- 1Department of Pathology, Chonnam National University Medical School and Hwasun Hospital, Hwasun-gun, South Korea
- 2Department of Biomedical Science and Engineering, Gwangju Institute of Science and Technology (GIST), Gwangju, South Korea
- 3Department of Parasitology and Tropical Medicine, Chonnam National University Medical School, Hwasun-gun, South Korea
- 4Department of Hepatobiliary Pancreas Surgery, Chonnam National University Medical School and Hwasun Hospital, Hwasun-gun, South Korea
- 5Department of Neurosurgery, Brain Tumor Clinic & Gamma Knife Center, Chonnam National University Hwasun Hospital and Medical School, Hwasun-gun, South Korea
The molecular profile of cholangiocarcinoma (CC) remains elusive. The prognostic value of isocitrate dehydrogenase (IDH) mutations in CC is controversial, and there have been few relevant studies in Asian populations. In the present study, we investigated the frequency and prognostic significance of IDH mutations in Korean patients with CC. CC specimens were collected from patients who underwent surgical liver resection between 2004 and 2019. Clinical and pathological data were retrospectively reviewed from medical records. Mutational IDH profiling was performed by peptide nucleic acid-mediated PCR clamping in 206 surgical specimens; IDH-mutant samples were confirmed by next-generation sequencing (NGS). Of the 195 patients with CC, six (3.13%) were found to exhibit IDH1 (n = 5) or IDH2 (n = 1) mutations. Among patients with IDH1 mutations, four had R132C (c.394C>T) and one had R132G (c.394C>G) mutations. One patient had R172W (c.514A>T) mutations in IDH2. All IDH-mutant samples were of intrahepatic origin, and patients with IDH mutations had physiological to low serum levels of carbohydrate antigen 19-9 (CA19-9). No association between IDH mutation status and long-term survival outcomes was observed. The frequency of IDH mutations was considerably lower than the 10–20% reported in previous studies. The frequency and pattern of IDH mutations in CC are likely to vary among patients with different ethnicities. These findings suggest that characterization of the oncogenic mutation profile in different populations is of high clinical importance.
Introduction
Cholangiocarcinoma (CC) is a heterogeneous group of malignancies, which can be classified as intrahepatic, perihilar, and distal, according to their anatomic location. Several risk factors have been identified for certain types of CC, including chronic viral hepatitis, bile duct stones, and primary sclerosing cholangitis. The incidence rates of CC vary considerably among different geographical locations; its prevalence is markedly higher in Southeast Asia than in other parts of the world, due to liver fluke infestation. Although tumor resection can provide curative treatment of CC, surgery is only available for 10–20% of patients who are diagnosed at early stages. For patients diagnosed with advanced disease, treatment strategies include combination chemotherapy and targeted molecular therapy. However, patients with advanced CC have a poor prognosis, with a median overall survival of <1 year (1, 2).
Several studies have been performed to identify novel molecular targets in CC. A recent study suggested that patients with intrahepatic CC frequently have inactivating mutations in multiple chromatin-remodeling genes, including BAP1, ARID1A, and PBRM1 (3). Additionally, somatic mutations in isocitrate dehydrogenase 1 (IDH1) and IDH2 have been observed in 10–20% of patients with CC (4–7). However, the relationships of IDH mutations to clinicopathologic features and prognosis among patients with CC remains controversial (3–9). IDH mutations in CC have been associated with long-term improvement, worsening, or no impact (3, 5, 6, 8, 9). Moreover, while some studies showed that IDH mutations were associated with poorly differentiated CC and clear-cell histology (7), others showed no association with histological grade (3). Significant differences in IDH mutations between certain types of parasite-associated CC have also been reported (10).
However, most studies have focused on IDH mutations in intrahepatic CC and have involved non-Asian cohorts. Therefore, the aim of this study was to assess IDH mutations in a large cohort of Korean patients with CC at various anatomic locations, including intrahepatic and extrahepatic lesions. Furthermore, this study explored the associations of IDH mutations with clinicopathological features and long-term outcomes in patients with resected CC. In addition, this study used next-generation sequencing (NGS) analysis to investigate differences in IDH mutation patterns among patients with CC of different ethnicities.
Materials and Methods
Patients and Clinicopathological Data
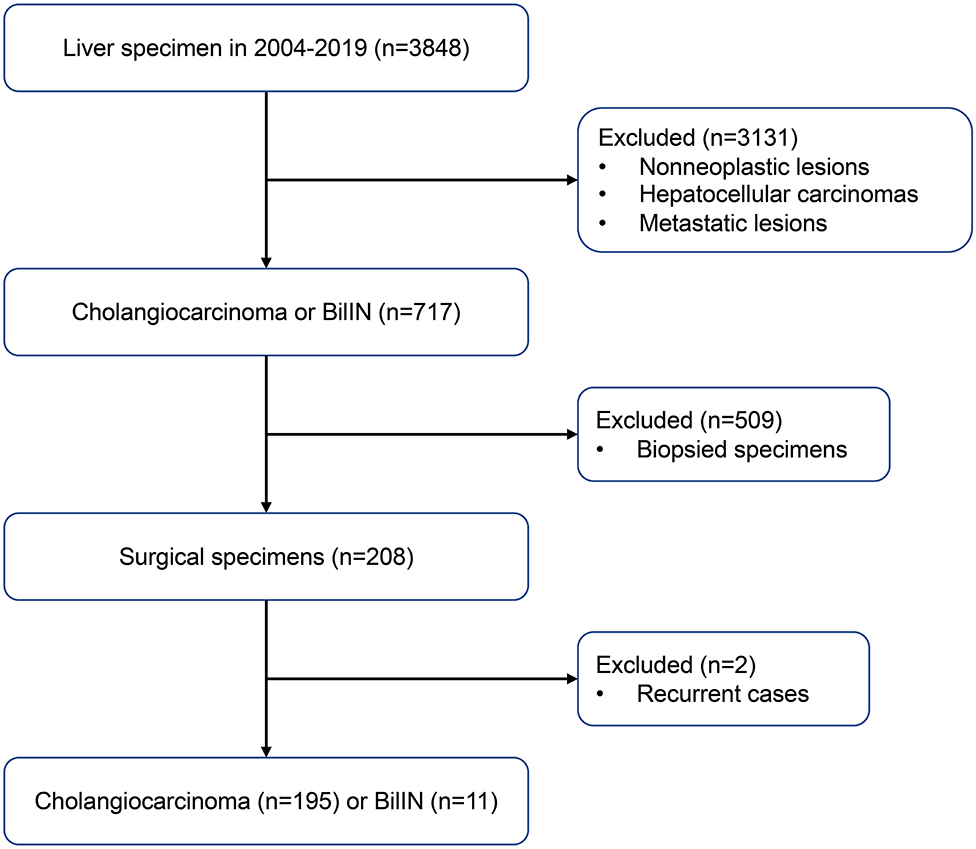
Samples from 195 patients with CC and 11 patients with biliary intraepithelial neoplasia (BilIN) were obtained during surgical resection of the liver at the Chonnam National University Hwasun Hospital between 2004 and 2019. A flow chart of case selection is presented in Figure 1. Clinical data were retrospectively reviewed from the patients' medical records. To investigate relationships between IDH mutations and clinicopathological parameters, the following variables were evaluated: age, sex, tumor size, tumor localization, presence of parasites or hepatitis, serum CA 19-9 levels, T stage, lymph node metastasis, distant metastasis, and surgical margin involvement. The pathological stage was determined in accordance with the American Joint Committee on Cancer (AJCC) staging system, 7th edition (11). Surgical margin involvement was defined as the presence of tumor within <1 mm from the excision margin. Disease-free survival (DFS) was calculated from the date of surgery to the date of recurrence, progression, or death. Overall survival (OS) was calculated from the date of surgery until death or the last follow-up visit.
All available tumor tissue slides were independently reviewed by two pathologists (KNI and LKH) to evaluate histopathological features and select representative tissue blocks. Tumors were classified and subtyped according to the World Health Organization classification of tumors of the digestive system (12). The following histopathological features were evaluated: histologic type, histologic grade, presence of mucinous or signet ring cell component, presence of sarcomatoid component, coexistence of BilIN, association with intraductal papillary neoplasm, bile duct stone, periductal inflammation, and presence of cirrhosis. Well-differentiated and moderately differentiated tumors were defined as low grade; poorly differentiated tumors, as well as carcinosarcomas and combined hepatocellular-CCs, were defined as high grade. Periductal inflammation was graded as minimal to mild, or moderate to severe, according to the degree of necroinflammatory activity. This study was approved by the Institutional Review Board of the Chonnam National University Hwasun Hospital (CNUHH-2019-213).
Detection of IDH Mutations by Peptide Nucleic Acid (PNA)-Mediated Real-Time PCR Clamping
Formalin-fixed paraffin-embedded tissue blocks from 206 patients were used to prepare 10-μm-thick sections. Genomic tumor DNA was extracted using the Maxwell 16 MDx Instrument (Promega, Madison, WI, USA), in accordance with the manufacturer's instructions. The tumor tissues were macro-dissected, in accordance with a previously described method (13). IDH1 and IDH2 mutations were analyzed using genomic DNA from 195 patients with CC and 11 patients with BilIN. The extracted genomic DNA was subjected to mutation analysis using the PNAClamp™ IDH Mutation detection kit (Panagene, Daejeon, Korea), as previously described (14). The mutation spots of IDH1 and IDH2 that are detected with the PNAClamp™ kit are listed in Supplementary Table 1. The PNA clamping probes were complementary to the wild-type sequence and competitively inhibited the binding of the DNA primers. Consequently, they preferentially amplified the mutant alleles. The reported sensitivity of the PNAClamp™ kit for IDH1/2 mutation testing is 1% with cloned DNA, according to the manufacturer's verification. Real-time PCR reactions were performed using a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
Next-Generation Sequencing (NGS) Analysis
NGS analysis was performed for the validation of IDH1 and IDH2 mutations. Genomic DNA was extracted from tumors after macro-dissection using the Gene-Read™ DNA FFPE Kit (Qiagen, Hilden, Germany), in accordance with the manufacturer's protocol. Targeted sequencing for 90 cancer-related genes was performed using the SureSelect targeted panel (Agilent Technologies, Santa Clara, CA, USA), as previously described (15). The processed libraries were loaded onto the MiSeqDx instrument (Illumina, San Diego, CA, USA), in accordance with the manufacturer's protocol. Sequenced reads were aligned to the human reference genome (GRCh37/hg19) using the BWA-MEM (0.7.15); common germline variants were distinguished from somatic variant candidates in accordance with a previously described method (15).
Parasite Detection Using PCR
Specimens with parasites were selected after observation under the microscope. Detection and identification of parasites (Opisthorchis viverrini and Clonorchis sinensis) were achieved using a PCR-based method. Two pairs of species-specific primers were designed to bind to the mitochondrial NADH dehydrogenase subunit 2 (nad2) genes of O. viverrini and C. sinensis. The primer sequences were OV-F (5′-ATG TAG TGT TGG TTG GAG TT-3′) and OV-R (5′-CAC AAT TAC CGC CGT AGC-3′) for O. viverrini, and CS-F (5′-GTC TGT TGA GCT TTC TCC T-3′) and CS-R (5′-TAA AGA CCC TGG AAA CGA GAT-3′) for C. sinensis. The PCR mixture contained 10 × reaction buffer, 10 mM dNTPs Mixture, Prime Taq DNA polymerase, and each of the four primers (OV-F, OV-R, CS-F, and CS-R), in a total reaction volume of 50 μL. The PCR cycling included one cycle at 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 55°C for 8 s, and 72°C for 15 s. PCR products were confirmed by sequencing using the same primers.
Immunohistochemistry
Immunohistochemistry for CD56 was performed on tissues found to exhibit IDH mutations. Tissue sections (3 μm thick) were prepared from paraffin-embedded tissue blocks. Tissue slides were subjected to immunohistochemistry staining using an automated immunostainer (Bond-MaX DC2002, Leica Biosystems, Bannockburn, IL, USA). Tissue sections were pretreated with bond epitope retrieval solution 1 (containing citrate buffer, pH 6.0), followed by incubation with CD56 antibody (1:400 dilution; cat. no.: M7304, DAKO, Glostrup, Denmark). Unstained tissues served as negative controls. Immunostained tissues were evaluated independently by two pathologists (KNI and LKH).
Statistical Analysis
Statistical analysis was performed using SPSS Statistics, version 25.0 (IBM Corp., Armonk, NY, USA) for Windows. To evaluate relationships between IDH mutations and clinicopathological parameters, the Pearson chi-squared test or Wilcoxon signed-rank test was used as appropriate. The effects of individual variables on survival were determined by univariate and multivariate analyses. For multivariate analysis, independent prognostic factors were determined using Cox proportional hazards models. Survival rates were calculated using the Kaplan–Meier method, and survival curves were compared using the log-rank test. P < 0.05 were considered statistically significant.
Results
Clinical Characteristics of Patients With CC
This study involved 195 patients with CC; 138 (70.8%) were men, and the remaining 57 (29.2%) were women. The mean age of patients at diagnosis was 63.1 years (range, 35–80 years). The mean tumor size was 4.38 cm. The majority of tumors were located in the intrahepatic area (168/195; 86.2%); adenocarcinoma (168/195; 86.2%) was the most common histologic type. T2 was the most common stage (81/195, 41.5%), according to the AJCC criteria. Forty-six patients had lymph node metastasis, while four exhibited distant metastasis at the time of diagnosis. Parasites were detected in eight patients by histological examination or gross detection in the operating field. Further PCR analysis using genomic DNA from the tumors and organism-specific primers did not reveal O. viverrini or C. sinensis infestations in other patients. The clinicopathological features of the patients in this study are summarized in Table 1.
Relationships Between IDH Mutations and Clinicopathological Features
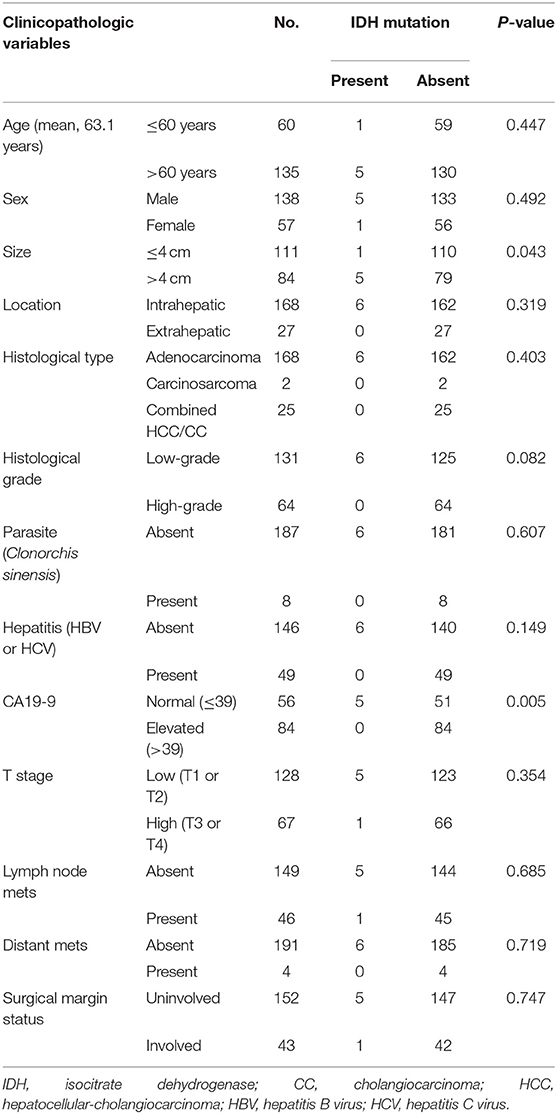
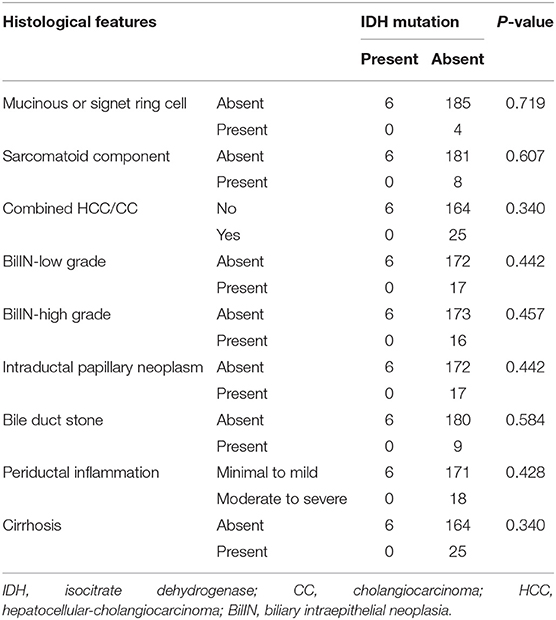
In total, samples from 195 patients with CC and 11 patients with BilIN were screened for the presence of IDH mutations. IDH mutations were detected in six patients with CC by PNA clamping; further validation with NGS confirmed the presence of these mutations. Five patients harbored mutations in codon 132 of IDH1 (R132C in four patients and R132G in one patient), while one patient had mutations in codon 172 of IDH2 (R172W) (Figure 2). No IDH mutations were identified in patients with BilIN. In patients with CC, the presence of IDH mutations was significantly associated with larger tumor size (>4 cm; P = 0.043). Although patients harboring IDH mutations tended to have histologically low-grade tumors, the association between IDH mutations and tumor grade was not statistically significant (P = 0.082). Furthermore, IDH mutations were significantly associated with normal CA19-9 serum levels (P = 0.005). There were no significant relationships between IDH mutations and the following factors: age, sex, tumor location, histologic type, presence of parasites or hepatitis, pathologic stage, metastasis, or surgical margin status (Table 1). Additionally, there were no significant association between IDH mutations and these additional factors: the presence of bile duct stones, inflammation, cirrhosis, BilIN, or intraductal papillary neoplasms. This absence of an association was also true regarding histopathological features of the tumor, including histologic components of mucinous, signet ring, or sarcomatoid cells (all P > 0.05, Table 2). In addition, CD56 immunohistochemistry was performed using tissues from six patients with IDH mutations; patchy and focal CD56 expression was observed in all tumors with IDH mutations (Figure 3). There were no notable histopathological differences among different IDH1 or IDH2 mutations.
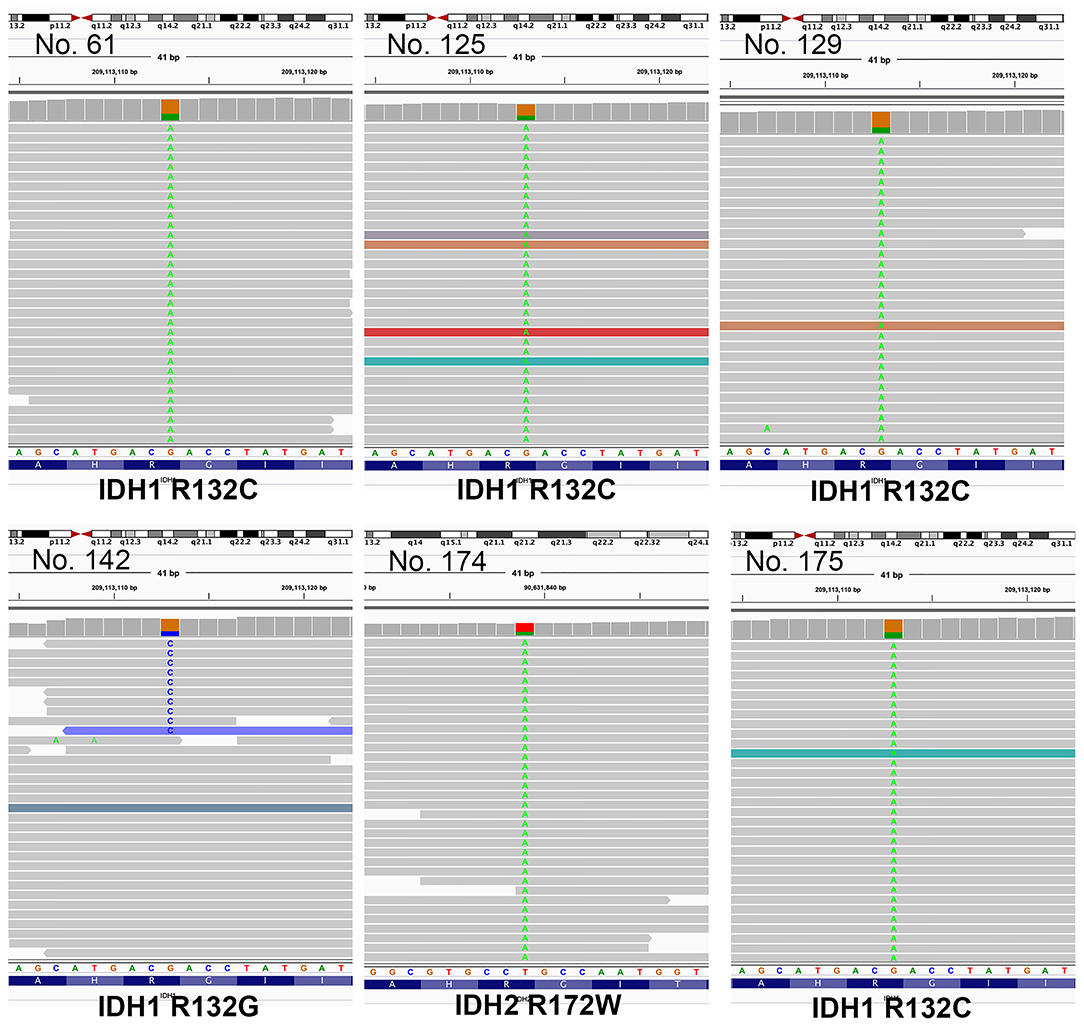
Figure 2. Illustration of mutations identified by next-generation sequencing (NGS), visualized in Integrative Genome Viewer (IGV). Patients 61, 125, 129, and 175 had mutations in codon 132 of IDH1 (R132C). Patient 142 had R132G mutations, while patient 172 had IDH2 mutations in codon 172 (R172W).
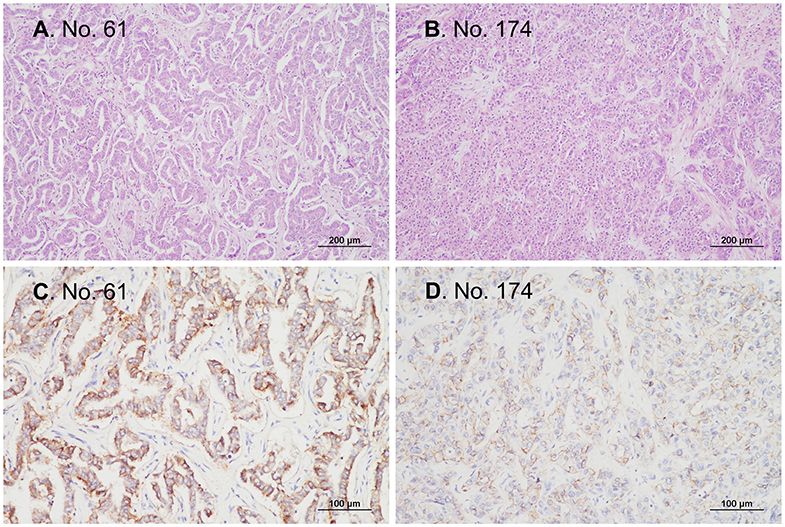
Figure 3. Representative microphotographs of tissues from patients with IDH-mutant cholangiocarcinoma. All patients with IDH-mutant cholangiocarcinoma in the current study were patients with conventional adenocarcinomas that were immunopositive for CD56. (A,B) H&E staining, original magnification ×100. (C,D) Immunohistochemistry, original magnification ×200.
Identification of DFS and OS Predictors in Patients With CC
DFS was analyzed in relation to clinical variables. Univariate and multivariate analyses revealed significant effects of the presence of cirrhosis, lymph node metastasis, surgical margin status, or serum CA 19-9 levels on patient survival (all P < 0.05; Figure 4). Although univariate analysis revealed that smaller tumor size (P = 0.001) and low pathological T stage (P < 0.001) were significantly associated with prolonged DFS, neither of these was identified as an independent prognostic factor by multivariate analysis (P = 0.291 and P = 0.355, respectively). Moreover, sex and enhanced periductal inflammation were significantly associated with DFS in multivariate analysis (P = 0.013 and P = 0.001, respectively). Neither univariate nor multivariate analysis identified IDH mutations as significant prognostic factors in CC (both P > 0.05, Table 3).
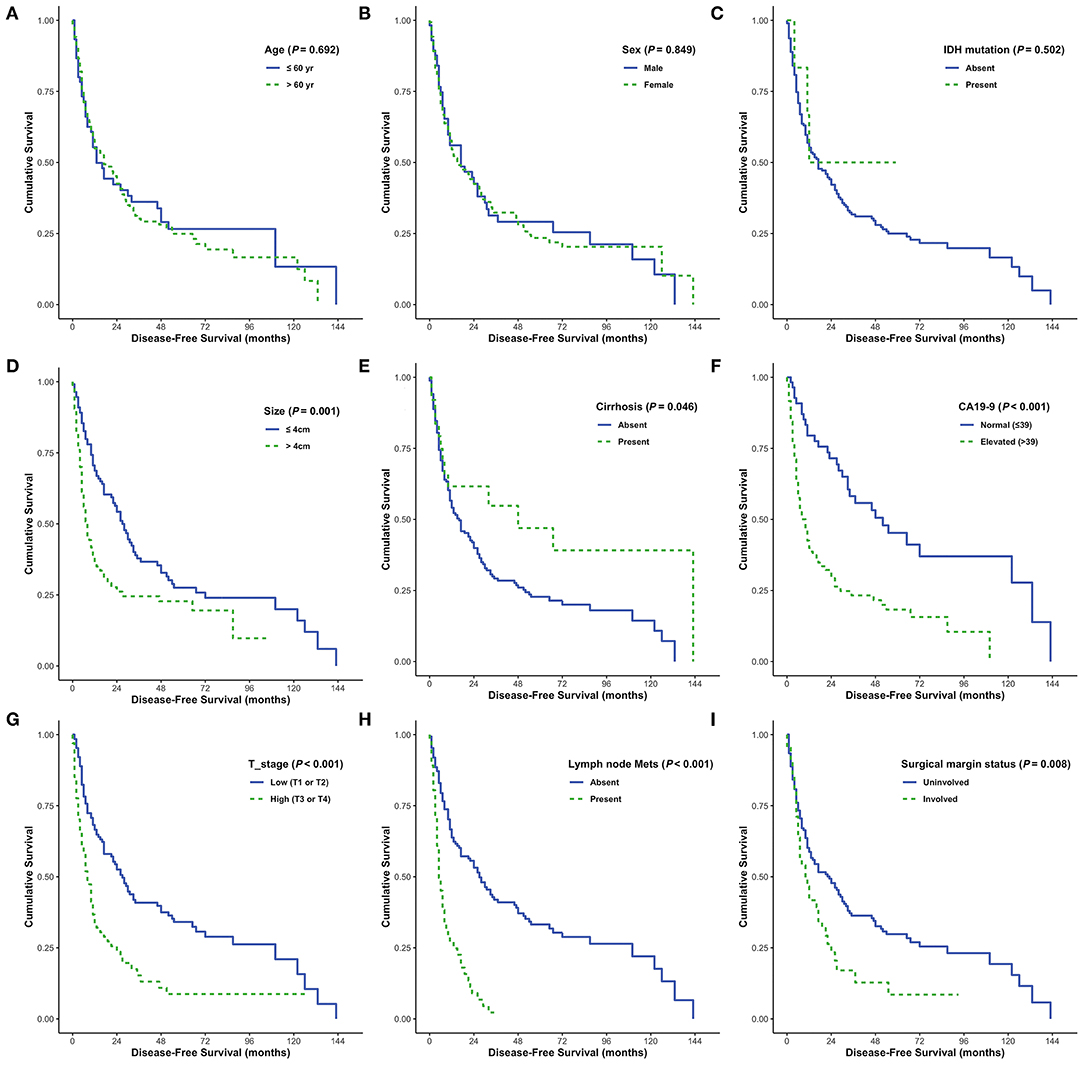
Figure 4. Disease-free survival (DFS) analyses using the Kaplan-Meier estimator and log-rank test were performed according to age (A), sex (B), IDH mutations (C), tumor size (D), cirrhosis (E), CA19-9 (F), T stage (G), lymph node metastasis (H), and surgical margin status (I).
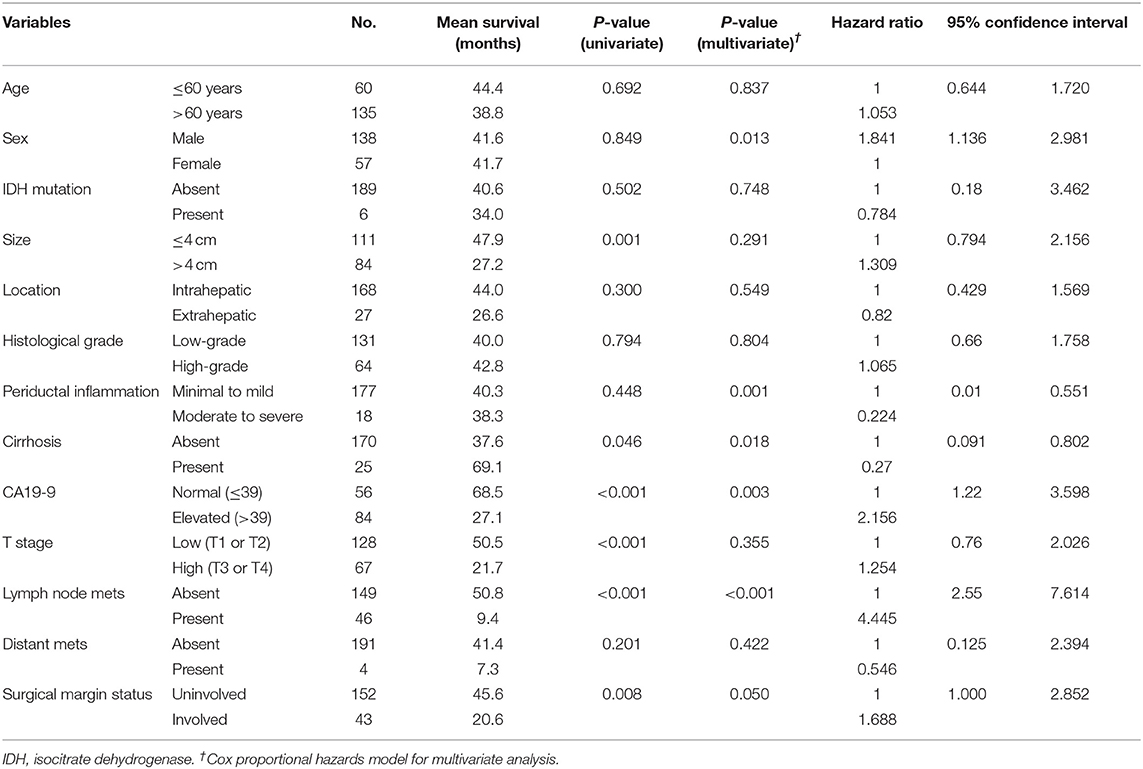
Table 3. Univariate and multivariate analysis for progression-free survival predictors in 195 patients with cholangiocarcinoma.
Elevated serum CA19-9 levels, lymph node metastasis, and positive surgical margins were independent predictors of poor OS in patients with CC (all P < 0.05; Figure 5). Moreover, cirrhosis was associated with prolonged survival in both univariate and multivariate analyses (P = 0.009 and P = 0.072, respectively). Intrahepatic CC and absence of distant metastases were associated with prolonged survival in univariate analysis (P = 0.064 and P = 0.069, respectively); however, no significant association was identified by multivariate analysis. Enhanced periductal inflammation showed a significant association with improved OS in multivariate analysis (P = 0.012). Age, sex, tumor size, and histological grade were not significantly associated with OS. Furthermore, no significant associations were observed between the presence of IDH mutations and OS (both P > 0.05, Table 4).
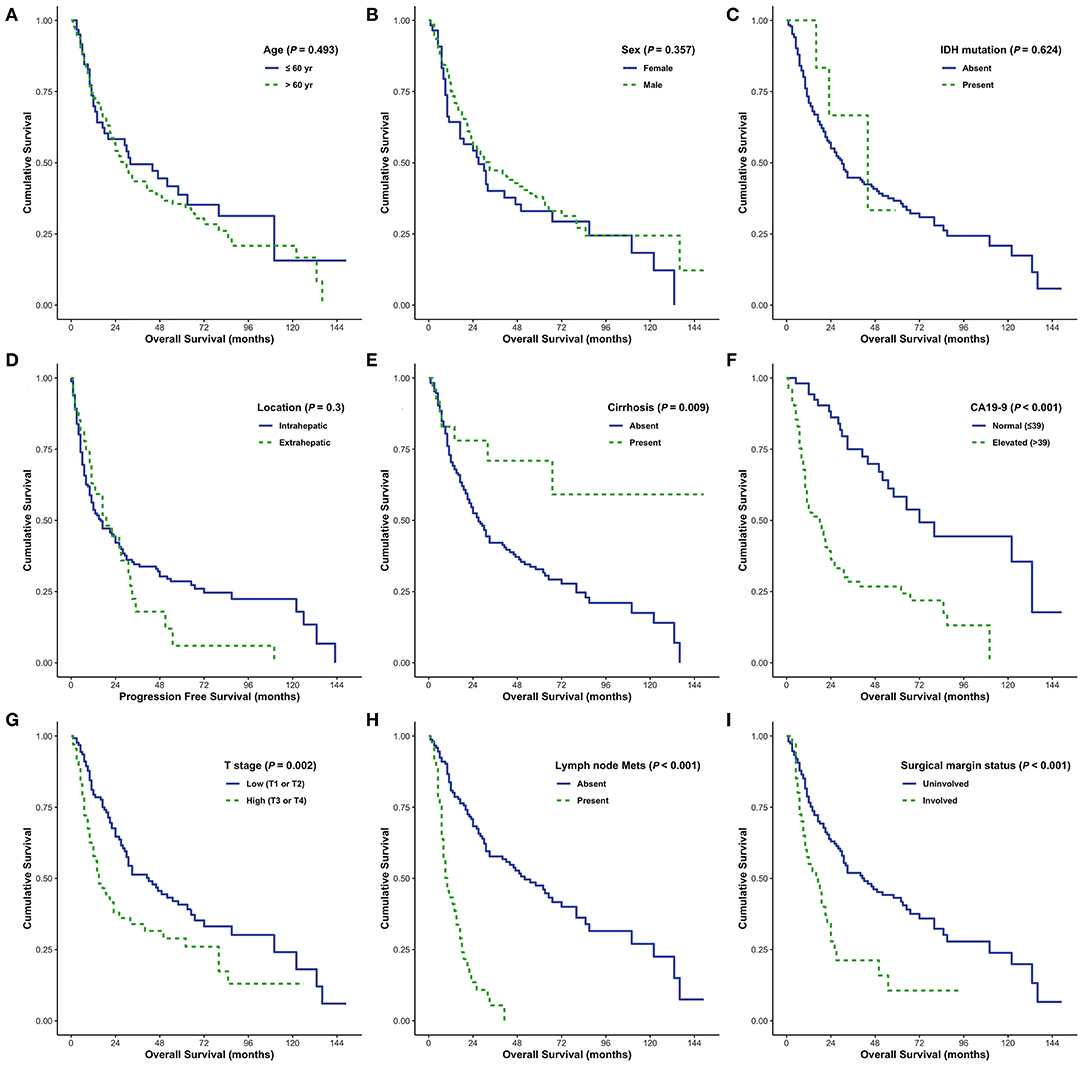
Figure 5. Overall survival (OS) analyses using the Kaplan–Meier estimator and log-rank test were performed according to age (A), sex (B), IDH mutations (C), tumor size (D), cirrhosis (E), CA19-9 (F), T stage (G), lymph node metastasis (H), and surgical margin status (I).
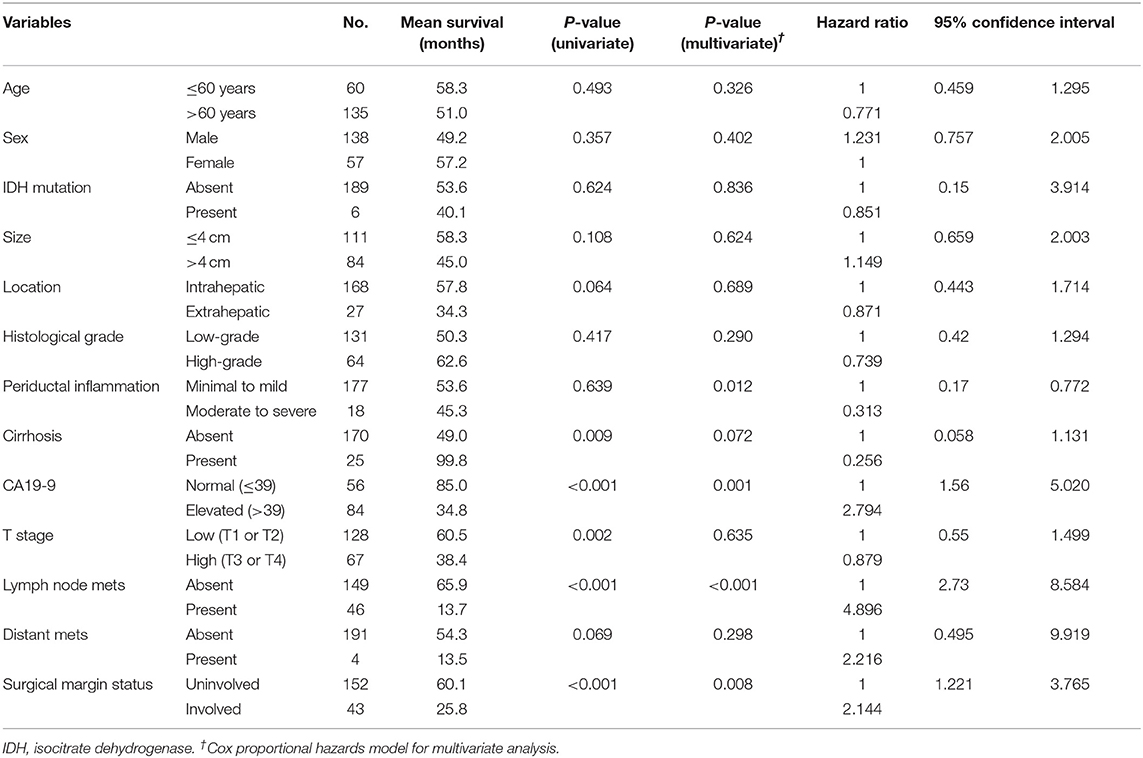
Table 4. Univariate and multivariate analysis for overall survival predictors in 195 patients with cholangiocarcinoma.
Discussion
Despite recent improvements in understanding the molecular mechanisms underlying cancer, little is known regarding genomics in CC. The genomic profiling of CC and analysis of IDH mutations, in particular, have gained increasing interest in recent years. IDH1 and IDH2 exhibit high sequence similarity and are often present in various malignancies, including glioma, leukemia, and cartilaginous tumors (16). IDH1 mutations have been identified in 9.2% (191 of 2,079) of biliary tract tumors in the COSMIC database (as of March 2020). In the present study, we identified IDH1 and IDH2 mutations in 3.1% (six of 195) of patients with CC; hence, the incidence of IDH mutations in our cohort was relatively low, compared to the incidences in previous reports (5–7, 9, 17). However, these results were consistent with the findings of previous studies in Asian populations, which reported low frequencies of IDH mutations. Because the vast majority of data regarding IDH mutations in CC are derived from studies in non-Asian populations, differences in patient ethnicities among studies may contribute to discrepancies in IDH mutation frequencies. Risk factors associated with CC include liver flukes and chronic viral hepatitis, which are prevalent in Asian countries; thus, differences in risk factor distributions might partly explain the differences in IDH mutation prevalences in CC among individuals of different ethnicities.
Moreover, C. sinensis and O. viverrini infestations are strongly associated with CC development. Chan-On et al. reported that IDH1 and IDH2 mutations were more frequent in non-O. viverrini CC, compared to O. viverrini-associated CC (10). However, we were unable to reproduce the previously reported associations between IDH mutations and parasite infestation. Future large cohort and multi-ethnic studies are needed to confirm the relationships between liver fluke infestation and IDH mutations.
Types of IDH mutations vary significantly among tumor types. IDH1 mutations in codon R132 are the predominant IDH mutations in brain tumors; these mutations are functionally similar to R172 mutations in IDH2. IDH1 R132H mutations represent ~90% of all glioma-associated IDH mutations; IDH1 R132C, R132S, R132G, and R132L, as well as IDH2 R172K, R172M, and R172W represent the remaining 10%. Unlike glioma, most previously reported IDH mutations in CC occurred in codon R132C of IDH1. Our study confirmed the presence of IDH1 and IDH2 mutations in CC. Among the six resected CC specimens harboring IDH1 or IDH2 mutations, mutations in codon R132C and R132G of IDH1 were detected in specimens from five patients, while a specimen from the remaining patient had mutations in codon R172W. IDH1 mutations were more frequent than IDH2 mutations in CC, and the majority of IDH1 mutations were present in codon R132C, consistent with the findings of previous studies. However, unlike glioma, R132H mutations were not observed in our cohort. IDH mutations in glioma typically accumulate in lower-grade gliomas early during tumor initiation and are maintained throughout progression to high-grade malignancy (18). To assess the prevalence of IDH mutations early during CC development, we analyzed IDH mutations in samples from patients with BilIN, which constitute precursor lesions of CC. However, no IDH mutations were detected in BilIN specimens.
IDH mutations are found in ~20% of patients with intrahepatic CC. Borger et al. presumed that IDH1 mutations represented a molecular feature of CC of intrahepatic origin (4). Kipp et al. also found that IDH mutations were more frequent in intrahepatic CC than in extrahepatic CC; moreover, IDH mutations were associated with clear cell lesions and poorly differentiated histology (7). In the present study, we found that all IDH-mutant CC samples were CD56-positive; thus, it is likely that these tumors represented the small duct type of intrahepatic CC (19). In CC, conflicting data exist regarding the prevalence and clinical significance of IDH mutations. Most previous studies focused on IDH1 genetic profiling in patients with intrahepatic CC; conversely, the present study involved analysis of mutations in both IDH1 and IDH2 in a large cohort of patients with diverse types of CC. We found that IDH mutations tended to be associated with low-grade histology, although this relationship was not statistically significant (P = 0.082). No other histological types or features were associated with IDH1 or IDH2 mutations. Consistent with the results of prior studies, we found that all instances of IDH-mutant CC originated in the intrahepatic area. Given that the majority of CC samples were of intrahepatic origin and were acquired during surgical resection of the liver, the frequency of IDH mutations might have been skewed.
The relationships between IDH mutations and prognosis have been investigated in various tumors. IDH mutations were associated with favorable outcomes in patients with glioma (20). In contrast, worse OS or no impact on prognosis was observed in patients with acute myeloid leukemia (21). In a cohort of 326 patients with resected intrahepatic CC, Wang et al. found that patients with IDH mutations exhibited prolonged DFS and OS (5). Another study showed that IDH1 mutations in intrahepatic CC were associated with favorable prognosis, smaller tumor size, lower serum CA19-9 levels, and lower TNM stage. Furthermore, IDH1 mutations reportedly affected growth inhibition in intrahepatic CC by suppressing AKT signaling, both in vivo and in vitro (8). In contrast, analysis of a cohort of 32 patients with IDH mutant or IDH wild-type intrahepatic CC revealed shorter median OS in patients with IDH mutations (3). However, more patients with IDH mutations had stage IV disease (50%), compared to patients with IDH wild-type (15%); this difference might have affected the survival rate and prognosis. Zhu et al. detected IDH mutations in 20% of the patients (15.5% IDH1 and 4.5% IDH2) in their study of 200 patients with resected intrahepatic CC (6); no significant differences in DFS or OS were reported among patients with different IDH mutation statuses. Similarly, Goyal et al. found no association between IDH mutations and prognosis in patients with intrahepatic CC; however, patients with IDH mutations had lower serum CA19-9 levels at presentation (9). We found that IDH mutations were more frequent (5/6) in patients with larger tumor size (>4 cm). This finding was consistent with the results described by Goyal et al., who reported that IDH mutations were significantly associated with physiological serum CA19-9 levels. In addition, they observed no associations between mutational status and long-term outcomes. We also found no significant associations between IDH mutations and OS or DFS in patients with CC.
There were some limitations in this study. First, the number of extrahepatic cases was relatively small because patients who underwent liver resection were enrolled in this study. Second, the study cohort was from a single institution, and the results might have been influenced by the regional characteristics of the patients. Nevertheless, this study included a large Asian cohort. Data regarding genetic variation in CC are primarily derived from studies in Western cohorts, which have a relatively low incidence of hepatobiliary cancer. Therefore, analysis of genetic variation in Asian patients with CC is crucial. The identification of genetic characteristics associated with risk factors for CC, one of the most intractable cancers with poor prognosis, will foster the development of more effective treatment strategies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena/browser/view/PRJEB38130.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of the Chonnam National University Hwasun Hospital (CNUHH-2019-213). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
K-HL and J-HL designed this study. EW and YL performed the experiments. NK and M-GN drafted the manuscript. M-GN and J-HK performed data analysis. YH and K-SM collected clinical data. NK and K-HL performed the pathological examination. K-HL and M-GN performed statistical analyses. K-SM and J-HL assisted with manuscript preparation and data analysis. All authors read and approved the final manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), which was funded by the Ministry of Science and ICT (2018R1A5A2024181 and 2019R1A2B5B01070598) and the Chonnam National University Hospital Biomedical Research Institute (HCRI19030 for K-HL and K-SM). The funding bodies had no involvement in the study design; collection, analysis, and interpretation of data; or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to express our gratitude to Wun-Cheol Kim, for his technical support for PNA clamping PCR and NGS analysis. The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/index/jsvw93.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01514/full#supplementary-material
References
1. Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. (2004) 24:115–25. doi: 10.1055/s-2004-828889
2. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. (2010) 362:1273–81. doi: 10.1056/NEJMoa0908721
3. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. (2013) 45:1470–3. doi: 10.1038/ng.2813
4. Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. (2012) 17:72–9. doi: 10.1634/theoncologist.2011-0386
5. Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. (2013) 32:3091–100. doi: 10.1038/onc.2012.315
6. Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, et al. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Ann Surg Oncol. (2014) 21:3827–34. doi: 10.1245/s10434-014-3828-x
7. Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, Zhang L, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. (2012) 43:1552–8. doi: 10.1016/j.humpath.2011.12.007
8. Wang J, Zhang ZG, Ding ZY, Dong W, Liang HF, Chu L, et al. IDH1 mutation correlates with a beneficial prognosis and suppresses tumor growth in IHCC. J Surg Res. (2018) 231:116–25. doi: 10.1016/j.jss.2018.04.056
9. Goyal L, Govindan A, Sheth RA, Nardi V, Blaszkowsky LS, Faris JE, et al. Prognosis and clinicopathologic features of patients with advanced stage isocitrate dehydrogenase (IDH) mutant and IDH wild-type intrahepatic cholangiocarcinoma. Oncologist. (2015) 20:1019–27. doi: 10.1634/theoncologist.2015-0210
10. Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. (2013) 45:1474–8. doi: 10.1038/ng.2806
11. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. New York, NY: Springer International Publishing (2010).
12. Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. 4th ed. Bosman FT, Jaffe ES, Lakhani SR, Ohgaki H, editors. Lyon: France International Agency for Research on Cancer (2010).
13. Na JI, Kim JH, Kim HJ, Kim HK, Moon KS, Lee JS, et al. VE1 immunohistochemical detection of the BRAF V600E mutation in thyroid carcinoma: a review of its usefulness and limitations. Virchows Arch. (2015) 467:155–68. doi: 10.1007/s00428-015-1773-0
14. Lee D, Suh YL, Kang SY, Park TI, Jeong JY, Kim SH. IDH1 mutations in oligodendroglial tumors: comparative analysis of direct sequencing, pyrosequencing, immunohistochemistry, nested PCR and PNA-mediated clamping PCR. Brain Pathol. (2013) 23:285–93. doi: 10.1111/bpa.12000
15. Kim SS, Cho YM, Kim GH, Kee KH, Kim HS, Kim KM, et al. Recurrent KRAS mutations identified in papillary renal neoplasm with reverse polarity-a comparative study with papillary renal cell carcinoma. Mod Pathol. (2020) 33:690–9. doi: 10.1038/s41379-019-0420-8
16. Schaap FG, French PJ, Bovee JV. Mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in tumors. Adv Anat Pathol. (2013) 20:32–8. doi: 10.1097/PAP.0b013e31827b654d
17. Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. (2010) 28:3717–23. doi: 10.1200/JCO.2010.28.2285
18. Kim JH, Jang WY, Jung TY, Jung S, Kim KK, Kim HS, et al. Recurrent glioma with lineage conversion from oligodendroglioma to astrocytoma in two cases. Front Oncol. (2019) 9:828. doi: 10.3389/fonc.2019.00828
19. Sigel CS, Drill E, Zhou Y, Basturk O, Askan G, Pak LM, et al. Intrahepatic cholangiocarcinomas have histologically and immunophenotypically distinct small and large duct patterns. Am J Surg Pathol. (2018) 42:1334–45. doi: 10.1097/PAS.0000000000001118
20. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. (2009) 360:765–73. doi: 10.1056/NEJMoa0808710
Keywords: cholangiocarcinoma, isocitrate dehydrogenase, mutation, high-throughput nucleotide sequencing, survival rate
Citation: Kim NI, Noh M-G, Kim J-H, Won EJ, Lee YJ, Hur Y, Moon K-S, Lee K-H and Lee J-H (2020) Frequency and Prognostic Value of IDH Mutations in Korean Patients With Cholangiocarcinoma. Front. Oncol. 10:1514. doi: 10.3389/fonc.2020.01514
Received: 04 May 2020; Accepted: 14 July 2020;
Published: 18 August 2020.
Edited by:
Qingfeng Zhu, Johns Hopkins Medicine, United StatesReviewed by:
Ding Ding, Johns Hopkins Medicine, United StatesZach Reitman, Duke Cancer Institute, United States
Copyright © 2020 Kim, Noh, Kim, Won, Lee, Hur, Moon, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyung-Hwa Lee, mdkaylee@jnu.ac.kr; mdkaylee@gmail.com; Jae-Hyuk Lee, jhlee@chonnam.ac.kr
†These authors have contributed equally to this work
 Nah Ihm Kim1†
Nah Ihm Kim1† Myung-Giun Noh
Myung-Giun Noh Eun Jeong Won
Eun Jeong Won Kyung-Sub Moon
Kyung-Sub Moon Kyung-Hwa Lee
Kyung-Hwa Lee