
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 12 August 2020
Sec. Cancer Genetics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.01627
 Xiaohan Dong1†
Xiaohan Dong1† Shanshan Ding1†
Shanshan Ding1† Miao Yu2
Miao Yu2 Limin Niu1
Limin Niu1 Linlin Xue1
Linlin Xue1 Yajing Zhao1
Yajing Zhao1 Li Xie1
Li Xie1 Xingguo Song3*
Xingguo Song3* Xianrang Song1*
Xianrang Song1*Background: Small nuclear RNA (snRNA) levels are extremely variable across a wide range of biological conditions. SnRNAs could potentially regulate alternative splicing to drive genetic, dysplastic and neoplastic disease, which might be the main reason for mRNA profile alteration in tumor educated platelets (TEPs).
Methods: Platelets were isolated from the plasma of lung cancer patients and healthy donors by low-speed centrifugation and subjected to RNA isolation. SnRNA U1, U2, U5 levels were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Exosomes were isolated by ultracentrifugation and identified by qNano.
Results: TEP U1, U2, U5 levels were significantly decreased in patients with lung cancer as well as with early stage patients, their downregulation was correlated with lung cancer progression, possessing favorable diagnostic efficiency. More importantly, TEP U1, U2 and U5 levels were closely correlated between paired exosomes and TEP from treated patients but not from untreated ones, and U1, U5 but not U2 in platelets were elevated by apo-exosomes.
Conclusion: Tumor educated platelet small nuclear RNAs are downregulated and act as promising biomarkers in lung cancer.
Lung cancer is the most common cause of cancer-related death worldwide, with an average 5-year survival rate of 15% (1). Unfortunately, symptoms of lung cancer are obscured until the patients reach the end stage of the disease, and approximately two−thirds of patients present with metastatic tumors at the time of diagnosis (2), reflecting the urgent need to develop the reliable biomarkers for the early diagnosis of lung cancer and the monitoring of its spatial and temporal progression.
Platelets, originating as anucleate cytoplasts from megakaryocytes (3), play crucial roles not only in hemostasis, but also in systemic and local responses to the presence of cancer (4), thereby sequestering tumor-specified biomolecules including RNA transcripts and proteins, as well as altering their spliced RNA profiles, called “tumor educated platelets (TEPs)” (5). Recently, TEPs emerged as non-invasive biomarker source for cancer detection and progression monitoring including colorectal carcinoma (CRC) (6), glioblastoma (7), non–small cell lung cancer (NSCLC) (8), prostate cancer (9) and etc., due to their significantly altered RNA profiles. Moreover, RNA profiles in TEPs enabled to discriminate patients with localized and metastasized cancer from healthy controls, even precisely pinpoint the primary origin of pan-cancer, as well as predict the oncogenic status including MET or HER2 positivity and the existence of KRAS, EGFR, or PIK3CA mutations (10). Due to the lifespan of 7–10 days and the structure of platelet membrane, tumor-specified biosources and biomolecules are enriched in the TEPs and protected from circulating RNAs, and thus TEP RNA analysis is capable to reflect tumor bioactivity up-to-date, intensively, and dynamically.
Platelet lacks a nucleus, thus no genomic DNA is available for transcription of new RNA molecules, but endogenous pre-mRNAs can be spliced into mature mRNAs (11). Specific splice events in TEPs may be induced by external stimuli such as activation of platelet surface receptors, or in response to signals released by cancer cells and the tumor microenvironment, endowing TEPs with a highly dynamic mRNA repertoire with potential applicability to cancer diagnostics (12). It has been reported mRNA profiles were significantly altered in TEPs of lung cancer, with specific spliced mRNA signature (8, 13). Spliceosome is a complex of small ribonucleoproteins (snRNPs) composed of splicing proteins and snRNAs including U1, U2, U4, U5, and U6, all of which are present in platelet (14). SnRNAs of the spliceosome are not merely the basal factors as has long been assumed, ubiquitously expressed in all cells since they are required for splicing, whereas snRNA levels are extremely variable across a wide range of biological conditions, even across all samples taken from breast ductal carcinoma (15). Endogenous variation in snRNAs could potentially enable these RNAs to regulate alternative splicing to drive genetic, dysplastic and neoplastic disease (15, 16). Therefore, we supposed snRNA levels in TEPs of lung cancer were varied from those in healthy individuals, which might the main reason for alteration of TEP mRNA profiles, exerting the potential as the biomarker for lung cancer diagnostics.
In the present study, we aimed to validate whether TEP snRNAs served as the potential biomarkers for lung cancer. We found that TEP U1, U2, U5 were significantly downregulated in lung cancer patients compared with those in healthy controls, possessing with the favorable diagnostic efficiency. Moreover, we observed that relative snRNAs expression was elevated after anticancer therapy, which seemed to attribute to apo-exosomes (apoptotic exosomes)-dependent horizontal transmission, suggesting that aberrant expression of TEP U1, U2, U5 act as novel biomarkers for lung cancer.
A total of 382 lung cancer patients without any anticancer treatment and 23 patients with initial treatment from Shandong Cancer Hospital and Institute, were enrolled between January 2019 to November 2019. Cancer diagnosis was determined through a histological examination of tumor specimens, and the tumor stage was determined according to the 8th edition of the lung cancer TNM staging standards formulated by IASLC; 156 healthy volunteers from the above hospital and 205 from Shandong Provincial Third Hospital were excluded from any malignant tumor after examination and enrolled in this study. All participants gave their informed consents for specimen collection and clinical information collection.
The human NSCLC-derived cell line A549 was obtained from China Center for Type Culture Collection (Wuhan, China), and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 U ml–1 penicillin and streptomycin (Gibco) at 37°C in a humidified 5% CO2 incubator.
Platelet isolation was performed as described previously (17). In brief, peripheral blood was collected in EDTAK2-coated purple-cap Vacutainer tubes (BD, Franklin Lakes, NJ, United States), and centrifuged at 120 × g for 10 min twice to separated platelet-rich plasma (PRP) from nucleated blood cells, followed by another 360 × g centrifugation for 20 min to pellet platelets at room temperature.
Total RNA was extracted with the TRIzol reagent and reverse−transcribed into complementary DNA (cDNA) using the Takara PrimeScript RT reagent Kit (Takara Bio, Kusatsu, Japan) in 20 μl reaction according to the manufacturer’s instructions. The qRT-PCR was performed using UltraSYBR Mixture (CWBio, Beijing, China) in a volume of 25 μl (12.5 μl of 2 × UltraSYBR Mixture, 0.5 μl of forward premier, 0.5 μl of reverse premier, 1 μl cDNA and 10.5 μl water). The reactions were performed with LightCycler 480 qPCR system (Roche Diagnostics, Germany). 18S acted as control as described previously (18). The relative expression of snRNAs was evaluated by the comparative cycle threshold (ΔCt) method: (ΔCt = CtsnRNA – Ct18S) as described previously (19). The qPCR primers are listed in Table 1.
Exosomes isolation was performed as described previously (20). Briefly, the platelet-free plasma was centrifuged at 12,000 g for 30 min at 4°C to exclude the cell debris, then subjected to a 0.22 μm pore filter (Millipore), followed by ultracentrifugation at 100,000 × g for 120 min at 4°C to harvest exosomes (Beckman Coulter, Brea, CA, United States). Isolated exosomes were resuspended with cold phosphate-buffered saline (PBS) and identified using qNano (Izon Science Ltd., Christchurch, New Zealand).
A549 cells were seeded in 100 mm dishes in exosome-depleted DMEM with 10% exosome-depleted FBS, treated with or without 50 μM cisplatin (Sigma-Aldrich, Shanghai, China) for 24 h. Then total 50 ml contained media were collected for exosome isolation. The amount of exosomes was measured by the BCA Protein Assay kit (Thermo Scientific, CA, United States), 100 μg exosomes were collected for qPCR or for coincubation with platelets.
Platelets were isolated from healthy volunteers and transferred to a 15 ml centrifugal tube and counted under a microscopy after washing by 0.8% EDTA-PBS. Then, 1 × 109 platelets were suspended with 1 ml exosome- depleted medium and cultured with above 100 μg exosomes in EDTAK2-coated purple-cap Vacutainer tubes on a Platelet Oscillometer (L-Y70, Suzhou, China) for 4 h. After washing twice by 0.8% EDTA-PBS, platelets were collected and subjected to qPCR.
The data analyses were performed with GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA, United States) and SPSS 22.0 software (IBM, Ehningen, Germany). The Kolmogorov–Smirnov test was carried out to check the normality of the distribution. If the data followed normal analysis, unpaired t-test would be used; if not, Mann-Whitney test would be used. Chi-square test was used for analyzing categorical variables. Multi-group analysis was tested by Kruskal-Wallis test. In paired data, the normally distributed numeric variables were evaluated by paired t-test, whereas non-normally distributed variables were analyzed by Wilcoxon rank-test. Receiving operating characteristic (ROC) curve was used to evaluate diagnostic efficiency. For correlation, if the data followed normal analysis, Pearson correlation test would be used; if not, Spearman correlation test would be used. All the values were represented as mean ± interquartile range and p values <0.05 were defined as statistically significance, and all tests were set as double-tailed.
To screen out the differential snRNAs expression levels in lung cancer, 10 snRNAs expression including U1, U2, U4, U5, U6, U7, U8, U12, U4ATAC and U6ATAC was detected in TEPs from 48 healthy donors and 48 lung cancer patients (data not shown). U1, U2, U5 were identified due to their remarkable differences and then subjected to a larger scale validation cohort including 361 healthy donors and 382 lung cancer patients naive to any anticancer treatment.
First, Mann-Whitney test was performed to analyze the differential expression between lung cancer patients (n = 382) and healthy controls (n = 361). TEP U1, U2, U5 were significantly decreased in lung cancer patients compared with those in healthy controls (p < 0.0001, p < 0.0001 and p < 0.0001, respectively) (Figure 1A), indicting the variable expression of TEP snRNAs in lung cancer. The correlation between U1, U2, U5 expression and clinicopathological characteristics was also evaluated. As shown in Table 2, U1 and U5 were obviously associated with gender and smoking and T stage, whereas U1 was associated with drinking, U5 was related to histology type and tumor size; Moreover, all these three snRNAs were closely relevant to TNM stage and lymph node metastasis, both U1 and U2 were significantly associated with distant metastasis.
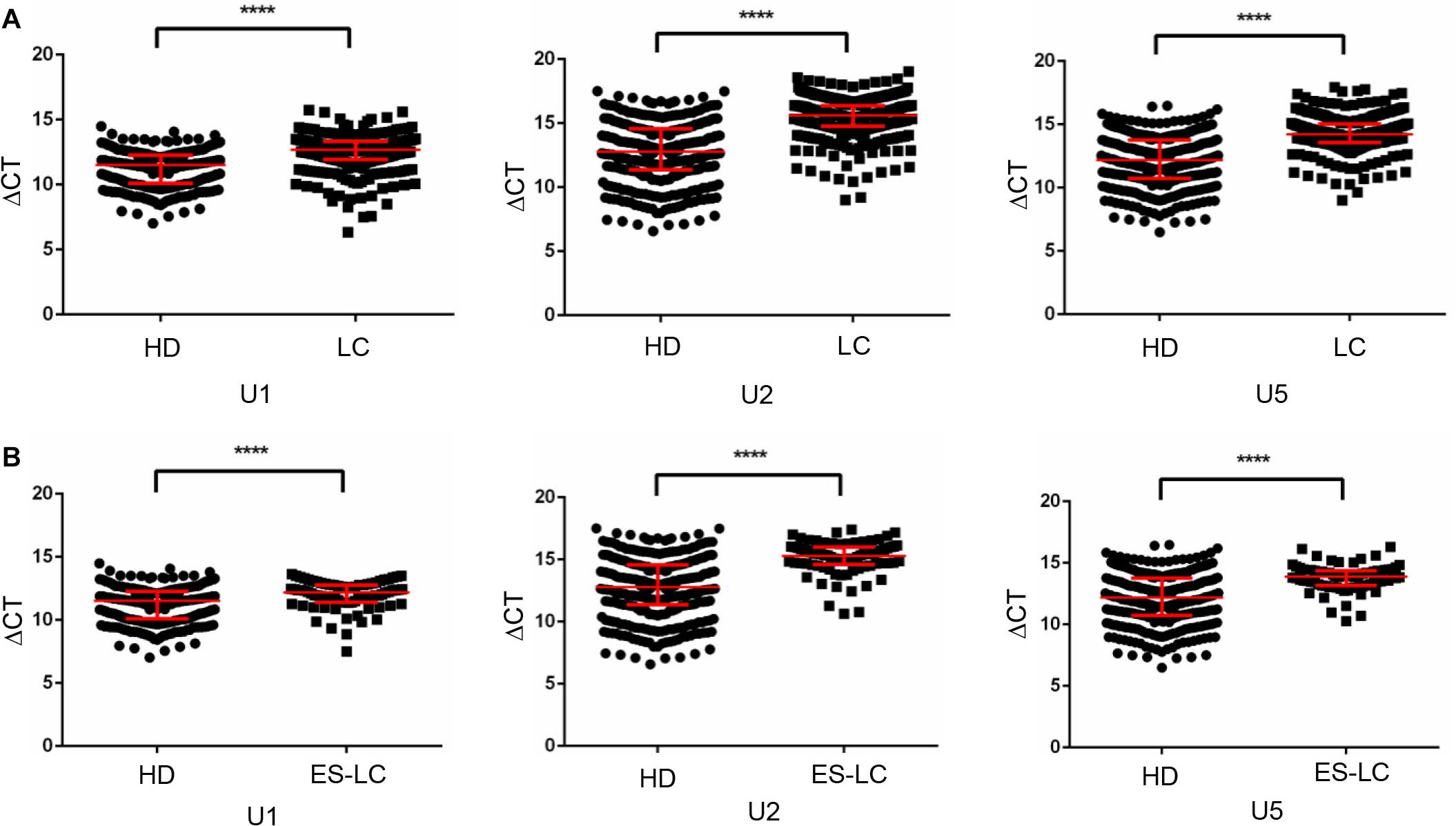
Figure 1. TEP U1, U2 and U5 were downregulated in lung cancer. (A) TEP U1, U2, U5 were down-regulated in lung cancer patients (n = 382) compared with healthy controls (n = 361); (B) TEP U1, U2, U5 were down-regulated in early stage lung cancer patients (n = 80) compared with healthy controls (n = 361). HD, healthy donors; LC, lung cancer patients; ES-LC, early stage lung cancer patients. ****p < 0.0001.
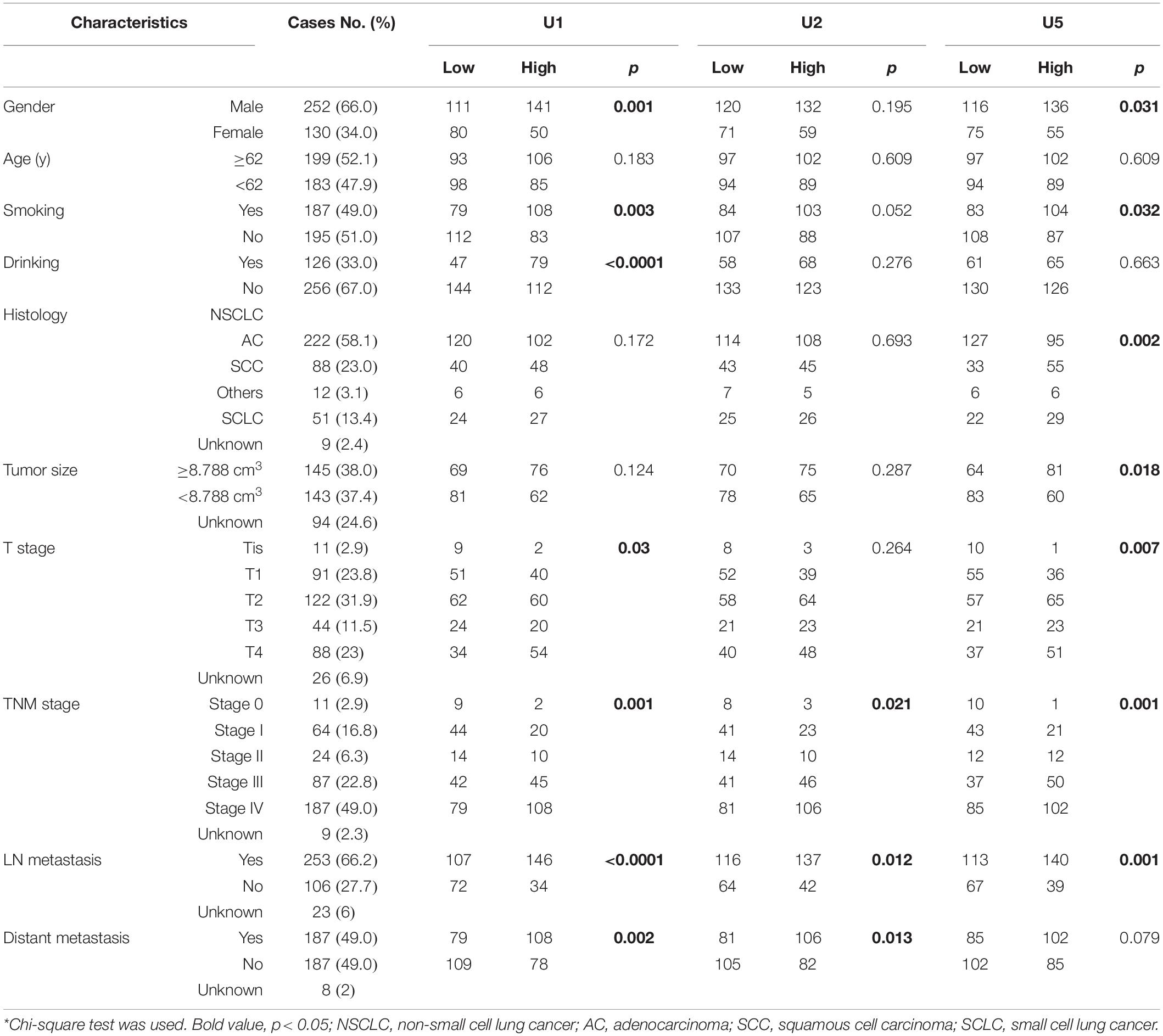
Table 2. Correlation between TEP U1, U2, U5 expression and clinicopathologic characteristics of lung cancer patients.
Then, we analyzed the differential expression of TEP U1, U2, U5 in 80 early stage lung cancer patients (Tis stage = 11, I stage = 64, IIA stage = 5) as well as in 361 healthy donors. Consistently, TEP U1, U2, U5 were also dramatically decreased in early stage lung cancer patients (p < 0.0001, p < 0.0001 and p < 0.0001, respectively) compared with those in healthy subjects (Figure 1B). Taken together, these data imply that TEP U1, U2, U5 might be involved in carcinogenesis and metastasis of lung cancer.
Furthermore, we analyzed the expression levels of TEP U1, U2, U5 among different stages of lung cancer patients and healthy donors. As shown in Figure 2A, when compared with the healthy donors, the TEP U1, U2, U5 expression was significantly down-regulated in various TNM stage. Next, we retrospectively analyzed the differential expression of TEP U1, U2, U5 in 80 early stage lung cancer patients (Tis stage = 11, I stage = 64, IIA stage = 5) and 293 advanced-stage lung cancer patients (IIB stage = 19, III stage = 87, IV stage = 187). As shown in Figure 2B, their expression was also significantly downregulated in advanced-stage lung cancer patients (p < 0.0001, p = 0.0013 and p < 0.0001, respectively) compared with those early stage lung cancer patients. Similarly, TEP U1, U2, U5 levels were much lower in lymph node metastasis while U1, U5 levels were lower in distant metastasis group than those in control group, respectively (Figures 2C,D). Collectively, our data support TEP U1, U2, U5 downregulation correlates with lung cancer progression.
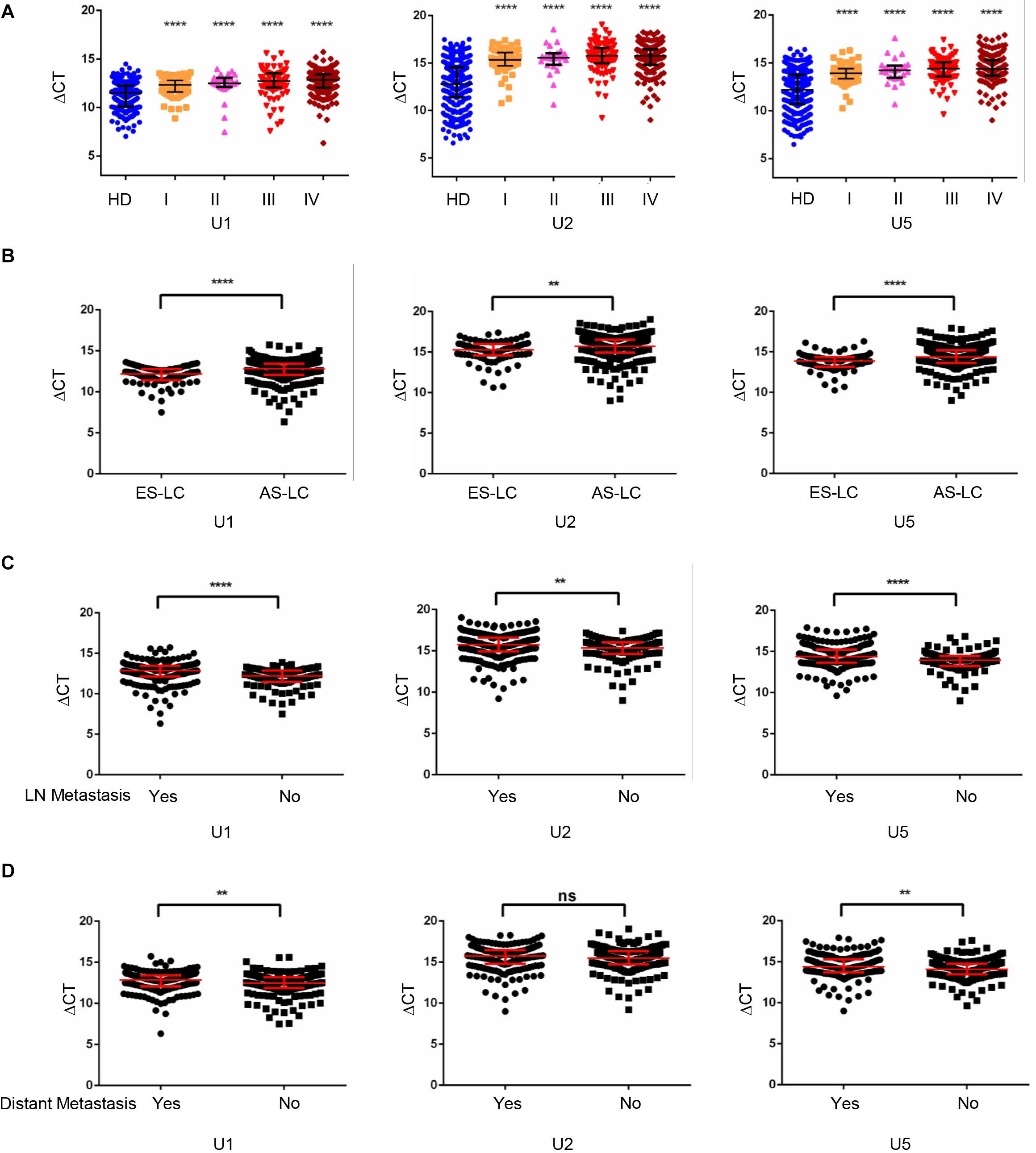
Figure 2. TEP U1, U2, U5 downregulation correlated with lung cancer progression. (A) TEP U1, U2, U5 were down-regulated in in various TNM stage; (B) TEP U1, U2, U5 were down-regulated in advanced-stage lung cancer patients (n = 274) compared with early-stage patients (n = 80); (C,D) TEP U1, U2, U5 expression was lower in lymph node metastasis (C) and distant metastasis (D) group than in control group. AS-LC, advanced-stage lung cancer patients; **p < 0.005; ****p < 0.0001; ns, no significance.
To explore the potentiality of TEP U1, U2, U5 as circulating diagnostic markers for lung cancer, ROC curves were employed. As shown in Figure 3A, the areas under the curve (AUCs) of TEP U1, U2 and U5 were 0.769 with 74.6% sensitivity and 66.5% specificity, 0.840 with 81.4% sensitivity and 74.2% specificity, and 0.809 with 90.1% sensitivity and 63.7% specificity, respectively. Moreover, the diagnostic efficiency of their combination was also calculated, possessing AUC of 0.840 with a relative sensitivity of 85.9% and a relative specificity of 70.1%, indicating TEP U1, U2 and U5 potentially act as the non-invasive circulating biomarkers for lung cancer.
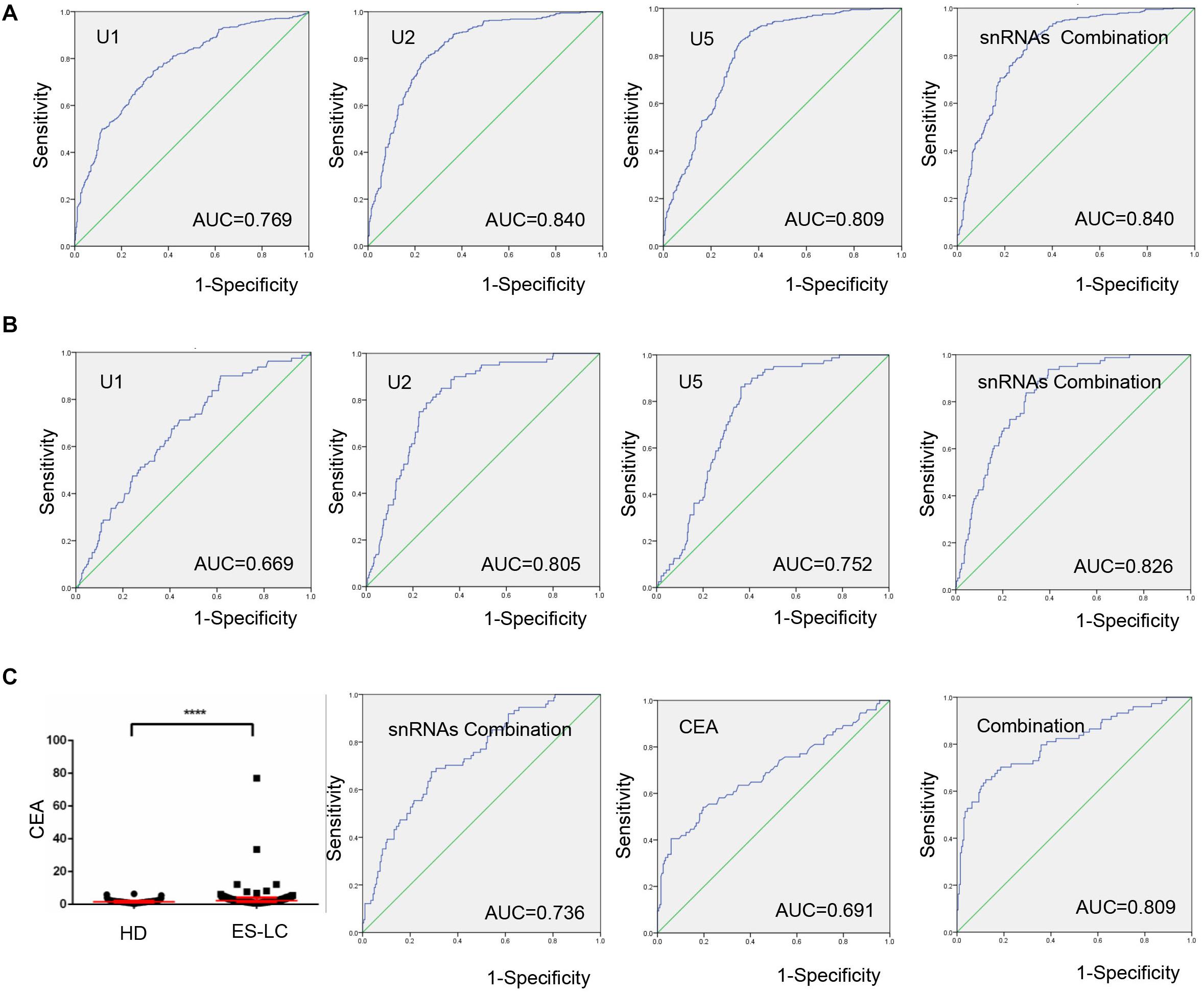
Figure 3. TEP U1, U2 and U5 as biomarkers for lung cancer diagnosis. (A) The ROC curve analysis of U1, U2, U5 and their combination for lung cancer; (B) The ROC curve analysis of U1, U2, U5 and their combination for early stage lung cancer; (C) The levels of CEA were increased in early stage lung cancer patients (n = 74) compared with healthy controls (n = 204). The ROC curve analysis of snRNAs combination, CEA and the combination of snRNAs and CEA (AUC = 0.809) for early stage lung cancer. AUC, area under curve; ****P < 0.0001.
Subsequently, when comparing the patients with early stage lung cancer to healthy controls, ROC curves demonstrated favorable diagnostic efficiencies of TEP U1, U2, U5, possessing AUCs of 0.669 with 90.0% sensitivity and 38.5% specificity, 0.805 with 78.8% sensitivity and 74.2% specificity, and 0.752 with 86.3% sensitivity and 63.7% specificity, respectively. Moreover, the diagnostic performance for their combination demonstrated the AUC of 0.826 with a relative sensitivity of 93.8% and a relative specificity of 60.7% (Figure 3B).
Carcinoembryonic antigen (CEA) is a widely used blood biomarker for lung cancer diagnosis since 1980 (21), but possesses the poor clinic diagnostic efficiency at the early stage of cancer development (22). We detected the CEA levels in 74 early stage lung cancer patients and 204 healthy subjects, which were obviously higher in early stage lung cancer patients (p < 0.0001) than those in healthy controls (Figure 3C). Importantly, combination of TEP U1, U2, U5 significantly facilitated the diagnostic efficiency for lung cancer of CEA, the AUC of CEA was elevated from 0.691 to 0.809 with a sensitivity of 64.9% and specificity of 87.7%. In summary, TEP U1, U2, U5 potentially act as biomarkers for lung cancer diagnosis.
Next, we investigated whether TEP U1, U2, U5 could serve as biomarkers to monitor lung cancer progression, and ROC curve was applied to analyze the diagnostic efficiency between early-stage and advanced-stage lung cancer patients. As shown in Figure 4A, AUCs of TEP U1, U2, U5 and their combination were 0.677 with 51.9% sensitivity and 76.25%specificity, 0.617 with 59.7% sensitivity and 62.5% specificity, and 0.655 with 44.7% sensitivity and 83.75% specificity, as well as 0.702 with 60.4% sensitivity of and 73.75% specificity, respectively, indicating that TEP U1, U2, U5 levels might be used to monitor lung cancer progression.
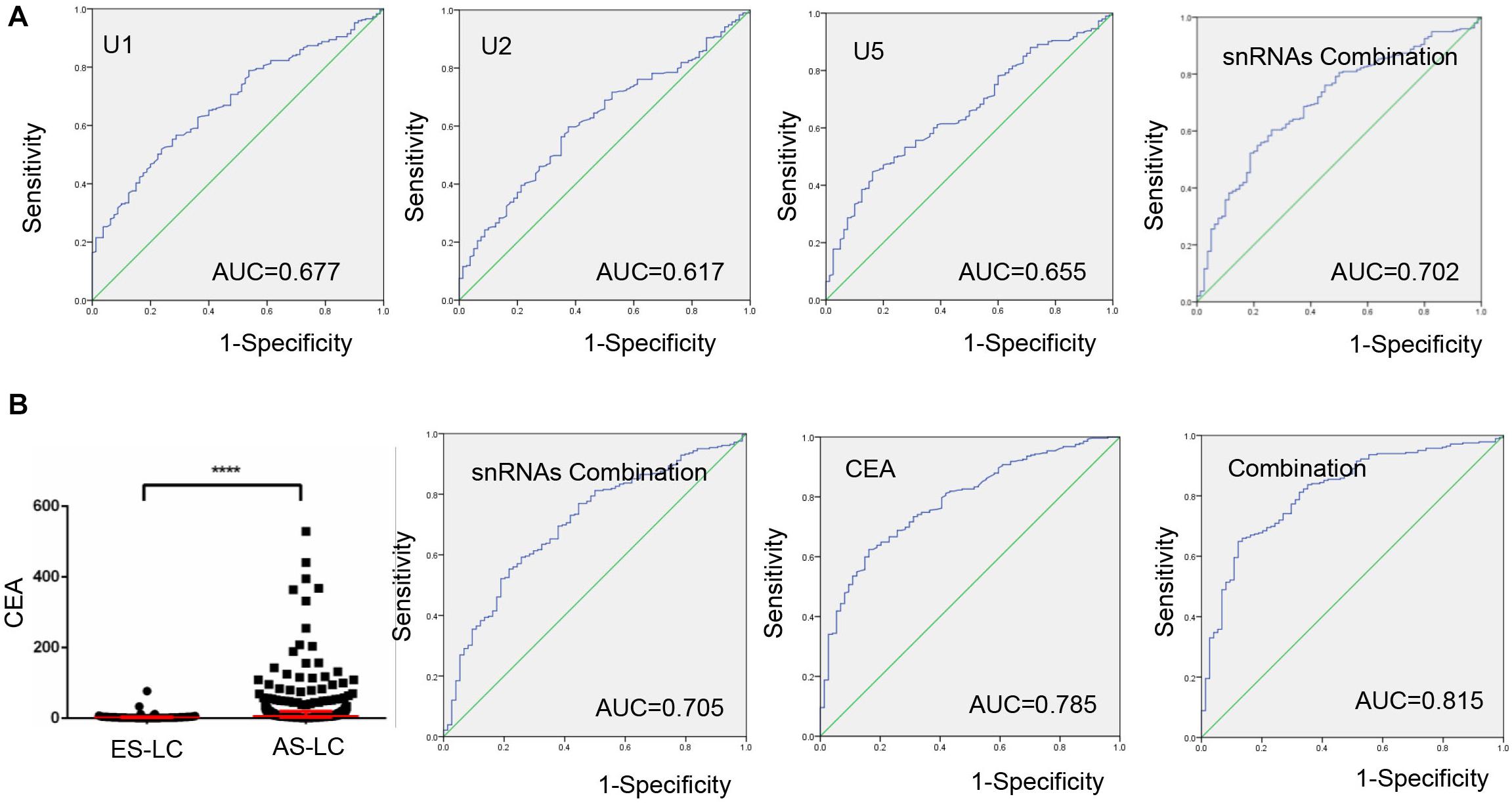
Figure 4. TEP U1, U2, U5 as biomarkers for lung cancer progression monitoring. (A) The ROC curve analysis of U1, U2, U5 and their combination for lung cancer progression; (B) The ROC curve analysis of CEA for lung cancer progression combined with U1, U2, U5; ****P < 0.0001.
We also combined TEP U1, U2, U5 with CEA to evaluate their roles in monitoring cancer progression. Consistent with previous reports, CEA level was upregulated in 282 advantaged-stage compared with that in 74 early-stage lung cancer (Figure 4B). Besides, we demonstrated that the combination of TEP U1, U2, U5 and CEA improved the diagnostic efficiency of cancer progression, the AUC was 0.815 with 64.9% sensitivity and 87.8% specificity, higher than that for CEA alone (AUC = 0.785, with 62.4% sensitivity and 83.8% specificity), indicating the potential roles of TEP U1, U2, U5 in lung cancer progression monitoring.
Increased platelet counts could accompany various cancers including lung cancer (3), revealed as a predictor for prognosis of lung cancer (23). Therefore, in current study, we divided lung cancer patients into high (n = 192) and low (n = 189) level groups based on their median value 266 × 109/L of platelet counts. As shown in Figure 5A, TEP U1, U2, U5 levels were dramatically downregulated in the patients with higher platelet counts (p < 0.0001, p < 0.0001 and p < 0.0001, respectively).
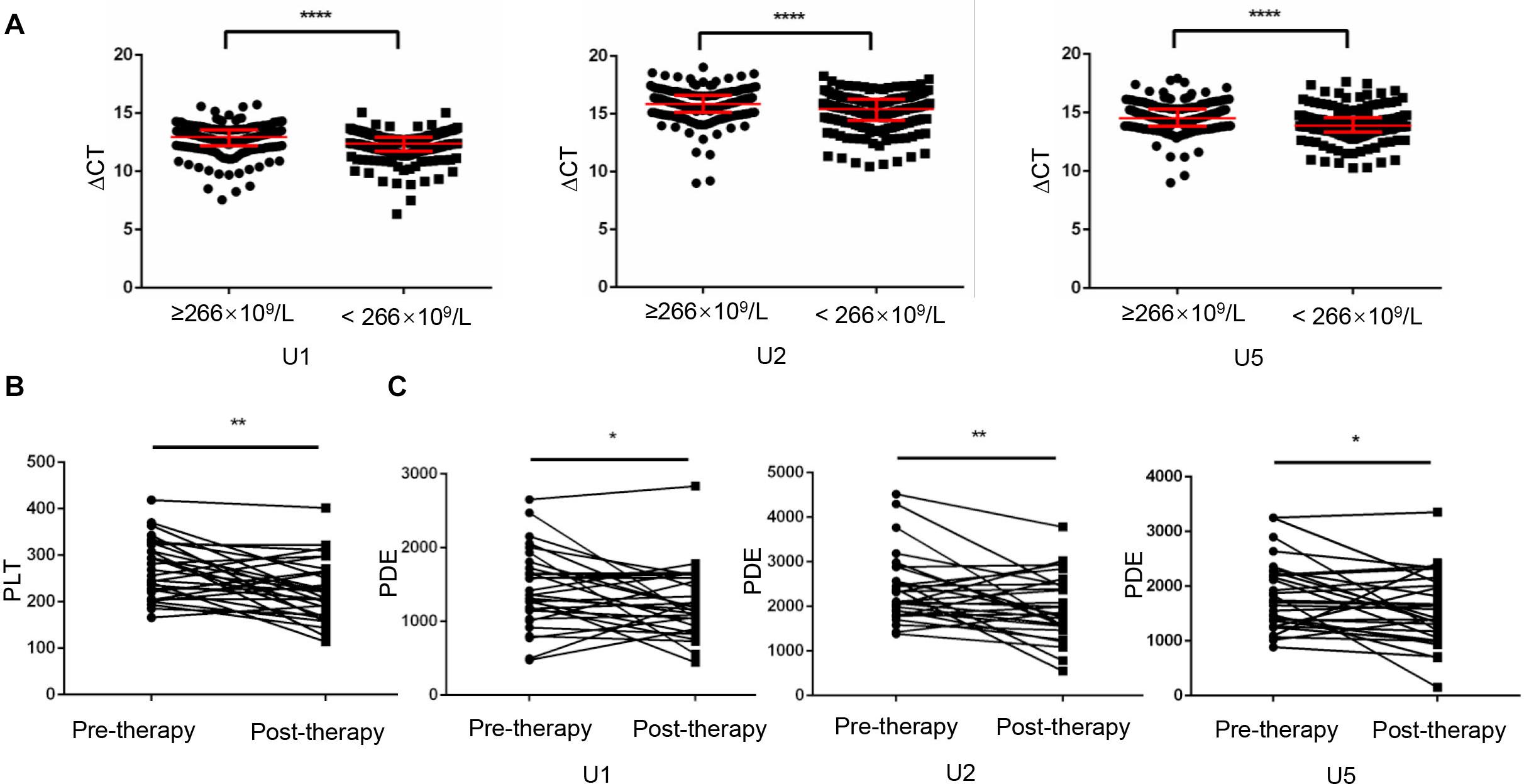
Figure 5. Platelet-count dependent expression of U1, U2 and U5 was elevated by anticancer therapy. (A) TEP U1, U2, U5 expression was down-regulated in high platelet counts patients (n = 192) compared with that in low platelet counts patients (n = 189); (B) The platelet count levels were significantly lower in the post-therapy patients than in paired pre-therapy patients (n = 30); (C) The PDE levels were significantly lower in the post-therapy patients than in paired pre-therapy patients; *P < 0.05; **P < 0.005; ****P < 0.0001.
Given that anticancer therapy was capable to induce a decrease (p = 0.0016) in platelet counts (Figure 5B), snRNAs expression per platelet was elevated by anticancer therapy much more significantly, as evidence from that platelet-count dependent expression (PDE, ΔCT × platelet counts) of U1, U2 and U5 was downregulated (Figure 5C) after anticancer therapy (p = 0.0495, p = 0.0098 and p = 0.0276, respectively).
Previous study had been reported chemotherapy-induced apoptosis might lead to significant release of apoptotic extracellular vesicles (apoEVs), in which snRNAs specifically were enriched (24, 25). We supposed apoEVs could be captured by TEPs, thus causing an increase of TEP snRNAs expression. Hence, we hypothesized a correlation of snRNAs level between in exosome and in TEP. To prove this hypothesis, 58 lung cancer patients before receiving any anticancer treatment (untreated) and 23 patients with initial anti-cancer therapy (treated) were enrolled, then the paired TEPs and exosomes from the same donor were collected. First, the exosomes were identified by qNano, the diameters of most exosomes concentrated on the 50–150 nm (Figure 6A). Then these three snRNAs were detected in paired TEPs and exosomes. As show in Figure 6B, all TEP U1, U2 and U5 levels were closely correlated between exosomes and TEPs in treated patients (R = 0.781, p < 0.0001; R = 0.467, p = 0.025; R = 0.467, p = 0.025, respectively), but not in untreated ones unexpectedly. These data support anti-cancer therapy induced apo-exosomes might transmit snRNAs to TEPs.
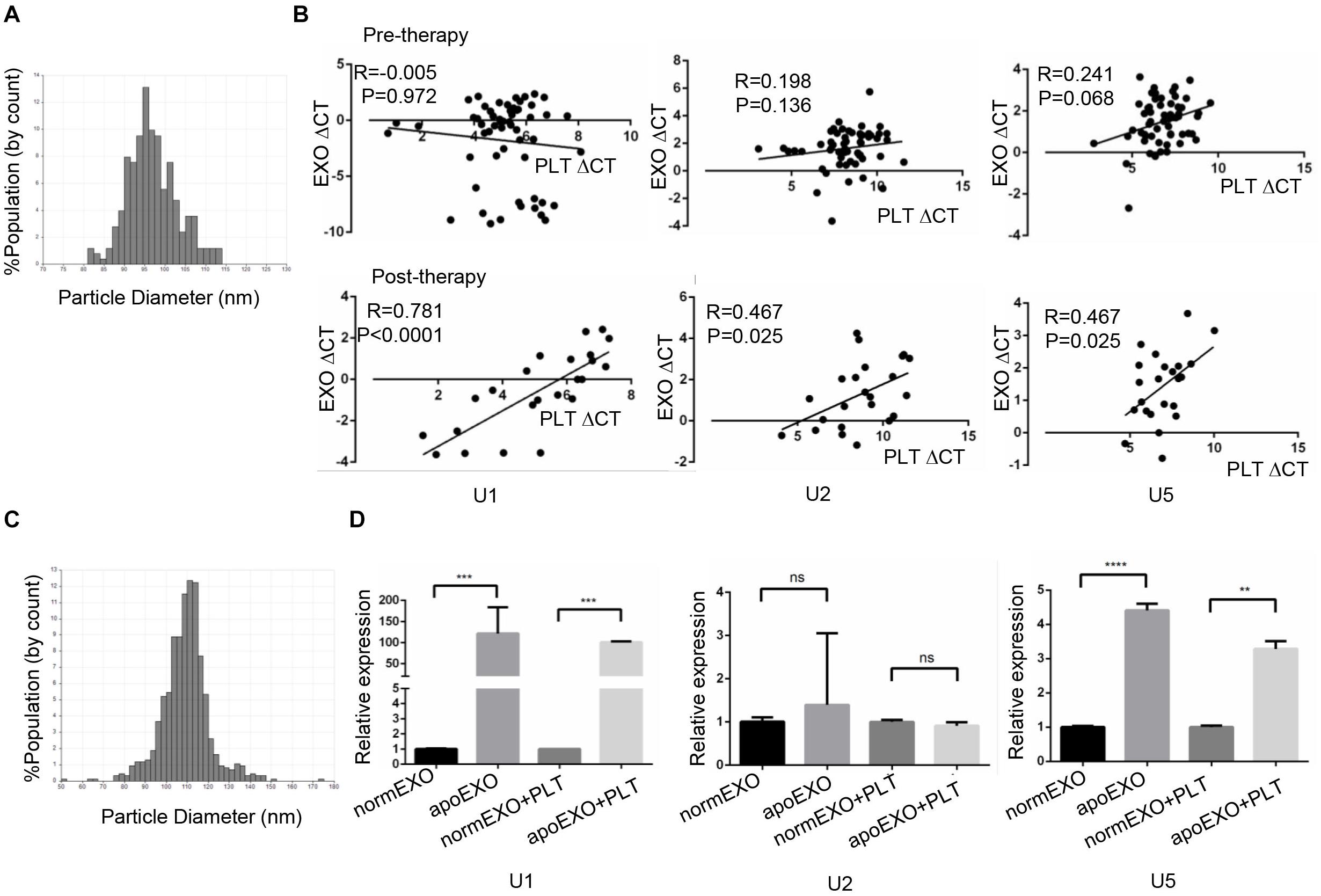
Figure 6. TEP U1, U5 but not U2 expression was affected by apo-exosomes. (A) Distribution of exosomes of 50–150 nm diameter in the samples from lung cancer patients’ plasma based on the qNano system; (B) The correlations between U1, U2, U5 expression and platelet counts in 58 untreated patients (upper) and 20 treated lung cancer patients (down); (C) Distribution of exosomes of 50–150 nm diameter in the samples from A549 cells based on the qNano system; (D) Exosomal U1, U5 from apoptotic A549 cells (apoEXO) were significantly increased compared to those derived from normal A549 cells. normEXO/apoEXO, exosomes derived from normal/apoptotic A549 cells. **p < 0.005; ***p < 0.001; ****p < 0.0001; ns, no significance.
To confirm the transmission of snRNAs to platelets by apo-exosomes, A549 cells were employed and treated with or without 50 μm cisplatin for 24 h to induce apoptosis. A549 derived exosomes were isolated and identified using qNano system (Figure 6C), followed by co-culture with platelets from healthy volunteers for 4 h. As shown in Figure 6D, exosomal U1, U5 from apoptotic A549 cells (apoExo) were significantly increased (p = 0.0008 and p < 0.0001, respectively) compared to those from normal A549 cells (normEXO). In consistence, U1, U5 in platelets were also significantly upregulated (p = 0.0333 and p = 0.005, respectively) after coincubation with exosomes from apoptotic A549 cells (apoEXO + PLT) than that from normal A549 cells (normEXO + PLT). Unexpectedly, no significant differences of U2 was observed neither in normEXO nor in apoEXO, as well as neither in normEXO + PLT nor in apoEXO + PLT.
Despite continuous improvements of therapy strategies, the mortality rate of lung cancer patients still remains high, mainly since most patients were diagnosed at an advanced-stage (24). Thus, sensitive and specific biomarkers to identify lung cancer patients are urgently needed. Platelets, “educated” by their environment, contain a rich and dynamical repertoire of RNA varieties, including mRNAs and small non-coding RNAs, thus providing the biosources and biomolecules as diagnostic, prognostic, predictive or monitoring biomarkers. In our study, we demonstrated TEP U1, U2, U5 served as potential biomarkers for diagnosis and progression monitoring for lung cancer.
Splicing usually occurs within nuclei, yet platelets devoid of a nucleus do possess essential splicing factors (25), including all of the five snRNAs (U1, U2, U4, U5 and U6), which were detected in nucleus and cytoplasm of megakaryocytes, and in proplatelets that extend from megakaryocytes, as well as in primary circulating platelets (26) implementing signal-dependent splicing, a unique splicing mechanism upon platelet activation by tissue factor (27) or clooxygenase-2 (Cox-2) (28), providing platelets with a novel mechanism to alter their mRNA profile. The U1, approximately 165 nucleotides in length, participates in splicing by recognizing and binding to 5′ splice sites (SS) at pre-mRNA exon/intron boundaries to form the E complex (29); U2 forms part of the RNA network to bring the reactive sites into approach of pre-mRNA at the catalytic core of spliceosome; U5 snRNA interacting with the 5′ exon is thought to assist in docking the 5′ SS into catalytic core of the spliceosome (30). Our data validated TEP U1, U2, U5 as potential biomarkers for lung cancer. Firstly, TEP U1, U2, U5 expression levels were decreased significantly in patients with lung cancer or early stage lung cancer. The ROC curve analysis showed TEP U1, U2, U5 could distinguish patients from healthy donors, possessing favorable diagnostic efficiencies. Then, TEP U1, U2, U5 downregulation was correlated with lung cancer progression. They were significantly downregulated in advanced-stage, distant metastasis or lymph node metastasis lung cancer patients compared with those in control groups, respectively. The ROC curve analysis demonstrated TEP U1, U2, U5 could distinguish advanced-stage patients from early-stage ones with favorable diagnostic efficiencies.
The snRNA U4 and U6 also play crucial roles in splicing behavior of nucleated cell. During splicing, U4 and U6 interact with one another by RNA-RNA base pairs, at the time U4/U6 and U5 snRNPs are recruited as a preassembled U4/U6-U5 tri-snRNP, and interact with the pre-mRNA (31). In our study, we also analyzed the differential expression of TEP U4 and U6 between healthy donors and lung cancer patients, but no significance was observed (data not shown), indicating the different roles of U4 and U6 in platelet. In addition, second minor snRNAs including U11, U12, U4ATAC and U6ATAC which represent functional counterparts of the major U1, U2, U4, and U6 (29), were also detected in our research. However, most of them expressed very low in platelets, which are inappropriate to act as the potential biomarkers.
Interestingly, snRNAs U1, U2 and U5 levels were closely correlated between paired exosomes and TEP from treated patients but not from untreated ones, this was probably because apo-exosomes were accumulated during anticancer therapy, in which splicing messages were enriched, and then captured by TEPs, thus causing increases of TEP U1 and U5 expression. Nevertheless, U2 seemed not to be assembled in platelets as well as in apo-exosomes, implying another mechanism might be involved.
Accumulating evidences have highlighted that platelets acted as active players in all steps of tumorigenesis, and coordinated with tumor cells by both local and distant crosstalk. One of the mechanisms by which platelets are educated by tumor cells is receptor-ligand interaction. For example, pdoplanin, expressed on cancer associated fibroblasts and tumor cells, is a major ligand to attract platelets by binding to C-type lectin-like receptor 2 (CLEC-2), which has been identified as a receptor for the platelet activating snake venom protein rhodocytin, then facilitates hematogenous cancer metastasis and cancer associated thrombosis (32). In current study, we proved that horizontal transmission by apo-exosomes played a crucial role in the process that tumor educated platelets. However, we can’t exclude the possibility that direct interaction of tumor-platelet induced TEP U1, U2, U5 downregulation, which need more exploration.
However, several limitations should be carefully considered in the present study. First, our results included 382 lung cancer patients, the total sample sizes were small and might lack statistically vigorous power. Long-term clinical follow-up data were also absent, which currently limit the ability to explore the prognostic values of TEP U1, U2, U5. Moreover, the mechanisms of enrichment and transmission of exosomal snRNAs were still unclear, so the detailed process that cancer educated platelets still need more investigation.
In summary, our findings reveal that TEP U1, U2, U5 were significantly downregulated in lung cancer, and their downregulations were correlated with lung cancer progression, possessing favorable diagnostic efficiencies. More importantly, TEP U1, U2 and U5 levels were closely correlated between paired exosomes and TEP from treated patients but not from untreated ones, and U1, U5 but not U2 in platelets were elevated by apo-exosomes, thus providing the evidence TEP U1, U2, U5 as biomarkers for lung cancer.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Shandong Cancer Hospital Affiliated to Shandong First Medical University and Shandong Academy of Medical Sciences of Committee. The patients/participants provided their written informed consent to participate in this study.
XRS and XGS designed the experiments. XD and SD carried out the experiments. MY provided the blood samples. XD and XGS wrote the manuscript and prepared the figures. LXi, LN, LXu, and YZ contributed to analysis the experimental data. All authors reviewed the manuscript read and approved the final manuscript.
This work was supported by the Shandong Provincial Key Research and Development Program (2017CXGC1207), National Natural Science Foundation of China (81972014, 81672104, and 8150111724) and the Shandong Provincial Natural Science Foundation (ZR2019MH004, ZR2019LZL016, and ZR2019LZL015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Δ Ct, comparative cycle threshold; ApoEVs, apoptotic extracellular vesicles; Apo-exosomes, apoptotic exosomes; AUC, area under the curve; cDNA, complementary DNA; CEA, carcinoembryonic antigen; CRC, colorectal carcinoma; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; NSCLC, non-small cell lung cancer; PBS, phosphate-buffered saline; PDE, platelet-count dependent expression; PRP, platelet rich plasma; qRT-PCR, quantitative real-time polymerase chain reaction; ROC, receiving operating characteristic; SnRNA, small nuclear RNA; SnRNPs, small nuclear ribonucleoproteins; SS, splice site; TEP, tumor educated platelet.
1. Cheung CHY, Juan HF. Quantitative proteomics in lung cancer. J Biomed Sci. (2017) 24:37. doi: 10.1186/s12929-017-0343-y
2. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. (2019) 103:463–73. doi: 10.1016/j.mcna.2018.12.006
3. Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, Sood AK. The platelet lifeline to cancer: challenges and opportunities. Cancer Cell. (2018) 33:965–83. doi: 10.1016/j.ccell.2018.03.002
4. McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. (2014) 16:717–27. doi: 10.1038/ncb3015
5. Best MG, Wesseling P, Wurdinger T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. (2018) 78:3407–12. doi: 10.1158/0008-5472.CAN-18-0887
6. Yang L, Jiang Q, Li D-Z, Zhou X, Yu D-S, Zhong J, et al. TIMP1 mRNA in tumor-educated platelets is diagnostic biomarker for colorectal cancer. Aging (Albany NY). (2019) 11:8998–9012. doi: 10.18632/aging.102366
7. Nilsson RJ, Balaj L, Hulleman E, Van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. (2011) 118:3680–3. doi: 10.1182/blood-2011-03-344408
8. Calverley DC, Phang TL, Choudhury QG, Gao B, Oton AB, Weyant MJ, et al. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci. (2010) 3:227–32. doi: 10.1111/j.1752-8062.2010.00226.x
9. Tjon-Kon-Fat LA, Lundholm M, Schröder M, Wurdinger T, Thellenberg-Karlsson C, Widmark A, et al. Platelets harbor prostate cancer biomarkers and the ability to predict therapeutic response to abiraterone in castration resistant patients. Prostate. (2018) 78:48–53. doi: 10.1002/pros.23443
10. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
11. Best MG, Vancura A, Wurdinger T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost. (2017) 15:1295–306. doi: 10.1111/jth.13720
12. Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. (2017) 36:263–72. doi: 10.1007/s10555-017-9674-0
13. Best MG, Sol N, In ‘t Veld SGJG, Vancura A, Muller M, Anna-Larissa NN, et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell. (2017) 32:238–52.e9.
14. Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. (2001) 13:302–9. doi: 10.1016/S0955-0674(00)00212-X
15. Dvinge H, Guenthoe J, Porter PL, Bradley RK. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. (2019) 29:1591–604. doi: 10.1101/gr.246678.118
16. Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. (2016) 16:413–30. doi: 10.1038/nrc.2016.51
17. Liu L, Song X, Li X, Xue L, Ding S, Niu L, et al. A three-platelet mRNA set: MAX, MTURN and HLA-B as biomarker for lung cancer. J Cancer Res Clin Oncol. (2019) 145:2713–23. doi: 10.1007/s00432-019-03032-9
18. Ficko T, Cernelc P. Real-time quantitative PCR assay for analysis of platelet glycoprotein IIIa gene expression. J Biochem Biophys Methods. (2005) 62:241–50. doi: 10.1016/j.jbbm.2004.12.002
19. Ko HH, Lee J-J, Chen H-M, Kok S-H, Kuo MY-P, Cheng S-J, et al. Upregulation of vascular endothelial growth factor mRNA level is significantly related to progression and prognosis of oral squamous cell carcinomas. J Formos Med Assoc. (2015) 114:605–11. doi: 10.1016/j.jfma.2015.05.005
20. Perez-Callejo D, Romero A, Provencio M, Torrente M. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res. (2016) 5:455–65. doi: 10.21037/tlcr.2016.10.07
21. Ferrigno D, Buccheri G, Biggi A. Serum tumour markers in lung cancer: history, biology and clinical applications. Eur Respir J. (1994) 7:186–97. doi: 10.1183/09031936.94.07010186
22. Molina R, Filella X, Augé JM, Fuentes R, Bover I, Rifa J, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. (2003) 24:209–18.
23. Pedersen LM, Milman N. Prognostic significance of thrombocytosis in patients with primary lung cancer. Eur Respir J. (1996) 9:1826–30. doi: 10.1183/09031936.96.09091826
24. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
25. Zucker M, Hauschner H, Seligsohn U, Rosenberg N. Platelet factor XI: intracellular localization and mRNA splicing following platelet activation. Blood Cells Mol Dis. (2018) 69:30–7. doi: 10.1016/j.bcmd.2017.04.006
26. Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. (2005) 122:379–91. doi: 10.1016/j.cell.2005.06.015
27. Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med. (2006) 203:2433–40. doi: 10.1084/jem.20061302
28. Shashkin PN, Brown GT, Ghosh A, Marathe GK, McIntyre TM. Lipopolysaccharide is a direct agonist for platelet RNA splicing. J Immunol. (2008) 181:3495–502. doi: 10.4049/jimmunol.181.5.3495
29. Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. (2009) 136:701–18. doi: 10.1016/j.cell.2009.02.009
30. MacRae AJ, Mayerle M, Hrabeta-Robinson E, Chalkley RJ, Guthrie C, Burlingame AL, et al. Prp8 positioning of U5 snRNA is linked to 5’ splice site recognition. RNA. (2018) 24:769–77. doi: 10.1261/rna.065458.117
31. Schultz A, Nottrott S, Hartmuth K, Lührmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem. (2006) 281:28278–86. doi: 10.1074/jbc.M603350200
Keywords: small nuclear RNA (snRNA U1, U2, U5), tumor educated platelet, lung cancer, biomarkers, alternative splicing
Citation: Dong X, Ding S, Yu M, Niu L, Xue L, Zhao Y, Xie L, Song X and Song X (2020) Small Nuclear RNAs (U1, U2, U5) in Tumor-Educated Platelets Are Downregulated and Act as Promising Biomarkers in Lung Cancer. Front. Oncol. 10:1627. doi: 10.3389/fonc.2020.01627
Received: 27 May 2020; Accepted: 27 July 2020;
Published: 12 August 2020.
Edited by:
Rengyun Liu, The First Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Rafael Rosell, Catalan Institute of Oncology, SpainCopyright © 2020 Dong, Ding, Yu, Niu, Xue, Zhao, Xie, Song and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingguo Song, c3Quc2FtdWVsQDE2My5jb20=; Xianrang Song, YmFzaWNsYWJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.