- Hepatic Surgery Center, Tongji Medical College, Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China
Circular RNAs (circRNAs), which act as initiators and promoters of various diseases, were thought to be mostly noncoding RNAs (ncRNAs) in eukaryotes, until recent studies confirmed that some circRNAs have the function of encoding proteins. Accumulating research findings have proved that dysregulation of circRNAs is associated with the developmental process of multiple cancers. circHIPK3, an example of circRNA, is frequently expressed in many diseases, such as diabetes, age-related cataract, idiopathic pulmonary fibrosis, preeclampsia, osteoblasts, and retinal vascular dysfunction, leading to disease development and progression. In addition, circHIPK3 may also serve as a potential biomarker, to help us know more about the rules of occurrence and development of cancers. In recent studies, many circHIPK3-related cancers have been identified, including nasopharyngeal carcinoma, gallbladder cancer, lung cancer, hepatocellular carcinoma, osteosarcoma, glioma, colorectal cancer, ovarian cancer, bladder cancer, prostate cancer, gastric cancer, oral squamous cell carcinoma, and chronic myeloid leukemia. This review summarizes recent studies on the biological mechanisms of circHIPK3 and expounds the molecular mechanisms of circHIPK3 in these malignant tumors.
Introduction
The proportion of protein-coding genes in the human genome is ~2%. Non-protein-coding sequences of the human genome account for over 70% of the noncoding RNAs (ncRNAs) (1–3). Recent studies have shown that ncRNAs act as a necessary element in many kinds of biological processes, including post-transcriptional regulation, epigenetics, chromatin modification, and the cell cycle (4, 5). It is increasingly being reported that ncRNAs also affect human cancers and disease progression (6). ncRNAs are subdivided into small (≤ 200 nucleotides) and long (> 200 nucleotides) (7, 8).
The lengths of circular RNAs (circRNAs) are ~200–2,000 bp, with most being around 500 bp. circRNAs are a class of endogenous RNAs that are transcribed by RNA polymerase II (9). They are divided into noncoding and coding circRNAs. Coding circRNAs were found to have an open reading box, which generally contains at least one internal ribosome entry site (10). The closed loop structure can be formed by a back-splicing mechanism, in which an upstream 5′ splice site is linked with a downstream 3' splice site, in a linear mRNA; thus, the regulation of circRNA biogenesis may be influenced by canonical spliceosomal signals (11–14) (Figure 1). circRNAs can be produced from many genomic positions, such as exons, 3′ UTRs, 5′ UTRs, antisense RNAs, intergenic regions, and intronic regions (15, 16). Exonic circRNAs (ecircRNAs), which make up 80% of identified circRNAs, are mainly derived from one or more exons (14, 17, 18). Lariat-driven circularization is one proposed hypothesis for the formation of ecircRNA. The precursor mRNA (pre-mRNA) forms a lariat intermediate, composed of numerous exons and introns (19, 20). The introns are excised, and then the closed loop structure is generated through the “head-to-tail” joining of the upstream 3′ splice site to the downstream 5′ splice site (21, 22). The generation of exon-intron circRNAs (EIciRNAs) occurs, in some cases, when the introns between exons are not spliced out completely (21, 23, 24). Another mechanism of formation of circRNA is intron pairing-driven circularization, which can form a circular structure via the base-pairing of introns flanking inverted repeats. Transfer RNA (tRNA) intronic circular RNAs (tricRNAs) can be generated after removing the excised tRNA introns from tRNA precursors by splicing enzymes and ligating the intron termini (25–27). In addition, RNA-binding proteins (RBPs) cause a difference in circRNA biogenesis, by acting as trans-factors (28). Circular intronic RNAs (ciRNAs) depend on conserved sequences at both ends of the intron and are generated from intron lariats. These conserved sequences can help introns avoid branching, and form circular structures (29).
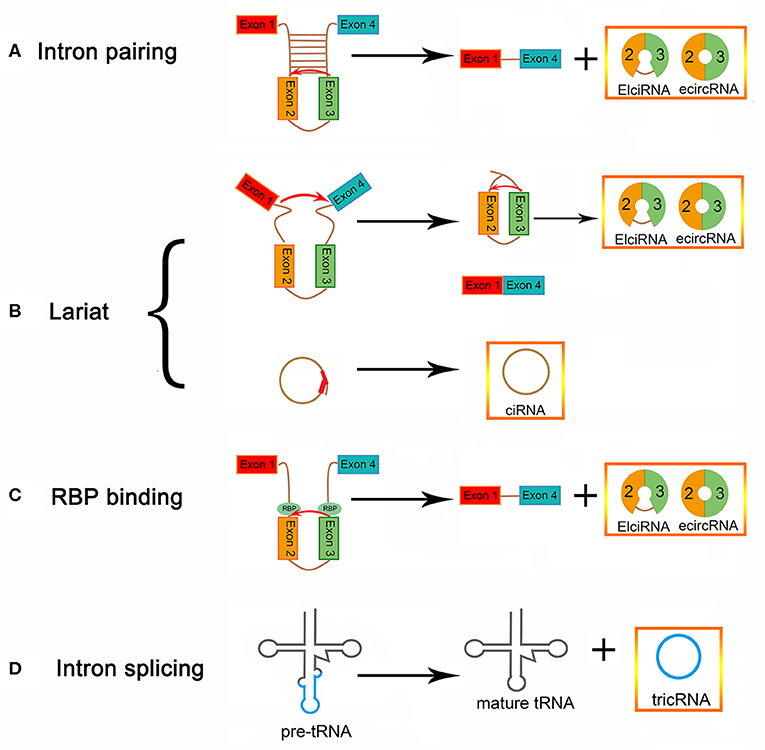
Figure 1. circRNA biogenesis (A) EIciRNA biogenesis and ecircRNA biogenesis via intron pairing-driven circularization. (B) EIciRNA, ecircRNA, and ciRNA biogenesis via lariat-driven circularization. (C) EIciRNA biogenesis via RBP-associated pairing-driven circularization. (D) TricRNA biogenesis via intron splicing from pre-tRNA.
Multiple studies have revealed that RNA interference could target circRNAs; thus, circRNAs might guide the specific regulation of miRNA-targeted genes by acting as competitive endogenous RNAs (ceRNAs) and miRNA sponges (30, 31). As shown in previous studies, some ecircRNAs are distributed in the cytoplasm and combine with miRNAs to behave as miRNA sponges, to control their function, and the others can interfere with the RNA replication of viruses and viroids (32). However, ciRNAs and EIciRNAs located in the nucleus cannot act as miRNA sponges but can influence gene expression and transcription, by such as cis- or trans-activation (33). Thus, circRNA may regulate protein production and function by binding, storing, sorting, and sequestering proteins to appropriate subcellular locations and may modulate protein-protein interactions (34). In addition, circRNAs can bind to RNA-binding proteins (RBP), in competition with their parent mRNAs, to affect the translation process. Many recent studies have used RNA-seq to analyze the biological functions of circRNAs and have identified various circRNAs, in many model organisms, with different cell types (35, 36). RNase R is the exonuclease of circRNAs, but circRNAs lacking 3′ termini cannot usually be degraded by RNase R, and possess greater stability than the related linear mRNAs. Thus, the concentration of circRNAs in quiescent and postmitotic cells is higher than that of linear mRNAs (37). In addition, genome-wide analyses suggest that circRNAs are abundant in human body fluids, with a highly conserved evolution, and the expression of cell-type-specific circRNAs indicates that they may be regarded as potential biomarkers for predicting disease progression and prognosis (38, 39).
In this review, we characterize one abundant circRNA, produced from exon 2 of the homeodomain-interacting protein kinase 3 (HIPK3) gene, circHIPK3, which has been reported in many recent studies (40). HIPK3, Gen Bank Accession ID NM_005734.5, is located on chromosome 11p13 and comprises 7,551 base pairs (Figure 2). The genome sequence indicates that the second exon (1,099 bp) from HIPK3, plus the long introns at both ends of the gene combine to make the structure of circHIPK3 (41). circHIPK3 is the dominant circRNA isoform, as evidenced by a mass of back-spliced unique reads (14, 41). The formation of circHIPK3 requires long flanking introns with complementary Alu repeats (19, 42). circHIPK3 is mostly located in the cytoplasm and is commonly expressed in diverse tissues, such as the lung, heart, stomach, colon, and brain (41). Several biological and pathological processes, beyond cancer, are related to the dysregulated expression of circHIPK3. This review summarizes the aberrant expression and molecular mechanism of circHIPK3 in human cancers, and addresses its clinical significance.
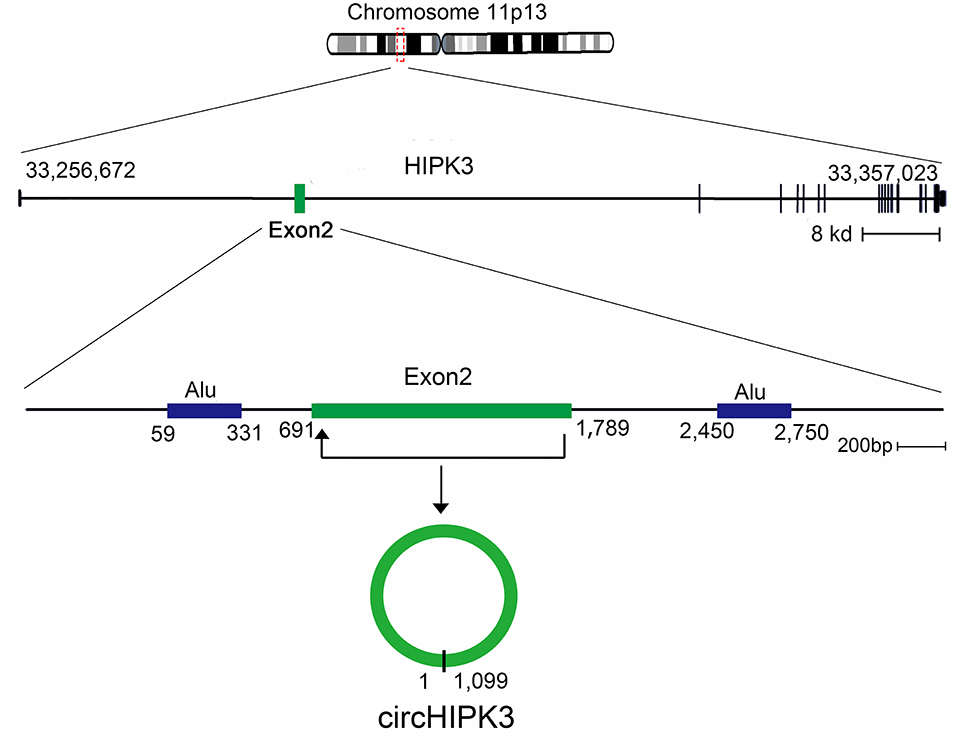
Figure 2. Generation of circHIPK3. HIPK3 is located on chromosome 11p13, and the genomic regions of HIPK3 exon 2 contain flanking Alu repeats and long introns. HIPK3 yields an abundant 1,099 nt circular RNA comprised solely of exon 2.
The Function of circHIPK3 in Cancers
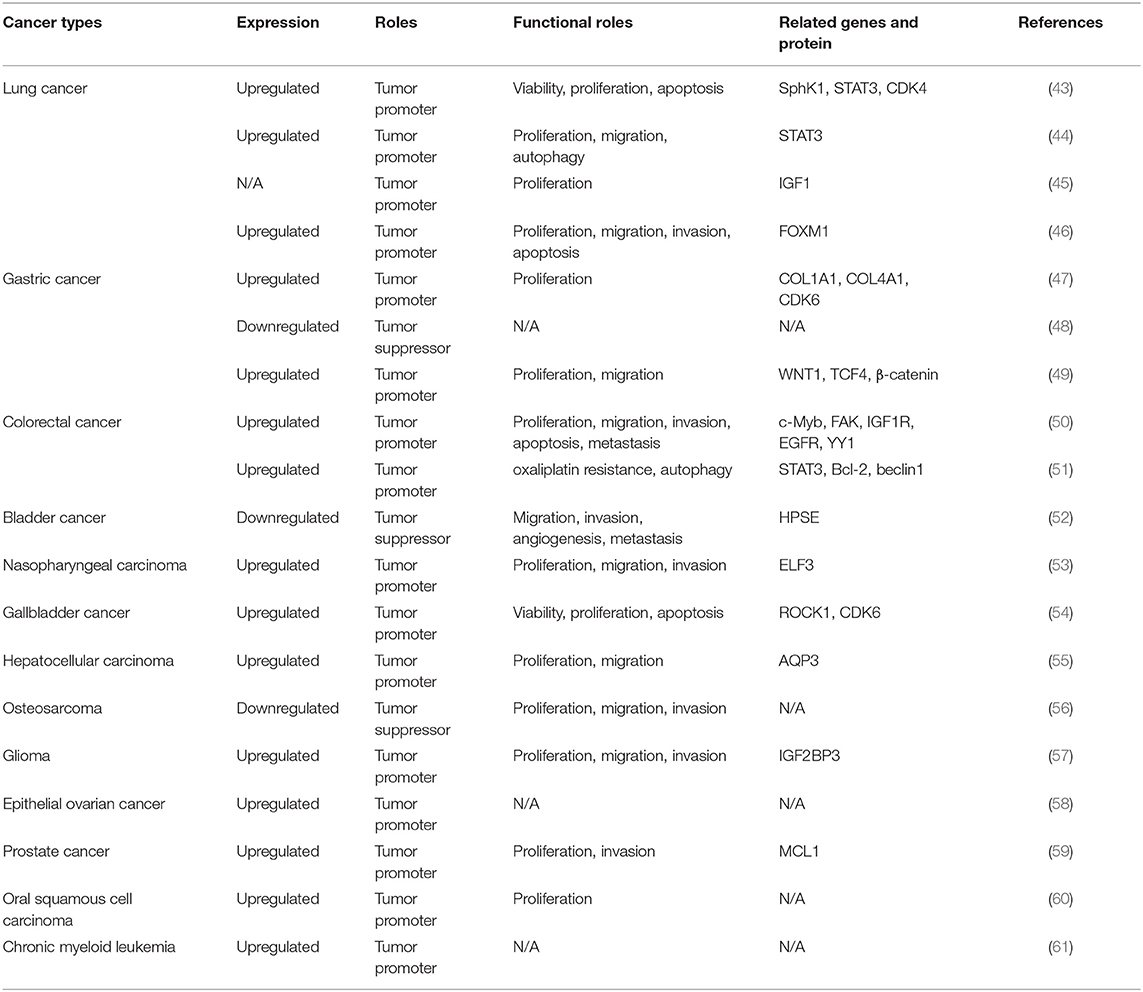
Lung Cancer
According to the morphology of tumor cells, lung cancer is classified into two categories: small cell lung cancer (SCLC) and non-SCLC (NSCLC). Chen et al. noted that circHIPK3 is an important autophagy regulator in STK11 mutant lung cancer, by regulating miR-124-3p expression (Figure 3, Table 1) (44). Knockdown of circHIPK3 significantly induced cell autophagy, while cell proliferation, migration, and invasion were impaired. In addition, transfection with an miR-124-3p mimic or abrogation of STAT3 also induced autophagy. They demonstrated that STAT3 is the downstream target of miR-124-3p, which influences cell autophagy via the PRKAA/AMPKa pathway. The researchers observed that a high ratio of circHIPK3 to linHIPK3 was a sign of poor prognosis, especially in patients with advanced NSCLC (44). The proliferation of NSCLC cells was investigated by Tian et al. (45). Significantly, overexpression of circHIPK3 accelerated the proliferation of NSCLC cells, and interference of circHIPK3 inhibited it. Moreover, miR-379 can be directly combined with circHIPK3 and IGF1 mRNA. Overexpression of circHIPK3 increased the expression of IGF1, and downregulation of IGF1 was observed with interference of circHIPK3 (45). Furthermore, miR-379 mimics restored the upregulated expression and cell proliferation phenotype induced by IGF1, in a cell line stably expressing circHIPK3. Thus, circHIPK3 increased the expression of IGF1 by reducing miR-379, to accelerate NSCLC cell proliferation (45). In addition to these findings, Lu et al. indicated that miR-149 expression was increased by circHIPK3 silencing. Viability and proliferation of NSCLC cells decreased with miR-149 overexpression. The function of NSCLC cells was regulated by miR-149-mediated FOXM1 expression (46). In the study of Yu et al., circHIPK3 overexpression promoted proliferation in lung cancer cell line A549. The researchers reported that the expression level of circHIPK3 was higher in human lung cancer cells and tissues than in normal tissues. They showed that miR-124 was accumulated upon silencing circHIPK3, induced tumor cell apoptosis, and alleviated tumor cell proliferation. Further studies in tumor tissues demonstrated that downstream target genes (Sphk1, STAT3, and CDK4) were upregulated, suggesting the possibility of identifying therapeutic targets from this axis (43).
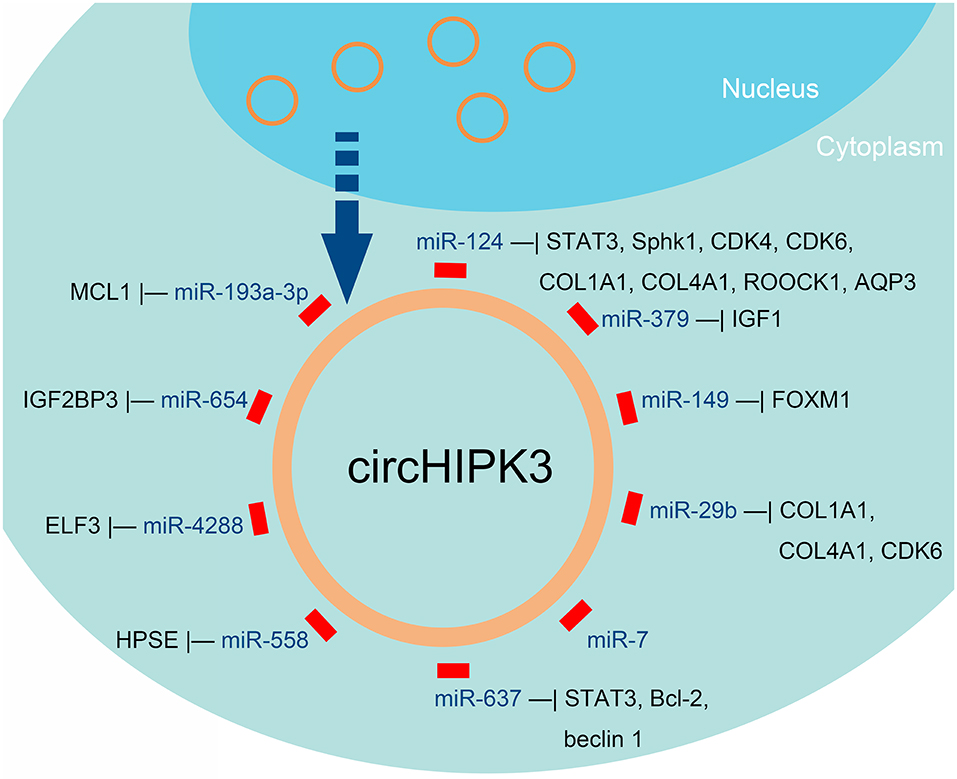
Figure 3. miRNAs that can be sponged by circHIPK3, according to published reports about human cancers.
Gastric Cancer
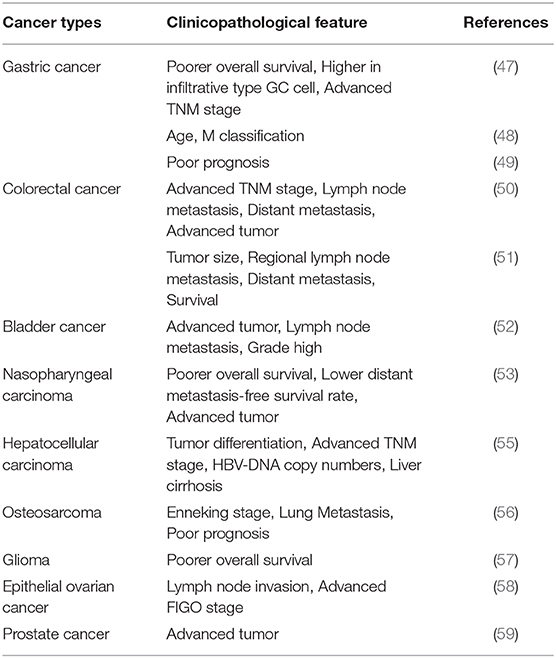
The function of circHIPK3 in gastric cancer (GC) was investigated by Cheng et al. (47). The level of circHIPK3 was significantly higher in GC tissues than in paired, adjacent normal tissues. In addition, the expression of circHIPK3 in infiltrative-type GC cells was higher than that in expanding-type GC cells, suggesting that circHIPK3 expression is related to aggressive clinical factors in GC. GC cell proliferation was inhibited by circHIPK3 knockdown and promoted by circHIPK3 overexpression. The researchers performed bioinformatic analyses and identified COL1A1, COL4A1, and CDK6 as common targets in the circHIPK3-miR-124/miR-29b axes (47). Ghasemi et al. investigated the clinical significance of circHIPK3 (48). Its expression was downregulated in GC tumoral tissues and was significantly correlated with various clinicopathological parameters. However, Liu et al. found that circHIPK3 levels were higher in GC tissues than in paracancerous tissues (49). Similarly, its level was higher in GC cells than in normal cells. In addition, an inverse correlation between circHIPK3 expression level and overall survival of GC patients was found. Silencing of circHIPK3 weakened the proliferative and migratory abilities of GC cells (49). Additionally, circHIPK3 knockdown markedly downregulated the levels of WNT1, TCF4, and β-catenin (49). circHIPK3 promoted tumor cell proliferation and migration through the Wnt/β-catenin signaling pathway, which is a classic proliferation-related pathway (62). Their results also showed that upregulation of circHIPK3 indicated poor prognosis in patients with GC (49).
Colorectal Cancer
As a malignancy of the digestive tract, colorectal cancer (CRC) is a widespread disease with high incidence and fatality rate and is a significant threat to human health (63). Experimental induction of highly expressed circHIPK3 in vivo and in vitro was first carried out in 2018 (50). These data indicated that the c-Myb/circHIPK3/miR-7 axis might be a therapeutic target against CRC. The results proved that circHIPK3 expression in CRC was significantly higher than in normal tissues and that increased circHIPK3 expression indicates poor prognosis. Downregulation of circHIPK3 suppressed cell invasion, proliferation, and migration, and induced apoptosis in CRC cells. Moreover, c-Myb, an upstream transcription factor, elevated circHIPK3 expression by directly binding to its promoter region. Furthermore, circHIPK3 can sponge miR-7 in CRC cell lines and upregulate the expression of oncogenes (FAK, IGF1R, EGFR, and YY1) to promote CRC progression (50). Recent studies have found that the main cause of recurrence in CRC patients is resistance to oxaliplatin-based chemotherapy (51, 64). Aiming at the miR-637/STAT3/Bcl-2/beclin1 axis, circHIPK3 in CRC cells functioned as a chemoresistant gene. Therefore, circHIPK3 might be used as a prognostic prediction for oxaliplatin-based chemotherapy in CRC patients (51). The transcriptomic data from the Gene Expression Omnibus database (GSE116589) revealed that circHIPK3 is highly expressed in CRC tissues and exosomes from CRC patients (65, 66).
Bladder Cancer
circHIPK3, also known as bladder cancer-related circular RNA-2 (BCRC-2), was found to be downregulated in bladder cancer and negatively correlated with cancer grade, invasion, and lymph node metastasis (52, 67). circHIPK3 overexpression efficiently inhibited cell migration, invasion, and angiogenesis in vitro, while cell growth and metastasis were suppressed in vivo. miR-558 can bind with circHIPK3 at two critical sites and is sponged abundantly to suppress heparanase (HPSE) expression (52).
Nasopharyngeal Cancer
Ke et al. demonstrated that the expression of circHIPK3 was higher in nasopharyngeal cancer (NPC) tissues and cells, a common malignant tumor occurring in the head and neck region, than in normal tissues and cells (53). circHIPK3 overexpression enhanced tumor cell proliferation and increased colony numbers, and circHIPK3 silencing dramatically inhibited the migration and invasion of NPC cells, suggesting that circHIPK3 exerts an oncogenic role. The effect of miR-4288 on the expression of E74-like ETS transcription factor 3 (ELF3) was counteracted by overexpression of circHIPK3 overexpression, but not by mutant circHIPK3. In addition, the level of ELF3 in NPC tissues increased with an increase in circHIPK3 expression. This study demonstrated that circHIPK3 upregulated ELF3 expression by reducing the levels of miR-4288, and the regulatory circHIPK3-miR-4288-ELF3 axis plays a critical role in NPC progression (53).
Gallbladder Cancer
Kai et al. showed that the level of circHIPK3 in human gallbladder cancer (GBC) was higher than that in gallbladder epithelial cells (54). Cell apoptosis was induced when circHIPK3 silencing, by siRNA, downscaled the survival and proliferation of GBC cells. They found that the levels of miR-124 targets (ROCK1 and CDK6) increased upon circHIPK3 inhibition of the activity of miR-124 in GBC cells. Moreover, the upregulation of ROCK1-CDK6 and downregulation of miR-124 were correlated with circHIPK3 in GBC tissues (54). Notably, CDK4 and CDK6 are involved in the Ras/Raf/MAPK pathway, which transfer signals from the extracellular environment to the nucleus. This signaling pathway plays a key role in the occurrence and development of malignant tumors, such as by stimulating angiogenesis via changing the expression of genes involved in neovascularization, cell cycle regulation, integrin signaling, and cell migration (68). Hence, circHIPK3 may affect the progression of GBC through the Ras/Raf/MAPK pathway (54).
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is a fatal carcinoma, with nearly 6 × 105 deaths each year (69). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analyses validated that the expression of miR-124 was decreased in HCC tissues, whereas adjacent nontumor liver tissues had normal miR-124 expression (55). Chen et al. (55) demonstrated that miR-124 inhibited the proliferation and migration of HCC cells. In addition, the 3′ UTR of aquaporin 3 (AQP3) contained an miR-124 binding motif. Elevating the expression of AQP3 promoted tumor proliferation and migration. circHIPK3 knockdown decreased HCC cell proliferation and migration rate, which was rescued by an miR-124 inhibitor. Also, inhibition of miR-124 overcame the inhibition of AQP3 by circHIPK3. circHIPK3 knockdown suppressed Huh7 xenograft tumor growth and results indicated that circHIPK3 sponged miR-124 to regulate AQP3 expression in HCC (55).
Osteosarcoma
Studies of circHIPK3 in bone cancers have focused on osteosarcoma (OS), which is more common in children and adolescents (70). Ma et al. (56) found that circHIPK3 expression was downregulated in OS tissues and plasma samples, which is associated with poor prognosis. Functional analysis revealed that, in OS cells, proliferation, migration, and invasion were significantly suppressed when circHIPK3 was overexpressed in vitro (56).
Glioma
Glioma originates from the neuroepithelium of the brain, accounting for the majority of intracranial tumors (71). Jin et al. found that circHIPK3 was upregulated in glioma tissues, and higher levels of circHIPK3 were associated with worse prognosis. A series of experiments showed that circHIPK3 accelerated tumor cell proliferation, invasion, and metastasis in vivo. Moreover, circHIPK3 promoted IGF2BP3 expression by targeting miR-654 in glioma cells. Overexpression of IGF2BP3 reversed the effects of circHIPK3 depletion (57).
Ovarian Cancer
Ovarian cancer (OC) is a type of gynecological malignancy that causes massive mortality worldwide, and the pathology of most OC is epithelial ovarian cancer (EOC) (72). It was demonstrated using qRT-PCR analysis that the expression level of circHIPK3 is significantly increased in EOC tissue samples over matched, adjacent noncancerous tissues (58). The experimental results indicated that higher lymph invasion rates and shorter survival times were positively correlated with circHIPK3 expression in EOC patients (58).
Prostate Cancer
Chen et al. indicated that circHIPK3 overexpression was associated with prostate cancer (PCa) malignancy and poor prognosis (59). They noted that downregulation of circHIPK3 significantly inhibited malignant phenotypes, including proliferation, migration, and invasion in vitro, and tumor growth in vivo. They found that miR-193a-3p functioned as a tumor suppressor in PCa cells, through bioinformatic prediction and biotin-coupled miRNA capture studies. Functional studies showed that miR-193a-3p reduced MCL1 expression, to suppress proliferation, migration, and invasion in cancer cells (59).
Oral Squamous Cell Carcinoma
Wang et al. explored the expression and clinical applications of circHIPK3 in oral squamous cell carcinoma (OSCC) and the effect of miR-124 expression, regulated by circHIPK3, on OSCC cell proliferation (60). The circHIPK3 expression in OSCC tissues was significantly higher than that in paracancerous tissues. The expression of circHIPK3 in cancer tissues at different clinical stages and with varying degrees of cell differentiation was observed to be statistically different. The proliferation ability of OSCC cells was significantly inhibited by reducing the expression of circHIPK3. Moreover, miR-124 expression in OSCC tissues was significantly lower than that in paracancerous tissues. There was a negative correlation between circHIPK3 and miR-124 expression in OSCC tissues. Taken together, downregulating the expression level of circHIPK3, which is upregulated in OSCC, inhibited the proliferation of OSCC cells, and circHIPK3 contributed to the occurrence and development of OSCC by regulating the expression of miR-124 (60).
Chronic Myeloid Leukemia
Feng et al. investigated the expression of circHIPK3 in chronic myeloid leukemia (CML). They found that circHIPK3 was significantly upregulated in peripheral blood mononuclear cells and serum samples from patients with CML compared with those from healthy controls. High circHIPK3 expression predicted poor outcomes in patients with CML. Further loss-of-function experiments suggested an oncogenic role of circHIPK3 in CML (61).
The Function of circHIPK3 in Cancers
Clinical Features of circHIPK3
As mentioned above, circHIPK3 is able to block circularization, like other circRNAs, which allows it to escape the actions of exonucleases. Therefore, circHIPK3 can exist stably in cells for a long time (6). It is precisely because of its stability that we can easily determine whether circHIPK3 can be a potential biomarker molecule for clinical diagnosis (Table 2).
circHIPK3 has been shown to be closely related to multiple clinical features, through the study of bioinformatics, statistics, and various types of cancer patient. In solid tumors, high expression of circHIPK3 often represents larger tumor mass and volume. More advanced tumor morphology is often found in cancer tissues that highly expressed circHIPK3. Distant metastasis and lymph node metastasis often represent poor prognosis, which is usually observed in patients with high circHIPK3 expression, suggesting that reduced overall survival is related to circHIPK3 in gastric cancer, colorectal cancer, bladder cancer, nasopharyngeal carcinoma, hepatocellular carcinoma, osteosarcoma, glioma, epithelial ovarian cancer, prostate cancer, and so on. At the same time, increased differentiation and advanced TNM stages are associated with abnormal expression of circHIPK3. In addition, Zhang et al. also noted a relationship between circHIPK3 and resistance to chemotherapy drugs, such as the relationship between oxaliplatin resistance and circHIPK3, thus establishing a causative link. These findings suggest that circHIPK3 is a promising target in cancer therapy (51).
The Function of circHIPK3 in Other Diseases
The function of circHIPK3 has also been reported in many other diseases. circHIPK3's role in diabetes was the first to be investigated. Stoll et al. suggested that circHIPK3 is a novel regulator of β-cell activities, and that β-cell dysfunction is an essential hallmark of diabetic conditions (73). They showed that circHIPK3 acts by sequestering miR-124-3p and miR-338-3p to regulate the expression of crucial target genes, including Slc2a2, Akt1, and Mtpn. Cao et al. showed that the circHIPK3-miR-124 pathway participated in high glucose (HG)-induced endothelial cell injury (74). circHIPK3 was downregulated by the accumulation of miR-124 in both HG-treated human umbilical vein endothelial cells and primary aortic endothelial cells, from diabetic patients. According to research on type 2 diabetes patients, by Wang et al., a positive association was shown between circHIPK3 expression and the grade of neuropathic pain (75). They demonstrated that neuropathic pain could be alleviated in diabetic rats when circHIPK3 was silenced. Further mechanistic studies illustrated that circHIPK3 negatively regulated the expression of miR-124. As additional proof, in diabetic rats with circHIPK3 knockdown, inhibition of miR-124 would mediate the moderation of neuropathic pain, thereby inhibiting neuroinflammation (75).
George et al. (76) suggested that circHIPK3 may regulate human lens epithelial cell (HLEC) function through miR-193a-mediated crystallin alpha A (CRYAA) expression. They found that circHIPK3 was significantly downregulated in all subtypes of age-related cataract. The silencing of circHIPK3 significantly accelerated apoptosis upon oxidative stress and decreased cell viability and proliferation (76). Meanwhile, circHIPK3 knockdown inhibited epithelial-mesenchymal transition and increased miR-193a expression. miR-193a reduced the mRNA and protein levels of CRYAA by targeting the binding sites in its 3′ UTR. Furthermore, miR-193a decreased the viability, promoted proliferation, and increased the apoptosis of HLECs under conditions of oxidative stress (77).
Zhang et al. illustrated that circHIPK3 silencing could improve idiopathic pulmonary fibrosis and inhibit fibroblasts (78). Substantial evidence was found that circHIPK3 increases SOX4 and COL1A1 expression by inhibiting miR-338-3p activity, as an endogenous sponge. Furthermore, these results revealed that, in vitro, circHIPK3 plays a critical role in the emergence of TGF-β-induced myofibroblasts, predominantly through the fibroblast-to-myofibroblast transition (FMT). From this research, the correlation between the circHIPK3-miR-338-3p/SOX4/COL1A1 network and FMT regulation was revealed (78).
Zhang et al. found that circHIPK3 was decreased in patients with preeclampsia and affected biological processes such as the migration and invasion of human trophoblast cells (79). The levels of circHIPK3 detected in preeclampsia were significantly lower than those in the normal pregnancy group. Silencing circHIPK3 inhibited cell migration, invasion, proliferation, and tubular formation, but circHIPK3 overexpression effectively promoted these abilities in addition to promoting cell proliferation (79).
Dexamethasone (DEX) exerts cytotoxicity to osteoblasts by inducing apoptosis and necrosis. Zhu et al. found that circHIPK3 expression was downregulated in DEX-treated human osteoblasts and necrotic femoral head tissues (80). Overexpression of circHIPK3 suppressed DEX-induced apoptosis and programmed necrosis, and knockdown of circHIPK3 enhanced DEX-induced cytotoxicity in osteoblasts. They found that miR-124 could induce cytotoxicity by DEX in human osteoblasts. Conversely, the DEX-induced actions were attenuated when miR-124 was sponged by circHIPK3 (80).
Conclusions and Prospects for Future Research
Several studies have validated that circRNAs regulate gene expression and are closely related to tumorigenesis and tumor progression, in a complicated gene regulatory network. In this review, we provide an overview of many new characterizations of circHIPK3 that may deepen our understanding of how it regulates the process of tumor development. In addition, the relationship between circHIPK3 and clinical features was also examined. Various experiments have demonstrated that the expression of circHIPK3 might be a marker of accelerated tumor progression, both in vitro and in vivo. circHIPK3 expression was related to increased tumor volume of tumors, advanced TNM stage, shortened survival, and metastasis. Meanwhile, circHIPK3 has potent effects in controlling the cell cycle, influencing cell proliferation, regulating apoptosis, promoting invasion, and managing migration in cancers. A new target could be provided by circHIPK3 for diagnosing and treating different types of cancer. Moreover, a growing number of studies have demonstrated the mechanisms of circHIPK3 in several pathways, such as the Wnt/β-catenin signaling pathway. It is well known that there is competition between circHIPK3 and miRNAs, which results in the inhibition of the downstream genes of those miRNAs.
However, findings on the role of circHIPK3 are limited; further experiments are needed to explore the function of circHIPK3 and its pathway, in a range of human malignancies. Due to technical limitations, research has focused on the role of circHIPK3 as a miRNA sponge, and its other functions need further study. In addition, the mechanism of circHIPK3 in other diseases may provide new ideas and considerations for future research on malignant cancers. In addition to C-MYB, there may be other significant genes regulating the expression of circHIPK3, which represents a future direction of study. It is worth noting that, because of the complex mechanisms associated with circHIPK3, it may play an opposite role in different cancers, and within the same tumor, with circHIPK3 promoting the development of some tumors, while inhibiting the proliferation and growth
Author Contributions
JW and JLiao collected the related paper. JW, JLiao, JLian, and LC drafted and revised the manuscript. JW and LC designed the review. BZ and XC participated in the design of the review and helped to draft and revise the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Fund (31671348, 31301064).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All authors thank Dr. Zhao Huang for his valuable suggestions.
Abbreviations
ncRNAs, Noncoding RNAs; circRNAs, circular RNAs; ecirRNAs, exonic circRNAs; ElciRNAs, exon-intron circRNAs; tRNA, transfer RNA; tricRNAs, tRNA intronic circRNAs; RBPs, RNA-binding proteins; ciRNAs, circular intronic RNAs; ceRNA, Competitive endogenous RNAs; HIPK3, Homeodomain-interacting protein kinase3; NSCLL, Non-small-cell lung cancer; SCLL, Small-cell-lung cancer; STAT, Signal transducer and activator of transcription 3; PRKAA, Protein kinase AMP-activated catalytic subunit alpha 2; AMPKa, AMP-activated protein kinase alpha subunit; IGF1, Insulin like growth factor 1; FOXM1, Forkhead box M1; SPHK1, Sphingosine kinase 1; CDK4, Cyclin-dependent kinase4; GC, gastric cancer; COL1A1, collagen type I alpha 1 chain; COL4A1, collagen type IV alpha 1 chain; CDK6, Cyclin-dependent kinase6; WNT1, Wnt family member 1; TCF4, Transcription factor 4; CRC, Colorectal cancer; c-Myb, MYB proto-oncogene; FAK, Focal adhesion kinase; IGF1R, Insulin like growth factor 1 receptor; YY1, YY1 transcription factor; Bcl-2, B cell leukemia/lymphoma 2; GEO, Gene Expression Omnibus; BCRC-2, Bladder cancer-related circular RNA-2; HPSE, Heparinase; BC, Bladder cancer; NPC, Nasopharyngeal carcinoma; ELF3, E74-like ETS transcription factor 3; GBC, Gallbladder cancer; ROCK1, Rho associated coiled-coil containing protein kinase 1; MAPK, Mitogen activated kinase-like protein; HCC, Hepatocellular carcinoma; qRT-PCR, quantitative reverse transcription polymerase chain reaction; AQP3, 3′-UTR of aquaporin3; OS, Osteosarcoma; IGF2BP3, Insulin like growth factor 2 mRNA binding protein 3; CCK8, Cell counting kit-8; OC, Ovarian cancer; EOC, Epithelial ovarian cancer; Pca, Prostate cancer; MCL1, Myeloid cell leukemia sequence 1; OSCC, Oral squamous cell carcinoma; CML, Chronic myeloid leukemia; SLc2a2, Solute carrier family 2 member 2; AKT1, AKT serine/threonine kinase 1; Mtpn, Myotrophin; HG, High glucose; HLEC, Human lens epithelial cell; CRYAA, Crystallin alpha A; 3′-UTR, 3′-untranslated region; SOX4, SRY-box transcription factor 4; TGF-β, Transforming growth factor-β; FMT, Fibroblast-to-myofibroblast transition; DEX, Dexamethasone; OS, Overall Survival.
References
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
2. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. doi: 10.1038/nrg3074
3. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. (2019) 18:6. doi: 10.1186/s12943-018-0934-6
4. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
5. Fan H, Tian H, Cheng X, Chen Y, Liang S, Zhang Z, et al. Aberrant Kank1 expression regulates YAP to promote apoptosis and inhibit proliferation in OSCC. J Cell Physiol. (2020) 235:1850–65. doi: 10.1002/jcp.29102
6. Liang H, Yu T, Han Y, Jiang H, Wang C, You T, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. (2018) 17:119. doi: 10.1186/s12943-018-0870-5
7. He X, Bao W, Li X, Chen Z, Che Q, Wang H, et al. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. (2014) 33:325–32. doi: 10.3892/ijmm.2013.1570
8. Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. (2014) 9:3–12. doi: 10.4161/epi.27473
9. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
10. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. (2019) 110:3630–8. doi: 10.1111/cas.14211
11. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. (2016) 17:205–11. doi: 10.1038/nrm.2015.32
12. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
13. Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. (2019) 18:35. doi: 10.1186/s12943-019-0996-0
14. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
15. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. (2016) 26:1277–87. doi: 10.1101/gr.202895.115
16. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
17. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
18. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
19. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
20. Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. (2018) 48:173–84. doi: 10.1159/000491716
21. Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers. (2018) 10:258. doi: 10.3390/cancers10080258
22. Xu S, Zhou L, Ponnusamy M, Zhang L, Dong Y, Zhang Y, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. (2018) 6:e5503. doi: 10.7717/peerj.5503
23. Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. (2017) 404:8–18. doi: 10.1016/j.canlet.2017.07.002
24. Cao LH, Li HT, Lin WQ, Tan HY, Xie L, Zhong ZJ, et al. Morphine, a potential antagonist of cisplatin cytotoxicity, inhibits cisplatin-induced apoptosis and suppression of tumor growth in nasopharyngeal carcinoma xenografts. Sci Rep. (2016) 6:18706. doi: 10.1038/srep18706
25. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
26. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. (2016) 1859:1245–51. doi: 10.1016/j.bbagrm.2016.07.009
27. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
28. Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. (2012) 149:1393–406. doi: 10.1016/j.cell.2012.04.031
29. Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. (2006) 40:47–76. doi: 10.1146/annurev.genet.40.110405.090625
30. Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. (2017) 18:547–57. doi: 10.1093/bib/bbw045
31. Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. (2017) 10:2. doi: 10.1186/s13045-016-0370-2
32. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
33. Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. (2014) 24:1774–86. doi: 10.1101/gr.177790.114
34. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. (2017) 7:4183–91. doi: 10.7150/thno.21299
35. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. (2016) 17:679–92. doi: 10.1038/nrg.2016.114
36. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
37. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. (2016) 143:1838–47. doi: 10.1242/dev.128074
38. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. (2017) 8:10.1002/wrna.1386. doi: 10.1002/wrna.1386
39. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. (2019) 18:90. doi: 10.1186/s12943-019-1002-6
40. Xie Y, Yuan X, Zhou W, Kosiba AA, Shi H, Gu J, et al. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: review. Life Sci. (2020) 1:117252. doi: 10.1016/j.lfs.2019.117252
41. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. doi: 10.1038/ncomms11215
42. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. (2014) 28:2233–47. doi: 10.1101/gad.251926.114
43. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. (2018) 506:455–62. doi: 10.1016/j.bbrc.2018.10.087
44. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. (2019) 16:659–71. doi: 10.1080/15548627.2019.1634945
45. Tian F, Wang Y, Xiao Z, Zhu X. [Circular RNA CircHIPK3 promotes NCI-H1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1]. Zhongguo Fei Ai Za Zhi. (2017) 20:459–67. doi: 10.3779/j.issn.1009-3419.2017.07.04
46. Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. (2019) 21:113–21. doi: 10.1080/15384047.2019.1669995
47. Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. (2018) 16:216. doi: 10.1186/s12967-018-1582-8
48. Ghasemi S, Emadi-Baygi M, Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. (2019) 49:687–95. doi: 10.3906/sag-1806-50
49. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. (2019) 23:7905–12. doi: 10.26355/eurrev_201909_19004
50. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. (2018) 9:417. doi: 10.1038/s41419-018-0454-8
51. Hao S, Lv J, Yang Q, Wang A, Li Z, Guo Y, et al. Identification of key genes and circular RNAs in human gastric cancer. Med Sci Monit. (2019) 25:2488–504. doi: 10.12659/MSM.915382
52. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. (2017) 18:1646–59. doi: 10.15252/embr.201643581
53. Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. (2019) 234:1699–706. doi: 10.1002/jcp.27041
54. Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. (2018) 503:863–9. doi: 10.1016/j.bbrc.2018.06.088
55. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. (2018) 9:175. doi: 10.1038/s41419-017-0204-3
56. Ma X-L, Zhu K-P, Zhang C-L. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. (2018) 9:1856–62. doi: 10.7150/jca.24619
57. Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. (2018) 503:1570–4. doi: 10.1016/j.bbrc.2018.07.081
58. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. (2018) 22:3713–8. doi: 10.26355/eurrev_201806_15250
59. Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. (2019) 11:1415–23. doi: 10.2147/CMAR.S190669
60. Wang J, Zhao S, Ouyang S, Huang Z, Luo Q, Liao L. Circular RNA circHIPK3 acts as thespongeof microRNA‘124 topromotehuman oralsquamouscell carcmoma cellsproliferation. Chin J Stomatol. (2018) 53:546–51. doi: 10.3760/cma.j.issn.1002-0098.2018.08.009
61. Feng XQ, Nie SM, Huang JX, Li TL, Zhou JJ, Wang W, et al. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. (2020) 67:171–7. doi: 10.4149/neo_2018_181129N908
62. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. (2017) 169:985–99. doi: 10.1016/j.cell.2017.05.016
63. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. (2014) 383:1490–502. doi: 10.1016/S0140-6736(13)61649-9
64. Harper KL, McDonnell E, Whitehouse A. CircRNAs: from anonymity to novel regulators of gene expression in cancer (Review). Int J Oncol. (2019) 55:1183–93. doi: 10.3892/ijo.2019.4904
65. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. (2002) 30:207–10. doi: 10.1093/nar/30.1.207
66. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. (2013) 41(Database issue):D991–5. doi: 10.1093/nar/gks1193
67. Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. (2018) 414:301–9. doi: 10.1016/j.canlet.2017.11.022
68. Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. (2006) 1:7–9. doi: 10.1097/01243894-200601000-00004
69. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
70. Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. (2017) 8:59417–34. doi: 10.18632/oncotarget.19727
71. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
72. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. (2016) 66:7–30. doi: 10.3322/caac.21332
73. Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab. (2018) 9:69–83. doi: 10.1016/j.molmet.2018.01.010
74. Cao Y, Yuan G, Zhang Y, Lu R. High glucose-induced circHIPK3 downregulation mediates endothelial cell injury. Biochem Biophys Res Commun. (2018) 507:362–8. doi: 10.1016/j.bbrc.2018.11.041
75. Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun. (2018) 505:644–50. doi: 10.1016/j.bbrc.2018.09.158
76. George AK, Master K, Majumder A, Homme RP, Laha A, Sandhu HS, et al. Circular RNAs constitute an inherent gene regulatory axis in the mammalian eye and brain (1). Can J Physiol Pharmacol. (2019) 97:463–72. doi: 10.1139/cjpp-2018-0505
77. Liu X, Liu B, Zhou M, Fan F, Yu M, Gao C, et al. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/CRYAA axis. Biochem Biophys Res Commun. (2018) 503:2277–85. doi: 10.1016/j.bbrc.2018.06.149
78. Zhang JX, Lu J, Xie H, Wang DP, Ni HE, Zhu Y, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. (2019) 10:182. doi: 10.1038/s41419-019-1430-7
79. Zhang Y, Cao L, Jia J, Ye L, Wang Y, Zhou B, et al. CircHIPK3 is decreased in preeclampsia and affects migration, invasion, proliferation, and tube formation of human trophoblast cells. Placenta. (2019) 85:1–8. doi: 10.1016/j.placenta.2019.07.010
Keywords: circular RNA, cancer, circHIPK3, mRNA splicing, biomarker, miRNA sponge
Citation: Wen J, Liao J, Liang J, Chen X, Zhang B and Chu L (2020) Circular RNA HIPK3: A Key Circular RNA in a Variety of Human Cancers. Front. Oncol. 10:773. doi: 10.3389/fonc.2020.00773
Received: 31 January 2020; Accepted: 21 April 2020;
Published: 15 May 2020.
Edited by:
Kyle Schachtschneider, University of Illinois at Chicago, United StatesReviewed by:
Junming Guo, Ningbo University, ChinaQi Zhao, Sun Yat-sen University Cancer Center (SYSUCC), China
Copyright © 2020 Wen, Liao, Liang, Chen, Zhang and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Chu, liangchu@tjh.tjmu.edu.cn
 Jingyuan Wen
Jingyuan Wen Jingyu Liao
Jingyu Liao Junnan Liang
Junnan Liang Xiao-ping Chen
Xiao-ping Chen Bixiang Zhang
Bixiang Zhang Liang Chu
Liang Chu