- 1Department of Urology, Yijishan Affiliated Hospital, Wannan Medical College, Wuhu, China
- 2Department of Oncology and Hematology, Hospital (T.C.M) Affiliated to Southwest Medical University, Luzhou, China
- 3Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, China
- 4South Sichuan Institute of Translational Medicine, Luzhou, China
- 5Department of Pharmacy, Yijishan Affiliated Hospital, Wannan Medical College, Wuhu, China
- 6Department of Oncology, The Affiliated Hospital of Southwest Medical University, Luzhou, China
Docetaxel is a major treatment for advanced prostate cancer (PCa); however, its resistance compromises clinical effectiveness. Estrogen receptor-related receptor alpha (ERRα) belongs to an orphan nuclear receptor superfamily and was recently found to be closely involved in cancer. In the present study, we found that ERRα was involved in docetaxel resistance in PCa. Overexpression of ERRα conferred docetaxel resistance in PCa cell lines, and cells with ERRα downregulation were more sensitive to docetaxel. Among the drug resistance-related genes, ABCC4 demonstrated synchronous expression after ERRα manipulation in cells. Moreover, both ERRα and ABCC4 were overexpressed in the docetaxel-resistant cell, which could be reversed by ERRα knockdown. The knockdown of ERRα also reversed the reduced drug accumulation in the docetaxel-resistant cell. We also demonstrated for the first time that ABCC4 was a direct target of ERRα as determined by the CHIP and luciferase assays. Bioinformatics analysis revealed high expression of ERRα and ABCC4 in PCa patients, and a number of potential ERRα/ABCC4 targets were predicted. In conclusion, our study demonstrated a critical role for ERRα in docetaxel resistance by directly targeting ABCC4 and stressed the importance of ERRα as a potential therapeutic target for drug-resistant PCa.
Introduction
Prostate cancer (PCa) is one of the most common and heritable malignancies and the third leading cause of cancer death in men, especially in the western population (1). Docetaxel provides effective control for most of PCa, including high-risk or localized advanced PCa, metastatic hormone-sensitive PCa, metastatic castrate-sensitive, and castration-resistant PCa (2–5). However, docetaxel chemotherapy and docetaxel-based combination therapy (docetaxel with androgen deprivation therapy) also have certain limitations and problems clinically. Docetaxel can moderately prolong the overall survival (OS) of patients but gradually lead to disease progression on account of inherent or acquired docetaxel resistance (6, 7). In addition, a combination treatment of docetaxel and other medicines (like abiraterone, Enzalutamide) even can give rise to clinical cross-resistance (8, 9). Multiple mechanisms, including reduced intracellular concentrations of drug (increased drug efflux), androgen-receptor (AR) activation, central transcription factor, changes in B-tubulin isotype expression, apoptotic pathways, angiogenesis, mutations in tumor suppressor proteins, and so on, have been demonstrated to be involved in the development of docetaxel resistance (7, 10, 11). Docetaxel resistance is one of the major hurdles that must be overcome in order to achieve successful treatment of PCa with chemotherapy and docetaxel-based combination therapy.
Estrogen receptor-related receptor alpha (ERRα) is one of the three subtypes of ERRs (ERRα/NR3B1, ERRβ/NR3B2, ERRγ/NR3B3) (12) and belongs to an orphan nuclear receptor super family of DNA-binding transcription factors. Increasing scientific evidence has indicated that ERRα is closely involved in carcinogenesis and tumor progression, including breast cancer (13), prostate cancer (14), and bone tumor progression which are related to advanced prostate cancer (15). Indeed, in PCa, ERRα is detected in cancer cell lines, xenografts, and cancerous lesions (16), and it is regarded as a negative prognostic predictor (17). However, thorough knowledge about the functional role of ERRα in PCa is lacking.
The protein encoded by ABCC4 is an important member of the ATP-binding cassette (ABC) membrane transporter family which can transport various molecules across extra/intra cellular membranes, and is involved in the export of endogenous signaling molecules and chemotherapeutic agents (18, 19) and the transport of steroid hormones (20). The inhibition of ABCC4 has been shown to be beneficial for atherothrombotic disease due to its involvement in vascular biology and in platelet functions (21–23). Importantly, the broad-spectrum resistance in cancer cells is often caused by the overexpression of ABC transporters (24), and upregulation of ABCC4 transcription is correlated with multidrug resistance in various kinds of cancer (25, 26). Hence, ABCC4 is also known as the multidrug resistance protein 4 (MRP4). In PCa, higher-level expression of ABCC4 is reported in malignant prostate tissues when compared to benign prostate tissues (27). Importantly, ABCC4 is considered as a key determinant of docetaxel resistance in PCa cells because of reversed drug resistance by inhibiting ABCC4 expression (28).
A growing body of scientific evidence has demonstrated that both ERRα and ABCC4 are highly expressed in PCa and relate to tumor progression. It is also notable that ABCC4 is crucial for multi-drug resistance, especially docetaxel resistance. This study aimed at discovering whether ERRα could regulate drug resistance-related genes and revealing the relationship between ERRα and ABCC4.
Materials and Methods
Cell Culture and Reagents
Human epithelial PCa cells (PC3) were cultured in F-12K medium with 10% fetal bovine serum, and C4-2B was cultured in 4:1 ratio of DMEM and F12 medium with 11% fetal bovine serum and 1.1% T-Media. To establish docetaxel-resistant cells including PC3/DR and C4-2B/DR, we used the increasing concentration method. Firstly, we confirmed the IC50 of PC3 and C4-2B for docetaxel, respectively. We used the IC50 as a starting concentration. Cells were cultured in increasing concentrations of docetaxel for about 2 months and were subsequently stimulated with docetaxel once in a while. Docetaxel was purchased from Sigma (USA). The antibodies used were ERRa (ab76228, Abcam) for Western blots, ERRa (PA5-28390, Thermo Fisher) for ChIP, Rabbit monoclonal for MRP4 (ab233382, Abcam), and beta-actin (ab115777, Abcam) and HRP-conjugated secondary antibodies (A0208, Beyotime).
MTT Assay
Cell viability was measured by the 3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay as previously described (29).
Total RNA and Protein Extraction
Total RNA was isolated from the PCa cell by Trizol (Invitrogen) in accordance with the instructions. Total protein was lysed from the PCa cell in RIPA buffer with complete protease inhibitor and phosphatase inhibitor. Both RNA and protein were stored at −80°C.
Plasmid Construction
Full-length human ERRα cDNA was synthetized and inserted into plenti-puro vector using homologous recombination. The plasmid was named plenti-ERRα (Genscript Co. Ltd., Nan Jing, China). Cells were transfected with plenti-ERRα using Lipofectamine 2000 (Thermo Fisher, USA), and a stable clone was established by puromycin selection. The sh-ERRα sequence which was well-identified before was synthetized and inserted into a pLKO.1-TRC vector by homologous recombination. The knockdown plasmid was named pLKO-shERRα (Genscript Co. Ltd., Nan Jing, China). Lentiviral shRNA production, infection, and stable cell selection were performed as previously described (30).
Quantitative Real-Time PCR Analysis
Reverse transcription was done using the FastKing RT kit (with gDNase). Real-time qPCR was performed by using FastFire qPCR PreMix. Both RT and qPCR kits were purchased from TIANGEN (China). Primers for qPCR were as follows: ERRα Forward: CCACTATGGTGTGGCATCCTGT, Reverse: GGTGATCTCACACTCGTTGGAG; ABCC1 Forward: CCGTGTACTCCAACGCTGACAT, Reverse: ATGCTGTGCGTGACCAAGATCC; ABCC2 Forward: GCCAACTTGTGGCTGTGATAGG, Reverse: ATCCAGGACTGCTGTGGGACAT; ABCC3 Forward: GAGGAGAAAGCAGCCATTGGCA, Reverse: TCCAATGGCAGCCGCACTTTGA; ABCC4 Forward: CTGTTGGAGGATGGTGATCTGAC, Reverse: CTGCTAACTTCCGCATCTACTGC; ABCC5 Forward: GGCTGTATTACGGAAAGAGGCAC, Reverse: TCTTCTGTGAACCACTGGTTTCC; ABCG2 For-ward: GTTCTCAGCAGCTCTTCGGCTT, Reverse: TCCTCCAGACACACCACGGATA; MDR1 Forward: GCTGTCAAGGAAGCCAATGCCT, Reverse: TGCAATGGCGATCCTCTGCTTC; β-actin Forward: CACCATTGGCAATGAGCGGTTC, Reverse: AGGTCTTTGCGGATGTCCACGT.
Western Blot
Electrophoretic analysis of different proteins was carried out on SDS–PAGE and transferred to a PVDF membrane. Experiment was performed following the standard laboratory protocol as previously reported (29).
Chromatin Immunoprecipitation Assay (ChIP)
The ChIP assay was performed as described previously (29). Cross-linked chromatin was incubated overnight with anti-ERRα. The precipitated DNA was analyzed by qPCR. Primers used were as follows: ABCC4 P1 Forward: TTACCCGGCTTTCTTGAGGT, Reverse: GGTTTGGGAAGACTGGGAGA; ABCC4 P2 Forward: GGGTGGATATGAAGAGCAGC, Reverse: TCTAAGCATGGCCTGTCTCC; ABCC4 P3 Forward: GGTGACAGAGCAAGACCCTA, Reverse: ACTCTTGTCTTAGGGTCTTGTCA.
Luciferase Reporter Assay
The wild-type or ERRα-binding site-deleted ABCC4 promoter region was subcloned into the pMIR–REPORT reporter (Life Technologies). ERRα binding sites P1 and P2 were deleted separately (Genscript Co. Ltd., Nan Jing, China). The luciferase activity was determined as previously described (31).
High Performance Liquid Chromatography (HPLC)
Chromatographic separation was performed on an Agilent 1,290 LC system using a ZORBAX Eclipse Plus C18 column (2.1 × 100 mm, 1.7 μm, Agilent Technologies, USA). The column temperature was set at 35°C, and the flow rate was 0.30 mL/min. The temperature of the autosampler was maintained at 4°C. The mobile phases consisted of A (Acetonitrile) and B (Distilled water with 0.1% formic acid). The gradient elution program was as follows: 0–1.5 min, A, 10–30%; 1.5–5 min, A, 30–95%; 5–7 min, A, 95%; and return to the initial condition for a 3-min equilibration. The injection volume was 20 μL. The UV absorbance was monitored at 227 nm.
Accumulation of Docetaxel in C4-2B/DR Cells
All the cells were seeded into 24-well plates and allowed to grow overnight. To study the intracellular accumulation of docetaxel, the cells were incubated with docetaxel in complete culture medium at 37°C for 30 min. Afterwards, the cells were washed twice with ice-cold PBS. The intracellular docetaxel was determined by HPLC after cell lysis. The values were normalized to the protein content.
cBioPortal Database Analysis
The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) is one of the major and authoritative sources of cancer genomics data which is crucial for our research. Relevant information about ERRα and ABCC4 in PCa including mRNA expression levels, association with clinical parameters, genetic alteration, protein expression, interaction network, and co-expression genes was analyzed by cBioPortal.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7. All data are expressed as mean ± SD from three separate experiments performed in triplicate. The co-expression level was analyzed by Spearman's correlation test, and a cutoff of 0.3 was used. The Pearson test and Spearman rank test were used for correlation analysis. The Kaplan-Meier survival curve by using the log-rank test was used for overall survival analysis. p < 0.5 was considered statistically significant.
Results
The Influence of ERRα Manipulation on Docetaxel Sensitivity
In order to assess the influence of ERRα expression on docetaxel sensitivity in PCa, we chose two PCa cells (C4-2B and PC3) treated with different concentrations of docetaxel (ranging from 10 to 50 nM) following the overexpression and knockdown of ERRα. Results demonstrated that C4-2B and PC3 cells with elevated expression of ERRα displayed significant resistance to docetaxel treatment when compared with the control group (Figures 1A,B). At the same time, C4-2B cells with ERRα knockdown were more sensitive to docetaxel (Figure 1C). However, there is no significant difference in PC3 cells with or without ERRα knockdown to docetaxel treatment (Figure 1D). We also confirmed the result with the colony formation assay (Supplementary Figure 1). The above data suggested that ERRα up-expression was linked to docetaxel resistance in PCa cell lines.
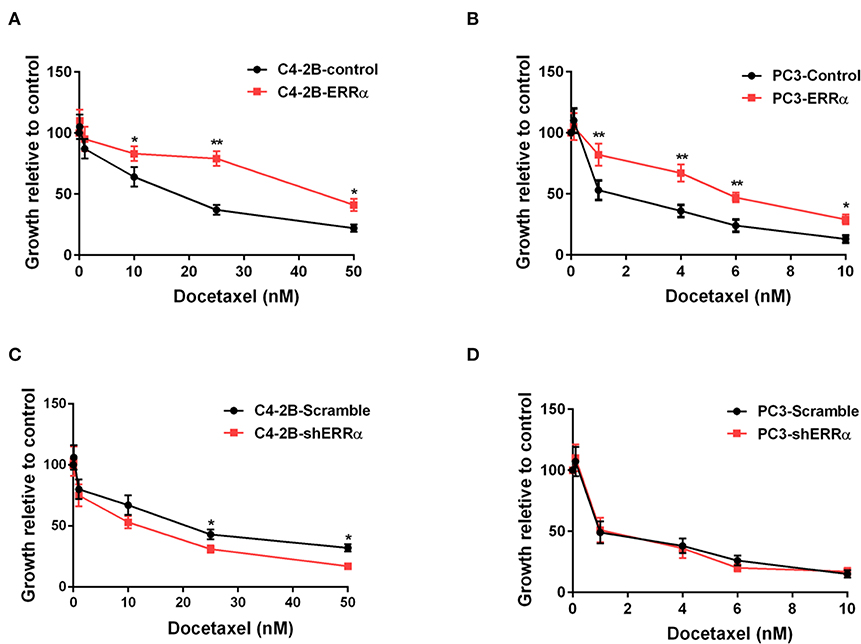
Figure 1. Effects of ERRα overexpression and knockdown on docetaxel sensitivity. (A,B) Effect of ERRα overexpression on docetaxel sensitivity in C4-2B and PC3 cells was determined by MTT assay at 24 h. (C,D) Effect of ERRα knockdown on docetaxel sensitivity in C4-2B and PC3 cells was determined by MTT assay at 24 h. *p < 0.05, **p < 0.01 compared with control.
The Influence of ERRα Manipulation on Drug Resistance-Related Genes
We then determined the expression of major drug resistance-related genes, including ABCC1-5, ABCG2, and MDR1, after ERRα manipulation. Overexpression of ERRα in both C4-2B and PC3 cells was accompanied by a significant upregulation of ABCC4 at the mRNA and protein levels (Figures 2A–D). Similarly, the knockdown of ERRα in C4-2B resulted in significant downregulation of ABCC4 (Figures 2E,F). These data indicated that ERRα may influence docetaxel sensitivity by regulating ABCC4 expression in PCa. Docetaxel works as a microtubule stabilizer, and overexpression of βIII-tubulin (TUBB3) was often found in docetaxel resistance in different cancers (32). Therefore, we also determined the relationship of ERRα and TUBB3 by bioinformatics analysis using TCGA prostate cancer data containing 551 samples (Supplementary Figure 2). Their expression showed some but not very strong positive correlation.
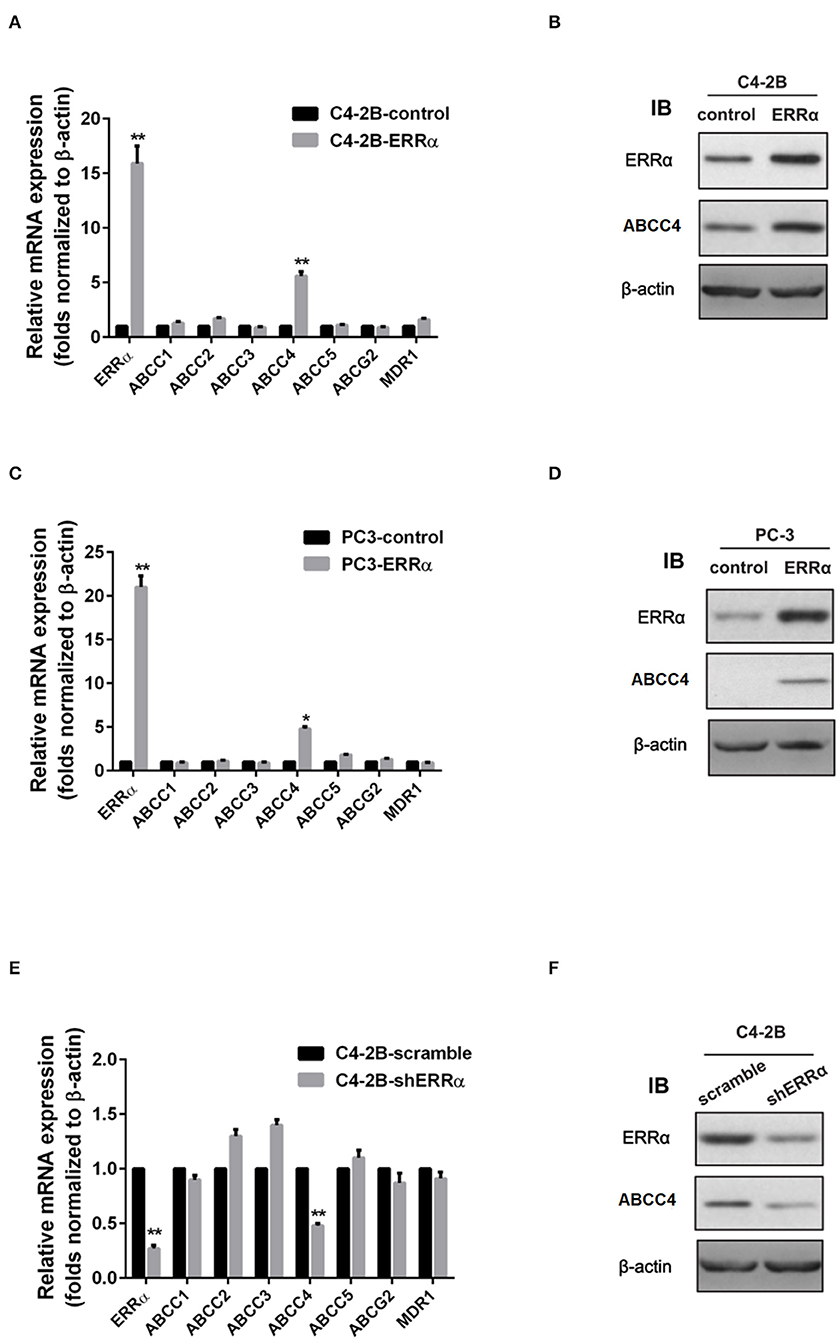
Figure 2. Regulation of ERRα on drug resistance-related genes. (A,C) mRNA expression of drug resistance-related genes after ERRα overexpression in C4-2B and PC3 cells. (B,D) Protein expression of ERRα and ABCC4 (MRP) after ERRα overexpression in C4-2B and PC3 cells. (E) mRNA expression of drug resistance-related genes after ERRα knockdown in the C4-2B cell. (F) Protein expression of ERRα and ABCC4 (MRP) after ERRα knockdown in the C4-2B cell. *p < 0.05, **p < 0.01 compared with respective control.
Expression of ERRα and ABCC4 in Docetaxel-Resistant PCa Cells
To further elucidate the role of the ERRα and ABCC4 in docetaxel resistance, we determined the sensitivity of C4-2B and PC3 for docetaxel and established docetaxel-resistant C4-2B and PC3 cell lines (Figure 3A). PC3 was more sensitive to docetaxel than C4-2B. Consistent with our hypothesis that ERRα overexpression is involved in docetaxel resistance, higher expression of ERRα is found in docetaxel-resistant C4-2B and PC3 (Figures 3B,C). At the same time, ABCC4 was also highly expressed in docetaxel-resistant C4-2B and PC3 compared with their respective parental cells (Figures 3B,C). Moreover, the knockdown of ERRα by shRNA in the resistant cell lines reversed the high expression of ABCC4 (Figures 3D,E). Based on the above findings, we suspect that ERRα might exert a regulatory role on ABCC4.
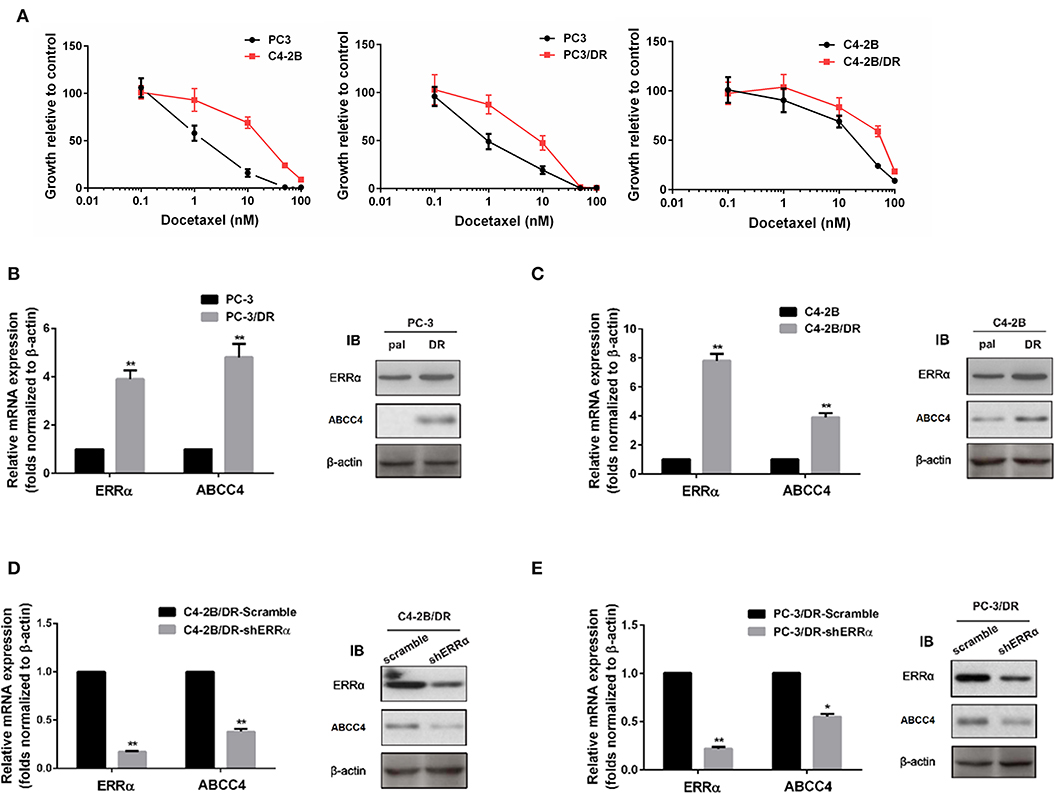
Figure 3. Expression of ERRα and ABCC4 in docetaxel-resistant cells. (A) Sensitivity of C4-2B, PC3, and their drug-resistant cells to docetaxel was determined by MTT assay. (B,C) Expression of ERRα and ABCC4 in docetaxel-resistant C4-2B and PC3 cells was increased compared with parental cells. (D,E) Expression of ERRα and ABCC4 was decreased after ERRα knockdown in drug-resistant C4-2B and PC3 cells. Pal, parental cell; DR, drug-resistant cell. *p < 0.05, **p < 0.01 compared with respective control.
Intracellular Docetaxel Accumulation After ERRα Manipulation
Since ABCC4 confers drug resistance through the efflux of anticancer drugs from cancer cells (33), we next determined whether intracellular docetaxel quantity was changed after ERRα manipulation. In drug-resistant C4-2B, there was a significantly lower intracellular docetaxel level due to the efflux of drugs (Figures 4A,B). Interestingly, after the knockdown of ERRα, the intracellular docetaxel level was significantly elevated, indicating that inhibition of ERRα could inhibit drug efflux associated with ABCC4.
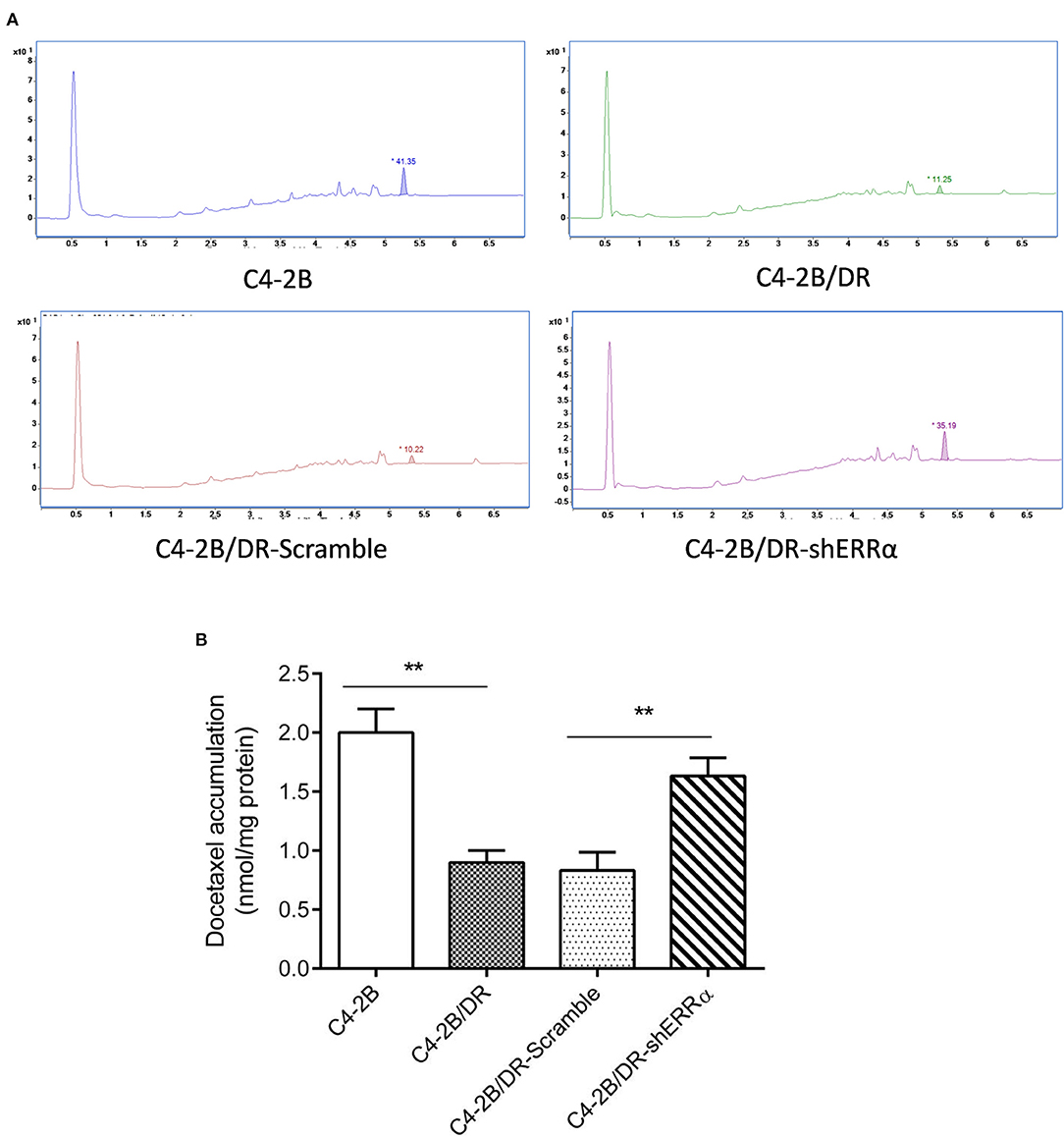
Figure 4. Knockdown of ERRα increased drug accumulation in docetaxel-resistant cell. (A) Representative HPLC figure of docetaxel accumulation in C4-2B and drug-resistant cells. (B) Statistical analysis of HPLC results. Intracellular docetaxel was significantly lowered in the drug-resistant C4-2B cell and the decrease was rescued by ERRα knockdown. **p < 0.01.
Interaction Between ERRα and ABCC4
To verify whether ERRα directly regulates ABCC4, CHIP and luciferase assays were performed. Three potential ERRα binding sites (namely P1, P2, P3) were predicted in the ABCC4 gene promoter region (Figure 5A). The CHIP assay revealed that the ERRα occupancy was significantly elevated in the P1 and P3 regions on the ABCC4 promoter (Figure 5B). Subsequently, we constructed ABCC4 promoter luciferase reporters expressing promoter regions around P1 and P3 (ABCC4-I-Luc and ABCC4-III-Luc) and their respective mutant expressing the same region but with P1 or P3 deletion (Figures 5C,D). Results showed that after transfection with ERRα-overexpressing plasmids, both ABCC4-I-Luc and ABCC4-III-Luc exhibited enhanced luciferase signal, while their mutant counterparts did not show such enhancement (Figures 5C,D), indicating a direct binding of ERRα and the ABCC4 gene promoter regions P1 and P3. Taken together, our results demonstrated that ABCC4 is a direct target of ERRα.
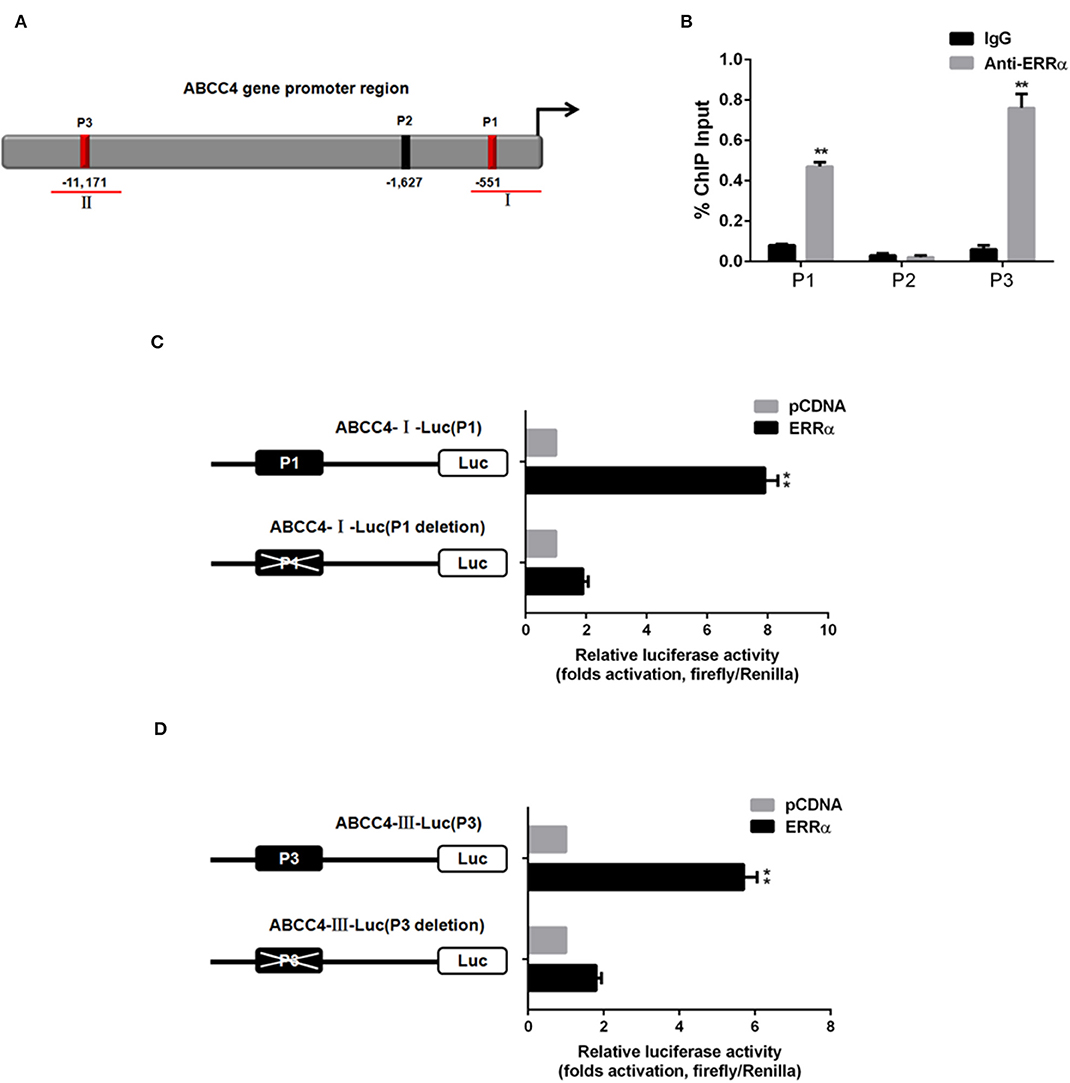
Figure 5. Direct interaction between ERRα and ABCC4. (A) Representative graph of ABCC4 promoter regions containing putative ERRα binding sites. (B) P1 and P3 regions were pulled down by ERRα antibody by CHIP assay. (C) Luciferase signal for plasmid-containing P1 region was increased by ERRα. (D) Luciferase signal for plasmid-containing P3 region was increased by ERRα. **p < 0.01 compared with respective control.
Bioinformatics Analysis and Validation
We analyzed the expression level of ERRα and ABCC4 in cBioPortal. ERRα and ABCC4 were significantly upregulated in PCa in a data set consisting of 496 tumor vs. 53 normal samples (Figure 6A). To find out the potential targets of ERRα/ABCC4, we investigated genes co-expressed with ERRα and ABCC4 at mRNA level in cBioPortal using TCGA data consisting of two independent data sets (TCGA cell 2015, 333 samples and TCGA provisional, 499 samples downloaded on January 5, 2018). As shown in Figure 6B, 36 genes were co-expressed with ERRα and ABCC4 from both data sets. Among them, 13 genes showed a significant association with ERRα and ABCC4 (Figure 6C). Three genes (GNL3, SDND1, and VPS37C) were positively associated with ERRα and ABCC4 and others showed a negative correlation. The expression of these genes after ERRα manipulation was validated (Figure 6D). In accordance with the bioinformatics finding, GNL3 and VPS37C were positively correlated with ERRα and ABCC4, and FAXDC2, PDPN, PLEKHA2, and POP2 were negatively correlated with them.
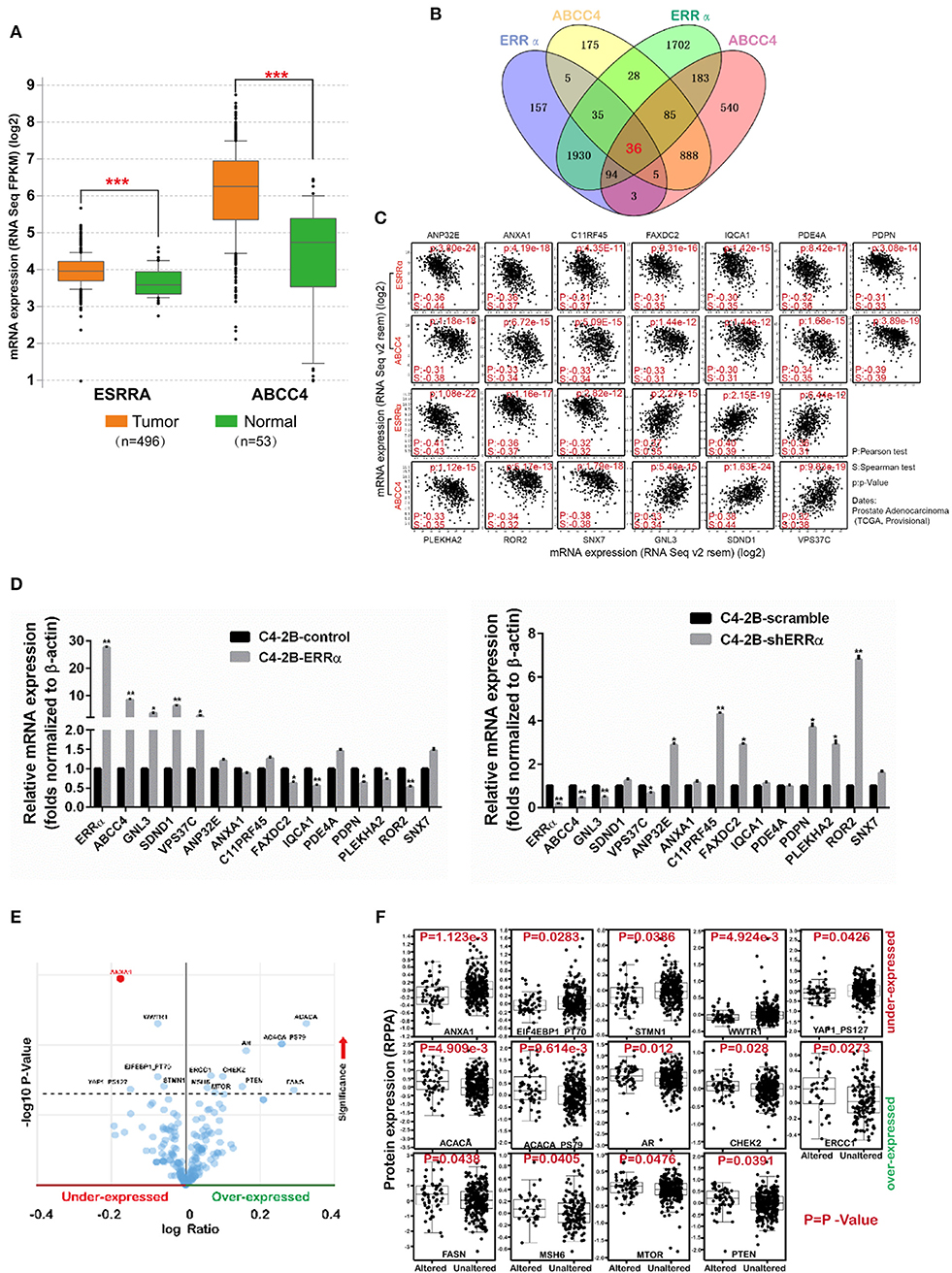
Figure 6. The expression level and potential targets of ERRα and ABCC4 from bioinformatics analysis. (A) The expression of ERRα and ABCC4 were elevated in prostate cancer using data from TCGA. (B) Genes co-expressed with ERRα and ABCC4 at mRNA level; 36 genes were co-expressed with both ERRα and ABCC4 from two independent data sets. Co-expression level was analyzed by Spearman's correlation test, and 0.3 cutoff was used. (C) Thirteen genes from (B) were significantly co-expressed with ERRα and ABCC4. (D) The 13 genes were validated by qPCR. (E) Genes with significant protein level change upon ERRα and ABCC4 gene alteration. (F) The expression level of 14 proteins was significantly affected by ERRα and ABCC4 gene alteration. *p < 0.01, **p < 0.001, ***p < 0.001 compared with normal sample.
We next analyzed proteins whose expressions were significantly affected by ERRα and ABCC4 gene alteration by protein enrichment analysis in cBioPortal. Results showed that 14 proteins were significantly influenced by ERRα and ABCC4 (Figures 6E,F), among which five (ANXA1, EIF4EBP1_PT70, WWTR1, STMN1, YAP1_PS127) were downregulated and accompanied by ERRα and ABCC4 alteration, and 9 (ACACA, ACACA PS79, AR, CHEK2, ERCC1, FASN, MSH6, MTOR, and PTEN) were upregulated (Figures 6E,F). Interestingly, the ANXA1 protein was decreased with ERRα and ABCC4 alteration (Figure 6F), and its mRNA was also negatively correlated with ERRα and ABCC4 (Figure 6C).
Discussion
Docetaxel is effective against most of PCa; however, its resistance hampers the successful clinical use of docetaxel-based therapy. According to previous studies, docetaxel resistance has occurred in different kinds of PCa in clinic, such as hormone-refractory prostate cancer (HRPC), castration-resistant PCa, metastatic PCa, and so on. Recently, accumulating studies have focused on the mechanisms of docetaxel resistance in PCa. For example, ASC-J9, an AR degradation enhancer, combined with docetaxel can restore the docetaxel sensitivity and suppress castration-resistant PCa (34). ERRα, an orphan nuclear receptor, is implicated as an important regulator in drug resistance by many studies. It has been reported that in endometrial cancer, silencing ERRα can make HEC-1A cells more sensitive to paclitaxel and lead to a decreased tumor growth and angiogenesis (35). Pharmacological inhibition of ERRα could restore sensitivity to anti-cancer treatment in breast cancer (36, 37). In the current study, we sought to determine whether ERRα is involved in docetaxel resistance and whether targeting ERRα could overcome docetaxel resistance in PCa.
ERRα was first shown to be upregulated in prostate cancer in 2007, and its elevated expression was found to be associated with poor patient survival (17). It has been demonstrated that ERRα expression is correlated with castration-resistant prostate cancer, and it could promote tumor progression by targeting a number of cancer-related genes, including VEGF-A, WNT5A and TGFβ1, and WNT11 and HIF-1α (15, 38, 39). Recently, it has been shown that ERRα could promote the castration-resistant growth of PCa by regulating intratumoral androgen biosynthesis (40). However, the role of ERRα in the drug resistance of PCa is not well-elucidated. We found that overexpression of ERRα conferred tolerance to docetaxel and improved the survival of PCa cells (Figures 1A,B). Moreover, downregulation of ERRα sensitized a PCa cell to docetaxel treatment (Figure 1C), indicating ERRα was involved in docetaxel resistance. To elucidate the target of ERRα in regulating docetaxel resistance, we determined the impact of ERRα on major drug resistance-related genes. As shown in Figure 2, overexpression of ERRα led to a significantly elevated level of ABCC4, and the knockdown of ERRα was accompanied by a decrease in ABCC4 level, suggesting that ERRα contributed to docetaxel resistance probably through ABCC4. It has been reported that ABCC4 mRNA/protein expression was upregulated, and it was an important determinant of docetaxel resistance in PCa (17, 27, 41, 42). Recently, the inhibition of ABCC4 is confirmed as a potential cure for neuroblastoma by inhibiting tumor proliferation and sensitizing to chemotherapeutic drug (18). Notably, increased ABCC4 expression has a strong correlation with multidrug resistance in PCa (28, 43). Docetaxel works as a microtubule stabilizer, and it has been reported that docetaxel resistance was also related to the overexpression of βIII-tubulin (TUBB3) (32). Our bioinformatics analysis showed that the expression of ERRα and TUBB3 exhibited some positive correlation (Supplementary Figure 1). Whether ERRα could regulate TUBB3 expression in PCa warrants further study.
The involvement of ERRα in drug resistance has been widely studied in breast cancer and osteosarcoma (44–48). However, its role in PCa drug resistance was rarely examined. In our study, we established docetaxel-resistant PCa cell lines and found that both ERRα and ABCC4 were highly expressed in drug-resistant cells compared with their parental cells (Figures 3B,C). The knockdown of ERRα in resistant cells was accompanied by a decrease in ABCC4 (Figures 3D,E), suggesting that ERRα and ABCC4 were involved in docetaxel resistance in PCa. The importance of ERRα in drug resistance was functionally tested by docetaxel accumulation inside the drug-resistant cells after ERRα manipulation (Figure 4). The docetaxel level was significantly lower in resistant cells due to drug efflux, which could be reversed by ERRα knockdown, suggesting the potential clinical benefit of inhibiting ERRα to treat docetaxel-resistant PCa (Figure 4). Subsequently, the CHIP and luciferase assays were performed to assess the direct regulation of ERRα on ABCC4. Results showed that ERRα could directly bind two regions in the ABCC4 promoter (Figure 5). This is the first report on the direct regulation of ERRα on ABCC4.
We further confirmed our finding using publicly available data from PCa patients and discovered that the expression level of both ERRα and ABCC4 was significantly higher in the prostate tumor vs. the normal samples (Figure 6A). Next, in order to explore other potential mechanisms, we tried to find out the potential targets of the ERRα/ABCC4 axis by bioinformatics analysis. We first looked at genes that are co-expressed with both ERRα and ABCC4 (Figures 6C,D). Thirteen genes were significantly co-expressed with ERRα and ABCC4 using data from two patient cohorts (Figure 6D). Among them, ANP32E (49), ANXA1(50), PDE4A (51), PDPN (52), and ROR2 (53) play an oncogenic role according to the literature, which is contrary to their negative correlation with ERRα and ABCC4. Therefore, they might not be a direct target of ERRα/ABCC4, and their function is irrelevant to their co-expression with ERRα/ABCC4. GNL3 was one of the genes that were positively associated with ERRα and ABCC4. Earlier studies have reported that the elevated expression of GNL3 is related to cancer proliferation and metastasis (54). Recently, GNL3 is also considered as a susceptibility gene for PCa metastasis due to its expression involved in aggressive human PCa multiplication, migration, and invasion (55). Moreover, its positive correlation with ERRα and ABCC4 was validated by qPCR (Figure 6D). Thus, GNL3 can be considered as a potential target for ERRα/ABCC4 and awaits further validation.
Moreover, we analyzed protein expression of potential ERRα/ABCC4 targets (Figures 6E,F). In total, 14 proteins were significantly affected by ERRα/ABCC4 mutation. Of these, five were significantly decreased with ERRα/ABCC4 alteration, and nine were increased with ERRα/ABCC4 alteration (Figure 6F). Consistent with previous results that ANXA1 mRNA is negatively correlated with ERRα/ABCC4, its protein is decreased accompanying ERRα/ABCC4 alteration. Among other decreased proteins, STMN1 has been reported as an oncogene (56). Interestingly, WWTR1 and YAP1_PS127 both belong to the Hippo signaling pathway, and they are frequently hyperactivated in cancer (57). For the proteins that increased with ERRα/ABCC4 alteration, AR (androgen receptor) is expressed in primary and metastatic PCa, and downregulation of AR is considered a potential therapy for PCa (58, 59). CHEK2 is a gene involved in DNA repair and its mutation is associated with PCa (60). ERCC1 has been demonstrated to be an independent prognostic marker in PCa and a therapeutic target to sensitize cancer cells to chemotherapy (61, 62). FASN (fatty acid synthase), the key enzyme in the control of fatty acid synthesis, has received considerable attention as a therapeutic target in cancer including PCa (63, 64). MSH6, one of DNA mismatch repair genes, is overexpressed in PCa and is linked to genetic instability and tumor aggressiveness (65). mTOR belongs to the PI3K-AKT-mTOR signaling pathway, which is strongly involved in many cancers. It interplays with androgen receptor (AR) in PCa (66) and thus represents a potential target for PCa (67). PTEN is a well-known tumor suppressor and is also frequently lost in PCa (68). Taken together, many of the proteins that increased with ERRα/ABCC4 alterations are strongly involved in PCa and awaits further investigation, including AR, CHEK2, ERCC1, FASN, MSH6, and MTOR.
In summary, our findings reveal for the first time the involvement of ERRα in docetaxel resistance in PCa by directly regulating ABCC4. With the pharmacological inhibitors for ERRα available, it appears to be a promising potential adjuvant therapy for docetaxel-resistant PCa and warrants further investigation.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
HH, JL, and JS conducted the experiments and drafted the manuscript. LL, XW, and YL collected and processed the data. YX, QZ, and ML analyzed and interpreted the data. PK and YZ prepared the figures. XL and SX revised the article critically for important intellectual content and approved the final version to be published. QW, WW, and ZX managed the project, designed the study, and provided the funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant nos. 81770562, 81503093, 81602166, and 81672444), the Joint Funds of the Southwest Medical University & Luzhou (2016LZXNYD-T01, 2017LZXNYD-Z05 and 2017LZXNYD-J09), the Wuhu Science and Technology Benefit People Project (2016hm13), and the Clinical Research Project of Shenzhen Municipal Health and Family Planning Commission (SZLY2017027).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01474/full#supplementary-material
Supplementary Figure 1. Colony formation assay in PC-3 cell after ERRα manipulation. **p < 0.01, ***p < 0.001 compared with respective control.
Supplementary Figure 2. Correlation of ERRα and TUBB3 in prostate cancer. Data were extracted from TCGA (n = 551).
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
2. Davda R, Hughes S, Jones R, Crabb SJ, Troup J, Payne H. Chemotherapy at first diagnosis of advanced prostate cancer - revolution or evolution? Findings from a British Uro-oncology group UK survey to evaluate oncologists' views on first-line docetaxel in combination with androgen deprivation therapy in castrate-sensitive metastatic and high-risk/locally advanced prostate cancer. Clin Oncol. (2016) 28:376–85. doi: 10.1016/j.clon.2016.01.006
3. Vale CL, Burdett S, Rydzewska LHM, Albiges L, Clarke NW, Fisher D, et al. Addition of docetaxel or bisphosphonates to standard of care in men with localised or metastatic, hormone-sensitive prostate cancer: a systematic review and meta-analyses of aggregate data. Lancet Oncol. (2016) 17:243–56. doi: 10.1016/S1470-2045(15)00489-1
4. Puente J, Grande E, Medina A, Maroto P, Lainez N, Arranz JA. Docetaxel in prostate cancer: a familiar face as the new standard in a hormone-sensitive setting. Ther Adv Med Oncol. (2017) 9:307–18. doi: 10.1177/1758834017692779
5. Miller RE, Sweeney CJ. Chemotherapy for metastatic castrate-sensitive prostate cancer. Prostate Cancer Prostatic Dis. (2016) 19:139–44. doi: 10.1038/pcan.2016.10
6. Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. (2004) 351:1502–12. doi: 10.1056/NEJMoa040720
7. O'Neill AJ, Prencipe M, Dowling C, Fan Y, Mulrane L, Gallagher WM, et al. Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol Cancer. (2011) 10:126. doi: 10.1186/1476-4598-10-126
8. Schweizer MT, Zhou XC, Wang H, Bassi S, Carducci MA, Eisenberger MA, et al. The influence of prior abiraterone treatment on the clinical activity of docetaxel in men with metastatic castration-resistant prostate cancer. Eur Urol. (2014) 66:646–52. doi: 10.1016/j.eururo.2014.01.018
9. van Soest RJ, de Morree ES, Kweldam CF, de Ridder CMA, Wiemer EAC, Mathijssen RHJ, et al. Targeting the androgen receptor confers in vivo cross-resistance between enzalutamide and docetaxel, but not cabazitaxel, in castration-resistant prostate cancer. Eur Urol. (2015) 67:981–5. doi: 10.1016/j.eururo.2014.11.033
10. Mahon KL, Henshall SM, Sutherland RL, Horvath LG. Pathways of chemotherapy resistance in castration-resistant prostate cancer. Endocr Relat Cancer. (2011) 18:R103–23. doi: 10.1530/ERC-10-0343
11. Ganju A, Yallapu MM, Khan S, Behrman SW, Chauhan SC, Jaggi M. Nanoways to overcome docetaxel resistance in prostate cancer. Drug Resist Updat. (2014) 17:13–23. doi: 10.1016/j.drup.2014.04.001
12. Wu D, Cheung A, Wang Y, Yu S, Chan FL. The emerging roles of orphan nuclear receptors in prostate cancer. Biochim Biophys Acta. (2016) 1866:23–36. doi: 10.1016/j.bbcan.2016.06.001
13. Chang CY, McDonnell DP. Molecular pathways: the metabolic regulator estrogen-related receptor alpha as a therapeutic target in cancer. Clin Cancer Res. (2012) 18:6089–95. doi: 10.1158/1078-0432.CCR-11-3221
14. Bonkhoff H. Estrogen receptor signaling in prostate cancer: implications for carcinogenesis and tumor progression. The Prostate. (2018) 78:2–10. doi: 10.1002/pros.23446
15. Fradet A, Bouchet M, Delliaux C, Gervais M, Kan C, Benetollo C, et al. Estrogen related receptor alpha in castration-resistant prostate cancer cells promotes tumor progression in bone. Oncotarget. (2016) 7:77071–86. doi: 10.18632/oncotarget.12787
16. Cheung CP, Yu S, Wong KB, Chan LW, Lai FM, Wang X, et al. Expression and functional study of estrogen receptor-related receptors in human prostatic cells and tissues. J Clin Endocrinol Metab. (2005) 90:1830–44. doi: 10.1210/jc.2004-1421
17. Fujimura T, Takahashi S, Urano T, Kumagai J, Ogushi T, Horie-Inoue K, et al. Increased expression of estrogen-related receptor alpha (ERRalpha) is a negative prognostic predictor in human prostate cancer. Int J Cancer. (2007) 120:2325–30. doi: 10.1002/ijc.22363
18. Murray J, Valli E, Yu DMT, Truong AM, Gifford AJ, Eden GL, et al. Suppression of the ATP-binding cassette transporter ABCC4 impairs neuroblastoma tumour growth and sensitises to irinotecan in vivo. Eur J Cancer. (2017) 83:132–41. doi: 10.1016/j.ejca.2017.06.024
19. Russel FG, Koenderink JB, Masereeuw R. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol Sci. (2008) 29:200–7. doi: 10.1016/j.tips.2008.01.006
20. Cho E, Montgomery RB, Mostaghel EA. Minireview: SLCO and ABC transporters: a role for steroid transport in prostate cancer progression. Endocrinology. (2014) 155:4124–32. doi: 10.1210/en.2014-1337
21. Vogt K, Mahajan-Thakur S, Wolf R, Broderdorf S, Vogel C, Bohm A, et al. Release of platelet-derived sphingosine-1-phosphate involves multidrug resistance protein 4 (MRP4/ABCC4) and is inhibited by statins. Thromb Haemost. (2018) 118:132–42. doi: 10.1160/TH17-04-0291
22. Belleville-Rolland T, Sassi Y, Decouture B, Dreano E, Hulot JS, Gaussem P, et al. MRP4 (ABCC4) as a potential pharmacologic target for cardiovascular disease. Pharmacol Res. (2016) 107:381–9. doi: 10.1016/j.phrs.2016.04.002
23. Marcantoni E, Allen N, Cambria MR, Dann R, Cammer M, Lhakhang T, et al. Platelet transcriptome profiling in HIV and ATP-binding cassette subfamily C member 4 (ABCC4) as a mediator of platelet activity. JACC Basic Transl Sci. (2018) 3:9–22. doi: 10.1016/j.jacbts.2017.10.005
24. Tsukamoto M, Sato S, Satake K, Miyake M, Nakagawa H. Quantitative evaluation of drug resistance profile of cells expressing wild-type or genetic polymorphic variants of the human ABC transporter ABCC4. Int J Mol Sci. (2017) 18:1435. doi: 10.3390/ijms18071435
25. Zhang G, Wang Z, Qian F, Zhao C, Sun C. Silencing of the ABCC4 gene by RNA interference reverses multidrug resistance in human gastric cancer. Oncol Rep. (2015) 33:1147–54. doi: 10.3892/or.2014.3702
26. Zhu X, Xue L, Yao Y, Wang K, Tan C, Zhuang M, et al. The FoxM1-ABCC4 axis mediates carboplatin resistance in human retinoblastoma Y-79 cells. Acta Biochim Biophys Sin. (2018) 50:914–20. doi: 10.1093/abbs/gmy080
27. Ho LL, Kench JG, Handelsman DJ, Scheffer GL, Stricker PD, Grygiel JG, et al. Androgen regulation of multidrug resistance-associated protein 4 (MRP4/ABCC4) in prostate cancer. Prostate. (2008) 68:1421–9. doi: 10.1002/pros.20809
28. Oprea-Lager DE, Bijnsdorp IV, Van Moorselaar RJA, Van Den Eertwegh AJM, Hoekstra OS, Geldof AA. ABCC4 Decreases docetaxel and not cabazitaxel efficacy in prostate cancer cells in vitro. Anticancer Res. (2013) 33:387–91.
29. Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang Y, et al. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res. (2014) 74:6236–47. doi: 10.1158/0008-5472.CAN-14-0855
30. Zhou J, Guo X, Chen W, Wang L, Jin Y. Targeting survivin sensitizes cervical cancer cells to radiation treatment. Bioengineered. (2020) 11:130–40. doi: 10.1080/21655979.2020.1717297
31. Shen J, Xiao Z, Wu WK, Wang MH, To KF, Chen Y, et al. Epigenetic silencing of miR-490–3p reactivates the chromatin remodeler SMARCD1 to promote Helicobacter pylori-induced gastric carcinogenesis. Cancer Res. (2015) 75:754–65. doi: 10.1158/0008-5472.CAN-14-1301
32. Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. (2010) 10:194–204. doi: 10.1038/nrc2803
33. Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. (2010) 10:147–56. doi: 10.1038/nrc2789
34. Luo J, Tian J, Chou F, Lin C, Xing EZ, Zuo L, et al. Targeting the androgen receptor (AR) with AR degradation enhancer ASC-J9 led to increase docetaxel sensitivity via suppressing the p21 expression. Cancer Lett. (2018) 444:35–44. doi: 10.1016/j.canlet.2018.09.025
35. Matsushima H, Mori T, Ito F, Yamamoto T, Akiyama M, Kokabu T, et al. Anti-tumor effect of estrogen-related receptor alpha knockdown on uterine endometrial cancer. Oncotarget. (2016) 7:34131–48. doi: 10.18632/oncotarget.9151
36. Park S, Chang CY, Safi R, Liu X, Baldi R, Jasper JS, et al. ERRalpha-regulated lactate metabolism contributes to resistance to targeted therapies in breast cancer. Cell Rep. (2016) 15:323–35. doi: 10.1016/j.celrep.2016.03.026
37. Thewes V, Simon R, Schroeter P, Schlotter M, Anzeneder T, Buttner R, et al. Reprogramming of the ERRα and ERα target gene landscape triggers tamoxifen resistance in breast cancer. Cancer Res. (2015) 75:720–31. doi: 10.1158/0008-5472.CAN-14-0652
38. Dwyer MA, Joseph JD, Wade HE, Eaton ML, Kunder RS, Kazmin D, et al. WNT11 expression is induced by estrogen-related receptor alpha and beta-catenin and acts in an autocrine manner to increase cancer cell migration. Cancer Res. (2010) 70:9298–308. doi: 10.1158/0008-5472.CAN-10-0226
39. Zou C, Yu S, Xu Z, Wu D, Ng CF, Yao X, et al. ERRα augments HIF-1 signalling by directly interacting with HIF-1α in normoxic and hypoxic prostate cancer cells. J Pathol. (2014) 233:61–73. doi: 10.1002/path.4329
40. Xu Z, Ma T, Zhou J, Gao W, Li Y, Yu S, et al. Nuclear receptor ERRα contributes to castration-resistant growth of prostate cancer via its regulation of intratumoral androgen biosynthesis. Theranostics. (2020) 10:4201–16. doi: 10.7150/thno.35589
41. Montani M, Hermanns T, Muntener M, Wild P, Sulser T, Kristiansen G. Multidrug resistance protein 4 (MRP4) expression in prostate cancer is associated with androgen signaling and decreases with tumor progression. Virchows Arch. (2013) 462:437–43. doi: 10.1007/s00428-013-1390-8
42. Cai C, Omwancha J, Hsieh CL, Shemshedini L. Androgen induces expression of the multidrug resistance protein gene MRP4 in prostate cancer cells. Prostate Cancer Prostatic Dis. (2007) 10:39–45. doi: 10.1038/sj.pcan.4500912
43. Li YF, Ji HH, Zhang ZL, Zhang TT, Gan W, Zhang SF. Targeting MRP4 expression by anti-androgen treatment reverses MRP4-mediated docetaxel resistance in castration-resistant prostate cancer. Oncol Lett. (2017) 14:1748–56. doi: 10.3892/ol.2017.6357
44. Li X, Zhang K, Hu Y, Luo N. ERRalpha activates SHMT2 transcription to enhance the resistance of breast cancer to lapatinib via modulating the mitochondrial metabolic adaption. Biosci Rep. (2020) 40:BSR20192465. doi: 10.1042/BSR20192465
45. Zhang W, Wu M, Chong QY, Zhang M, Zhang X, Hu L, et al. Loss of estrogen-regulated MIR135A1 at 3p21.1 promotes tamoxifen resistance in breast cancer. Cancer Res. (2018) 78:4915–28. doi: 10.1158/0008-5472.CAN-18-0069
46. Deblois G, Smith HW, Tam IS, Gravel SP, Caron M, Savage P, et al. ERRalpha mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat Commun. (2016) 7:12156. doi: 10.1038/ncomms12156
47. Chen P, Wang H, Duan Z, Zou JX, Chen H, He W, et al. Estrogen-related receptor alpha confers methotrexate resistance via attenuation of reactive oxygen species production and P53 mediated apoptosis in osteosarcoma cells. Biomed Res Int. (2014) 2014:616025. doi: 10.1155/2014/616025
48. Chen Y, Zhang K, Li Y, Guo R, Zhang K, Zhong G, et al. Oestrogen-related receptor alpha mediates chemotherapy resistance of osteosarcoma cells via regulation of ABCB1. J Cell Mol Med. (2019) 23:2115–24. doi: 10.1111/jcmm.14123
49. Xiong Z, Ye L, Zhenyu H, Li F, Xiong Y, Lin C, et al. ANP32E induces tumorigenesis of triple-negative breast cancer cells by upregulating E2F1. Mol Oncol. (2018) 12:896–912. doi: 10.1002/1878-0261.12202
50. Bizzarro V, Belvedere R, Milone MR, Pucci B, Lombardi R, Bruzzese F, et al. Annexin A1 is involved in the acquisition and maintenance of a stem cell-like/aggressive phenotype in prostate cancer cells with acquired resistance to zoledronic acid. Oncotarget. (2015) 6:25076–92. doi: 10.18632/oncotarget.4725
51. Peng Y, Li Y, Tian Y, Ao G. PDE4a predicts poor prognosis and promotes metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. J Cancer. (2018) 9:2389–96. doi: 10.7150/jca.24079
52. Retzbach EP, Sheehan SA, Nevel EM, Batra A, Phi T, Nguyen ATP, et al. Podoplanin emerges as a functionally relevant oral cancer biomarker and therapeutic target. Oral Oncol. (2018) 78:126–36. doi: 10.1016/j.oraloncology.2018.01.011
53. Debebe Z, Rathmell WK. Ror2 as a therapeutic target in cancer. Pharmacol Ther. (2015) 150:143–8. doi: 10.1016/j.pharmthera.2015.01.010
54. Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci USA. (2011) 108:20388–93. doi: 10.1073/pnas.1015171108
55. Lee M, Williams KA, Hu Y, Andreas J, Patel SJ, Zhang S, et al. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. (2015) 32:769–82. doi: 10.1007/s10585-015-9745-y
56. Li M, Yang J, Zhou W, Ren Y, Wang X, Chen H, et al. Activation of an AKT/FOXM1/STMN1 pathway drives resistance to tyrosine kinase inhibitors in lung cancer. Br J Cancer. (2017) 117:974–83. doi: 10.1038/bjc.2017.292
57. Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nature Rev Cancer. (2015) 15:73–9. doi: 10.1038/nrc3876
58. Olson BM, Gamat M, Seliski J, Sawicki T, Jeffery J, Ellis L, et al. Prostate cancer cells express more androgen receptor (AR) following androgen deprivation, improving recognition by AR-specific T cells. Cancer Immunol Res. (2017) 5:1074–85. doi: 10.1158/2326-6066.CIR-16-0390
59. Culig Z, Santer FR. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. (2014) 33:413–27. doi: 10.1007/s10555-013-9474-0
60. Wang Y, Dai B, Ye D. CHEK2 mutation and risk of prostate cancer: a systematic review and meta-analysis. Int J Clin Exp Med. (2015) 8:15708–15.
61. Cummings M, Higginbottom K, McGurk CJ, Wong OG, Koberle B, Oliver RT, et al. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem Pharmacol. (2006) 72:166–75. doi: 10.1016/j.bcp.2006.04.025
62. Jacobsen F, Taskin B, Melling N, Sauer C, Wittmer C, Hube-Magg C, et al. Increased ERCC1 expression is linked to chromosomal aberrations and adverse tumor biology in prostate cancer. BMC Cancer. (2017) 17:504. doi: 10.1186/s12885-017-3489-9
63. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Rev Cancer. (2007) 7:763–77. doi: 10.1038/nrc2222
64. Wen S, Niu Y, Lee SO, Yeh S, Shang Z, Gao H, et al. Targeting fatty acid synthase with ASC-J9 suppresses proliferation and invasion of prostate cancer cells. Mol Carcinog. (2016) 55:2278–90. doi: 10.1002/mc.22468
65. Wilczak W, Rashed S, Hube-Magg C, Kluth M, Simon R, Buscheck F, et al. Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. Carcinogenesis. (2017) 38:19–27. doi: 10.1093/carcin/bgw116
66. Audet-Walsh E, Dufour CR, Yee T, Zouanat FZ, Yan M, Kalloghlian G, et al. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev. (2017) 31:1228–42. doi: 10.1101/gad.299958.117
67. Edlind MP, Hsieh AC. PI3K-AKT-mTOR signaling in prostate cancer progression and androgen deprivation therapy resistance. Asian J Androl. (2014) 16:378–86. doi: 10.4103/1008-682X.122876
Keywords: prostate cancer, docetaxel resistance, ERRα, ABCC4, bioinformatics
Citation: Huang H, Li J, Shen J, Lin L, Wu X, Xiang S, Li Y, Xu Y, Zhao Q, Zhao Y, Kaboli PJ, Li M, Li X, Wang W, Wen Q and Xiao Z (2020) Increased ABCC4 Expression Induced by ERRα Leads to Docetaxel Resistance via Efflux of Docetaxel in Prostate Cancer. Front. Oncol. 10:1474. doi: 10.3389/fonc.2020.01474
Received: 02 January 2020; Accepted: 10 July 2020;
Published: 28 August 2020.
Edited by:
Eva Martinez-Balibrea, Catalan Institute of Oncology, SpainReviewed by:
Kenneth K. W. To, The Chinese University of Hong Kong, ChinaVicenç Ruiz De Porras, Germans Trias i Pujol Health Science Research Institute (IGTP), Spain
Copyright © 2020 Huang, Li, Shen, Lin, Wu, Xiang, Li, Xu, Zhao, Zhao, Kaboli, Li, Li, Wang, Wen and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiping Wang, d3dwOTk4MjYmI3gwMDA0MDsxNjMuY29t; Qinglian Wen, d3FsNzMxMTUmI3gwMDA0MDsxNjMuY29t; Zhangang Xiao, emhhbmdhbmd4aWFvJiN4MDAwNDA7c3dtdS5lZHUuY24=
†These authors have contributed equally to this work
 Houbao Huang1†
Houbao Huang1† Jing Shen
Jing Shen Xu Wu
Xu Wu Yawei Li
Yawei Li Qijie Zhao
Qijie Zhao Yueshui Zhao
Yueshui Zhao Zhangang Xiao
Zhangang Xiao