
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 03 September 2020
Sec. Cancer Immunity and Immunotherapy
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.01632
This article is part of the Research Topic Targeting Indoleamine 2,3-dioxygenases and Tryptophan Dioxygenase for Cancer Immunotherapy View all 9 articles
 Raghavan Chinnadurai1*
Raghavan Chinnadurai1* Rafaela Scandolara1
Rafaela Scandolara1 Olatunji B. Alese2
Olatunji B. Alese2 Dalia Arafat3
Dalia Arafat3 Deepak Ravindranathan2
Deepak Ravindranathan2 Alton B. Farris4
Alton B. Farris4 Bassel F. El-Rayes2
Bassel F. El-Rayes2 Greg Gibson3
Greg Gibson3Mechanisms of dysfunctional T cell immunity in Hepatocellular Carcinoma (HCC) need to be well defined. B7 family molecules provide both co-stimulatory and co-inhibitory signals to T cells while tryptophan degrading enzymes like Indoleamine 2,3 dioxygenase (IDO) and Tryptophan 2,3 Dioxygenase (TDO) mediate tumor immune tolerance. It is necessary to identify their in situ correlative expression, which informs targets for combined immunotherapy approaches. We investigated B7 family molecules, IDO, TDO and immune responsive effectors in the tumor tissues of patients with HCC (n = 28) using a pathway-focused quantitative nanoscale chip real-time PCR. Four best correlative expressions, namely (1) B7-1 & PD-L2, (2) B7-H2 & B7-H3, (3) B7-2 & PD-L1, (4) PD-L1 & PD-L2, were identified among B7 family ligands, albeit they express at different levels. Although TDO expression is higher than IDO, PD-L1 correlates only with IDO but not TDO. Immune effector (Granzyme B) and suppressive (PD-1 and TGF-β) genes correlate with IDO and B7-1, B7-H5, PD-L2. Identification of the in situ correlation of PD-L1, PD-L2 and IDO suggest their cumulative immuno suppressive role in HCC. The distinct correlations among B7-1, B7-2, B7-H2, and B7-H3, correlation of PD-1 with non-cognate ligands such as B7-1 and B7-H5, and correlation of tumor lytic enzyme Granzyme B with IDO and PD-L2 suggest that HCC microenvironment is complexly orchestrated with both stimulatory and inhibitory molecules which together neutralize and blunt anti-HCC immunity. Functional assays demonstrate that both PDL-1 and IDO synergistically inhibit T cell responses. Altogether, the present data suggest the usage of combined immune checkpoint blocking strategies targeting co-inhibitory B7 molecules and IDO for HCC management.
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related mortalities (1). Defining the immunological mechanisms contributing to HCC pathogenesis in the intrahepatic tumor microenvironment is of translational interest (2). Co-stimulatory, co-inhibitory (B7 family) pathways and enzymes of tryptophan degradation predominantly attenuate anti-tumor T-cell immunity. FDA-approved inhibitors of PD-L1 (B7-H1) & PD-L2 (B7-DC)-PD-1 interaction to revive exhausted anti-tumor T-cell immunity have shown benefit to cancer patients (3, 4). In addition, clinical trials are ongoing to test the efficacy of inhibitors of tryptophan degrading enzymes for cancer treatment (NCT03695250) (5–7). Early phase clinical trials are ongoing to test the effect of immunotherapy agents targeting B7 family and tryptophan degradation pathways on HCC (8–10). Importantly, inhibitors of PD-L1 (B7-H1) & PD-L2 (B7-DC)-PD-1 pathways are promising for anti-HCC management (11, 12). Defining the complex interaction among B7 family ligands and enzymes of tryptophan degradation pathways in the HCC microenvironment will inform additional checkpoint immune blocking strategies.
B7 family ligands are the dominant family of molecules providing co-stimulation and co-inhibition to T-cells. Accumulated evidence demonstrates that there are many members present in the B7 family, namely, B7-1, B7-2, PD-L1 (B7-H1), PD-L2 (B7-DC), B7-H2, B7-H3, B7-H4, B7-H5, B7-H6 and B7-H7 (13, 14). Receptors of the majority of the B7 family ligands have been defined on T-cells while some ambiguity exists in identifying definitive receptors of new members B7-H3, B7-H4, B7-H5, and B7-H7 (Table 1). Nevertheless, dysfunctional and exhausted T-cells co-express receptors of B7-family ligands, which suggest that a complex co-receptor and ligand interaction occur in a tumor microenvironment and confer dysfunctional immune response (15–18). The interaction of PD-L1/PD-L2 with PD-1 on T cells plays an important role in modulating tumor immunity (19). B7-H2 has been characterized as a co-stimulatory ligand for Inducible Costimulator (ICOS) and skews T-cell differentiation toward Th2 responses (20, 21). In contrast, B7-H3 is a negative regulator by preferentially affecting Th1 responses (22, 23). B7-H4 has been shown to play a major role in the negative regulation of T cell immunity (24, 25). B7-H5 or V-domain Ig Suppressor of T cell Activation (VISTA) is identified as both co-stimulatory (26) and co-inhibitory molecule (27). B7-H6 is highly expressed on many cancer conditions and interacts with NK-cell receptor NKP30 to induce activation while its significance on tumor immunity is yet to be defined (28). B7-H7 is a co-inhibitory molecule and inhibits CD4+ and CD8+ T cell functions (29). The pattern of correlative expressions of B7 family molecules and their interaction with the immune effectors of HCC microenvironment is currently unknown.
Indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) deplete tryptophan by converting it into the immunosuppressive catabolite, kynurenine (30–32). Although inconsistent results emerge in clinical trials targeting IDO (33), it needs to be further clarified on how IDO and/or TDO operate with B7 family ligands in modulating T-cell responses in the liver of patients with HCC. Important questions are: How are B7 family ligands expressed relative to each other in controlling T cell fate? How do B7 family ligands correlate among themselves in the tumor microenvironment of HCC? Do B7 family ligands co-express with IDO and TDO in an intricate intrahepatic tumor microenvironment? How do B7 family ligands, IDO and TDO correlate with immune responsive effector molecules in HCC? Better understanding of the underlying in situ associations of IDO/TDO and B7 family members will inform the rationale of translational development of specific checkpoint inhibitors for primary liver cancer management.
The Emory University Institutional Review Board (IRB) approved the study. All consecutive cases of patients with HCC who were older than 18 years of age and treated between 2013 and 2015 at Winship Cancer Institute/Emory University Hospital system were identified and enrolled. All the patients (n = 28) who were included in the study had active HCC (Table 2).
Paraffin embedded tissue blocks from HCC patients were subjected to hematoxylin and eosin (H&E) staining to ensure that the tissue slices subjected to RNA analysis have adequate (minimum 30–40%) tumor volume. % Tumor volume and % leukocytes in tissue sections are given in Table 2. High Pure FFPE (formalin-fixed, paraffin-embedded) RNA Micro Kit was used to isolate RNA from the dissected paraffin slices according to the manufacturer instructions (Roche GmbH, Germany). Hundred to five-hundred nanogram of isolated RNA was used for total cDNA preparation (QuantiTect Reverse Transcription Kit, Qiagen, USA).
Quantitative RT-PCR was performed using Fluidigm 48 × 48 nanofluidic arrays. Briefly, cDNA samples were pre amplified with 14-cycle PCR reaction for each sample with the combination of 100 ng cDNA and pooled primers as described by TaqMan Pre-Amp Mastermix (Fluidigm BioMark™) manufacturer's protocols. Two thousand three hundred four parallel qRT-PCR reactions were performed for each primer pair on each sample on a 48 × 48 chip array. Human cDNA library was used as a positive control. Amplification was detected in Eva Green detection assay on a Biomark I machine based on standard Fluidigm protocols as described previously (34, 35). Two independent primer pairs were used for each target (Table 3). Cycle of Threshold (CT) values were normalized based on the endogenous GAPDH & beta actin controls. CT value of each target was subtracted with the CT value of endogenous GAPDH & beta actin (Control CT values) of the respective samples to get delta CT values. The delta CT values were subjected to a second standardization by adding all the delta CT values with a constant CT value, which was derived from the total average of control CT values from all the samples. In the case of Osteopontin or Glypican normalization, GAPDH & beta actin normalized CT values were further normalized with Osteopontin or Glypican-3. The normalized CT values are expressed as an inverse (CT−1) to better graphically represent the increase or decrease in expression as described previously (36). Inverse CT values were obtained by dividing 1 with the appropriate normalized CT value. High or low inverse CT values represent high or low expression of target genes, respectively.
Peripheral Blood Mononuclear Cells (PBMCs) were isolated from heparin-anticoagulated whole blood using a Ficoll-Paque PLUS density gradient (GE Healthcare Biosciences, Sweden), and cryopreserved in medium containing 90% fetal calf serum (HyClone) and 10% dimethyl sulfoxide (Corning, USA). Prior to the experiment, cells were thawed and rested overnight in a 37°C CO2 incubator. For proliferation assays, Carboxyfluorescein succinimidyl ester (CFSE) (Biolegend USA) labeled PBMC were stimulated for 4 days with plate bound 1 μg/ml anti-CD3 and anti-CD28 (Biolegend, USA) antibodies and 10ug/ml of PD-L1 Ig or Control Ig protein (Biolegend, USA) with varying concentrations of Kynurenine (Tocris, USA). Four days later, cells were stained with CD3-APCCy7 antibody (Biolegend, USA) and acquired in BD FACSAria instrument. Results were analyzed in Flow Jo software. Supernatants were analyzed for IFNγ using the kit, Human IFN gamma ELISA Ready-SET-Go according to the manufacture's instruction.
Data were analyzed with GraphPad Prism 5.0 software for statistics. An unpaired two-sided t-test was used to determine significance between the means of two groups, and a one-way ANOVA with Tukey's multiple comparison tests was used to compare multiple groups simultaneously. P-value < 0.05 was considered statistically significant. Linear regression analysis was performed using normalized CT values to obtain R2 and P-values. R2 = Goodness of Fit; P = Significance of the slope deviation from Zero. Correlations with R2 values above 0.5 with the P-value of < 0.05 were considered as the best correlation. Correlation with R2 values between 0.3 and 0.5 with the P-value of < 0.05 were considered as moderate correlations/statistical trend toward a correlation. Correlations with R2 values below 0.3 were considered relatively less significant.
B7 family molecules are either co-stimulatory or co-inhibitory in stimulating and negating T-cell responses (Table 1). We have evaluated their relative expression in the hepatic tumor microenvironment in patients with HCC. Surgically resected liver tumor tissues of the patients with HCC (n = 28) were fixed with formalin and embedded in paraffin. Hematoxylin and Eosin (H&E) staining was performed for the inclusion criteria that the tissue slices subjected to RNA analysis have at least 30–40% tumor volume (Table 2). % Tumor volume and % leukocytes in tissue sections are given in Table 2. Patient #1 had a minute focus of cholangiocarcinoma on morphology which was excluded on histologic analysis. Total RNA was extracted from tissue sections, which was then converted into cDNA. cDNA samples were then used to perform Fluidigm nanoscale quantitative real-time chip PCR with two independent primer pairs for each target. Human cDNA library was used as a positive control to ensure the effectiveness of the primer pairs. The cycle of threshold (CT) values were normalized based on endogenous GAPDH & beta actin controls. Hierarchical organization of the cumulative median CT−1 values (inverse CT values were used to depict the expression) identified the ranking of expression as values identified the ranking of expression from high to low as B7-H5, B7-H3, B7-H2, B7-1, PD-L1, B7-2, PD-L2, B7-H6, B7-H4, and B7-H7 (Figure 1A). B7-H5 and B7-H3 showed statistically significant enhanced expression compared to all other B7 family molecules. B7-H2, PD-L1, PD-L2, B7-1, and B7-2 showed comparable expression among themselves while B7-H4, B7-H6, and B7-H7 are expressed at low levels (Figures 1A,B). The immunosuppressive tryptophan degradation pathway (IDO, TDO) is an immunotherapy target to revive anti-tumor immunity (30–32). We set out to identify the mRNA expression levels of IDO and TDO in HCC and our results show that TDO expression is higher than IDO (Figure 1C).
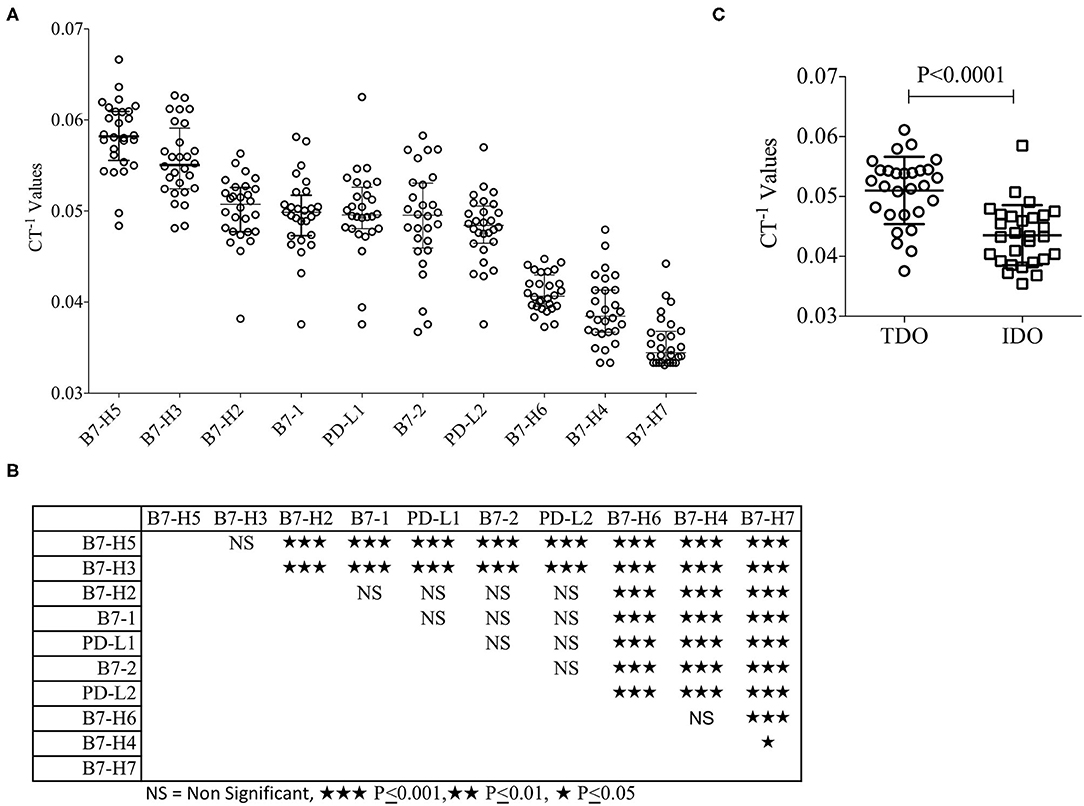
Figure 1. Relative expression of B7 familiy ligands and enzymes of Tryptophan degradation in HCC. Paraffin embedded liver tissue (10 micron) sections were collected in tubes and Hematoxylin and Eosin stained sections were reviewed to ensure tumor content. RNA was extracted and converted into cDNA which was used for the B7 Family ligand amplification in FluidigmTM nanoscale quantitative RT PCR. Each target is validated with two independent primer sets with the amplicon length of <100 base pairs. (A) Inverse CT values (CT−1) of B7 Family ligands derived from each patient were plotted with median and range. Median values were organized in hierarchical order of high to low expression. (B) One-way ANOVA and Tukey's multiple comparison test was performed to obtain P-values. Table shows the statistical significance of the median of CT−1 values of each B7 family ligand in comparison to others. (C) Inverse CT values (CT−1) of TDO and IDO derived from each patient were plotted with the median and range. CT values were normalized based on GAPDH & beta actin controls.
To identify the correlation among B7 family ligands, we subjected the CT values of each B7 family ligands in combinations through linear regression analysis. The degree of R2 correlation values (Example: R2 = 1 and R2 = 0 represent the best and no correlation between two molecules, respectively) determines the correlation between two B7 family molecules. Fifty-five combinations tested among the B7 family members and the R2 values were subjected to hierarchical ranking analysis ranging from best to no correlation. B7-1 expression showed the highest correlation with PD-L2 (R2 = 0.7174; P < 0.0001) (Figures 2A,B). We also identified moderate correlations among three B7 combinations such as (1) B7-2 and PD-L1 (R2 = 0.4244; P = 0.0002) (Figures 2A,C), (2) B7-H2 and B7-H3 (R2 = 0.4090; P = 0.0002) (Figures 2A,D), (3) PD-L1 and PD-L2 (R2 = 0.3863; P = 0.0004) (Figures 2A,E). All other combinations did not show a significant correlation. Altogether, the present analysis identified that B7-1, B7-2, PD-L1, PD-L2, and B7-H3 are correlated in a distinct combinatorial expression pattern.
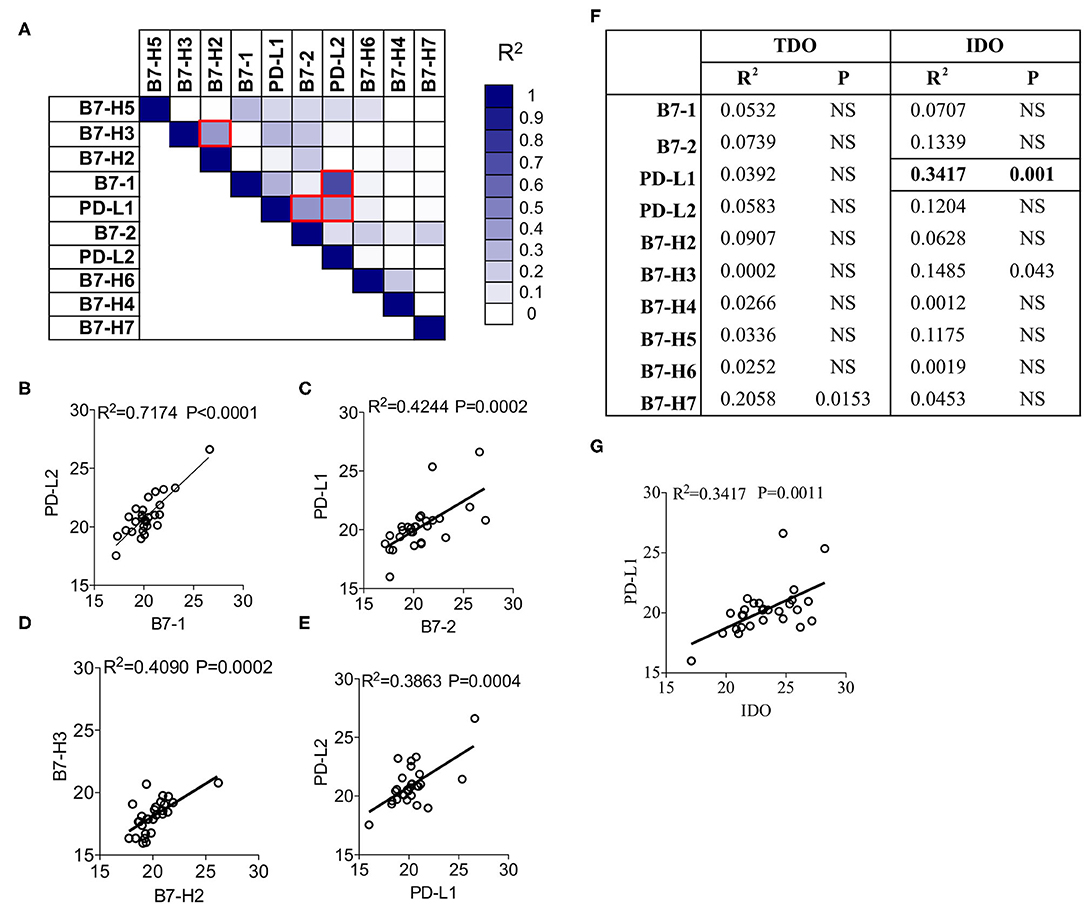
Figure 2. Correlation patterns among B7-Familiy ligands and IDO in the liver of HCC patients. (A) CT Values of B7 Family ligands from HCC patients (n = 28) were subjected to linear regression analysis among each other. R2 values were color-coded and red boxes indicate the best correlations based on the hierarchical ranking. Correlative plot with R2 values above 0.3 are shown for the combination (B) B7-1&PD-L2, (C) B7-2&PD-L1, (D) B7-H2&B7-H3, and (E) PD-L2&PD-L1. (F) CT values of IDO and TDO were subjected to linear regression analysis with B7 family ligands. Correlative plot with the R2 values above 0.3 is shown for the combination (G) IDO & PD-L1. Linear regression analysis was performed in GraphPad Prism to get R2 and P-values. R2, Goodness of Fit; P, Significance of the slope deviation from Zero.
To investigate the effect of the expression of IDO and TDO on B7 family ligands, we performed a linear regression analysis between the CT values of IDO, TDO and B7 family ligands. Twenty combinations were investigated to identify R2 values. Although TDO expression is superior to IDO, it does not correlate with any of the B7 family ligands (Figure 2F). However, IDO shows a trend toward a correlation with PD-L1 (R2 = 0.3417; P = 0.001) (Figures 2F,G). We did not observe any associations between IDO and other B7 family molecules. Altogether, these results demonstrate the correlation between IDO and PD-L1.
Tumor microenvironment is not only occupied with the co-stimulatory and co-inhibitory ligands but also the immune responsive effector molecules that actually facilitate immune suppression or stimulation. We analyzed the expression of TGF-β, IL-10 (immune suppression), IFNγ, TNFα, IL-2, Perforin, Granzyme B (immune stimulation), and PD-1 (immune exhaustion). We categorized the expression levels of these effector molecules relative to each other by descending hierarchical organization of median CT−1 values. Our results demonstrate that TGF-β, PD-1, and IL-10 are expressed at high levels, TNFα and Granzyme B expressed at moderate levels, while IFNγ, IL-2, and Perforin are expressed at very low levels (Figure 3A). Descending hierarchical organization of median CT−1 values identified the ranking as TGF-β, PD-1, IL-10, GranzymeB, TNFα, IFNγ, IL-2 and Perforin (Figures 3A,B). Altogether these results demonstrate the dominant expression of immunosuppressive molecules (TGF-β, PD-1, IL-10) in HCC microenvironment.
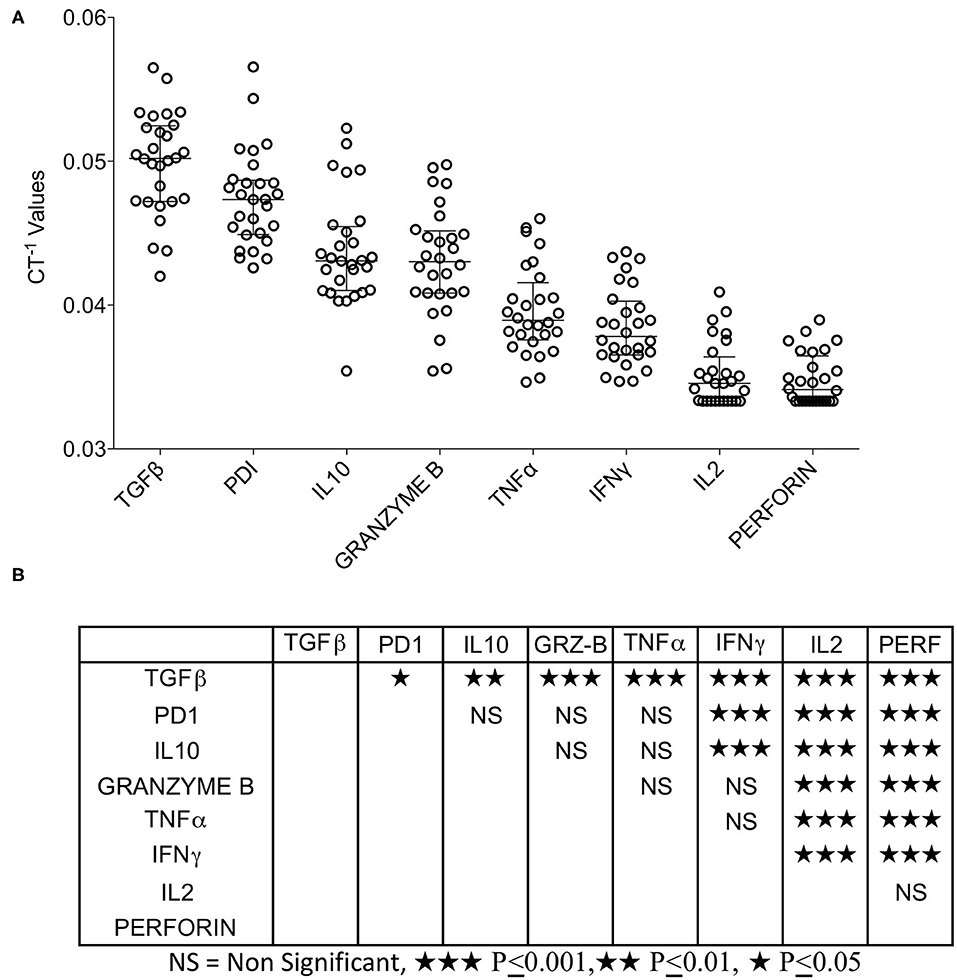
Figure 3. Expression pattern of immune responsive effector molecules in HCC. RNA transcripts of immune responsive effector molecules TGFβ, PD-1, IL-10, IFNγ, TNFα, IL-2, Granzyme B and Perforin were analyzed in FluidigmTM nanoscale quantitative RT-PCR. (A) Inverse CT values (CT−1) of immune responsive effector molecules derived from each patient were plotted with the median and range. Median values were organized in hierarchical order of high to low expression. (B) One-way ANOVA and Tukey's multiple comparison test was performed to obtain P-values. Table shows the statistical significance of the median of CT−1 values of each effector molecule in comparison to others. CT values were normalized based on GAPDH & beta actin controls.
Next we aim to determine the effect of the expression of individual B7 family ligands on the expression of immune responsive effector molecules within HCC microenvironment. CT values of the immune responsive effector molecules were subjected to linear regression analysis with the CT values of B7 family ligands. Eighty combinations were tested to identify R2 values, which determine the correlation of immune responsive effector molecules with B7 family ligands. Our results identified the best correlation between TGF-β and B7-H5 (R2 = 0.6765; P < 0.0001) (Figures 4A,B). In addition, a statistical trend toward the best correlation was identified with four other combinations. (1) PD-1 and B7-H5 (R2 = 0.48; P < 0.0001), (Figures 4A,C), (2) PD-L2 and Granzyme B (R2 = 0.46; P < 0.0001) (Figures 4A,D), (3) PD-1 and B7-1 (R2 = 0.4224; P = 0.0002) (Figures 4A,E). (4) IFNγ and PD-L2 (R2 = 0.3174; P = 0.0018) (Figures 4A,F).
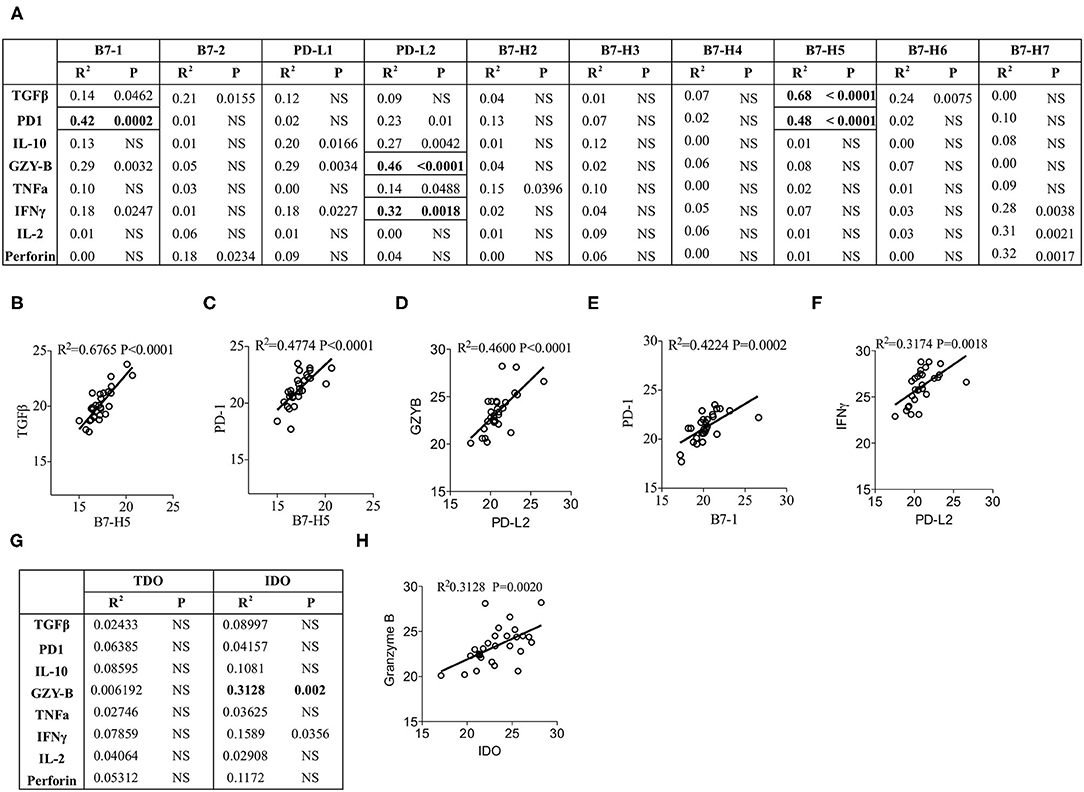
Figure 4. Correlation of B7 family ligands and IDO with immune responsive effector molecules. CT values of each immune responsive effector molecules were subjected to linear regression analysis with the B7 family. (A) Correlation B7 family ligands with immune responsive effector molecules is shown in table format. Correlative plots with R2 values above 0.3 are shown as (B) B7-H5&TGFβ, (C) PD-1&B7-H5, (D) PD-L2&GranzymeB, (E) B7-1&PD-1 and (F) PDL2&IFNγ. (G) IDO and TDO were subjected to linear regression analysis with immune responsive effector molecules to get R2 and p-values. Correlative plots with the R2 values above 0.3 is shown for the combination (H) IDO & Granzyme B. Linear regression analysis was performed in GraphPad Prism to get R2 and P-values. R2, Goodness of Fit; P, Significance of the slope deviation from Zero; NS, Non-significant.
None of the other combinations of B7 family ligands and immune responsive molecules showed a significant correlation values above 0.3 (Figure 4A). Correlations such as B7-H7 and IL-2, B7-H7 and Perforin were disregarded due to their very low expression levels (Figures 1A, 3A). Altogether, these results demonstrate that although PD-1 is the cognate receptor for the ligands PD-L1 and PD-L2, its expression correlates with other B7 family ligands B7-1 and B7-H5. In addition, the immunosuppressive cytokine TGF-β correlates with B7-H5 while IFNγ and tumorlytic enzyme Granzyme B correlates with PD-L2.
Next we investigated the correlation of IDO and TDO with immune responsive effector molecules. Linear regression analysis was performed between these two families of molecules with 16 combinations (Figure 4G). Identification of R2 values demonstrates that TDO expression does not show any correlation with immune responsive effector molecules (Figure 4G). However, IDO expression shows a trend toward correlation with Granzyme B but not with any other immune responsive effector molecules (R2 = 0.3128; P = 0.002) (Figures 4G,H). Altogether these results demonstrate that although TDO is dominant, it does not correlate with immune responsive effector molecules. In contrast, IDO expression correlates with Granzyme B.
Glypican-3 and Osteopontin are over expressed in HCC and studies have demonstrated that both of these molecules could serve as the biomarker of HCC progression and could be targeted (37–39). Hence, we determined the mRNA expression levels of Glypican-3 and Osteopontin in HCC samples. Our results show that Glypican-3 and Osteopontin expressions are not significantly different from each other (Figure 5A). In addition, Glypican-3 and Osteopontin did not show significant levels of correlation with B7 family ligands, immune response genes and IDO, except Osteopontin showing a moderate correlation with B7-2 (Figures 5B,C). Next, we normalized CT values based on Osteopontin and Glypican-3 expression and validated the key identified correlation (as identified in Figures 2, 4) that were determined based on GAPDH and beta-actin normalization. We recapitulated and confirmed the identified correlations among B7 family ligands, immune response genes and IDO based on the normalization with HCC associated biomarkers Glypican-3 and Osteopontin (Figures 5D,E).
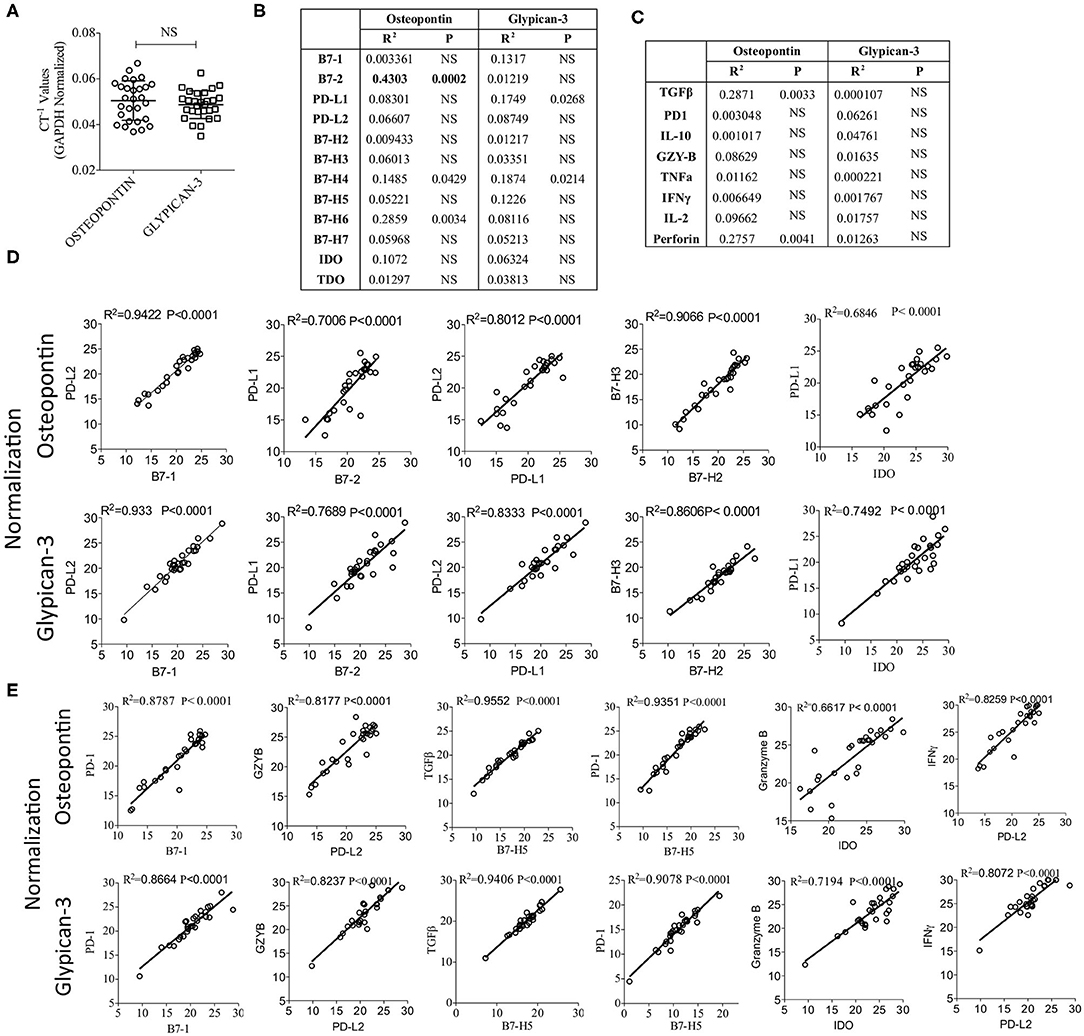
Figure 5. Validation of the identified correlations using HCC biomarkers Osteopontin and Glypican-3. (A) Inverse CT values (CT−1) of Osteopontin and Glypican-3 derived from each patient, after GAPDH & beta actin normalization, were plotted with median and range. CT values of Osteopontin and Glypican-3 were subjected to linear regression analysis with (B) B7 Family ligands, Tryptophan Degrading enzymes and (C) immune response genes. Correlation R2 values and statistical significance are shown in a table format. (D,E) GAPDH & beta actin normalized CT values were further normalized based on the expression of Osteopontin or Glypican-3. Correlations that are identified in Figures 2, 4 were further confirmed using Osteopontin and Glypican-3 normalization. (D) Correlations between B7 family ligands and IDO. (E) Correlations between B7 family ligands, IDO and immune response genes. Linear regression analysis was performed in GraphPad Prism to get R2 and P-values. R2, Goodness of Fit; P, Significance of the slope deviation from Zero; NS, Non-significant.
IDO correlates only with PD-L1 but not with other B family ligands, which suggests that this correlation is significant. Hence, to investigate the synergistic functional effect of IDO and PD-L1, we tested the effect of PD-L1 Ig/Control Ig and Kynurenine (IDO metabolite) on human peripheral blood mononuclear cells (PBMCs). We stimulated CFSE labeled human PBMCs with plate-bound anti-CD3 & anti-CD28 in the presence of PD-L1 Ig or Control Ig along with the dose escalating concentrations of Kynurenine. Four days later, T cell proliferation was determined by analyzing the percentage of CFSE dilution using flow cytometry. In addition, we also determined the concentrations of secreted Interferon γ (IFNg) in the supernatants of the cultures by Enzyme Linked Immunosorbent Assay (ELISA). Our results demonstrate that PD-L1 Ig inhibits T cell proliferation in the absence of Kynurenine. Similarly, Kynurenine inhibits T cell proliferation in the absence of PD-L1 Ig (Figures 6A,B). In the cultures with both PD-L1 and Kynurenine, the inhibitory effect on T cell proliferation is superior over their individual counterparts (Figures 6A,B). Similarly, both PD-L1 Ig and Kynurenine synergistically inhibit Interferon γ (IFNg) secretion (Figure 6C). Altogether these results demonstrate the synergistic inhibition of PD-L1 and IDO on T cell responses.
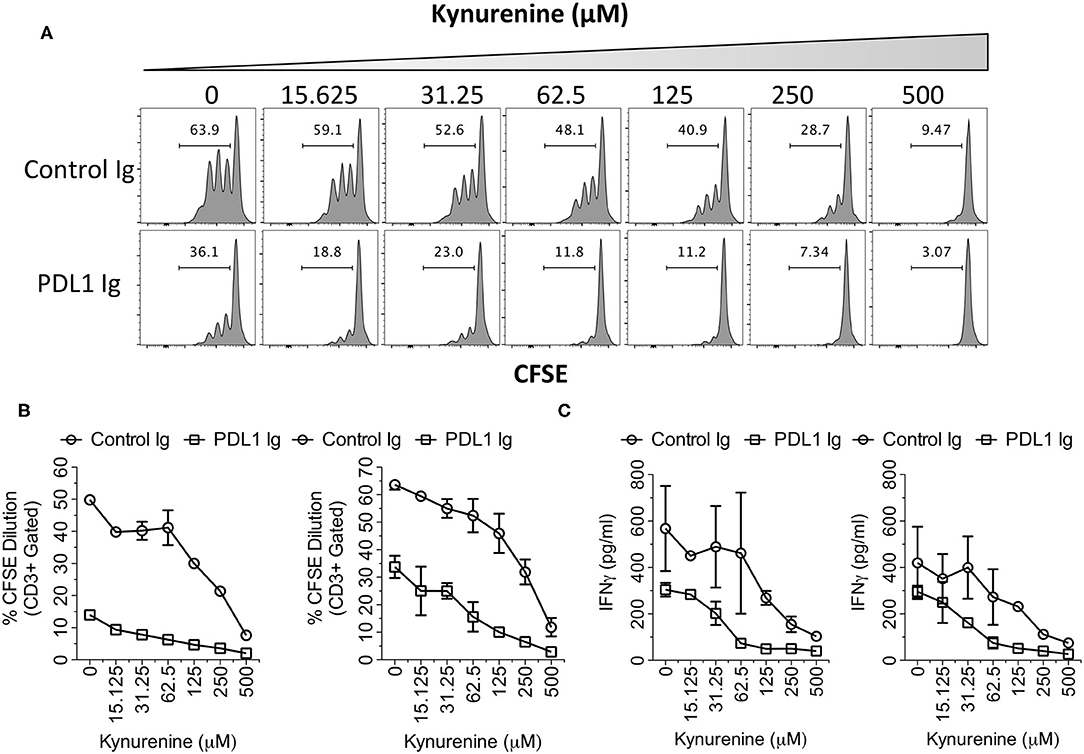
Figure 6. Synergistic inhibitory effects of PD-L1 and Kynurenine on T cell responses. CFSE labeled PBMCs were stimulated on the plates coated with anti-CD3 and anti-CD28 (1 μg/ml) alone or in combination with PD-L1 Ig or Control Ig (10 μg/ml). Subsequently, Kynurenine was added to the culture at a final concentration as indicated. 4 days later, T cell proliferation analysis was performed using flow cytometry. (A) Representative CFSE dilution plot is shown. CFSE plots were derived from Forward Scatter, Side Scatter, CD3+ parental gates. (B) % CD3+ CFSE dilution is plotted with two independent donors. (C) Supernatants of the cultures were analyzed for human IFNγ using ELISA. Concentrations of the IFNγ are shown from two independent experiments using independent donor PBMCs.
Reviving the immune system with immunotherapy is becoming an attractive strategy to treat HCC (40, 41). Immunotherapy agents such as immune checkpoint blocking agents targeting single co-inhibitory molecule show some benefits to patients with HCC (11). These prompted to use second-generation combined immune blocking strategies targeting additional co-inhibitory molecules of the tumor microenvironment. However, practically it is not feasible to test combined immune blocking strategies targeting an array of immunomodulatory molecules with all the combinations in clinical trials or immunotherapy animal model studies. Hence, it is necessary to identify the in situ correlation and interaction of immunomodulatory molecules in the tumor microenvironment. We hypothesized that immunomodulatory molecules express in a specific combinatorial pattern in the HCC microenvironment and identification of such in situ combinations not only inform the targets for combined immunotherapy approaches but also unveil the immuno physiology of the tumor microenvironment.
Herein, we analyzed the correlative expressions of B7 family molecules and enzymes of tryptophan degradation in the tumor microenvironment of HCC. Consistent with other tumor types (42), we identified the differential expressions of B7 family molecules within the hepatic tumor microenvironment. Using a correlative hierarchical ranking analysis, we identified four best correlations out of 55 combinations tested among B7 family molecules. They are (1) B7-1 and PD-L2, (2) B7-2 and PD-L1 (3) PD-L1 and PD-L2, (4) B7-H2 and B7-H3. Identification of the correlative expression of PD-L1 and PD-L2 suggests that both of these ligands may function together in dampening T cell responses through its receptor PD-1. Clinical trials targeting PD-1 or PD-L1 with monoclonal antibodies showed encouraging results. FDA approved this immunotherapy for the patients who show advanced stage HCC and sorafenib resistance. Although FDA approved this therapy with the contingency that advanced phase clinical trials should demonstrate efficacy, the results are both positive and negative (40, 41). Thus, combined immunotherapy approach may be necessary to demonstrate consistent clinical efficacy. In addition, it has also been demonstrated that the binding structure, kinetics and affinity of PD-L1 and PD-L2 with PD1 are different (43, 44). These structural insights and our observation of the correlation of PD-L1 and PD-L2 suggest that the combined immunotherapy approaches targeting PD-L1, PD-L2 and PD-1 are warranted for advanced HCC management.
We also observed correlations such as (1) B7-1 and PD-L2, (2) B7-2 and PD-L1, (3) PD-1 and B7-1 which are intriguing since PD-L1, PD-L2, PD-1 are co-inhibitory ligands/receptor while B7-1 and B7-2 are dimorphic ligands since they can interact with CD28 or CTLA4 to promote co-stimulation or co-inhibition, respectively. It has been recently demonstrated that B7-1, B7-2 and CD28 co-stimulation is necessary to revive exhausted T cells using anti-PD-1 therapy in animal models and lung cancer patients (45). Thus, both of these two co-stimulatory and co-inhibitory pathways, (1) B7-1, B7-2-CD28 and (2) PD-L1, PD-L2-PD1 may function together in promoting T-cell exhaustion. Our results further support this hypothesis that in the liver tumor microenvironment, B7-1, B7-2, PD-L1, PD-L2 and PD-1 interactions occur extensively, which cumulatively confer dysfunctional anti-tumor immunity. In addition, our data shows the correlation of PD-1 with its non-cognate ligands B7-1 and B7-H5 which further suggests that PD-1 should be targeted in conjunction with other B7 family molecules.
B7-H2 and B7-H3 are co-stimulatory and co-inhibitory molecules, respectively (20, 22). The identification of their correlation suggests that neutralization/dampening of immune activation ensue as a result of the interaction of these molecules. In addition, it is also entirely possible that in the liver tumor microenvironment both of these pathways cumulatively promote T-cell exhaustion/tolerance similar to CD28 and PD-1 (45). Hence, these observations propose that blocking of the inhibitory pathways in HCC microenvironment will not only negate the immunosuppressive signals but also boost the functions of endogenous co-stimulatory molecules, which collectively revive exhausted/tolerized T cells.
B7-H5 plays an important role in the generation of inducible FoxP3+ regulatory T cells through TGF-β (46). Similarly, PD-1 and B7-H5 cumulatively attenuate T cell responses (47). Recent evidences suggest that HCC tumor microenvironment is enriched with Foxp3+CD4+ and PD-1+CD8+ T cells which together provide an extensive immunosuppressive microenvironment (48, 49). In addition, reduction in PD-1 and Foxp3 has been shown to be a predictor of the survival of HCC patients (50). Our observation of the correlation of B7-H5 with TGF-β and PD-1 add to these previous findings that B7-H5 plays an important role in the origin of intrahepatic regulatory T cells and in association with PD1+ T cells, it dampens anti-HCC T cell immunity.
IDO upregulation and its role in immunosuppression have been well established in generic and intrahepatic tumor immunity (51–53). TDO has a predominant homeostatic housekeeping role specific to liver (54), and the observation that TDO overexpression above IDO is due to the indigenous nature of the liver microenvironment. However, none of the B7 family molecules correlate with TDO expression in the liver while IDO shows a correlation with PD-L1 stronger than any other B7 family molecules. Correlation of IDO and PD-L1 has been shown in other tumors (55–57). Preclinical animal model studies also demonstrated that immune checkpoint blocking strategies with IDO targeting show an enhanced anti-tumor responses (58, 59). Our functional investigation on the effect of PD-L1 and IDO metabolite Kynurenine on T cell responses has demonstrated that both Kynurenine and PD-L1 exhibit synergistic inhibitory effect on T cells. Thus, co-blocking of IDO and PD-L1 is a promising strategy to promote anti-HCC T-cell immunity within the liver. However, although several clinical trials are aiming to test combined immune checkpoint blockade agents to revive anti-tumor immunity, some did not show efficacy. For example, a large phase III clinical trial investigating the combined checkpoint blockade of IDO and PD-L1 did not show efficacy in melanoma patients (60). This suggests that additional immune suppression persists beyond IDO and PD-L1 which neutralizes anti-tumor immunity. We have identified additional immunomodulatory pathways that coexist beyond IDO and PD-L1 suggesting that HCC tumor microenvironment is complex. Further mechanistic and functional studies are warranted to identify which pathways are dominant and superior over others in reviving anti-HCC immunity.
Granzyme B is a tumorlytic effector enzyme, which is predominantly used by cytotoxic lymphocytes and natural killer cells to lyse tumor cells (61, 62). In addition, Granzyme B+ lymphocytes were shown to be significantly associated with improved survival of HCC patients (63). We observed a correlation of Granzyme B with PD-L2 and IDO. IDO plays an important role in conferring adaptive tumor resistance to immune lysis since animal model studies have demonstrated that sole blocking of PD-1 or CTLA4 pathway enhances the IDO mediated tumor resistance. However, co-blocking of IDO and PD-1/CTLA4 significantly abolish tumor progression (58, 59). Similarly HCC patients treated with single immune checkpoint blocking antibodies to PD-1 or PDL-1 are always not responsive to therapy (40, 41). Thus, we speculate that although Granzyme B is tumorlytic, both PD-L2 and IDO may override and neutralize Granzyme B+ lymphocyte activity. This also proposes that both PDL1 and IDO may need to be targeted to enhance Granzyme B tumorlytic activity. Altogether, our data suggest that co-blocking of IDO and B7 family is a promising strategy to promote anti-HCC T-cell immunity without adaptive tumor resistance.
A limitation of our present study is the investigation of the correlation of B7 family molecules, Tryptophan degrading enzymes and immune effectors only at RNA but not protein expression levels. Although significance of our approach is the identification of correlation pattern by combination analysis, such an approach at protein levels is laborious and often limited with the sensitivity to demonstrate correlation. Nevertheless, our results were supported by previous studies which demonstrated that some B7 family molecules such as PD-L1 (64, 65), PD-L2 (66), PD-1 (67), B7-H2 (68), B7-H3 (69) and IDO (70) play a significant functional role in modulating anti-HCC immunity. Although we have demonstrated the synergistic functions of IDO and PD-L1, future studies are necessary to determine if the other identified correlation of costimulatory and coinhibitory molecules play a functional synergistic role in modulating anti-HCC immunity.
Liver is a tolerogenic organ rich in parenchymal and non-parenchymal cell types such as Hepatocytes, Kupffer Cells, Liver Sinusoidal Endothelial Cells, Hepatic Dendritic Cells, Hepatic Stellate Cells, Mesenchymal Stromal Cells and Hepatic B cells that constantly interact with T cells in order to execute immune tolerance. Upon injury, the resting status of these intrahepatic antigen presenting cells gets compromised leading to inflammation followed by fibrosis/cirrhosis. Eventually, immune suppression prevails that leads to HCC and significance of these cell types in HCC progression is important. Our earlier study has demonstrated that Hepatic Stellate Cells upregulate PD-L1 and IDO by IFNγ while blocking of IDO activity completely abolishes their immunosuppressive potential (71). Similarly, Kupffer cells in HCC were shown to execute immune suppression through PD-L1-PD-1 pathways (72). In the present study, we observed a correlation between PD-L2 and IFNγ. Future studies are warranted to define the coexpression, regulation and functions of B7 family molecules and Tryptophan degrading enzymes in intrahepatic APCs derived from HCC microenvironment. In conclusion, our study provided evidence that co-stimulatory and co-inhibitory molecules exhibit specific correlation pattern among themselves, with IDO and immune responsive effector molecules. This information is important to inform second-generation immunotherapy approaches for liver cancer management.
All datasets generated for this study are included in the article/supplementary material.
The studies involving human participants were reviewed and approved by Emory University IRB. The ethics committee waived the requirement of written informed consent for participation.
RC conceived, designed the research plan, performed most experiments, analyzed results, and wrote the manuscript. RS performed experiments related to flow cytometer and ELISA. OA, DR, AF, and BE provided patient materials. DA and GG helped with qPCR array. All authors contributed to the article and approved the submitted version.
This study was supported by the Winship Cancer Institute's Gala Scholar Award to RC. RC was partly supported by the WES Leukemia Research Foundation. This research was also supported by Mercer University School of Medicine's research funds (RC) and generous support from the Landings Women's Golf Association (Savannah, GA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Suresh Ramalingam (Professor, Emory University) for advice in this project. We also thank Dr. Jacques Galipeau (Professor, University of Wisconsin-Madison) and Dr. Devi Rajan (Instructor, Mercer University School of Medicine, Georgia) for critically reading the manuscript.
1. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. (2016) 2:16018. doi: 10.1038/nrdp.2016.18
2. Kudo M. Immuno-oncology in hepatocellular carcinoma: 2017 update. Oncology. (2017) 93 (Suppl. 1):147–59. doi: 10.1159/000481245
3. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. (2012) 366:2455–65. doi: 10.1056/NEJMoa1200694
4. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. (2015) 27:450–61. doi: 10.1016/j.ccell.2015.03.001
5. Komiya T, Huang CH. Updates in the clinical development of epacadostat and other indoleamine 2,3-dioxygenase 1 inhibitors (IDO1) for human cancers. Front Oncol. (2018) 8:423. doi: 10.3389/fonc.2018.00423
6. Gunther J, Dabritz J, Wirthgen E. Limitations and off-target effects of tryptophan-related IDO inhibitors in cancer treatment. Front Immunol. (2019) 10:1801. doi: 10.3389/fimmu.2019.01801
7. Labadie BW, Bao R, Luke JJ. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the tryptophan-kynurenine-aryl hydrocarbon axis. Clin Cancer Res. (2019) 25:1462–71. doi: 10.1158/1078-0432.CCR-18-2882
8. Vacchelli E, Aranda F, Eggermont A, Sautes-Fridman C, Tartour E, Kennedy EP, et al. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. (2014) 3:e957994. doi: 10.4161/21624011.2014.957994
9. El Dika I, Khalil DN, Abou-Alfa GK. Immune checkpoint inhibitors for hepatocellular carcinoma. Cancer. (2019) 125:3312–9. doi: 10.1002/cncr.32076
10. Keenan BP, Fong L, Kelley RK. Immunotherapy in hepatocellular carcinoma: the complex interface between inflammation, fibrosis, and the immune response. J Immunother Cancer. (2019) 7:267. doi: 10.1186/s40425-019-0749-z
11. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. (2017) 389:2492–502. doi: 10.1016/S0140-6736(17)31046-2
12. Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. (2018) 19:940–52. doi: 10.1016/S1470-2045(18)30351-6
13. Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity. (2016) 44:955–72. doi: 10.1016/j.immuni.2016.05.002
14. Ni L, Dong C. New B7 family checkpoints in human cancers. Mol Cancer Ther. (2017) 16:1203–11. doi: 10.1158/1535-7163.MCT-16-0761
15. Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. (2009) 10:29–37. doi: 10.1038/ni.1679
16. Popovic A, Jaffee EM, Zaidi N. Emerging strategies for combination checkpoint modulators in cancer immunotherapy. J Clin Invest. (2018) 128:3209–18. doi: 10.1172/JCI120775
17. Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining 'T cell exhaustion'. Nat Rev Immunol. (2019) 19:665–74. doi: 10.1038/s41577-019-0221-9
18. Kurachi M. CD8(+) T cell exhaustion. Semin Immunopathol. (2019) 41:327–37. doi: 10.1007/s00281-019-00744-5
19. Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. (2016) 375:1767–78. doi: 10.1056/NEJMra1514296
20. Mak TW, Shahinian A, Yoshinaga SK, Wakeham A, Boucher LM, Pintilie M, et al. Costimulation through the inducible costimulator ligand is essential for both T helper and B cell functions in T cell-dependent B cell responses. Nat Immunol. (2003) 4:765–72. doi: 10.1038/ni947
21. Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol. (2018) 11:39. doi: 10.1186/s13045-018-0582-8
22. Prasad DV, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. (2003) 18:863–73. doi: 10.1016/S1074-7613(03)00147-X
23. Flem-Karlsen K, Fodstad O, Tan M, Nunes-Xavier CE. B7-H3 in cancer - beyond immune regulation. Trends Cancer. (2018) 4:401–4. doi: 10.1016/j.trecan.2018.03.010
24. Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. (2003) 18:849–61. doi: 10.1016/S1074-7613(03)00152-3
25. Macgregor HL, Ohashi PS. Molecular pathways: evaluating the potential for B7-H4 as an immunoregulatory target. Clin Cancer Res. (2017) 23:2934–41. doi: 10.1158/1078-0432.CCR-15-2440
26. Zhu Y, Yao S, Iliopoulou BP, Han X, Augustine MM, Xu H, et al. B7-H5 costimulates human T cells via CD28H. Nat Commun. (2013) 4:2043. doi: 10.1038/ncomms3043
27. Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O'connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. (2014) 74:1924–32. doi: 10.1158/0008-5472.CAN-13-1504
28. Schlecker E, Fiegler N, Arnold A, Altevogt P, Rose-John S, Moldenhauer G, et al. Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res. (2014) 74:3429–40. doi: 10.1158/0008-5472.CAN-13-3017
29. Janakiram M, Chinai JM, Fineberg S, Fiser A, Montagna C, Medavarapu R, et al. Expression, clinical significance, and receptor identification of the newest B7 family member HHLA2 protein. Clin Cancer Res. (2015) 21:2359–66. doi: 10.1158/1078-0432.CCR-14-1495
30. Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. (2013) 34:137–43. doi: 10.1016/j.it.2012.10.001
31. Prendergast GC, Mondal A, Dey S, Laury-Kleintop LD, Muller AJ. Inflammatory reprogramming with IDO1 inhibitors: turning immunologically unresponsive ‘cold' tumors ‘hot'. Trends Cancer. (2018) 4:38–58. doi: 10.1016/j.trecan.2017.11.005
32. Lemos H, Huang L, Prendergast GC, Mellor AL. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat Rev Cancer. (2019) 19:162–75. doi: 10.1038/s41568-019-0106-z
33. Garber K. A new cancer immunotherapy suffers a setback. Science. (2018) 360:588. doi: 10.1126/science.360.6389.588
34. Chinnadurai R, Copland IB, Ng S, Garcia M, Prasad M, Arafat D, et al. Mesenchymal stromal cells derived from Crohn's patients deploy indoleamine 2,3-dioxygenase-mediated immune suppression, independent of autophagy. Mol Ther. (2015) 23:1248–61. doi: 10.1038/mt.2015.67
35. Chinnadurai R, Rajan D, Qayed M, Arafat D, Garcia M, Liu Y, et al. Potency analysis of mesenchymal stromal cells using a combinatorial assay matrix approach. Cell Rep. (2018) 22:2504–17. doi: 10.1016/j.celrep.2018.02.013
36. Chinnadurai R, Garcia MA, Sakurai Y, Lam WA, Kirk AD, Galipeau J, et al. Actin cytoskeletal disruption following cryopreservation alters the biodistribution of human mesenchymal stromal cells in vivo. Stem Cell Reports. (2014) 3:60–72. doi: 10.1016/j.stemcr.2014.05.003
37. Sun T, Li P, Sun D, Bu Q, Li G. Prognostic value of osteopontin in patients with hepatocellular carcinoma: A systematic review and meta-analysis. Medicine. (2018) 97:e12954. doi: 10.1097/MD.0000000000012954
38. Nishida T, Kataoka H. Glypican 3-targeted therapy in hepatocellular carcinoma. Cancers. (2019) 11:1339. doi: 10.3390/cancers11091339
39. Guo M, Zhang H, Zheng J, Liu Y. Glypican-3: a new target for diagnosis and treatment of hepatocellular carcinoma. J Cancer. (2020) 11:2008–21. doi: 10.7150/jca.39972
40. Giannini EG, Aglitti A, Borzio M, Gambato M, Guarino M, Iavarone M, et al.). Overview of immune checkpoint inhibitors therapy for hepatocellular carcinoma, and the ITA.LI.CA cohort derived estimate of amenability rate to immune checkpoint inhibitors in clinical practice. Cancers. (2019) 11:1689. doi: 10.3390/cancers11111689
41. Mahipal A, Tella SH, Kommalapati A, Lim A, Kim R. Immunotherapy in hepatocellular carcinoma: is there a light at the end of the tunnel? Cancers. (2019) 11:1078. doi: 10.3390/cancers11081078
42. Xu Z, Shen J, Wang MH, Yi T, Yu Y, Zhu Y, et al. Comprehensive molecular profiling of the B7 family of immune-regulatory ligands in breast cancer. Oncoimmunology. (2016) 5:e1207841. doi: 10.1080/2162402X.2016.1207841
43. Ghiotto M, Gauthier L, Serriari N, Pastor S, Truneh A, Nunes JA, et al. PD-L1 and PD-L2 differ in their molecular mechanisms of interaction with PD-1. Int Immunol. (2010) 22:651–60. doi: 10.1093/intimm/dxq049
44. Zak KM, Grudnik P, Magiera K, Domling A, Dubin G, Holak TA. Structural biology of the immune checkpoint receptor PD-1 and its ligands PD-L1/PD-L2. Structure. (2017) 25:1163–74. doi: 10.1016/j.str.2017.06.011
45. Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. (2017) 355:1423–7. doi: 10.1126/science.aaf0683
46. Wang Q, He J, Flies DB, Luo L, Chen L. Programmed death one homolog maintains the pool size of regulatory T cells by promoting their differentiation and stability. Sci Rep. (2017) 7:6086. doi: 10.1038/s41598-017-06410-w
47. Liu J, Yuan Y, Chen W, Putra J, Suriawinata AA, Schenk AD, et al. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc Natl Acad Sci USA. (2015) 112:6682–7. doi: 10.1073/pnas.1420370112
48. Chew V, Lai L, Pan L, Lim CJ, Li J, Ong R, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci USA. (2017) 114:E5900–9. doi: 10.1073/pnas.1706559114
49. Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. (2017) 169:1342–56 e1316. doi: 10.1016/j.cell.2017.05.035
50. Kalathil SG, Lugade AA, Miller A, Iyer R, Thanavala Y. PD-1(+) and Foxp3(+) T cell reduction correlates with survival of HCC patients after sorafenib therapy. JCI Insight. (2016) 2016:e86182. doi: 10.1172/jci.insight.86182
51. Johnson TS, Munn DH. Host indoleamine 2,3-dioxygenase: contribution to systemic acquired tumor tolerance. Immunol Invest. (2012) 41:765–97. doi: 10.3109/08820139.2012.689405
52. Han Y, Chen Z, Yang Y, Jiang Z, Gu Y, Liu Y, et al. Human CD14+ CTLA-4+ regulatory dendritic cells suppress T-cell response by cytotoxic T-lymphocyte antigen-4-dependent IL-10 and indoleamine-2,3-dioxygenase production in hepatocellular carcinoma. Hepatology. (2014) 59:567–79. doi: 10.1002/hep.26694
53. Mukhopadhyay B, Schuebel K, Mukhopadhyay P, Cinar R, Godlewski G, Xiong K, et al. Cannabinoid receptor 1 promotes hepatocellular carcinoma initiation and progression through multiple mechanisms. Hepatology. (2015) 61:1615–26. doi: 10.1002/hep.27686
54. Platten M, Wick W, Van Den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. (2012) 72:5435–40. doi: 10.1158/0008-5472.CAN-12-0569
55. Dill EA, Dillon PM, Bullock TN, Mills AM. IDO expression in breast cancer: an assessment of 281 primary and metastatic cases with comparison to PD-L1. Mod Pathol. (2018) 31:1513–22. doi: 10.1038/s41379-018-0061-3
56. Mills AM, Peres LC, Meiss A, Ring KL, Modesitt SC, Abbott SE, et al. Targetable immune regulatory molecule expression in high-grade serous ovarian carcinomas in african american women: a study of PD-L1 and IDO in 112 cases from the African American Cancer Epidemiology Study (AACES). Int J Gynecol Pathol. (2018) 38:157–70. doi: 10.1097/PGP.0000000000000494
57. Rosenberg AJ, Wainwright DA, Rademaker A, Galvez C, Genet M, Zhai L, et al. Indoleamine 2,3-dioxygenase 1 and overall survival of patients diagnosed with esophageal cancer. Oncotarget. (2018) 9:23482–93. doi: 10.18632/oncotarget.25235
58. Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J Exp Med. (2013) 210:1389–402. doi: 10.1084/jem.20130066
59. Brown ZJ, Yu SJ, Heinrich B, Ma C, Fu Q, Sandhu M, et al. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol Immunother. (2018) 67:1305–15. doi: 10.1007/s00262-018-2190-4
60. Long GV, Dummer R, Hamid O, Gajewski T, Caglevic C, Dalle S, et al. Epacadostat. (E) plus pembrolizumab. (P) versus pembrolizumab alone in patients. (pts) with unresectable or metastatic melanoma: Results of the phase 3 ECHO-301/KEYNOTE-252 study. J Clin Oncol. (2018) 36:108–108. doi: 10.1016/S1470-2045(19)30274-8
61. Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. (2015) 15:388–400. doi: 10.1038/nri3839
62. Durgeau A, Virk Y, Corgnac S, Mami-Chouaib F. Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front Immunol. (2018) 9:14. doi: 10.3389/fimmu.2018.00014
63. Ding W, Xu X, Qian Y, Xue W, Wang Y, Du J, et al. Prognostic value of tumor-infiltrating lymphocytes in hepatocellular carcinoma: a meta-analysis. Medicine. (2018) 97:e13301. doi: 10.1097/MD.0000000000013301
64. Long J, Qu T, Pan XF, Tang X, Wan HH, Qiu P, et al. Expression of programmed death ligand-1 and programmed death 1 in hepatocellular carcinoma and its clinical significance. J Cancer Res Ther. (2018) 14:S1188–92. doi: 10.4103/0973-1482.204850
65. Nelson A. The social life of DNA: racial reconciliation and institutional morality after the genome - a response. Br J Sociol. (2018) 69:575–9. doi: 10.1111/1468-4446.12612
66. Liao H, Chen W, Dai Y, Richardson JJ, Guo J, Yuan K, et al. Expression of programmed cell death-ligands in hepatocellular carcinoma: correlation with immune microenvironment and survival outcomes. Front Oncol. (2019) 9:883. doi: 10.3389/fonc.2019.00883
67. Kim HD, Song GW, Park S, Jung MK, Kim MH, Kang HJ, et al. Association between expression level of PD1 by tumor-infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. (2018) 155:1936–50 e1917. doi: 10.1053/j.gastro.2018.08.030
68. Zheng Y, Liao N, Wu Y, Gao J, Li Z, Liu W, et al. High expression of B7H2 or B7H3 is associated with poor prognosis in hepatocellular carcinoma. Mol Med Rep. (2019) 19:4315–25. doi: 10.3892/mmr.2019.10080
69. Sun TW, Gao Q, Qiu SJ, Zhou J, Wang XY, Yi Y, et al. B7-H3 is expressed in human hepatocellular carcinoma and is associated with tumor aggressiveness and postoperative recurrence. Cancer Immunol Immunother. (2012) 61:2171–82. doi: 10.1007/s00262-012-1278-5
70. Wang Y, Yao R, Zhang L, Xie X, Chen R, Ren Z. IDO and intra-tumoral neutrophils were independent prognostic factors for overall survival for hepatocellular carcinoma. J Clin Lab Anal. (2019) 33:e22872. doi: 10.1002/jcla.22872
71. Chinnadurai R, Sands J, Rajan D, Liu X, Arafat D, Das R, et al. Molecular genetic and immune functional responses distinguish bone marrow mesenchymal stromal cells from hepatic stellate cells. Stem Cells. (2019) 37:1075–82. doi: 10.1002/stem.3028
Keywords: B7 family molecules, indoleamine 2,3 dioxygenase, hepatocellular carcinoma, T cell immunity, co-stimulation, co-inhibition, PD-L1
Citation: Chinnadurai R, Scandolara R, Alese OB, Arafat D, Ravindranathan D, Farris AB, El-Rayes BF and Gibson G (2020) Correlation Patterns Among B7 Family Ligands and Tryptophan Degrading Enzymes in Hepatocellular Carcinoma. Front. Oncol. 10:1632. doi: 10.3389/fonc.2020.01632
Received: 25 November 2019; Accepted: 27 July 2020;
Published: 03 September 2020.
Edited by:
Lieve Brochez, Cancer Research Institute, Ghent University, BelgiumReviewed by:
William J. Magner, University at Buffalo, United StatesCopyright © 2020 Chinnadurai, Scandolara, Alese, Arafat, Ravindranathan, Farris, El-Rayes and Gibson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raghavan Chinnadurai, Y2hpbm5hZHVyYWlfckBtZXJjZXIuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.