- 1Research Center for Functional Genomics, Biomedicine and Translational Medicine, “Iuliu-Hatieganu” University of Medicine and Pharmacy Cluj-Napoca, Cluj-Napoca, Romania
- 2Department of Psychology, Babes-Bolyai University, Cluj-Napoca, Romania
- 3Research Center for Advanced Medicine MedFuture, “Iuliu Hatieganu” University of Medicine and Pharmacy Cluj-Napoca, Cluj-Napoca, Romania
- 4Pathology Unit, Department of Experimental, Diagnostic and Specialty Medicine, DIMES, University of Bologna, Bologna, Italy
- 5Department of Functional Genomics and Experimental Pathology, The Oncology Institute “Prof. Dr. Ion Chiricută”, Cluj-Napoca, Romania
Non-small cell lung cancer (NSCLC) remains a problem worldwide due to its rapid progression and low rate of response to treatment. The heterogeneity of these tumors observed in histopathology exam but also in the mutational status and gene expression pattern makes this malignancy difficult to treat in clinic. The present study investigated the effect of miR-21 and let-7 family members as prognostic biomarkers in NSCLC patients based on the results published in different studies regarding this subject until March 2019. The analysis revealed that these two transcripts are steady biomarkers for prediction of patient outcome or survival. Upregulated expression of miR-21 is associated with poor outcome of patients with NSCLC [HR = 1.87, 95% CI = (1.41, 2.47), p < 0.001]. The analysis regarding let-7 family, specifically let-7a/b/e/f, revealed that downregulated expression of these transcripts predicts poor outcome for NSCLC patients [HR = 2.61, 95% CI = (1.58, 4.30), p < 0.001]. Besides, the reliability of these microRNAs is reflected in the fact that their prognostic significance is constant given the different sample types (tissue, FFPE tissue, serum, serum/plasma or exosomes) used in the selected studies.
Introduction
Lung cancer represents the leading cause of cancer related deaths, with more than 1.7 million deaths in 2018, being also one of the most diagnosed oncogenic pathology worldwide, alongside with breast cancer (1). Non-small cell lung cancer (NSCLC) accounts for more than 85% of the lung cancers, having two major histological types—adenocarcinoma and squamous cell carcinoma (2). According to the reports of World Health Organization (WHO), smoking habits in both sexes appear to indicate a decrease, from 22.1% in 2010 to an expected 17.3% in 2025 (3). Nevertheless, lung cancer incidence in patients increased, raising serious concerns in terms of prevention and response to therapy (1). The explanation would be that, independent to exogenous carcinogens, lung tumorigenesis is driven by genetic dysregulations of coding and non-codingRNAs (4, 5).
Understanding the molecular imbalance that underlies a malignancy is the key to develop and apply an efficient therapy scheme (6). In NSCLC, precisely adenocarcinoma, there are several targeted therapies developed to overcome the disequilibrium that leads to the transformation and abnormal behavior of cells. Mutations in epidermal growth factor receptor (EGFR), or other recombinant DNA processes in anaplastic lymphoma kinase (ALK) or c-ros oncogene 1 (ROS1) represent the main alterations that occur in patients diagnosed with NSCLC (7). Another important determinant in the development of the malignant phenotype in NSCLCs is represented by the non-coding part of the genome (8). Over the last two decades, microRNAs (miRNAs) gained the attention of the medical researchers, as they were proved to be important players in cancer development (9–11). MiRNAs represent short RNAs that do not undergo the translation into proteins, but regulate the expression of many protein coding genes, by complementary binding (12). Considering the dysregulation of miRNAs, these molecules can be categorized as tumor suppressor or oncogenic miRNAs (13, 14).
Intensively studied let-7 family is known to have regulatory function in several cancers, such as pancreatic cancer (15), thyroid cancer (16), and colorectal cancer (17), with differences in the expression profile of different members of let-7 family. In thyroid cancer, let-7b appears to be downregulated, while let-7f is overexpressed in tissue samples collected from patients with follicular thyroid cancer (16). Target genes of let-7 include c-Myc (18), high-mobility group A (HMGA) (19–21) or signal transducer and activator of transcription 3 (STAT3) and Janus-activated kinase 2 (JAK2) (15), genes involved in cell proliferation and cell cycle. In NSCLC cells, let-7 family transfection considerably reduced proliferation rate, hence acting as tumor suppressor gene (22). By regulating the Ras family, let-7 takes part in controlling cellular proliferation in in vivo models, although treatment with this miRNA revealed that there are additional mechanisms with downstream activation that lead to resistance of cancer cells to the tumor suppressive functions of this transcript (23). In NSCLC, let-7 underexpression was reported as significantly correlated with patient outcome (24–26), but there are studies that were not successful in establishing a direct link between low expression of let-7 and prognostic of NSCLC patients (27, 28).
The oncogenic role of miR-21 in patients with NSCLC was reported in a substantial number of publications (24–27, 29–47). MiR-21 appears to be upregulated in many malignancies, such as breast cancer, head and neck cancers, non-small cell lung cancer, melanoma, glioblastoma, confirming its oncogenic role. Its function is exerted by downregulating the expression of suppressor of cytokine signaling 1 and 6 (SOCS1/6) and phosphatase and tensin homolog (PTEN), thus distorting the apoptotic mechanism, cellular growth and proliferation of NSCLC cells (48–50). Further information regarding the involvement of miR-21 in different cellular processes and pathways in cell lines/animal models and in NSCLC patients' outcome is systematically formulated in a previous review (51).
Given the differences of the results obtained by various groups regarding both miR-21 and let-7, the focus of this study was to evaluate the prognostic significance of the expression levels of miR-21 (as oncogene) and let-7 (as tumor suppressor gene) in patients with NSCLC. Therefore, a comprehensive literature search was assessed to perform a meta-analysis on these two molecules and their implication in patient outcome.
Methods
Search Procedure and Study Selection
In order to perform this meta-analysis, the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) protocol was followed (52, 53). The comprehensive search for eligible studies began by interrogating the PubMed database, as it provided the literature most consistent with the topic. The database was scrutinized using the following terms: “let-7*” OR “let7*” AND “lung cancer” for let-7 family, and “miR-21” OR “microRNA 21” AND “lung cancer” AND “patients” for miR-21. The searching terms for let-7 family were simplified due to the poor results generated using the same search scheme that was applied for miR-21 in lung cancer.
The articles published until 20 March 2019 were assessed for eligibility, reviewing the titles and abstracts, and full-text publications where the abstract text was inconclusive. Publications listed that were written in another language than English, review/meta-analysis/letters/erratum papers were all ruled out from the meta-analysis. Publications that did not assess the association between the expression levels of miR-21/let-7 with the survival in patients with lung cancer, specifically NSCLC, were not selected for the analysis.
Publications had several inclusion criteria, as follows: (i) studies that characterized patients with NSCLC, including sample size, sample type, and histological type; (ii) miR-21/let-7 evaluation of expression in human samples; (iii) Hazard ratios (HR) calculated in relation with specific microRNA expression for overall survival (OS), progression-free survival (PFS), or recurrence free survival (RFS), along with the p-values and 95% confidence intervals (95% CIs).
Each set of publications (for miR-21 and let-7) was evaluated and verified by two independent reviewers (C.P-B. and L.M). Publications that included any data regarding the investigation of the correlation between the expression levels of the selected microRNAs and survival time of patients suffering from NSCLC were organized in an excel worksheet.
The following parameters were marked from the eligible studies for this meta-analysis: (i) reference data (authors, year); (ii) demographic characteristics: population, male/female ratio, ethnicity, smoking status; (iii) pathology data: histology, stage, TNM status; (iv) experiment design: sample type, techniques, microRNA expression; (v) statistical data: survival type, hazard ratio (HR), 95% confidence interval (CI), p-value, statistical tests/methods.
Data Set and Coding Procedure
In order to standardize the way results were reported across studies and compare survival between miR-21 and let-7, we applied the following procedure. All hazard ratios (HR) reported for miR-21 were computed and introduced in our database as high/low expression, while for let-7 results were computed as low/high. As a consequence, for miR-21, HR > 1 indicates higher mortality for high expression, while for let-7 HR > 1 indicates higher mortality for low expression.
Data Analysis
Analyses were conducted by using Comprehensive Meta-Analysis software, version 2.2.050 (Biostat Inc., Englewood, NJ, USA). As an indicator of effect sizes, the hazard ratio was used. Given the heterogeneity of the studies, all analyses were based on a random effects model.
Publication Bias Analysis
To assess the risk of publication bias for the results we calculated the Begg and Mazumdar's rank correlation test according to the recommendations of Kepes et al. (2012) (54). This test computes the rank order correlation (Kendall's tau b) between the effect size and the standard error (which is driven primarily by sample size). This test identifies if large studies tend to be included in the analysis regardless of their effect size, whereas small studies are more likely to be included when they show a relatively large effect size.
TCGA Analysis
Data analysis was also performed using The Cancer Genome Atlas (TCGA) data available for lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) and downloaded from FireBrowse as one data matrix including the expression levels of the mature miRNA as normalized and log2 transformed values. Clinical data and follow-up data for these patients were downloaded from the UCSC (University of California Santa Cruz) Xena browser, and then matched with the expression data. Kaplan-Meier survival analysis was carried out only on samples which included complete survival data. Therefore, the patients were separated based on the miRNA expression levels as high-expression and low-expression according to the median value in the group. This analysis was initially performed on the entire cohort from the TCGA set, and then the cohort was split into males and females, and the same investigation was performed.
Results
Qualified Studies
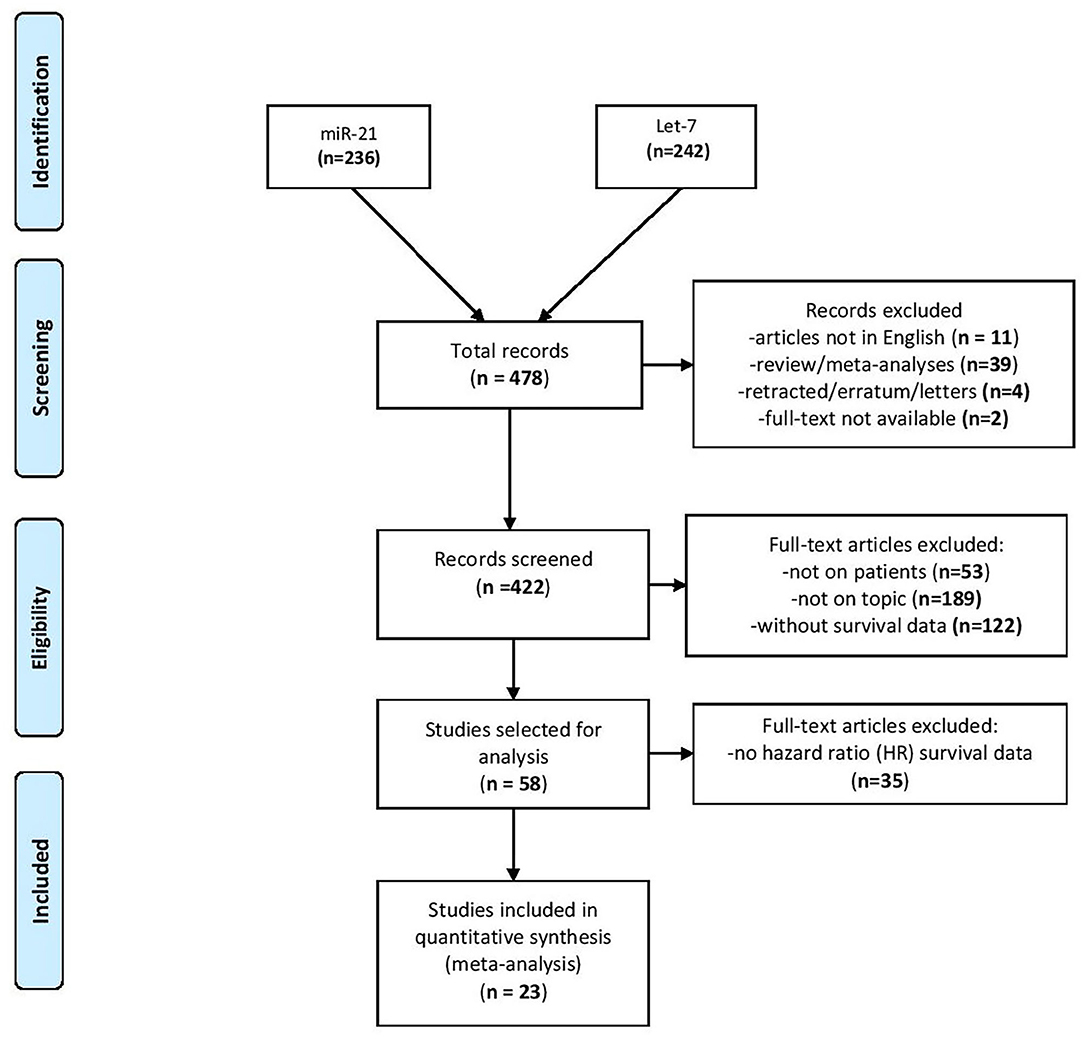
After interrogating the PubMed database, the search disclosed 236 publications for miR-21 and 242 publications for let-7 family (from now on reported for simplicity as “let-7”) for lung cancer topics. From the total number of 478 articles revealed after the search, 56 publications were expelled as by the criteria presented in the Methods section. The abstracts and full-text articles of 422 studies were selected for a more exhaustive assessment. Another 364 studies were excluded as they were not on topic or the expression of miRNAs was not evaluated in patients or was not precisely linked to the effect on patient outcome. Therefore, 58 articles were selected as candidate studies for further analysis. From these, 35 studies did not provide HR data in the survival analysis or the effect of miR-21/let-7 on patient outcome was not evaluated alone. Finally, 23 articles were identified as eligible for the present meta-analysis (24–27, 29–47), of which two studies provided HR survival data for both miR-21 and let-7 (Figure 1) (27, 32).
Study Characteristics
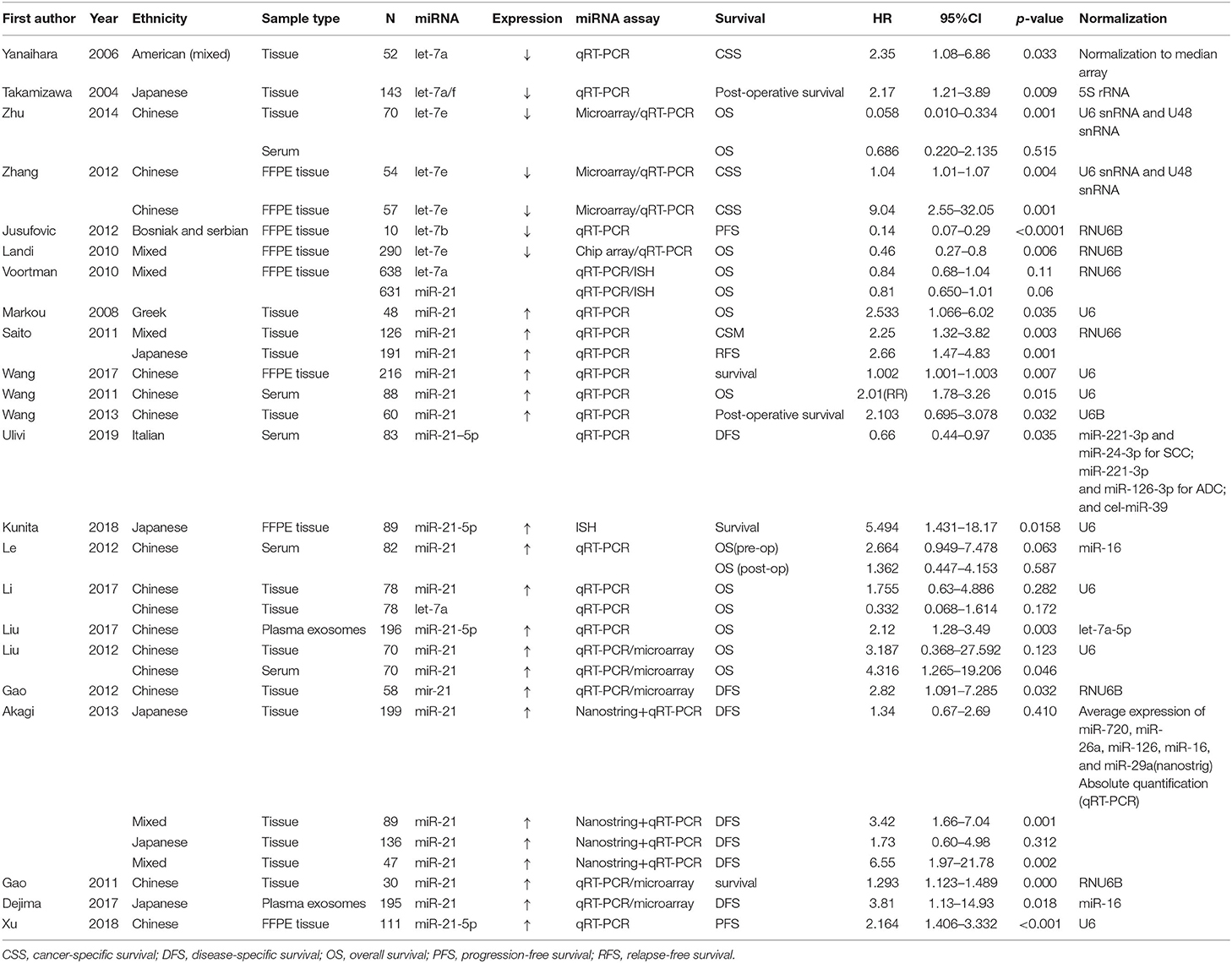
From the 23 studies selected for the meta-analysis, sample size ranged between 10 and 638 patients, with a median study size of 81.5, categorized according to ethnicity: Chinese, Bosnian and Serbian, Greek, Japanese, Italian, and mixed population. The studies used as sample fresh frozen tissue (26, 29, 30, 32–34, 37, 43–46), formalin-fixed paraffin embedded (FFPE) tissue (24, 25, 27, 31, 35, 38, 40), serum (30, 36, 39, 41, 43) or plasma exosomes (42, 47) (two studies used both tissue and serum) (30, 43). For miRNA quantification, microarray and qRT-PCR were used in six studies (30, 31, 43, 44, 46, 47), in situ hybridization (ISH) and qRT-PCR or Nanostring were used in one study, and in rest of the studies quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was mainly used. The prognostic significance of miR-21/let-7 was reported in all 23 studies listed in Table 1.
Prognostic Significance of miR-21/let-7 in NSCLC
All 23 selected studies reported a direct connection between the expression levels of miR-21/let-7 and the outcome of lung cancer patients. The outcome is reported in these publications as: overall survival (OS), progression-free survival (PFS), relapse-free survival (RFS), disease-free survival (DFS), cancer-specific survival (CSS), post-operative survival or survival.
Overall HR for miR-21
Figure 2 describes the forest plot for miR-21 with the hazard ratio for each study included in the meta-analysis. The overall corrected hazard ratio for miR-21 is HR = 1.87, 95% CI = (1.41, 2.47) and statistically significant (Z = 4.43, p < 0.001), indicating that a high expression of miR-21 is associated with poor survival.
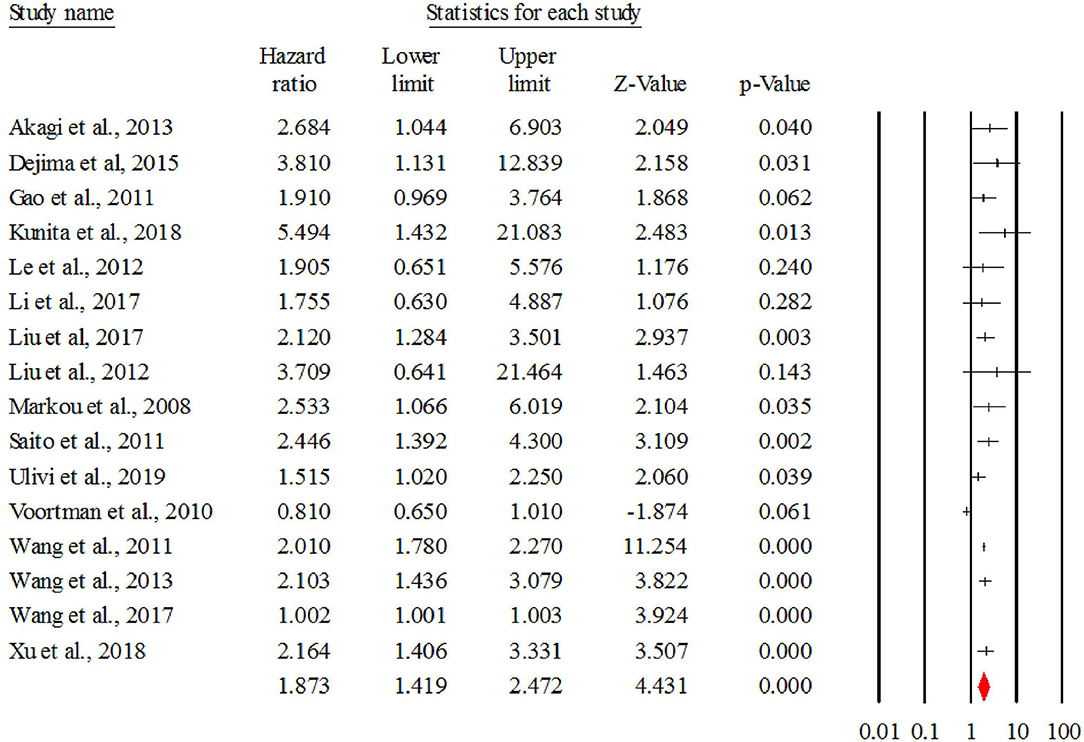
Figure 2. The forest plot of HR from all studies assessing the prognostic significance of miR-21 in NSCLC patients and summary.
The data downloaded from TCGA revealed 776 patients with LUAD/LUSC with complete clinical and survival data available. Kaplan-Meier survival analysis was performed on these samples organized as high-expression and low-expression compared to the median value. The evaluation revealed that overexpression of miR-21 in NSCLC patients was associated with poor overall survival (p = 0.008) (Figure 3).
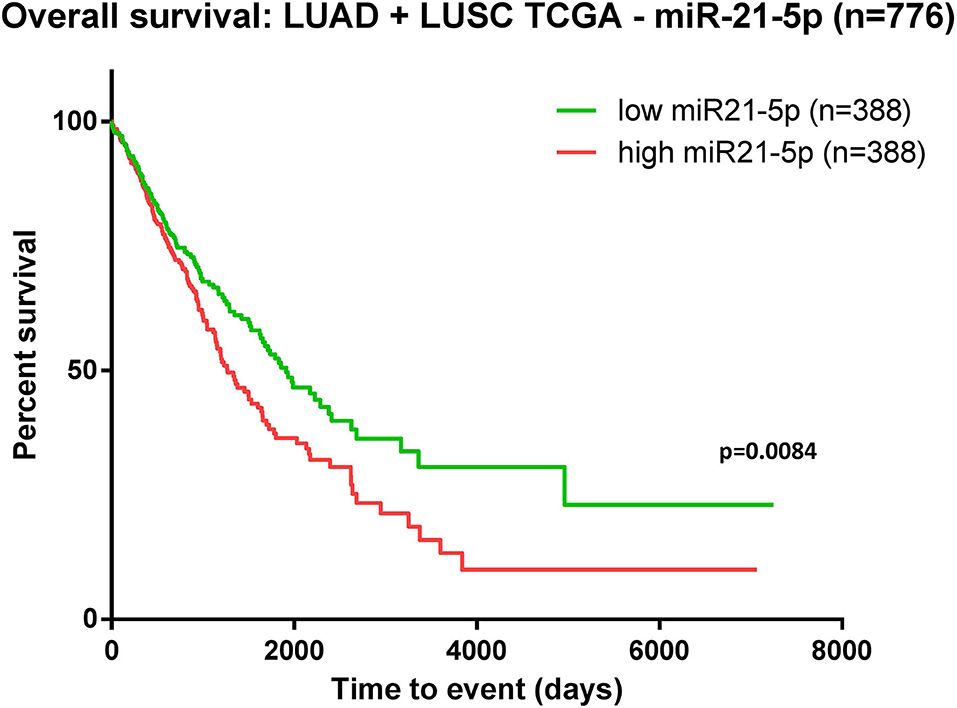
Figure 3. Kaplan-Meier curve using TCGA data analysis regarding the expression of miR-21 in patients diagnosed with lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (n = 776). Patients were classified as high-/low-expression according to the median value. Upregulated expression of miR-21 is correlated with poor survival (p = 0.0084).
Overall HR for let-7
Figure 4 describes the forest plot for let-7 with the hazard ratio for each study included in the meta-analysis. The overall corrected hazard ratio for let-7 is HR = 2.61, 95% CI = (1.58, 4.30) and statistically significant (Z = 3.75, p < 0.001), indicating that a low expression of let-7 is associated with poor survival.
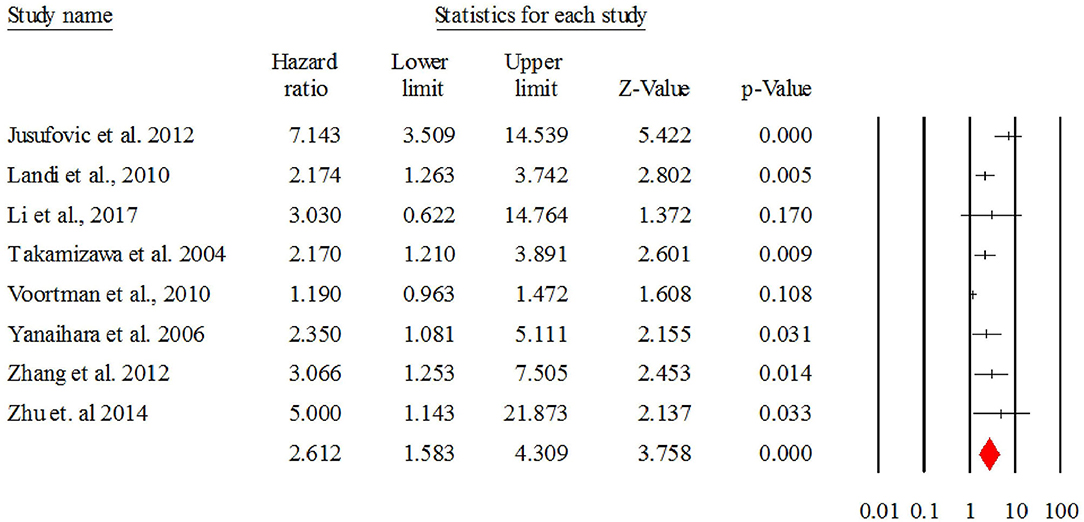
Figure 4. The forest plot of HR from all studies assessing the prognostic significance of let-7 in NSCLC patients and summary.
MiR-21 vs. let-7
The comparison between the hazard ratios generated by miR-21 (high vs. low) vs. let-7 (low vs. high) indicated that there were no significant differences, Q(1) = 1.30, p = 0.253.
Publication Bias Analysis
For miR-21, the publication bias analysis using the Begg and Mazumdar's rank correlation test revealed a non-significant Kendall's tau b of −0.14, with a two-tailed p-value of 0.444 (based on continuity-corrected normal approximation), suggesting there is no tendency for studies which are more precise (and implicitly larger) to generate larger effect sizes.
The same analysis performed for let-7 revealed a non-significant Kendall's tau b of 0.10, with a two-tailed p-value of 0.710 (based on continuity-corrected normal approximation), suggesting there is no tendency for studies which are more precise (and implicitly larger) to generate larger effect sizes.
Heterogeneity Analysis
For miR-21, the distribution of effects proved to be significantly heterogeneous, Q(15) = 206.17, p < 0.001, leading us to perform the moderators' analysis to test several explanations for this heterogeneity.
In the case of let-7, the distribution of effects proved to be non-significantly heterogeneous, Q(7) = 32.58, p < 0.001. As a consequence, we performed the moderators' analysis to test several explanations for this heterogeneity.
Also, because of this heterogeneity, all analysis were performed under the random effects model.
Moderators' Analysis for miR-21
Categorical Moderators
Population
As Table 2 shows, the hazard ratios obtained by studies performed on Chinese and Japanese population proved to be statistically significant, HR = 1.85, 95% CI = (1.25, 2.73), p = 0.002 for Chinese samples, and HR = 2.78, 95% CI = (1.39, 5.55), p = 0.004 for Japanese samples. The hazard ratio obtained on Greek and Italian population seems to be not significant, but these results should be interpreted with caution since they are represented in our meta-analysis by only one study each.
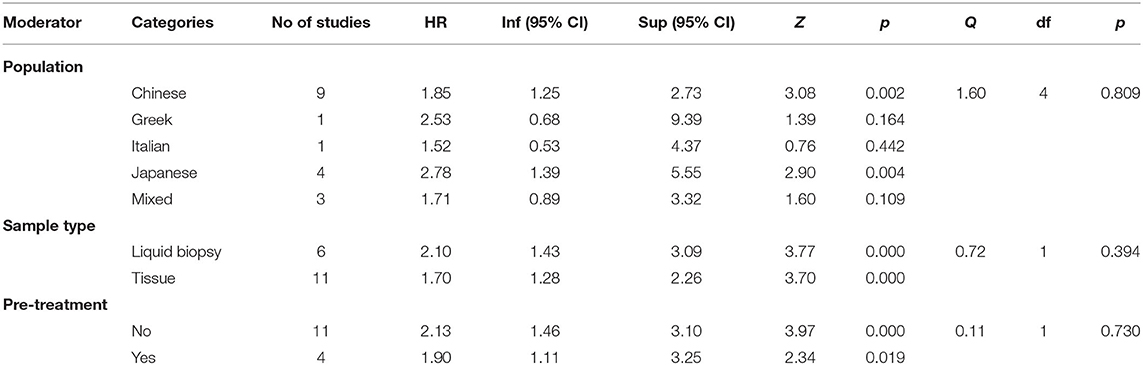
Table 2. The analysis of HR (miR-21) as a function of population, sample type, and existence of pre-treatment.
Sample type
The analysis of hazard rations as a function of sample type revealed significant HR for both type of samples analyzed, HR = 2.10, 95% CI = (1.43, 3.09), p < 0.001 for liquid biopsy and HR = 1.70, 95% CI = (1.28, 2.26), with no significant differences between those two categories, Q(1) = 0.72, p = 0.394.
Pre-treatment
Our analysis indicated that the existence of pre-treatment did not make any difference regarding the hazard ratio obtained, Q(1) = 0.11, p = 0.730, both categories of studies (with vs. without pre-treatment) yielding significant hazard ratios.
Continuous Moderators
We used percentage of women in the samples, percentage of smokers, and percentage of patients with lymph node metastasis. In order to test continuous moderators, we performed a meta-regression in which percentage (%) of women, percentage of smokers and percentage of patients with lymph node metastasis (LNM) were treated as predictors of hazard ratios.
Percentage of women
Among the studies analyzed, 14 reported the proportion of women. We performed a meta-regression analysis in order to test the predictive value of the proportion of women in the samples upon the hazard ratios. The results revealed that this variable had a significant positive predictive value, B = 0.016, p < 0.001. In other words, larger proportions of women in the samples are associated with higher levels of hazard ratios obtained.
Using the TCGA data available, we performed the survival analysis separating the patients according to their gender. Consistent with the above-mentioned findings, the separate analysis—on women and men—revealed that, the correlation between overexpression of miR-21 and worse outcome is statistically significant in females, but not in males (Figure 5).
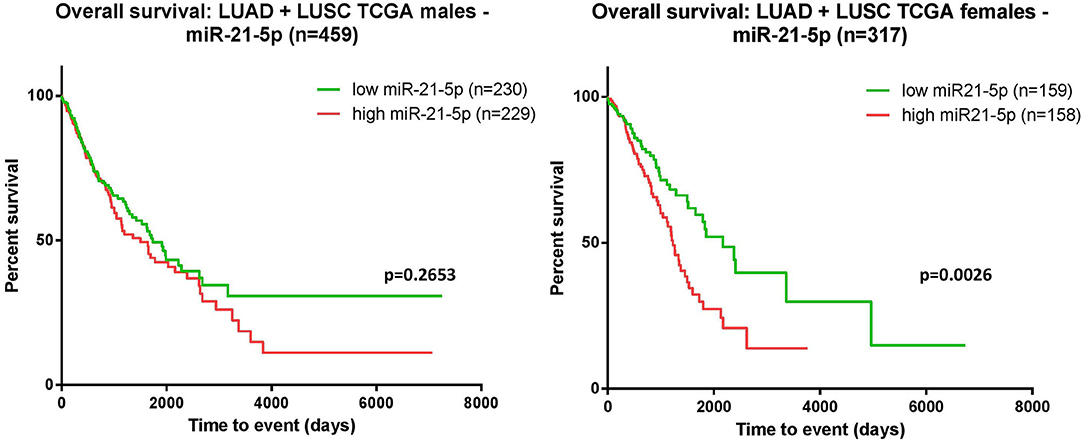
Figure 5. Kaplan-Meier curves. The survival analysis was assessed for the male cohort indicated that the overexpression of miR-21 and poor survival did not reach a statistic significance p = 0.2653 (left); in females, overexpression of miR-21 correlated with poor survival p = 0.0026 (right).
Percentage of smokers
Among the selected studies, only 12 reported the proportion of smokers. The analysis of the predictive value of the proportion of smokers in each sample revealed that this variable had a significant negative predictive value, B = −0.009, p = 0.002, which means that as the proportion of smokers increases, the hazard ratio decreases (gets closer to 1).
Percentage of patients with LNM
The meta-analytical regression performed on the 10 studies which reported the percentage of patients with LNM, proved that this percentage is a significant negative predictor of the hazard ratio, B = −0.017, p < 0.001. In other words, as the percentage of patients with LNM increases, the hazard ratio decreases (gets closer to 1).
Moderators' Analysis for let-7
Categorical Moderators
We used population and pre-treatment as moderators. The type of sample (liquid biopsy vs. tissue) could not be tested as potential moderator because all studies used tissue as sample for analysis.
Population
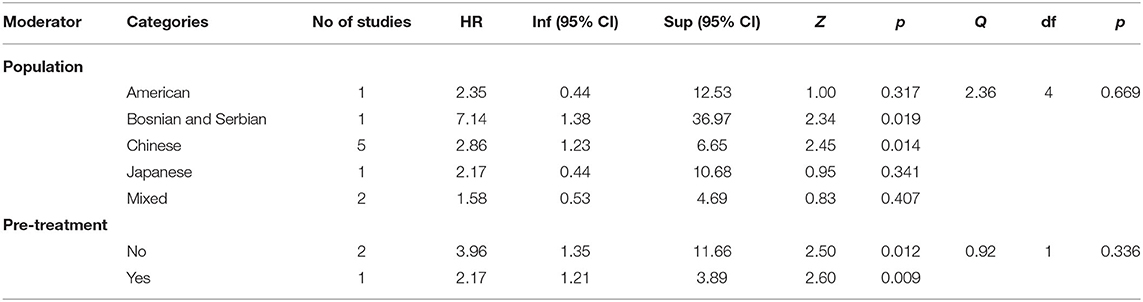
As Table 3 shows, the hazard ratios obtained by studies performed on Chinese and Bosnian/Serbian populations proved to be statistically significant, HR = 2.86, 95% CI = (1.23, 6.65), p = 0.014 for Chinese samples, and HR = 7.14, 95% CI = (1.38, 36.97), p = 0.019 for Bosnian/Serbian population. The last result should be taken with reserve since it has been obtained by analyzing only one study. The hazard ratios obtained on American, Japanese and mixed populations seem to be not significant, but these results should be interpreted with caution due to the availability of one or two studies.
Pre-treatment
Our analysis indicated that the existence of pre-treatment did not make any difference regarding the hazard ratio obtained, Q(1) = 0.92, p = 0.336, both categories of studies (with vs. without pre-treatment) yielding significant hazard ratios. Being performed on a small number of studies, this analysis should be also interpreted with precaution.
Continuous Moderators
We used percentage of women in the samples, percentage of smokers, and percentage of patients with lymph node metastasis.
Percentage of women
Among the studies analyzed, eight reported the proportion of women. We performed a meta-regression analysis in order to test the predictive value of the proportion of women in the samples upon the hazard ratios. The results revealed that this variable had a significant positive predictive value, B = 0.014, p = 0.008. In other words, larger proportions of women in the samples are associated with higher levels of hazard ratios obtained.
Percentage of smokers
Among the selected studies, only three reported the proportion of smokers. The analysis of the predictive value of the proportion of smokers in each sample revealed that this variable had a non-significant predictive value, B = 0.024, p = 0.115. Being performed on a small number of studies, this analysis should be regarded with discretion.
Percentage of patients with LNM
The meta-analytical regression performed on the four studies which reported the percentage of patients with LNM, proved that this percentage is a significant negative predictor of the hazard ratio, B = −0.037, p = 0.009. In other words, as the percentage of patients with LNM increases, the hazard ratio decreases (gets closer to 1).
Discussion
Since the recognition of miRNAs as relevant players in cancer development and progression, many studies reported the aberrant expression (overexpression or underexpression) of these RNAs in several cancers (55). This meta-analysis reports that the expression levels of both miR-21 and let-7 family are highly correlated with the outcome of NSCLC patients.
The analysis for miR-21 revealed that overexpression of miR-21 in NSCLC patients resulted in shortened survival time (p < 0.001), bringing new evidence of the oncogenic nature of this miRNA in lung cancer. In accordance with this result, TCGA data analysis on patients with lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) indicated a significant association between upregulated levels of miR-21 and poor survival (Figure 3). The moderators' analysis on miR-21 disclosed the evidence that regardless of sample type (liquid biopsy or tissue) the analysis showed no significant differences in HR for each category. This observation supports the idea that the abnormal upregulation of miR-21 in tumor tissue sample is also mirrored in plasma/serum samples of these patients. This consideration is not necessary valid for other cancers or other miRNAs, as there are recent publications with examples of molecules reported up-/downregulated in tissue, and opposite regulation is observed in liquid biopsies of the same patient (56, 57). One important highlight is that the pre-treatment does not affect the HRs, suggesting that in this particular case, miR-21 could be considered a stable biomarker for the prognosis of NSCLC patients. When analyzing how the proportion of women is reflected in the HRs obtained, the results showed that the accuracy of the HR calculated as high vs. low is higher when the proportion of woman increases. This means that in woman, the aberrant expression of miR-21 seems to be more consistent with its impact on patient survival. This result is also emphasized in the TCGA survival evaluation, as the survival analysis on LUAD and LUSC patients revealed that upregulated expression of miR-21 indicated a significant correlation with worse survival in female patients, but not in males (Figure 5). This result is not completely new, in a previous review published by our group, the prognostic value of miR-21 in LUAD patients from TCGA was already assessed, with the confirmation that upregulation of this transcript predicts poor outcome of LUAD patients (51) and supported by other groups' research in NSCLC TCGA datasets (58).
Another moderator investigated in this meta-analysis was smoking status. The analysis disclosed that the HR tends to get closer to 1, meaning that the differences between the outcomes of patients do not differ substantially with the proportion of smokers in the analyzed sample. This observation is not unexpected, as the effects of smoking are already notorious in prompting lung cancer (59), and the survival rates in smoking patients tend to be similar, despite their categorization as high or low expressing miR-21. One similar impact appears to have the presence of lymph node metastasis in NSCLC patients, as previously reported (60). Our results imply that patients with LNM tend to have comparable survival rates and the differences in survival rates decrease when LNM are present. All these findings, specifically the fact that the prognostic value of miR-21 appears to be more accurate in patients who are female, non-smoking or Asian, are consistent with previous studies regarding biomarkers in NSCLC patients (61, 62). Our results suggest the fact that in the mentioned categories miR-21 expression is a valuable prognosis biomarker in NSCLC patients and that in males, smokers and patients with LNM, the examination of other biomarkers should be taken into consideration when assessing the prognostic of a patient.
Analysis of the impact of let-7 family members in patients diagnosed with NSCLC indicates that low expression of this miRNA is a predictor for shortened survival rates. The inquiry comprised eight studies and the summary of the HR is statistically significant, emphasizing the importance of let-7's expression in NSCLC. The TCGA data analysis did not confirm this finding (Supplementary Material), although this is not unexpected, given the fact that although the members of let-7 family possess similar functions in humans (63), there are definite differences between the representatives of this family and their role in mediating particular aspects of malignant development and progression. As the TCGA survival analysis performed separately on LUAD and LUSC datasets for the transcripts (miR-21, let-7a/b/e/f) indicated (Supplementary Figure 1), in LUAD, the expression of these microRNAs appears to have a significant correlation with the outcome of these patients. This may be regarded as a consequence of the fact that even though the two classes of cancers—lung adenocarcinoma and squamous cell carcinoma—are part of the same pathology—NSCLC-, the differences in the genomic mutational landscape have consequences observed at molecular level. In this respect, for LUAD patients' mutations in TP53, STK11, ALK, EGFR genes are more common, unlike LUSC patients, which have more characteristic mutations in RB1, KEAP, NFE2L2 or NF1 genes (64–68). The literature mentions the efficacy of let-7b in NSCLC cell lines with TP53 and EGFR mutations as an adjuvant in the therapy with erlotinib in order to potentiate the effect of this therapeutic compound (69). Besides, In NSCLC miR-21 was reported as being regulated by EGFR in non-smoker patients, its upregulation being correlated with EGFR mutations (70). Given the fact that EGFR and TP53 are genes more commonly mutated in LUAD patients, this would bring an explanation regarding the results of the TCGA survival analysis. When considering pre-treatment in these patients, the examination indicated that HR is not influenced by a previous chemo-/radiotherapy. Given the small number of studies in which treatment information was available or patients had undergone treatment before sample collection, this aspect should be further investigated for solid evidences. As was the case for miR-21, the percentage of women positively influenced the HR, suggesting that the expression profile of let-7's as survival predictor is more stable in women. As expected, the presence of LNM appears as a negative predictor for HR, the survival rates of high/low expressing let-7 patients tend to be similar in patients who already developed LNM, as the spreading of the tumor cells—leading to the development of new tumors—is harder to control and eventually leads to decease. One interesting observation is that smoking does not affect HRs and smoking patient's survival rates differ according to their high/low expression levels of let-7.This observation has to be confirmed in additional studies, as the number of present studies reporting the smoking status of the patients is rather small (three studies). As in miR-21, expression levels of let-7 have a more accurate prognostic value in females, rather than in males and patients presenting LNM. The present meta-analysis proved that miR-21 and let-7 can serve as prognostic biomarkers in NSCLC patients, although there are several drawbacks that need to be addressed. Since smokers represent an important part of patients with NSCLC, it would argue against the fact that these microRNAs alone could be used as prognostic biomarkers in NSCLC. There is a clear need to further investigate other molecules or expression patterns in a group of molecules in order to establish a definite prognostic biomarker/panel in smokers. Besides, in the absence of consistency among studies regarding patient characteristics, clinical or methodological settings, the constructive heterogeneity hindered us the possibility to investigate the influence of other moderators on miR-21 and let-7 stability as prognostic biomarkers in NSCLC patients. In order to reinforce the importance and the utility of these two transcripts in non-smoker NSCLC patients, future studies focused exclusively on non-smoker patients should be conducted to investigate their roles and/or correlation with common mutations encountered also in non-smoking patients diagnosed with NSCLC, such as EGFR, ALK, KRAS, RET, etc. (71).In this respect, a meta-analysis published in 2016 revealed that in the genes most frequently mutated in NSCLC, such as EGFR, KRAS, and ALK mutations are associated with patient characteristics, such gender, smoking status and histological type (72). As already stated, in non-smokers, miR-21 appears to be regulated by EGFR, miR-21 overexpression being associated with EGFR mutations (70).
The impact of the anomalous expression of these two microRNAs is reflected in the cluttering of different molecular pathway, by targeting genes encoding proteins essential in these pathways. In NSCLC cell lines, miR-21 is reported to directly target phosphatase and tensin homolog (PTEN) a tumor suppressor gene (73), with effects on cell differentiation, tumor growth, apoptosis and cell proliferation (74). Also, it was reported that miR-21 targets transforming growth factor b-induced gene (TGFB1), which encodes the same protein with recognized functions in cell proliferation. Specifically, in A549 cells TGFB1 overexpression reduces cell proliferation and overexpression of miR-21 decreases TGFB1 levels, thus promoting aberrant cell proliferation and highlighting the function of miR-21 as an oncomiR in NSCLC (75). Also, there is solid evidence that miR-21 regulates radiosensitivity in lung cancer cells, and the mechanism reported by Jiang et al. indicate the interference with PI3K/AKT/mTOR signaling by inhibiting the transcription of pathway programmed cell death 4 (PDCD4) gene (76) (Figure 6).
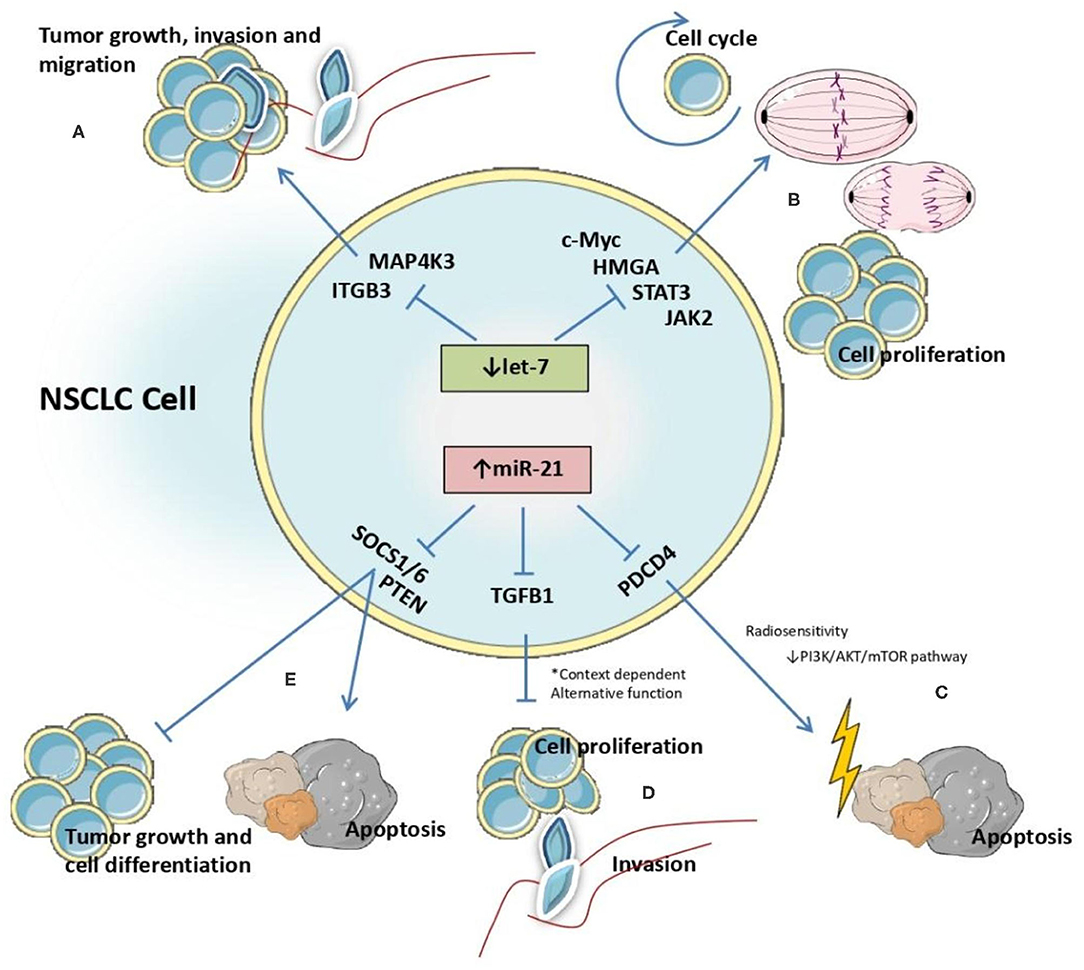
Figure 6. Identified interactions between let-7 and miR-21 microRNAs and target genes affecting molecular processes involved in NSCLC malignant transformation.
The role of let-7 family members as tumor suppressors in NSCLC cells was evaluated in several publications that focused on the mechanism by which this molecule interacts with other genes. In SKMES-1 cells, the effect of let-7c as inhibitor of cellular growth, invasion and migration was demonstrated via downregulation of integrin beta-3 (ITGB3) and mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3). Let-7c reduces cell motility by binding to the 3'-UTR region of these genes and promoting their degradation (77).
In conclusion, our meta-analysis confirmed the predictive value of both miR-21 and let-7 for the outcome of patients with NSCLC. Besides, this investigation revealed that the impact of smoking is reflected in the survival rates of these patients, smoking patients having similar outcomes in both low expression/high expression groups for miR-21. One interesting finding refers to the fact that the accuracy of these molecules as predictors increases with the proportion of women in the sample. Future studies to evaluate the prognostic significance of other microRNAs may offer a complete understanding of the mechanisms of action of microRNAs in NSCLC and therefore would provide the basis to establish a miRNA/gene lung cancer panel that would help in assessing the outcome of NSCLC patients and therefore choose the right treatment course according to the molecular mechanisms affected reflected from the customized miRNA/gene panel. Given the evidence that miR-21 and let-7 expression remain statistically significant in assessing the outcome of NSCLC patients disregarding the type of biological sample used, these microRNAs appear to be valuable molecules to be taken into consideration for the set up of a miRNA/gene NSCLC cancer panel for prognostic.
Conclusion
The present study revealed the applicability and stability of miR-21 and let-7 as prognostic biomarkers in NSCLC, disregarding the sample type—tissue sample or liquid biopsy. Our results highlight the importance of these two transcripts in lung cancer biology, particularly NSCLC, by interfering with crucial pathways associated with induction of apoptosis, cell cycle and cell proliferation. All these findings can represent valuable information for the management of NSCLC patients, with therapeutic significance, as this can count as a stepping stone for further studies focused on a better management of these patients.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://pubmed.ncbi.nlm.nih.gov/–the meta-analysis was made by searching Pubmed database for relevant articles that were included in the analysis.
Author Contributions
CP-B and LM: conceptualization, methodology, and writing. SP and RC: software and formal analysis. DG and MF: review and editing. IB-N: supervision and project administration. All authors contributed to the article and approved the submitted version.
Funding
This work was granted by Competitiveness Operational Program, 2014–2020, entitled Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance—CANTEMIR, no. 35/01.09.2016, MySMIS 103375, project PNCDI III 2015-2020 entitled Increasing the performance of scientific research and technology transfer in translational medicine through the formation of a new generation of young researchers—ECHITAS, no. 29PFE/18.10.2018 and project CNFIS-FDI-2019-0666 entitled Sustenance and valorification of research of excellence in the domain of personalized medicine by internationalization and increasement of research activities visibility.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.516850/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 world health organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. (2015) 10:1240–2. doi: 10.1097/JTO.0000000000000663
3. Commar APV, Tursan d'Espaignet E, Wolfenden L. WHO Global Report on Trends in Prevalence of Tobacco Smoking 2000–2025. World Health Organization (2018).
4. Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet. (2010) 70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6
5. Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. (2009) 23:3243–50. doi: 10.1096/fj.09-135251
6. Calin GA, Trapasso F, Shimizu M, Dumitru CD, Yendamuri S, Godwin AK, et al. Familial cancer associated with a polymorphism in ARLTS1. N Engl J Med. (2005) 352:1667–76. doi: 10.1056/NEJMoa042280
7. Desai A, Menon SP, Dy GK. Alterations in genes other than EGFR/ALK/ROS1 in non-small cell lung cancer: trials and treatment options. Cancer Biol Med. (2016) 13:77–86. doi: 10.20892/j.issn.2095-3941.2016.0008
8. Ling H, Girnita L, Buda O, Calin GA. Non-coding RNAs: the cancer genome dark matter that matters! Clin Chem Lab Med. (2017) 55:705–14. doi: 10.1515/cclm-2016-0740
9. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. (2013) 12:847–65. doi: 10.1038/nrd4140
10. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. (2007) 318:1931–4. doi: 10.1126/science.1149460
11. Gulei D, Raduly L, Berindan-Neagoe I, Calin GA. CRISPR-based RNA editing: diagnostic applications and therapeutic options. Expert Rev Mol Diagn. (2019) 19:83–8. doi: 10.1080/14737159.2019.1568242
12. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
13. Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer-an emerging concept. EBioMedicine. (2016) 12:34–42. doi: 10.1016/j.ebiom.2016.09.017
14. Berindan-Neagoe I, Calin GA. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin Cancer Res. (2014) 20:6247–53. doi: 10.1158/1078-0432.CCR-13-2500
15. Patel K, Kollory A, Takashima A, Sarkar S, Faller DV, Ghosh SK. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. (2014) 347:54–64. doi: 10.1016/j.canlet.2014.01.020
16. Damanakis AI, Eckhardt S, Wunderlich A, Roth S, Wissniowski TT, Bartsch DK, et al. MicroRNAs let7 expression in thyroid cancer: correlation with their deputed targets HMGA2 and SLC5A5. J Cancer Res Clin Oncol. (2016) 142:1213–20. doi: 10.1007/s00432-016-2138-z
17. Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, et al. A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. (2011) 22:104–9. doi: 10.1093/annonc/mdq315
18. Yang X, Cai H, Liang Y, Chen L, Wang X, Si R, et al. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncol Rep. (2015) 33:1723–30. doi: 10.3892/or.2015.3757
19. Liu Q, Liu T, Zheng S, Gao X, Lu M, Sheyhidin I, et al. HMGA2 is down-regulated by microRNA let-7 and associated with epithelial-mesenchymal transition in oesophageal squamous cell carcinomas of Kazakhs. Histopathology. (2014) 65:408–17. doi: 10.1111/his.12401
20. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. (2007) 315:1576–9. doi: 10.1126/science.1137999
21. Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. (2007) 21:1025–30. doi: 10.1101/gad.1540407
22. Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. (2008) 7:759–64. doi: 10.4161/cc.7.6.5834
23. Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. (2008) 105:3903–8. doi: 10.1073/pnas.0712321105
24. Jusufovic E, Rijavec M, Keser D, Korosec P, Sodja E, Iljazovic E, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non–small–cell lung cancer. PLoS ONE. (2012) 7:e45577. doi: 10.1371/journal.pone.0045577
25. Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. (2010) 16:430–41. doi: 10.1158/1078-0432.CCR-09-1736
26. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. (2006) 9:189–98. doi: 10.1016/j.ccr.2006.01.025
27. Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. (2010) 70:8288–98. doi: 10.1158/0008-5472.CAN-10-1348
28. Silva J, Garcia V, Zaballos A, Provencio M, Lombardia L, Almonacid L, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. (2011) 37:617–23. doi: 10.1183/09031936.00029610
29. Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. (2004) 64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637
30. Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY, et al. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest. (2014) 32:394–401. doi: 10.3109/07357907.2014.922569
31. Zhang YK, Zhu WY, He JY, Chen DD, Huang YY, Le HB, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. (2012) 138:1641–50. doi: 10.1007/s00432-012-1240-0
32. Li Y, Zhang H, Dong Y, Fan Y, Li Y, Zhao C, et al. MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer. J Cancer. (2017) 8:1704–16. doi: 10.7150/jca.16961
33. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. (2008) 54:1696–704. doi: 10.1373/clinchem.2007.101741
34. Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. (2011) 17:1875–82. doi: 10.1158/1078-0432.CCR-10-2961
35. Wang X, Zhang Y, Zhi X. Correlation between microRNA expression, clinicopathological characteristics, and prognosis in patients with non-small cell Lung Cancer: a retrospective study. Thorac Cancer. (2017) 8:511–6. doi: 10.1111/1759-7714.12480
36. Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, De W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol. (2011) 104:847–51. doi: 10.1002/jso.22008
37. Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol. (2013) 8:146. doi: 10.1186/1748-717X-8-146
38. Xu X, Yu S, Sun W, Qin X, Chen Y, Zhou L, et al. MiRNA signature predicts the response of patients with advanced lung adenocarcinoma to platinum-based treatment. J Cancer Res Clin Oncol. (2018) 144:431–8. doi: 10.1007/s00432-017-2562-8
39. Ulivi P, Petracci E, Marisi G, Baglivo S, Chiari R, Billi M, et al. Prognostic role of circulating miRNAs in early-stage non-small cell lung cancer. J Clin Med. (2019) 8:131. doi: 10.3390/jcm8020131
40. Kunita A, Morita S, Irisa TU, Goto A, Niki T, Takai D, et al. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci Rep. (2018) 8:8838. doi: 10.1038/s41598-018-27128-3
41. Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, et al. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. (2012) 29:3190–7. doi: 10.1007/s12032-012-0303-z
42. Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. (2017) 8:13048–58. doi: 10.18632/oncotarget.14369
43. Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. (2012) 29:618–26. doi: 10.1007/s12032-011-9923-y
44. Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. (2012) 13:330–40. doi: 10.4161/cbt.19073
45. Akagi I, Okayama H, Schetter AJ, Robles AI, Kohno T, Bowman ED, et al. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res. (2013) 73:3821–32. doi: 10.1158/0008-5472.CAN-13-0031
46. Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. (2011) 137:557–66. doi: 10.1007/s00432-010-0918-4
47. Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett. (2017) 13:1256–63. doi: 10.3892/ol.2017.5569
48. Xue X, Liu Y, Wang Y, Meng M, Wang K, Zang X, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. (2016) 7:84508–19. doi: 10.18632/oncotarget.13022
49. Wan J, Ma J, Mei J, Shan G. The effects of HIF-1alpha on gene expression profiles of NCI-H446 human small cell lung cancer cells. J Exp Clin Cancer Res. (2009) 28:150. doi: 10.1186/1756-9966-28-150
50. Perez-Ramirez C, Canadas-Garre M, Molina MA, Faus-Dader MJ, Calleja-Hernandez MA. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics. (2015) 16:1843–62. doi: 10.2217/pgs.15.122
51. Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. (2018) 75:3539–51. doi: 10.1007/s00018-018-2877-x
52. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. (2010) 8:336–41. doi: 10.1016/j.ijsu.2010.02.007
53. Pop-Bica C, Pintea S, Cojocneanu-Petric R, Del Sal G, Piazza S, Wu ZH, et al. MiR-181 family-specific behavior in different cancers: a meta-analysis view. Cancer Metastasis Rev. (2018) 37:17–32. doi: 10.1007/s10555-017-9714-9
54. Kepes S, Banks GC, McDaniel M, Whetzel DL. Publication bias in the organizational sciences. Organ Res Methods. (2012) 15:624–62. doi: 10.1177/1094428112452760
55. Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci. (2011) 121:141–58. doi: 10.1042/CS20110005
56. Nagy ZB, Bartak BK, Kalmar A, Galamb O, Wichmann B, Dank M, et al. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol Oncol Res. (2019) 25:97–105. doi: 10.1007/s12253-017-0308-1
57. Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. (2018) 12:229–41. doi: 10.1016/j.omtn.2018.05.009
58. Li C, Yin Y, Liu X, Xi X, Xue W, Qu Y. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget. (2017) 8:24564–78. doi: 10.18632/oncotarget.15596
59. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. (2008) 83:584–94. doi: 10.4065/83.5.584
60. Ding N, Pang Z, Zhang X, Huang C, Yang Y, Liu Q, et al. Prognostic and predictive effects of positive lymph node number or ratio in NSCLC. Sci Rep. (2017) 7:584. doi: 10.1038/s41598-017-00619-5
61. Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR, Threapleton D, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. (2016) 7:78985–93. doi: 10.18632/oncotarget.12587
62. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. (2004) 64:8919–23. doi: 10.1158/0008-5472.CAN-04-2818
63. Su JL, Chen PS, Johansson G, Kuo ML. Function and regulation of let-7 family microRNAs. MicroRNA. (2012) 1:34–9. doi: 10.2174/2211536611201010034
64. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. (2012) 489:519–25. doi: 10.1038/nature11404
65. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. (2014) 511:543–50. doi: 10.1038/nature13385
66. Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. (2010) 16:4864–75. doi: 10.1158/1078-0432.CCR-10-0199
67. Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS ONE. (2012) 7:e36530. doi: 10.1371/journal.pone.0036530
68. Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. (2006) 24:5079–90. doi: 10.1200/JCO.2005.05.1748
69. Stahlhut C, Slack FJ. Combinatorial action of MicroRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. (2015) 14:2171–80. doi: 10.1080/15384101.2014.1003008
70. Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. (2009) 106:12085–90. doi: 10.1073/pnas.0905234106
71. Gou LY, Niu FY, Wu YL, Zhong WZ. Differences in driver genes between smoking-related and non-smoking-related lung cancer in the Chinese population. Cancer. (2015) 121(Suppl. 17):3069–79. doi: 10.1002/cncr.29531
72. Chapman AM, Sun KY, Ruestow P, Cowan DM, Madl AK. Lung cancer mutation profile of EGFR, ALK, and KRAS: meta-analysis and comparison of never and ever smokers. Lung Cancer. (2016) 102:122–34. doi: 10.1016/j.lungcan.2016.10.010
73. Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. (2013) 372:35–45. doi: 10.1007/s11010-012-1443-3
74. Yamada KM, Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci. (2001) 114(Pt 13):2375–82.
75. Yan L, Ma J, Wang Y, Zan J, Wang Z, Zhu Y, et al. miR-21-5p induces cell proliferation by targeting TGFBI in non-small cell lung cancer cells. Exp Ther Med. (2018) 16:4655–63. doi: 10.3892/etm.2018.6752
76. Jiang LP, He CY, Zhu ZT. Role of microRNA-21 in radiosensitivity in non-small cell lung cancer cells by targeting PDCD4 gene. Oncotarget. (2017) 8:23675–89. doi: 10.18632/oncotarget.15644
Keywords: prognostic, biomarker, NSCLC, miR-21, let-7, patients
Citation: Pop-Bica C, Pintea S, Magdo L, Cojocneanu R, Gulei D, Ferracin M and Berindan-Neagoe I (2020) The Clinical Utility of miR-21 and let-7 in Non-small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front. Oncol. 10:516850. doi: 10.3389/fonc.2020.516850
Received: 08 January 2020; Accepted: 21 September 2020;
Published: 19 October 2020.
Edited by:
Rengyun Liu, The First Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Vincenzo Graziano, University of Cambridge, United KingdomSimone Anfossi, University of Texas MD Anderson Cancer Center, United States
Copyright © 2020 Pop-Bica, Pintea, Magdo, Cojocneanu, Gulei, Ferracin and Berindan-Neagoe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ioana Berindan-Neagoe, aW9hbmEubmVhZ29lQHVtZmNsdWoucm8=
†These authors have contributed equally to this work
 Cecilia Pop-Bica
Cecilia Pop-Bica Sebastian Pintea
Sebastian Pintea Lorand Magdo
Lorand Magdo Roxana Cojocneanu
Roxana Cojocneanu Diana Gulei
Diana Gulei Manuela Ferracin
Manuela Ferracin Ioana Berindan-Neagoe
Ioana Berindan-Neagoe