonucleoprotein complex biogenesis to promote malignant behaviors in HBV-HCC. CHST4 may also recruit CD4+ T cells, macrophages, dendritic cells, and neutrophils into the tumor microenvironment to inhibit the progression of HBV-HCC. Overall, our findings suggest that CHST4 acts as a tumor suppressor in HCC-HBV and represents a potential diagnostic and therapeutic target.
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer deaths worldwide (1–3). Although there has been considerable progress in the treatment of HCC, patient prognosis remains bleak (4). Chronic hepatitis B virus (HBV) infection is a major risk factor for HCC (5) and is responsible for about 50% of HCC cases and almost all childhood liver cancer (6). Indeed, in China, most HCC is related to HBV infection (7–9). Many biomarkers have been used to monitor at-risk populations, such as alpha-fetoprotein L3 (AFP-L3), des gamma carboxyprothrombin (DCP), glypican 3 (GPC3), and Golgi membrane protein 73 (GP73), yet these serum markers are not elevated in all HCC subtypes (10, 11). Therefore, effective and reliable biomarkers for the early detection of HCC are in high demand.
According to the Human Protein Atlas, CHST4 (carbohydrate sulfotransferase 4, GlcNAc6ST2) is expressed in the gallbladder, fallopian tube, liver, and pancreas. CHST4 is a member of the GlcNAc6ST family of proteins and catalyzes the transfer of sulfate to position 6 of non-reducing GlcNAc residues within mucin-associated glycans that ultimately serve as L-selectin ligands (12, 13). These L-selectin ligands are present in high endothelial cells (HEVs) and contribute to lymphocyte homing (14–16). Although previous researches showed that CHST4 was specifically expressed in HEVs (17), follow-up studies have revealed that CHST4 is abnormally expressed in a variety of solid tumors. For example, CHST4 is highly expressed in mucinous adenocarcinomas (18, 19), where it adds sulfate to extended core 1- and core 2-based-glycans (20). Additionally, in those with early-stage uterine corpus and cervical cancer, serum CHST4 expression was higher than that of cancer antigen 125 (CA125) and squamous cell carcinoma antigen (SCC) (21). CHST4 expression is also elevated in gastric cancer tissues (22), urothelial bladder carcinoma (23), cholangiolocellular carcinoma (24), and Opisthorchis viverrini (OV)-related intrahepatic cholangiocarcinoma (25), and the CHST4 promoter is hypermethylated in HBV-HCC (26). In gliomas, CHST4 was shown to have a high frequency of mutant alleles and amplifications and was related to poor prognosis (27). Overall, the current evidence suggests that CHST4 is a cancer-related enzyme and is a potential biomarker in some tumor subtypes. However, the specific functions and roles of CHST4 in tumor progression are largely unknown.
Although CHST4 has been implicated in HBV-HCC (26), its precise function and prognostic value remain unclear. Therefore, this study aimed to clarify the function and clinical value of CHST4 in cancer by performing expression profiling in mice and human tumors. The function of CHST4 in HBV-HCC was also analyzed both in vitro and in vivo.
Four male BALB/c mice (4 weeks old) were obtained from the Animal Science Centre Laboratory of Southern University (Guangzhou). All animals were housed in a specific pathogen-free (SPF)-class and temperature-controlled sterile room. All mice were sacrificed by cervical dislocation, and different organs and tissues were obtained. All collected tissues were stored at −80°C for subsequent quantitative real-time-PCR (qRT-PCR) and immunohistochemistry examination. The BALB/c mice were obtained from the Animal Science Centre Laboratory of Southern University (Guangzhou) with the approval of the Institutional Animal Care Committee of Southern Medical University.
Eight paired tumors and normal tissues were collected from 76 patients who underwent surgical resection at the Nanfang Hospital (Guangzhou, China) from January 2019 to October 2019, with over three specimens collected for each patient. Meanwhile, 40 pairs of freshly resected HCC and adjacent non-neoplastic liver tissues were collected from patients who had undergone hepatectomies for the curative treatment.
All patients had not received chemotherapy, radiotherapy, multiple operation, and biological immunotherapy before surgery. HCC patients were HBV positive, while recurrent HCC and incomplete follow-up data were excluded. The BCLC staging system was used for staging. All collected tissues were stored at -80°C until needed. All patients provided informed consent, and the study was approved by the Institutional Review Board of Nanfang Hospital.
Four-micrometer-thick sections from each sample were cut, deparaffinized using xylene, hydrated through graded alcohol, and incubated with 3% hydrogen peroxide for endogenous peroxidase blockade, and processed for antigen retrieval by microwave heating for 15 min in 1 mM Tris–EDTA buffer (pH 8.0). The primary anti-CHST4 antibody for mice (ARP35787_T100, Aviva Systems Biology) was diluted 1:50 in PBS containing 1% BSA, and the anti-CHST4 antibody for humans (66623-1-Ig, Proteintech) was diluted 1:200, prior to incubation with the sections (overnight at 4°C). As a negative control, a section on each slide was incubated with rabbit serum (provided by Aviva Systems Biology) for mice and mouse monoclonal IgG (B900620, Proteintech) for humans. Subsequently, the sections were incubated with horseradish-peroxidase-conjugated anti-rabbit secondary antibody (DakoCytomation, Glostrup, Denmark) for 1 h at 37°C and then developed with diaminobenzidine. All sections were counterstained with hematoxylin, visualized under a microscope (Olympus, Japan; four images per section). Two pathologists, who were blind to clinical data, calculated CHST4 expression as the sum of the percent positivity of stained tumor cells and the staining intensity (28), and they independently scored CHST4 expression in human tumors as negative, low, moderate, or high. ImageJ software (National Institutes of Health, Bethesda, MD, United States) was used for quantitative analysis of the immunohistochemical staining (29).
Total RNA was isolated from tissues or cells using TRIzol (Invitrogen, United States) and reverse transcribed using the PrimeScript RT Reagent Kit (Perfect Real Time, Takara). qRT-PCR was performed with TB GreenTM Premix Ex TaqTM II (Tli RNaseH Plus, Takara) using the ABI 7500 fast Real-Time PCR System (Applied Biosystems 7500 Fast), as previously described (30). The primer sequences used for CHST4 (human) were: forward: 5′-CCTGCTGTTTCTGGTTTCCCA-3′, reverse: 5′-TGCCCCACAAAAGAAGAGCC-3′; and those for CHST4 (mice) were: forward: 5′-TCCATACTAACGCCAGGAACG-3′, reverse: 5′- TGGTGACTAAGGCTGGAACC-3′. The relative CHST4 level was calculated using the comparative Ct method (ΔΔCt), and normalized to the reference gene, GAPDH.
All cell lines were obtained from the Shanghai Institute for Biological Sciences (Shanghai, China). The human hepatoma cell lines (HCCL-M3, MHCC-97h, Huh7, SMMC-7721, HepG2, and HepG1), and the immortalized normal hepatocyte cell line (LO2) were cultured in high glucose DMEM (Biological Industries, Cronwell, CT, United States) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Cronwell, CT, United States). All media were supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (GibcoTM, Thermo Fisher Scientific, Waltham, MA, United States) and incubated at 37°C in a humidified chamber containing 5% CO2.
Cells were transfected with CHST4 plasmid (GeneCopoeia, United States) or human siRNA1, siRNA2, and siRNA3 sequences (sense: 5′-CGTCAGATCTGAACAAGAA-3′; sense: 5′-GGAGGACCAACCCTACTAT-3′; sense: 5′-GGAGATCTCATGATTGACA-3′; synthesized by RiboBio) by Lipofectamine®2000, as per manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States). RNA and protein were collected at 48 and 72 h after transfection, and CHST4 expression was detected by qRT-PCR and Western blot to verify the transfection efficiency, as previously described (31).
Western blotting was performed according to standard methods, and antibodies were used against CHST4 (1:500, 66623-1-Ig, Proteintech) and GAPDH (1:1,000, 60004-1-Ig, Proteintech).
Cells were seeded into 96-well plates and incubated at 37°C. A cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) was used to detect viable cells over 7 days. Briefly, 10 μl of CCK-8 solution was added, and the absorbance at 450 nm was measured after 1 h (three duplicate wells/sample). Additionally, in vitro Transwell assays were performed to assess cell migration ability. At 24 h after siRNA or plasmid transfection, 5 × 104 cells (cultured with serum-free medium) were placed in the upper insert dish of a 24-well plate chamber filled with DMEM containing 10% FBS (Corning, United States). After incubation for an appropriate period of time at 37°C, the cells were fixed with 100% methanol for 40 min and stained with crystal violet solution for 15 min. Migrated cells were photographed and counted in five random regions with an inverted microscope.
The chromosome location and gene structure of CHST4 were analyzed by GeneCards1 and Uniprot2. The protein sequences of CHST4 from human and other species were compared using DNAMAN software (Lynnon Biosoft, United States). The expression profiles of CHST4 in various human normal and cancer tissues were examined using the TIMER platform3. The CHST4 mRNA expression and copy number variation (CNV) in human cancer cell lines were analyzed using the CCLE database4. CHST4 mutations in human tumors were evaluated via the cBioPortal database5. The clinical prognosis of CHST4 in human tissues was analyzed using the GEPIA database6, which is based on The Cancer Genome Atlas (TCGA).
DIANA, miRDB, mirDIP, and miRwalk databases were used to predict microRNAs (miRNAs) that target CHST4, and the DIANA LncBase V2 database was used to predict long non-coding RNAs (lncRNAs). The competing endogenous RNA (ceRNA) network was constructed using the Cytoscape software (28). The heatmaps of differentially expressed miRNAs and lncRNAs were produced using GraphPad Prism (v8). In addition, the Gene Ontology (GO) enrichment analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were generated by ImageGP7.
We screened for genes that interact with CHST4 using the STRING database8. The expressions of CHST4, miRNAs, and lncRNAs were obtained from TCGA9. CHST4 gene expression and corresponding clinical data were obtained from the GSE14520 dataset of HBV-HCC and downloaded from the Gene Expression Omnibus (GEO) database10. The X-tile program was used to select the cut-off value of CHST4 expression in GSE14520. To investigate the difference of biological functions and pathways between high and low expression groups of CHST4 in HBV-HCC survival, gene set enrichment analysis (GSEA) was used to investigate potential mechanisms in the Molecular Signatures Database (MSigDB) of c2 (c2.cp.kegg.v7.1.symbols) and c5 (c5.all.v7.1.symbols) (32). False discovery rate (FDR) <0.25 and the nominal P-value < 0.05 were considered statistically significant. We examined the correlation between the expression level of CHST4 and other genes using two-side Pearson correlation coefficients and the z-test (33). The “cor” function of the R software (34) (R version 3.5.1) was used in this study (| Pearson correlation coefficient| > 0.5 and P < 0.01). The degree of immune infiltration in HBV-HCC was evaluated by the TIMER platform (TIMER, QUANTISEQ, MCPCOUNTER, and EPIC databases) (35, 36) and R-ssGSEA algorithm (37).
All statistical analyses were carried out using SPSS version 22.0 (IBM, United States). All results are presented as mean ± SD. All experiments were performed in triplicate unless otherwise specified. The comparison between the two groups was performed using the Student’s t-test. The associations between the clinicopathological characteristics of the patients and CHST4 expression were analyzed using the Chi-squared test. Univariate and multivariate Cox regression analyses were used to evaluate survival data. The receiver-operating characteristic (ROC) curve was plotted by SPSS, and the area under the ROC curve (AUC) was used for the predicted value. In all tests, a two-tailed P < 0.05 was considered statistically significant.
For research involving biohazards, correct standard procedures have been carried out.
The CHST4 gene is located at 16q22.2 and includes a 5′UTR exon, one CDS exon, a 3′UTR exon, and two introns (Figure 1A). Human CHST4 shares 78, 78, 79, 72, and 79% sequence identity to Bos taurus, Capra hircus, Felis catus, Mus musculus, and Sus scrofa CHST4, respectively, suggesting that CHST4 is highly conserved in mammals (Supplementary Figure S1).
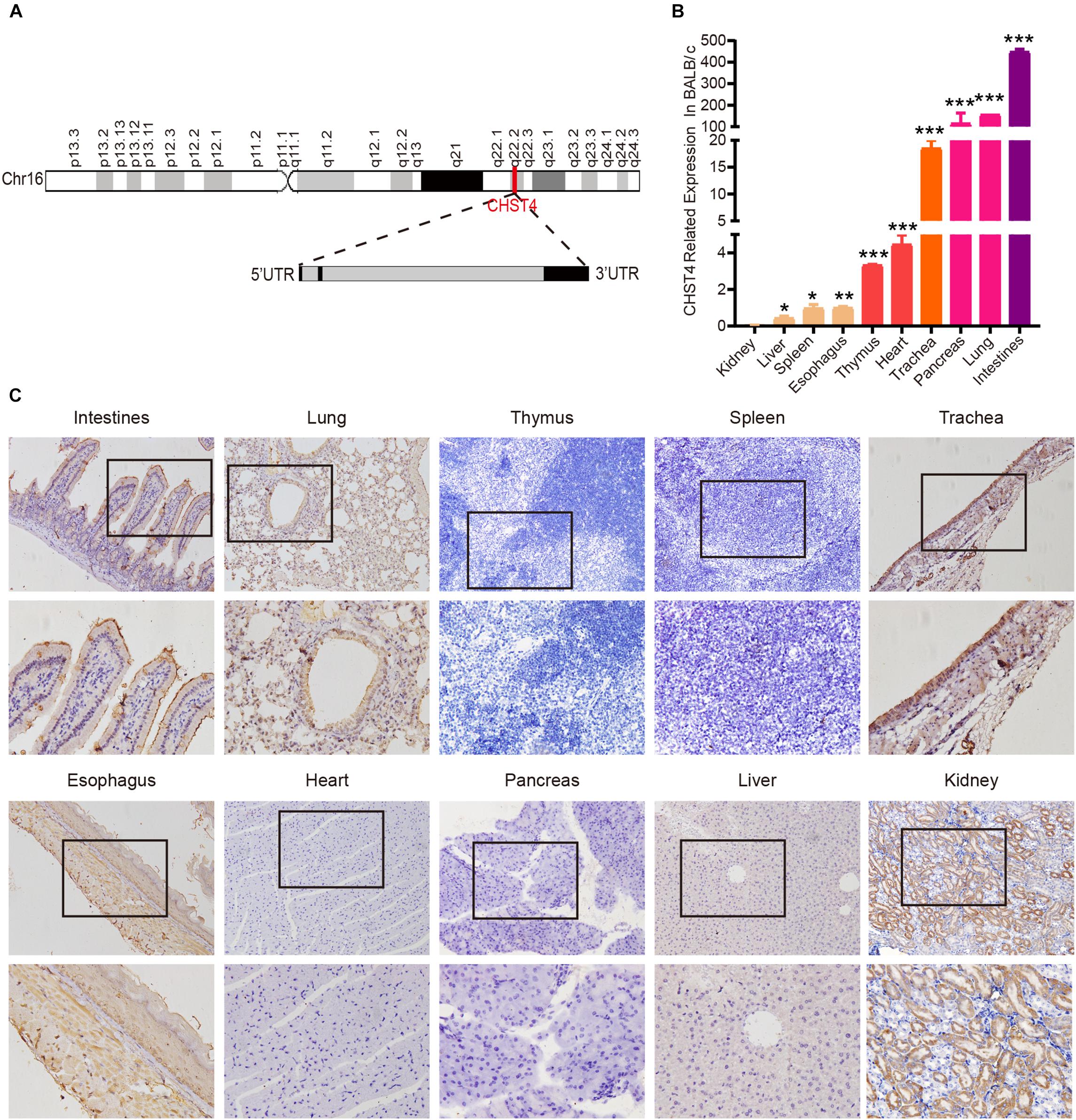
Figure 1. Structure of the carbohydrate sulfotransferase 4 (CHST4) gene and its distribution in BALB/c mice tissues. (A) Chromosome location (red line; top) and gene structure of CHST4 (bottom; black boxes represent exons and gray boxes represent introns). (B) Relative expression of CHST4 mRNA in various tissues of BALB/c mice by quantitative real-time (qRT)-PCR. GAPDH was used as the loading control. Bars represent the mean ± SD (n = 4); *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test). All samples were tested in triplicate. (C) Immunohistochemical analysis of the expression and distribution of CHST4 protein in BALB/c mice tissues (100×; 400×; four samples in each group).
We used BALB/c mice, the most commonly used laboratory animal, to examine the expression of CHST4 in 10 different tissue types by qRT-PCR. As shown in Figure 1B, mRNA expression of CHST4 was highest in the intestine, followed by the lung and pancreas, then the trachea, and finally, the heart and thymus tissues. Meanwhile, there was little mRNA expression of CHST4 in the kidney, liver, spleen, and esophagus tissues. In terms of protein expression (Figure 1C and Supplementary Figure S2A), low levels of CHST4 were observed in intestinal epithelial cells, and strong expression was found in goblet cells. In the respiratory system, CHST4 was low expressed in the bronchial and alveolar epithelial cells of the lung and moderately expressed in the tracheal epithelial cells. CHST4 was also moderately expressed in the epithelial cells of renal tubules. Esophageal epithelial cells and smooth muscle cells showed low expression. However, there was no protein expression of CHST4 in the thymus, spleen, myocardium, pancreas, or liver. The corresponding negative control is shown in Supplementary Figure S2B.
We used the TIMER platform to analyze the expression of CHST4 mRNA in 34 types of human tumors, either alone or with their paired normal tissues. As shown in Figure 2A, CHST4 was highly expressed in cholangiocarcinoma (CHOL), mesothelioma (MESO), and prostate adenocarcinoma (PRAD), and was only lowly expressed in skin cutaneous melanoma (SKCM) and uveal melanoma (UVM). Among the subtypes of breast invasive carcinoma (BRCA), the CHST4 expression in BRCA-basal was significantly higher than in BRCA-Her2 and BRCA-luminal. Interestingly, there was a higher expression level of CHST4 in metastatic SKCM than in non-metastatic SKCM. In head and neck squamous cell carcinoma (HNSCC), CHST4 expression was higher in human papillomavirus (HPV)-positive subtypes than those that were HPV-negative. Five types of human tumors showed higher mRNA expression of CHST4 in the tumor compared to the adjacent normal tissue, including colon adenocarcinoma (COAD), stomach adenocarcinoma (STAD), and kidney renal papillary cell carcinoma (KIRP). Meanwhile, five types of human tumors showed lower CHST4 expression than the adjacent normal tissue, especially BRCA and liver hepatocellular carcinoma (LIHC).
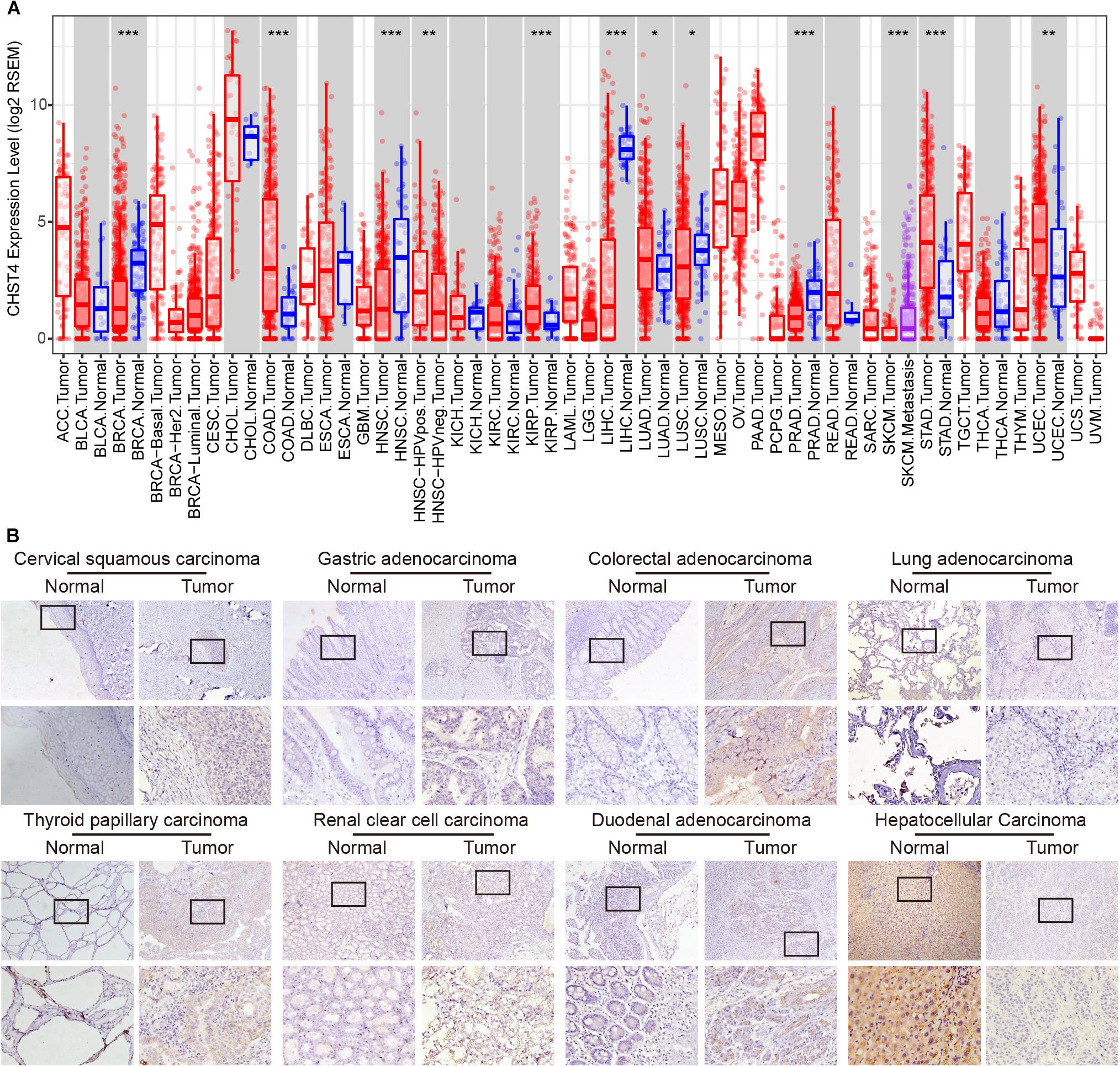
Figure 2. Expression, copy number, and gene alteration of CHST4 in human tumors. (A) mRNA expression of CHST4 in 34 types of human tumors, alone or with their paired normal tissues from the TIMER platform. Bars represent mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).588896101588896101 (B) Immunohistochemical analysis of the expression and distribution of CHST4 proteins in human tissues (100×; 400×; more than three samples in each group).
We further detected the protein expression of CHST4 in eight types of human tumors and paired normal tissues by immunohistochemistry. As shown in Figure 2B and Supplementary Figure S3A, the CHST4 was weakly expressed in cervical squamous carcinoma, gastric adenocarcinoma, thyroid papillary carcinoma, renal clear cell carcinoma, and duodenal adenocarcinoma compared to the matched normal tissues. Additionally, CHST4 was strongly expressed in LIHC compared to the paired normal tissues. Moreover, CHST4 was moderately expressed in colorectal adenocarcinoma but not expressed in paired normal tissues. Meanwhile, there was no obvious protein expression of CHST4 in either normal or cancerous tissues of the lung. CHST4 was distributed in the nuclei in LIHC and gastric adenocarcinoma; in all other tissues, it was distributed in the cytoplasm. The corresponding negative control is shown in Supplementary Figure S3B.
We used the Cancer Cell Line Encyclopedia (CCLE) database to explore the role of CHST4 in human tumor cell lines. Both mRNA expression and copy number of CHST4 were highest in the pancreas, colon, lung non-small-cell, and lymphoma Burkitt cell lines (Supplementary Figures S4A,B). We also used the cBioPortal database to analyze CHST4 gene alterations in 32 human tumor types (Supplementary Figure S4C). The change rate of CHST4 (primarily gene amplification) was highest in cholangiocarcinoma, followed by melanoma and uterine cancer (mainly gene mutation).
We used the GEPIA database to analyze the correlation between CHST4 expression and the prognosis of patients with tumors. As shown in Figure 3, in adrenocortical carcinoma (ACC), patients with high CHST4 expression had longer overall survival (OS) and disease-free survival (DFS) than those with low CHST4 expression. However, in kidney renal clear cell carcinoma (KIRC) and LIHC, the opposite was observed: patients with high CHST4 expression had shorter OS and DFS. In SKCM and MESO, patients with high CHST4 expression had longer OS than those with low expression. Additionally, in uterine corpus endometrial carcinoma (UCEC), those with high CHST4 expression had longer DFS than those with low expression. In KIRP, patients with high CHST4 expression had short OS. Finally, in bladder urothelial carcinoma (BLCA) and glioblastoma multiforme (GBM), those with high CHST4 expression had shorter DFS than those with low expression. Therefore, the relationship between CHST4 expression and prognosis varies according to the tumor type.
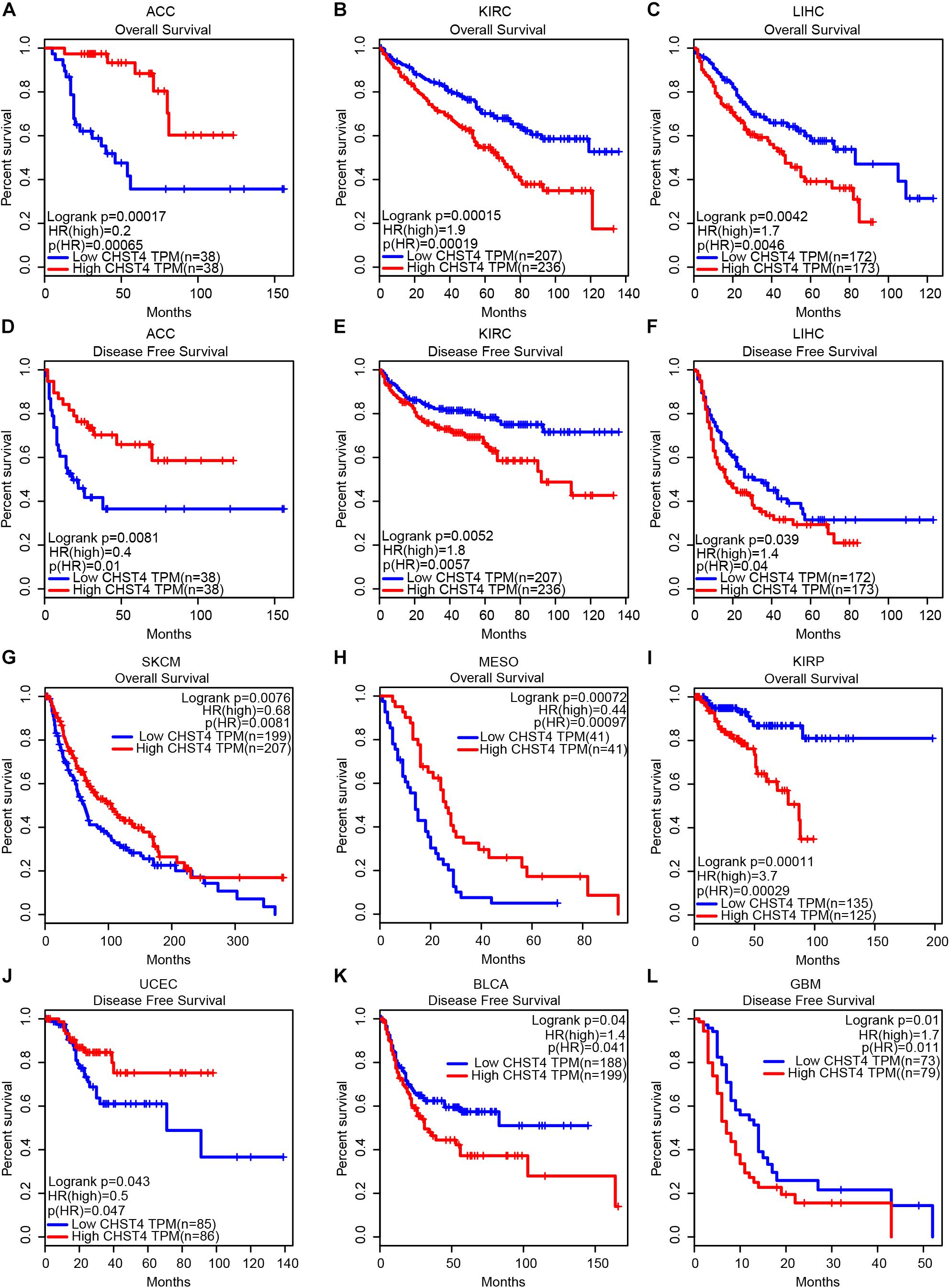
Figure 3. The significance of CHST4 in the clinical prognosis of human tumors. (A–L) Kaplan–Meier analysis of the overall survival and disease-free survival in patients with ACC, adrenocortical carcinoma; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; SKCM, skin cutaneous melanoma; MESO, mesothelioma; KIRP, kidney renal papillary cell carcinoma; UCEC, uterine corpus endometrial carcinoma; BLCA, bladder urothelial carcinoma, and GBM, glioblastoma multiforme using the GEPIA database. The division of CHST4 high and low expression is based on the median (50%) of CHST4 expression in each tumor type.
In HCC, patients with high CHST4 expression had shorter OS and DFS, which suggests that CHST4 plays a tumor-promoting role in HCC. As most HCC is related to HBV in China (7, 8), we sought to determine CHST4 expression in Chinese patients with HBV-HCC using the GEO gene chip. We found that CHST4 expression was lower in tumor tissues than in normal tissues, and HBV-HCC patients with high CHST4 expression had longer OS and DFS. This finding indicates that CHST4 may inhibit the development or progression of HBV-HCC.
After excluding the patients without HBV infection reports and complete survival information from the GSE14520 dataset, we analyzed a total of 242 HBV-HCC tumor tissues and 239 adjacent normal liver tissues. As shown in Figure 4A, CHST4 expression was significantly lower in tumors than in normal tissues (P < 0.0001). This finding was verified in our collected HBV-HCC samples (P < 0.001) (Figure 4B). However, we found that the CHST4 expression levels varied among different HBV-HCC specimens (Figure 4C). Therefore, we focused on the aberrant expression of CHST4 in tumor tissues.
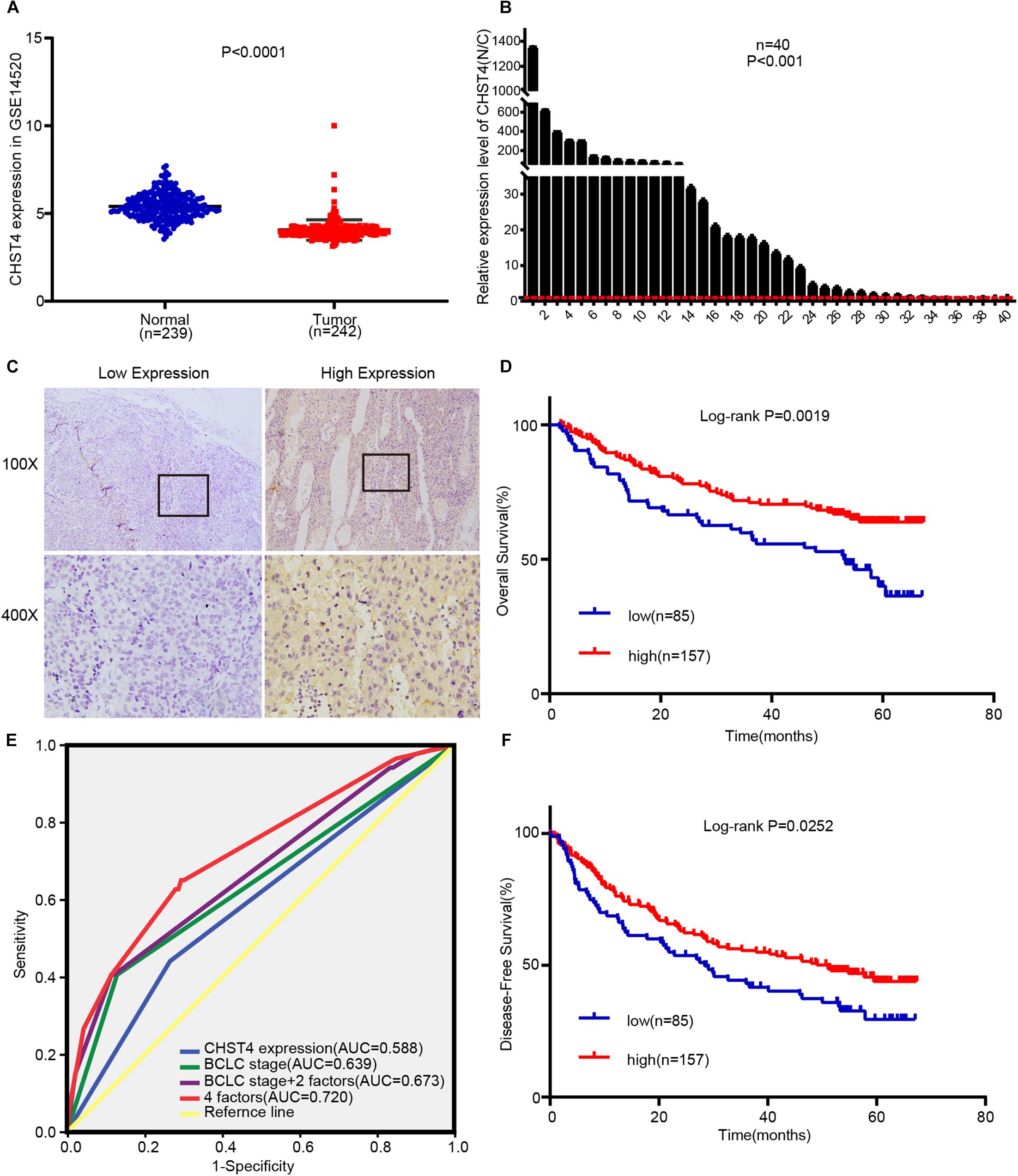
Figure 4. High CHST4 expression is related to better prognoses in patients with hepatitis B virus-related hepatocellular carcinoma (HBV-HCC). (A) Expression of CHST4 in patients from the GSE14520 dataset. (B) mRNA expression of CHST4 in 40 paired HCC and normal tissue samples (N/C) by qRT-PCR assays. All samples were tested in triplicate. The statistical analysis is paired-samples t-test. Bars represent the mean ± SD. (C) Representative staining of low and high CHST4 expression in the tumor cell cytoplasm. (D,F). Kaplan–Meier analysis of overall survival and disease-free survival in those with high CHST4 expression in the GSE14520 dataset versus low expression. (E) Receiver-operating characteristic curves (ROC) of the prognostic accuracy of CHST4 expression and clinicopathological risk factors of patients in the GSE14520 dataset. Comparative ROC curves for CHST4 expression, Barcelona Clinic Liver Cancer (BCLC) stage, and BCLC stage with two factors (multinodular and cirrhosis) or with four factors (CHST4 expression, BCLC stage, multinodular, and cirrhosis) are shown (AUC, area under the curve).
We used X-tile analysis (38, 39) to divide patients in the GSE14520 dataset into two groups: those with high or low expression of CHST4 (Supplementary Figure S5). We then analyzed the relationship between CHST4 expression and clinicopathological characteristics. As shown in Table 1, strong associations were observed between CHST4 expression and alanine transaminase levels (P = 0.026) and TNM staging (P = 0.042). However, the expression of CHST4 was not associated with age, gender, tumor size, multinodular, cirrhosis, BCLC stage, or serum AFP (P > 0.05).
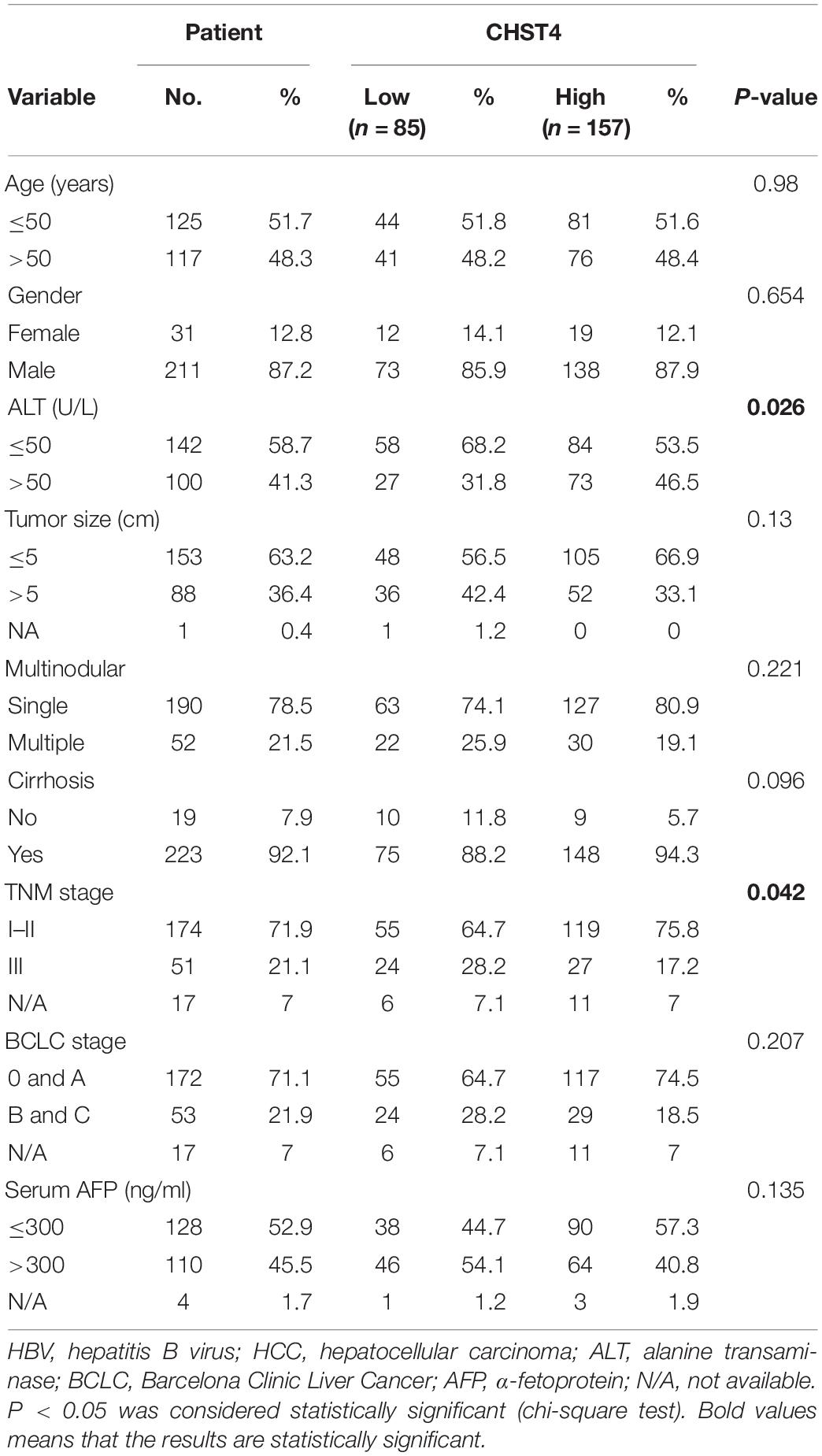
Table 1. Correlation between carbohydrate sulfotransferase 4 (CHST4) expression and clinicopathologic characteristics of HBV-HCC patients from the GSE14520 dataset.
To determine whether CHST4 expression was an independent prognostic factor in HBV-HCC, we performed Cox regression analysis. As shown in Table 2, high expression of CHST4 was associated with a significantly lower risk of death in HBV-HCC patients (P = 0.013) compared to those with low CHST4 expression. Moreover, we found that CHST4 expression level could be used to predict survival (P = 0.001). We also found that multinodular (P = 0.017), cirrhosis (P = 0.019), and BCLC stage (P = 0.001) were independent prognostic factors for survival in HBV-HCC. When these four parameters were considered together (Figure 4E), the area under the curve (AUC) increased to 0.72 (95% CI 0.651–0.79), which was higher than using the BCLC stage alone (0.639, 95% CI 0.562–0.716) (Supplementary Table S1). We also proved high CHST4 expression correlated with a significantly decreased risk of recurrence in HBV-HCC patients (P = 0.026). Furthermore, CHST4 expression (P = 0.027), gender (P = 0.016), and BCLC stage (P = 0.044) were independent prognostic factors for recurrence in HBV-HCC (Table 2). Kaplan–Meier survival curves also demonstrated that patients with higher expression of CHST4 had better OS (P = 0.0019) and longer time to recurrence (P = 0.0252) (Figures 4D,F). Together, these data suggest that increased CHST4 expression in HBV-HCC correlates with a better prognosis.
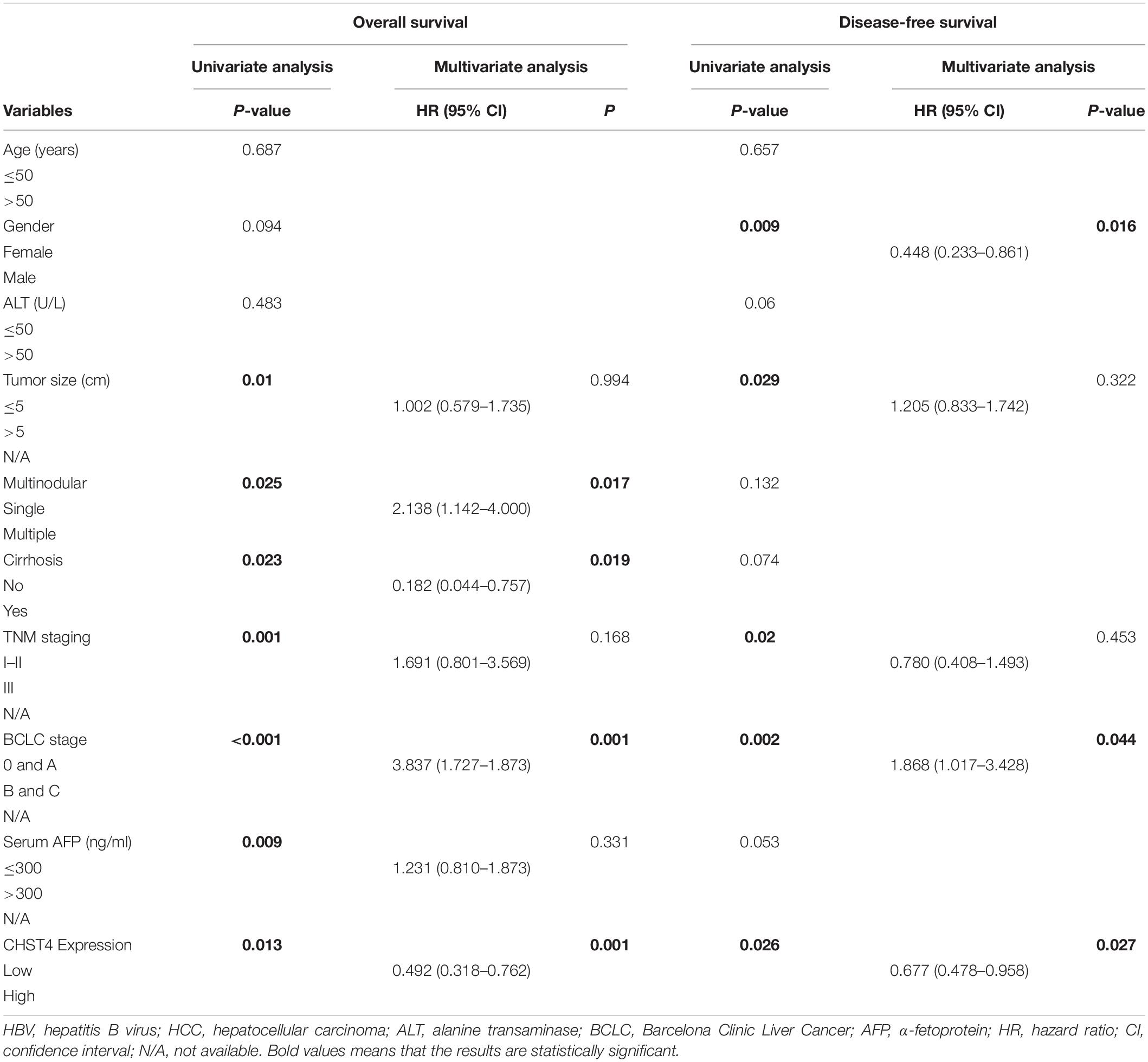
Table 2. Univariate and multivariate analyses of prognostic factors of HBV-HCC patients from the GSE14520 dataset.
As low CHST4 expression in HBV-HCC tumor tissues was associated with malignant clinicopathological features, we explored the biological function of CHST4 in HCC cells. First, we examined the CHST4 expression pattern in HCC cell lines (HCCL-M3, MHCC-97h, Huh7, SMMC-7721, hepG2, and HepG1) and normal liver cells (L02) (Figures 5A,B). Next, according to the endogenous level of CHST4, we silenced CHST4 in HepG2 and Huh7 cells using a CHST4 siRNA and overexpressed CHST4 in MHCC-97h and HCCL-M3 cell lines using a CHST4 plasmid. The efficiencies of CHST4 knockdown and overexpression were validated by qRT-PCR and Western blot (Figure 5C and Supplementary Figures S6A,B): siRNA2 targeting of CHST4 showed satisfactory knockdown efficiency and was used in the subsequent in vitro experiments (Supplementary Figure S6C).
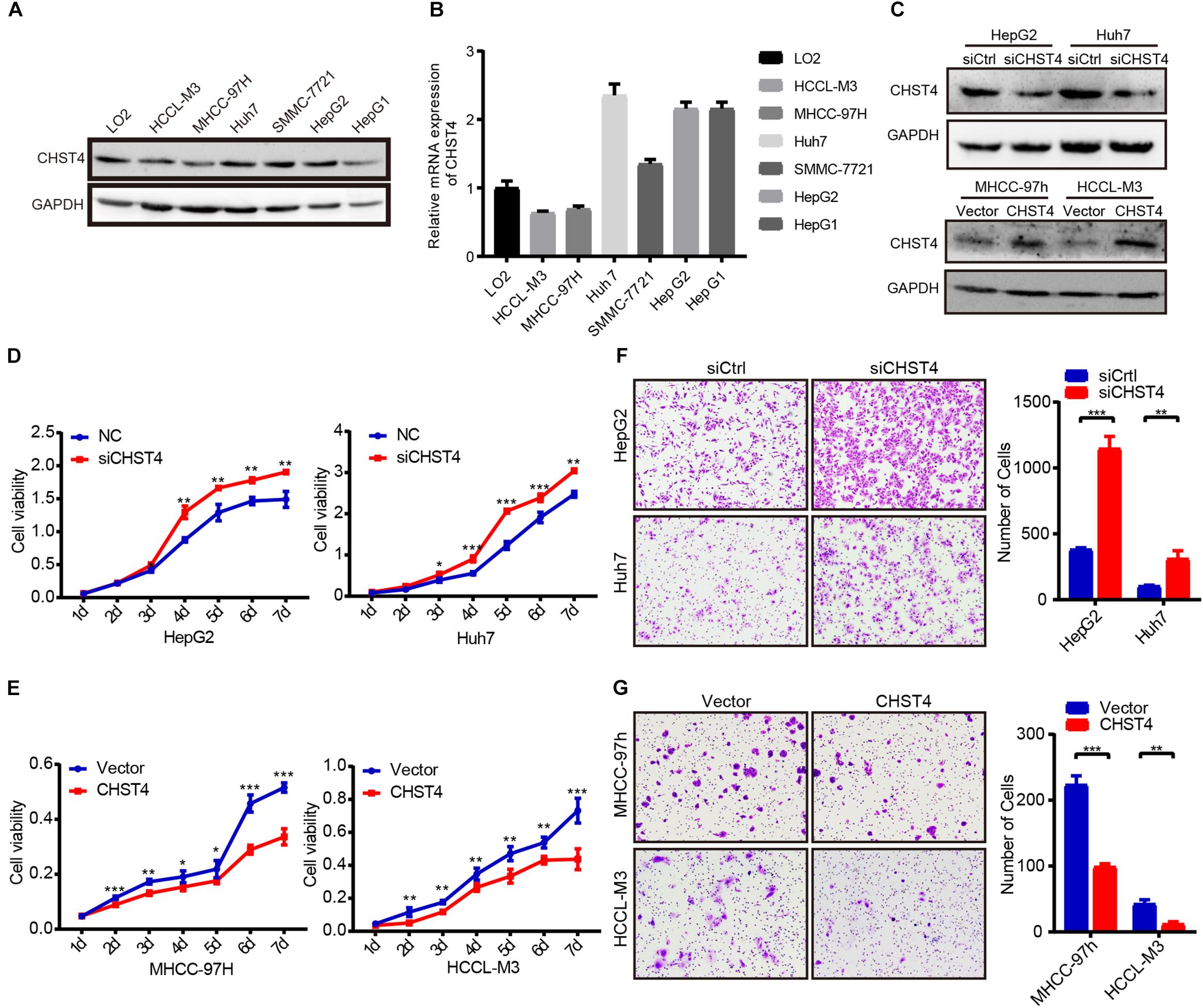
Figure 5. CHST4 inhibits the proliferation and migration of HCC cells. (A,B) Protein and mRNA expression of CHST4 in various HCC cell lines. (C) Knockdown efficiencies were validated in HepG2 and Huh7 cells, and overexpression efficiencies were validated in MHCC-97h and MCCL-M3 cells by Western blot. (D,E) Effects of CHST4 knockdown and overexpression on HCC proliferation determined by CCK-8 assays. Bars represent the mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).588896105588896105 (F,G) Effects of CHST4 knockdown and overexpression on HCC migration were determined by Transwell assays. The right panels show the respective quantitative histograms. All experiments were performed in triplicate. Bars represent the mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).588896108588896108.
We then investigated the role of CHST4 in the pathogenesis of HCC in vitro. CHST4 knockdown resulted in elevated cell viability in HepG2 and Huh7 cells than controls. Conversely, CHST4-overexpressing MHCC-97h and HCCL-M3 cells showed decreased cell viability compared to in control cells (all P < 0.05) Figures 5D,E). Transwell migration assays showed that CHST4 knockdown greatly increased the migration abilities of HepG2 and Huh7 cells (Figure 5F). In contrast, CHST4 overexpression hindered the migration capacities of HCC cells (Figure 5G). Together, our data demonstrate that CHST4 is critical for the proliferation and migration of HCC cells.
There are currently three known regulatory gene expression pathways: DNA methylation (40), RNA splicing (41), and translation blocking (42). Previous studies showed that the promoter of CHST4 is hypermethylated in HBV-HCC (26), which may lead to the downregulation of CHST4 expression. However, intracellular miRNA may also regulate the expression of CHST4 through translation inhibition. Therefore, to explore the upstream regulation of CHST4 expression in HBV-HCC, we constructed a ceRNA network (43).
Using the DIANA, miRDB, mirDIP, and miRwalk databases to predict upstream miRNAs of CHST4, we identified miR-10b-5p and miR-10a-5p as potential regulators (Figure 6A). As miR-10a-5p shares the same seed sequence with miR-10b-5p, they may target the same lncRNAs. Therefore, we used DIANA LncBase V2 to predict the lncRNAs that are targeted by these miRNAs in the liver and constructed a potential lncRNA–miRNA–CHST4 ceRNA network (Figure 6B).
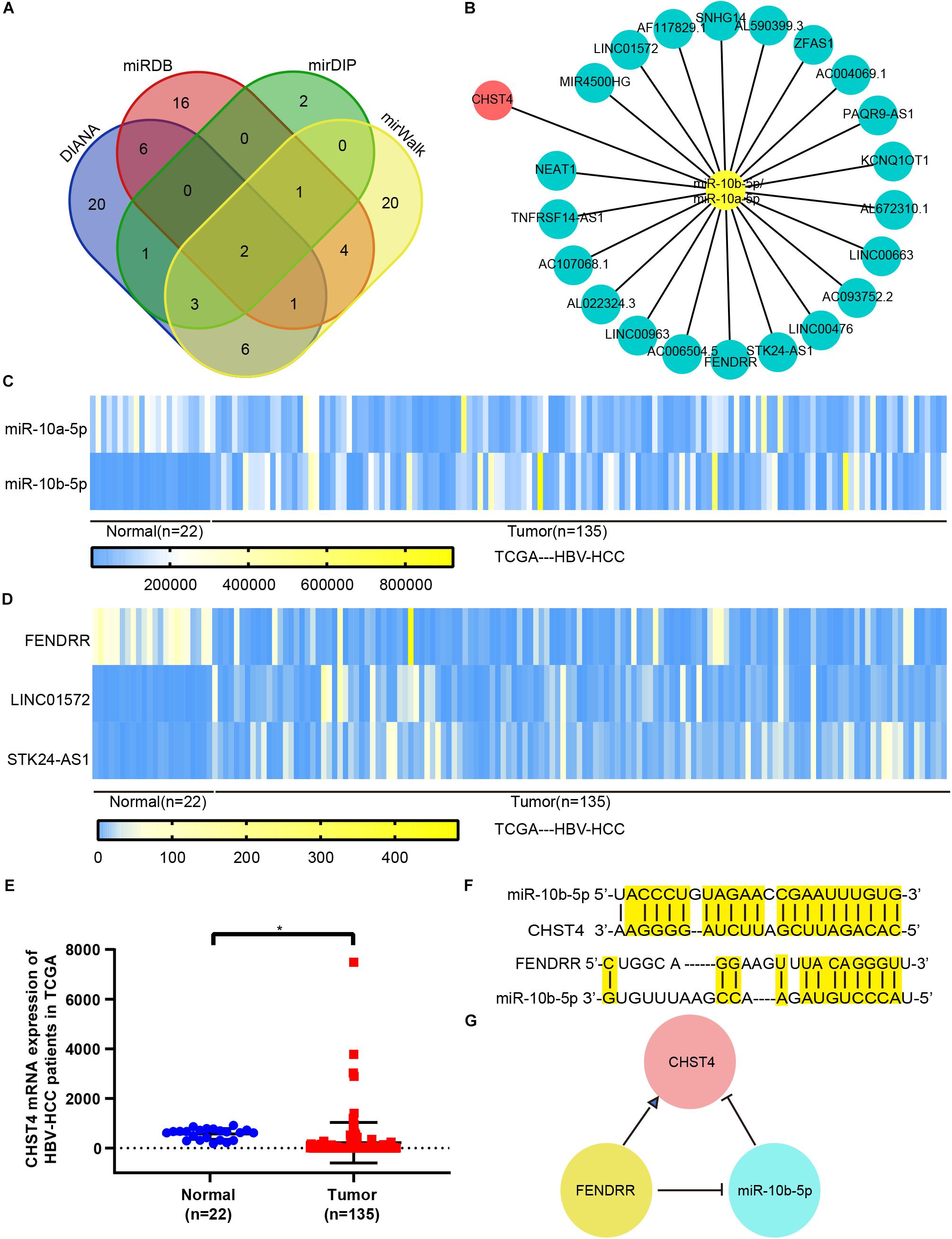
Figure 6. The competing endogenous RNA (ceRNA) network for CHST4 in HBV-HCC. (A) Results of the predicted miRNAs using four different databases: miR-10b-5p and miR-10a-5p were finally screened. (B) Construction of the lncRNA–miRNA–CHST4 network. (C–E) The expression of mRNA, miRNA, and lncRNA in the network of HBV-HCC patients from The Cancer Genome Atlas. Bar represents the mean ± SD; *P < 0.05 (Student’s t-test).588896110588896110 (F) Complementary sequences between CHST4 and miR-10b-5p, and miR-10b-5p with FENDRR. (G) A ceRNA network constructed using FENDRR, miR-10b-5p, and CHST4 in HBV-HCC.
We compared the expression levels of CHST4 and differentially expressed miRNAs and lncRNAs between normal and tumor tissues in human HBV-HCC from TCGA. We screened HBV-HCC patients (i.e., those who were positive for the hepatitis B surface antigen) with complete clinical data in TCGA, and identified 135 HBV-HCC tumor tissues and 22 normal liver tissues. CHST4 expression was significantly lower in HBV-HCC tumor tissues than the corresponding normal tissue, while only miR-10b-5p was more highly expressed (P < 0.01) (Figures 6C,E). Therefore, miR-10b-5p is likely the miRNA target of CHST4 in HBV-HCC. We also found that the lncRNA FENDRR was highly expressed in tumors (P < 0.01) (Figure 6D). Therefore, we analyzed any complementary sequences between CHST4 and miR-10b-5p, miR-10b-5p and FENDRR, respectively (Figure 6F). Together, our findings suggest that the lncRNA FENDRR inhibits the expression of miR-10b-5p, which, in turn, increases the expression of CHST4 in HBV-HCC (Figure 6G).
Next, we performed GSEA to investigate the potential biological roles of CHST4 using the Molecular Signatures Database (MSigDB) for GSE14520. GO analysis showed that CHST4 participates in ribonucleoprotein complex biogenesis, RNA splicing, and mRNA metabolic process, and was also closely related to viral gene expression (Figures 7A–C,E). Furthermore, the KEGG pathway analysis showed that CHST4 regulates the ubiquitin-mediated proteolysis signaling pathway and takes part in nucleotide excision repair, DNA replication, and aminoacyl tRNA biosynthesis (Figure 7D). We also identified the co-expressed genes of CHST4 in GSE14520 (Figure 7F and Supplementary Table S2).
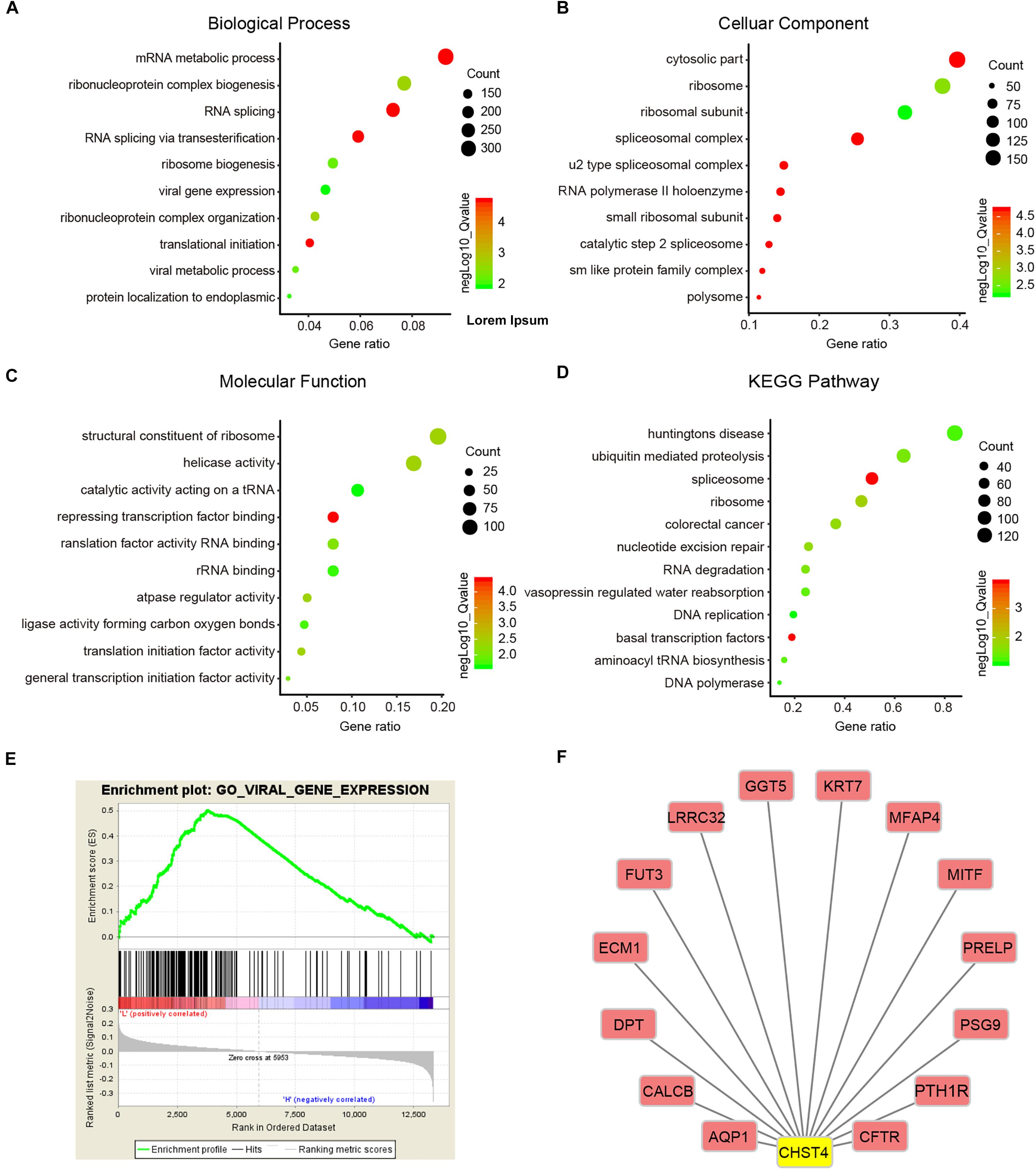
Figure 7. Gene function and downstream pathway analysis for CHST4. (A–D) Gene ontology (GO) analysis of CHST4 for HBV-HCC patients in the GSEA14520 dataset by gene set enrichment analysis (GSEA). (E) Relationship between CHST4 and viral gene expression by GO analysis. (F) The co-expressed genes of CHST4 in GSE14520.
We then constructed a protein–protein interaction (PPI) network to identify 40 significant interacting proteins of CHST4 from the String database (Supplementary Figure S7A) and used the DAVID database to explore the potential roles of CHST4 and these interacting proteins. GO analysis showed these genes were mainly located in the Golgi, several biofilms, and extracellular exosomes. The genes participate in multiple biological metabolic processes, such as sulfur compound metabolism, glycoprotein biosynthesis, and oligosaccharide metabolism. They also regulate the immune response, as well as leukocyte migration, tethering, or rolling (Supplementary Figures S7B–D). The KEGG analysis showed that these genes regulate metabolic pathways and proteoglycans in cancer. They are also involved in the interleukin (IL)-17 signaling pathway and the intestinal immune network for IgA production, and are related to the hematopoietic cell lineage (Supplementary Figure S7E), which indicates that CHST4 may participate in immune regulation of the tumor microenvironment.
We further evaluated the immune cell infiltration score in HBV-HCC using TIMER platform (TIMER, QUANTISEQ, MCPCOUNTER, and EPIC databases) and R-ssGSEA. We found that those with high expression of CHST4 had higher scores of B cells, CD4+ T cells, macrophages, dendritic cells, and neutrophils (Figures 8A,B). Therefore, overall findings indicate that CHST4 can recruit immune cells into the tumor microenvironment in HBV-HCC.
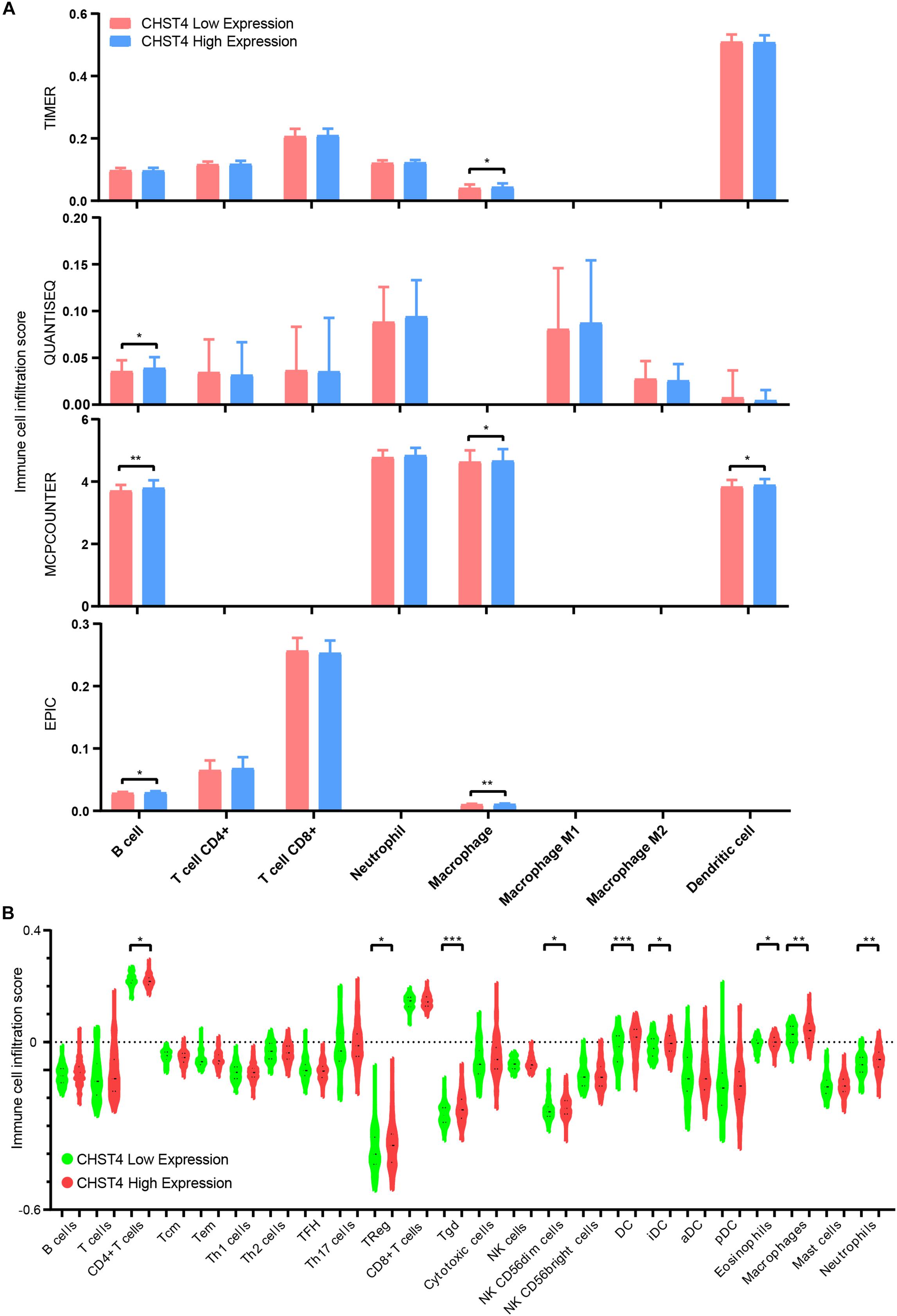
Figure 8. The correlation between CHST4 and immune cell infiltration in HBV-HCC. (A) The degree of immune infiltration in HBV-HCC was evaluated by TIMER platform (TIMER, QUANTISEQ, MCPCOUNTER, and EPIC databases). Bar represents the mean ± SD; *P < 0.05, **P < 0.01 (Student’s t-test). (B) The immune cell infiltration score in HBV-HCC using R-ssGSEA (Tcm, T central memory cells; Tem, T effector memory cells; Th1 cells, T helper 1 cells; Th2 cells, T helper 2 cells; TFH, T follicular helper cells; Th17 cells, T helper 17 cells; Treg, regulatory T cells; Tgd, T gamma delta cells; NK cells, natural killer cells; DC, dendritic cell; iDC, immature DC; aDC, activated DC; pDC, plasmacytoid DC). Bars represent the mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).
In this study, we showed that CHST4 is highly conserved in mammals, indicating that it has a potential role in species evolution. We found that both protein and mRNA levels of CHST4 were high in the intestines and the lung in BALB/c mice. Similarly, previous studies showed that CHST4 was highly expressed in the mouse colon (44). We also found that protein and mRNA levels of CHST4 were low in the liver in BALB/c mice, while high mRNA levels and low protein levels were observed in the pancreas. In contrast, the mRNA levels of CHST4 were high in the pancreas and liver in C57BL/6 mice (12), which may be related to the different strains of mice (45). The difference in protein and mRNA levels may be due to translation or posttranslational modifications (46) of CHST4, although this has not yet been verified.
We also analyzed the mRNA levels of CHST4 in 34 different types of human tumors. Consistent with previous studies, we found that the expression of CHST4 mRNA was higher in cholangiocarcinoma (24), colon adenocarcinoma (18), and gastric adenocarcinoma (22), and we partially verified this conclusion immunohistochemically. We also found that tumor expression of CHST4 at the mRNA and protein level is only partially consistent. We speculate this difference is partly because the TIMER data from the TCGA database contains ethnic differences, which could result in tumor heterogeneity. Additionally, our specimen size is small, which could also lead to deviations in tumor characteristics. We also found significant differences in the expression of CHST4 among different subtypes of the same tumor. For example, the expression of CHST4 in BRCA-basal was higher than that in BRCA-Her2 and BRCA-luminal. There was also a higher expression of CHST4 in metastatic in SKCM compared to non-metastatic cancer, and the expression of CHST4 was higher in the HPV-positive subtype of HNSCC than in the HPV-negative one. Similarly, differences in CHST4 expression were previously observed among the various subtypes of colon adenocarcinoma (18) and ovarian cancer (19). Therefore, we speculate that CHST4 may be a potential biomarker for specific tumor subtypes, and this hypothesis should be validated in future studies.
We also showed that CHST4 expression was downregulated in HBV-HCC. Moreover, patients from the GSE14520 dataset of HBV-HCC with higher CHST4 expression had longer OS and DFS. Further analysis showed that CHST4 expression was an independent prognostic indicator among patients with HBV-HCC. Moreover, when we combined CHST4 with three other independent prognostic factors (multinodular, cirrhosis, and BCLC stage), the predictive value for prognosis significantly improved. Thus, potentially, these four factors could be useful survival predictors for patients with HBV-HCC.
Additionally, in vitro experiments confirmed that ectopic overexpression of CHST4 inhibited the proliferation and metastasis of HCC, while silencing of CHST4 expression promoted tumor progression. These findings indicate that CHST4 has an important biological function and prognostic value in HBV-HCC. Indeed, previous studies show that CHST4 can flow in the bloodstream in soluble form (21); however, whether CHST4 can be used as a serological indicator of HBV-HCC requires further research. Moreover, as the relationship between the expression level of CHST4 and HBV-HCC prognosis was contradictory to the prognosis of HCC from TCGA, the role of CHST4 in other subtypes of HCC should also be further investigated.
Our analysis of the upstream regulatory network of CHST4 in HBV-HCC revealed both FENDRR and miR-10b-5p are most likely involved. FENDRR has previously been shown to play an important role in tumor suppression. For example, in NSCLC, lncRNA FENDRR was lowly expressed and inhibited tumor cell growth and cisplatin resistance by regulating ABCC10 (3). Also, in colorectal cancer, FENDRR interacts with miRNA-18a-5p to upregulate the expression of growth inhibitor 4, thus inhibiting the proliferation, migration, and invasion of colorectal cancer cells (47). In breast cancer, FENDRR inhibits tumor cell proliferation, promotes cell apoptosis, and is related to good prognosis (48). Furthermore, miR-10b-5p is overexpressed in HCC tissues and is associated with poor prognosis (49). Therefore, based on this data from previous studies (combined with the complementary sequences and expressions of miR-10b-5p, CHST4, and FENDRR in HBV-HCC), we speculate that the FENDRR–miR-10b-5p–CHST4 network plays an important role in HBV-HCC, although further experiments are necessary to prove this hypothesis.
CHST4 expression was found to be related to viral gene expression, and thus, low CHST4 expression may promote HBV expression and replication in HCC, thereby increasing chromosomal instability and tumor cell proliferation, as reported previously (6). We also discovered that CHST4 participates in ribonucleoprotein complex biogenesis, RNA splicing, and mRNA metabolic process, and these biological processes have previously been implicated in HCC development (50, 51). Moreover, ribosome biogenesis and mutations of ribosome genes were previously shown to be related to progression of various tumors (52, 53). CHST4 may also regulate nucleotide excision repair, which is involved in every step of the DNA recognition–unwinding–incision process, which affects HCC risk (54). We also found that downregulation of CHST4 may impact the immune response and the IL-17 signaling pathway. IL-17 recruits neutrophils to the peritumor matrix of HCC to produce matrix metalloproteinase-9, which stimulates angiogenesis (55). Overall, these findings suggest that the low expression of CHST4 in HBV-HCC may upregulate HBV expression and regulate spliceosome and ribonucleoprotein complex biogenesis to promote malignant behaviors.
Finally, we showed that CHST4 expression positively correlates with the score of infiltration of B cells, CD4 + T cells (56), macrophages (57, 58), dendritic cells (59), and neutrophils (60–62). Therefore, CHST4 might recruit immune cells with tumor-killing activity into the tumor microenvironment, thus inhibiting the occurrence and development of HBV-HCC.
Our study provides evidence that CHST4 expression may relate to tumor heterogeneity in several types of cancers. In particular, we showed that CHST4 inhibits HCC progression, likely through recruiting immune cells to the tumor microenvironment. We also showed that CHST4 is a prognostic candidate for HBV-HCC, particularly if its expression is combined with multinodular, cirrhosis, and BCLC stage. The function and mechanism of CHST4 in various tumors and their subtypes, and its role as a potential biomarker, should be further explored.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Institutional Review Board of Nanfang hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care Committee of Southern Medical University.
JG and LC contributed to the study design and draft revision. LZ completed most of the cytologic experiments, drafted the manuscript, and coordinated the data collection and analysis. YF and XWa contributed to data collection and interpretation of bioinformatics results. JL, MY, and XWu contributed to the human specimen collection and analyzed the clinicopathological characteristics of the patients. WenH, LL, and WeiH contributed to implementation and analysis of qRT-PCR and immunohistochemical assay. LY, HP, and YW contributed to mice and tissue preparation. All authors contributed to the article and approved the submitted version.
This project was supported by the National Natural Science Foundation of China (Nos. 81672992 and 81602685) and the Natural Science Foundation of Guangdong Province (No. 2017A030313486).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the State Key Laboratory of Oncology in Southern Medical University for providing the experimental platform.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.554331/full#supplementary-material
1. Beste LA, Leipertz SL, Green PK, Dominitz JA, Ross D, Ioannou GN. Trends in burden of cirrhosis and hepatocellular carcinoma by underlying liver disease in US veterans, 2001-2013. Gastroenterology. (2015) 149:1471–82.e5. doi: 10.1053/j.gastro.2015.07.056
2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. (2011) 61:69–90. doi: 10.3322/caac.20107
3. Xu R, Han Y. Long non-coding RNA FOXF1 adjacent non-coding developmental regulatory RNA inhibits growth and chemotherapy resistance in non-small cell lung cancer. Arch Med Sci. (2019) 15:1539–46. doi: 10.5114/aoms.2019.86707
4. Villanueva A. Hepatocellular carcinoma. Reply. N Engl J Med. (2019) 381:e2. doi: 10.1056/NEJMc1906565
5. Lee TY, Hsu YC, Tseng HC, Yu SH, Lin JT, Wu MS, et al. Association of daily aspirin therapy with risk of hepatocellular carcinoma in patients with chronic hepatitis B. JAMA Intern Med. (2019) 179:633–40. doi: 10.1001/jamainternmed.2018.8342
6. Amaddeo G, Cao Q, Ladeiro Y, Imbeaud S, Nault JC, Jaoui D, et al. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut. (2015) 64:820–9. doi: 10.1136/gutjnl-2013-306228
7. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
9. Lei Y, Liu H, Yang Y, Wang X, Ren N, Li B, et al. Interaction of LHBs with C53 promotes hepatocyte mitotic entry: a novel mechanism for HBV-induced hepatocellular carcinoma. Oncol Rep. (2012) 27:151–9. doi: 10.3892/or.2011.1489
10. Yoshida K. New biomarkers for early detection of hepatocellular carcinoma. Ebiomedicine. (2015) 2:370–1. doi: 10.1016/j.ebiom.2015.04.001
11. Yu JP, Xu XG, Ma RJ, Qin SN, Wang CR, Wang XB, et al. Development of a clinical chemiluminescent immunoassay for serum GPC3 and simultaneous measurements alone with AFP and CK19 in diagnosis of hepatocellular carcinoma. J Clin Lab Anal. (2015) 29:85–93. doi: 10.1002/jcla.21733
12. Bistrup A, Bhakta S, Lee JK, Belov YY, Gunn MD, Zuo FR, et al. Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J Cell Biol. (1999) 145:899–910. doi: 10.1083/jcb.145.4.899
13. Uchimura K, Gauguet JM, Singer MS, Tsay D, Kannagi R, Muramatsu T, et al. major class of L-selectin ligands is eliminated in mice deficient in two sulfotransferases expressed in high endothelial venules. Nat Immunol. (2005) 6:1105–13. doi: 10.1038/ni1258
14. Kawashima H, Petryniak B, Hiraoka N, Mitoma J, Huckaby V, Nakayama J, et al. acetylglucosamine-6-O-sulfotransferases 1 and 2 cooperatively control lymphocyte homing through L-selectin ligand biosynthesis in high endothelial venules. Nat Immunol. (2005) 6:1096–104. doi: 10.1038/ni1259
15. Tsuboi K, Hirakawa J, Seki E, Imai Y, Yamaguchi Y, Fukuda M, et al. Role of high endothelial venule-expressed heparan sulfate in chemokine presentation and lymphocyte homing. J Immunol. (2013) 191:448–55. doi: 10.4049/jimmunol.1203061
16. Ohmichi Y, Hirakawa J, Imai Y, Fukuda M, Kawashima H. Essential role of peripheral node addressin in lymphocyte homing to nasal-associated lymphoid tissues and allergic immune responses. J Exp Med. (2011) 208:1015–25. doi: 10.1084/jem.20101786
17. Li X, Tu L, Murphy PG, Kadono T, Steeber DA, Tedder TF. CHST1 and CHST2 sulfotransferase expression by vascular endothelial cells regulates shear-resistant leukocyte rolling via L-selectin. J Leukoc Biol. (2001) 69:565–74.
18. Seko A, Nagata K, Yonezawa S, Yamashita K. Ectopic expression of a GlcNAc 6-O-sulfotransferase, GlcNAc6ST-2, in colonic mucinous adenocarcinoma. Glycobiology. (2002) 12:379–88. doi: 10.1093/glycob/12.6.379
19. Kanoh A, Seko A, Ideo H, Yoshida M, Nomoto M, Yonezawa S, et al. Ectopic expression of N-acetylglucosamine 6-O-sulfotransferase 2 in chemotherapy-resistant ovarian adenocarcinomas. Glycoconj J. (2006) 23:453–60. doi: 10.1007/s10719-006-6979-6
20. Yu SY, Hsiao CT, Izawa M, Yusa A, Ishida H, Nakamura S, et al. Distinct substrate specificities of human GlcNAc-6-sulfotransferases revealed by mass spectrometry-based sulfoglycomic analysis. J Biol Chem. (2018) 293:15163–77. doi: 10.1074/jbc.RA118.001937
21. Seko A, Kataoka F, Aoki D, Sakamoto M, Nakamura T, Hatae M, et al. Acetylglucosamine 6-O-sulfotransferase-2 as a tumor marker for uterine cervical and corpus cancer. Glycoconj J. (2009) 26:1065–73. doi: 10.1007/s10719-008-9227-4
22. Okayama H, Kumamoto K, Saitou K, Hayase S, Kofunato Y, Sato Y, et al. Ectopic expression of MECA-79 as a novel prognostic indicator in gastric cancer. Cancer Sci. (2011) 102:1088–94. doi: 10.1111/j.1349-7006.2011.01895.x
23. Taga M, Hoshino H, Low S, Imamura Y, Ito H, Yokoyama O, et al. A potential role for 6-sulfo sialyl Lewis X in metastasis of bladder urothelial carcinoma. Urol Oncol. (2015) 33:496.e1–9. doi: 10.1016/j.urolonc.2015.05.026.
24. Hoshino H, Ohta M, Ito M, Uchimura K, Sakai Y, Uehara T, et al. Apical membrane expression of distinct sulfated glycans represents a novel marker of cholangiolocellular carcinoma. Lab Invest. (2016) 96:1246–55. doi: 10.1038/labinvest.2016.104
25. Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, et al. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. (2006) 44:1025–38. doi: 10.1002/hep.21330
26. Gao F, Liang H, Lu H, Wang J, Xia M, Yuan Z, et al. Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin Epigenetics. (2015) 7:86. doi: 10.1186/s13148-015-0121-1
27. Subbarayan K, Seliger B. Tumor-dependent effects of proteoglycans and various glycosaminoglycan synthesizing enzymes and sulfotransferases on patients’, outcome. Curr Cancer Drug Targets. (2019) 19:210–21. doi: 10.2174/1568009618666180706165845
28. Yang M, Huang W, Sun Y, Liang H, Chen M, Wu X, et al. Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis. Br J Cancer. (2019) 121:699–709. doi: 10.1038/s41416-019-0571-x
29. Laurent V, Guerard A, Mazerolles C, Le Gonidec S, Toulet A, Nieto L, et al. Periprostatic adipocytes act as a driving force for prostate cancer progression in obesity. Nat Commun. (2016) 7:10230. doi: 10.1038/ncomms10230
30. Fan Y, Zhang L, Sun Y, Yang M, Wang X, Wu X, et al. Expression profile and bioinformatics analysis of COMMD10 in BALB/C mice and human. Cancer Gene Ther. (2020) 27:216–25. doi: 10.1038/s41417-019-0087-9
31. Shen Z, Li Y, Fang Y, Lin M, Feng X, Li Z, et al. SNX16 activates c-Myc signaling by inhibiting ubiquitin-mediated proteasomal degradation of eEF1A2 in colorectal cancer development. Mol Oncol. (2020) 14:387–406. doi: 10.1002/1878-0261.12626
32. Liao X, Liu X, Yang C, Wang X, Yu T, Han C, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma. J Cancer. (2018) 9:2357–73. doi: 10.7150/jca.25221
33. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. (2017) 16:129. doi: 10.1186/s12943-017-0696-6
34. Wu J, Chen L, Zheng C, Xu S, Gao Y, Wang J. Co-expression network analysis revealing the potential regulatory roles of lncRNAs in Alzheimer’s disease. Interdiscip Sci. (2019) 11:645–54. doi: 10.1007/s12539-019-00319-w
35. Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. (2019) 35:i436–45. doi: 10.1093/bioinformatics/btz363
36. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
37. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. (2013) 39:782–95. doi: 10.1016/j.immuni.2013.10.003
38. Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang LP, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. (2011) 475:226–30. doi: 10.1038/nature10169
39. Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. (2006) 66:5487–94. doi: 10.1158/0008-5472.CAN-06-0100
40. Greenberg M, Bourc’His D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. (2019) 20:590–607. doi: 10.1038/s41580-019-0159-6
41. Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. (2019) 9:1493–510. doi: 10.1158/2159-8290.CD-19-0399
42. Meins FJ, Si-Ammour A, Blevins T. RNA silencing systems and their relevance to plant development. Annu Rev Cell Dev Biol. (2005) 21:297–318. doi: 10.1146/annurev.cellbio.21.122303.114706
43. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
44. Kawashima H, Hirakawa J, Tobisawa Y, Fukuda M, Saga Y. Conditional gene targeting in mouse high endothelial venules. J Immunol. (2009) 182:5461–8. doi: 10.4049/jimmunol.0802327
45. Zhong Y, Li S, Chen L, Liu Z, Luo X, Xu P, et al. In vivo toxicity of solasonine and its effects on cyp450 family gene expression in the livers of male mice from four strains. Toxins (Basel). (2018) 10:487. doi: 10.3390/toxins10120487
46. Tolsma TO, Hansen JC. Post-translational modifications and chromatin dynamics. Essays Biochem. (2019) 63:89–96. doi: 10.1042/EBC20180067
47. Yin SL, Xiao F, Liu YF, Chen H, Guo GC. Long non-coding RNA FENDRR restrains the aggressiveness of CRC via regulating miR-18a-5p/ING4 axis. J Cell Biochem. (2019) 121:3973–85. doi: 10.1002/jcb.29555
48. Li Y, Zhang W, Liu P, Xu Y, Tang L, Chen W, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. (2018) 11:1403–12. doi: 10.2147/OTT.S149511
49. Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, et al. Serum exosomal MicroRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J Clin Med. (2020) 9:281. doi: 10.3390/jcm9010281
50. Ke RS, Zhang K, Lv LZ, Dong YP, Pan F, Yang F, et al. Prognostic value and oncogene function of heterogeneous nuclear ribonucleoprotein A1 overexpression in HBV-related hepatocellular carcinoma. Int J Biol Macromol. (2019) 129:140–51. doi: 10.1016/j.ijbiomac.2019.02.012
51. Jin YJ, Byun S, Han S, Chamberlin J, Kim D, Kim MJ, et al. Differential alternative splicing regulation among hepatocellular carcinoma with different risk factors. BMC Med Genomics. (2019) 12:175. doi: 10.1186/s12920-019-0635-z
52. Nieminen TT, O’Donohue MF, Wu Y, Lohi H, Scherer SW, Paterson AD, et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology. (2014) 147:595–8.e5. doi: 10.1053/j.gastro.2014.06.009
53. De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. (2013) 45:186–90. doi: 10.1038/ng.2508
54. Wang B, Xu Q, Yang HW, Sun LP, Yuan Y. The association of six polymorphisms of five genes involved in three steps of nucleotide excision repair pathways with hepatocellular cancer risk. Oncotarget. (2016) 7:20357–67. doi: 10.18632/oncotarget.7952
55. Kuang DM, Zhao Q, Wu Y, Peng C, Wang J, Xu Z, et al. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. J Hepatol. (2011) 54:948–55. doi: 10.1016/j.jhep.2010.08.041
56. Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, et al. CD4(+) T cell help is required for the formation of a cytolytic CD8(+) T cell subset that protects against chronic infection and cancer. Immunity. (2019) 51:1028–42.e4. doi: 10.1016/j.immuni.2019.10.009
57. Li J, Li N, Wang J. M1 macrophage-derived exosome-encapsulated cisplatin can enhance its anti-lung cancer effect. Minerva Med. (2020). doi: 10.23736/S0026-4806.20.06564-7 [Epub ahead of print].
58. Guerra AD, Yeung O, Qi X, Kao WJ, Man K. The anti-tumor effects of M1 macrophage-loaded poly (ethylene glycol) and gelatin-based hydrogels on hepatocellular carcinoma. Theranostics. (2017) 7:3732–44. doi: 10.7150/thno.20251
59. Hansen M, Andersen MH. The role of dendritic cells in cancer. Semin Immunopathol. (2017) 39:307–16. doi: 10.1007/s00281-016-0592-y
60. Masucci MT, Minopoli M, Carriero MV. Tumor associated neutrophils, their role in tumorigenesis, metastasis, prognosis and therapy. Front Oncol. (2019) 9:1146. doi: 10.3389/fonc.2019.01146
61. Li SC, Xu Z, Deng YL, Wang YN, Jia YM. Higher neutrophil-lymphocyte ratio is associated with better prognosis of hepatocellular carcinoma. Medicine (Baltimore). (2020) 99:e20919. doi: 10.1097/MD.0000000000020919
Keywords: HBV-related hepatocellular carcinoma, CHST4, prognostic biomarker, expression profiling, competing endogenous RNA
Citation: Zhang L, Fan Y, Wang X, Yang M, Wu X, Huang W, Lan J, Liao L, Huang W, Yuan L, Pan H, Wu Y, Chen L and Guan J (2020) Carbohydrate Sulfotransferase 4 Inhibits the Progression of Hepatitis B Virus-Related Hepatocellular Carcinoma and Is a Potential Prognostic Marker in Several Tumors. Front. Oncol. 10:554331. doi: 10.3389/fonc.2020.554331
Received: 21 April 2020; Accepted: 15 September 2020;
Published: 15 October 2020.
Edited by:
Giuseppe Giaccone, Cornell University, United StatesReviewed by:
Christine Fillmore Brainson, University of Kentucky, United StatesCopyright © 2020 Zhang, Fan, Wang, Yang, Wu, Huang, Lan, Liao, Huang, Yuan, Pan, Wu, Chen and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Guan, Z3VhbmppYW41NDYxQDE2My5jb20=; Longhua Chen, Y2hlbmxoNTQ2MUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.