- 1Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, United States
- 2Department of Pathology, Kangnam Sacred Heart Hospital, Hallym University College of Medicine, Seoul, South Korea
- 3Department of Pathology, Incheon St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, South Korea
- 4Department of Pathology, Kosin University College of Medicine, Busan, South Korea
- 5Department of Pathology, Hanyang University College of Medicine, Seoul, South Korea
- 6Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea
Objective: Hairy and enhancer of split-1 (HES-1), which is a downstream target of the Notch signaling pathway, has been linked to KRAS mutations. HES-1 has been proposed as harboring oncogenic activity in colorectal cancer but has not been investigated in adenocarcinoma of the small intestine, where the drivers of oncogenesis are not as well-understood.
Materials and Methods: To investigate the clinicopathologic and prognostic implications of HES-1, HES-1 immunohistochemical expression was analyzed in digital images along with clinicopathological variables, including survival and KRAS genotype, in 185 small intestinal adenocarcinomas.
Results: The loss of HES-1 expression (HES-1Loss) was observed in 38.4% (71/185) of the patients, and was associated with higher pT category (P = 0.018), pancreatic invasion (P = 0.005), high grade (P = 0.043), and non-tubular histology (P = 0.004). Specifically, in tumors with mutant KRAS (KRASMT), HES-1Loss was related to proximal location (P = 0.024), high T and N categories (P = 0.005 and 0.047, respectively), and pancreatic invasion (P = 0.004). Patients with HES-1Loss showed worse overall survival compared to those with intact HES-1 (HES-1Intact) (P = 0.013). Patients with HES-1Loss/KRASMT (median, 17.3 months) had significantly worse outcomes than those with HES-1Intact/KRASWT (39.9 months), HES-1Intact/KRASMT (47.6 month), and HES-1Loss/KRASWT (36.2 months; P = 0.010). By multivariate analysis, HES-1Loss (hazard ratio = 1.55, 95% confidence interval (CI), 1.07–2.26; P = 0.022) remained an independent prognostic factor.
Conclusion: HES-1expression can be used as a potential prognostic marker and may aid in the management of patients with small intestinal adenocarcinomas.
Introduction
Small intestinal adenocarcinoma is rare cancer that is clinically distinct from colorectal cancer but managed similarly due to the lack of prospective data necessary for establishing optimal management. However, recent studies demonstrated that the clinicopathologic and molecular features of small intestinal adenocarcinoma differed from those of colorectal cancer (1, 2). The majority of small intestinal adenocarcinoma patients are diagnosed at an advanced disease stage because of a lack of early detection tools, low incidence, and non-specific clinical symptoms (1, 3). A prospective ARCAD-NADEGE cohort study found that 35.6% of patients with small intestinal adenocarcinomas were diagnosed with metastatic disease, compared to 15.6% of those with colorectal cancer (4). The incidence of small intestinal adenocarcinoma is increasing while that of colorectal cancer is declining (5). Thus, there is an urgent need for research efforts on prognostic predictors and/or guides for the treatment of small intestinal adenocarcinoma.
The hairy and enhancer of split (HES) family of proteins consists of seven members that share a highly conserved tetrapeptide domain (Trp-Arg-Pro-Trp) at the C-terminus (6). Within this family, HES-1 is a transcriptional factor that plays an important role in intracellular processes, such as cell cycle arrest, differentiation, and apoptosis (7, 8). HES-1 is a downstream target of the Notch signaling pathway and is regulated by the Hedgehog and Wnt signaling pathways (9–11). It is expressed in the intestine along with HES-3, HES-5, HES-6, and HES-7 (12). Among the HESs, HES-1 is crucial for the normal development of the small intestine because it regulates the differentiation of Paneth cells (13). Prior studies suggested that HES-1 might play an oncogenic role in colorectal cancer. However, its role in tumorigenesis or prognosis remains unclear (11, 14–17). The clinicopathologic and prognostic significance of HES-1 expression has not been elucidated in small intestinal adenocarcinoma.
KRAS is the most frequently altered gene among the three human RAS isoforms and is mutated in approximately 30–50% of colorectal cancers (18–20). The significance of KRAS mutations has been demonstrated in a genetic colorectal cancer model, in which mutant KRAS harboring adenomatous polyposis coli (APC) mutations induced tumorigenesis and metastasis (21, 22). Schrock et al. (2) recently reported that KRAS was mutated in 53.6% (170/317) of the small intestinal adenocarcinomas. Abnormalities in KRAS-mediated differentiation and proliferation were linked to activation of the HES-1 transcription factor in colorectal carcinomas (23).
In this study, we investigated the clinicopathologic and prognostic significance of HES-1 expression in small intestinal adenocarcinomas by utilizing combined immunohistochemistry (IHC) and digital image analysis. In addition, we examined the potential clinical significance of HES-1 expression in patients with small intestinal adenocarcinomas harboring mutant KRAS.
Materials and Methods
Tissue Samples and Clinicopathological Data
This study was approved by the Institutional Review Board of Incheon St. Mary's Hospital (Seoul, Republic of Korea; OC14OIMI0133). A cohort of 197 patients who underwent surgical resections for primary small intestinal adenocarcinomas from the surgical pathology archives of 22 South Korean institutions were examined, as reported previously (24). Patients with primary carcinomas in the duodenum, jejunum, and ileum were included in the study. Patients with carcinomas grossly involving the stomach, the ampulla of Vater, pancreas, cecum, or appendix were excluded from the study.
The clinical and pathological data collected in a previous study were used in this study. The patient's gender, age, tumor location, and survival data were included as clinical data. Duodenal adenocarcinomas were defined as proximal tumors, whereas jejunal and/or ileal adenocarcinomas were considered distal tumors. Pathological data included histological type, differentiation, pathologic tumor-node-metastasis (pTNM) stage, lymph node metastasis, pancreatic invasion, and perineural and lymphovascular invasion. Histologic types and tumor grading were classified according to the 2019 World Health Organization (WHO) classification (25). All cases were staged according to the eighth edition of the American Joint Committee on Cancer (AJCC) cancer staging system (26).
HES-1 Expression
Immunohistochemical staining was performed on tissue microarrays (TMAs), which were constructed as part of a previous study (27). Briefly, the representative areas of each sample were selected and marked on the corresponding hematoxylin and eosin-stained slides. Three tissue cylinders with 1-mm tumor diameter each and one matched core from normal mucosa were punched from each formalin-fixed, paraffin-embedded (FFPE) tissue block and transplanted into recipient blocks using a tissue arrayer (Beecher Instruments, Inc., Silver Spring, MD, USA).
For IHC, TMA sections with 5-μm thickness were deparaffinized in xylene and rehydrated in a graded ethanol series. The endogenous peroxidase activity of the samples was quenched with 3% H2O2 solution (Dako, Carpinteria, CA, USA) for 15 min at room temperature. Heat-induced antigen retrieval was performed for 20 min in a target retrieval buffer at pH 6.0 (Dako). The slides were then incubated with rabbit monoclonal anti-HES-1 antibody (Cell Signaling Technology, Danvers, MA, USA; clone D6P2U; cat# 11988) at 1:500 for 1 h at room temperature in a Dako Autostainer Plus Slide Stainer (Dako). Subsequently, the slides were incubated with Envision+Rb HRP dual-link secondary (Dako) and visualized with 3,3′-diaminobenzidine (Dako), and counterstained with hematoxylin. The primary antibody and rabbit immunoglobulin were omitted in the negative control, and human placenta was used as a positive control (28).
All immunostained slides were digitalized using an Aperio AT2 digital scanner (Leica Biosystems, Vista, CA, USA) at 40 × objective magnification and the images were automatically analyzed using Visiopharm software v6.9.1 (Visiopharm, Hørsholm, Denmark). In brief, screenshots of single relevant regions of interest were generated by a single pathologist (JWK) who was blinded to the clinical and pathological data. Blue-colored (hematoxylin) tumor nuclei were initially defined, and then brown-colored (DAB) nuclei and cytoplasm were separated spectrally. Subsequently, the brown nuclear staining intensity (0 = negative, 1 = weak, 2 = moderate, and 3 = strong) and the percentage of nuclear-stained tumor cells (range, 0–100) were obtained using a predefined algorithm and optimized settings (Figure 1). Histoscores were calculated by multiplying the intensity score and proportion score and ranged from 0 to 300 (Supplementary Figure S1A). For the statistical analyses, the values were dichotomized using the cutoff value showing the most discriminative power. The samples with histoscores of 40.0 or lower were classified as loss of HES-1 expression (HES-1Loss), while cases with a histoscore higher than 40.0 were classified as intact HES-1 expression (HES-1Intact). There was no significant intra-tumor heterogeneity in HES-1 expression (Supplementary Figure S1B).
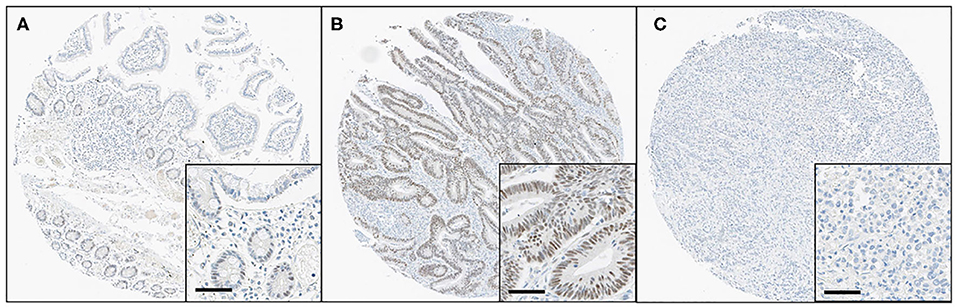
Figure 1. Immunohistochemical findings for HES-1 in small intestinal adenocarcinomas. (A) Some epithelial cells of basal crypts in normal mucosa show immunoreactivity for HES-1. (B) The tumor cells of low-grade small intestinal adenocarcinomas show diffuse, strong positivity for HES-1, corresponding to HES-1Intact. (C) In contrast, the high-grade tumor cells are completely negative for HES-1, indicating HES-1Loss (Original magnification, ×8; inset, ×40; scale bar, 50 μm).
KRAS Mutation
We used previously reported KRAS mutation data in the same cohort (29). In brief, genomic DNA was extracted from the FFPE tissue blocks by a QIAmp DNA Mini Kit (Qiagen, Valencia, CA, USA) as previously described (29). The KRAS genes were amplified using the following primers: forward, 5′-TGACATGTTCTAATATAGTCAC-3′, and reverse, 5′-ACAAGATTTACCTCTATTGTT-3′). The PCR reaction volume was 25 μl, including 0.3 μM of each primer and AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The cycling conditions were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturing at 95°C for 50 s, annealing for 50 s, elongation at 72°C for 1 min, and a final elongation at 72°C for 7 min. The PCR amplicons were purified by a QIAquick PCR Purification Kit (Qiagen) and sequencing reactions were performed in the forward and reverse directions using the BigDye Terminator Cycle Sequencing Kit, version 1.1 (Applied Biosystems).
Statistical Analysis
Unpaired Student's t-test was applied to compare the continuous variables. The relationships between the categorical variables were analyzed by the chi-squared test or Fisher's exact test. All survival analyses used an overall survival (OS) model, which captured all patient deaths as events and censored other patients at their last visit dates. The Kaplan–Meier method was used to compare survival between the groups and survival was analyzed by the log-rank test using a cutoff histoscore of 40.0. A Cox proportional hazards model was used to estimate the hazard ratios (HRs) and confidence intervals (CIs) in both the univariate and multivariate models. In all statistical analyses, a P-value of < 0.05 was considered statistically significant. Data analysis was performed using SPSS Statistics for Windows, version 23 (IBM Corp., Armonk, NY, USA).
Results
Clinicopathological Characteristics
The TMA contained 197 small intestinal adenocarcinoma samples. However, due to tissue loss and folding during sectioning and staining, along with sample heterogeneity, only 185 samples could be interpreted and included in this study. One hundred sixteen patients were male (62.7%) and 69 were female (37.3%), with a mean age of 58.9 years (range, 23 to 86 years). The most common tumor location was the duodenum in 103 (55.7%) patients, followed by the jejunum in 54 (29.2%), and the ileum in 28 (15.1%) patients. The clinicopathological characteristics of the study are summarized in Supplementary Table S1. The patients were followed-up for a median of 28.8 months, ranging from 0.3 to 168.4 months.
HES-1 Expression
The histoscores of the nuclear HES-1 expression ranged from 0 to 290.7, with a median of 62.2. Of the cancer specimens, 114 (61.6%) of the185 cases exhibited HES-1Intact, whereas 71 (38.4%) cases showed HES-1Loss. As summarized in Table 1, HES-1Loss was significantly associated with younger age (<60 years; P = 0.023). In terms of histologic subtype, non-tubular adenocarcinomas, including mucinous, signet ring cell, and undifferentiated carcinomas, frequently showed HES-1Loss, whereas tubular adenocarcinomas tended to have HES-1Intact (P = 0.004). HES-1Loss was more frequent in carcinomas with extended T category (P = 0.018), high grade (P = 0.043), and pancreatic invasion (P = 0.005). No significant association was identified between HES-1 expression and other clinicopathological variables, including sex, tumor location, type of growth, lymphovascular and perineural invasion, pN category, stage group, and KRAS genotype.
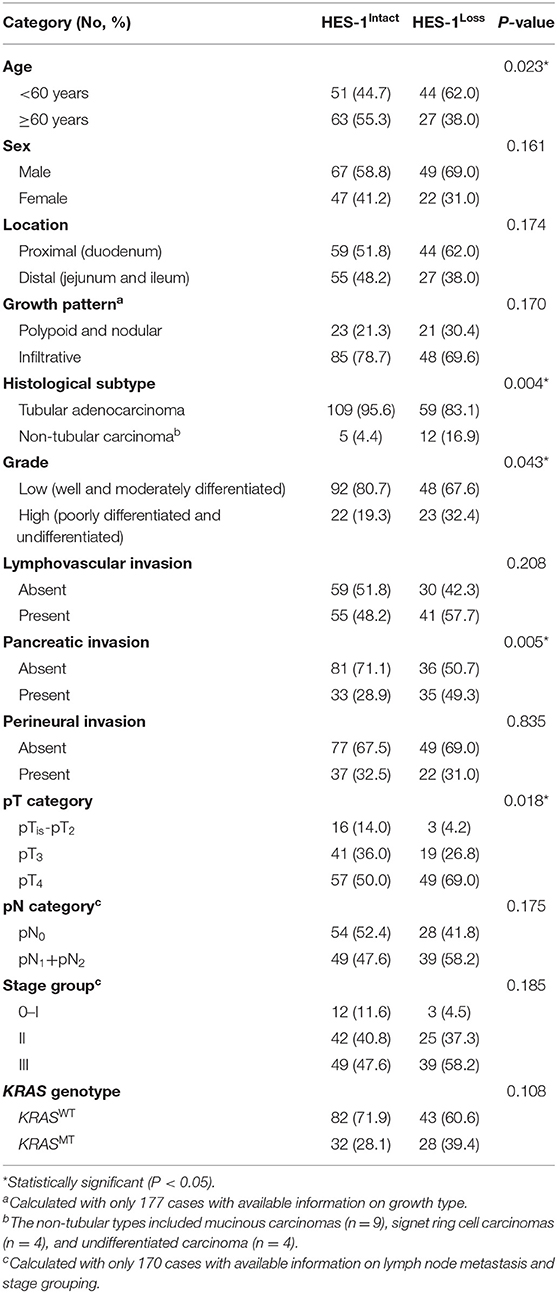
Table 1. Correlation between clinicopathologic factors and HES-1 expression of small intestinal adenocarcinoma patients.
KRAS Mutation
KRAS mutations (KRASMT) were found in 32.4% (60/185) of the patients. Among the small intestinal adenocarcinomas with KRASMT, 81.7% (49/60) of the mutations were detected in codon 12 and 18.3% (11/60) were identified in codon 13. The main type of KRASMT was p.G12D (30/60 cases, 50.0%), followed by p.G13D (11/60, 18.3%), p.G12C (7/60, 11.7%), p.G12V (6/60, 10.0%), p.G12A (4/60, 6.6%), p.G12R (1/60, 1.7%), and p.G12S (1/60, 1.7%) (Figure 2A).
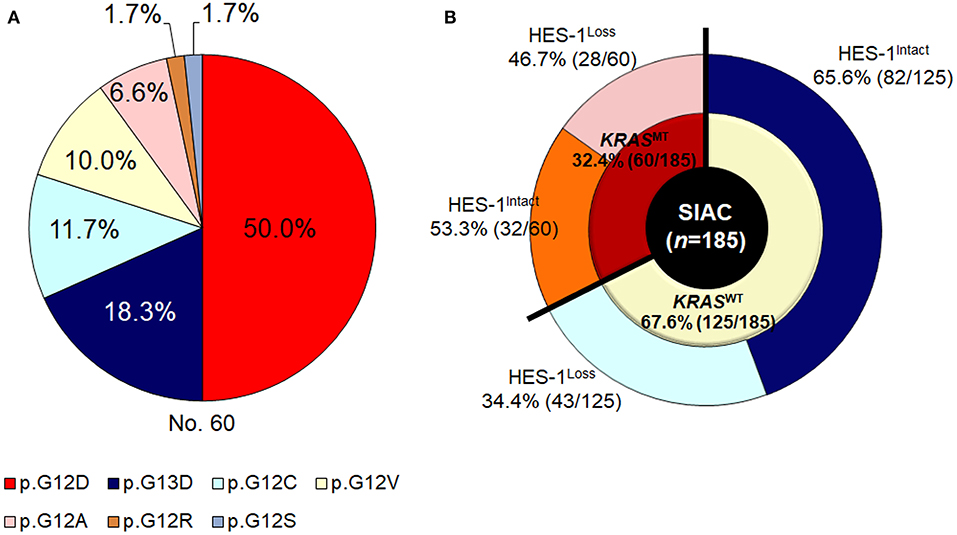
Figure 2. KRAS genotypes of small intestinal adenocarcinomas. (A) Frequency of KRASMT and (B) HES-1 expression according to KRAS genotypes.
HES-1 Expression and KRAS Genotypes
As described in Figure 2B, in the KRASWT group (n = 125), 43 (34.4%) cases exhibited HES-1Loss. In contrast, in tumors with KRASMT (n = 60), 28 (46.8%) had HES-1Loss. The relationship between HES-1 expression and clinicopathologic factors according to KRAS mutation status are summarized in Table 2. In the KRASMT group, HES-1Loss was significantly associated with higher pT category (P = 0.005), proximal location (P = 0.024), pancreatic invasion (P = 0.004), and nodal metastasis (P = 0.047). In the KRASWT group, HES-1Loss was only correlated with non-tubular types of small intestinal adenocarcinomas (P = 0.028).
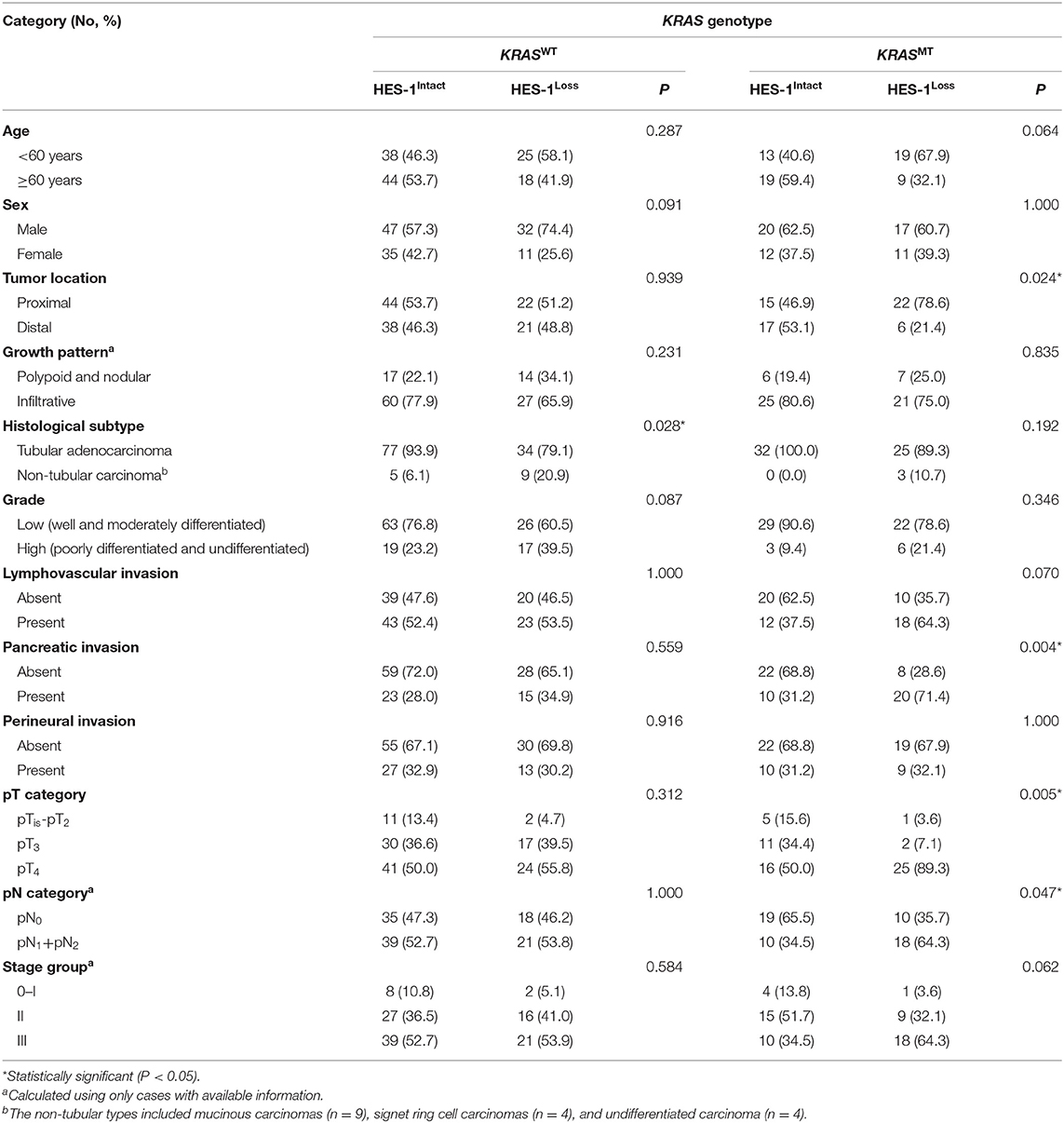
Table 2. Correlation between clinicopathologic factors and HES-1 expression based on KRAS genotype in small intestinal adenocarcinoma patients.
Survival Analysis
The relationship between HES-1 expression and OS is described in Figure 3. Patients with HES-1Loss (median, 26.3 months) had significantly shorter OS times than those with HES-1Intact (41.7 months; P = 0.013) (Figure 3A). The median OS of the patients with KRASMT tended to be shorter than that of the patients with KRASWT (18.7 vs. 38.5 months), but it did not reach statistical significance (P = 0.063, Figure 3B). In the KRASMT subgroup, patients with HES-1Loss (median, 17.3 months) had worse OS than those with HES-1Intact (47.6 months; P = 0.027), whereas there was no significant survival difference in the KRASWT subgroup based on HES-1 expression status (Supplementary Figure S2).
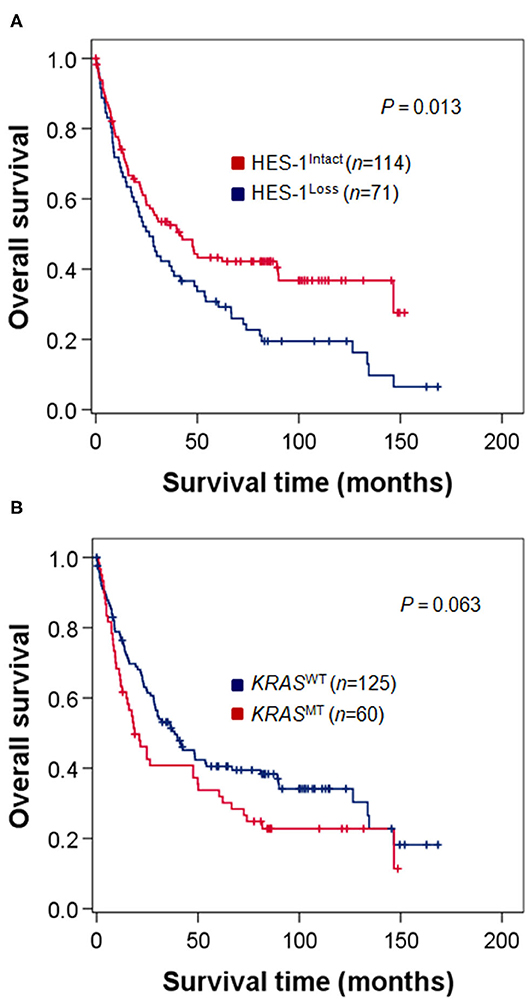
Figure 3. Survival analysis of patients with small intestinal adenocarcinomas. (A) Patients with HES-1Loss show poor OS compared to those with HES-1Intact (median, 26.3 vs. 41.7 months, P = 0.013). (B) Patients with KRASMT exhibit a tendency toward worse OS than those with KRASWT, but the difference was not statistically significant (18.7 vs. 38.5 months, P = 0.063).
Survival Analysis Based on HES-1 Expression and KRAS Genotypes
Furthermore, we analyzed the OS of patients in the four groups, which were classified according to the combined patterns of HES-1 expression and KRAS genotypes: HES-1Loss/KRASMT (28 cases, 15.2%), HES-1Loss/KRASWT (43, 23.2%), HES-1Intact/KRASMT (32, 17.3%), and HES-1Intact/KRASWT (82, 44.3%). Patients with HES-1Loss/KRASMT (median, 17.3 months) had significantly worse outcomes than those with HES-1Intact/KRASWT (39.9 months), HES-1Intact/KRASMT (47.6 month), and HES-1Loss/KRASWT (36.2 months) (P = 0.010; Figure 4). Significant differences in survival rates were observed between the groups with HES-1Loss/KRASMT and HES-1Intact/KRASWT (P = 0.001), and HES-1Loss/KRASWT and HES-1Loss/KRASMT (P = 0.001) in pair-wise comparisons. However, there were no significant differences between the HES-1Intact/KRASWT and HES-1Intact/KRASMT (P = 0.252), and HES-1Intact/KRASMT and HES-1Loss/KRASWT (P = 0.533) groups.
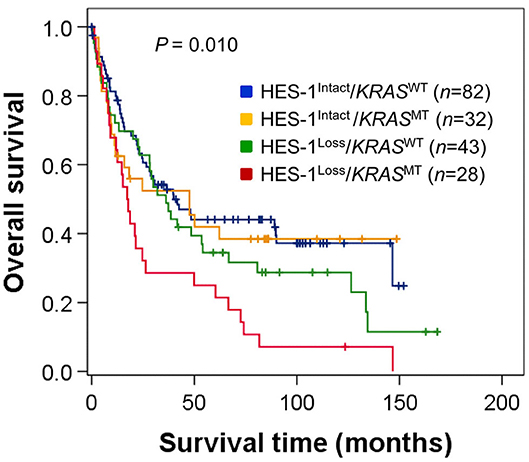
Figure 4. Survival analysis of patients with small intestinal adenocarcinomas according to the combined patterns of HES-1 expression and KRAS genotypes. Survival differences are observed among four groups classified according to HES-1 expression and KRAS genotype (log-rank, P = 0.010).
Multivariate Analysis
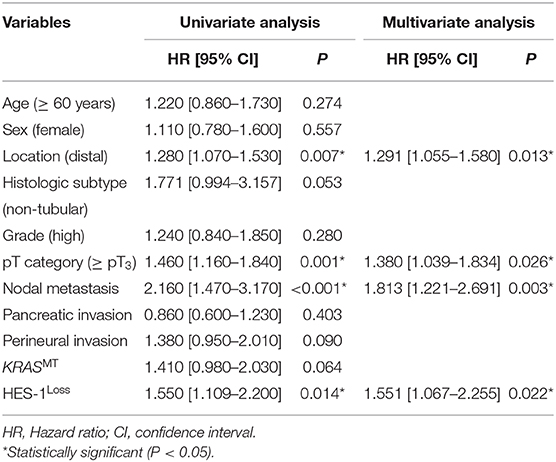
Cox multivariate proportional hazard analyses were performed using HES-1 expression and other factors considered significant by univariate analysis (Table 3). Non-tubular histology (HR = 1.771, 95% CI: 0.994–3.157, P = 0.053) and KRASMT (HR = 1.410, 95% CI: 0.980–2.030, P = 0.064) were exhibited a tendency of shorter patient survival by univariate survival analysis. Distal location (HR = 1.291, 95% CI: 1.055–1.580, P = 0.013), high pT category (≥pT3) (HR = 1.380, 95% CI: 1.039–1.834, P = 0.026), lymph node metastasis (HR = 1.813, 95% CI: 1.221–2.691, P = 0.003), and HES-1Loss (HR = 1.551, 95% CI: 1.067–2.255, P = 0.022) were revealed as independent negative prognostic factors. Notably, dual HES-1Loss and KRASMT is also an independent prognostic factor for poor OS in small intestinal adenocarcinoma patients (HR = 1.312 [95% CI: 1.125–1.529], P = 0.001; Supplementary Table S2).
Discussion
Notch signaling not only affects cell differentiation, proliferation, and apoptosis but controls the expression of HES-1 (30, 31). In general, Notch signaling is known to suppress squamous cancers of the skin, but stimulate hematologic malignancies and adenocarcinomas of the stomach, colon, and pancreas (30). A previous study has reported that Notch3 expression is correlated with lower T stage and the absence of lymphovascular invasion in small intestinal adenocarcinomas (32). HES-1 is known as a transcriptional inhibitor. However, recent studies showed that HES-1 was more than a repressor and contributed to cancer stem cell maintenance, cancer metastasis, and tumor multidrug resistance (33). The regulation of HES-1 expression is mediated by not only the canonical Notch signaling pathway, but also other signaling pathways, such as Hedgehog, c-Jun N-terminal kinase, Wnt, and TGF-a/Ras/mitogen-activated protein kinase (MAPK) (10, 33, 34). To our knowledge, this is the first study to assess the prognostic value of HES-1 expression, alone and in combination with the KRAS genotype in patients with small intestinal adenocarcinomas.
We found HES-1Loss to be strongly associated with tumor aggressiveness, indicated by high T category, high grade, pancreatic invasion, and carcinoma showing non-tubular histology. Moreover, HES-1Loss was an independent poor prognostic factor of small intestinal adenocarcinomas for OS. In contrast to our results, some studies have demonstrated that increased HES-1 expression may be an adverse prognostic factor in colorectal cancer (15, 35). We noted that those studies evaluated HES-1 expression via mRNA rather than IHC. Since HES-1 expression is sometimes preserved in non-neoplastic stromal cells of the colorectal mucosa, mRNA expression assays would have measured both tumor and stromal HES-1 expression (16). To accurately assess the nuclear expression of HES-1 in tumor cells via IHC, we selected regions only composed of tumor cell nests and analyzed them using digital image analysis. Consistent with our findings, a recent study performed by Ahadi et al. reported that the loss of HES-1 nuclear expression in colorectal carcinomas was significantly associated with mucinous or medullary histology, higher histological grade, and worse survival (16). With regards to tumor histology, Vanoli et al. demonstrated that non-glandular histology type is an independent prognostic factor for poor outcome in small intestinal adenocarcinoma patients (36). In this study, patients with non-tubular small intestinal adenocarcinomas had a tendency of worse OS, but it was not statistically significant (Table 3). This discrepancy might be due to the small proportion of non-tubular type of tumors (9.2%, 17/185) in this study, comparing to higher proportion of them (44.7%, 34/76) in the study of Vanoli et al. Further studies utilizing large numbers of non-tubular type of small intestinal adenocarcinomas are needed to establish the prognostic power of different histologic features.
It has been hypothesized that the pathogenesis of small intestinal adenocarcinoma varies depending upon the tumor location (4). Proximal small intestinal carcinomas are sometimes accompanied by background gastric metaplasia, suggesting a gastric metaplasia-dysplasia-carcinoma sequence, or pancreaticobiliary differentiation. In contrast, distal small intestine carcinomas are significantly associated with Crohn's disease (4).
Studies on the interactions between Notch signaling, HES-1 expression, and KRAS mutations in gastrointestinal cancers have been limited and contradictory. Nishikawa et al. (37) suggested that mutant KRAS-induced HES-1 played an essential role in the initiation and progression of pancreatic ductal adenocarcinoma by regulating acinar-to-ductal reprogramming-related genes. Feng et al. (23) also reported that in colorectal carcinomas, abnormalities in KRAS-mediated differentiation and proliferation required MAPK signaling and were linked to activation of the HES-1 transcription factor. Meanwhile, Chung et al. (38) suggested that downregulation of Notch signaling occurs during the initiation of KRAS-driven gastric carcinogenesis. Based on these findings, we hypothesized that the KRAS mutation status and HES-1 expression may be associated with tumor location in small intestinal adenocarcinomas, and Notch-independent HES-1 expression may be linked to mutated KRAS. In this study, we found that the survival rates of patients with KRASMT were significantly reduced only in the loss of HES-1 expression (HES-1Loss/KRASWT and HES-1Loss/KRASMT, P = 0.001), while the survival rate of patients with HES-1Intact tumors was not dependent on KRAS mutation status (HES-1Intact/KRASWT and HES-1Intact/KRASMT, P = 0.252) (Figure 4). Thus, we investigated HES-1Loss/KRASMT and found that it could be an independent prognostic marker for poor OS time in small intestinal adenocarcinoma patients (Supplementary Table S2). HES-1Loss harboring KRASMT was more frequently detected in proximal location compared to distal location, which suggests a link between HES-1Loss/KRASMT and tumor location. These findings suggest that intact HES-1 expression, independent of KRAS genotype, prolonged the survival rate of small intestinal adenocarcinoma patients. Furthermore, HES-1Loss was revealed as an independent prognostic factor for poor outcome. Further studies are needed to clarify the relationship between HES-1 and KRAS genotype in small intestinal adenocarcinomas.
The Notch signaling pathway is a promising target for anti-cancer therapy (30). However, activation of this pathway can lead to tumor-suppressive or oncogenic effects, and nonspecific inhibition of the Notch pathways has been toxic (30). In metastatic colon cancer, a phase II clinical trial of RO-4929097 targeting cleavage mediated by γ-secretase, which is a crucial step in Notch activation, was evaluated. However, there was no evidence of objective radiographic response and survival increase (39). As γ-secretase inhibitor (GSI) nonspecifically inhibits the Notch target gene, it causes a rapid differentiation of intestinal progenitor cells into goblet cells and this may be the primary cause of gastrointestinal toxicities associated with GSI (33, 40). Therefore, aiming at HES-1 may result in fewer side effects because many other Notch target genes will be unaffected (33). Moreover, since HES-1 lies at the crossroads of multiple signaling pathways, the co-inhibition of these pathways through targeting HES-1 might represent a new strategy for cancer therapy (33). It is also notable that the regulation of HES-1 expression and Notch pathway activity is dependent upon tissue, spatial, and temporal factors and the proteins with which they interact (10, 34). Therefore, we proposed that a more sophisticated approach is needed for tailored therapy targeting the Notch pathway in small intestinal adenocarcinomas.
Akce et al. demonstrated that duodenal localization tends to have worse patients' survival than jejunal/ileal adenocarcinomas (1). In contrast, we found that patients with distal (jejunal/ileal) adenocarcinomas had significantly shorter OS times than those with proximal (duodenal) adenocarcinomas. The present study only included surgically resected cases without stage IV disease, while the study by Akce et al. using a cohort derived from the National Cancer Data Base (n = 7,954) included inoperable stage IV cases (n = 2,889) (1). In addition, in the study of Akce et al., 37.6% of patients had duodenal adenocarcinomas presented as stage IV disease. Thus, this discrepancy may be resulted from differences in percentage of patients with surgical resection, ethnicity, and lifestyle.
In this study, we revealed that HES-1Loss was associated with tumor aggressiveness, including high T category, high grade, and pancreatic invasion in small intestinal adenocarcinomas. Moreover, HES-1Loss could predict a worse prognosis in patients with small intestinal adenocarcinomas. Further elucidation of the underlying molecular mechanism of HES-1 will contribute to development of new therapeutic targets in patients with small intestinal adenocarcinomas.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Incheon St. Mary's Hospital (OC14OIMI0133). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JK, S-YJ, J-YC, and SH designed the study. JK, S-YJ, KY, H-KC, Y-HO, S-MH, and J-YC collected the experimental or clinical data. JK, J-YC, and SH analyzed the data. JK, S-YJ, and J-YC drafted and edited the manuscript. J-YC and SH reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (2017R1D1A1B03031817, awarded to S-YJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the members of the Korean Small Intestinal Cancer Study Group: Dr. Eun Sun Jung, The Catholic University of Korea College of Medicine, Seoul; Dr. Young Kyung Bae, Yeungnam University College of Medicine, Daegu; Dr. Joon Mee Kim, Inha University College of Medicine, Incheon; Dr. Gwang Il Kim, CHA Bundang Medical Center, CHA University, Seongnam; Dr. Kee-Taek Jang, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul; Dr. Jung Yeon Kim, Inje University Sanggye Paik Hospital, Seoul; Dr. Soo Jin Jung, Inje University College of Medicine, Busan; Dr. Ghilsuk Yoon, Kyungpook National University School of Medicine, Daegu; Dr. Kyu Yun Jang, Chonbuk National University Medical School, Jeonju; Dr. Jinwon Seo, Hallym University Sacred Heart Hospital, Anyang; Dr. Tae Jung Kim, Yeouido St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul; Dr. Eun Kyoung Kwak, Fatima Hospital, Daegu; Dr. Dae Woon Eom, University of Ulsan College of Medicine, Gangneung Asan Hospital, Gangneung; Dr. Hee Kyung Kim, Soonchunhyang University Bucheon Hospital, Bucheon; Dr. Hee Sung Park, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul; and Dr. Ji Shin Lee, Chonnam National University Medical School, Gwangju, Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01427/full#supplementary-material
References
1. Akce M, Jiang R, Zakka K, Wu C, Alese OB, Shaib WL, et al. Clinical outcomes of small bowel adenocarcinoma. Clin Colorectal Cancer. (2019) 18:257–68. doi: 10.1016/j.clcc.2019.08.002
2. Schrock AB, Devoe CE, McWilliams R, Sun J, Aparicio T, Stephens PJ, et al. Genomic profiling of small-bowel adenocarcinoma. JAMA Oncol. (2017) 3:1546–53. doi: 10.1001/jamaoncol.2017.1051
3. Hong SH, Koh YH, Rho SY, Byun JH, Oh ST, Im KW, et al. Primary adenocarcinoma of the small intestine: presentation, prognostic factors and clinical outcome. Jpn J Clin Oncol. (2009) 39:54–61. doi: 10.1093/jjco/hyn122
4. Aparicio T, Henriques J, Manfredi S, Tougeron D, Bouche O, Pezet D, et al. Small bowel adenocarcinoma: results from a nationwide prospective ARCAD-NADEGE cohort study of 347 patients. Int J Cancer. (2020) 147:967–77. doi: 10.1002/ijc.32860
5. Overman MJ, Hu CY, Kopetz S, Abbruzzese JL, Wolff RA, Chang GJ. A population-based comparison of adenocarcinoma of the large and small intestine: insights into a rare disease. Ann Surg Oncol. (2012) 19:1439–45. doi: 10.1245/s10434-011-2173-6
6. Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. (1996) 16:2670–7. doi: 10.1128/MCB.16.6.2670
7. Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. (2006) 298:22–31. doi: 10.1016/j.ydbio.2006.05.036
8. Kimura H, Kawasaki H, Taira K. Mouse microRNA-23b regulates expression of Hes1 gene in P19 cells. Nucleic Acids Symp Ser (Oxf). (2004) 48:213–4. doi: 10.1093/nass/48.1.213
9. Rani A, Greenlaw R, Smith RA, Galustian C. HES1 in immunity and cancer. Cytokine Growth Factor Rev. (2016) 30:113–7. doi: 10.1016/j.cytogfr.2016.03.010
10. Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. (2008) 27:1489–500. doi: 10.1038/sj.onc.1210767
11. Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. (2009) 106:6309–14. doi: 10.1073/pnas.0900427106
12. Liang SJ, Li XG, Wang XQ. Notch signaling in mammalian intestinal stem cells: determining cell fate and maintaining homeostasis. Curr Stem Cell Res Ther. (2019) 14:583–90. doi: 10.2174/1574888X14666190429143734
13. Suzuki K, Fukui H, Kayahara T, Sawada M, Seno H, Hiai H, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. (2005) 328:348–52. doi: 10.1016/j.bbrc.2004.12.174
14. Gao F, Huang W, Zhang Y, Tang S, Zheng L, Ma F, et al. Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3β pathway in human colon cancer. Oncotarget. (2015) 6:38667–80. doi: 10.18632/oncotarget.5484
15. Weng MT, Tsao PN, Lin HL, Tung CC, Change MC, Chang YT, et al. Hes1 Increases the invasion ability of colorectal cancer cells via the STAT3-MMP14 pathway. PLoS One. (2015) 10:e0144322. doi: 10.1371/journal.pone.0144322
16. Ahadi M, Andrici J, Sioson L, Sheen A, Clarkson A, Gill AJ. Loss of Hes1 expression is associated with poor prognosis in colorectal adenocarcinoma. Hum Pathol. (2016) 57:91–7. doi: 10.1016/j.humpath.2016.07.010
17. Candy PA, Phillips MR, Redfern AD, Colley SM, Davidson JA, Stuart LM, et al. Notch-induced transcription factors are predictive of survival and 5-fluorouracil response in colorectal cancer patients. Br J Cancer. (2013) 109:1023–30. doi: 10.1038/bjc.2013.431
18. Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II' study. Br J Cancer. (2001) 85:692–6. doi: 10.1054/bjoc.2001.1964
19. Zlobec I, Kovac M, Erzberger P, Molinari F, Bihl MP, Rufle A, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. (2010) 127:2569–75. doi: 10.1002/ijc.25265
20. Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, et al. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. (2012) 13:12153–68. doi: 10.3390/ijms131012153
21. Nash GM, Gimbel M, Shia J, Nathanson DR, Ndubuisi MI, Zeng ZS, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol. (2010) 17:572–8. doi: 10.1245/s10434-009-0605-3
22. Jeong WJ, Yoon J, Park JC, Lee SH, Lee SH, Kaduwal S, et al. Ras stabilization through aberrant activation of Wnt/beta-catenin signaling promotes intestinal tumorigenesis. Sci Signal. (2012) 5:ra30. doi: 10.1126/scisignal.2002242
23. Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, et al. Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology. (2011) 141:1003–13.e1–10. doi: 10.1053/j.gastro.2011.05.007
24. Chang HK, Yu E, Kim J, Bae YK, Jang KT, Jung ES, et al. Adenocarcinoma of the small intestine: a multi-institutional study of 197 surgically resected cases. Hum Pathol. (2010) 41:1087–96. doi: 10.1016/j.humpath.2010.01.006
25. Nagtegaal ID, Arends MJ, Salto-Tellez M editors Colorectal Adenocarcinoma. In: WHO Classification of Tumors Editorial Board. Digestive System Tumours. 5th ed. Lyon: International Agency for Research on Cancer (2019). p. 177–87.
26. Coit DG, Kelsen D, Tang LH, Erasmus JJ, Gerdes H, Hofstetter WL. Small intestine. In: Amin MB, Edge SB, Greene FL, Byrd DR, Brookland RK, Washington MK, et al. editors. AJCC Cancer Staging Manual. 8th ed. Chicago, IL: American College of Surgeons (2017). p. 221–34.
27. Lee HJ, Lee OJ, Jang KT, Bae YK, Chung JY, Eom DW, et al. Combined loss of E-cadherin and aberrant beta-catenin protein expression correlates with a poor prognosis for small intestinal adenocarcinomas. Am J Clin Pathol. (2013) 139:167–76. doi: 10.1309/AJCPS54RTFCTHGWX
28. Dailey DD, Anfinsen KP, Pfaff LE, Ehrhart EJ, Charles JB, Bonsdorff TB, et al. HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC Vet Res. (2013) 9:130. doi: 10.1186/1746-6148-9-130
29. Jun SY, Kim M, Jin Gu M, Kyung Bae Y, Chang HK, Sun Jung E, et al. Clinicopathologic and prognostic associations of KRAS and BRAF mutations in small intestinal adenocarcinoma. Mod Pathol. (2016) 29:402–15. doi: 10.1038/modpathol.2016.40
30. Previs RA, Coleman RL, Harris AL, Sood AK. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. (2015) 21:955–61. doi: 10.1158/1078-0432.CCR-14-0809
31. Huang T, Zhou Y, Cheng AS, Yu J, To KF, Kang W. NOTCH receptors in gastric and other gastrointestinal cancers: oncogenes or tumor suppressors? Mol Cancer. (2016) 15:80. doi: 10.1186/s12943-016-0566-7
32. Eom DW, Hong SM, Kim J, Kim G, Bae YK, Jang KT, et al. Notch3 signaling is associated with MUC5AC expression and favorable prognosis in patients with small intestinal adenocarcinomas. Pathol Res Pract. (2014) 210:501–7. doi: 10.1016/j.prp.2014.04.003
33. Liu ZH, Dai XM, Du B. Hes1: a key role in stemness, metastasis and multidrug resistance. Cancer Biol Ther. (2015) 16:353–9. doi: 10.1080/15384047.2015.1016662
34. Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. (2003) 194:237–55. doi: 10.1002/jcp.10208
35. Yuan R, Ke J, Sun L, He Z, Zou Y, He X, et al. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin Exp Metastasis. (2015) 32:169–79. doi: 10.1007/s10585-015-9700-y
36. Vanoli A, Di Sabatino A, Martino M, Klersy C, Grillo F, Mescoli C, et al. Small bowel carcinomas in celiac or Crohn's disease: distinctive histophenotypic, molecular and histogenetic patterns. Mod Pathol. (2017) 30:1453–66. doi: 10.1038/modpathol.2017.40
37. Nishikawa Y, Kodama Y, Shiokawa M, Matsumori T, Marui S, Kuriyama K, et al. Hes1 plays an essential role in Kras-driven pancreatic tumorigenesis. Oncogene. (2019) 38:4283–96. doi: 10.1038/s41388-019-0718-5
38. Chung WC, Zhou Y, Atfi A, Xu K. Downregulation of notch signaling in Kras-induced gastric metaplasia. Neoplasia. (2019) 21:810–21. doi: 10.1016/j.neo.2019.06.003
39. Strosberg JR, Yeatman T, Weber J, Coppola D, Schell MJ, Han G, et al. A phase II study of RO4929097 in metastatic colorectal cancer. Eur J Cancer. (2012) 48:997–1003. doi: 10.1016/j.ejca.2012.02.056
Keywords: HES-1, KRAS, prognosis, small intestine, adenocarcinoma
Citation: Kim JW, Jun S-Y, Ylaya K, Chang H-K, Oh Y-H, Hong S-M, Chung J-Y and Hewitt SM (2020) Loss of HES-1 Expression Predicts a Poor Prognosis for Small Intestinal Adenocarcinoma Patients. Front. Oncol. 10:1427. doi: 10.3389/fonc.2020.01427
Received: 27 May 2020; Accepted: 06 July 2020;
Published: 19 August 2020.
Edited by:
Alessandro Vanoli, University of Pavia, ItalyCopyright © 2020 Kim, Jun, Ylaya, Chang, Oh, Hong, Chung and Hewitt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joon-Yong Chung, Y2h1bmdqb0BtYWlsLm5paC5nb3Y=; Stephen M. Hewitt, Z2VuZWpvY2tAaGVsaXgubmloLmdvdg==
†These authors have contributed equally to this work
‡ORCID: Joon-Yong Chung orcid.org/0000-0001-5041-5982
Stephen M. Hewitt orcid.org/0000-0001-8283-1788
 Jeong Won Kim
Jeong Won Kim Sun-Young Jun
Sun-Young Jun Kris Ylaya1
Kris Ylaya1 Young-Ha Oh
Young-Ha Oh Joon-Yong Chung
Joon-Yong Chung Stephen M. Hewitt
Stephen M. Hewitt