- Department of Chronic-Degenerative Diseases, National Institute of Respiratory Diseases “Ismael Cosío Villegas”, Mexico City, Mexico
Despite the recent advances in chemotherapeutic treatments against cancer, some types of highly aggressive and invasive cancer develop drug resistance against conventional therapies, which continues to be a major problem in the fight against cancer. In recent years, studies of alterations of DNA methylome have given us a better understanding of the role of DNA methylation in the development of tumors. DNA methylation (DNAm) is an epigenetic change that promotes the covalent transfer of methyl groups to DNA. This process suppresses gene expression through the modulation of the transcription machinery access to the chromatin or through the recruitment of methyl binding proteins. DNAm is regulated mainly by DNA methyltransferases. Aberrant DNAm contributes to tumor progression, metastasis, and resistance to current anti-tumoral therapies. Aberrant DNAm may occur through hypermethylation in the promoter regions of tumor suppressor genes, which leads to their silencing, while hypomethylation in the promoter regions of oncogenes can activate them. In this review, we discuss the impact of dysregulated methylation in certain genes, which impact signaling pathways associated with apoptosis avoidance, metastasis, and resistance to therapy. The analysis of methylome has revealed patterns of global methylation, which regulate important signaling pathways involved in therapy resistance in different cancer types, such as breast, colon, and lung cancer, among other solid tumors. This analysis has provided gene-expression signatures of methylated region-specific DNA that can be used to predict the treatment outcome in response to anti-cancer therapy. Additionally, changes in cancer methylome have been associated with the acquisition of drug resistance. We also review treatments with demethylating agents that, in combination with standard therapies, seem to be encouraging, as tumors that are in early stages can be successfully treated. On the other hand, tumors that are in advanced stages can be treated with these combination schemes, which could sensitize tumor cells that are resistant to the therapy. We propose that rational strategies, which combine specific demethylating agents with conventional treatment, may improve overall survival in cancer patients.
Introduction
During carcinogenesis, genetic and epigenetic alterations lead to dysregulated expression of genes associated with cellular pathways that regulate processes such as cell proliferation, cell differentiation, cell death, and cell cycle, among others. Epigenetic alterations that include DNA methylation (DNAm), histone modifications, aberrant expression of microRNAs (miRNAs), and long non-coding RNA (lncRNA) are common in several types of cancer. These epigenetic changes are hereditary, transient, and reversible and do not cause modification in the DNA sequence (1). Cancer can be treated by resection, chemotherapeutic agents, radiation, and immunotherapy, among others, as well as any combination of the aforementioned therapies. However, the 5-year survival rate remains low in many solid tumors due to tumor intrinsic or acquired resistance (2).
DNAm is a pivotal mechanism in normal cell development, which plays an important role in the regulation of gene expression, as well as chromatin stability, genetic imprinting, X-chromosome inactivation, the suppression of repetitive element transcription, and transposition. In mammals, DNAm involves the covalent transfer of methyl groups (-CH3) from S-adenosyl-1-methionine (SAM) to cytosine in the CpG islands (2). CpG islands are characterized by a length longer than 200 bp. They possess a GC content >50% and present a ratio of observed to expected CpG dinucleotides >0.6. Moreover, CpG islands have been located in or near ~50% of human promoters (3). DNAm is catalyzed by three DNA methyltransferases (DNMTs), which have been identified in mammals: DNMT1, DNMT3A, and DNMT3B. DNMT1 maintains hemimethylated DNA patterns during DNA replication, while DNMT3A and DNMT3B establish new patterns of methylation in early embryonic development (4).
Here, we review several DNAm alterations in cancer that have been associated with carcinogenesis, apoptosis avoidance, migration, invasion, and metastasis. Several studies have found that some of these DNAm alterations may be associated with tumor clinical features such as disease risk, TNM (tumor, node and metastasis)-stage, prognosis, diagnosis, survival, and response to treatment. We also discuss several DNAm alterations in genes and some pathways that have been reported to promote tumor resistance to therapeutic agents. Additionally, we argue that the promotion or inhibition of DNAm in a non-specific way should be carefully revised because of their side effects. In contrast, more extensive studies should be further developed by considering the targeting specific alterations in DNAm or editing the epigenome by CRISPR-Cas9 technology.
DNA Methylation Regulates Gene Expression
Genetic mechanisms and epigenetic modifications such as DNAm, histone modifications, and non-coding RNAs (including microRNAs) regulate gene expression, which is a fundamental process that maintains cellular homeostasis. Each cell type possesses its own gene expression pattern, driven by a specific epigenetic signature, which may also produce cell heritable characteristics (5).
DNAm is a covalent modification in which a methyl group is linked to the cytosine in the dinucleotides cytosine-guanine (CpG), which is often located in “CpG islands” in the gene promoters; as a consequence, DNAm can modify gene expression. The CpG island is a short sequence of DNA in which the frequency of the CpG sequence is higher than that in other regions. Hypomethylation of promoter regions allows gene expression machinery to access the promoters of target genes. Hypermethylation, on the other hand, can suppress gene expression through the modulation of the transcription machinery access to the chromatin or through the recruitment of methyl binding proteins (5). In addition to promoters, other upstream DNA regions are rich in CpG sequences, up to 2 Kb distant to CpG islands, which are named “CpG island shores.” These CpG island shores have been observed in colon and breast cancers (6, 7), or up to 700 bp in prostate cancer (8). Methylation of these CpG island shores also regulates gene expression. This epigenetic regulation was confirmed with the reactivation of downregulated genes in colon cancer, by demethylation of hypermethylated CpG island shores, using 5-aza2′-deoxycytidine (a DNA methyltransferase inhibitor) and DNA methyltransferase knockout (6).
Throughout the human lifespan, epigenetic patterns may change and constitute an important component of the aging process. Studies of human DNA methylome have revealed that one-third of 476,366 DNAm sites are affected by age. When age increases from 14 to 94 years, 60.5% of these affected DNAm sites become hypomethylated, and 39.5% become hypermethylated (9).
DNAm is a dynamic process that can also be affected by environmental factors, diet, and exercise habits, which can induce particular gene-expression signatures. For instance, the presence of short-chain fatty acids, such as butyric acid, can induce changes in DNAm patterns in normal and cancer cells; diets deficient in methyl-donor folic acid also promote dynamic changes in DNAm (5). Exercise favors hypomethylation of peroxisome proliferator-activated receptor gamma coactivator 1-alfa and delta (PGC-1α, PPAR-δ), pyruvate dehydrogenase kinase-4 (PDK4), which regulate mitochondrial function and fuel usage. mRNA upregulation of these genes during acute exercise is correlated with a transient but marked hypomethylation on each respective promoter (5). Interestingly, the change in the DNAm patterns associated with exercise is stronger among older people. The decreased DNAm associated with exercise habits among older people has been associated with cancer prevention, rewinding the “epigenetic clock” as people age (10).
Cancer Modifies Gene Expression Through DNA Methylation
Studies of global DNA methylation have found methylation patterns or signatures that have been associated with different cancer hallmarks, such as cell proliferation, migration, invasion, and metastasis, and also with clinical features such as disease stage, prognosis, survival, and response to treatment (2). Many regulatory regions of tumor suppressor genes and oncogenes present an altered methylation pattern (6). Aberrant DNAm, mediated by the overexpression of DNMTs, affects tumor suppressor genes through their hypermethylation, leading to transcriptional silencing of these genes. On the other hand, transcriptional activation by hypomethylation is observed in proto-oncogenes. Hypomethylation may be detected in early and late stages of the tumorigenesis in several cancer types, such as lung cancer, breast cancer, prostate cancer, gastric cancer (GC), and hepatocellular cancer, among others (6).
Neoplastic transformation, carcinogenesis, and cancer progression may be led by DNAm disruption, given that epigenetic changes have been demonstrated in multiple cancers (11). Most DNAm changes in cancer occur in both CpG islands and CpG island shores, affecting the expression of tumor suppressors and oncogenes (6). For instance, it has been found that ~7,000 CpG islands are altered in the genome of human bladder cancer (12).
Analysis of methylation patterns in genomes of normal breast tissue indicates that the 5' end of highly expressed genes presents enriched sites of hypomethylation. This 5' end region includes the promoter, first intron, and first exon. In contrast, the methylome's analysis of genomes of the breast tumor cell lines (MDA-MB-231 and MCF-7) shows extensive hypomethylation in the intergenic and intragenic regions. These tumor cell lines present megabase-sized hypomethylated zones, which are associated with gene-poor regions containing tissue-specific gene clusters, fragile sites, chromosomal rearrangement breakpoints, and large genes. This suggests that hypomethylation is involved in genome instability. Interestingly, the extensively hypomethylated genes are all silenced. Also, primary breast tumors exhibit a methylation pattern that is between those of the cell lines and the normal tissue (13). It is well-documented that inactivation of tumor-suppressor genes can also be caused by deletions, point mutations, or allelic loss. Marsit et al. speculated that there might be a mutual relationship between the predisposition to promoter hypermethylation and genetic deletion in non-small cell lung carcinomas (NSCLCs). Interestingly, tumors that exhibit a high loss of heterozygosity show a reduced propensity for hypermethylation. The authors conclude that tumor suppressor gene silencing might be caused by allele loss events or epigenetic silencing events, occurring in a roughly dichotomous fashion, which would promote different molecular phenotypes in lung cancer (14).
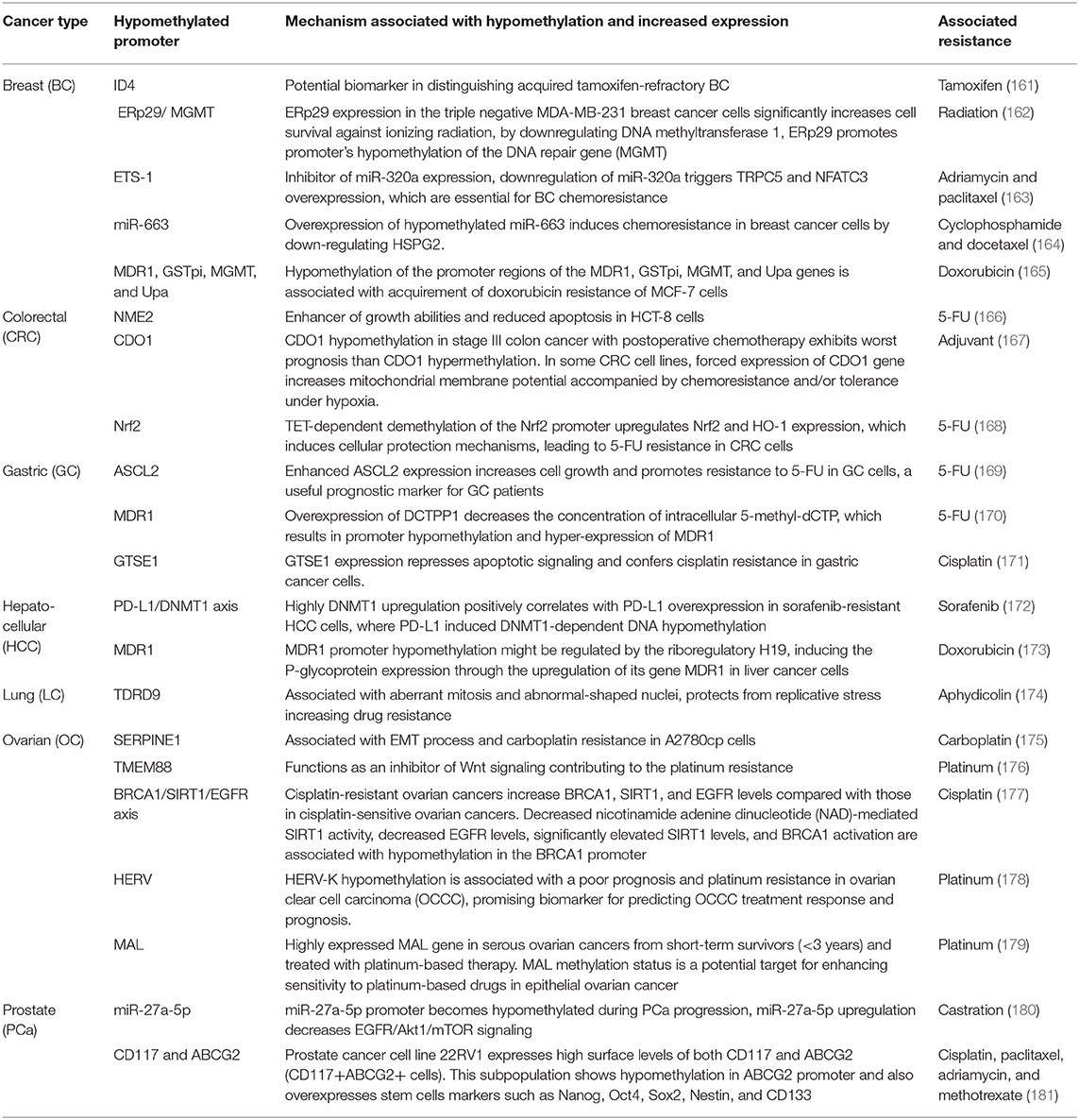
Table 1 (15–85) summarizes genes with decreased expression in cancer as a consequence of hypermethylation of their promoter regions. The absence or reduction of the protein function associated with these genes has been implicated in the development, progression, invasion, and metastasis of many cancer types. Moreover, many hypermethylated genes included in Table 1 participate in pathways involved with cell death processes. On the other hand, Table 2 (86–120) summarizes epigenetically regulated genes by hypomethylation of their promoter regions, which have been found to be highly expressed in cancer. The expression or increased protein function associated with some of these genes has been shown to support cell proliferation, migration, and invasion of many cancer types. Many hypomethylated genes are highly expressed and participate in pathways involved in proliferation and evasion of the immune system (see Table 2). Both the hypermethylation and hypomethylation status of the regulatory regions of tumor suppressor genes and oncogenes have been tested as possible biomarkers for evaluating several parameters, such as cancer risk, diagnosis, and prognosis. Furthermore, analysis of the methylation status of certain genes may be useful for chemotherapy selection for cancer patients, and even for immunotherapy or target therapy (see Tables 1, 2).

Table 1. Hypermethylated promoters of genes associated with tumor suppression, prognosis, response to treatment, or as potential biomarkers.
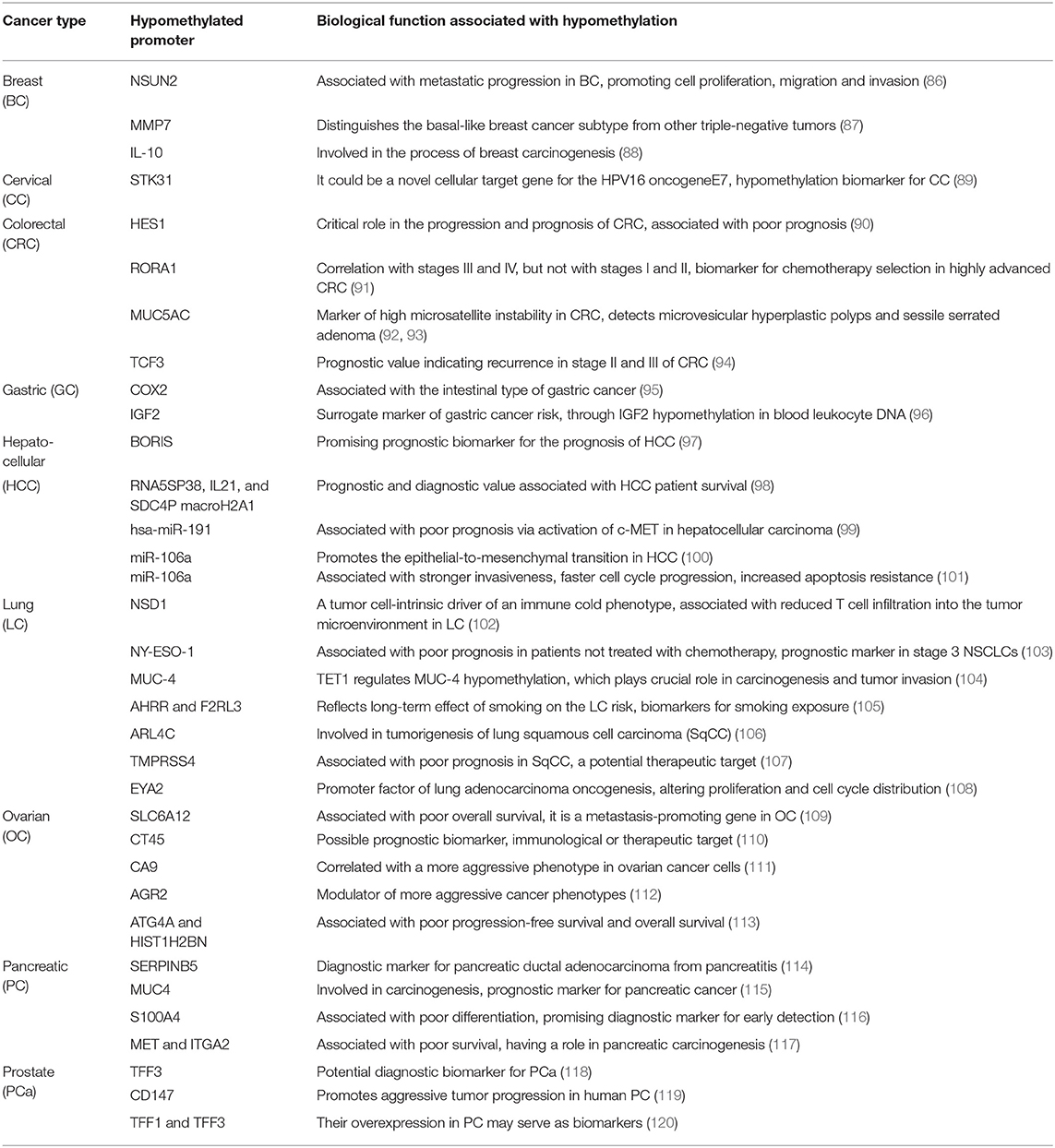
Table 2. Hypomethylated promoters of genes involved in tumor progression, prognosis, or potential therapeutic targets.
Cancer Cells Present Therapy Resistance by Changing Their DNA Patterns
One of the major reasons for the failure of cancer chemotherapy is multidrug resistance (MDR). MDR is divided into the categories of primary drug resistance, which already existed prior to chemotherapy treatment (intrinsic resistance), and acquired drug resistance, which develops during the administration of chemotherapy. This MDR is associated with the regulation and function of apoptotic pathways, intracellular pH, drug pumps, DNA damage repair ability, and drug detoxification. All of these mechanisms reduce the concentration of chemotherapeutic drugs inside the cell, and hyper- and hypomethylation of certain genes appear to play a role (121).
Multiple changes in the methylation of CpG islands and CpG island shores have been found following the acquisition of drug resistance in different cancers. Studies of global DNA methylation profiling have identified different proportions of hypermethylated genes against hypomethylated ones. Baharudin et al. performed DNAm profiling on five recurrent and 43 non-recurrent patients with colorectal cancer (CRC) with 5-fluorouracil (5-FU) treatment (122). The researchers identified 4,787 significantly differentially methylated genes in the recurrent group of CRC compared to the non-recurrent group; 3,112 genes were hypermethylated, and 1,675 genes were hypomethylated. Interestingly, many hypermethylated genes were associated with the MAPK signaling pathway, which is implicated in apoptosis regulation. Conversely, many hypomethylated genes were associated with the PI3K-AKT signaling pathway and the promotion of proliferation (122). In another study, Guo et al. compared the methylation of promoters in a genome-wide study of human lung adenocarcinoma A549 cells resistant to cisplatin (A549/CDDP) with its progenitor A549 cells. The study identified 3,617 genes with differentially methylated promoters; 2,036 were hypomethylated, and 1,581 were hypermethylated. The promoters of RAS association domain family gene 1 (RASSF1), metallothionein 1G (MT1G), and G protein-coupled receptor 56 isoform 3 (GPR56) showed significantly higher hypermethylation in A549/CDDP cells compared to the progenitor A549 cells (123). Thus, increasing evidence supports the notion that epigenetic changes are a driving force behind the acquisition of drug resistance.
Hypermethylation of Key Genes Associated With Therapy Resistance in Cancer
Downregulation of specific genes by hypermethylation of their promoters may lead to MDR. Table 3 (124–153) shows genes whose promoters may suffer hypermethylation and have been associated with resistance to antitumoral therapy in several types of cancer. The products of some of these genes are associated with signaling pathways, such as JAK-STAT, Wnt/β-catenin, MAPK/mTOR, and FAK/Ekt. The promoter hypermethylation pattern or the downregulated gene expression are promising biomarkers for early detection of intrinsic or acquired MRD (Table 3).
The transcriptional silencing mediated by hypermethylation can be used as a therapeutic strategy to diminish the expression of genes associated with drug resistance. For instance, it has been shown that the transmembrane ectoenzyme CD13 endows GC patients with insensitivity to CDDP and that expression of this molecule predicts a poor prognosis in CDDP-treated GC patients. CD13 functions upstream of the epithelial membrane protein 3 (EMP3) to induce its expression. The optimal phosphorylation of PI3K is facilitated by EMP3 upregulation. Phosphorylated PI3K activates the PI3K/AKT/NF-κB pathway suppressing autophagy and epithelial-mesenchymal transition (EMT) and overcoming CDDP resistance in GC cells. Ubenimex, a CD13 inhibitor, induces transcriptional silencing of EMP3 that is mediated by hypermethylation. Therefore, to overcome CDDP resistance in GC cells, ubenimex epigenetically inhibits the activation of the CD13/EMP3/PI3K/AKT/NF-κB pathway (154). Additionally, the NFκB pathway participates in the acquisition of resistance to tyrosine kinase inhibitor (TKI) treatment in lung cancer. Although it is a rare event, methylated cytosine may be converted to thymine by deamination. As a result, the methylated CG sequence could be converted into the TG sequence. Treatment with EGFR TKIs leads to activation of the NFκB pathway and also induces the activation-induced cytidine deaminase (AICDA) expression. AICDA deaminates the 5-methylcytosine, resulting in thymine to generate a T790M mutation. Hence, this is also a methylation-associated mechanism behind the acquisition of a mutation that provides resistance to TKI treatment in lung cancer (155).
DNAm-induced silencing of tumor suppressors is common in cancer. The hypermethylation can be reverted using the FDA-approved DNMT inhibitor 5-aza-2'-deoxycytidine, also named 5-azacytidine or decitabine. 5-Azacytidine has proven to be effective in the treatment of hematological neoplasms. However, its antitumor effect varies in solid tumors (156). The inhibition of methylation has presented good results in GC; Zhang et al. reported that growth arrest-specific transcript 5 (GAS5), which is a tumor suppressor lncRNA, is downregulated in GC. Adriamycin (ADM)-resistant cells (SGC-7901/ADM) have significantly higher levels of hypermethylation in the GAS5 promoter than GC SGC-7901 cells. The authors enforced GAS5 expression, provoking a significant reduction in tumor growth rate and apoptosis after Adriamycin treatment (157). Additionally, Wu et al. have shown that hypermethylation of miR-129-5p CpG island promotes miR-129-5p downregulation, favoring chemoresistance in GC cells. In the GC MDR cell line (SGC7901/VCR), the expression of miR-129-5p was restored through the use of 5-azacytidine, which reduced the chemo-resistance to 5-FU, vincristine, and cisplatin in this cell line. When the authors downregulated miR-129-5p, the chemoresistance was recovered. Furthermore, three members of ABC transporters (ABCG1, ABCC5, and ABCB1), which are associated with MDR, are direct targets of miR-129-5p regulation (158). In contrast, the demethylation of regulatory regions can induce chemoresistance in cervical cancer. Sensitivity to DNA topoisomerase I inhibitors in cancer therapy can be affected by DNA hypermethylation of the Werner (WRN) gene that reduces WRN expression. The WRN gene codes for a DNA helicase that contributes to genomic stability. Masuda et al. reported that cervical cancer-derived cell lines and primary cervical cancer that presented decreased WRN expression due to DNA hypermethylation showed high sensitivity to the topoisomerase I inhibitor (CPT-11). After treatment with 5-azacytidine, the tumor cells became resistant to CPT-11. To confirm this result, they transfected with a siRNA against WRN in tumor cells. These cells increased the sensitivity to CPT-11 (159). Therefore, treatment with demethylating drugs may have unforeseen and opposing results in cancer patients.
Silencing mechanisms to prevent the expression of tumor suppressor genes may also be induced by hypermethylation in cancer. For instance, potassium (K+) channels are dysregulated in different tumors and contribute significantly to the malignant phenotypes, such as chemoresistance, proliferation, and migration. KCNQ1 (potassium channel) can interact with β-catenin to affect its subcellular distribution. The interaction of KCNQ1 with β-catenin reduces Wnt/β-catenin signaling, which consequently blocks the expression of its downstream targets, such as MMP7, CCND1, and c-Myc. As a result, proliferation and cell migration are inhibited. DNA hypermethylation of KCNQ1 promoter has been shown to downregulate KCNQ1 expression in hepatocellular carcinoma (HCC). Downregulation of KCNQ1 is found in HCC cell lines and tissues and is associated with a poor prognosis (138). Additionally, the KCNQ1 Opposite Strand/Antisense Transcript 1 (KCNQ1OT1) gene is a lncRNA, which has been reported to be highly expressed in colorectal and lung cancers. High KCNQ1OT1 expression is correlated with malignant phenotypes in lung cancer. The transfection of si-KCNQ1OT1 can effectively knock down the expression of KCNQ1OT1, increasing KCNQ1 levels and, thus, inhibiting the malignancy and chemoresistance of lung cancer cells to paclitaxel (160). Accordingly, treatments that focus on recovering KCNQ1 expression must consider both the hypermethylation of regulatory regions and the expression of lncRNA. Another example of multiple mechanisms for silencing gene expression is the downregulation of BCL2 interacting protein 3 (BNIP3), which is a proapoptotic member of the BCL-2 family that induces necrotic-like cell death. Loss of BNIP3 expression in pancreatic cancer is correlated with methylation of the BNIP3 promoter. Mahon et al. showed an association between the decreased expression of BNIP3 and chemoresistance to gemcitabine in pancreatic ductal adenocarcinoma (PDAC) cell lines. Besides promoter hypermethylation, S100A4 overexpression, which belongs to the S100 calcium-binding protein family, represents an alternative mechanism for inhibiting BNIP3 function in PDAC. S100A4 knockdown, mediated by RNA interference, upregulated the expression of BNIP3 in PDAC cell lines that have an unmethylated BNIP3 promoter, which led to an increased sensitivity to gemcitabine in PDAC cell lines (150). Consequently, it is important to keep in mind that hypermethylation is one of several mechanisms that inhibits tumor suppressor genes, and anti-cancer treatments must consider this fact.
Hypomethylation of Key Genes Is Associated With Therapy Resistance in Cancer
Hypomethylation of promoters for a certain type of gene may also function as tumor mechanisms for acquiring resistance to drug therapy. Table 4 (161–181) shows some genes whose promoters are hypomethylated in several types of cancer. Hypomethylation of the promoters of these genes leads to their upregulation. The product of some of these genes supports mechanisms involved in MDR, proliferation, the repression of apoptotic signaling, mitochondrial function, and DNA repair. For instance, Luzhna et al. found that diminished radiation responsiveness was correlated with significant global DNA hypomethylation in radiation-resistant cells (MCF-7/DOX). This radiation resistance can be reversed by an epigenetic treatment, which is the use of SAM, a methyl donor. The radiation sensitivity in MCF-7/DOX cells was promoted through use of the SAM-mediated reversal of DNA methylation. However, the researchers found that SAM should be carefully used because the SAM application decreased responsiveness to radiation on MCF-7 cells that were originally radiation-sensitive and highly methylated. Remarkably, the authors concluded that a fine balance of DNA methylation is needed to ensure proper drug and radiation responsiveness (182).
Activation of drug-resistance-associated genes, besides the hypomethylation of their promoters, can be caused by de novo gene fusions. In the case of breast cancer, BRCA1-deficient tumors are extremely sensitive to DNA-damaging drugs and poly(ADP-ribose) polymerase (PARP) inhibitors. However, BRCA1 protein was detected in 31 of 42 drug-resistant cases, despite presenting a hypermethylated promoter. BRCA1-intragenic deletions and the loss of BRCA1 promoter hypermethylation have been shown to occur, and de novo gene fusions take place, where BRCA1 expression can be under the transcriptional control of a heterologous promoter (183).
Therefore, targeting methylation should be carefully evaluated because most compounds that promote or inhibit this process are not gene-specific, which may lead to undesirable effects.
Regulation of Wnt Canonical and NON-Canonical Pathways by DNA Methylation that Supports Cancer Development and Therapy Resistance
In this section, we include potential mechanisms by which differentially methylated genes take part in the development of cancer, by integrating protein interactions and pathways regulated by methylation (11). Tumors present a dysregulated pattern of methylation in genes that impact in pathways like the Wnt canonical pathway and PI3K/AKT/mTOR (e.g., DKK, SFRPs, WIF1, DVL, APC, PTEN, SALL2, and IGFBP-3), which support resistance to specific inhibitors as well as conventional chemotherapeutic agents.
The Wnt/beta-catenin signaling pathway plays important roles in carcinogenesis and therapy resistance. Wnt is a large family of secreted lipoproteins that can join to receptors and co-receptors at the cell surface and activate a complex signaling network. This pathway participates in a wide range of physiological cellular processes like embryonic development, tissue homeostasis, tissue regeneration, cell polarity, cell proliferation, cell migration, and apoptosis (184, 185). The Wnt signaling pathway may be activated by the Wnt/ß-catenin pathway (also known as the canonical pathway (Figure 1) (185, 186) or non-canonical Wnt signaling. Non-canonical Wnt signaling, which is independent of β-catenin, is activated by the pathways Wnt/planar cell polarity and Wnt/Ca2+ (187, 188).
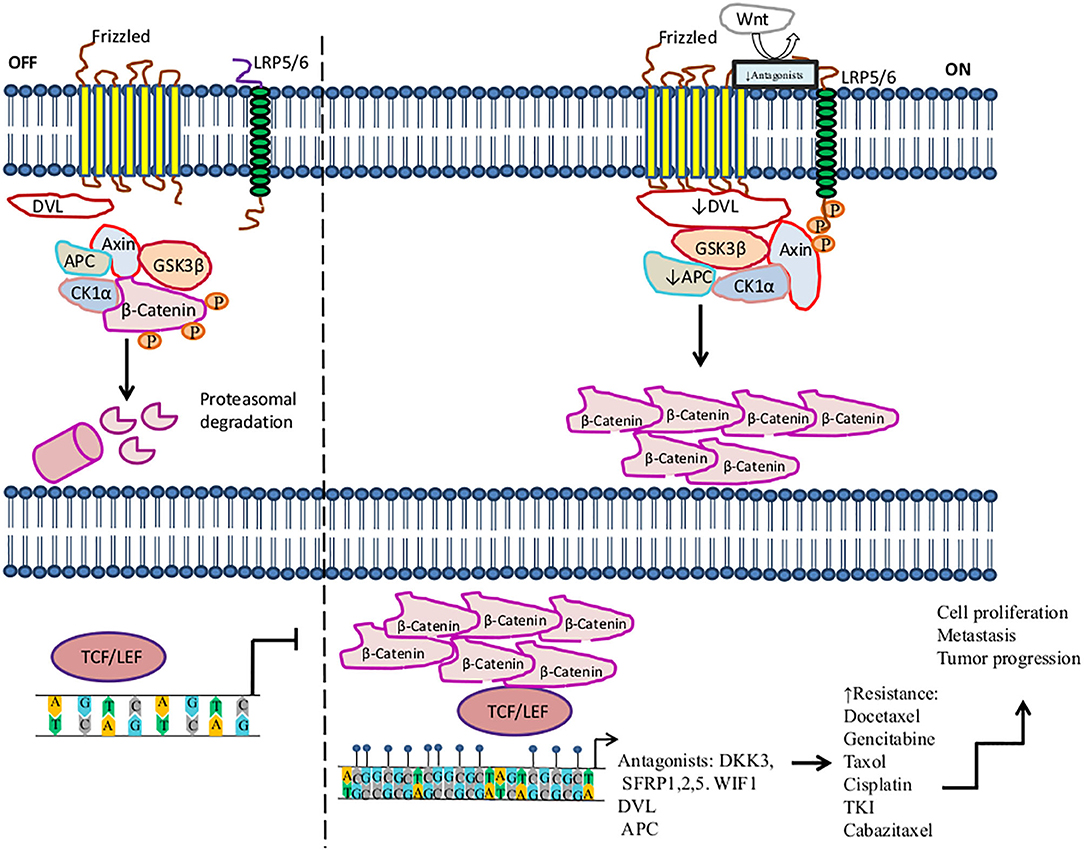
Figure 1. Activation of the Wnt/β-catenin signaling pathway in the resistance to therapy in cancer by methylation. The canonical Wnt/ß-catenin pathway is activated by the binding of Wnt to the Frizzled receptor (Fzd). Then LRP is phosphorylated by casein kinase 1 (CK1α) and glycogen synthase kinase 3 beta (GSK3β) associated with tumor suppressor adenomatous polyposis coli (APC). LRP phosphorylation promotes the recruitment of disheveled proteins (DVL) to the plasma membrane, where they are polymerized and activated. DVL complex interacts with Axin, which inhibits the degradation of β-catenin and leads to its accumulation in the cytoplasm and translocation into the nucleus, where β-catenin promotes the activation of LEF/TCF transcription factors inducing the transcription of several genes. In the absence of Wnt, β-catenin is a target of the destruction complex conformed by Axin, CK1α, APC, and GSK-3β. CK1α and GSK-3β phosphorylate β-catenin, promoting its ubiquitination by the β-TrCP ubiquitin ligase and degradation through the proteasome. On the other hand, this figure shows some aberrantly methylated key genes that increase resistance to therapeutic agents and the dysregulation of the Wnt/β-catenin signaling pathway. These genes play an antagonist role in the Wnt pathway, for instance, Dickkopf-related protein (DKK3), secreted frizzled-related proteins (SFRP1, SFRP2), and WNT inhibitory factor-1 (WIF1), which are tumor suppressor genes and inhibit the signaling of Wnt to bind LRP5/6 receptors.  , phosphorylation; ↑, increase; ↓, decrease,
, phosphorylation; ↑, increase; ↓, decrease, , methylation.
, methylation.
Methylation of genes involved in the Wnt pathway plays a crucial role in regulating the development and progression of tumors, as well as metastasis, diagnosis, and treatment. Several tumors, such as lung, breast, prostate, colon, gastric, and ovarian cancers, among others, exhibit a pattern of deregulated methylation in this pathway (184, 189, 190). For instance, Dickkopf-related protein (DKK3), secreted frizzled-related protein 1 (SFRP1), SFRP2, and Wnt inhibitory factor-1 (WIF1), which are tumor suppressor genes, prevent LRP5/6 receptors from interacting with their ligands, consequently inhibiting the signaling of the Wnt pathway (191, 192). In the context of methylation, it has been said that several tumors show the downregulation of DKK3, SFRP1, SFRP2, and WIF1 by hypermethylation in their promoters (193). The hypermethylation of DKK3 has been associated with docetaxel (DTX) resistance in the lung cancer H1299/DTX cell line. Moreover, treatment with 5-azacytidine on the H1299/DTX cell line upregulates DKK3 expression at both the mRNA and protein levels, which inhibits colony formation and induces apoptosis due to recovered sensitivity to DTX. Additionally, P-glycoprotein is a drug efflux pump associated with MDR, encoded by the MDR-1 gene (MDR-1). MDR-1 overexpression is associated with DTX resistance in lung cancer. Restored expression of DKK3 leads to the downregulation of MDR-1 and P-glycoprotein, thus increasing sensitivity to DTX. This is a mechanism of regulation in lung cancer therapy. Therefore, DKK3 may be a therapeutic target that may help tumor cells recover sensitivity to DTX (194). Another study found that the decreased expression of DKK3 is associated with hypermethylation in pancreatic cancer biopsies in comparison to non-tumor tissue. In this study, DKK3 expression was not detected in three pancreatic cancer cell lines (Aspc-1, Bxpc-3, and CFPAC-1). DKK3 overexpression by DKK3 transfection in the Bxpc-3 pancreatic cell line promotes the inhibition of β-catenin translocation to the nucleus, as well as its transcriptional role under conditions of hypoxia or normoxia. Furthermore, DKK3 repressed the EMT and migration of Bxpc-3 cells, mediated by the inhibition of β-catenin. These effects improved the response to gemcitabine in Bxpc-3 tumor cells, suggesting that DKK3 may be a potential target for therapy (195).
In advanced stages of lung cancer, treatment based on taxanes is one treatment option, such as paclitaxel and DTX; however, resistance to therapy is presented in some patients (196). Ren et al. showed that hypermethylated SFRP1 regulates the chemotherapy resistance of taxanes and DTX in A-549 and SPC-A1 lung adenocarcinomas cell lines. The resistance was mediated by Wnt pathway activation because SFRP1 reduces β-catenin stability, leading to cell death, whereas SFRP2 promotes β-catenin accumulation, inducing resistance to apoptosis. Moreover, 5-azacytidine treatment restored the SFRP1 expression level, inducing the inhibition of the Wnt pathway and promoting drug sensitivity in resistant cell lines. Thus, the overexpression of SFRP1 can improve patients' responses to taxanes and DTX therapies (196). Zhu et al. also found that SFRP1 and SFRP5 were hypermethylated in NSCLC. Furthermore, the hypermethylation of SFRP5 predicted a poor response to TKI therapy; hence, SFRP5 methylation could be associated with TKI resistance (197). Additionally, higher levels of the hypermethylation of SFRP1, SFRP2, and WIF1 genes were found in colon cancer compared to non-tumoral tissues. Thus, the hypermethylation of one or both SFRP1and SFRP2 genes is a promising prognostic marker for predicting survival in patients who receive post-operative chemotherapy (198–200). Su et al. found that SFRP5 was hypermethylated in 44.4% of ovarian cancer tissues, as well as in SKOV3 and A2780 tumor cell lines. The SFRP5 low expression promotes EMT, tumor growth, invasion, tumor progression, and cisplatin resistance. Restored expression of SFRP5 reduces Wnt non-canonical signaling, promoting sensitivity to cisplatin in a mouse model of ovarian cancer (201).
Several genes whose protein products participate in the Wnt transduction signaling cascade are regulated by hypermethylation. Hence, the deregulation of this process during carcinogenesis may contribute to drug resistance. For instance, hypermethylation of DVL in prostate cancer has been suggested to favor resistance to cabazitaxel in DU145 cells. Reactivation of DVL by 5-azacytidine treatment in DU145 10DRCR cells restores sensitivity to cabazitaxel in this prostate tumor cell line (202). Hypermethylation of adenoma polyposis coli (APC), a tumor suppressor gene, inhibits the Wnt pathway, promoting tumorigenesis and tumor progression in CRC (203, 204). In breast cancer, Matuschek et al. reported that hypermethylated APC promotes tumor aggressiveness in circulating tumor cells. Additionally, 70% of breast cancer tissues presented hypermethylation in the APC gene (205). By the same token, loss of APC inactivates the repair of double-stranded breaks mediated by ATM, Chk1, and Chk2, which induces doxorubicin resistance (205, 206). Thus, we suggest that the methylation status of several key genes involved in the Wnt/canonical signaling pathway may be used as predictive markers of tumor progression and therapy response.
Regulation of the PI3K/PTEN/AKT/mTOR Signaling Pathway by DNA Methylation Supports Cancer Development and Therapy Resistance
AKT is also recognized as protein kinase B (Serine/Threonine Kinase 1, or Protein Kinase B), which participates in several processes such as cell metabolism, cell proliferation, angiogenesis, apoptosis, motility, and cell survival (207). Aberrant DNAm of key genes in PI3K/PTEN/AKT/mTOR signaling pathway promotes therapy resistance in solid tumors (Figure 2).
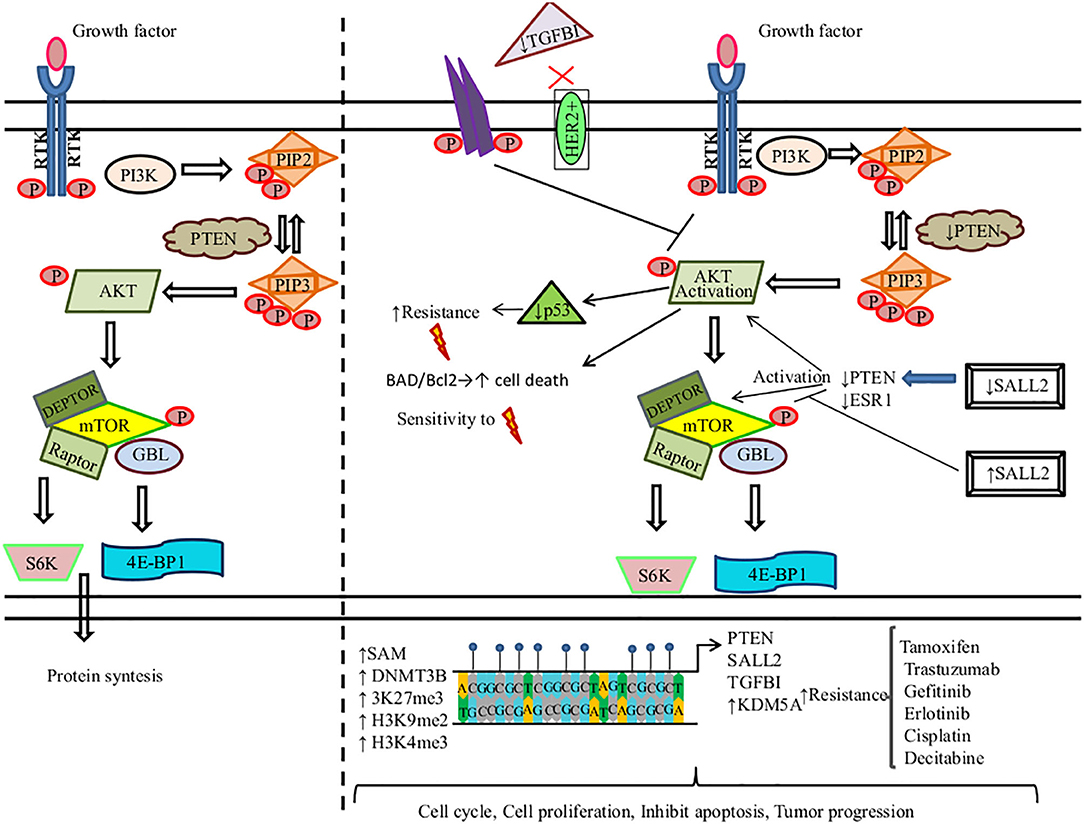
Figure 2. DNA methylation regulates the PI3K/PTEN/AKT/mTOR signaling pathway in the resistance to therapy in cancer. (Left) PI3K induces the phosphorylation and activation of AKT/mTOR. This transduction signal begins with the activation of the membrane tyrosine kinase receptors (RTKs) or G-protein-coupled receptors, which promotes the change of phosphatidylinositol (4,5)-bisphosphate (PIP2) in phosphatidylinositol (3-5)-trisphosphate (PIP3). The activation of PI3K (phosphoinositide-3-kinase) is regulated by the phosphatase and tensin homolog (PTEN) by dephosphorylating PIP3 into PIP2. (Right) We show the aberrant methylation of the PTEN, Spalt-like transcription factor 2 (SALL2), transforming growth factor beta-induced protein (TGFB1), and Lysine (K)-specific demethylase 5A (KDM5A) genes through the high expression of methyltransferase (DNMT3B), s-adenosylmethionine (SAM), H3K27me3, H3K9me2, and H3K4me3, promoting a continued activation of the PI3K/AKT/mTOR signaling pathway associated with resistance therapy in solid tumors.  , phosphorylation; ↑, increase; ↓, decrease;
, phosphorylation; ↑, increase; ↓, decrease;  , methylation,
, methylation,  , radiotherapy.
, radiotherapy.
Tamoxifen (TAM) is the first line of therapy for the treatment of estrogen-receptor-positive breast cancer. This type of breast cancer develops TAM resistance, promoting tumor relapse (208). Phuong et al. found that the MCF-7 breast cancer cell line showed TAM resistance (TAMR/MCF-7). This resistance is mediated by a high expression of DNMT1. DNMT1 along with SAM induce the hypermethylation of PTEN in amplicon A and amplicon B sites, leading to its downregulation and the constitutive activation and phosphorylation of PI3K/AKT. 5-Azacytidine treatment inhibits DNMT1 in TAMR/MCF-7 cells, restoring PTEN expression, suppressing cell proliferation, and promoting cell death by apoptosis (209). Spalt-like transcription factor 2 (SALL2) functions as a tumor suppressor, which regulates the AKT/mTOR pathway (Figure 2). Hypermethylated SALL2 is found in the TAM-resistant ER+ TAMR/MCF-7 breast cancer cell line, which leads to SALL2 downregulation. SALL2 decreased expression promotes decreased expression levels of estrogen receptor-alpha (ERα) and PTEN, which causes the continued activation of AKT/mTOR. In addition, hypomethylation of SALL2 increases its expression, leading to the upregulation of ER and PTEN and the further inhibition of the AKT/mTOR signaling pathway, which consequently leads to TAM sensitivity (210).
Among breast cancers, 15–20% are human epidermal growth factor receptor 2 positive (HER2+) and may develop resistance to trastuzumab. Palomeras et al. reported that primary breast tumors that developed trastuzumab resistance present a loss of expression of transforming growth factor beta-induced protein (TGFBI) by hypermethylation. TGFBI inhibits the HER2 receptor and AKT (124). Abnormal activation of PI3K/AKT is common in breast cancer. Among the most frequent causes is constitutive signaling through mutational activation of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), which is mutated in 45% of luminal breast cancers. Thus, a promising therapeutic strategy is to develop PI3K/AKT inhibitors. KDM5A lysine demethylase can remove tri- and dimethyl marks on histone H3 (H3K4me3), leading to tumor progression and drug tolerance. KDM5A is a target of AKT and, together, they regulate certain cell-cycle genes. AKT phosphorylates KDM5A, thus promoting the subcellular localization of KDM5A from the chromatin-bound regions and nucleus to the cytoplasm. As a result, KDM5A is rendered unable to demethylate H3K4me2/3. PI3K/AKT inhibition decreases KDM5A phosphorylation, promoting the low expression of cell-cycle promoting genes. Additionally, KDM5A regulates resistance to PI3K/AKT inhibitors (211).
On the other hand, some mutated EGFR lung cancers induce resistance to EGFR-TKIs (gefitinib and erlotinib). Two gefitinib-resistant cell lines (GEF1-1 and GEF2-1) derived from the PC-9 cell line were treated with 5-azacytidine. This treatment restored PTEN expression and promoted sensitivity to gefitinib and erlotinib in GEF1-1 and GEF2-1 cell lines. Nonetheless, the parental cell line (PC-9 cells) did not show this sensitivity due to the hypermethylation of PTEN and hyperactivation of AKT (212). Furthermore, the hypermethylation of insulin-like growth factor-binding protein-3 (IGFBP-3) promotes cisplatin resistance in lung cancer. Downregulation of IGFBP-3 induces PI3K/AKT activation by specific de-repression of insulin-like growth factor-I receptor (IGFIR) signaling (213).
Methylation of PTEN promoter is an alternative mechanism to PTEN downregulation that induces drug resistance. For instance, lung cancer cells develop radioresistance due to hypermethylated PTEN that induces low expression of pAKT and downregulates p53 expression (214). In similar research, Pappas et al. showed that restoring PTEN expression in the human lung cancer cell line H1299 by the use of the adenovirus expression vector (Ad-PTEN) increases sensitivity to ionizing therapy. Of note, PTEN promoter is methylated in H1299 cells. The phosphorylation of BAD, a proapoptotic molecule regulated by AKT, inhibits its binding to Bcl-2, leading to apoptosis. Thus, restoring PTEN induces lower levels of phosphorylated AKT and BAD, which sensitizes to apoptosis. Also, Ad-PTEN regulates the DNA repair of double-strand breaks, mediated by the activation of H2AX (215).
Differential DNA methylation profiles are found in prostate cancer samples. These tumors show hypermethylated PTEN and hemi- and homozygous PTEN loss. The latter has been associated with poor prognosis, recurrence, and tumor progression (216).
Qian et al. found that DNMTs were highly expressed in nasopharyngeal carcinoma resistant cells; consequently, PTEN and PPP2R2B promoter hypermethylation is induced. DNMT upregulation activates two important signaling pathways, PI3K/mTOR and PDK1/MYC, favoring survival, proliferation, and resistance to the BEZ235 inhibitor (217). Treatment with BEZ235 and the inhibition of DNMT expression with 5-azacytidine induces drug sensitivity in resistant tumor cells. Furthermore, 5-azacytidine dephosphorylates the AKT, GSK3β, MYC, P70, and 4EBP-1 proteins involved in the AKT/mTOR and PDK1/MYC pathways. The combination of decitabine and BEZ235 upregulates PTEN protein expression, inhibiting cell growth. Hence, new combinations of chemotherapeutic agents with inhibitors against components of the PI3K/AKT/mTOR signaling pathway should be tested to increase tumor chemotherapy sensitivity.
Conclusions and Future Directions
Several studies have shown that modifications in DNAm patterns may support cancer development, invasion, and metastasis. Global methylation analyses suggest that CpG islands tend to be a highly altered methylation status that depends on the cancer type (122, 123). Importantly, several studies have reported that DNAm patterns of drug-treated tumor cells can change and support the acquisition of resistance to treatments, such as radiotherapy and chemotherapy. Some of these DNAm changes have been proposed as promising biomarkers whose presence would be indicative that therapy must be replaced (Tables 3, 4).
Additionally, the study of the DNAm patterns and their involvement in the regulation of several signaling pathways in cancer has provided significant insight into the molecular mechanisms underlying the development of cancer. For instance, dysregulation of Wnt canonical and PI3K/AKT/mTOR signaling pathways, caused by an altered methylation status in a variety of genes, has been associated with resistance to current treatments (taxanes, DTX, cisplatin, TKI, etc.) in many types of cancer.
Some studies have tried to change drug-resistance-associated DNAm patterns using SAM and DNMTs to increase the methylation grade or TET-dependent demethylation to diminish DNAm. Although they have changed the DNAm pattern of the target gene, these treatments also modify the DNAm patterns of other genes, causing undesirable secondary effects. For this reason, a fine balance of DNAm is needed to ensure proper drug responsiveness.
Recent advances in applied genetic engineering and genome editing may provide new tools for targeting methylation status in cancer patients. In particular, the clustered regularly interspaced short palindromic repeats/associated protein 9 (CRISPR/Cas9) system allows for the addition or removal of DNA from the genome in a specific manner. Genetic engineering has produced a new version of Cas9 (dCas9), in which the activity of endonuclease has been removed but in which the DNA binding activity is maintained. dCas9 can also be linked to DNMT3A or ten-eleven translocation-1 (TET1) enzymes, generating the systems dCas9-DNMT3A or dCas9-TET1, respectively (218, 219). These systems can induce the methylation and demethylation of target genes in a specific-sequence manner, which makes them promising tools in the fight against cancer or the acquisition of therapy resistance.
Author Contributions
SR-G and AC-R designed the study. SR-G, HP-G, and AC-R wrote the manuscript and integrated the tables. AC-R designed and elaborated the figures. All authors contributed to the critical revision of the manuscript, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge to Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas for supporting the publication of this manuscript.
References
1. Ilango S, Paital B, Jayachandran P, Padma PR, Nirmaladevi R. Epigenetic alterations in cancer. Front Biosci. (2020) 25:4847. doi: 10.2741/4847
2. Fouad MA, Salem SE, Hussein MM, Zekri ARN, Hafez HF, El Desouky ED, et al. Impact of global DNA methylation in treatment outcome of colorectal cancer patients. Front Pharmacol. (2018) 9:1173. doi: 10.3389/fphar.2018.01173
3. Janitz KJM. Assessing epigenetic information. In: Handbook of Epigenetics, 1st ed. Amsterdam: Elsevier (2011). p. 173–81. doi: 10.1016/B978-0-12-375709-8.00012-5
4. Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol Cell Biol. (2007) 27:3891–9. doi: 10.1128/MCB.00036-07
5. Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. (2012) 15:405–11. doi: 10.1016/j.cmet.2012.01.001
6. Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. (2009) 41:178–86. doi: 10.1038/ng.298
7. Tsuboi K, Nagatomo T, Gohno T, Higuchi T, Sasaki S, Fujiki N, et al. Single CpG site methylation controls estrogen receptor gene transcription and correlates with hormone therapy resistance. J Steroid Biochem Mol Biol. (2017) 171:209–17. doi: 10.1016/j.jsbmb.2017.04.001
8. Ramachandran K, Gopisetty G, Gordian E, Navarro L, Hader C, Reis IM, et al. Methylation-mediated repression of GADD45alpha in prostate cancer and its role as a potential therapeutic target. Cancer Res. (2009) 69:1527–35. doi: 10.1158/0008-5472.CAN-08-3609
9. Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS ONE. (2013) 8:e67378. doi: 10.1371/journal.pone.0067378
10. Brown WM. Exercise-associated DNA methylation change in skeletal muscle and the importance of imprinted genes: a bioinformatics meta-analysis. Br J Sports Med. (2015) 49:1567–78. doi: 10.1136/bjsports-2014-094073
11. Aref-Eshghi E, Schenkel LC, Ainsworth P, Lin H, Rodenhiser DI, Cutz JC, et al. Genomic DNA methylation-derived algorithm enables accurate detection of malignant prostate tissues. Front Oncol. (2018) 8:100. doi: 10.3389/fonc.2018.00100
12. Markl ID, Cheng J, Liang G, Shibata D, Laird PW, Jones PA. Global and gene-specific epigenetic patterns in human bladder cancer genomes are relatively stable in vivo and in vitro over time. Cancer Res. (2001) 61:5875–84.
13. Shann YJ, Cheng C, Chiao CH, Chen DT, Li PH, Hsu MT. Genome-wide mapping and characterization of hypomethylated sites in human tissues and breast cancer cell lines. Genome Res. (2008) 18:791–801. doi: 10.1101/gr.070961.107
14. Marsit CJ, Houseman EA, Nelson HH, Kelsey KT. Genetic and epigenetic tumor suppressor gene silencing are distinct molecular phenotypes driven by growth promoting mutations in nonsmall cell lung cancer. J Cancer Epidemiol. (2008) 2008:215809. doi: 10.1155/2008/215809
15. Yadav P, Masroor M, Nandi K, Kaza RCM, Jain SK, Khurana N, et al. Promoter methylation of BRCA1, DAPK1 and RASSF1A is associated with increased mortality among indian women with breast cancer. Asian Pac J Cancer Prev. (2018) 19:443–8. doi: 10.22034/APJCP.2018.19.2.443
16. Guo L, Wang X, Yang Y, Xu H, Zhang Z, Yin L, et al. Methylation of DACT2 contributes to the progression of breast cancer through activating WNT signaling pathway. Oncol Lett. (2018) 15:3287–94. doi: 10.3892/ol.2017.7633
17. Begam N, Jamil K, Raju SG. Promoter hypermethylation of the ATM gene as a novel biomarker for breast cancer. Asian Pac J Cancer Prev. (2017) 18:3003–9. doi: 10.22034/APJCP.2017.18.11.3003
18. Espinal AC, Buas MF, Wang D, Cheng DT, Sucheston-Campbell L, Hu Q, et al. FOXA1 hypermethylation: link between parity and ER-negative breast cancer in African American women? Breast Cancer Res Treat. (2017) 166:559–68. doi: 10.1007/s10549-017-4418-y
19. Guerrero-Setas D, Perez-Janices N, Blanco-Fernandez L, Ojer A, Cambra K, Berdasco M, et al. RASSF2 hypermethylation is present and related to shorter survival in squamous cervical cancer. Mod Pathol. (2013) 26:1111–22. doi: 10.1038/modpathol.2013.32
20. Li JY, Huang T, Zhang C, Jiang DJ, Hong QX, Ji HH, et al. Association between RASSF1A promoter hypermethylation and oncogenic HPV infection status in invasive cervical cancer: a meta-analysis. Asian Pac J Cancer Prev. (2015) 16:5749–54. doi: 10.7314/APJCP.2015.16.14.5749
21. Dong Y, Tan Q, Tao L, Pan X, Pang L, Liang W, et al. Hypermethylation of TFPI2 correlates with cervical cancer incidence in the uygur and han populations of Xinjiang, China. Int J Clin Exp Pathol. (2015) 8:1844–54.
22. Kim HJ, Kim CY, Jin J, Bae MK, Kim YH, Ju W, et al. Aberrant single-minded homolog 1 methylation as a potential biomarker for cervical cancer. Diagn Cytopathol. (2018) 46:15–21. doi: 10.1002/dc.23838
23. Zhang J, Yao T, Lin Z, Gao Y. Aberrant methylation of MEG3 functions as a potential plasma-based biomarker for cervical cancer. Sci Rep. (2017) 7:6271. doi: 10.1038/s41598-017-06502-7
24. Han YD, Wang XB, Cui NH, Zhang S, Wang C, Zheng F. Associations of P16INK4a promoter hypermethylation with squamous intra-epithelial lesion, cervical cancer and their clinicopathological features: a meta-analysis. Oncotarget. (2017) 8:1871–83. doi: 10.18632/oncotarget.12202
25. Wei X, Zhang S, Cao D, Zhao M, Zhang Q, Zhao J, et al. Aberrant hypermethylation of SALL3 with HPV involvement contributes to the carcinogenesis of cervical cancer. PLoS ONE. (2015) 10:e0145700. doi: 10.1371/journal.pone.0145700
26. Ma D, Jiang C, Hu X, Liu H, Li Q, Li T, et al. Methylation patterns of the IFN-gamma gene in cervical cancer tissues. Sci Rep. (2014) 4:6331. doi: 10.1038/srep06331
27. Yang WT, Zheng PS. Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis. PLoS ONE. (2014) 9:e88827. doi: 10.1371/journal.pone.0088827
28. Paulikova S, Chmelarova M, Petera J, Palicka V, Paulik A. Hypermethylation of RAD51L3 and XRCC2 genes to predict late toxicity in chemoradiotherapy-treated cervical cancer patients. Folia Biol. (2013) 59:240–5.
29. Chen H, Xu Z, Yang B, Zhou X, Kong H. RASGRF1 hypermethylation, a putative biomarker of colorectal cancer. Ann Clin Lab Sci. (2018) 48:3–10.
30. Zhu Y, Lu H, Zhang D, Li M, Sun X, Wan L, et al. Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer. Clin Epigenetics. (2018) 10:30. doi: 10.1186/s13148-018-0458-3
31. Barati Bagerabad M, Tavakolian S, Abbaszadegan MR, Kerachian MA. Promoter hypermethylation of the eyes absent 4 gene is a tumor-specific epigenetic biomarker in iranian colorectal cancer patients. Acta Med Iran. (2018) 56:21–7. doi: 10.18502/acta.v57i5.1874
32. Yin MD, Ma SP, Liu F, Chen YZ. Role of serine/threonine kinase 33 methylation in colorectal cancer and its clinical significance. Oncol Lett. (2018) 15:2153–60. doi: 10.3892/ol.2017.7614
33. Lin RK, Hung WY, Huang YF, Chang YJ, Lin CH, Chen WY, et al. Hypermethylation of BEND5 contributes to cell proliferation and is a prognostic marker of colorectal cancer. Oncotarget. (2017) 8:113431–43. doi: 10.18632/oncotarget.22266
34. Islam F, Gopalan V, Pillai S, Lu CT, Kasem K, Lam AK. Promoter hypermethylation inactivate tumor suppressor FAM134B and is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. (2018) 57:240–51. doi: 10.1002/gcc.22525
35. Sun Z, Liu J, Jing H, Dong SX, Wu J. The diagnostic and prognostic value of CHFR hypermethylation in colorectal cancer, a meta-analysis and literature review. Oncotarget. (2017) 8:89142–8. doi: 10.18632/oncotarget.19408
36. Barrow TM, Klett H, Toth R, Bohm J, Gigic B, Habermann N, et al. Smoking is associated with hypermethylation of the APC 1A promoter in colorectal cancer: the colocare study. J Pathol. (2017) 243:366–75. doi: 10.1002/path.4955
37. Hu YH, Chen Q, Lu YX, Zhang JM, Lin C, Zhang F, et al. Hypermethylation of NDN promotes cell proliferation by activating the Wnt signaling pathway in colorectal cancer. Oncotarget. (2017) 8:46191–203. doi: 10.18632/oncotarget.17580
38. Zhang HF, Lu YW, Xie ZR, Wang KH. Relationship between human mutL homolog 1 (hMLH1) hypermethylation and colorectal cancer: a meta-analysis. Med Sci Monit. (2017) 23:3026–38. doi: 10.12659/MSM.895643
39. Ge Y, Wu Q, Ma G, Shao W, Liu H, Zhang Q, et al. Hypermethylation of EIF4E promoter is associated with early onset of gastric cancer. Carcinogenesis. (2018) 39:66–71. doi: 10.1093/carcin/bgx110
40. Chen Z, Hu T, Zhu S, Mukaisho K, El-Rifai W, Peng DF. Glutathione peroxidase 7 suppresses cancer cell growth and is hypermethylated in gastric cancer. Oncotarget. (2017) 8:54345–56. doi: 10.18632/oncotarget.17527
41. Tahara T, Tahara S, Horiguchi N, Kawamura T, Okubo M, Yamada H, et al. Methylation status of IGF2 DMR and LINE1 in leukocyte DNA provides distinct clinicopathological features of gastric cancer patients. Clin Exp Med. (2018) 18:215–20. doi: 10.1007/s10238-017-0471-4
42. Mohsenzadeh M, Sadeghi RN, Vahedi M, Kamani F, Hashemi M, Asadzadeh H, et al. Promoter hypermethylation of RAR-beta tumor suppressor gene in gastric carcinoma: association with histological type and clinical outcomes. Cancer Biomark. (2017) 20:7–15. doi: 10.3233/CBM-160331
43. Liu L, Liu C, Fotouhi O, Fan Y, Wang K, Xia C, et al. TERT promoter hypermethylation in gastrointestinal cancer: a potential stool biomarker. Oncologist. (2017) 22:1178–88. doi: 10.1634/theoncologist.2017-0064
44. Zhang Z, Xin S, Gao M, Cai Y. Promoter hypermethylation of MGMT gene may contribute to the pathogenesis of gastric cancer: a PRISMA-compliant meta-analysis. Medicine. (2017) 96:e6708. doi: 10.1097/MD.0000000000006708
45. Pei YF, Zhang YJ, Lei Y, Wu WD, Ma TH, Liu XQ. Hypermethylation of the CHRDL1 promoter induces proliferation and metastasis by activating Akt and Erk in gastric cancer. Oncotarget. (2017) 8:23155–66. doi: 10.18632/oncotarget.15513
46. Guo L, Huang C, Ji QJ. Aberrant promoter hypermethylation of p16, survivin, and retinoblastoma in gastric cancer. Bratisl Lek Listy. (2017) 118:164–8. doi: 10.4149/BLL_2017_033
47. Zhang JK, Li YS, Zhang CD, Dai DQ. Up-regulation of CRKL by microRNA-335 methylation is associated with poor prognosis in gastric cancer. Cancer Cell Int. (2017) 17:28. doi: 10.1186/s12935-017-0387-9
48. Wang H, Duan XL, Qi XL, Meng L, Xu YS, Wu T, et al. Concurrent hypermethylation of SFRP2 and DKK2 activates the Wnt/beta-catenin pathway and is associated with poor prognosis in patients with gastric cancer. Mol Cells. (2017) 40:45–53. doi: 10.14348/molcells.2017.2245
49. Chen X, Yang Y, Liu J, Li B, Xu Y, Li C, et al. NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population. Oncotarget. (2017) 8:8105–19. doi: 10.18632/oncotarget.14099
50. Wang N, Sui F, Ma J, Su X, Liu J, Yao D, et al. Site-specific hypermethylation of RUNX3 predicts poor prognosis in gastric cancer. Arch Med Res. (2016) 47:285–92. doi: 10.1016/j.arcmed.2016.07.011
51. Chen J, Zhang J, Li X, Zhang C, Zhang H, Jin J, et al. Downregulation of ADAMTS8 by DNA hypermethylation in gastric cancer and its clinical significance. Biomed Res Int. (2016) 2016:5083841. doi: 10.1155/2016/5083841
52. Wang H, Xu M, Cui X, Liu Y, Zhang Y, Sui Y, et al. Aberrant expression of the candidate tumor suppressor gene DAL-1 due to hypermethylation in gastric cancer. Sci Rep. (2016) 6:21755. doi: 10.1038/srep21755
53. Ng PKS, Lau CPY, Lam EKY, Li SSK, Lui VWY, Yeo W, et al. Hypermethylation of NF-kappaB-activating protein-like (NKAPL) promoter in hepatocellular carcinoma suppresses its expression and predicts a poor prognosis. Dig Dis Sci. (2018) 63:676–86. doi: 10.1007/s10620-018-4929-3
54. Guo Y, Peng Y, Gao D, Zhang M, Yang W, Linghu E, et al. Silencing HOXD10 by promoter region hypermethylation activates ERK signaling in hepatocellular carcinoma. Clin Epigenetics. (2017) 9:116. doi: 10.1186/s13148-017-0412-9
55. Zhang Y, Xu X, Chen Z, Zhao Z. Association of FHIT expression and FHIT gene hypermethylation with liver cancer risk: a PRISMA-compliant meta-analysis. Onco Targets Ther. (2017) 10:3083–93. doi: 10.2147/OTT.S138036
56. Mansour LA, El Raziky M, Mohamed AA, Mahmoud EH, Hamdy S, El Sayed EH. Circulating hypermethylated RASSF1A as a molecular biomarker for diagnosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. (2017) 18:1637–43. doi: 10.22034/APJCP.2017.18.6.1637
57. Tian MM, Fan YC, Zhao J, Gao S, Zhao ZH, Chen LY, et al. Hepatocellular carcinoma suppressor 1 promoter hypermethylation in serum. A diagnostic and prognostic study in hepatitis B. Clin Res Hepatol Gastroenterol. (2017) 41:171–80. doi: 10.1016/j.clinre.2016.10.003
58. Hoan NX, Van Tong H, Giang DP, Cuong BK, Toan NL, Wedemeyer H, et al. SOCS3 genetic variants and promoter hypermethylation in patients with chronic hepatitis B. Oncotarget. (2017) 8:17127–39. doi: 10.18632/oncotarget.15083
59. Yu Q, Xiang L, Yin L, Liu X, Yang D, Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-beta-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. (2017) 50:e12384. doi: 10.1111/cpr.12384
60. Hu H, Chen X, Zhou C, Li B, Yang Y, Ying X, et al. Aberrant methylation of mutL homolog 1 is associated with increased risk of non-small cell lung cancer. J Clin Lab Anal. (2018) 32:e22370. doi: 10.1002/jcla.22370
61. Gao C, Xing X, He Z, Chen S, Wang S, Li Q, et al. Hypermethylation of PGCP gene is associated with human bronchial epithelial cells immortalization. Gene. (2018) 642:505–12. doi: 10.1016/j.gene.2017.11.063
62. Chen R, Hong Q, Jiang J, Chen X, Jiang Z, Wang J, et al. AGTR1 promoter hypermethylation in lung squamous cell carcinoma but not in lung adenocarcinoma. Oncol Lett. (2017) 14:4989–94. doi: 10.3892/ol.2017.6824
63. El-Sherif WT, Sayed SK, Galal SH, Makhlouf HA, Hassan AT, Yousef HA. Diagnostic role of RASSF1A and p16INK4a promoter gene hypermethylation in serum DNA of lung cancer patients: clinicopathological significance. Egypt J Immunol. (2016) 23:1–16.
64. Zheng Y, Li X, Jiang Y, Xu Y, Song B, Zhou Q, et al. Promoter hypermethylation of Wnt inhibitory factor-1 in patients with lung cancer: a systematic meta-analysis. Medicine. (2016) 95:e5433. doi: 10.1097/MD.0000000000005433
65. Rezk NA, Mohamed RH, Alnemr AA, Harira M. Promoter methylation of RASSF1A gene in egyptian patients with ovarian cancer. Appl Biochem Biotechnol. (2018) 185:153–62. doi: 10.1007/s12010-017-2648-4
66. Kim JY, Do SI, Bae GE, Kim HS. B-cell translocation gene 1 is downregulated by promoter methylation in ovarian carcinoma. J Cancer. (2017) 8:2669–75. doi: 10.7150/jca.21037
67. Shen C, Sheng Q, Zhang X, Fu Y, Zhu K. Hypermethylated APC in serous carcinoma based on a meta-analysis of ovarian cancer. J Ovarian Res. (2016) 9:60. doi: 10.1186/s13048-016-0271-6
68. Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer. (2016) 16:102. doi: 10.1186/s12885-016-2135-2
69. Ding JJ, Wang G, Shi WX, Zhou HH, Zhao EF. Promoter hypermethylation of FANCF and susceptibility and prognosis of epithelial ovarian cancer. Reprod Sci. (2016) 23:24–30. doi: 10.1177/1933719115612136
70. Hafner N, Steinbach D, Jansen L, Diebolder H, Durst M, Runnebaum IB. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int J Cancer. (2016) 138:217–28. doi: 10.1002/ijc.29690
71. Chou JL, Huang RL, Shay J, Chen LY, Lin SJ, Yan PS, et al. Hypermethylation of the TGF-beta target, ABCA1 is associated with poor prognosis in ovarian cancer patients. Clin Epigenetics. (2015) 7:1. doi: 10.1186/s13148-014-0036-2
72. Sheng X, Li J, Yang L, Chen Z, Zhao Q, Tan L, et al. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. (2014) 32:277–85. doi: 10.3892/or.2014.3208
73. Faleiro I, Apolonio JD, Price AJ, De Mello RA, Roberto VP, Tabori U, et al. The TERT hypermethylated oncologic region predicts recurrence and survival in pancreatic cancer. Future Oncol. (2017) 13:2045–51. doi: 10.2217/fon-2017-0167
74. Wang L, Wang M, Hu C, Li P, Qiao Y, Xia Y, et al. Protein salvador homolog 1 acts as a tumor suppressor and is modulated by hypermethylation in pancreatic ductal adenocarcinoma. Oncotarget. (2017) 8:62953–61. doi: 10.18632/oncotarget.17972
75. Ushiku H, Yamashita K, Kawamata H, Waraya M, Katoh H, Yokoi K, et al. Homeobox-only protein expression is a critical prognostic indicator of pancreatic neuroendocrine tumor and is regulated by promoter DNA hypermethylation. Pancreas. (2016) 45:1255–62. doi: 10.1097/MPA.0000000000000646
76. Tang B, Li Y, Qi G, Yuan S, Wang Z, Yu S, et al. Clinicopathological significance of CDKN2A promoter hypermethylation frequency with pancreatic cancer. Sci Rep. (2015) 5:13563. doi: 10.1038/srep13563
77. Haldrup C, Pedersen AL, Ogaard N, Strand SH, Hoyer S, Borre M, et al. Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol Oncol. (2018) 12:545–60. doi: 10.1002/1878-0261.12183
78. Pugongchai A, Bychkov A, Sampatanukul P. Promoter hypermethylation of SOX11 correlates with adverse clinicopathological features of human prostate cancer. Int J Exp Pathol. (2017) 98:341–6. doi: 10.1111/iep.12257
79. Kuffer S, Gutting T, Belharazem D, Sauer C, Michel MS, Marx A, et al. Insulin-like growth factor 2 expression in prostate cancer is regulated by promoter-specific methylation. Mol Oncol. (2018) 12:256–66. doi: 10.1002/1878-0261.12164
80. Liu T, Qiu X, Zhao X, Yang R, Lian H, Qu F, et al. Hypermethylation of the SPARC promoter and its prognostic value for prostate cancer. Oncol Rep. (2018) 39:659–66. doi: 10.3892/or.2017.6121
81. Lounglaithong K, Bychkov A, Sampatanukul P. Aberrant promoter methylation of the PAQR3 gene is associated with prostate cancer. Pathol Res Pract. (2018) 214:126–9. doi: 10.1016/j.prp.2017.10.010
82. Zhang P, Wang H, Wang J, Liu Q, Wang Y, Feng F, et al. Association between protocadherin 8 promoter hypermethylation and the pathological status of prostate cancer. Oncol Lett. (2017) 14:1657–64. doi: 10.3892/ol.2017.6282
83. Strand SH, Switnicki M, Moller M, Haldrup C, Storebjerg TM, Hedegaard J, et al. RHCG and TCAF1 promoter hypermethylation predicts biochemical recurrence in prostate cancer patients treated by radical prostatectomy. Oncotarget. (2017) 8:5774–88. doi: 10.18632/oncotarget.14391
84. Castelo-Branco P, Leao R, Lipman T, Campbell B, Lee D, Price A, et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: a retrospective cohort study. Oncotarget. (2016) 7:57726–36. doi: 10.18632/oncotarget.10639
85. Zelic R, Fiano V, Zugna D, Grasso C, Delsedime L, Daniele L, et al. Global hypomethylation (LINE-1) and gene-specific hypermethylation (GSTP1) on initial negative prostate biopsy as markers of prostate cancer on a rebiopsy. Clin Cancer Res. (2016) 22:984–92. doi: 10.1158/1078-0432.CCR-15-0606
86. Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang H, et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. (2017) 8:20751–65. doi: 10.18632/oncotarget.10612
87. Sizemore ST, Sizemore GM, Booth CN, Thompson CL, Silverman P, Bebek G, et al. Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast Cancer Res Treat. (2014) 146:25–40. doi: 10.1007/s10549-014-2989-4
88. Son KS, Kang HS, Kim SJ, Jung SY, Min SY, Lee SY, et al. Hypomethylation of the interleukin-10 gene in breast cancer tissues. Breast. (2010) 19:484–8. doi: 10.1016/j.breast.2010.05.011
89. Yin FF, Wang N, Bi XN, Yu X, Xu XH, Wang YL, et al. Serine/threonine kinases 31(STK31) may be a novel cellular target gene for the HPV16 oncogene E7 with potential as a DNA hypomethylation biomarker in cervical cancer. Virol J. (2016) 13:60. doi: 10.1186/s12985-016-0515-5
90. Wu Y, Gong L, Xu J, Mou Y, Xu X, Qian Z. The clinicopathological significance of HES1 promoter hypomethylation in patients with colorectal cancer. Onco Targets Ther. (2017) 10:5827–34. doi: 10.2147/OTT.S151857
91. Kano H, Takayama T, Midorikawa Y, Nagase H. Promoter hypomethylation of RAR-related orphan receptor alpha 1 is correlated with unfavorable clinicopathological features in patients with colorectal cancer. Biosci Trends. (2016) 10:202–9. doi: 10.5582/bst.2016.01097
92. Renaud F, Mariette C, Vincent A, Wacrenier A, Maunoury V, Leclerc J, et al. The serrated neoplasia pathway of colorectal tumors: identification of MUC5AC hypomethylation as an early marker of polyps with malignant potential. Int J Cancer. (2016) 138:1472–81. doi: 10.1002/ijc.29891
93. Renaud F, Vincent A, Mariette C, Crepin M, Stechly L, Truant S, et al. MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int J Cancer. (2015) 136:2811–21. doi: 10.1002/ijc.29342
94. Li C, Cai S, Wang X, Jiang Z. Hypomethylation-associated up-regulation of TCF3 expression and recurrence in stage II and III colorectal cancer. PLoS ONE. (2014) 9:e112005. doi: 10.1371/journal.pone.0112005
95. de Melo CF, Gigek CO, da Silva JN, Cardoso Smith Mde A, de Araujo RM, Burbano RR, et al. Association of COX2 gene hypomethylation with intestinal type gastric cancer in samples of patients from northern Brazil. Tumour Biol. (2014) 35:1107–11. doi: 10.1007/s13277-013-1148-6
96. Yuasa Y, Nagasaki H, Oze I, Akiyama Y, Yoshida S, Shitara K, et al. Insulin-like growth factor 2 hypomethylation of blood leukocyte DNA is associated with gastric cancer risk. Int J Cancer. (2012) 131:2596–603. doi: 10.1002/ijc.27554
97. He J, Huang Y, Liu Z, Zhao R, Liu Q, Wei L, et al. Hypomethylation of BORIS is a promising prognostic biomarker in hepatocellular carcinoma. Gene. (2017) 629:29–34. doi: 10.1016/j.gene.2017.07.077
98. Zhong D, Cen H. Aberrant promoter methylation profiles and association with survival in patients with hepatocellular carcinoma. Onco Targets Ther. (2017) 10:2501–9. doi: 10.2147/OTT.S128058
99. Zhu C, Utsunomiya T, Ikemoto T, Yamada S, Morine Y, Imura S, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol. (2014) 21(Suppl 4):S729–35. doi: 10.1245/s10434-014-3874-4
100. He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, et al. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. (2011) 13:841–53. doi: 10.1593/neo.11698
101. Yuan R, Zhi Q, Zhao H, Han Y, Gao L, Wang B, et al. Upregulated expression of miR-106a by DNA hypomethylation plays an oncogenic role in hepatocellular carcinoma. Tumour Biol. (2015) 36:3093–100. doi: 10.1007/s13277-014-2945-2
102. Brennan K, Shin JH, Tay JK, Prunello M, Gentles AJ, Sunwoo JB, et al. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep. (2017) 7:17064. doi: 10.1038/s41598-017-17298-x
103. Chueh AC, Liew MS, Russell PA, Walkiewicz M, Jayachandran A, Starmans MHW, et al. Promoter hypomethylation of NY-ESO-1, association with clinicopathological features and PD-L1 expression in non-small cell lung cancer. Oncotarget. (2017) 8:74036–48. doi: 10.18632/oncotarget.18198
104. Yokoyama S, Higashi M, Tsutsumida H, Wakimoto J, Hamada T, Wiest E, et al. TET1-mediated DNA hypomethylation regulates the expression of MUC4 in lung cancer. Genes Cancer. (2017) 8:517–27. doi: 10.18632/genesandcancer.139
105. Lee DH, Hwang SH, Lim MK, Oh JK, Song DY, Yun EH, et al. Performance of urine cotinine and hypomethylation of AHRR and F2RL3 as biomarkers for smoking exposure in a population-based cohort. PLoS ONE. (2017) 12:e0176783. doi: 10.1371/journal.pone.0176783
106. Fujii S, Shinjo K, Matsumoto S, Harada T, Nojima S, Sato S, et al. Epigenetic upregulation of ARL4C, due to DNA hypomethylation in the 3'-untranslated region, promotes tumorigenesis of lung squamous cell carcinoma. Oncotarget. (2016) 7:81571–87. doi: 10.18632/oncotarget.13147
107. Villalba M, Diaz-Lagares A, Redrado M, de Aberasturi AL, Segura V, Bodegas ME, et al. Epigenetic alterations leading to TMPRSS4 promoter hypomethylation and protein overexpression predict poor prognosis in squamous lung cancer patients. Oncotarget. (2016) 7:22752–69. doi: 10.18632/oncotarget.8045
108. Gao T, Zheng S, Li Q, Ran P, Sun L, Yuan Y, et al. Aberrant hypomethylation and overexpression of the eyes absent homologue 2 suppresses tumor cell growth of human lung adenocarcinoma cells. Oncol Rep. (2015) 34:2333–42. doi: 10.3892/or.2015.4245
109. Sung HY, Yang SD, Park AK, Ju W, Ahn JH. Aberrant hypomethylation of solute carrier family 6 member 12 promoter induces metastasis of ovarian cancer. Yonsei Med J. (2017) 58:27–34. doi: 10.3349/ymj.2017.58.1.27
110. Zhang W, Barger CJ, Link PA, Mhawech-Fauceglia P, Miller A, Akers SN, et al. DNA hypomethylation-mediated activation of cancer/testis antigen 45 (CT45) genes is associated with disease progression and reduced survival in epithelial ovarian cancer. Epigenetics. (2015) 10:736–48. doi: 10.1080/15592294.2015.1062206
111. Sung HY, Ju W, Ahn JH. DNA hypomethylation-mediated overexpression of carbonic anhydrase 9 induces an aggressive phenotype in ovarian cancer cells. Yonsei Med J. (2014) 55:1656–63. doi: 10.3349/ymj.2014.55.6.1656
112. Sung HY, Choi EN, Lyu D, Park AK, Ju W, Ahn JH. Aberrant hypomethylation-mediated AGR2 overexpression induces an aggressive phenotype in ovarian cancer cells. Oncol Rep. (2014) 32:815–20. doi: 10.3892/or.2014.3243
113. Liao YP, Chen LY, Huang RL, Su PH, Chan MW, Chang CC, et al. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum Mol Genet. (2014) 23:1894–906. doi: 10.1093/hmg/ddt583
114. Mardin WA, Ntalos D, Mees ST, Spieker T, Senninger N, Haier J, et al. SERPINB5 promoter hypomethylation differentiates pancreatic ductal adenocarcinoma from pancreatitis. Pancreas. (2016) 45:743–7. doi: 10.1097/MPA.0000000000000526
115. Zhu Y, Zhang JJ, Zhu R, Zhu Y, Liang WB, Gao WT, et al. The increase in the expression and hypomethylation of MUC4 gene with the progression of pancreatic ductal adenocarcinoma. Med Oncol. (2011) 28(Suppl. 1):S175–84. doi: 10.1007/s12032-010-9683-0
116. Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, et al. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. (2002) 160:45–50. doi: 10.1016/S0002-9440(10)64347-7
117. Nones K, Waddell N, Song S, Patch AM, Miller D, Johns A, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. (2014) 135:1110–8. doi: 10.1002/ijc.28765
118. Norgaard M, Haldrup C, Storebjerg TM, Vestergaard EM, Wild PJ, Hoyer S, et al. Comprehensive evaluation of TFF3 promoter hypomethylation and molecular biomarker potential for prostate cancer diagnosis and prognosis. Int J Mol Sci. (2017) 18:2017. doi: 10.3390/ijms18092017
119. Liang YX, Mo RJ, He HC, Chen JH, Zou J, Han ZD, et al. Aberrant hypomethylation-mediated CD147 overexpression promotes aggressive tumor progression in human prostate cancer. Oncol Rep. (2015) 33:2648–54. doi: 10.3892/or.2015.3870
120. Vestergaard EM, Nexo E, Torring N, Borre M, Orntoft TF, Sorensen KD. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int J Cancer. (2010) 127:1857–65. doi: 10.1002/ijc.25209
121. Gao B, Yang F, Chen W, Li R, Hu X, Liang Y, et al. Multidrug resistance affects the prognosis of primary epithelial ovarian cancer. Oncol Lett. (2019) 18:4262–9. doi: 10.3892/ol.2019.10745
122. Baharudin R, Ab Mutalib NS, Othman SN, Sagap I, Rose IM, Mohd Mokhtar N, et al. Identification of predictive DNA methylation biomarkers for chemotherapy response in colorectal cancer. Front Pharmacol. (2017) 8:47. doi: 10.3389/fphar.2017.00047
123. Guo R, Wu G, Li H, Qian P, Han J, Pan F, et al. Promoter methylation profiles between human lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP) cells and its progenitor A549 cells. Biol Pharm Bull. (2013) 36:1310–6. doi: 10.1248/bpb.b13-00153
124. Palomeras S, Diaz-Lagares A, Vinas G, Setien F, Ferreira HJ, Oliveras G, et al. Epigenetic silencing of TGFBI confers resistance to trastuzumab in human breast cancer. Breast Cancer Res. (2019) 21:79. doi: 10.1186/s13058-019-1160-x
125. Zhang J, Zhou C, Jiang H, Liang L, Shi W, Zhang Q, et al. ZEB1 induces ER-alpha promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. (2017) 8:e2732. doi: 10.1038/cddis.2017.154
126. Ponnusamy L, Mahalingaiah PKS, Chang YW, Singh KP. Reversal of epigenetic aberrations associated with the acquisition of doxorubicin resistance restores drug sensitivity in breast cancer cells. Eur J Pharm Sci. (2018) 123:56–69. doi: 10.1016/j.ejps.2018.07.028
127. Tuo YL, Ye YF. MGP is downregulated due to promoter methylation in chemoresistant ER+ breast cancer and high MGP expression predicts better survival outcomes. Eur Rev Med Pharmacol Sci. (2017) 21:3871–8.
128. De Marchi T, Timmermans MA, Sieuwerts AM, Smid M, Look MP, Grebenchtchikov N, et al. Phosphoserine aminotransferase 1 is associated to poor outcome on tamoxifen therapy in recurrent breast cancer. Sci Rep. (2017) 7:2099. doi: 10.1038/s41598-017-02296-w
129. Kim MH, Kim MS, Kim W, Kang MA, Cacalano NA, Kang SB, et al. Suppressor of cytokine signaling (SOCS) genes are silenced by DNA hypermethylation and histone deacetylation and regulate response to radiotherapy in cervical cancer cells. PLoS ONE. (2015) 10:e0123133. doi: 10.1371/journal.pone.0123133
130. Wu NY, Zhang X, Chu T, Zhu S, Deng Y, Zhou Y, et al. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med. (2019) 7:328. doi: 10.21037/atm.2019.06.15
131. Chang SY, Kuo CC, Wu CC, Hsiao CW, Hu JM, Hsu CH, et al. NKX6.1 hypermethylation predicts the outcome of stage II colorectal cancer patients undergoing chemotherapy. Genes Chromosomes Cancer. (2018) 57:268–77. doi: 10.1002/gcc.22529
132. Bosch LJW, Trooskens G, Snaebjornsson P, Coupe VMH, Mongera S, Haan JC, et al. Decoy receptor 1 (DCR1) promoter hypermethylation and response to irinotecan in metastatic colorectal cancer. Oncotarget. (2017) 8:63140–54. doi: 10.18632/oncotarget.18702
133. Wang X, Ghareeb WM, Zhang Y, Yu Q, Lu X, Huang Y, et al. Hypermethylated and downregulated MEIS2 are involved in stemness properties and oxaliplatin-based chemotherapy resistance of colorectal cancer. J Cell Physiol. (2019) 234:18180–91. doi: 10.1002/jcp.28451
134. Wang B, Lu FY, Shi RH, Feng YD, Zhao XD, Lu ZP, et al. MiR-26b regulates 5-FU-resistance in human colorectal cancer via down-regulation of Pgp. Am J Cancer Res. (2018) 8:2518–27.
135. Jingyue S, Xiao W, Juanmin Z, Wei L, Daoming L, Hong X. TFAP2E methylation promotes 5fluorouracil resistance via exosomal miR106a5p and miR421 in gastric cancer MGC803 cells. Mol Med Rep. (2019) 20:323–31. doi: 10.3892/mmr.2019.10237
136. Sun J, Du N, Li J, Zhou J, Tao G, Sun S, et al. Transcription factor AP2epsilon: a potential predictor of chemoresistance in patients with gastric cancer. Technol Cancer Res Treat. (2016) 15:285–95. doi: 10.1177/1533034615577028
137. Fujiyoshi S, Honda S, Minato M, Ara M, Suzuki H, Hiyama E, et al. Hypermethylation of CSF3R is a novel cisplatin resistance marker and predictor of response to postoperative chemotherapy in hepatoblastoma. Hepatol Res. (2020) 50:598–606. doi: 10.1111/hepr.13479
138. Fan H, Zhang M, Liu W. Hypermethylated KCNQ1 acts as a tumor suppressor in hepatocellular carcinoma. Biochem Biophys Res Commun. (2018) 503:3100–7. doi: 10.1016/j.bbrc.2018.08.099
139. Tian FM, Zhong CY, Wang XN, Meng Y. PDE3A is hypermethylated in cisplatin resistant non-small cell lung cancer cells and is a modulator of chemotherapy response. Eur Rev Med Pharmacol Sci. (2017) 21:2635–41.
140. Grasse S, Lienhard M, Frese S, Kerick M, Steinbach A, Grimm C, et al. Epigenomic profiling of non-small cell lung cancer xenografts uncover LRP12 DNA methylation as predictive biomarker for carboplatin resistance. Genome Med. (2018) 10:55. doi: 10.1186/s13073-018-0562-1
141. Yue J, Lv D, Wang C, Li L, Zhao Q, Chen H, et al. Epigenetic silencing of miR-483-3p promotes acquired gefitinib resistance and EMT in EGFR-mutant NSCLC by targeting integrin beta3. Oncogene. (2018) 37:4300–12. doi: 10.1038/s41388-018-0276-2
142. Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N, et al. UCHL1 is a putative tumor suppressor in ovarian cancer cells and contributes to cisplatin resistance. J Cancer. (2013) 4:662–70. doi: 10.7150/jca.6641
143. Yang SD, Ahn SH, Kim JI. 3-Oxoacid CoA transferase 1 as a therapeutic target gene for cisplatin-resistant ovarian cancer. Oncol Lett. (2018) 15:2611–8. doi: 10.3892/ol.2017.7560
144. Prieske K, Prieske S, Joosse SA, Trillsch F, Grimm D, Burandt E, et al. Loss of BRCA1 promotor hypermethylation in recurrent high-grade ovarian cancer. Oncotarget. (2017) 8:83063–74. doi: 10.18632/oncotarget.20945
145. Deng Y, Zhao F, Hui L, Li X, Zhang D, Lin W, et al. Suppressing miR-199a-3p by promoter methylation contributes to tumor aggressiveness and cisplatin resistance of ovarian cancer through promoting DDR1 expression. J Ovarian Res. (2017) 10:50. doi: 10.1186/s13048-017-0333-4
146. Tian H, Yan L, Xiao-Fei L, Hai-Yan S, Juan C, Shan K. Hypermethylation of mismatch repair gene hMSH2 associates with platinum-resistant disease in epithelial ovarian cancer. Clin Epigenetics. (2019) 11:153. doi: 10.1186/s13148-019-0748-4
147. Ha YN, Sung HY, Yang SD, Chae YJ, Ju W, Ahn JH. Epigenetic modification of alpha-N-acetylgalactosaminidase enhances cisplatin resistance in ovarian cancer. Korean J Physiol Pharmacol. (2018) 22:43–51. doi: 10.4196/kjpp.2018.22.1.43
148. Kritsch D, Hoffmann F, Steinbach D, Jansen L, Mary Photini S, Gajda M, et al. Tribbles 2 mediates cisplatin sensitivity and DNA damage response in epithelial ovarian cancer. Int J Cancer. (2017) 141:1600–14. doi: 10.1002/ijc.30860
149. Tian J, Xu YY, Li L, Hao Q. MiR-490-3p sensitizes ovarian cancer cells to cisplatin by directly targeting ABCC2. Am J Transl Res. (2017) 9:1127–38.
150. Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, Harada T, et al. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res. (2007) 67:6786–95. doi: 10.1158/0008-5472.CAN-07-0440
151. Abukiwan A, Nwaeburu CC, Bauer N, Zhao Z, Liu L, Gladkich J, et al. Dexamethasone-induced inhibition of miR-132 via methylation promotes TGF-beta-driven progression of pancreatic cancer. Int J Oncol. (2019) 54:53–64. doi: 10.3892/ijo.2018.4616
152. Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu L, et al. Methylation-induced silencing of miR-34a enhances chemoresistance by directly upregulating ATG4B-induced autophagy through AMPK/mTOR pathway in prostate cancer. Oncol Rep. (2016) 35:64–72. doi: 10.3892/or.2015.4331
153. Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. (2010) 1:e105. doi: 10.1038/cddis.2010.85
154. Guo Q, Jing FJ, Xu W, Li X, Li X, Sun JL, et al. Ubenimex induces autophagy inhibition and EMT suppression to overcome cisplatin resistance in GC cells by perturbing the CD13/EMP3/PI3K/AKT/NF-kappaB axis. Aging. (2019) 12:80–105. doi: 10.18632/aging.102598
155. El Kadi N, Wang L, Davis A, Korkaya H, Cooke A, Vadnala V, et al. The EGFR T790M mutation is acquired through AICDA-mediated deamination of 5-methylcytosine following TKI treatment in lung cancer. Cancer Res. (2018) 78:6728–35. doi: 10.1158/0008-5472.CAN-17-3370
156. Yu J, Qin B, Moyer AM, Nowsheen S, Liu T, Qin S, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. (2018) 128:2376–88. doi: 10.1172/JCI97924
157. Zhang N, Wang AY, Wang XK, Sun XM, Xue HZ. GAS5 is downregulated in gastric cancer cells by promoter hypermethylation and regulates adriamycin sensitivity. Eur Rev Med Pharmacol Sci. (2016) 20:3199–205.
158. Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y, et al. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget. (2014) 5:11552–63. doi: 10.18632/oncotarget.2594
159. Masuda K, Banno K, Yanokura M, Tsuji K, Kobayashi Y, Kisu I, et al. Association of epigenetic inactivation of the WRN gene with anticancer drug sensitivity in cervical cancer cells. Oncol Rep. (2012) 28:1146–52. doi: 10.3892/or.2012.1912
160. Ren K, Xu R, Huang J, Zhao J, Shi W. Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol. (2017) 80:243–50. doi: 10.1007/s00280-017-3356-z
161. Zhang Y, Zhang B, Fang J, Cao X. Hypomethylation of DNA-binding inhibitor 4 serves as a potential biomarker in distinguishing acquired tamoxifen-refractory breast cancer. Int J Clin Exp Pathol. (2015) 8:9500–5.
162. Chen S, Zhang Y, Zhang D. Endoplasmic reticulum protein 29 (ERp29) confers radioresistance through the DNA repair gene, O(6)-methylguanine DNA-methyltransferase, in breast cancer cells. Sci Rep. (2015) 5:14723. doi: 10.1038/srep14723
163. He DX, Gu XT, Jiang L, Jin J, Ma X. A methylation-based regulatory network for microRNA 320a in chemoresistant breast cancer. Mol Pharmacol. (2014) 86:536–47. doi: 10.1124/mol.114.092759
164. Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2). J Biol Chem. (2013) 288:10973–85. doi: 10.1074/jbc.M112.434340
165. Chekhun VF, Kulik GI, Yurchenko OV, Tryndyak VP, Todor IN, Luniv LS, et al. Role of DNA hypomethylation in the development of the resistance to doxorubicin in human MCF-7 breast adenocarcinoma cells. Cancer Lett. (2006) 231:87–93. doi: 10.1016/j.canlet.2005.01.038
166. Wen S, Wang X, Wang Y, Shen J, Pu J, Liang H, et al. Nucleoside diphosphate kinase 2 confers acquired 5-fluorouracil resistance in colorectal cancer cells. Artif Cells Nanomed Biotechnol. (2018) 46:896–905. doi: 10.1080/21691401.2018.1439835
167. Yokoi K, Harada H, Yokota K, Ishii S, Tanaka T, Nishizawa N, et al. Epigenetic status of CDO1 gene may reflect chemosensitivity in colon cancer with postoperative adjuvant chemotherapy. Ann Surg Oncol. (2019) 26:406–14. doi: 10.1245/s10434-018-6865-z
168. Zhao XQ, Zhang YF, Xia YF, Zhou ZM, Cao YQ. Promoter demethylation of nuclear factor-erythroid 2-related factor 2 gene in drug-resistant colon cancer cells. Oncol Lett. (2015) 10:1287–92. doi: 10.3892/ol.2015.3468
169. Kwon OH, Park JL, Baek SJ, Noh SM, Song KS, Kim SY, et al. Aberrant upregulation of ASCL2 by promoter demethylation promotes the growth and resistance to 5-fluorouracil of gastric cancer cells. Cancer Sci. (2013) 104:391–7. doi: 10.1111/cas.12076
170. Xia LL, Tang YB, Song FF, Xu L, Ji P, Wang SJ, et al. DCTPP1 attenuates the sensitivity of human gastric cancer cells to 5-fluorouracil by up-regulating MDR1 expression epigenetically. Oncotarget. (2016) 7:68623–37. doi: 10.18632/oncotarget.11864
171. Subhash VV, Tan SH, Tan WL, Yeo MS, Xie C, Wong FY, et al. GTSE1 expression represses apoptotic signaling and confers cisplatin resistance in gastric cancer cells. BMC Cancer. (2015) 15:550. doi: 10.1186/s12885-015-1550-0
172. Liu J, Liu Y, Meng L, Liu K, Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. (2017) 38:899–907. doi: 10.3892/or.2017.5722
173. Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. (2007) 26:4877–81. doi: 10.1038/sj.onc.1210266
174. Guijo M, Ceballos-Chavez M, Gomez-Marin E, Basurto-Cayuela L, Reyes JC. Expression of TDRD9 in a subset of lung carcinomas by CpG island hypomethylation protects from DNA damage. Oncotarget. (2018) 9:9618–31. doi: 10.18632/oncotarget.22709
175. Pan JX, Qu F, Wang FF, Xu J, Mu LS, Ye LY, et al. Aberrant SERPINE1 DNA methylation is involved in carboplatin induced epithelial-mesenchymal transition in epithelial ovarian cancer. Arch Gynecol Obstet. (2017) 296:1145–52. doi: 10.1007/s00404-017-4547-x
176. de Leon M, Cardenas H, Vieth E, Emerson R, Segar M, Liu Y, et al. Transmembrane protein 88 (TMEM88) promoter hypomethylation is associated with platinum resistance in ovarian cancer. Gynecol Oncol. (2016) 142:539–47. doi: 10.1016/j.ygyno.2016.06.017
177. Li D, Wu QJ, Bi FF, Chen SL, Zhou YM, Zhao Y, et al. Effect of the BRCA1-SIRT1-EGFR axis on cisplatin sensitivity in ovarian cancer. Am J Transl Res. (2016) 8:1601–8.
178. Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer. (2011) 21:51–7. doi: 10.1097/IGC.0b013e3182021c1a
179. Lee PS, Teaberry VS, Bland AE, Huang Z, Whitaker RS, Baba T, et al. Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer. (2010) 126:1378–89. doi: 10.1002/ijc.24797
180. Barros-Silva D, Costa-Pinheiro P, Duarte H, Sousa EJ, Evangelista AF, Graca I, et al. MicroRNA-27a-5p regulation by promoter methylation and MYC signaling in prostate carcinogenesis. Cell Death Dis. (2018) 9:167. doi: 10.1038/s41419-017-0241-y
181. Liu T, Xu F, Du X, Lai D, Liu T, Zhao Y, et al. Establishment and characterization of multi-drug resistant, prostate carcinoma-initiating stem-like cells from human prostate cancer cell lines 22RV1. Mol Cell Biochem. (2010) 340:265–73. doi: 10.1007/s11010-010-0426-5
182. Luzhna L, Kovalchuk O. Modulation of DNA methylation levels sensitizes doxorubicin-resistant breast adenocarcinoma cells to radiation-induced apoptosis. Biochem Biophys Res Commun. (2010) 392:113–7. doi: 10.1016/j.bbrc.2009.12.093
183. Ter Brugge P, Kristel P, van der Burg E, Boon U, de Maaker M, Lips E, et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst. (2016) 108:djw148. doi: 10.1093/jnci/djw148
184. Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. (2008) 4:68–75. doi: 10.4161/org.4.2.5851
185. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. (2017) 169:985–99. doi: 10.1016/j.cell.2017.05.016
186. MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. (2009) 17:9–26. doi: 10.1016/j.devcel.2009.06.016
187. Shi J, Chi S, Xue J, Yang J, Li F, Liu X. Emerging role and therapeutic implication of wnt signaling pathways in autoimmune diseases. J Immunol Res. (2016) 2016:9392132. doi: 10.1155/2016/9392132
188. Lang CMR, Chan CK, Veltri A, Lien WH. Wnt signaling pathways in keratinocyte carcinomas. Cancers. (2019) 11:1216. doi: 10.3390/cancers11091216
189. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. (2017) 36:1461–73. doi: 10.1038/onc.2016.304
190. Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. (2009) 4:307–12. doi: 10.4161/epi.4.5.9371
191. Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. (2013) 5:a015081. doi: 10.1101/cshperspect.a015081
192. Choi HJ, Park H, Lee HW, Kwon YG. The Wnt pathway and the roles for its antagonists, DKKS, in angiogenesis. IUBMB Life. (2012) 64:724–31. doi: 10.1002/iub.1062
193. Ghosh N, Kazi M, Vora H, Gajjar K, Kobawala T. Frequent methylation of genes encoding wnt pathway antagonists: Secreted frizzled-related protein 1 and dickkopf 3 in invasive breast cancer. Clin Cancer Invest J. (2019) 8:106–13. doi: 10.4103/ccij.ccij_14_19
194. Tao L, Huang G, Chen Y, Chen L. DNA methylation of DKK3 modulates docetaxel chemoresistance in human nonsmall cell lung cancer cell. Cancer Biother Radiopharm. (2015) 30:100–6. doi: 10.1089/cbr.2014.1797
195. Guo Q, Qin W. DKK3 blocked translocation of beta-catenin/EMT induced by hypoxia and improved gemcitabine therapeutic effect in pancreatic cancer Bxpc-3 cell. J Cell Mol Med. (2015) 19:2832–41. doi: 10.1111/jcmm.12675
196. Ren J, Wang R, Song H, Huang G, Chen L. Secreted frizzled related protein 1 modulates taxane resistance of human lung adenocarcinoma. Mol Med. (2014) 20:164–78. doi: 10.2119/molmed.2013.00149
197. Zhu J, Wang Y, Duan J, Bai H, Wang Z, Wei L, et al. DNA Methylation status of Wnt antagonist SFRP5 can predict the response to the EGFR-tyrosine kinase inhibitor therapy in non-small cell lung cancer. J Exp Clin Cancer Res. (2012) 31:80. doi: 10.1186/1756-9966-31-80
198. Liu S, Chen X, Chen R, Wang J, Zhu G, Jiang J, et al. Diagnostic role of Wnt pathway gene promoter methylation in non small cell lung cancer. Oncotarget. (2017) 8:36354–67. doi: 10.18632/oncotarget.16754
199. Hu H, Wang T, Pan R, Yang Y, Li B, Zhou C, et al. Hypermethylated promoters of secreted frizzled-related protein genes are associated with colorectal cancer. Pathol Oncol Res. (2019) 25:567–75. doi: 10.1007/s12253-018-0505-6
200. Liu X, Fu J, Bi H, Ge A, Xia T, Liu Y, et al. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer. (2019) 19:1212. doi: 10.1186/s12885-019-6436-0
201. Su HY, Lai HC, Lin YW, Liu CY, Chen CK, Chou YC, et al. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer. (2010) 127:555–67. doi: 10.1002/ijc.25083
202. Ramachandran K, Speer C, Nathanson L, Claros M, Singal R. Role of DNA methylation in cabazitaxel resistance in prostate cancer. Anticancer Res. (2016) 36:161–8.
203. Hiltunen MO, Alhonen L, Koistinaho J, Myohanen S, Paakkonen M, Marin S, et al. Hypermethylation of the APC (Adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int J Cancer. (1997) 70:644–8. doi: 10.1002/(SICI)1097-0215(19970317)70:6<644::AID-IJC3>3.0.CO;2-V
204. Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. (2007) 120:3327–35. doi: 10.1242/jcs.03485
205. Matuschek C, Bolke E, Lammering G, Gerber PA, Peiper M, Budach W, et al. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res. (2010) 15:277–86. doi: 10.1186/2047-783X-15-7-277
206. Stefanski CD, Keffler K, McClintock S, Milac L, Prosperi JR. APC loss affects DNA damage repair causing doxorubicin resistance in breast cancer cells. Neoplasia. (2019) 21:1143–50. doi: 10.1016/j.neo.2019.09.002
207. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. (2017) 170:605–35. doi: 10.1016/j.cell.2017.07.029
208. Tancredi R, Furlanetto J, Loibl S. Endocrine therapy in premenopausal hormone receptor positive/human epidermal growth receptor 2 negative metastatic breast cancer: between guidelines and literature. Oncologist. (2018) 23:974–81. doi: 10.1634/theoncologist.2018-0077
209. Phuong NT, Kim SK, Lim SC, Kim HS, Kim TH, Lee KY, et al. Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Res Treat. (2011) 130:73–83. doi: 10.1007/s10549-010-1304-2
210. Ye L, Lin C, Wang X, Li Q, Li Y, Wang M, et al. Epigenetic silencing of SALL2 confers tamoxifen resistance in breast cancer. EMBO Mol Med. (2019) 11:e10638. doi: 10.15252/emmm.201910638
211. Spangle JM, Dreijerink KM, Groner AC, Cheng H, Ohlson CE, Reyes J, et al. PI3K/AKT signaling regulates H3K4 methylation in breast cancer. Cell Rep. (2016) 15:2692–704. doi: 10.1016/j.celrep.2016.05.046
212. Maeda M, Murakami Y, Watari K, Kuwano M, Izumi H, Ono M. CpG hypermethylation contributes to decreased expression of PTEN during acquired resistance to gefitinib in human lung cancer cell lines. Lung Cancer. (2015) 87:265–71. doi: 10.1016/j.lungcan.2015.01.009
213. Cortes-Sempere M, de Miguel MP, Pernia O, Rodriguez C, de Castro Carpeno J, Nistal M, et al. IGFBP-3 methylation-derived deficiency mediates the resistance to cisplatin through the activation of the IGFIR/Akt pathway in non-small cell lung cancer. Oncogene. (2013) 32:1274–83. doi: 10.1038/onc.2012.146
214. Jung IL, Kang HJ, Kim KC, Kim IG. PTEN/pAkt/p53 signaling pathway correlates with the radioresponse of non-small cell lung cancer. Int J Mol Med. (2010) 25:517–23. doi: 10.3892/ijmm_00000372
215. Pappas G, Zumstein LA, Munshi A, Hobbs M, Meyn RE. Adenoviral-mediated PTEN expression radiosensitizes non-small cell lung cancer cells by suppressing DNA repair capacity. Cancer Gene Ther. (2007) 14:543–9. doi: 10.1038/sj.cgt.7701050
216. Geybels MS, Fang M, Wright JL, Qu X, Bibikova M, Klotzle B, et al. PTEN loss is associated with prostate cancer recurrence and alterations in tumor DNA methylation profiles. Oncotarget. (2017) 8:84338–48. doi: 10.18632/oncotarget.20940
217. Qian XJ, Li YT, Yu Y, Yang F, Deng R, Ji J, et al. Inhibition of DNA methyltransferase as a novel therapeutic strategy to overcome acquired resistance to dual PI3K/mTOR inhibitors. Oncotarget. (2015) 6:5134–46. doi: 10.18632/oncotarget.3016
218. Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. (2016) 44:5615–28. doi: 10.1093/nar/gkw159
Keywords: DNA methylation, tumor suppressor genes, oncogenes, DNMTs, therapeutic targets, biomarkers, solid tumors, chemotherapy
Citation: Romero-Garcia S, Prado-Garcia H and Carlos-Reyes A (2020) Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front. Oncol. 10:1152. doi: 10.3389/fonc.2020.01152
Received: 15 March 2020; Accepted: 08 June 2020;
Published: 07 August 2020.
Edited by:
Atsushi Fujimura, Okayama University, JapanReviewed by:
Prasanna Ekambaram, University of Pittsburgh, United StatesPatrizia Zavattari, University of Cagliari, Italy
Copyright © 2020 Romero-Garcia, Prado-Garcia and Carlos-Reyes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angeles Carlos-Reyes, cmV5ZXNfY2FyZG9zb0B5YWhvby5jb20=
 Susana Romero-Garcia
Susana Romero-Garcia Heriberto Prado-Garcia
Heriberto Prado-Garcia Angeles Carlos-Reyes
Angeles Carlos-Reyes