- 1Department of Urology, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang, China
- 2Department of Pathophysiology, College of Basic Medical Science, China Medical University, Shenyang, China
- 3School of Kinesiology, Shenyang Sport University, Shenyang, China
- 4Department of Biostatistics, Fairbanks School of Public Health, Indiana University, Indianapolis, IN, United States
- 5Division of Hematology-Oncology, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
- 6Cancer Therapeutics Program, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, United States
m6A, the main form of mRNA modification, participates in regulating multiple normal and pathological biological events, especially in tumorigenesis. However, there is little known about the association of m6A-related genes with prognosis of clear cell renal cell cancer (ccRCC). Therefore, the prognostic value of m6A-related genes was investigated using Kaplan–Meier curves of overall survival (OS) with the log-rank test and Cox regression analysis. The differential expression of YTHDF2 mRNA in ccRCC and tumor-adjacent normal tissues and associated with clinicopathological characteristics was also analyzed. The alteration of cancer signaling pathways was screened by Gene Set Enrichment Analysis (GSEA). Univariate analysis showed that 15 m6A-related genes (including YTHDF2) were closely related to prognosis. Multivariate analysis further confirmed that YTHDF2 could serve as an independent prognostic factor for the OS of ccRCC patients (P < 0.001). Low-level expression of YTHDF2 had poor prognosis in ccRCC patients with lower tumor–node–metastasis (TNM) stage, age > 61, non-distant metastasis, non-lymph node metastasis, female gender, and higher histological grade (P < 0.05). Moreover, YTHDF2 expression in ccRCC tissues (N = 529) is significantly lower than that of tumor-adjacent normal tissues (N = 72, P = 0.0086). Furthermore, GSEA demonstrated that AKT/mTOR/GSK3 pathway, EIF4 pathway, CHREBP2 pathway, MET pathway, NFAT pathway, FAS pathway, EDG1 pathway, and CTCF pathway are altered in tumors with high YTHDF2 expression. Taken together, our results demonstrated that YTHDF2 (an m6A-related gene) could serve as a potential prognostic biomarker of ccRCC, and targeting epigenetic modification may be a novel therapeutic strategy for the treatment of ccRCC.
Introduction
In 2018, 65,340 new cases of renal cell carcinoma (RCC) were diagnosed and 14,970 resulted deaths in the United States (1). Moreover, ~3/4 of RCC belongs to clear cell renal cell cancer (ccRCC) (2). There are three main treatment measures for ccRCC, including radiotherapy, chemotherapy, and surgical resection (3–5). Due to the developments in medical imaging, the accurate for diagnostic rate of ccRCC is increased. However, ~30% of patients have distant metastasis once diagnosed (6), and these patients cannot be suitable for resection. Currently, the primary therapeutic measure for metastatic RCC (mRCC) is antiangiogenic therapy-based targeting tyrosine kinase. Although this treatment is of benefit for mRCC patients, and the reason for limited efficacy is development of drug resistance (7, 8), this biochemical alteration leads to poorer prognosis (9). Therefore, understanding the precise mechanisms of mRCC and looking for the key clinical biomarker and therapeutic target for RCC metastasis will contribute to successful treatment ccRCC.
N6-Methyladenosine (m6A), a key modification event of RNA, manipulated a series of genes called “writers” (METTL3, METTL14, and WTAP), “readers” (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2), and “erasers” (FTO and ALKBH5) (10). In general, m6A is near the long internal exons and stop codons, located in 3′-UTRs (11), resulting in changes of RNA stability, splicing, intracellular distribution, and translation (12–14). Recently, several studies reported that modification of m6A exerted a key role in multiple tumorigenesis (15–17). Previous studies demonstrated that genetic alterations of m6A-mediated genes occurred in ccRCC. The alteration of m6A-mediated genes is closely associated with poor clinical characteristics, including overall survival (OS) (18). In addition, METTL3, as a tumor suppressor, plays a crucial role in process of proliferation, migration/invasion, and cell cycle regulation of ccRCC cells (19). To date, there is little known about the correlation of m6A-related genes profile and clinicopathological character of ccRCC. Thus, the prognostic value of m6A-related genes was investigated using Kaplan–Meier curve with Cox regression analysis and log-rank test. The differential YTHDF2 expression in ccRCC and tumor-adjacent normal tissues and associated with clinicopathological characteristics was analyzed by using mRNA expression data from The Cancer Genome Atlas (TCGA) ccRCC cohort. The alteration of multiple cancer signaling pathways were identified by Gene Set Enrichment Analysis (GSEA).
Materials and Methods
RNA-Seq Gene Expression Analysis in Clear Cell Renal Cell Cancer Patients
Based on the TCGA (https://tcga-data.nci.nih.gov/tcga/) data portal, m6A-related gene expression data (HTSeq-FPKM data) and clinicopathological features of 529 ccRCC patients and 72 tumor-adjacent normal renal samples were obtained. The clinicopathological characteristics of ccRCC patients are listed in Table 1, as follows: age, sex, tumor grade, tumor–node–metastasis (TNM) stage, cancer status, laterality, race, hemoglobin, platelet, serum calcium, and white blood cell (WBC) count. The patients without complete clinicopathological characteristics were excluded. The repeat gene expression data for the same patient were averaged. Finally, the correlation of RNA-Seq gene expression of 529 ccRCC patients and clinic information were investigated.
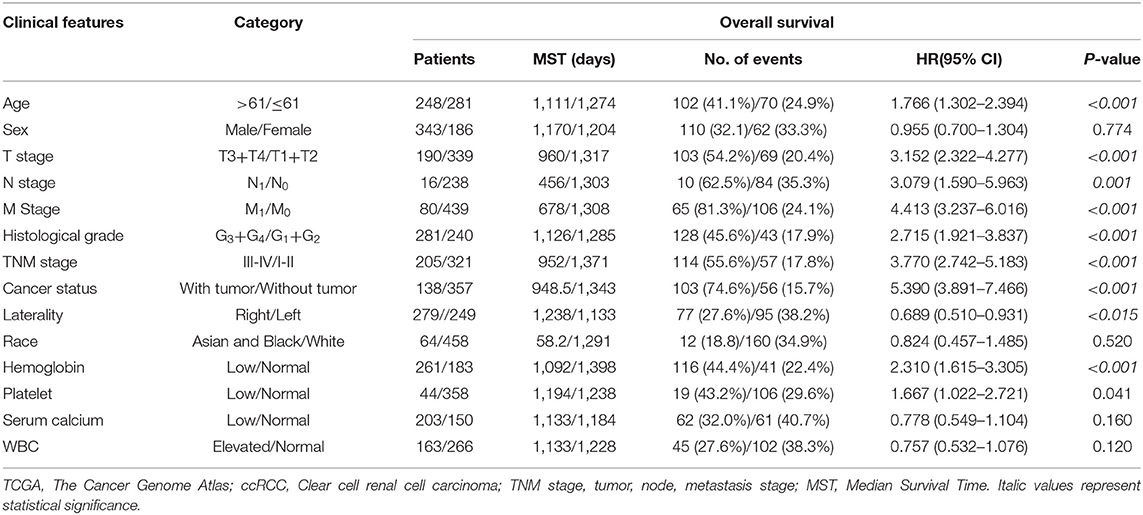
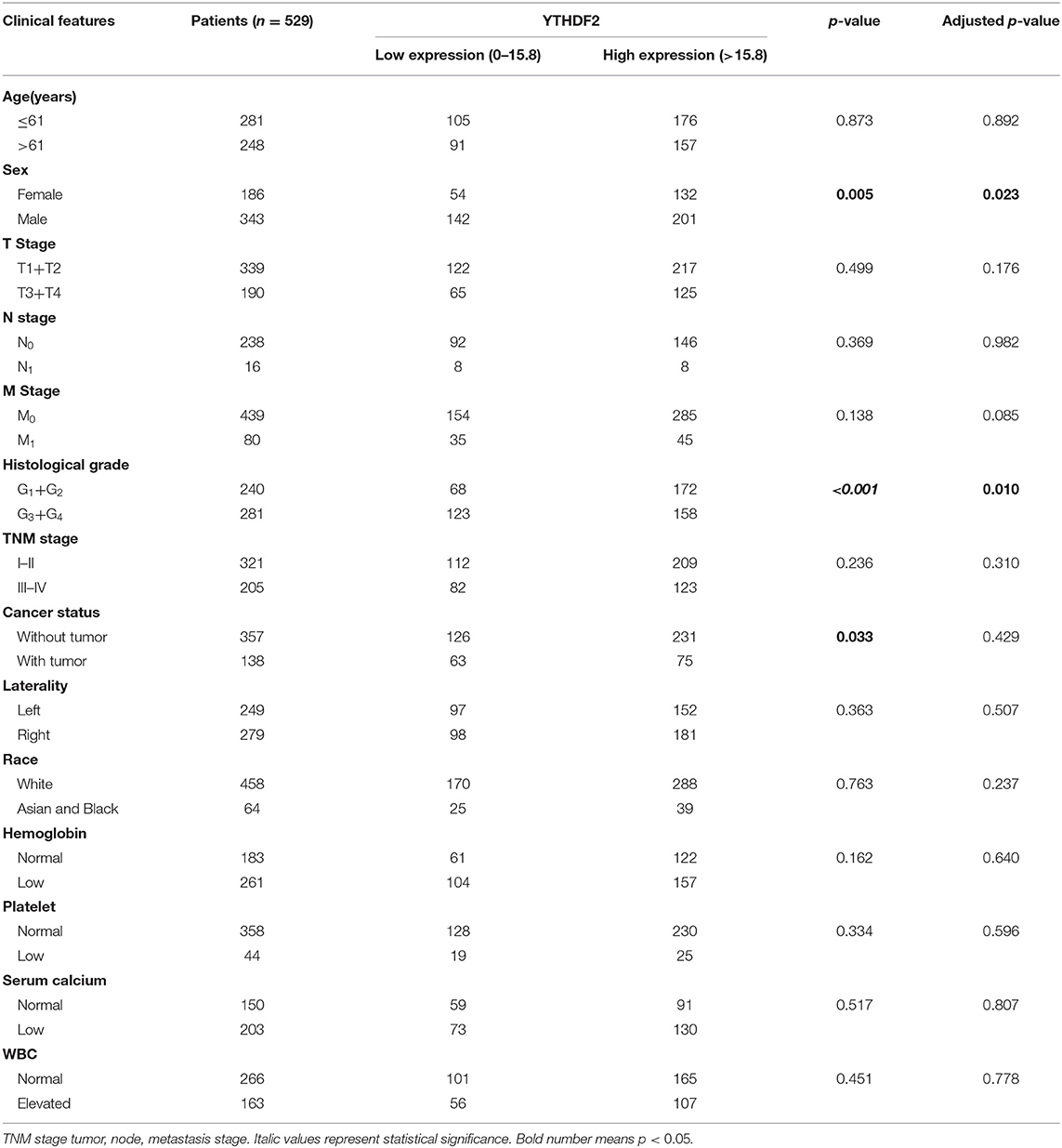
Table 1. Clinical characteristics of The Cancer Genome Atlas (TCGA) clear cell renal cell cancer (ccRCC) patients.
Gene Set Enrichment Analysis
According to the cutoff value of YTHDF2 gene, all the ccRCC samples were defined as high- and low-level expression groups. GSEA was obtained from the GSEA program from sangerbox software (http://sangerbox.com/) using the BioCarta gene profile. Meanwhile, three factors [normalized enrichment score (NES), nominal P-value (NOM P-val), and false discovery rate (FDR)] were evaluated for statistical significance and enrichment magnitude (20).
Statistical Analysis
X-tile software (Version 3.6.1) (21) was used to determine the optimal cutoff values for expression profile of m6A-related genes. Mann–Whitney test was used to analyze the expression difference between ccRCC samples and tumor-adjacent normal tissues. Chi-square test was used to evaluate the association between YTHDF2 and each clinicopathological characteristics of ccRCC patients, ignoring the effect of the other characteristics (shown as P-value). Multiple logistic regression model and Wald test were used to determine the association between YTHDF2 and each clinicopathological characteristic of ccRCC patients, adjusting for the effect of the other characteristics (shown as adjusted P-value). Log-rank test and Kaplan–Meier curve were used to compare the survival times. According to Cox hazards regression (HR) model, univariate and multivariate survival analyses were used to analyze the independent parameters associated with the OS. Prism software (Version 6.0), SPSS (Version 16.0), and SAS (Version 9.4) were used to perform data statistics. P < 0.05 was considered to be statistically significant.
Results
Clinical Profile and Prognosis of All Clear Cell Renal Cell Cancer Patients
The correlation of clinicopathological characteristic with the OS of 529 ccRCC patients was analyzed by univariate Cox proportion model. As shown in Table 1, age (>61/ ≤ 61), T stage (T3+T4/T1+T2), N stage (N1/N0), M stage (M1/M0, histological grade (G3+G4/G1+G2), cancer status (with/without tumor), TNM stage (III–IV/I–II), laterality (right/left), hemoglobin level (low/normal), and platelet level (low/normal) were closely related to the OS (P < 0.05), while sex (male/female), race (Asian and Black/White), serum calcium level (low/normal), and WBC (elevated/normal) were not significantly associated with the OS (P > 0.05) in patients with ccRCC.
Identification of m6A-Related Gene YTHDF2 as a Prognostic Factor in Clear Cell Renal Cell Cancer
Univariate Cox proportion model and calculation of hazard ratio were performed to screen prognostic factors from a total of 19 m6A-related genes. As shown in Table 2, 15 m6A-related genes (including ALKBH, FTO, METTL3, METTL14, YTHDF2, YTHDC1, YTHDC2, ZC3H13, METTL16, KIAA1429, CBLL1, IGF2BP1, IGF2BP2, IGF2BP3, and RBM15) were significantly correlated to prognosis of ccRCC (P < 0.05). Furthermore, multivariate Cox regression model revealed that YTHDF2 (HR = 0.471, 95% CI: 0.241–0.920; P = 0.028) as well as age (HR = 2.118, 95% CI: 1.142–3.931; P = 0.017) and cancer status (HR = 3.329, 95% CI: 1.608–6.526; P = 0.001) served as independent prognostic factors (P < 0.05).
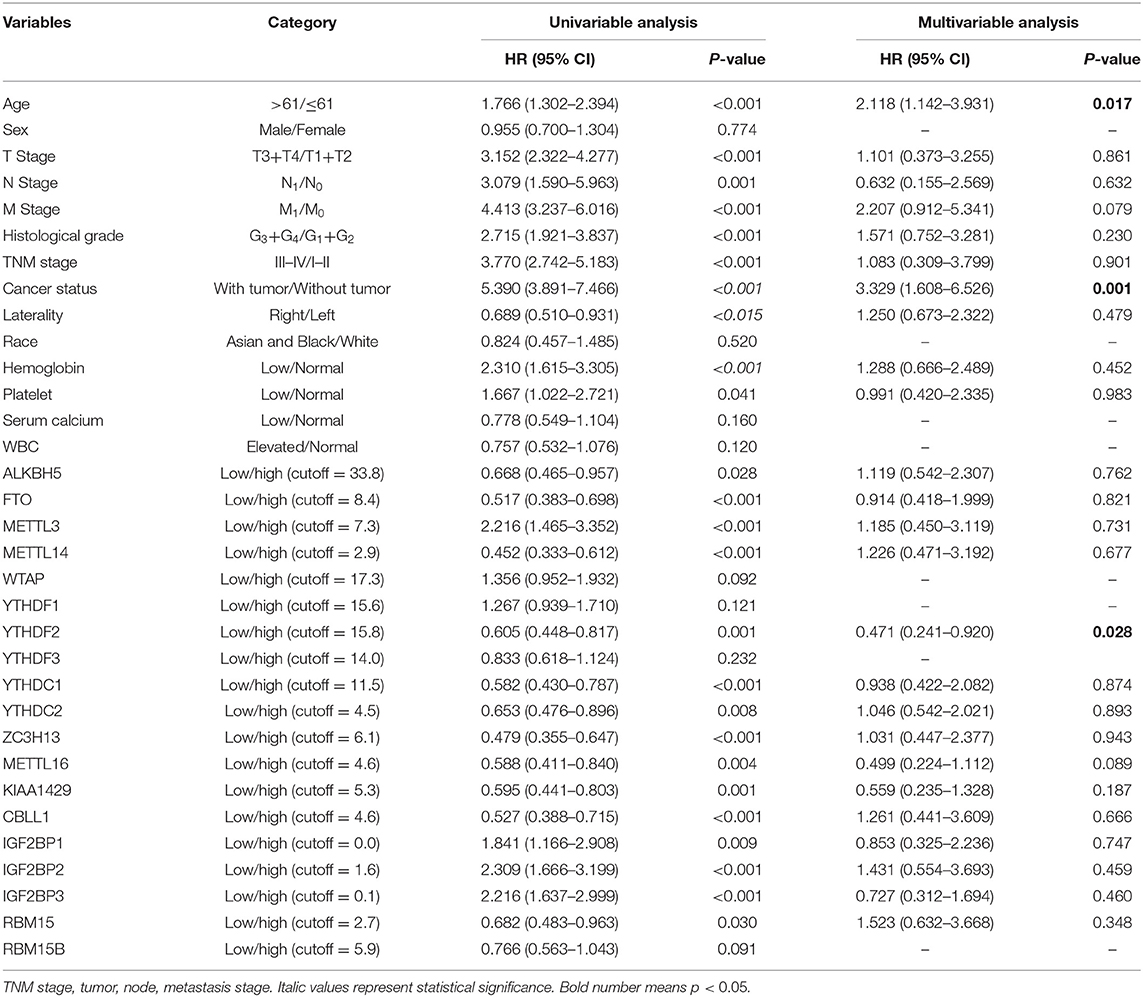
Table 2. Univariate and multivariate analysis of overall survival using the Cox proportional hazards regression model.
YTHDF2 Expression Is Associated With Overall Survival With Clinicopathological Characteristics in Clear Cell Renal Cell Cancer
Subsequently, the correlation of YTHDF2 mRNA expression and OS was evaluated by log-rank test and Kaplan–Meier survival analysis. As seen in Figure 1, the ccRCC patients with low-level mRNA expression of YTHDF2 presented shorter OS (P < 0.001). In stratified analysis (Figures 1B–O), lower YTHDF2 expression was significantly associated with poor prognosis of ccRCC patients with lower TNM stage (stage I–II, P = 0.013); elder age (>61, P = 0.005); non-distant metastasis (P = 0.002); late T stage (T3+T4, P = 0.043); non-lymph node metastasis (P < 0.001); female gender (P = 0.001); and higher histological grades (G3+G4, P = 0.011).
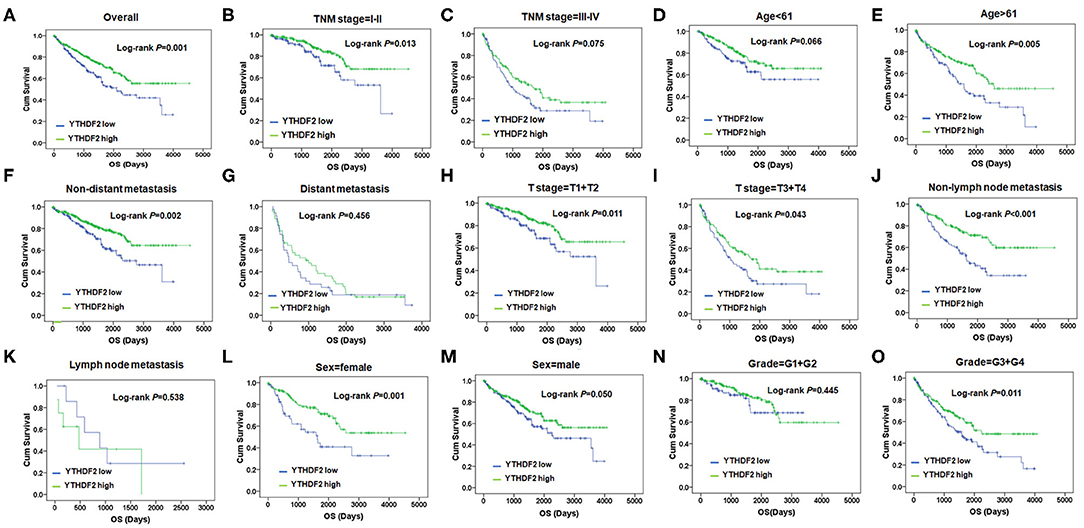
Figure 1. YTHDF2 expression is associated overall survival (OS) with clinicopathological characteristics in clear cell renal cell cancer (ccRCC). (A) Kaplan–Meier survival analysis and log-rank test were used to compare differences in OS between the groups classified using cutoff values determined by X-tile; Kaplan–Meier survival analysis and log-rank test were used to analyze the association of YTHDF2 expression and OS stratified by TNM stage (B,C); age (D,E); non-distant metastasis (F), distant metastasis (G); T-stage = T1+T2 (H); and T-stage = T1+T2 (I); non-lymph node metastasis (J) and lymph node metastasis (K), sex (L,M); and grade = G1+G2 (N) and grade = G3+G4 (O).
Expression Differences of YTHDF2 in Clear Cell Renal Cell Cancer Samples and Tumor-Adjacent Normal Tissues
To analyze YTHDF2 expression in ccRCC and tumor-adjacent normal tissues, we extracted and compared YTHDF2 gene expression from the TCGA database, including 529 ccRCC tumor tissues and 72 tumor-adjacent normal tissues. As shown in Figure 2A, the mRNA expression level of YTHDF2 was evidently decreased in ccRCC (N = 529) as compared with tumor-adjacent normal tissues (N = 72, P = 0.0086). To explore clinical significance of YTHDF2 expression, the correlation of YTHDF2 gene expression and various clinicopathological characteristics of ccRCC were further analyzed. As seen in Table 3 and Figures 2B,C, YTHDF2 expression was significantly lower in male patients (N = 186) than female patients (N = 343, P < 0.001). Meanwhile, YTHDF2 mRNA was significantly less in patients with high histological grade (G3+G4, N = 281) than in patients with low histological grade (G1+G2, N = 240, P < 0.001). After the effect of the other clinicopathological characteristics were adjusted for, histological grade (adjusted P = 0.010) and sex (adjusted P = 0.023) were still significantly associated with YTHDF2 expression in patients with ccRCC.
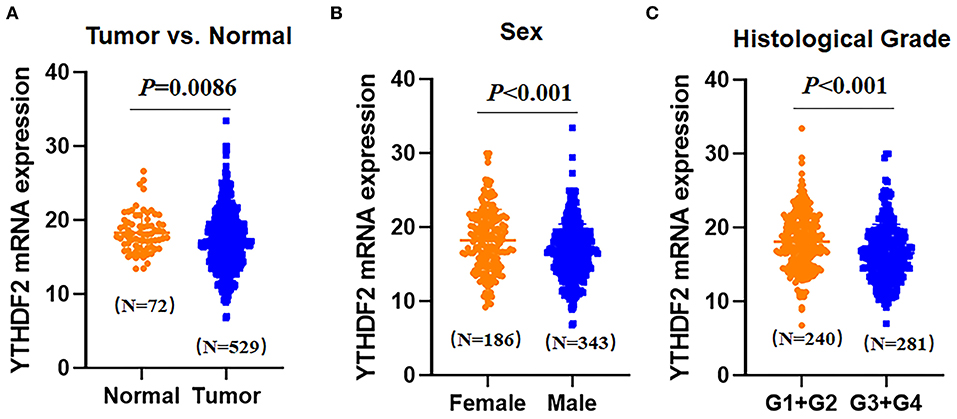
Figure 2. Expression differences of YTHDF2 in clear cell renal cell cancer (ccRCC) samples and tumor-adjacent normal tissues (A). Differential expression of YTHDF2 gene in ccRCC tissues (N = 529) and tumor-adjacent normal tissues (N = 72); (B) differential expression of YTHDF2 in female (N = 186) and male (N = 343) patients with ccRCC; and (C) differential expression of YTHDF2 gene between ccRCC tissues in lower histological grade (G1+G2, N = 240) and higher histological grade (G3+G4, N = 281).
Identification of YTHDF2-Regulated Multiple Pathways in Clear Cell Renal Cell Cancer by Gene Set Enrichment Analysis
To investigate the alteration of YTHDF2-related pathways in ccRCC, GSEA was performed between two data sets with low- and high-YTHDF2 expression. GSEA showed that there are significant differences in enrichment of MSigDB Collection (NOM P-val < 0.05, FDR < 0.05). According to their NES, the top 10 most evidently enriched pathways were screened (Figure 3). It shows that mTOR pathway, GSK3 pathway, EIF4 pathway, CHREBP2 pathway, MET pathway, NFAT pathway, FAS pathway, EDG1 pathway, AKT pathway, and CTCF pathway were significantly enriched in the YTHDF2-related phenotype.
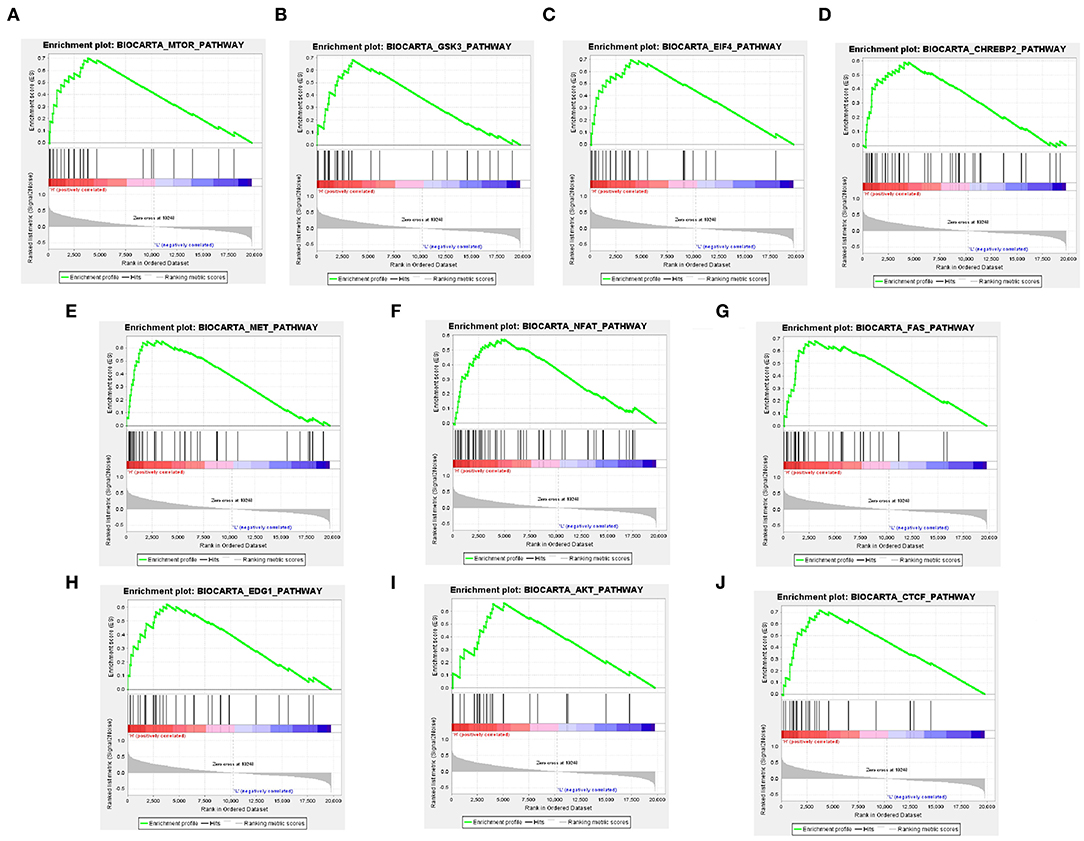
Figure 3. Identification of YTHDF2-regulated multiple pathways in clear cell renal cell cancer (ccRCC) by Gene Set Enrichment Analysis (GSEA). GSEA results showing mTOR pathway (A), GSK3 pathway (B), EIF4 pathway (C), CHREBP2 pathway (D), MET pathway (E), NFAT pathway (F), FAS pathway (G), EDG1 pathway (H), AKT pathway (I), and CTCF pathway (J) are differentially enriched in YTHDF2 increased expression phenotype. ES, enrichment score; NES, normalized ES; NOM, normalized P-value.
Discussion
Herein, we discovered that several specific m6A-related genes were closely related to distinct OS and that YTHDF2 can serve as an independent risk factor in ccRCC. What is more, our result showed that YTHDF2 mRNA expression significantly correlated with histological grade and sex. Therefore, YTHDF2, a key m6A-related gene, could serve as a prognostic biomarker and a therapeutic target of ccRCC.
m6A was initially reported by Ronald Desrosiers in 1974 (22), but the precise mechanism and regulatory function of the m6A modification remained largely unknown until recently (23). The m6A modifications were manipulated by precise interplay of recognition, removal, and deposition regulators, and increasing evidences indicated that m6A regulator contributed to initiation and progression of tumors (24). Noticeably, the biological role of these m6A regulators can be variable relying on the disease. For instance, Liu et al. (25) found that high METTL3 expression and decreased regulation of METTL14, METTL16, FTO, and ALKBH5 were positively correlated with poor prognosis in RCC patients. Zhou et al. (26) reported that YTHDF1 served as an independent poor prognostic factor in hepatocellular carcinoma (HCC). Wu et al. found that METTL3, METTL14, WTAP, and FTO present a valuable predictive strategy for breast cancer and contribute to the development of breast cancer (27). To date, the relationship of m6A-regulated genes and ccRCC prognosis is not clear. Herein, we provided an overall summary of the roles of m6A in tumorigenesis of ccRCC. Analysis of the TCGA-KIRC database suggested that elevation of METTL3, IGF2BP1, IGF2BP2, and IGF2BP3 and decreased expressions of ALKBH5, FTO, METTL14, YTHDF2, YTHDC1, YTHDC2, ZC3H13, METTL16, KIAA1429, CBLL1, and RBM15 in ccRCC were associated with poor OS probability. Furthermore, results showed that YTHDF2 could be an independent prognostic factor affecting OS. Similar results were previously reported. Specifically, Li et al. (19) observed that METTL3 exerted an oncogene role in RCC. Zhuang et al. (28) reported that FTO inhibits ccRCC via FTO-PGC-1α pathway. These m6A-related genes could be potential biomarkers utilized clinically in diagnostic and prognostic capacity for ccRCC.
YTHDF2 has been identified as an m6A-binding protein and regulating stability of mRNA (29). YTHDF2 accelerates degradation of target mRNAs through recognizing and binding with m6A sites in 3′-UTR (30, 31). Researchers demonstrated that YTHDF2 played key roles in the cancer progression. Zhong et al. (32) found that YTHDF2 targeting MEK/ERK pathway impacted on growth of HCC cells. Sheng et al. found that YTHDF2 caused tumor growth through altering 6PGD mRNA translation in lung cancer (33). Another study (34) found that YTHDF2 was overexpressed in pancreatic cancer and related to patients' poor prognosis. Kidney cancer is characterized by metabolic disorders (35). Several recent studies reported that YTHDF2 played an important role in the regulation of lipid metabolism (36, 37). However, the role of YTHDF2 in ccRCC metabolic disorders remains unknown. The present study analyzed the association of YTHDF2 with several metabolic-related factors by using TCGA-ccRCC data, including 6-phosphogluconate dehydrogenase (6-PGD) (33). As the result show in Figure S1, there is a weak positive correlation of YTHDF2 and 6-PGD in human ccRCC (Pearson correlation = 0.181, P < 0.001, N = 529). It is possible that alteration of YTHDF2 expression impacted on 6-PGD and cancer metabolic-related pathways in human ccRCC. These findings indicated that YTHDF2 may act as a potential diagnostic and prognostic biomarker for cancer. The present study found that decreased YTHDF2 expression had poor OS in ccRCC patients with lower TNM stage, higher age, non-distant metastasis, non-lymph node metastasis, female gender, and higher histological grade.
GSEA indicated that multiple cancer signaling pathways (mTOR pathway, GSK3 pathway, EIF4 pathway, CHREBP2 pathway, MET pathway, NFAT pathway, FAS pathway, EDG1 pathway, AKT pathway, and CTCF pathway) were differentially enriched in YTHDF2 increased expression phenotype. Previous studies reported that mTOR pathway (38), GSK3 pathway (39), AKT pathway (40), EIF4 pathway (41), MET pathway (42), NFAT pathway (43), and FAS pathway (44) acted as essential factors in the development of renal cancer and that CHREBP2 pathway (45), EDG1 pathway (46), and CTCF pathway (47) were also cancer-related pathways. Therefore, we supposed that poorer prognosis of ccRCC mediated by decreased YTHDF2 may be related to these pathways directly or indirectly, while the association YTHDF2 expression with these pathways and the precise mechanism need clarification.
There are still limitations in our study. First, the detailed reasons of death in patients cannot be obtained from the TCGA data portal; disease-free survival (DFS) was not considered. Second, the correlation between protein expression of m6A regulator and ccRCC prognosis is not clear. Third, the precise mechanism of the m6A regulator impact ccRCC patient prognosis has not been addressed. Thus, substantial proofs are needed to translate these results into clinical benefit.
In summary, our results demonstrated that downregulation of YTHDF2 (a key m6A-related gene) is associated with poor prognosis of ccRCC patients, suggesting that YTHDF2 can serve as a prognostic biomarker of ccRCC. In addition, GSEA showed that YTHDF2 impacted on multiple cancer signaling pathways, including mTOR pathway, GSK3 pathway, EIF4 pathway, and CHREBP2 pathway. Our findings provide a novel strategy for treatment of ccRCC through regulating epigenetic modification of target genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZM, DD, MS, LL, and BH analyzed and interpreted the data. BH and NW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was granted funding by the Natural Science Foundation of Liaoning Province (Grant No. 2020-BS-043), Natural Science Foundation of Liaoning Province (Grant No. 20170540560), Shenyang Science and Technology Innovation Special Plan (Grant No. 18-014-4-73), and the Shenyang Science & Technology Bureau (20-205-4-046).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01566/full#supplementary-material
Figure S1. The correlation of YTHDF2 and 6PGD in ccRCC.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
2. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. (2016) 70:93–105. doi: 10.1016/j.eururo.2016.02.029
3. Morris MR, Latif F. The epigenetic landscape of renal cancer. Nat Rev Nephrol. (2017) 13:47–60. doi: 10.1038/nrneph.2016.168
4. Cavaliere C, D'Aniello C, Pepa CD, Pisconti S, Berretta M, Facchini G. Current and emerging treatments for metastatic renal cell carcinoma. Curr Cancer Drug Targets. (2018) 18:468–79. doi: 10.2174/1568009617666170209094030
5. Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. (2018) 359:801–6. doi: 10.1126/science.aan5951
6. Gong J, Maia MC, Dizman N, Govindarajan A, Pal SK. Metastasis in renal cell carcinoma: biology and implications for therapy. Asian J Urol. (2016) 3:286–92. doi: 10.1016/j.ajur.2016.08.006
7. van der Mijn JC, Mier JW, Broxterman HJ, Verheul HM. Predictive biomarkers in renal cell cancer: insights in drug resistance mechanisms. Drug Resist Updat. (2014) 17:77–88. doi: 10.1016/j.drup.2014.10.003
8. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. (2016) 29:653–68. doi: 10.1016/j.ccell.2016.03.004
9. Kroeger N, Zimmermann U, Burchardt M, Pantuck AJ. Prognostication in localised renal cell carcinoma. Lancet Oncol. (2015) 16:603–4. doi: 10.1016/S1470-2045(15)70227-5
10. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. (2017) 27:444–7. doi: 10.1038/cr.2017.10
11. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. doi: 10.1038/nature11112
12. Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. (2016) 283:1607–30. doi: 10.1111/febs.13614
13. Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. (2013) 38:204–9. doi: 10.1016/j.tibs.2012.12.006
14. Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinform. (2013) 11:8–17. doi: 10.1016/j.gpb.2012.12.002
15. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
16. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. (2017) 65:529–43. doi: 10.1002/hep.28885
17. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. (2016) 113:E2047–56. doi: 10.1073/pnas.1602883113
18. Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging. (2019) 11:1633–47. doi: 10.18632/aging.101856
19. Li X, Tang J, Huang W, Wang F, Li P, Qin C, et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. (2017) 8:96103–16. doi: 10.18632/oncotarget.21726
20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
21. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. (2004) 10:7252–9. doi: 10.1158/1078-0432.CCR-04-0713
22. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
23. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
24. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. (2019) 18:103. doi: 10.1186/s12943-019-1033-z
25. Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, et al. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. (2019) 11:3972–91.
26. Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, et al. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manage Res. (2019) 11:3921–31. doi: 10.2147/CMAR.S191565
27. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. (2019) 19:326. doi: 10.1186/s12885-019-5538-z
28. Zhuang C, Zhuang C, Luo X, Huang X, Yao L, Li J, et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J Cell Mol Med. (2019) 23:2163–73. doi: 10.1111/jcmm.14128
29. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
30. Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. (2014) 24:1490–2. doi: 10.1038/cr.2014.153
31. Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. (2014) 24:1493–6. doi: 10.1038/cr.2014.152
32. Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. (2019) 442:252–61. doi: 10.1016/j.canlet.2018.11.006
33. Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. (2019) 41:541–50. doi: 10.1093/carcin/bgz152
34. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. (2017) 16:2259–71. doi: 10.1080/15384101.2017.1380125
35. Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. (2015) 75:2541–52. doi: 10.1158/0008-5472.CAN-14-1703
36. Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. (2018) 25:1816–28.e1814. doi: 10.1016/j.celrep.2018.10.068
37. Cai M, Liu Q, Jiang Q, Wu R, Wang X, Wang Y. Loss of m(6)A on FAM134B promotes adipogenesis in porcine adipocytes throughm(6) A-YTHDF2-dependent way. IUBMB Life. (2019) 71:580–6. doi: 10.1002/iub.1974
38. Ghidini M, Petrelli F, Ghidini A, Tomasello G, Hahne JC, Passalacqua R, et al. Clinical development of mTor inhibitors for renal cancer. Expert Opin Invest Drugs. (2017) 26:1229–37. doi: 10.1080/13543784.2017.1384813
39. Pal K, Cao Y, Gaisina IN, Bhattacharya S, Dutta SK, Wang E, et al. Inhibition of GSK-3 induces differentiation and impaired glucose metabolism in renal cancer. Mol Cancer Ther. (2014) 13:285–96. doi: 10.1158/1535-7163.MCT-13-0681
40. Mu Z, Dong D, Wei N, Sun M, Wang W, Shao Y, et al. Silencing of lncRNA AFAP1-AS1 inhibits cell growth and metastasis in clear cell renal cell carcinoma. Oncol Res. (2019) 27:653–61. doi: 10.3727/096504018X15420748671075
41. Nayak BK, Feliers D, Sudarshan S, Friedrichs WE, Day RT, New DD, et al. Stabilization of HIF-2alpha through redox regulation of mTORC2 activation and initiation of mRNA translation. Oncogene. (2013) 32:3147–55. doi: 10.1038/onc.2012.333
42. Nandagopal L, Sonpavde GP, Agarwal N. Investigational MET inhibitors to treat Renal cell carcinoma. Expert Opin Invest Drugs. (2019) 28:851–60. doi: 10.1080/13543784.2019.1673366
43. Kim JH, Hwang KH, Eom M, Kim M, Park EY, Jeong Y, et al. WNK1 promotes renal tumor progression by activating TRPC6-NFAT pathway. FASEB J. (2019) 33:8588–99. doi: 10.1096/fj.201802019RR
44. Kim YS, Kim KH, Choi JA, Lee JH, Kim HK, Won NH, et al. Fas (APO-1/CD95) ligand and Fas expression in renal cell carcinomas: correlation with the prognostic factors. Arch Pathol Lab Med. (2000) 124:687–93.
45. Iizuka K. The transcription factor carbohydrate-response element-binding protein (ChREBP): a possible link between metabolic disease and cancer. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:474–85. doi: 10.1016/j.bbadis.2016.11.029
46. Bao M, Chen Z, Xu Y, Zhao Y, Zha R, Huang S, et al. Sphingosine kinase 1 promotes tumour cell migration and invasion via the S1P/EDG1 axis in hepatocellular carcinoma. Liver Int. (2012) 32:331–8. doi: 10.1111/j.1478-3231.2011.02666.x
Keywords: m6A-related genes, The Cancer Genome Atlas, prognosis, YTHDF2, clear cell renal cell cancer
Citation: Mu Z, Dong D, Sun M, Li L, Wei N and Hu B (2020) Prognostic Value of YTHDF2 in Clear Cell Renal Cell Carcinoma. Front. Oncol. 10:1566. doi: 10.3389/fonc.2020.01566
Received: 01 April 2020; Accepted: 20 July 2020;
Published: 23 September 2020.
Edited by:
Ronald M. Bukowski, Cleveland Clinic, United StatesReviewed by:
Matteo Ferro, European Institute of Oncology (IEO), ItalyShomik Sengupta, Monash University, Australia
Copyright © 2020 Mu, Dong, Sun, Li, Wei and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Wei, Y2hpbmF3ZWluaW5nQHlhaG9vLmNvbQ==; Bin Hu, aHViaW4xMzQwQDEyNi5jb20=
†These authors have contributed equally to this work
 Zhongyi Mu
Zhongyi Mu Dan Dong2†
Dan Dong2† Ning Wei
Ning Wei Bin Hu
Bin Hu