- 1Department of Internal Medicine, University of South Florida, Tampa, FL, United States
- 2Department of Hematopathology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, United States
- 3Department of Malignant Hematology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, United States
Peripheral T-cell lymphoma (PTCL) is a rare, heterogenous group of mature T-cell neoplasms that comprise 10–15% of non-Hodgkin lymphoma cases in the United States. All subtypes of PTCL, except for ALK+ anaplastic T-cell lymphoma, are associated with poor prognosis, with median overall survival (OS) rates of 1–3 years. The diagnosis of PTCL is mainly based on clinical presentation, morphologic features, and immunophenotypes. Recent advances in genome sequencing and gene expression profiling have given new insights into the pathogenesis and molecular biology of PTCL. An enhanced understanding of its genomic landscape holds the promise of refining the diagnosis, prognosis, and management of PTCL. In this review, we examine recently discovered genetic abnormalities identified by molecular profiling in 3 of the most common types of PTCL: RHOAG17V and epigenetic regulator mutations in angioimmunoblastic T-cell lymphoma, ALK expression and JAK/STAT3 pathway mutations in anaplastic T-cell lymphoma, and T-follicular helper phenotype and GATA3/TBX21 expression in PTCL-not otherwise specified. We also discuss the implications of these abnormalities for clinical practice, new/potential targeted therapies, and the role of personalized medicine in the management of PTCL.
Introduction
Peripheral T-cell lymphoma (PTCL) is a rare and heterogeneous group of mature T-cell neoplasms, comprising 10–15% of all cases of non-Hodgkin lymphoma cases in the United States (1), with at least 29 subtypes recognized by the revised 2016 World Health Organization classification of lymphoid neoplasms (2). PTCL generally carries a poor prognosis. The complex pathobiology of these disorders is well-reflected in their heterogeneous clinical, histological, and immunophenotypic features. Advances in the genome sequencing and gene expression profiling (GEP) of PTCL have improved our understanding of its molecular pathobiology, and a precise definition of its molecular background has revealed novel therapeutic targets. In this review, we focus on the recently discovered somatic genetic abnormalities of and emerging therapies for 3 of the most common PTCL subtypes: angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), and PTCL-not otherwise specified (PTCL-NOS) (Supplementary Table 1). Germline mutations are beyond the scope of this review.
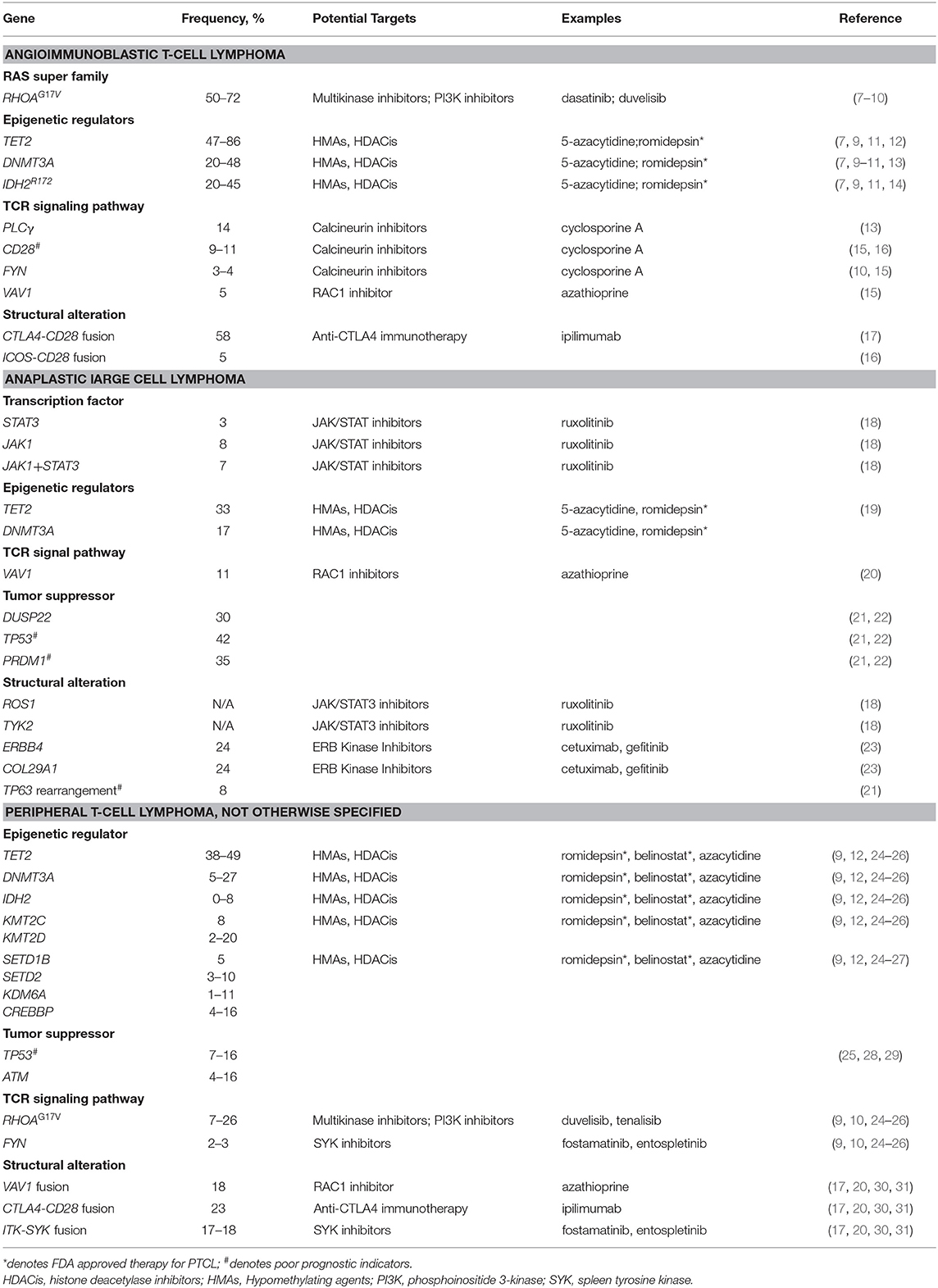
Angioimmunoblastic T-Cell Lymphoma and Other Nodal Lymphomas of Follicular T Helper Cell Origin
AITL is the one of the most common PTCL entities, accounting for ~20–30% of all PTCL cases in the United States (3). The AITL cell originates from CD4+ T follicular helper cells (TFHs) (4). AITL is associated with B-cell lymphoproliferative disorders and a constitutively activated immune system (5). The prognosis of patients with AITL is poor, with a 5-year overall survival (OS) rate of 33% (5). There has been no significant improvement in OS in the last 3 decades (6).
Recent advances in next-generation sequencing have led to the discovery of recurrent mutations in AITL. The most frequently reported somatic mutations include alterations in epigenetic regulators; ras homolog family member A (RHOA); and T-cell receptor (TCR) signaling pathway molecules (Table 1).
Epigenetic Regulators
Tet methylcytosine dioxygenase 2 (TET2), DNA methyltransferase 3α (DNMT3A), and mitochondrial isocitrate dehydrogenase 2 (IDH2) genes participate in the regulation of DNA methylation/hydromethylation. Mutations in TET2 and DNMT3A are associated with hypermethylation and dysregulated gene expression (11, 32), and the IDH2R172 mutant confers neo-enzymatic activity, producing oncometabolite D-2 hydroxyglutarate (D-2-HG). The accumulation of D-2-HG inhibits both histone lysine demethylase and the DNA hydroxylase in the TET family (33). Interestingly, the high co-occurrence of TET2 and IDH2R172 mutations in AITL suggests a synergistic effect, by which these genes upregulate follicular T helper–associated genes and downregulate genes associated with TH1, TH2, and TH17 cells (7, 19).
Epigenetic modulating agents are promising targets for patients with relapsed AITL. 5-azacytidine, has induced a sustained response in selected patients with TET2-mutated relapsed/refractory AITL (34). A similar response is reported with romidepsin (35).
TCR Signaling Pathway
The RHOAG17V mutation is common in AITL. RHOA is a small GTPase that mediates T-cell migration, polarity, and thymocyte development (36). Glycine at RHOA residue 17 is critical for GTP binding. Thus, the substitution of Valine leads to a loss of GTPase activity (8). It was initially believed that the RHOAG17V mutation played an oncogenic role by disrupting the classical RHOA signaling. However, a recently reported p.K18N mutant in AITL is associated with higher GTP binding capacity (15). This phenomenon is explained by the RHOA-VAV1 signaling pathway. VAV1, a guanine exchange factor protein, functions as an adaptor to facilitate and activate the TCR proximal signaling complex. The binding of G17V RHOA to VAV1 augments VAV1's adaptor function, resulting in an accelerated TCR signaling. An isolated VAV1 mutation has also been identified in AITL (37). Dasatinib blocked accelerated VAV1 phosphorylation and TCR signaling in vitro and improved the overall survival of the mice model (37).
In preclinical models, the expression of RHOAG17V induced TFH cell specification, upregulated the inducible co-stimulator (ICOS), and increased phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase signaling. PI3K inhibitors efficiently inhibited TET2-/-RHOA G17V tumor proliferation (38).
Other TCR-related mutations in AITL include PLCG1, CD28, and FYN. CD28 is the primary costimulatory receptor in T cells and induces sustained T-cell proliferation and cytokine production. The presence of CD28 mutations correlates with a poor prognosis (16). Cyclosporine A, a calcineurin inhibitor that blocks TCR signaling, effectively prevented the progression of AITL (39, 40). Two structural changes, CTLA4-CD28 (17) and ICOS-CD28 fusion genes (16), have also been described. Ipilimumab, an anti-CTLA4 immunotherapy, is a potential treatment for the CTLA4-CD28 fusion gene.
Multistep Tumorigenesis Model
To account for the complex genomic landscape of AITL, a multistep tumorigenesis model was proposed (41–43). The premalignant hematopoietic progenitor cells harboring mutations (e.g., TET2 and DNMT3A) are predisposed to the development of blood cancer, and the acquisition of second-hit mutations (e.g., RHOAG17V and IDH2R172) in a subclone of TFH cells eventually leads to AITL. This model is supported by the detection of TET2 and DNMT3A mutations in tumor-free peripheral blood cells, bone marrow cells, and hematopoietic progenitors, whereas RHOA and IDH mutations are specific to malignant cells from AITL tumors (13).
Nodal T-Cell Lymphomas With TFH Phenotype as a Newly Proposed Group of PTCL
Together with AITL, nodal PTCL with TFH phenotype and follicular T-cell lymphoma (F-PTCL) belong to a newly proposed group of PTCL called “nodal T-cell lymphomas with TFH phenotype,” described in the 2016 revised WHO classification (2, 44). This change reflects the observation that a subset of PTCLs expresses TFH-associated markers (45, 46). Interestingly, this subset shares common genetic abnormalities with AITL (9, 10, 12, 14, 24, 32). The analysis of 94 cases of AITL, 5 cases of F-PTCL, and 16 cases of nodal PTCL with TFH phenotype supported this grouping (13). These entities shared not only disease severity and prognosis, but also global and specific gene expression patterns. They had similar mutation frequencies in TET2, RHOA, DNMT3A, with the exception of IDH2R172 mutation, which was restricted to AITL.
We recommend routine screening of any PTCL-NOS for TFH markers and assigning them to this new category when at least 2 TFH markers are simultaneously detected on the neoplastic cells.
ALCL
ALCL accounts for ~12% of PTCL cases in the United States (3). ALCL is characterized by CD30 positivity. The 2016 revision of WHO classification system for lymphoid neoplasms recognizes 4 subtypes of ALCL: ALK+ ALCL, ALK− ALCL, primary cutaneous, and breast implant–associated ALCL (44, 47). The differentiation between ALK+ and ALK− subtypes has formed the backbone of the current classification system.
ALK+ ALCL
ALK+ ALCL commonly presents in young populations, generally within the first 3 decades of life and carries a significantly better prognosis (5-year OS, 70–85%) than ALK− ALCL (5-year OS, 30–49%) (48). The presence of ALK gene rearrangements in ALK+ ALCL, most commonly translocation t(2;5)(p23;q35), results in the fusion of nucleophosmin (NPM1) and ALK (49). Anti-ALK antibodies can identify the proteins produced by NPM1/ALK transcripts based on staining patterns. ALK+ ALCL expressed ALK in nucleus and cytoplasm; conversely, variant fusions lacked nuclear ALK-staining (50).
ALK gene rearrangements often occur within the intron, between exons 19 and 20, allowing the intracytoplasmic domain of ALK to fuse with NPM1. The dimerization domain auto-phosphorylates the ALK catalytic domain and activates multiple downstream signaling pathways, including PI3K/AKT, RAS/ERK, and JAK/STAT (51).
NPM1-ALK cell lines express STAT3 phosphorylated on serine residue 727 and tyrosine residue 705 and increase STAT3 expression at the transcriptional level. Although JAK3 is phosphorylated, its binding is not essential for STAT3 activity. NPM-ALK fusion transcripts could activate STAT3 directly (52). This activation is important, as STAT3 is integral to cell survival by controlling the transcription of numerous apoptosis-regulating proteins, such as cyclin D1, Bcl-X, Bcl-XL, and c-Myc (53). Although NPM1/ALK fusion transcripts are the most common rearrangements in ALK+ ALCL, other rearrangements, such as TPM3 (1q25), ATIC (2q35), TFG (3q21), TPM4 (19p13.1), MYH9 (22q11.2), RNF213 (17q25), TRAF1 (9q33.2), CLTC (17q23), and MSN (Xq11), have also been reported (54).
ALK− ALCL
ALK– ALCL was upgraded from a provisional to a definite entity in the revised 2016 WHO classification (55). It is difficult to differentiate between ALK– ALCL and PTCL-NOS based on CD30 positivity (20). To better define ALCL from PTCL-NOS, GEP of PTCL-NOS, and ALCL discovered a unique cluster of gene transcripts shared by ALK– and ALK+ALCLs (56). We can also distinguish ALK–ALCL from CD30+ PTCL-NOS through clinical outcomes (57, 58). CD30+ PTCL-NOS has a poorer prognosis and requires more aggressive treatment (59, 60). Based on GEP, a 3-gene model (TNFRSF8, BATF3, and TMOD1) was developed to separate ALK– ALCL from PTCL-NOS, with 97% accuracy (61).
Chromosomal Rearrangements of DUSP22 and TP63 as a Differentiating Factor
Two chromosomal rearrangements, DUSP22 and TP63, subclassify ALK−ALCL into 3 groups: DUSP22-rearranged, TP63-rearranged, and group without any rearrangement. DUSP22 rearrangement occurs in 30% of ALK− ALCL patients and is associated with a 5-year OS rate of 80–90%, similar to that of ALK+ ALCL (5-year OS, 85%) (62). It is associated with downregulation of DUSP22, which leads to the inhibition of TCR signaling and the promotion of apoptosis (21). Its unique immunogenic molecular signature, such as DNA hypomethylation, lower expression of PD-1, and higher expression of costimulatory gene CD58 and HLA Class II likely contributes to its favorable prognosis (63). Other clinical predictors, such as IPI risk factors, age, and CD3 positivity, also impact prognosis despite DUSP22 rearrangement (64, 65).
TP63 rearrangement, the fusion transcript of TBL1XR1/TP63, has similar structural homology to oncogenic deltaNp63 in p53 tumor suppressor pathway and is associated with inferior survival (66).
A third category, defined as triple-negative (ALK−, DUSP22−, and TP63−), harbors the remaining 62% of ALK− ALCL cases and has a 5-year OS rate of 42% (67). Although further validation of this model is needed, DUSP22 and TP63 rearrangements may serve as useful biomarkers in prognosis and direct therapy for patients with ALK− ALCL in the future.
Other Genetic Aberrations in ALK−ALCL
As in ALK+ ALCL, the JAK/STAT3 pathway is constitutively activated in ALK− ALCL (68). Recurrent single or convergent somatic mutations and translocations in the JAK1 and STAT3 genes are thought to upregulate the STAT3 pathway (18, 19). In addition, the gene fusions involving ROS1 and TYK2 in some ALK− ALCLs have led to the activation of STAT3 independent of JAK1 or STAT3 mutations. RNA sequencing has identified the co-expression of truncated ERBB4 and COL29A1 in 24% of patients with ALK− ALCL (23). These ERBB4-truncated forms are potentially oncogenic, and ERBB4 inhibition can partially arrest cell growth and stop disease progression. These transcripts were not observed in ALK+ ALCL or PTCL-NOS patients. More recently, losses at 6p21 and 17p13 were identified in ALK− ALCL using single nucleotide polymorphism arrays (22, 69). These losses correlated with the losses of TP53 and PRDM1 and poor prognoses.
PTCL-NOS
PTCL-NOS is the most common subtype of PTCL, accounting for 30–50% of PTCL cases in the United States (3, 70). Patients are diagnosed with PTCL-NOS if they do not meet the diagnostic criteria of other PTCL subtypes as per WHO 2016 revision (2, 70). As a diagnosis of exclusion, PTCL-NOS comprises a heterogeneous group of diseases with diverse cells of origin and presents with different cytogenic, molecular, and morphological phenotypes. This heterogeneity makes classification and treatment of the disease difficult. With the standard anthracycline-based chemotherapy, complete response rates range from 40 to 60%, and 5-year OS rates range from 30 to 40% (71, 72).
GATA3 and TBX21 Expression as a Differentiating Factor in PTCL-NOS
PTCL-NOS can be categorized based on GATA3 and TBX21 expression and T helper 1 and T helper 2 cell differentiation regulators (73, 74). PTCL-NOS cases with high expression of TBX21 have a tumor microenvironment gene signature, whereas those of GATA3 have a cytotoxic gene signature with poorer outcomes (75). The greater genomic complexity associated with GATA3 is characterized by frequent loss of tumor suppressor genes on the CDKN2A/B-TP3 axis and PTEN-PI3K pathways as well as genetic gains and amplification of STAT3 and MYC. Immunohistochemistry (IHC) algorithm can be used to identified the two subtypes and add in risk stratification for clinical trials (76).
Watatani's group studied the relation between PTCL-NOS with TFH phenotype and GATA3/TBX21 expression using GEP (25). PTCL-NOS without TFH phenotype often has mutations in TP53 and/or CDKNA2A genes, which can cause chromosomal instability and mediate immune escape and transcriptional regulations. Those mutations potentially explain the worse outcomes among patients with PTCL-NOS without TFH phenotype as compared with those with TFH phenotype. However, there was no difference in GATA3 and TBX21 expression in the TFH-related group and in the TP53/CDKN2A-altered group.
Other Genetic Aberrations in PTCL-NOS
The FYN gene encodes a tyrosinase kinase involved in T-cell activation and Src kinase inhibition. Dasatinib targeted the Src kinase in vitro, and could be a l target for patients with mutations in FYN genes (10). Recurrent loss at 9p21.3 decreases the expression of the cyclin-dependent kinase inhibitors 2A and 2B which are associated with a poorer prognosis (28). Guanine nucleotide exchange factor VAV1 encodes a critical component of TCR signaling, and recurrent gene fusion of VAV1 has also been identified (20). Recurrent genetic activating mutations and translocations of VAV1 gene in PTCL-NOS highlighted its role of a drive oncogene in catalytic-dependent (MAPK and JNK) and -independent (NFAT) VAV1 effector pathways (77). Azathioprine targets cells overexpressing the VAV1-GSS fusion protein (20). CTLA4-CD28 fusion and mutations in KMT2C and SET1B (histone methylation) have also been identified (17, 27).
FAT1 tumor suppressor binds to β-catenin and inhibits nuclear localization, thus inhibiting cell growth. The recurrent mutations in FAT1 tumor suppressor gene were seen in 39% cases of PTCL-NOS and is associated with inferior outcome (29).
With new differentiating factors such as TFH phenotype, GATA3/TBX21 expression, we may expect PTCL-NOS to be better categorized, which could provide more insight into defining targetable molecular pathways and developing novel therapeutic strategies for PTCL-NOS patients.
Clinical Implications and Personalized Medicine
Recent advances in genome sequencing and GEP have led to the identification of commonly dysregulated pathways, especially enhanced T-cell signaling pathways, in all of the 3 most common nodal subtypes of PTCL. Despite similarities in genomic profiles, the interaction between various pathways might play a role in determining divergent cell differentiation and tumorigenesis. For example, some mutations in the epigenetic modifier genes are similar between AITL and myeloid neoplasms; however, the mutation patterns are different. TET2 and IDH2 mutations are mutually exclusive in acute myeloid leukemia. In contrast, IDH2 mutations often cooccur with TET2 mutations in AITL (19, 78). These different mutation patterns suggest that the interaction between IDH2 and TET2 mutations possibly lead to the development of a TFH phenotype.
The different patterns of molecular signature profiles can help us to identify and to reclassify AITL and ALCL from PTCL-NOS in cases that do not meet morphological criteria (Figure 1). Iqbal et al. reclassified 14% of PTCL-NOS cases as AITL via GEP using the 3 prominent AITL signatures: B-cell signature, follicular dendritic-cell signature, and cytokine signature (79). This reclassification was then confirmed by the presence of the IDH2R172 mutation. The presence of the RHOAG17V mutation helped to identify nodal T-cell lymphomas with TFH phenotype, as this mutation key in TFH cell speciation and AITL transformation (38). ITK-SYK gene fusion could potentially differentiate a subset of PTCL-NOS patients with TFH phenotype from those with AITL (30, 31, 80, 81) (Figure 1).
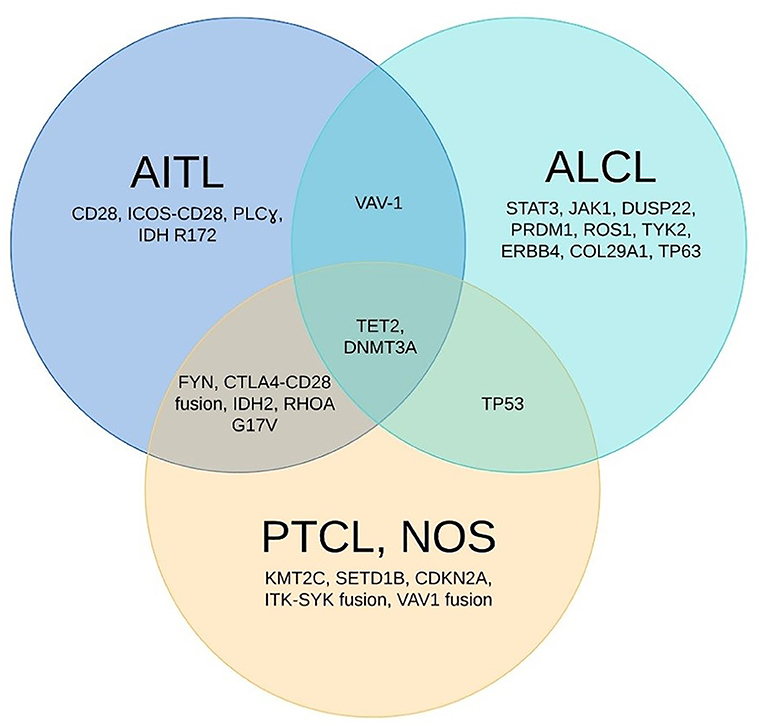
Figure 1. Unique and shared mutations identified in the 3 most common types of PTCL. The Venn diagram above showed the most frequently encountered genomic abnormalities in 3 most common types of peripheral T-cell lymphoma: angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), and peripheral T-cell lymphoma-not otherwise specified (PTCL-NOS). TET2 and DNMT3A mutations are seen in all 3 major subtypes.
Agnelli et al. reclassified 11% of PTCL-NOS cases as ALCL using a 3-gene model (TNFRSF8, BATF3, and TMOD1) with 97% accuracy (61). Similarly, lack of ERBB4 transcript in other PTCL subtypes could help to confirm the diagnosis of ALK− ALCL (23) (Figure 1).
Furthermore, many mutations have been associated with the prognosis of PTCL. The mutations associated with a poor prognosis include CD28 mutations in AITL (16); TP63 rearrangement (21), loss of TP53, and loss of PRDM1 (22) in ALK− ALCL; GATA3 (75), TP53, and/or CDKN2A (25) in ALK− ALCL; and alterations in histone methyltransferase genes KMT2A, KMT2B, or KDM6A (82) and FAT1 (29) in PTCL-NOS. The mutations associated with a favorable prognosis include the presence of DUSP22 in ALK− ALCL (21).
Potential Therapeutic Targets
Most PTCL subgroups have median OS rates of 1–3 years, except for ALK+ ALCL (3). Only a small portion (20%-40%) of patients with PTCL achieve long-term survival (6). During the past 3 decades, long-term survival has not been significantly improved by available therapies (83). Fortunately, a greater understanding of the pathogenesis of the major PTCL subtypes has led to the identification of potentially actionable biologic pathways, especially activating pathways. Novel targeting therapies are now available for clinical studies (Table 1).
Epigenetic modulators, such as romidepsin (NCT03141203, NCT00426764), chidamide (NCT03268889), and HBI-8000 (NCT02953652), are currently being investigated in clinical trials for PTCL, either as a monotherapy or in conjunction with other therapies.
The T-cell signaling pathway also has multiple candidate therapies: multikinase inhibitors, such as dasatinib (NCT01609816, NCT01643603), for PTCL; PI3K inhibitors, such as duvelisib (NCT03372057) and copanlisib (NCT03052933), for PTCL; an anti-ICOS monoclonal antibody in ICOS-PI3K pathways, MEDI−570 (NCT02520791), for the follicular variant of PTCL-NOS and AITL; and a CTLA-4 inhibitor, such as ipilimumab, for CLA-CD28 fusion-positive tumors.
The transcription factor NF-κB pathway is differentially activated in PTCL, especially in the AITL subtype (79). Bortezomib, a proteasome inhibitor with NF-κB inhibitory activity, has shown early promise in the treatment of adult T-cell lymphoma (84). An additional clinical study on bortezomib is currently underway (NCT04061772), as well as studies on ixazomib, a drug similar to bortezomib (NCT03547700).
The JAK-STAT3 pathways are sometimes aberrantly activated in PTCL, especially in ALCL and in some cases of PTCL-NOS. JAK inhibitors, such as ruxolitinib, were previously used extensively for myeloid disorder and are currently under investigation for treating patients with PTCL (NCT01431209).
Another promising target is SYK, a receptor-associated tyrosine kinase expressed in 94% of all PTCL patients (85). The SYK inhibitor R406 effectively caused apoptosis and inhibited cell growth in preclinical studies (86). SYK inhibitors, such as entospletinib, are promising potential agents.
The ALK-1 inhibitor crizotinib was used in patients with relapsed pediatric ALK+ ALCL, with a complete response rate of 83% (NCT00939770) (87). For ALK+ ALCL resistant to crizotinib, platelet-derived growth factor receptor-β (PDGFRB) blockade is potentially effective. Imatinib, acting on PDGFRA and PDGFRB blockade, induced a complete remission in a late stage NPM-ALK+ ALCL patient. As suggested in murine model, ALK promotes the expression of activator protein 1 family members JUN and JUNB, which subsequently promote tumor dissemination through PDGFRB regulation (88).
Other targeted agents tested in PTCL include BCL2 inhibitors (e.g., venetoclax [NCT03552692]); monoclonal antibodies targeting CD2 (e.g., siplizumab [NCT01445535]); CCR4 monoclonal antibodies (e.g., mogalizumab [NCT01611142]); FYN inhibitors; CD30 antibody drug conjugates (e.g., brentuximab [NCT01716806, NCT03496779]); ERB kinase inhibitors; VGEFR-2 inhibitors (e.g., apatinib [NCT03631862]); and PD-1 and PD-L1 inhibitors for immune modulation (e.g., pembrolizumab [NCT02535247], nivolumab [NCT03586999], durvalumab [NCT03161223], and avelumab [NCT03046953]).
Currently, novel therapies are being developed rapidly, and personalized medicine is made possible through commercial gene sequencing. The genetic heterogeneity in PTCL requires an individualized therapeutic approach that uses agents that specifically target genetic abnormalities or oncogenic pathways found in patients' tumors. More rationally designed clinical trials enrolling patients with specific genetic alterations are needed to provide higher response rates and more sustained responses. In the context of genome sequencing and GEP, targeted and personalized therapies will likely provide the best clinical outcomes in patients with PTCL in the near future.
Author Contributions
LS and MH conceptualized the idea for the manuscript and critically reviewed and edited the manuscript. YZ, DL, TB reviewed the relevant literature and collected the data, and wrote the original draft. YZ prepared the table and graph.
Conflict of Interest
LS and MH are associate guest editors of Frontiers in Oncology; during the course of writing and submitting this paper to the journal they were recused from the editorial decision-making process, and another associate editor handled the peer-review procedures independently.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Paul Fletcher and Daley Drucker (Moffitt Cancer Center) for editorial assistance. They were not compensated beyond their regular salaries.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00898/full#supplementary-material
References
1. Armitage JO. The aggressive peripheral T-cell lymphomas: 2012 update on diagnosis, risk stratification, and management. Am Hematol J. (2012) 87:511–9. doi: 10.1002/ajh.23144
2. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
3. Vose J, Armitage J, Weisenburger D, International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. (2008) 26:4124–30. doi: 10.1200/JCO.2008.16.4558
4. de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma. (AITL) and follicular helper T. (TFH) cells. Blood. (2007) 109:4952–63. doi: 10.1182/blood-2006-10-055145
5. Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. J Clin Oncol. (2013) 31:240–6. doi: 10.1200/JCO.2011.37.3647
6. Crozier JA, Sher T, Yang D, Swaika A, Foran J, Ghosh R, et al. Persistent disparities among patients with T-cell non-hodgkin lymphomas and B-cell diffuse large cell lymphomas over 40 years: a SEER database review. Clin Lymphoma Myeloma Leuk. (2015) 15:578–85. doi: 10.1016/j.clml.2015.06.005
7. Steinhilber J, Mederake M, Bonzheim I, Serinsoz-Linke E, Muller I, Fallier-Becker P, et al. The pathological features of angioimmunoblastic T-cell lymphomas with IDH2(R172) mutations. Mod Pathol. (2019) 32:1123–34. doi: 10.1038/s41379-019-0254-4
8. Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. (2014) 46:371–5. doi: 10.1038/ng.2916
9. Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. (2014) 46:171–5. doi: 10.1038/ng.2872
10. Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. (2014) 46:166–70. doi: 10.1038/ng.2873
11. Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. (2014) 123:1293–6. doi: 10.1182/blood-2013-10-531509
12. Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. (2012) 120:1466–9. doi: 10.1182/blood-2012-02-408542
13. Dobay MP, Lemonnier F, Missiaglia E, Bastard C, Vallois D, Jais JP, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. (2017) 102:e148–51. doi: 10.3324/haematol.2016.158428
14. Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. (2012) 119:1901–3. doi: 10.1182/blood-2011-11-391748
15. Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. (2016) 128:1490–502. doi: 10.1182/blood-2016-02-698977
16. Rohr J, Guo S, Huo J, Bouska A, Lachel C, Li Y, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia. (2016) 30:1062–70. doi: 10.1038/leu.2015.357
17. Yoo HY, Kim P, Kim WS, Lee SH, Kim S, Kang SY, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma. Haematologica. (2016) 101:757–63. doi: 10.3324/haematol.2015.139253
18. Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. (2015) 27:516–32. doi: 10.1016/j.ccell.2015.03.006
19. Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. (2015) 126:1741–52. doi: 10.1182/blood.V124.21.3580.3580
20. Boddicker RL, Razidlo GL, Dasari S, Zeng Y, Hu G, Knudson RA, et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood. (2016) 128:1234–45. doi: 10.1182/blood-2016-03-707141
21. Parrilla Castellar ER, Jaffe ES, Said JW, Swerdlow SH, Ketterling RP, Knudson RA, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. (2014) 124:1473–80. doi: 10.1182/blood-2014-04-571091
22. Boi M, Rinaldi A, Kwee I, Bonetti P, Todaro M, Tabbo F, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. (2013) 122:2683–93. doi: 10.1182/blood-2013-04-497933
23. Scarfo I, Pellegrino E, Mereu E, Kwee I, Agnelli L, Bergaggio E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood. (2016) 127:221–32. doi: 10.1182/blood-2014-12-614503
24. Manso R, Gonzalez-Rincon J, Rodriguez-Justo M, Roncador G, Gomez S, Sanchez-Beato M, et al. Overlap at the molecular and immunohistochemical levels between angioimmunoblastic T-cell lymphoma and a subgroup of peripheral T-cell lymphomas without specific morphological features. Oncotarget. (2018) 9:16124–33. doi: 10.18632/oncotarget.24592
25. Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. (2019) 33:2867–83. doi: 10.1038/s41375-019-0473-1
26. Laribi K, Alani M, Truong C, Baugier de Materre A. Recent advances in the treatment of peripheral T-cell lymphoma. Oncologist. (2018) 23:1039–53. doi: 10.1634/theoncologist.2017-0524
27. Ji MM, Huang YH, Huang JY, Wang ZF, Fu D, Liu H, et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica. (2018) 103:679–87. doi: 10.3324/haematol.2017.182444
28. Fujiwara SI, Yamashita Y, Nakamura N, Choi YL, Ueno T, Watanabe H, et al. High-resolution analysis of chromosome copy number alterations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified, with single nucleotide polymorphism-typing microarrays. Leukemia. (2008) 22:1891–8. doi: 10.1038/leu.2008.191
29. Laginestra MA, Cascione L, Motta G, Fuligni F, Agostinelli C, Rossi M, et al. Whole exome sequencing reveals mutations in FAT1 tumor suppressor gene clinically impacting on peripheral T-cell lymphoma not otherwise specified. Mod Pathol. (2020) 33:179–87. doi: 10.1038/s41379-019-0279-8
30. Huang Y, Moreau A, Dupuis J, Streubel B, Petit B, Le Gouill S, et al. Peripheral T-cell lymphomas with a follicular growth pattern are derived from follicular helper T cells. (TFH) and may show overlapping features with angioimmunoblastic T-cell lymphomas. Am J Surg Pathol. (2009) 33:682–90. doi: 10.1097/PAS.0b013e3181971591
31. Streubel B, Vinatzer U, Willheim M, Raderer M, Chott A. Novel t(5;9)(q33;q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia. (2006) 20:313–8. doi: 10.1038/sj.leu.2404045
32. Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. (2012) 366:95–6. doi: 10.1056/NEJMc1111708
33. Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. (2012) 18:5562–71. doi: 10.1158/1078-0432.CCR-12-1773
34. Lemonnier F, Dupuis J, Sujobert P, Tournillhac O, Cheminant M, Sarkozy C, et al. Treatment with 5-azacytidine induces a sustained response in patients with angioimmunoblastic T-cell lymphoma. Blood. (2018) 132:2305–9. doi: 10.1182/blood-2018-04-840538
35. Pro B, Horwitz SM, Prince HM, Foss FM, Sokol L, Greenwood M, et al. Romidepsin induces durable responses in patients with relapsed or refractory angioimmunoblastic T-cell lymphoma. Hematol Oncol. (2017) 35:914–7. doi: 10.1002/hon.2320
36. Zhang S, Konstantinidis DG, Yang JQ, Mizukawa B, Kalim K, Lang RA, et al. Gene targeting RhoA reveals its essential role in coordinating mitochondrial function and thymocyte development. Immunol J. (2014) 193:5973–82. doi: 10.4049/jimmunol.1400839
37. Fujisawa M, Sakata-Yanagimoto M, Nishizawa S, Komori D, Gershon P, Kiryu M, et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia. (2018) 32:694–702. doi: 10.1038/leu.2017.273
38. Cortes JR, Ambesi-Impiombato A, Couronne L, Quinn SA, Kim CS, da Silva Almeida AC, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. (2018) 33:259–73.e7. doi: 10.1016/j.ccell.2018.01.001
39. Advani R, Horwitz S, Zelenetz A, Horning SJ. Angioimmunoblastic T cell lymphoma: treatment experience with cyclosporine. Leuk Lymphoma. (2007) 48:521–5. doi: 10.1080/10428190601137658
40. Chen XG, Huang H, Tian Y, Guo CC, Liang CY, Gong YL, et al. Cyclosporine, prednisone, and high-dose immunoglobulin treatment of angioimmunoblastic T-cell lymphoma refractory to prior CHOP or CHOP-like regimen. Chin Cancer J. (2011) 30:731–8. doi: 10.5732/cjc.011.10071
41. Sakata-Yanagimoto M. Multistep tumorigenesis in peripheral T cell lymphoma. Int Hematol J. (2015) 102:523–7. doi: 10.1007/s12185-015-1738-8
42. Lemonnier F, Gaulard P, de Leval L. New insights in the pathogenesis of T-cell lymphomas. Curr Opin Oncol. (2018) 30:277–84. doi: 10.1097/CCO.0000000000000474
43. Fukumoto K, Nguyen TB, Chiba S, Sakata-Yanagimoto M. Review of the biologic and clinical significance of genetic mutations in angioimmunoblastic T-cell lymphoma. Cancer Sci. (2018) 109:490–6. doi: 10.1111/cas.13393
45. Agostinelli C, Hartmann S, Klapper W, Korkolopoulou P, Righi S, Marafioti T, et al. Peripheral T cell lymphomas with follicular T helper phenotype: a new basket or a distinct entity? Revising Karl Lennert's personal archive. Histopathology. (2011) 59:679–91. doi: 10.1111/j.1365-2559.2011.03981.x
46. Pileri SA. Follicular helper T-cell-related lymphomas. Blood. (2015) 126:1733–4. doi: 10.1182/blood-2015-08-665075
47. Stein H, Foss HD, Durkop H, Marafioti T, Delsol G, Pulford K, Pileri S, et al. CD30(+) anaplastic large cell lymphoma: a review of its histopathologic, genetic, clinical features. Blood. (2000) 96:3681–95. doi: 10.1182/blood.V96.12.3681
48. Moffitt AB, Dave SS. Clinical applications of the genomic landscape of aggressive non-hodgkin lymphoma. J Clin Oncol. (2017) 35:955–962. doi: 10.1200/JCO.2016.71.7603
49. Abate F, Zairis S, Ficarra E, Acquaviva A, Wiggins CH, Frattini V, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. (2014) 8:97. doi: 10.1186/s12918-014-0097-z
50. Damm-Welk C, Klapper W, Oschlies I, Gesk S, Rottgers S, Bradtke J, et al. Distribution of NPM1-ALK and X-ALK fusion transcripts in paediatric anaplastic large cell lymphoma: a molecular-histological correlation. Br Haematol J. (2009) 146:306–9. doi: 10.1111/j.1365-2141.2009.07754.x
51. Velusamy T, Kiel MJ, Sahasrabuddhe AA, Rolland D, Dixon CA, Bailey NG, et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood. (2014) 124:3768–71. doi: 10.1182/blood-2014-07-588434
52. Ducray SP, Natarajan K, Garland GD, Turner SD, Egger G. The transcriptional roles of ALK fusion proteins in tumorigenesis. Cancers. (2019) 11:E1074. doi: 10.3390/cancers11081074
53. Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, et al. Anaplastic lymphoma kinase. (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. (2002) 21:1038–47. doi: 10.1038/sj.onc.1205152
54. Xing X, Feldman AL. Anaplastic large cell lymphomas: ALK positive, ALK negative, primary cutaneous. Adv Anat Pathol. (2015) 22:29–49. doi: 10.1097/PAP.0000000000000047
55. Sandell RF, Boddicker RL, Feldman AL. Genetic landscape and classification of peripheral T cell lymphomas. Curr Oncol Rep. (2017) 19:28. doi: 10.1007/s11912-017-0582-9
56. Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I, et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol. (2010) 28:1583–90. doi: 10.1200/JCO.2008.20.9759
57. Piccaluga PP, Fuligni F, De Leo A, Bertuzzi C, Rossi M, Bacci F, et al. Molecular profiling improves classification and prognostication of nodal peripheral T-cell lymphomas: results of a phase III diagnostic accuracy study. J Clin Oncol. (2013) 31:3019–25. doi: 10.1200/JCO.2012.42.5611
58. Savage KJ, Harris NL, Vose JM, Ullrich F, Jaffe ES, Connors JM, et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood. (2008) 111:5496–504. doi: 10.1182/blood-2008-01-134270
59. Mereu E, Pellegrino E, Scarfo I, Inghirami G, Piva R. The heterogeneous landscape of ALK negative ALCL. Oncotarget. (2017) 8:18525–36. doi: 10.18632/oncotarget.14503
60. Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. (2007) 117:823–34. doi: 10.1172/JCI26833
61. Agnelli L, Mereu E, Pellegrino E, Limongi T, Kwee I, Bergaggio E, et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood. (2012) 120:1274–81. doi: 10.1182/blood-2012-01-405555
62. Pedersen MB, Hamilton-Dutoit SJ, Bendix K, Ketterling RP, Bedroske PP, Luoma IM, et al. DUSP22 and TP63 rearrangements predict outcome of ALK-negative anaplastic large cell lymphoma: a Danish cohort study. Blood. (2017) 130:554–7. doi: 10.1182/blood-2016-12-755496
63. Luchtel RA, Dasari S, Oishi N, Pedersen MB, Hu G, Rech KL, et al. Molecular profiling reveals immunogenic cues in anaplastic large cell lymphomas with DUSP22 rearrangements. Blood. (2018) 132:1386–98. doi: 10.1182/blood-2018-03-838524
64. Sibon D, Nguyen DP, Schmitz N, Suzuki R, Feldman AL, Gressin R, et al. Systemic ALK-positive anaplastic large-cell lymphoma. (ALCL): final analysis of an international, individual patient data study of 263 adults. Blood. (2017) 130:1514. doi: 10.1182/blood.V130.Suppl_1.1514.1514
65. Hapgood G, Ben-Neriah S, Mottok A, Lee DG, Robert K, Villa D, et al. Identification of high-risk DUSP22-rearranged ALK-negative anaplastic large cell lymphoma. Br Haematol J. (2019) 186:e28–31. doi: 10.1111/bjh.15860
66. Vasmatzis G, Johnson SH, Knudson RA, Ketterling RP, Braggio E, Fonseca R, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. (2012) 120:2280–9. doi: 10.1182/blood-2012-03-419937
67. Melard P, Idrissi Y, Andrique L, Poglio S, Prochazkova-Carlotti M, Berhouet S, et al. Molecular alterations and tumor suppressive function of the DUSP22. (Dual Specificity Phosphatase 22) gene in peripheral T-cell lymphoma subtypes. Oncotarget. (2016) 7:68734–48. doi: 10.18632/oncotarget.11930
68. Chen J, Zhang Y, Petrus MN, Xiao W, Nicolae A, Raffeld M, et al. Cytokine receptor signaling is required for the survival of ALK- anaplastic large cell lymphoma, even in the presence of JAK1/STAT3 mutations. Proc Natl Acad Sci USA. (2017) 114:3975–80. doi: 10.1073/pnas.1700682114
69. Feldman AL, Dogan A, Smith DI, Law ME, Ansell SM, Johnson SH, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. (2011) 117:915–9. doi: 10.1182/blood-2010-08-303305
70. Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. (2011) 117:5019–32. doi: 10.1182/blood-2011-01-293050
71. Gallamini A, Stelitano C, Calvi R, Bellei M, Mattei D, Vitolo U, et al. Peripheral T-cell lymphoma unspecified. (PTCL-U): a new prognostic model from a retrospective multicentric clinical study. Blood. (2004) 103:2474–9. doi: 10.1182/blood-2003-09-3080
72. Weisenburger DD, Savage KJ, Harris NL, Gascoyne RD, Jaffe ES, MacLennan KA, et al. Peripheral T-cell lymphoma, not otherwise specified: a report of 340 cases from the International Peripheral T-cell Lymphoma Project. Blood. (2011) 117:3402–8. doi: 10.1182/blood-2010-09-310342
73. Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. (2014) 123:2915–23. doi: 10.1182/blood-2013-11-536359
74. Wang T, Feldman AL, Wada DA, Lu Y, Polk A, Briski R, et al. GATA-3 expression identifies a high-risk subset of PTCL, NOS with distinct molecular and clinical features. Blood. (2014) 123:3007–15. doi: 10.1182/blood-2013-12-544809
75. Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. (2019) 133:1664–76. doi: 10.1182/blood-2018-09-872549
76. Amador C, Greiner TC, Heavican TB, Smith LM, Galvis KT, Lone W, et al. Reproducing the molecular subclassification of peripheral T-cell lymphoma-NOS by immunohistochemistry. Blood. (2019) 134:2159–70. doi: 10.1182/blood.2019000779
77. Abate F, da Silva-Almeida AC, Zairis S, Robles-Valero J, Couronne L, Khiabanian H, Quinn SA, et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc Natl Acad Sci USA. (2017) 114:764–9. doi: 10.1073/pnas.1608839114
78. Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. (2013) 121:3563–72. doi: 10.1182/blood-2013-01-451781
79. Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. (2010) 115:1026–36. doi: 10.1182/blood-2009-06-227579
80. Dierks C, Adrian F, Fisch P, Ma H, Maurer H, Herchenbach D, et al. The ITK-SYK fusion oncogene induces a T-cell lymphoproliferative disease in mice mimicking human disease. Cancer Res. (2010) 70:6193–204. doi: 10.1158/0008-5472.CAN-08-3719
81. Attygalle AD, Feldman AL, Dogan A. ITK/SYK translocation in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. (2013) 37:1456–7. doi: 10.1097/PAS.0b013e3182991415
82. Schatz JH, Horwitz SM, Teruya-Feldstein J, Lunning MA, Viale A, Huberman K, et al. Targeted mutational profiling of peripheral T-cell lymphoma not otherwise specified highlights new mechanisms in a heterogeneous pathogenesis. Leukemia. (2015) 29:237–41. doi: 10.1038/leu.2014.261
83. Xu B, Liu P. No survival improvement for patients with angioimmunoblastic T-cell lymphoma over the past two decades: a population-based study of 1207 cases. PLoS ONE. (2014) 9:e92585. doi: 10.1371/journal.pone.0092585
84. Ishitsuka K, Utsunomiya A, Katsuya H, Takeuchi S, Takatsuka Y, Hidaka M, et al. A phase II study of bortezomib in patients with relapsed or refractory aggressive adult T-cell leukemia/lymphoma. Cancer Sci. (2015) 106:1219–23. doi: 10.1111/cas.12735
85. Feldman AL, Sun DX, Law ME, Novak AJ, Attygalle AD, Thorland EC, et al. Overexpression of Syk tyrosine kinase in peripheral T-cell lymphomas. Leukemia. (2008) 22:1139–43. doi: 10.1038/leu.2008.77
86. Wilcox RA, Sun DX, Novak A, Dogan A, Ansell SM, Feldman AL. Inhibition of Syk protein tyrosine kinase induces apoptosis and blocks proliferation in T-cell non-Hodgkin's lymphoma cell lines. Leukemia. (2010) 24:229–32. doi: 10.1038/leu.2009.198
87. Mosse YP, Voss SD, Lim MS, Rolland D, Minard CG, Fox E, et al. Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: a children's oncology group study. J Clin Oncol. (2017) 35:3215–21. doi: 10.1200/JCO.2017.73.4830
Keywords: PTCL, AITL, ALCL, PTCL-NOS, genomics, personalized medicine, diagnosis, management
Citation: Zhang Y, Lee D, Brimer T, Hussaini M and Sokol L (2020) Genomics of Peripheral T-Cell Lymphoma and Its Implications for Personalized Medicine. Front. Oncol. 10:898. doi: 10.3389/fonc.2020.00898
Received: 04 February 2020; Accepted: 07 May 2020;
Published: 19 June 2020.
Edited by:
Anjali Mishra, Sidney Kimmel Cancer Center, United StatesReviewed by:
Javeed Iqbal, University of Nebraska Medical Center, United StatesStefano Aldo Pileri, University of Bologna, Italy
Copyright © 2020 Zhang, Lee, Brimer, Hussaini and Sokol. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lubomir Sokol, bHVib21pci5zb2tvbEBtb2ZmaXR0Lm9yZw==
 Yumeng Zhang
Yumeng Zhang Dasom Lee1
Dasom Lee1 Thomas Brimer
Thomas Brimer Mohammad Hussaini
Mohammad Hussaini Lubomir Sokol
Lubomir Sokol