
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 05 May 2020
Sec. Women's Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00695
 Xiuming Liu1,2,3†
Xiuming Liu1,2,3† Yuee Teng1,2,3†
Yuee Teng1,2,3† Xin Wu1,2,3
Xin Wu1,2,3 Zhi Li1,2,3
Zhi Li1,2,3 Bowen Bao1,2,3
Bowen Bao1,2,3 Yunpeng Liu1,2,3
Yunpeng Liu1,2,3 Xiujuan Qu1,2,3*
Xiujuan Qu1,2,3* Lingyun Zhang1,2,3*
Lingyun Zhang1,2,3*Background: Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b) is an E3 ubiquitin-protein ligase and a signal-transducing adaptor protein involved in the development and progression of cancer. Despite the known functions of Cbl-b, its role in breast cancer remains unclear. The aim of this study is to explore the prognostic value of Cbl-b in breast cancer.
Methods: Cbl-b expression was analyzed by immunohistochemistry in 292 breast cancer patients from the First Hospital of China Medical University between 1999 and 2008. Kaplan-Meier curve and Cox proportional hazards regression were used to analyze the independent prognostic factors for overall survival (OS) and disease-free survival (DFS). Nomogram was constructed based on these prognostic factors.
Results: Cbl-b expression was detected in 54.1% (158/292) breast cancer tissue samples. Cbl-b expression was correlated with DFS (p = 0.033), but was not significantly associated with the known clinic-pathological factors in this study. Log-rank analysis indicated that Cbl-b expression was correlated with better OS (p = 0.013) and DFS (p = 0.016). Multivariate analysis showed that Cbl-b expression was an independent prognostic factor in breast cancer. The nomogram we built for predicting OS was integrated with Cbl-b expression, age, tumor size, lymph node metastasis and histological grade. Except tumor size, all the above factors and date of diagnosis were used to construct the DFS nomogram. The C-indexes of the nomograms were 0.735 and 0.678, respectively. Our new clinical model was superior to the TNM staging for prediction of OS.
Conclusion: Cbl-b expression independently predicts favorable prognosis in breast cancer. Cbl-b expression, combined with other variables could be more precise clinical predictive models for predicting OS and DFS in patients with breast cancer.
Breast cancer is the most common malignancy and the leading cause of cancer death amongst women worldwide. It was estimated that there were 2.1 million newly diagnosed female breast cancer cases worldwide in 2018, accounting for a quarter of female cancer cases (1). Exploring the prognostic elements of breast cancer may provide clues for developing potential therapeutic targets. Some clinic-pathological features and biological indicators have been reported as prognostic factors for breast cancer patients, including hormone receptor (HR) status, which refers to estrogen receptor (ER) and progesterone receptor (PR) status, human epidermal growth factor receptor2 (HER2) status, histological grade, pathological type, TNM staging, age at diagnosis, marital status and some tumor markers such as CEA, CA15-3 (2–5). However, exploring new prognostic markers and models has high potential to improve outcomes for patients with breast cancer. Our previous study (6) revealed that Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b) might be a prognostic indicator in breast cancer by regulating the signaling pathways and cell proliferation.
It has been proposed that Cbl-b functions as an E3 ubiquitin protein ligase, thereby negatively regulating receptor tyrosine kinase (RTK) signal-transduction pathways and inhibiting tumor growth (7, 8). In addition, Cbl-b protein could regulate cell activation by acting as an adaptor protein. In the immune system, Cbl-b can play key roles in anti-tumor immunity by regulating T (9), B (10), and NK cell (11) activation pathways.
The prognostic value of Cbl-b in different tumors remains controversial. It has been reported that Cbl-b expression can predict poor prognosis in resectable pancreatic ductal adenocarcinoma (12). Conversely, our team has reported that Cbl-b can act as a favorable prognostic factor in multiple drug resistance gastric cancer (13) and non-small cell lung cancer (14) by inhibiting cancer cell proliferation. Furthermore, a preclinical study (15) has indicated that Cbl-b expression can inhibit breast cancer cell proliferation and migration in vivo by decreasing the activation of AKT and ERK. It suggested that Cbl-b may be a favorable prognostic factor. However, the clinical prognostic value of Cbl-b in patients with breast cancer remains unclear.
In the current study, we aimed to explore the prognostic value of Cbl-b expression in breast cancer. Furthermore, we developed new prognostic nomograms based on Cbl-b and other variables for predicting overall survival (OS) and disease-free survival (DFS). This may improve our understanding of the prognostic value of Cbl-b in breast cancer and provide more accurate individual prognosis estimates.
This study included 292 female patients with breast cancer who received surgery from the First Hospital of China Medical University between January 1999 and December 2008. Inclusion criteria were as follows: clear diagnosis by pathology; complete clinical data and follow-up data; patients with breast cancer surgery; patients who received postoperative adjuvant therapy (including chemotherapy, radiotherapy or endocrine therapy). However, because of the inaccessibility of drugs or unreimbursment by medical insurance in China in 1999–2008, we did not include anti-HER2 targeted therapy as the inclusion criteria. Exclusion criteria included patients with de novo stage IV breast cancer; patients who were combined with other malignant tumors.
Tumor specimens were collected from the Department of Pathology at the First Hospital of China Medical University. Immunohistochemistry staining for Cbl-b was performed as described previously (16). The immunoreactivity of Cbl-b was scored based on both intensity of staining (negative = 0, weak = 1, moderate = 2, strong = 3) and percentage of positive tumor cells (<10% = 0, 10–50% = 1, >50% = 2). The final score was calculated by multiplying the single scores obtained from the intensity and percentage of positive cells (ranging from 0 to 6) (6). The median expression score of Cbl-b was 2, which could be used as a cut-off value. Then patients with a score of at least 2 being applicable to the Cbl-b positive study population. Two pathologists independently scored the slides.
Data analysis was performed using SPSS software version 19.0 and R 3.6.0. The correlation between Cbl-b expression and clinic-pathological variables of breast cancer patients was assessed using the Chi-square test. Kaplan-Meier method was adopted to map survival curves of patients with breast cancer. Univariate and multivariate analyses were performed according to the Cox proportional hazards model. Statistically significant variables (P < 0.05) in multivariate analysis were included in the final nomograms. The C-index was used to evaluate the discrimination of the nomogram. The higher the C-index, the more accurate the prognostic prediction was. The receiver operating characteristic (ROC) analysis was also used to verify the accuracy of our new prognostic model.
A total of 292 histologically confirmed breast cancer samples between January 1999 and December 2008 were obtained. The majority of patients (88.7%) were diagnosed with invasive ductal carcinoma (n = 259). The median age of patients was 51 years (ranging from 26 to 76 years), and the median follow-up time was 125.7 months (ranging from 15.7 to 208 months). During the follow-up period, 99 patients developed disease progression (33.9%), of which 69 patients died of disease progression (23.6%).
Immunohistochemistry was performed to evaluate Cbl-b expression. Positive Cbl-b expression was observed in 54.1% (158/292) breast cancer tissue samples, Cbl-b staining for each of the scores (0–3) was shown in Figure 1. Cbl-b expression was significantly correlated with DFS (p = 0.033), but was not associated with HR or HER2 status, and other clinic-pathological factors (p > 0.05, Table 1).
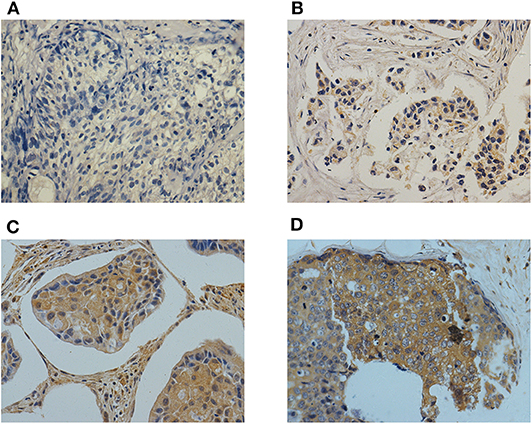
Figure 1. Representative images of Cbl-b immunohistochemical staining for each of the scores (0–3) in breast cancer tissues. Magnification × 400. (A) Negetive control (score = 0), (B) Cbl-b weak positive (score = 1), (C) Cbl-b moderate positive (score = 2), (D) Cbl-b strong positive (score = 3).

Table 1. Correlation of Cbl-b expression with clinic-pathological parameters in patients with breast cancer.
We evaluated the prognostic value of Cbl-b in patients with breast cancer through survival analyses. Kaplan–Meier survival analyses revealed that Cbl-b expression was significantly associated with better OS (p = 0.013) and DFS (p = 0.016) in patients with breast cancer, though the median OS and DFS were not reached at the last follow-up of November 2017 (Figures 2A,B).
The forest plot of subgroup analysis showed that positive Cbl-b expression was significantly associated with favorable prognosis in terms of OS in patients with HR-positive/HER2-positive, age > 35 years, histological grade 2-3, lymph nodes metastasis, and stage II-III breast cancer (Figure 3A). Moreover, positive Cbl-b expression indicated favorable prognosis in terms of DFS for patients with HR-positive/HER2-positive, age > 35 years, histological grade 2–3, tumor size > 2 cm, lymph node metastasis, stage II–III and premenopausal breast cancer (Figure 3B).
Univariate survival analysis showed that the patients with Cbl-b expression, age, tumor size, lymph node metastasis, histological grade, stage, radiotherapy and trastuzumab therapy were associated with prognosis (p < 0.2). In addition, HER2 status and neoadjuvant chemotherapy were also correlated with OS. And date of diagnosis was associated with DFS. Though HR status and HER2 status did not show statistical significance, many studies have confirmed their clinical importance. So we incorporated all the above factors into multivariate survival analysis. The results indicated that Cbl-b expression, age, tumor size, lymph node metastasis and histological grade were independent predictors for OS (Table 2A). Besides, Cbl-b expression, age, lymph node metastasis, histological grade and date of diagnosis were all independently associated with DFS (Table 2B).
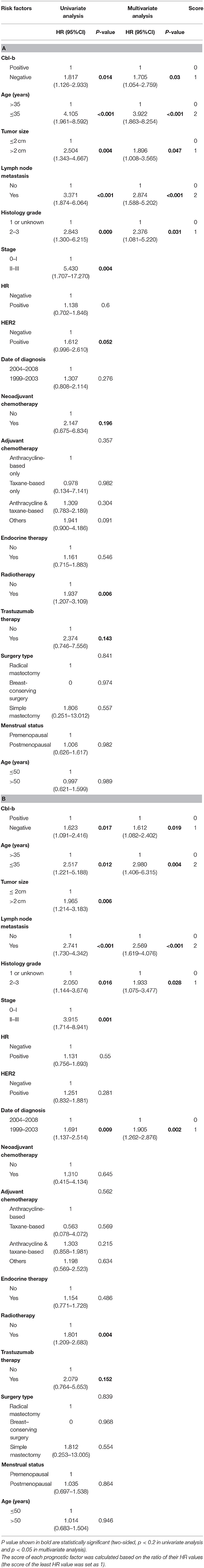
Table 2. Cox univariate and multivariate analysis of OS (A) and DFS (B) in patients with breast cancer.
We constructed predictive nomograms based on the multivariate analysis using a cut-off value of P < 0.05. According to the aforementioned multivariate survival analysis, the prognosis of patients with breast cancer could be predicted by using this model. Each variable in the nomogram was weighted based on some scores. We then obtained the total score of predicting 3- and 10-year OS, and DFS for each patient. The higher the scores patients obtained, the worse the predicted result. The nomogram was accurate for predicting OS, with Harrell's concordance index (C-index) of 0.735 [95% confidence interval (CI): 0.678–0.792] (Figure 4A). And the C-index of the nomogram for predicting DFS was 0.678 (95% CI:0.628–0.728) (Figure 5A). Calibration curves for predicting 3- and 10-year OS (Figures 4B,C) and DFS (Figures 5B,C) of the nomograms were consistent with the models.
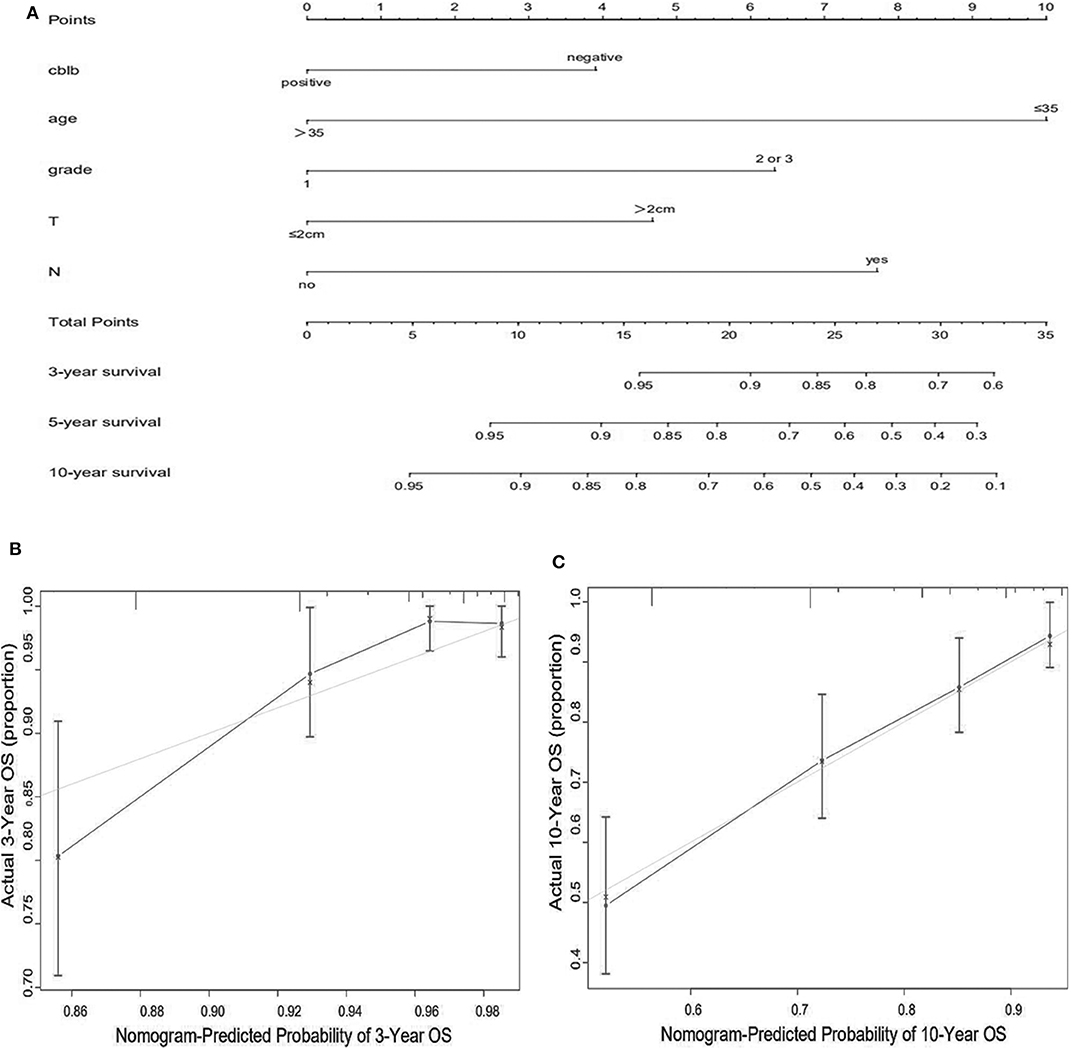
Figure 4. Prognostic nomogram (A) and calibration curve (B,C) for predicting OS in patients with breast cancer.
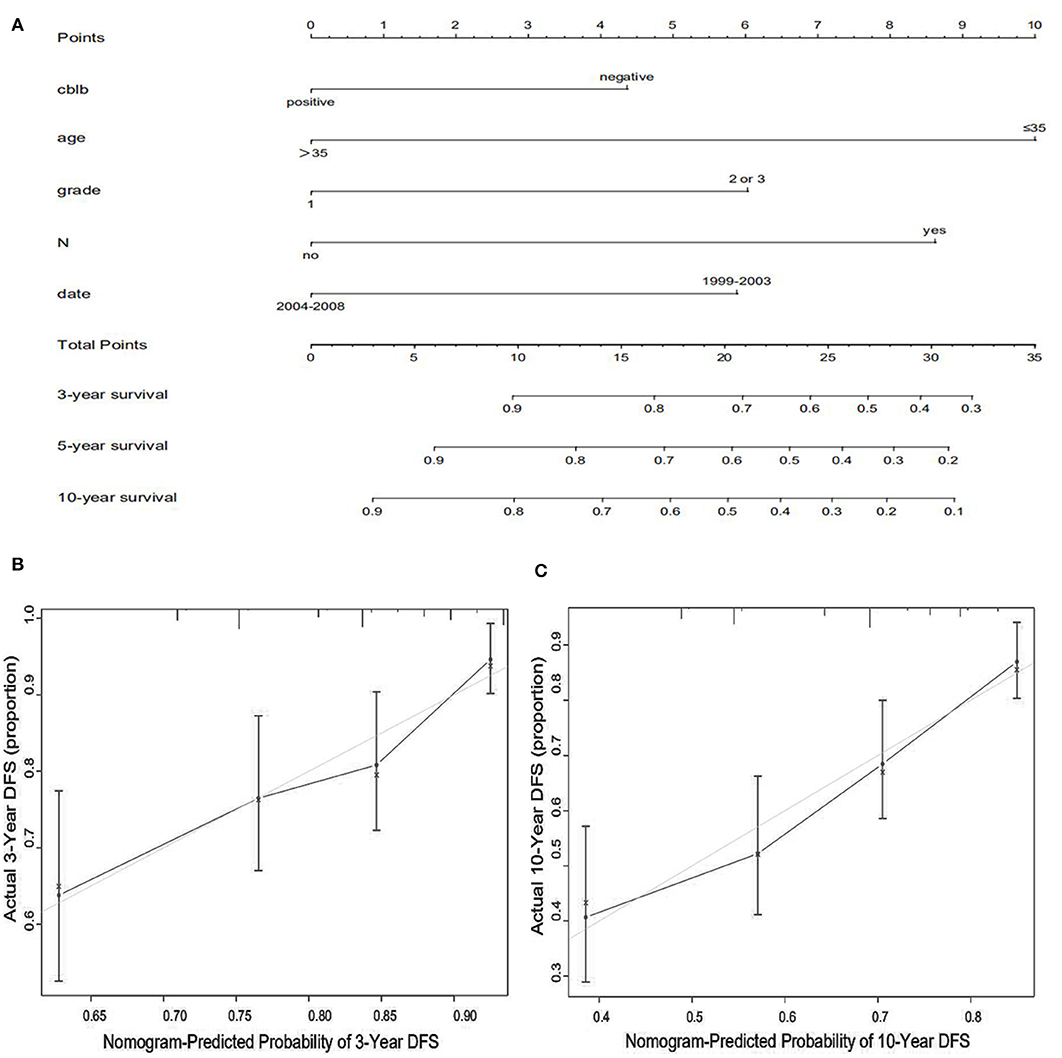
Figure 5. Prognostic nomogram (A) and calibration curve (B,C) for predicting DFS in patients with breast cancer.
We established a new prognostic score system to predict patient survival. The score of each prognostic factor was calculated based on the ratio of their HR values gained from the multivariate analysis (Tables 2A,B). Then, we summed the score of the above variables to develop a prognostic score index. Finally, Kaplan–Meier survival analysis of the prognostic score index showed that all the patients with breast cancer could be separated into different risk subgroups for OS with scores ranging from 0–1, 2–4, and 5–7 (Figure 6A). Similarly, the different risk subgroups for DFS were scored 0–1, 2–3, and 4–7 (Figure 6B). Patients with a higher score were predicted to have poorer prognosis.
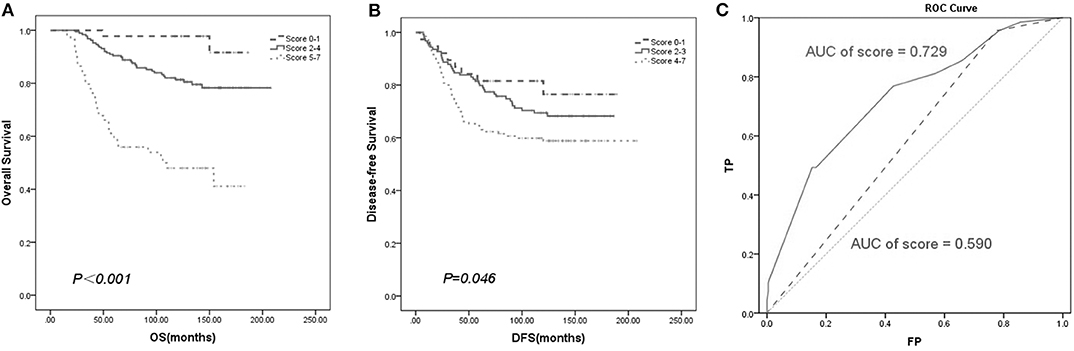
Figure 6. Survival curve based on prognostic score system. The new prognostic factors have separated the patients into different risk subgroups for OS (A) and DFS (B). (C) ROC analysis of overall survival rate based on established model (AUC of score = 0.729) and TNM stage (AUC of stage = 0.590) for breast cancer patients (n = 292).
To verify the accuracy of our new prognostic model compared with the TNM staging prognostic model, ROC analysis was performed. Our model yielded an area under the receiver operating characteristic curve (AUC) of 0.729 for predicting OS, which was superior to TNM staging according to the AJCC 8th with an AUC of 0.590 (Figure 6C).
Breast cancer is a highly heterogeneous disease. Several previous studies (17, 18) have summarized the prognostic factors and survival outcomes associated with breast cancer. Our results indicated that positive Cbl-b expression was an independent favorable prognostic factor in breast cancer. To the best of our knowledge, this is the first study to build nomograms based on Cbl-b and other variables that can be used to predict prognosis of patients with breast cancer. The novel prognostic model could improve the accuracy of traditional TNM staging for predicting OS.
In the present study, we firstly reported that Cbl-b was expressed in 54.1% (158/292) of breast cancer tissues. Our previous studies also showed that positive Cbl-b expression was observed in 69.5% (107/154) receptor activator of nuclear factor κ-B (RANK) positive breast cancer samples (6), and 60.3% (73/121) patients with P-glycoprotein positive multiple drug resistance breast cancer (16).
The E3 ubiquitin-protein ligase Cbl-b may have controversial functions in different tumor types. Cbl-b was reported to function as a negative regulator of growth factor receptor signaling and could suppress tumor cell proliferation (15). Therefore, Cbl-b might have anti-tumor activity. We have found that Cbl-b could inhibit cell proliferation by ubiquitinating the survival signal of the epidermal growth factor receptor (EGFR) pathway in lung (19, 20) and gastric cancer cells (13, 21, 22). Also, previous studies showed that Cbl-b could inhibit tumor growth in some breast cancer cells (6, 23). However, in other tumor types Cbl-b may promote the migration and proliferation of cancer cells. The expression of Cbl-b in skull base chordomas (24) and resectable pancreatic ductal adenocarcinoma (25) can predict tumor invasion and poor prognosis. Overall, the diversity of substrate proteins and cell type-dependent regulation of Cbl-b ubiquitination in tumor cells may explain why Cbl-b displays differential functions in different tumor cells. Our study suggested that Cbl-b expression was an independent prognostic factor. In addition, in the subgroup analysis we found that positive Cbl-b expression indicated favorable prognosis mainly in patients with histological grade 2–3, lymph node metastasis, and stage II–III. It was worth noting that all the above subgroups were relatively poor prognostic, indicating that the protective effect of Cbl-b might occur in clinically recognized subgroups of poor prognosis patients.
Recently it has been reported that a series of nomograms were built to predict prognosis of breast cancer. Xiong et al. (26) developed a nomogram to predict OS of metastatic breast cancer with the following factors: age, metastasis-free interval, metastasis location, and HR status. The C-index value of this model was 0.69. Prognostic factors used to create the nomograms of triple-negative breast cancer (27) and young women with breast cancer (28) included race, tumor size, number of positive lymph nodes, grade, and histological sub-type. The C-indexes for predicting OS were 0.779 and 0.724, respectively. To our knowledge, our study was the first to construct comprehensive nomograms for predicting both OS and DFS based on Cbl-b expression, age, lymph node metastasis, histological grade and other variables in breast cancer. The C-indexes of our nomograms were 0.735 for OS and 0.678 for DFS, indicting that our model showed good accuracy for predicting OS but was a weak model for DFS. Though the accuracy of DFS nomogram needed to be further improved, combining Cbl-b expression with other variables may provide a more precise prognostic model than traditional TNM stage system. Therefore, Cbl-b is still a critical prognostic factor in breast cancer.
The present study has some limitations. Firstly, we did not provide Ki67 and subsequent retesting or treatments. So we could not provide the precise luminal type, which is important for the choice of treatment and prognosis of breast cancer. Secondly, HR-positive patients accounted for only 55.8% of 292 patients with breast cancer, thus the distribution of tumor sub-type in our study could not stand for usual distribution. In addition, most HER2-positive patients did not receive trastuzumab due to the inaccessibility of drugs or unreimbursment by medical insurance in China in 1999–2008. Lastly, this study was performed retrospectively and requires larger multi-institutional prospective studies to further validate our findings in the future.
In conclusion, the expression of Cbl-b might be viewed as an independent prognostic factor for patients with breast cancer. Combining Cbl-b expression with other variables may provide a more precise prognostic model for patients with breast cancer. Although targeted drugs for Cbl-b have not yet been developed, the development of ubiquitin ligases (E3s) inhibitors are already being tested in clinical trials phase I, including inhibitors of MDM2, SPOP, and cIAP (29). Therefore, detection of Cbl-b expression may help clinicians make accurate individual prognosis estimates and provide clues for developing potential therapeutic targets for patients with breast cancer. However, our results are just hypothesis generating and further assessment in prospective trials are needed in order to evaluate the prognostic role of Cbl-b.
The datasets generated for this study are available on request to the corresponding author.
The studies involving human participants were reviewed and approved by Ethics Review Board of The First Hospital of China Medical University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
LZ and XL designed the study and collected data. XL wrote the manuscript. XL, XW, BB, and ZL contributed in the statistical analysis.YT, XQ, YL, and LZ supervised and approved the final draft.
This work was supported by The National Key Research and Development Program of China (2017YFC1308900); National Natural Science Foundation of China (No. 81672605); Science and Technology Plan Project of Liaoning Province (No. 2013225585); and The General Projects of Liaoning Province Colleges and Universities (LFWK201706). The funding source provided financial support for the study and did not have any other involvement in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge the support from the clinical lab team of the Department of Medical Oncology at The First Hospital of China Medical University for their excellent clinical and experimental work.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Leone JP, Leone J, Zwenger AO, Vallejo CT, Leone BA. Prognostic significance of tumor subtypes in women with breast cancer according to stage: a population-based study. Am J Clin Oncol. (2019) 42:588–95. doi: 10.1097/COC.0000000000000563
3. Martínez ME, Unkart JT, Tao L, Kroenke CH, Schwab R, Komenaka I, et al. Prognostic significance of marital status in breast cancer survival: a population-based study. PLoS ONE. (2017) 12:e0175515. doi: 10.1371/journal.pone.0175515
4. Pan X, Yang W, Chen Y, Tong L, Li C, Li H. Nomogram for predicting the overall survival of patients with inflammatory breast cancer: A SEER-based study. Breast. (2019) 47:56–61. doi: 10.1016/j.breast.2019.05.015
5. Nam Se, Lim W, Jeong J, Lee S, Choi J, Park H, et al. The prognostic significance of preoperative tumor marker (CEA, CA15-3) elevation in breast cancer patients: data from the Korean Breast Cancer Society Registry. Breast Cancer Res Treat. (2019) 177:669–78. doi: 10.1007/s10549-019-05357-y
6. Zhang L, Li C, Teng Y, Shi X, Fan Y, Qu X, et al. The E3 ubiquitin ligase Cbl-b improves the prognosis of RANK positive breast cancer patients by inhibiting RANKL-induced cell migration and metastasis. Oncotarget. (2015) 6:22918–33. doi: 10.18632/oncotarget.4382
7. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. (2010) 141:1117–34. doi: 10.1016/j.cell.2010.06.011
8. Liyasova MS, Ma K, Lipkowitz S. Molecular pathways: Cbl proteins in tumorigenesis and antitumor immunity—opportunities for cancer treatment. Clin Cancer Res. (2015) 21:1789–94. doi: 10.1158/1078-0432.CCR-13-2490
9. Lutz-Nicoladoni C, Wolf D, Sopper S. Modulation of immune cell functions by the E3 Ligase Cbl-b. Front Oncol. (2015) 5:58. doi: 10.3389/fonc.2015.00058
10. Tang N, Yang L, Li D, Liu R, Zhang J. Modulation of B cell activation threshold mediated by BCR/CD40 costimulation by targeting Cbl-b for ubiquitination. Biochem Biophys Rep. (2019) 18:100641. doi: 10.1016/j.bbrep.2019.100641
11. Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. (2014) 507:508–12. doi: 10.1038/nature12998
12. Dong Q, Li C, Ma Y, Li K, Zhang Y, Yang X, et al. Cbl-b predicts postoperative survival in patients with resectable pancreatic ductal adenocarcinoma. Oncotarget. (2017) 8:57163–73. doi: 10.18632/oncotarget.18714
13. Xu L, Zhang Y, Qu X, Che X, Guo T, Cai Y, et al. E3 ubiquitin ligase Cbl-b prevents tumor metastasis by maintaining the epithelial phenotype in multiple drug-resistant gastric and breast cancer cells. Neoplasia. (2017) 19:374–82. doi: 10.1016/j.neo.2017.01.011
14. Wang S, Xu L, Che X, Li C, Xu L, Hou K, et al. E3 ubiquitin ligases Cbl-b and c-Cbl downregulate PD-L1 in EGFR wild-type non-small cell lung cancer. FEBS Lett. (2018) 592:621–30. doi: 10.1002/1873-3468.12985
15. Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. (2015) 6:29209–23. doi: 10.18632/oncotarget.4976
16. Zhang Y, Qu X, Teng Y, Li Z, Xu L, Liu J, et al. Cbl-b inhibits P-gp transporter function by preventing its translocation into caveolae in multiple drug-resistant gastric and breast cancers. Oncotarget. (2015) 6:6737–48. doi: 10.18632/oncotarget.3253
17. Chen QF, Huang T, Shen L, Wu P, Huang ZL, Li W. Prognostic factors and survival according to tumor subtype in newly diagnosed breast cancer with liver metastases: a competing risk analysis. Mol Clin Oncol. (2019) 11:259–69. doi: 10.3892/mco.2019.1890
18. Kimbung S, Loman N, Hedenfalk I. Clinical and molecular complexity of breast cancer metastases. Semin Cancer Biol. (2015) 35:85–95. doi: 10.1016/j.semcancer.2015.08.009
19. Zhu T, An S, Choy MT, Zhou J, Wu S, Liu S, et al. Lnc RNA DUXAP 9-206 directly binds with Cbl-b to augment EGFR signaling and promotes non-small cell lung cancer progression. J Cell Mol Med. (2019) 23:1852–64. doi: 10.1111/jcmm.14085
20. Shtiegman K, Kochupurakkal B, Zwang Y, Pines G, Starr A, Vexler A, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. (2007) 26:6968–78. doi: 10.1038/sj.onc.1210503
21. Xu L, Zhang Y, Qu X, Che X, Guo T, Li C, et al. DR 5-Cbl-b/c-Cbl-TRAF 2 complex inhibits TRAIL-induced apoptosis by promoting TRAF 2-mediated polyubiquitination of caspase-8 in gastric cancer cells. Mol Oncol. (2017) 11:1733–51. doi: 10.1002/1878-0261.12140
22. Feng D, Ma Y, Liu J, Xu L, Zhang Y, Qu J, et al. Cbl-b enhances sensitivity to 5-fluorouracil via EGFR-and mitochondria-mediated pathways in gastric cancer cells. Int J Mol Sci. (2013) 14:24399–411. doi: 10.3390/ijms141224399
23. Che X, Zhang Y, Qu X, Guo T, Ma Y, Li C, et al. The E3 ubiquitin ligase Cbl-b inhibits tumor growth in multidrug-resistant gastric and breast cancer cells. Neoplasma. (2017) 64:887–92. doi: 10.4149/neo_2017_610
24. Luo P, Wang X, Zhou J, Li L, Jing Z. C-Cbl and Cbl-b expression in skull base chordomas is associated with tumor progression and poor prognosis. Hum Pathol. (2018) 74:129–34. doi: 10.1016/j.humpath.2017.12.019
25. Dong Q, Li C, Che X, Qu J, Fan Y, Li X, et al. MicroRNA-891b is an independent prognostic factor of pancreatic cancer by targeting Cbl-b to suppress the growth of pancreatic cancer cells. Oncotarget. (2016) 7:82338–53. doi: 10.18632/oncotarget.11001
26. Xiong Z, Deng G, Huang X, Li X, Xie X, Wang J, et al. Score for the survival probability in metastasis breast cancer: a nomogram-based risk assessment model. Cancer Res Treat. (2018) 50:1260–9. doi: 10.4143/crt.2017.443
27. Guo LW, Jiang LM, Gong Y, Zhang HH, Li XG, He M, et al. Development and validation of nomograms for predicting overall and breast cancer-specific survival among patients with triple-negative breast cancer. Cancer Manag Res. (2018) 10:5881–94. doi: 10.2147/CMAR.S178859
28. Gong Y, Ji P, Sun W, Jiang YZ, Hu X, Shao ZM. Development and validation of nomograms for predicting overall and breast cancer–specific survival in young women with breast cancer: a population-based study. Transl Oncol. (2018) 11:1334–42. doi: 10.1016/j.tranon.2018.08.008
Keywords: Cbl-b, breast cancer, overall survival, disease-free survival, prognosis, nomogram
Citation: Liu X, Teng Y, Wu X, Li Z, Bao B, Liu Y, Qu X and Zhang L (2020) The E3 Ubiquitin Ligase Cbl-b Predicts Favorable Prognosis in Breast Cancer. Front. Oncol. 10:695. doi: 10.3389/fonc.2020.00695
Received: 17 January 2020; Accepted: 14 April 2020;
Published: 05 May 2020.
Edited by:
Florian Clatot, Centre Henri Becquerel Rouen, FranceReviewed by:
Simona Ruxandra Volovat, Grigore T. Popa University of Medicine and Pharmacy, RomaniaCopyright © 2020 Liu, Teng, Wu, Li, Bao, Liu, Qu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiujuan Qu, cXVfeGl1anVhbkBob3RtYWlsLmNvbQ==; Lingyun Zhang, bHl6aGFuZ0BjbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.