- 1Department of Medical Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China
- 2State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Disease, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 3Department of Head and Neck/Thoracic Medical Oncology, The First People's Hospital of Foshan, Foshan, China
- 4Dongguan People's Hospital, Dongguan, China
- 5Department of Thoracic Surgery, Sun Yat-sen University Cancer Center, Guangzhou, China
Background: In patients with anaplastic lymphoma kinase (ALK) rearrangement-positive advanced non–small-cell lung cancer (NSCLC), ALK inhibitors are now the standard treatment, but their clinical efficacy varies widely for each patient. In this multicenter retrospective study, we evaluated the clinical efficacy of crizotinib according to the ALK rearrangement variants and concomitant mutations present.
Patients and Methods: A total 132 patients with ALK rearrangement advanced NSCLC from 4 centers in Guangdong province, China were evaluated. All patients received crizotinib treatment and their ALK rearrangement status was identified by next-generation sequencing (NGS).
Results: The median progression-free survival (PFS) in patients with EML4-ALK rearrangement (n = 121), non-EML4-ALK rearrangement (n = 5), and EML4-ALK arrangement accompanied by non-EML4-ALK rearrangement (n = 6) was 12.8, 7.5, and 7.4 months, respectively, with no significant difference between them (p = 0.1554). Similarly, among patients with various EML4-ALK variants (variant 1, variant 3a/b, and other variants), the median PFS values were again comparable. According to baseline NGS data, the median PFS in patients who had ALK rearrangement only, ALK rearrangement and concomitant tumor-suppressor gene mutations, and ALK rearrangement and concomitant oncogene mutations was 14.2, 10.9, and 4.9 months, respectively; (p = 0.0002). A multivariable analysis indicated that concomitant oncogene mutations and tumor-suppressor gene mutations were both negative factors influencing the efficacy of crizotinib in ALK rearrangement NSCLC.
Conclusion: Concomitant oncogene mutations and tumor-suppressor gene mutations had negative effects on the efficacy of crizotinib, while various ALK variants had a similar influence.
Introduction
Lung cancer remains the leading cause of cancer deaths in China. In patients with non–small-cell lung cancer (NSCLC), anaplastic lymphoma kinase (ALK) gene rearrangement is detected in approximately 3–7% of cases (1). In 2007, Soda et al. (2) first identified the echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion oncogene in NSCLC. Currently, more than 20 ALK rearrangement variants have been discovered, the most frequent among which are variant 1 (E13:A20) and variant 3a/b (E6a/b:A20) (3). All variants contain the ALK tyrosine kinase domain and an oligomerization domain in the N-terminal fusion partner gene, which activate downstream pathways to control the proliferation and apoptosis of carcinoma cells. In addition, more non-EML4 fusion variants have been discovered, including kinesin family member 5B (KIF5B) (4), kinesin light-chain 1 (KLC1) (5), cut-like homeobox 1 gene (CUX1) (6). Huntingtin-interacting protein 1(H1P1) (7), translocated promoter region (TPR) (8), baculoviral inhibition of apoptosis protein repeat-containing 6 (BIRC6) (9), and S1 RNA binding domain 1 (SRBD1) (10). These variants have all shown clinical responses to ALK inhibitors.
Since the first-generation ALK tyrosine kinase inhibitor (TKI) crizotinib (11, 12) was introduced, the development of targeted therapy has greatly improved the survival time and quality-of- life of patients with ALK rearrangement advanced NSCLC. In addition, second- and third-generation ALK TKIs, including ceritinib (13), alectinib (14), brigatinib (15), and lorlatinib (16), have also shown significant efficacy in these patients. However, despite their efficacy in patients with ALK rearrangement, all patients inevitably develop resistance to treatment and clinical efficacy varies widely for each patient. To date, a series of studies have investigated whether different ALK variants may affect the clinical response in patients who receive ALK inhibitors, and whether they are associated with resistance mechanisms. Lin et al. (17) have previously reported that ALK G1202R is significantly more common with variant 3 than variant 1 (57 vs. 30%; p = 0.023).
With the rapid development of next-generation sequencing (NGS), more and more ALK concomitant genes have been found. Epidermal growth factor receptor (EGFR) mutations are the most common mutations in NSCLC, there have been a series of studies showing that concomitant mutations are associated with inferior efficacy of EGFR TKI therapy (18, 19). In ALK rearrangement advanced NSCLC, it is still unclear whether concomitant mutations are negative predictive factors for ALK TKI therapy. Some retrospective studies and case reports have reported the poor efficacy of crizotinib treatment for ALK rearrangement NSCLC co-occurring with TP53, KRAS and EGFR mutations (20, 21). Therefore, we performed a retrospective multicenter study to explore the factors affecting the efficacy of crizotinib according to baseline next-generation sequencing data in patients with ALK rearrangement-positive advanced NSCLC.
Materials and Methods
Patients
Between January 2012 and June 2019, a total of 132 patients with ALK rearrangement advanced NSCLC from 4 medical centers across Guangdong province, China were evaluated. All patients had been histologically diagnosed with NSCLC, and with clinical stage IIIB, IV or recurrent disease according to the 7th American Joint Committee on Cancer (AJCC) staging system. The ALK rearrangement status was identified by next-generation sequencing. Clinicopathologic parameters including age, sex, histological type, clinical stage, ECOG performance status, smoking history, and gene status were collected prior to administering crizotinib therapy. The treatment progression-free survival (PFS) was defined as the time from initiation of crizotinib to the date of radiographically-confirmed progressive disease (PD) or death, whichever occurred first. The objective response rate (ORR) was defined as the percentage of patients with a complete response (CR) or partial response (PR), and the disease control rate (DCR) was defined as the percentage of patients with CR, PR, or stable disease (SD). The patients' clinical response was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1.
This study was approved by Guangdong Association of Thoracic Oncology (GASTO ID:1055). All patients signed informed consent to participate in the study.
Gene Analysis
All patient samples were identified as ALK rearrangement by next-generation sequencing. NGS-detected samples included formalin-fixed, paraffin-embedded (FFPE) tumor tissues (n = 100), malignant plural effusions (n = 10), or plasma (n = 22). Genomic DNA was extracted from FFPE samples, malignant plural effusions, or plasma samples, sheared into fragments and then subjected to end-repairing, A-tailing, and ligation with indexed adapters sequentially, followed by size selection using beads. Finally, libraries were amplified by PCR and purified for target enrichment. Libraries were sequenced on Illumina Hiseq platforms (425-gene panel or 1021-gene panel) and the Illumina NovaSeq6000 platform (543-gene panel). The sequencing depth was at least 500X mean coverage, and NGS detected genomic alterations included single-nucleotide variation (SNV), insertion/deletions (Indel), copy number variation (CNV), and gene rearrangement.
Statistical Analysis
The patients' baseline characteristics, concomitant mutations, and ALK variants were compared using χ2 or Fisher's exact test. PFS curves were estimated using the Kaplan-Meier method. Differences between ALK rearrangement variants and concomitant mutations were calculated with the log-rank test. Variables with p < 0.2 in the univariate Cox regression analysis were included in the multivariate Cox proportional hazards regression model to identify independent risk factors, which were expressed as hazards ratios (HR) with 95% confidence intervals (CI). All statistical analyses were performed using SAS™ 9.4 software, and R software (version 3.6.3). The statistical significance level was defined as a two-sides p < 0.05.
Results
Baseline Characteristics of the Patients
The baseline characteristics of the 132 patients with ALK rearranged NSCLC that were evaluated in this study are shown in Table 1. The patients' median age was 51 years (range 26–82 years), 55.3% were female, and 87.9% had adenocarcinoma. All patients received crizotinib therapy, of whom 95 patients (72.0%) received it as first-line treatment while 37 patients (28.0%) received it as second- or further-line treatment. In terms of clinical stage, 10 patients (7.6%) had stage IIIB disease, while 109 (82.6%) and 13 (9.8%) had stage IV or recurrent disease, respectively. Thirty-one patients (23.5%) had only lung or pleural metastasis (M1a). The most common distant metastatic site was bone (35.6% of patients), followed by brain (30.3%) and liver metastases (19.7%). At the end of the study, 71 patients (53.8%) had confirmed progressive disease (PD) or had died. Overall survival (OS) data are not yet mature.
ALK Rearrangement Variants and Clinical Efficacy of Crizotinib
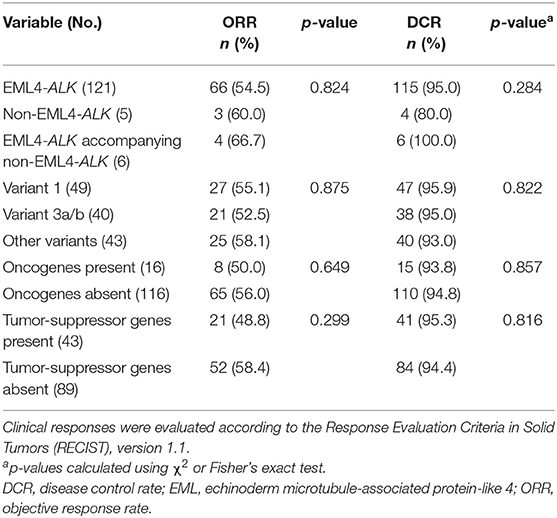
Among the 132 patients, 121 had EML4-ALK rearrangement, 5 patients had rare non-EML4-ALK rearrangement, including Lintergenic-ALK, KIF5B-ALK, ACTR3BP5-ALK, STRN-ALK and KLC1-ALK (one patient each), and 6 patients had EML4-ALK rearrangement accompanied by non-EML4-ALK rearrangement (detailed information on the non-EML4-ALK rearrangement and EML4-ALK rearrangement accompanied by non-EML4-ALK rearrangement variants and their best responses to crizotinib are shown in Supplementary Table 1 and Supplementary Figure 1). In terms of EML4-ALK rearrangement, the most common variant was variant 1 (E13:A20), which accounted for 37.1% of patients (49/132), followed by variant 3a/b (E6:A20) and variant 2 (E20:A20), which accounted for 30.3% (40/132) and 11.4% of patients (15/132), respectively (Figure 1A). The distant metastatic sites showed no significant correlation with the ALK variant type (Supplementary Table 2).
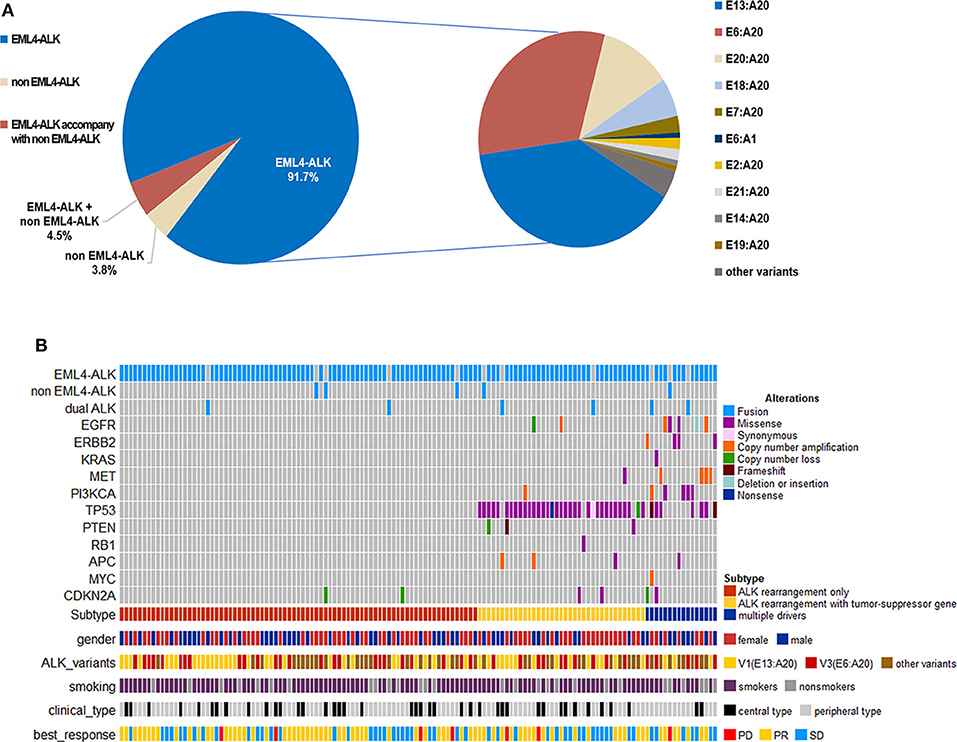
Figure 1. (A) Frequency of ALK variants in the study cohort (n = 132). (B) Distribution of concomitant mutations stratified by subgroups according to baseline NGS sequencing and their clinical features.
When comparing the efficacy of crizotinib, we considered two approaches. Firstly, we categorized patents into three subgroups: those with EML4-ALK rearrangement, non-EML4-ALK rearrangement, and EML4-ALK rearrangement accompanied by non-EML4-ALK rearrangement. The median PFS for patients with EML4-ALK rearrangement was 12.8 months (95% CI 11.2–16.8); for patients with non-EML4-ALK rearrangement the median PFS was 7.5 months (95% CI 1.0-NE), and for EML4-ALK rearrangement accompanied by non-EML4-ALK rearrangement, it was 7.4 months (95% CI 3.8–16.0), with no significant difference between them (P = 0.1554) (Figure 2A). The ORR in the three subgroups was 54.5, 60.0, and 66.7%, respectively, again with no significant differences between them (Table 2).
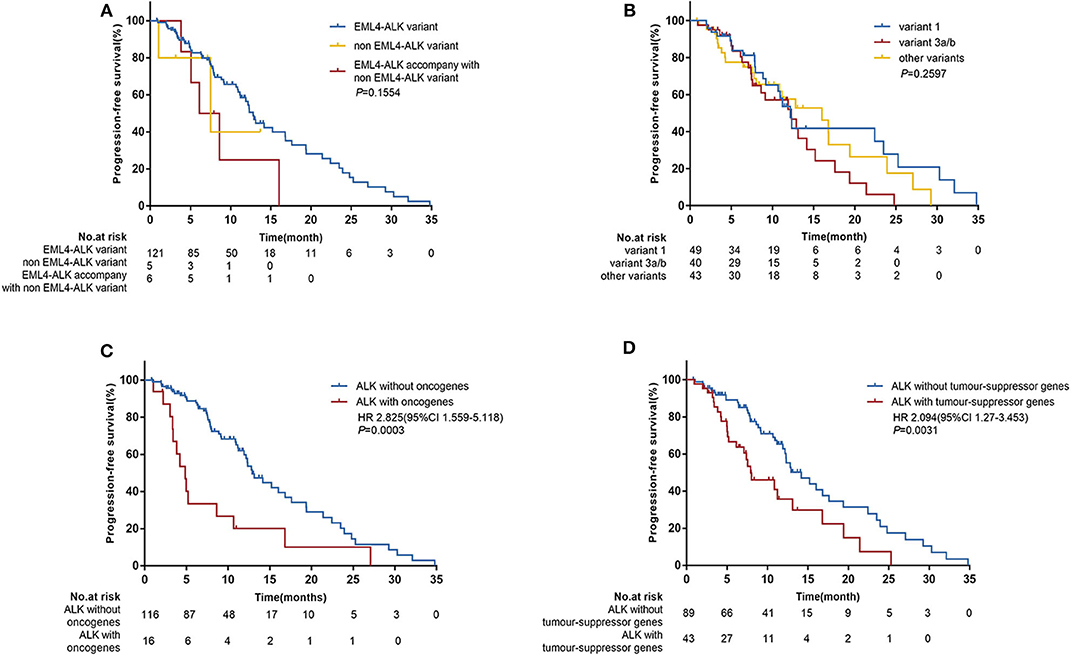
Figure 2. Progression-free survival (PFS) according to baseline next-generation sequencing (NGS) data. (A) Patients were categorized into three subgroups: EML4-ALK rearrangements (n = 121), non-EML4-ALK rearrangements (n = 5), and EML4-ALK rearrangements accompanied by non-EML4-ALK rearrangements (n = 6). (B) Patients with different EML4-ALK variants: variant 1 (n = 49), variant 3a/b (n = 40), and other variants (n = 43). (C) Patients with oncogene mutations (n = 16) vs. patients without oncogene mutations (n = 116). (D) Patients with tumor-suppressor gene mutations (n = 43) vs. patients without tumor-suppressor gene mutations (n = 89). HR, hazard ratios; CI, confidence interval; p-values were calculated using the log-rank test.
Secondly, according to the EML4-ALK rearrangement, we divided patients into variant 1, variant 3a/b, and other variant groups. The baseline characteristics of these three groups were well-balanced (Supplementary Table 2). The median PFS was similar in the three groups. In the variant 1 group the median PFS was 12.2 months (95% CI 9.2–23.5); in the variant 3a/b group, it was 12.3 months (95% CI 7.5–14.2); and in the group with other variants, it was 16.0 months (95% CI 8.0–19.4) (P = 0.2597) (Figure 2B). Similarly, no correlation was observed between EML4-ALK variants and the ORR with crizotinib treatment (Table 2). Similar results were observed in subgroups with baseline CNS metastases (Supplementary Figure 2A).
Prevalence and Clinical Impact of Concomitant Mutations
Among the 132 patients, 12.1% (16/132) patients had concomitant oncogene mutations (EGFR, ERBB2, KRAS, BRAF, MET, RET, ROS1, or PIK3CA), including 3 (2.3%) patients with EGFR mutations, 4 (3.0%) with ERBB2 mutations, 1 (0.76%) with KRAS mutations, 4 (3.0%) with MET amplification, and 4 (3.0%) with PI3KCA mutations. BRAF, RET, and ROS1 mutations were not found because of the limited sample size. In addition, we found that 32.6% of patients (43/132) had tumor-suppressor gene mutations (TP53, PTEN, APC, or RB1), the most common of which was TP53 mutation (39/132) (Figure 1B). There was no significant correlation between ALK rearrangement variants and concomitant mutations (Supplementary Table 2). However, concomitant mutations were significantly associated with poor efficacy of crizotinib. In patients with and without oncogene mutations, median PFS values were 4.9 months (95% CI 3.3–8.6) and 12.9 months (95% CI 11.9–16.8), respectively (HR 2.825; 95% CI 1.559–5.118; P = 0.0003) (Figure 2C). In patients with and without tumor-suppressor gene mutations, median PFS values were 7.9 months (95% CI 5.2–13.1) and 14.2 months (95% CI 11.9–17.6), respectively (HR 2.094; 95% CI 1.270–3.453; P = 0.0031) (Figure 2D). Similarly, in patients with baseline CNS metastases, concomitant mutations also had a negative effect on the clinical efficacy of crizotinib (Supplementary Figures 2B,C). No significant differences in the objective response rate (ORR) were observed according to concomitant mutations (Table 2).
In the univariate analysis which included age, gender, histological diagnosis, smoking status, ECOG, central nervous system metastases, clinical type, treatment line of crizotinib therapy, clinical stage, oncogenes, tumor-suppressor genes and ALK variants, we found that smoking status (P = 0.048), treatment line of crizotinib therapy (P = 0.047), oncogenes (P = 0.0003) and tumor-suppressor genes (P = 0.0031) were significantly associated with PFS. ALK variants tended to be associated with PFS (P = 0.254). When we included these factors in a multivariate Cox regression analysis, concomitant oncogene mutations (HR 2.615 [95% CI 1.398–4.889]; P = 0.0026) and tumor-suppressor gene mutations (HR 2.122 [95% CI 1.264–3.564]; P = 0.0044) both remained independent negative factors affecting the efficacy of crizotinib for patients with ALK rearrangement NSCLC (Figure 3B). However, the impacts of crizotinib treatment line and smoking status became less significant in the multivariate analysis.
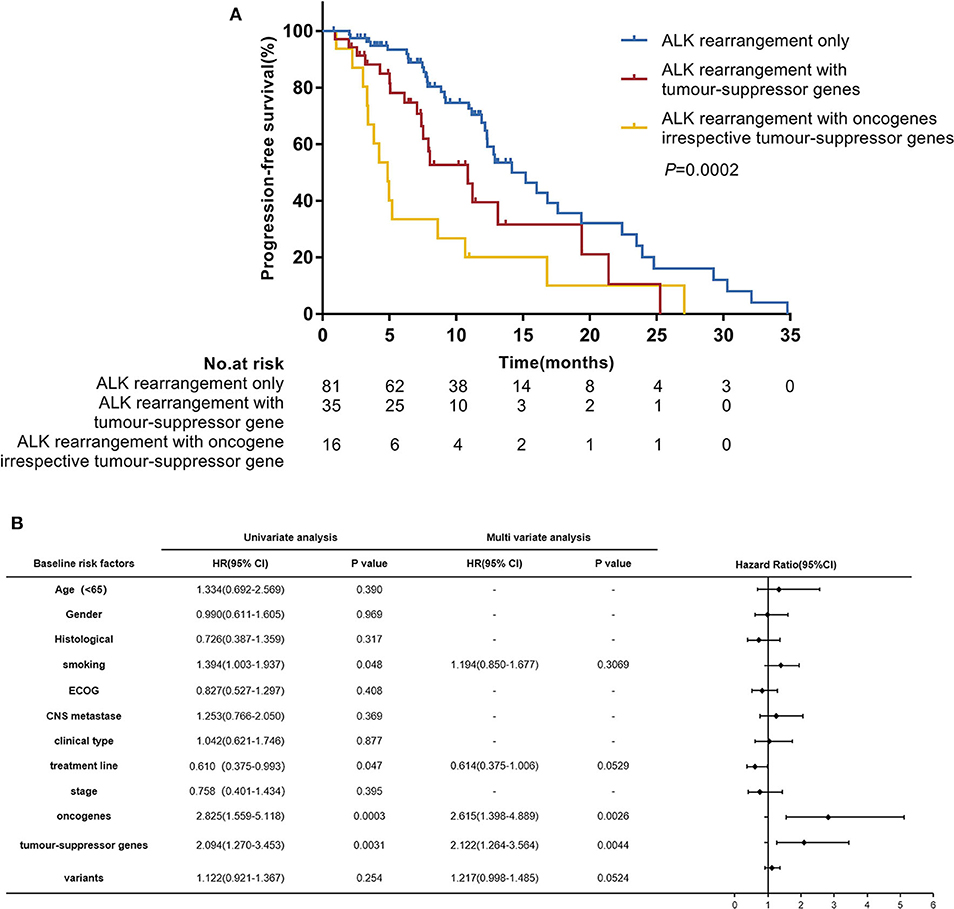
Figure 3. (A) Progression-free survival (PFS) according to concomitant mutations. (B) Hazard ratios (HR) were evaluated by Cox regression. CI, confidence interval; ECOG, Eastern Cooperative Oncology Group performance status.
In further analyses, we divided patients into three groups according to their concomitant mutations: patients with ALK-rearrangement only (n = 81), patients with ALK rearrangement and concomitant tumor-suppressor gene mutations (n = 35), and patients with ALK rearrangement and concomitant oncogene mutations irrespective of tumor-suppressor gene mutations (n = 16). The median PFS values in these three groups were 14.2 months (95% CI 12.2–19.4), 10.9 months (95% CI 7.4–19.4), and 4.9 months (95% CI 3.3–8.6), respectively; (P = 0.0002) (Figure 3A).
Progression Patterns and Resistance Mechanisms to Crizotinib
At the data cut-off time, a total of 71 patients (53.8%) had confirmed progressive disease (PD). Among these patients, 26 (36.6%) had isolated central nervous system (CNS) progression. Patients with CNS metastases at baseline were more likely to have isolated CNS progression compared with patients without CNS metastases at baseline (61.5 vs. 22.2%, respectively; P < 0.001). The patients with isolated CNS progression seemed to have inferior PFS values with crizotinib treatment compared with patients with progression at other sites; however, the difference between them was not significant (6.4 months [95% CI 4.9–10.9] vs. 9.2 months [95% CI 7.5–12.3], respectively; P = 0.5129) (Supplementary Figure 3). There was also no correlation between progression sites and different ALK rearrangement variants (Supplementary Table 2).
Twenty-five patients underwent repeat biopsies to detect acquired resistance mechanisms to crizotinib. Among these patients, 20 (80.0%) remained ALK rearrangement, but in 5 patients ALK rearrangement wasn't detected in their tissues. Secondary ALK mutations were identified in 8 (32.0%) patients. All secondary ALK mutations were detected in patients with ALK rearrangement present (Figure 4C) but there was no significant correlation for the ALK variants (variant3a/b, 25.0% vs. non-variant 3a/b, 35.3%; P = 0.607). The median PFS was significantly prolonged in patients with ALK rearrangement absent compared with patients with ALK rearrangement present (21.4 months [95% CI 6.3–34.8] vs. 10.8 months [95% CI 7.4–16.0]; P = 0.0453) (Figure 4A). Patients in whom secondary ALK mutations were detected showed inferior survival compared with those in whom secondary ALK mutations were not detected, although the difference was not statistically significant (PFS, 9.0 months [95% CI 4.9–16.0] vs. 12.9 months [95% CI 7.6–21.4]; P = 0.1063) (Figure 4B).
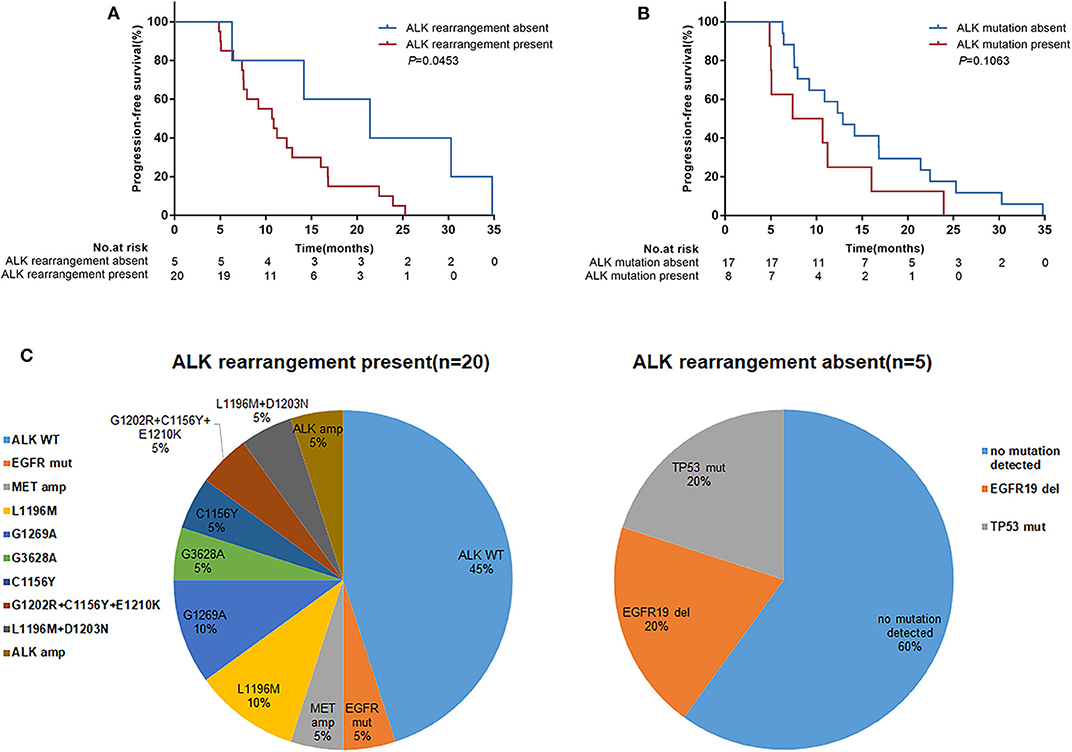
Figure 4. Progression-free survival (PFS) according to ALK resistance mechanisms detected in repeat tumor biopsies following disease progression on crizotinib treatment. (A) Patients with ALK rearrangement absent (n = 5) vs. ALK rearrangement present (n = 20); (B) Patients with ALK mutations absent (n = 17) vs. ALK mutations present (n = 8). p-values were calculated using the log-rank test. (C) Distribution of ALK resistance mutations after disease progression on crizotinib treatment by ALK rearrangement present or absent. WT, wild-type; amp, amplification; mut, mutation; del, deletion.
Discussion
To the best of our knowledge, the present study is the first large-sample size study to comprehensively investigate the correlation between concomitant mutations and the efficacy of ALK inhibitors according to next-generation sequencing data in patients with ALK rearrangement NSCLC. We showed that concomitant mutations, irrespective of oncogenes or tumor-suppressor genes, had a negative effect on the efficacy of crizotinib in patients with ALK rearrangement NSCLC. However, patients with different ALK variants had similar clinical responses to crizotinib.
In our study, we reported a relatively large data set in which the prevalence of different ALK variants was evaluated and we compared the clinical efficacy of crizotinib between the different ALK variants. Consistent with previous studies, EML4 was the most common fusion partner, but we also reported several rare fusion partners and dual fusion partners. When we evaluated clinical responses, patients with the rare fusion variants were found to have a similar median PFS with crizotinib treatment compared with EML4-ALK variants. In the case of dual fusion partners, all 6 patients had EML4-ALK rearrangement accompanied by a non-EML4-ALK rearrangement, and which was the major driver fusion gene was unclear. When we evaluated clinical responses, patients with the rare rearrangement variants or EML4-ALK accompanied by a non-EML4-ALK rearrangement were found to have a similar median PFS with crizotinib treatment compared with EML4-ALK variants. However, the sample size of these rare ALK variants was small, which limits conclusive data on the crizotinib sensitivity of rare ALK variants. Given the low occurrence rate of ALK in lung cancer, multicenter participation and predefined subgroup analysis of these rare ALK variants may be worth considering in future studies. In terms of EML4-ALK rearrangement, the most common variants were variant 1 (E13:A20), followed by variant 3a/b (E6:A20) and variant 2 (E20:A20), as has been reported in a series of other studies. Although the correlation between ALK variants and clinical efficacy has been investigated in several studies, a consensus has not yet been reached. Yoshida et al. (22) reported that variant 1 was associated with superior efficacy to crizotinib than other variant types, and Woo et al. (23) found that variant 3a/b, which has a stable EML4-ALK fusion protein, was associated with a significantly shorter PFS with ALK inhibitors than other variants. However, Mitiushkina et al. (24) found no difference in the treatment response between various ALK variants. Furthermore, in the prospective, phase III ALEX trial, there was a similar survival benefit with crizotinib and alectinib treatment for the different variants (25). In the present study, we found that various EML4-ALK variants had similar PFS values and response rates with crizotinib treatment, consistent with previous phase III ALEX study (25). The same results were observed in the subgroup with baseline CNS metastases.
For EGFR-mutated NSCLC, a series of studies have investigated the correlation of concomitant mutations and efficacy to EGFR TKIs. Hong et al. (19) reported that co-alteration mutations are associated with resistance to EGFR TKIs, and EGFR 21 L858R had a significantly higher incidence of co-alterations than EGFR 19 deletion. A prospective phase II study [the BENEFIT study (18)] also revealed that patients with an EGFR mutation only had superior responses to first-generation EGFR TKIs than those with oncogenes and tumor-suppressor genes present, or both (18). A similar conclusion was reported for ROS1 fusion in that concomitant mutations were observed to be frequent in patients with ROS1 fusion and these concomitant mutations had negative impacts on overall survival (26).
For ALK rearrangement NSCLC, several studies have found that ALK rearrangements are not absolutely exclusive with other driver mutations. Won et al. (27) reported that 4.4% of patients with ALK-positive NSCLC have EGFR concomitant mutation using Sanger sequencing, and this rose to 15.4% of patients when using NGS. Ulivi et al. (21) found that 1.6% and 2.5% of patients (n = 380) who harbor double EML4-ALK and EGFR mutations and EML4-ALK and KRAS mutations have a poor prognosis. Regarding the tumor-suppressor gene, Wang et al. (20) had previously reported that 38.1% of patients (8/22) with ALK rearrangement NSCLC had TP53 mutations, which reduced responsiveness to crizotinib and worsened the prognosis. However, all current studies of ALK-positive patients have been small-sample size and didn't use NGS to comprehensively investigate the baseline genetic mutations and the clinical response. In our study, we found concomitant mutations were common in patients with ALK rearrangement, and not related to ALK variants. Concomitant mutations are heterogeneous and may have different impacts on crizotinib efficacy. It seemed that concomitant oncogene mutations had a worse negative effect than concomitant tumor-suppressor gene mutations (HR 2.615 vs. 2.122, respectively), in multivariable analyses, and both remained poor independent factors for clinical efficacy of crizotinib after adjusting for ALK variants and patient characteristics.
In our study, the PFS was 4.9 months with crizotinib treatment in patients with concomitant oncogene mutations, which was inferior than that in previously reported phase III studies [7.7 months for chemotherapy-pretreated (11) and 10.9 months for treatment-naive patients (12)]. Our findings support previous views of high intratumor molecular heterogeneity in ALK rearrangement NSCLC, and the activation of bypass signaling pathways may induce the primary resistance to crizotinib in these patients. The status of these concomitant mutations should be considered when defining targeted treatments for ALK rearrangement patients as patients carrying these genomic aberrations may not benefit from crizotinib monotherapy. Our findings based on a small sample-size of patients with oncogene mutations remains to be verified and expanded in future studies. In EGFR mutation NSCLC, present studies have revealed that EGFR TKIs combined with chemotherapy (28, 29) or antiangiogenic (30) therapy may have better efficacy than monotherapy with EGFR TKIs. However, few studies have investigated the effectiveness and safety of combination therapies and it is not clear whether dual targeted TKI inhibitors for patients with concomitant oncogene mutations or combined with chemotherapy or antiangiogenic therapy may provide the better benefit for patients with concomitant tumor-suppressor gene mutations. In addition, there is a lack of evidence for first-line treatment with next-generation ALK inhibitors in patients with concomitant mutations. Kron et al. (31) found that patients with ALK/TP53 co-mutations had a worse PFS with next-generation ALK-inhibitors after crizotinib treatment compared with patients with TP53 with wild-type mutations (5.4 vs. 9.9 months, respectively; P = 0.039). The impact on efficacy of next-generation ALK inhibitors according to baseline NGS analysis needs to be further investigated in multicenter studies.
In our study, there were 25 patients who received repeat biopsies to detect the resistance mechanisms. It seemed that patients with ALK rearrangement absent have a longer PFS with crizotinib treatment than patients with ALK rearrangement present. This may be explained by tumor cells harboring ALK rearrangement decreasing or disappearing after effective therapy. Increasing evidence has shown that dynamic molecular changes are associated with clinical efficacy. The BENEFIT study (18) found that patients with clearance of an EGFR mutation after 8 weeks had a significantly prolonged PFS with first-line gefitinib treatment compared with patients with persisting EGFR mutations. Pailler et al. (32) also showed that a decrease in the number of circulating tumor cells (CTCs) and an ALK-copy number gain with crizotinib treatment was associated with a longer PFS (P = 0.025). The present study suggested that dynamic detection of ALK rearrangement may predict efficacy to crizotinib, but larger sample size prospective studies are needed for further analysis.
Our study has several limitations. Firstly, it was a retrospective study and still had a limited sample size, particularly for non-EML4-ALK rearrangement variants, dual ALK rearrangement variants and oncogene mutations, therefore, the results should be interpreted with caution. Multicenter studies based on next-generation ALK inhibitors will be conducted in future to validate and expand our findings. Secondly, we used three different gene panels in our studies, which were mainly based on patients' clinical characteristics and financial situation, although all contained lung cancer-related genes. The NGS-detected samples included tumor tissues and liquid biopsies, which may have different sensitivities for mutation detection. Recent studies have shown that sensitivity of EGFR ctDNA is lower for tumor tissues (33, 34), while the data for ALK rearrangement assessment using ctDNA is relatively limited compared with EGFR mutations. McCoach et al. (35) demonstrated that cfDNA NGS testing is a surrogate tool for detecting ALK alterations in newly diagnosed patients, as well as for resistant mutations in patients progressing on targeted therapy. Thirdly, the OS of patients according to ALK variants and concomitant mutations were not mature and further follow-up observation is required.
Conclusion
The present study found that concomitant mutations have a significant negative effect on the efficacy of crizotinib in patients with ALK rearrangement advanced NSCLC, but that various ALK variants may have a similar influence. The status of concomitant mutations should be considered when defining targeted treatment for ALK rearrangement patients. Our findings need further validation and expansion in future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The National Omics Data Encyclopedia [accession: OEP001055, https://www.biosino.org/node/project/detail/OEP001055].
Ethics Statement
The studies involving human participants were reviewed and approved by Guangdong Association of Thoracic Oncology (GASTO ID: 1055). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ML contributed to study conception and design, analysis of the data, and wrote the manuscript. XH, CZ, and WF contributed to study conception and design and acquisition of data. GJ, HL, SY, and JC contributed to acquisition of data. LC contributed to study conception and design and overall review. All authors reviewed the manuscript and approved the final version submitted for publication.
Funding
This study was supported by the Sun Yat-sen University Young Teacher Plan (19ykpy179) and Guangzhou Science and Technology Program (20200202007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the patients enrolled in this study, as well as the work of research staff. Editorial assistance with the manuscript was provided by Content Ed Net, Shanghai Co. Ltd. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01216/full#supplementary-material
References
1. Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res. (2015) 2227–35. doi: 10.1158/1078-0432.CCR-14-2791
2. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. (2007) 448:561. doi: 10.1038/nature05945
3. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers. (2017) 9:E118. doi: 10.3390/cancers9090118
4. Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. (2009) 15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248
5. Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS ONE. (2012) 7:e31323. doi: 10.1371/journal.pone.0031323
6. Zhang M, Wang Q, Ding Y, Wang G, Chu Y, He X, et al. CUX1-ALK, a novel ALK rearrangement that responds to crizotinib in non–small cell lung cancer. J Thorac Oncol. (2018) 13:1792–7. doi: 10.1016/j.jtho.2018.07.008
7. Hong M, Kim RN, Song JY, Choi SJ, Oh E, Lira ME, et al. HIP1–ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol. (2014) 9:419–22. doi: 10.1097/JTO.0000000000000061
8. Choi YL, Lira ME, Hong M, Kim RN, Choi SJ, Song JY, et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol. (2014) 9:563–6. doi: 10.1097/JTO.0000000000000093
9. Shan L, Jiang P, Xu F, Zhang W, Guo L, Wu J, et al. BIRC6-ALK, a novel fusion gene in ALK break-apart FISH-negative lung adenocarcinoma, responds to crizotinib. J Thorac Oncol. (2015) 10:e37–9. doi: 10.1097/JTO.0000000000000467
10. Hou X, Xu H, Chen L. SRBD1-ALK, a novel ALK fusion gene identified in an adenocarcinoma patient by next-generation sequencing. J Thorac Oncol. (2019) 14:e72–3. doi: 10.1016/j.jtho.2018.11.027
11. Shaw AT, Kim DW, Nakagawa K, Seto T, Crin L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. (2013) 368:2385–94. doi: 10.1056/NEJMoa1214886
12. Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. (2014) 371:2167–77. doi: 10.1056/NEJMoa1408440
13. Soria JC, Tan DSW, Chiari R, Wu YL, Paz-Ares L, Wolf J, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet. (2017) 389:917–29. doi: 10.1016/S0140-6736(17)30123-X
14. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med. (2017) 377:829–38. doi: 10.1056/NEJMoa1704795
15. Kim DW, Tiseo M, Ahn MJ, Reckamp KL, Hansen KH, Kim SW, et al. Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non-small-cell lung cancer: a randomized, multicenter phase II trial. J Clin Oncol. (2017) 35:2490–8. doi: 10.1200/JCO.2016.71.5904
16. Solomon BJ, Besse B, Bauer TM, Felip E, Soo RA, Camidge DR, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol. (2018) 19:1654–67. doi: 10.1016/S1470-2045(18)30649-1
17. Lin JJ, Zhu VW, Yoda S, Yeap BY, Schrock AB, Dagogo-Jack I, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol. (2018) 36:1199. doi: 10.1200/JCO.2017.76.2294
18. Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. (2018) 6:681–90. doi: 10.1016/S2213-2600(18)30264-9
19. Hong S, Gao F, Fu S, Wang Y, Fang W, Huang Y, et al. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non–small cell lung cancer. JAMA Oncol. (2018) 4:739–42. doi: 10.1001/jamaoncol.2018.0049
20. Wang WX, Xu CW, Chen YP, Liu W, Zhong LH, Chen FF, et al. TP53 mutations predict for poor survival in ALK rearrangement lung adenocarcinoma patients treated with crizotinib. J Thorac Dis. (2018). 10:2991–8. doi: 10.21037/jtd.2018.04.98
21. Ulivi P, Chiadini E, Dazzi C, Dubini A, Costantini M, Medri L, et al. Nonsquamous, non-small-cell lung cancer patients who carry a double mutation of EGFR, EML4-ALK or KRAS: frequency, clinical-pathological characteristics, and response to therapy. Clin Lung Cancer. (2016) 17:384–90. doi: 10.1016/j.cllc.2015.11.004
22. Yoshida T, Oya Y, Tanaka K, Shimizu J, Horio Y, Kuroda H, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non–small-cell lung cancer. J Clin Oncol. (2016) 34:3383–9. doi: 10.1200/JCO.2015.65.8732
23. Woo CG, Seo S, Kim SW, Jang SJ, Park KS, Song JY, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol. (2017) 28:791–7. doi: 10.1093/annonc/mdw693
24. Mitiushkina NV, Tiurin VI, Iyevleva AG, Kholmatov MM, Filippova EA, Moiseyenko FV, et al. Variability in lung cancer response to ALK inhibitors cannot be explained by the diversity of ALK fusion variants. Biochimie. (2018) 154:19–24. doi: 10.1016/j.biochi.2018.07.018
25. Camidge DR, Dziadziuszko R, Peters S, Mok T, Noe J, Nowicka M, et al. Updated efficacy and safety data and impact of the EML4-ALK fusion variant on the efficacy of alectinib in untreated ALK-positive advanced non–small-cell lung cancer in the global phase III ALEX study. J Thorac Oncol. (2019) 14:1233–43. doi: 10.1016/j.jtho.2019.03.007
26. Wiesweg M, Eberhardt WEE, Reis H, Ting S, Savvidou N, Skiba C, et al. High prevalence of concomitant oncogene mutations in prospectively identified patients with ROS1-positive metastatic lung cancer. J Thorac Oncol. (2017) 12:54–64. doi: 10.1016/j.jtho.2016.08.137
27. Won JK, Keam B, Koh J, Cho HJ, Jeon YK, Kim TM, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol. (2014) 26:348–54. doi: 10.1093/annonc/mdu530
28. Hosomi Y, Morita S, Sugawara S, Kato T, Fukuhara T, Gemma A, et al. Gefitinib alone versus gefitinib plus chemotherapy for non–small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 study. J Clin Oncol. (2020) 38:115–23. doi: 10.1200/JCO.19.01488
29. Yang JC, Mok T, Han B, Orlando M, Puri T, Park K. A review of regimens combining pemetrexed with an epidermal growth factor receptor tyrosine kinase inhibitor in the treatment of advanced nonsquamous non–small-cell lung cancer. Clin Lung Cancer. (2018) 19:27–34. doi: 10.1016/j.cllc.2017.06.013
30. Seto T, Kato T, Nishio M, Goto K, Atagi S, Hosomi Y, et al. Erlotinib alone or with bevacizumab as first-line therapy in patients with advanced non-squamous non-small-cell lung cancer harbouring EGFR mutations (JO25567): an open-label, randomised, multicentre, phase 2 study. Lancet Oncol. (2014) 15:1236–44. doi: 10.1016/S1470-2045(14)70381-X
31. Kron A, Alidousty C, Scheffler M, Merkelbach-Bruse S, Seidel D, Riedel R, et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol. (2018) 29:2068–75. doi: 10.1093/annonc/mdy333
32. Pailler E, Oulhen M, Borget I, Remon J, Ross K, Auger N, et al. Circulating tumor cells with aberrant ALK copy number predict progression-free survival during crizotinib treatment in ALK-rearranged non–small cell lung cancer patients. Cancer Res. (2017) 77:2222–30. doi: 10.1158/0008-5472.CAN-16-3072
33. Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. (2009) 27:2653–59. doi: 10.1200/JCO.2008.17.3930
34. Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib treatment in EGFRmutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. (2014) 9:1345–53. doi: 10.1097/JTO.0000000000000263
Keywords: ALK rearrangement, non–small-cell lung cancer, concomitant mutations, crizotinib, next-generation sequencing
Citation: Li M, Hou X, Zhou C, Feng W, Jiang G, Long H, Yang S, Chen J, Wang N, Wang K and Chen L (2020) Prevalence and Clinical Impact of Concomitant Mutations in Anaplastic Lymphoma Kinase Rearrangement Advanced Non-small-Cell Lung Cancer (Guangdong Association of Thoracic Oncology Study 1055). Front. Oncol. 10:1216. doi: 10.3389/fonc.2020.01216
Received: 11 April 2020; Accepted: 15 June 2020;
Published: 21 August 2020.
Edited by:
Alfredo Addeo, Geneva University Hospitals (HUG), SwitzerlandReviewed by:
Weiwei Tan, Pfizer, United StatesTommaso Martino De Pas, European Institute of Oncology (IEO), Italy
Copyright © 2020 Li, Hou, Zhou, Feng, Jiang, Long, Yang, Chen, Wang, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Likun Chen, Y2hlbmxrJiN4MDAwNDA7c3lzdWNjLm9yZy5jbg==
†These authors have contributed equally to this work
 Meichen Li
Meichen Li Xue Hou1†
Xue Hou1† Chengzhi Zhou
Chengzhi Zhou Weineng Feng
Weineng Feng Guanming Jiang
Guanming Jiang Jing Chen
Jing Chen Na Wang
Na Wang