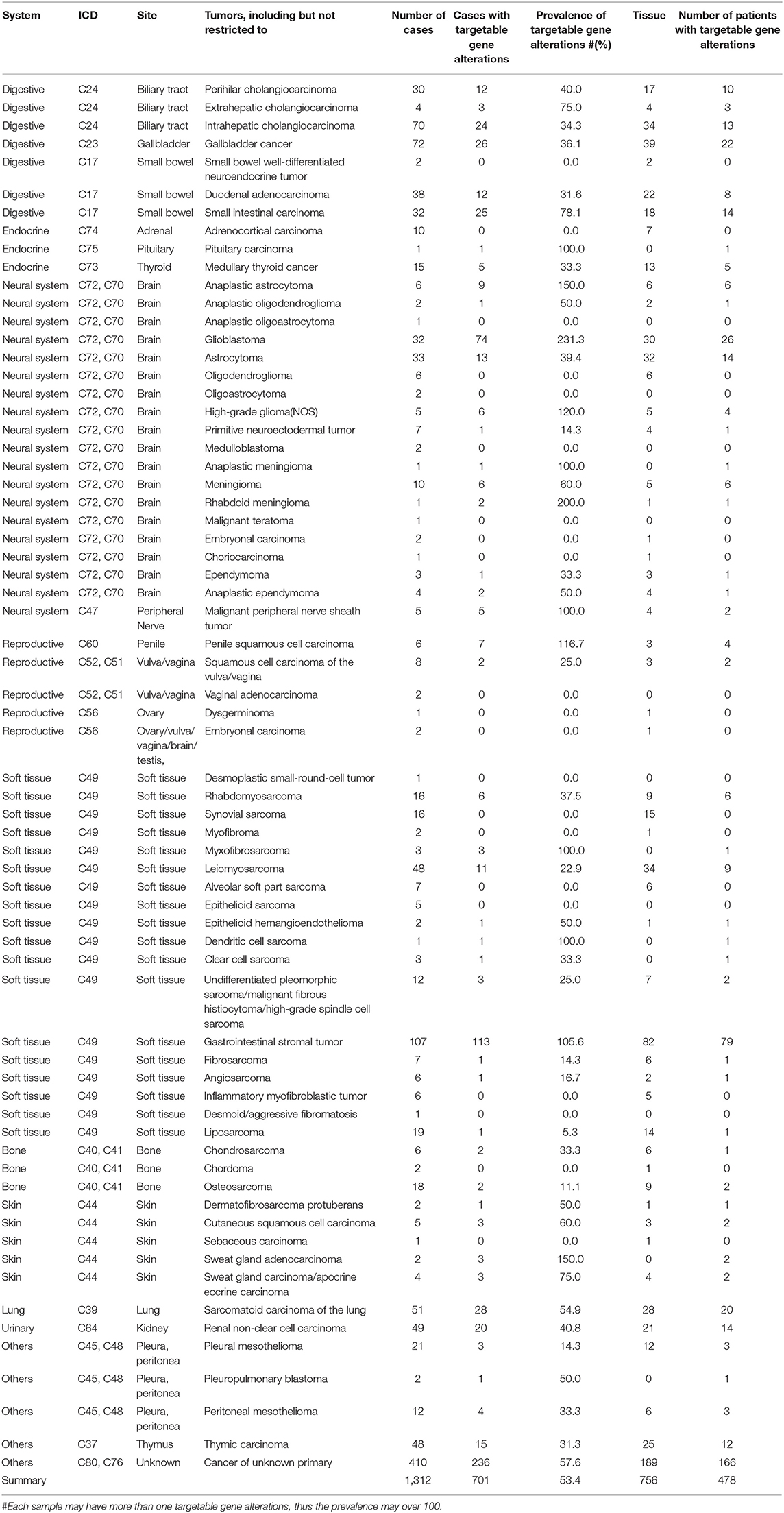
- 1Clinical Cancer Center, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of Medical Center, Geneplus-Beijing Institute, Beijing, China
Comprehensive Genomic Profiling may be informative for novel treatment strategies and to improve outcomes for patients with rare tumors. This study aims to discover opportunities for use of targeted therapies already approved for routine use in patients with rare tumors. Solid tumors with an incidence lower than 2.5/100,000 per year was defined as rare tumors in China after comprehensive analysis based on epidemiological data and current availability of standardized treatment. Genomic data of rare tumors from the public database cBioPortal were compared with that of the Chinese population for targetable genomic alterations (TGAs). TGAs were defined as mutations of ALK, ATM, BRAF, BRCA1, BRCA2, CDKN2A, EGFR, ERBB2, FGFR1,2,3, KIT, MET, NF1, NTRK1,2,3, PIK3CA, PTEN, RET, and ROS1 with level 1 to 4 of evidence according to the OncoKB knowledge database. Genomic data of 4,901 patients covering 63 subtypes of rare tumor from cBioPortal were used as the western cohort. The Chinese cohort was comprised of next generation sequencing (NGS) data of 1,312 patients from across China covering 67 subtypes. Forty-one subtypes were common between the two cohorts. The accumulative prevalence of TGAs was 20.40% (1000/4901) in cBioPortal cohort, and 53.43% (701/1312) in Chinese cohort (p < 0.001). Among those 41 overlapping subtypes, it was still significantly higher in Chinese cohort compared with cBioPortal cohort (54.1%% vs. 26.1%, p < 0.001). Generally, targetable mutations in BRAF, BRCA2, CDKN2A, EGFR, ERBB2, KIT, MET, NF1, ROS1 were ≥3 times more frequent in Chinese cohort compared with that of the cBioPortal cohort. Cancer of unknown primary tumor type, gastrointestinal stromal tumor, gallbladder cancer, intrahepatic cholangiocarcinoma, and sarcomatoid carcinoma of the lung were the top 5 tumor types with the highest number of TGAs per tumor. The incidence of TGAs in rare tumors was substantial worldwide and was even higher in our Chinese rare tumor population. Comprehensive genomic profiling may offer novel treatment paradigms to address the limited options for patients with rare tumors.
Introduction
Molecular profiling to identify potential therapeutic targets has been widely applied in common tumors such as lung cancer (1, 2), breast cancer (3, 4), melanoma (5), and colorectal cancer (6, 7). The use of targeted therapy in selected patients can significantly improve outcomes. Increasingly, clinical trials feature targeted therapeutic agents or require a specific biomarker for entry (8, 9). However, limited information is available regarding the utility of targeted therapy for rare tumors (10, 11). What's more, while rare individually, rare tumors cumulatively account for over 20% of adult malignant neoplasms in the United States (12, 13).
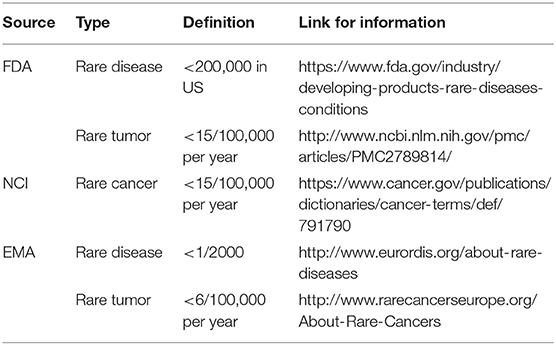
There is no universally applied definition for rare tumors (Table 1). The European Society for Medical Oncology (ESMO) defines a rare tumor as a tumor with an annual incidence of 6/10,000 (14) in Europe. The National Cancer Institute (NCI) (https://www.cancer.gov/publications/dictionaries/cancer-terms/def/791790) and Food and Drug Administration (FDA) (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2789814/) defines it as a tumor with an annual incidence of <15/10,000 in the US. According to the NCI definition, lung cancer, colon cancer, breast cancer, prostate cancer, endometrial carcinoma, rectal cancer, ovarian cancer, kidney cancer, melanoma, non-Hodgkin lymphoma, and gastric cancer belong to common cancers.
There is some discordance between these definitions and data specific to China. While esophageal cancer and hepatocellular carcinoma are rare tumors according to NCI definition, these are common in China based on annual incidence. On the other hand, skin tumors, especially basal cell carcinoma, are common tumors in the United States, with an incidence of 255.6/100,000 (15), but are relatively rare in China (14) (2.4/100,000 for all skin tumors). This suggests that the definitions from US and Europe were possibly not appropriate in China based on the different incidences and prevalence of tumors.
This study analyzed data from the National Cancer Registry office of the National Cancer Center (16) and integrated it with presently available treatment options to generate a definition of rare tumors specific to China. Subsequently, available data for targetable genomic alterations (TGAs) of two cohorts of rare tumors from the cBioPortal and Geneplus databases were collected and analyzed. Our work provides valuable knowledge to guide personalized, targeted therapy for rare tumors.
Methods
Definition of Rare Tumors in China
We consulted National Cancer Registry of the National Cancer Center, China (16) and generated an estimation of incidence of tumors in mainland China. Tumor types were classified according to the International Classification of Diseases (ICD), and we comprehensively synthesized the epidemiology data and availability of standard treatment in China as well as opinions of experts from National Cancer Center. We then defined rare tumors in China according to the following standardizations (Table 2):
1. First, we eliminated the tumors from systems or organs which have consensus or guidelines for treatment in China; an incidence of “2.5/100,000 per year” was selected as a cut-off value for “rare tumor” for tumors with unique ICD codes listed with systems or organs;
2. Secondly, we searched OncoTrees (http://oncotree.mskcc.org/) to further investigate the subtypes of those common tumors that (1) have a distinct ICD code and (2) exhibit an incidence >2.5/100,000 per year in China. We included subtypes of those tumors after further confirming that the incidence of which was ≤2.5/100,000 per year in China by searching Pubmed database (https://www.ncbi.nlm.nih.gov/pubmed/) and the China National Knowledge Infrastructure (CNKI) database;
3. Finally, we also included cancers of unknown primary (CUP) tumors, not only because the incidence of those tumors was ≤2.5/100,000 per year in China, but also because there were no consensus or guidelines for treatment of CUP in China.
Definition of Targetable Mutations According to the OncoKB Framework
The actionabilities of genetic alterations were mainly based on the OncoKB knowledge database (https://oncokb.org). Utilizing the OncoKB framework, mutations could be classified into 4 main levels of evidence for biomarker-guided therapy and those with unknown significance. OncoKB is a precision oncology knowledge base and contains information about the effects and treatment implications of specific cancer gene alterations. It is developed and maintained by the Knowledge Systems group in the Marie Josée and Henry R. Kravis Center for Molecular Oncology at Memorial Sloan Kettering Cancer Center (MSK) (17). Curated by a network of clinical fellows, research fellows, and faculty members at MSK, OncoKB contains detailed information about specific alterations in 668 cancer genes. The information is compiled from various sources, such as guidelines from the FDA, NCCN, or ASCO, ClinicalTrials.gov and the scientific literature. Level 1 is an FDA-recognized biomarker predictive of response to an FDA-approved drug in this indication. Level 2 is standard care biomarker predictive of response to an FDA-approved drug in this indication (2A) or in another indication, but not standard care in this indication (2B). Level 3 is compelling clinical evidence supports the biomarker as being predictive of response to a drug in this indication (3A) or in another indication (3B). Level 4 is compelling biological evidence supports the biomarker as being predictive of response to a drug (Supplementary Table 1).
cBioPortal
The cBioPortal for Cancer Genomics was originally developed at Memorial Sloan Kettering Cancer Center. The public cBioPortal site is hosted by the Center for Molecular Oncology at MSK. The cBioPortal currently hosts more than 40 datasets, including TCGA and other large-scale genomic studies, and makes them available for bulk download. Data from OCG's TARGET Initiative will be added to the database in the next year. The data types from the 13,000+ tumor samples include mutations, copy number alterations, mRNA expression changes, and DNA methylation values, as well as clinical parameters, such as disease-free survival.
(https://www.cbioportal.org/datasets).
Estimation of Targetable Mutations
To estimate the prevalence of targetable mutations in rare tumors, we queried the cBioPortal database using the genes listed in Supplementary Table 1 in a manually curated set of 175 non-redundant studies, including TCGA and non-TCGA studies, with no overlapping samples. Mutations of those genes were downloaded and filtered with the annotated oncoKB levels of evidence. Only mutations of level 1-4 were kept for further analysis. To calculate the prevalence, the cumulative number of targetable mutations in each cancer was divided by total numbers of samples for that cancer. The same criteria and workflow were used for the Chinese patient cohort.
Patient Recruitment
We retrospectively analyzed genomic profiling data of 1,312 patients with rare tumors from Geneplus database. This database contained patients enrolled from multiple hospitals of China from September 2015 to October 2019 (18, 19). All patients received next-generation sequencing (NGS) testing in Geneplus-Beijing Institute after obtaining written informed consent. Meanwhile, all the patients were stratified into different clinicopathological subgroups according to OncoTree system (http://oncotree.mskcc.org/).
All tissues samples included in this study underwent an onsite pathology review to confirm histologic classification and tumor tissue adequacy, which required a minimum of 20% of tumor cells. Genomic profiling was performed in a College of American Pathologists–accredited laboratory (Geneplus-Beijing) using the Illumina Nextseq CN 500 or Gene+Seq 2000 instrument (20, 21). Briefly, serial sections from formalin-fixed paraffin-embedded (FFPE) tumor tissues were used for genomic tumor DNA extraction using the QIAamp DNA mini kit (Qiagen, Valencia, CA). ctDNA was isolated from 4 to 5 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA). DNA from leukocytes was extracted using the DNeasy Blood Kit (Qiagen, Valencia, CA). Sequencing libraries were prepared from ctDNA using KAPA DNA Library Preparation Kits (Kapa Biosystems, Wilmington, MA, USA), and genomic DNA sequencing libraries were prepared with Illumina TruSeq DNA Library Preparation Kits (Illumina, San Diego, CA). Libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Roche NimbleGen, Madison, WI, USA) targeting 59-1021 genes (~1.4 Mbp genomic regions of 1021 cancer-related genes or ~230 Kbp genomic regions of 59 genes) (Supplementary Tables 2, 3). Prepared libraries were sequenced on using the Illumina Nextseq CN 500 (Illumina, San Diego, CA) or Gene+Seq 2000 (Geneplus-Beijing, China).
Sequencing data were analyzed using default parameters. Adaptor sequences and low-quality reads were removed. The clean reads were aligned to the reference human genome (hg19) using Burrows-Wheeler Aligner (BWA; version 0.7.12-r1039). Realignment and recalibration were performed using GATK (version 3.4-46-gbc02625). Single nucleotide variants (SNV) were called using MuTect (version 1.1.4) and NChot, a software developed in-house to review hotspot variants (22). Small insertions and deletions (InDels) were determined by GATK. Somatic copy number alterations were identified with CONTRA (v2.0.8). The final candidate variants were all manually verified using Integrative Genomics Viewer.
Targeted capture sequencing required a minimal mean effective depth of coverage of 300× in tissues and 1,000× in plasma samples. For the 1,312 patients included in our study, the mean effective depth of coverage is 1,295× in tissues and 2,014× in plasma samples and 299× in germline DNA samples (Supplementary Table 4).
TGAs simultaneously detected by this assay included base substitutions, short insertions and deletions, focal gene amplifications and homozygous deletions (copy number alterations) and select gene fusions and rearrangements. Variants were filtered to exclude synonymous variants, known germline variants in dbSNP, and variants that occur at a population frequency of >1% in the Exome Sequencing Project. Germline variants were interpreted following ACMG guidelines, and the variants were classified as pathogenic, likely pathogenic, unknown significance, likely benign, and benign.
Statistics
The Chi-square test or Fisher's exact test was performed to compare frequency targetable mutations between groups. All statistical analysis was performed with SPSS (v.23.0; STATA, College Station, TX, USA) or GraphPad Prism (v. 6.0; GraphPad Software, La Jolla, CA, USA) software. Statistical significance was defined as a two-sided P-value of < 0.05.
Results
Mutation Profiling of Rare Tumors in cBioPortal Database
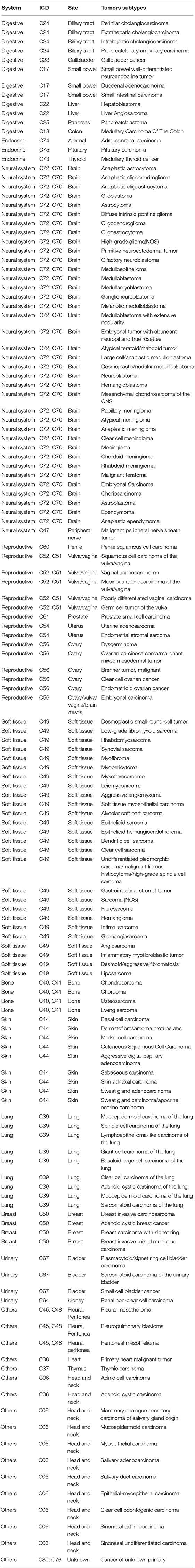
Rare tumors according to our China-specific definition included 141 tumor types. We analyzed a total of 45,666 samples from the cBioPortal database and identified 4,901 samples of rare tumors that matched our definition, representing 63 of the 141 possible tumor types. Neuroblastoma, adenoid cystic carcinoma, Ewing sarcoma, astrocytoma, and oligodendroglioma were the top 5 rare tumors, with 1321, 323, 263, 250, and 229 samples, respectively. One thousand (20.4%, 1000/4901) targetable mutations were identified in the 4901 samples, with PIK3CA, PTEN, KIT, CDKN2A, ATM, FGFR, BRAF, NF1, ALK, and BRCA2 as the top 10 genes with targetable mutations identified in 266, 149, 119, 112, 75, 66, 33, 33, 27, and 27 samples, respectively (Table 3 and Supplementary Table 5).
Mutation Profiling of Chinese Patients With Rare Tumors
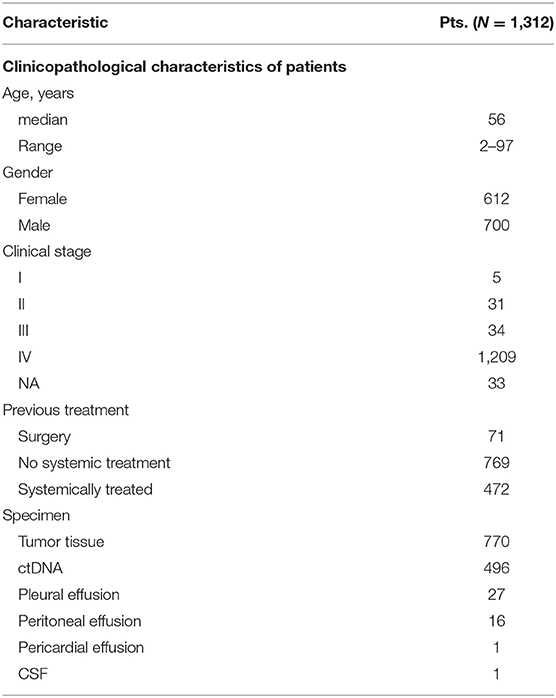
We recruited a second, independent patient cohort from another pan-China database, Geneplus. One thousand three hundred and twelve patients (1312) with rare tumors were included for the study. The clinicopathological characteristics of all the patients are summarized in Table 4. The median age was 56, and 53.4% (700/1312) of the cohort were male. Ninety two percent (92.1%, 1209/1312) of the patients were at stage IV, and 58.6% (769/1312) of the patient were systemic treatment-naïve while 36% (472/1312) had been systemically treated. Tumor tissue was available for genetic analysis in 770 of these patients, while 469, 27, 16, 1, and 1 patient, respectively, had ctDNA, pleural effusion, peritoneal effusion, pericardial effusion, and cerebrospinal fluid (CSF) available as an alternative.
These 1,312 cases included 67 tumor subtypes out of our defined rare tumor types, with cancer of unknown primary, gastrointestinal stromal tumor, gallbladder cancer, intrahepatic cholangiocarcinoma, and sarcomatoid carcinoma of the lung as the top 5 tumors including 410, 107, 72, 70, and 51 patients, respectively.
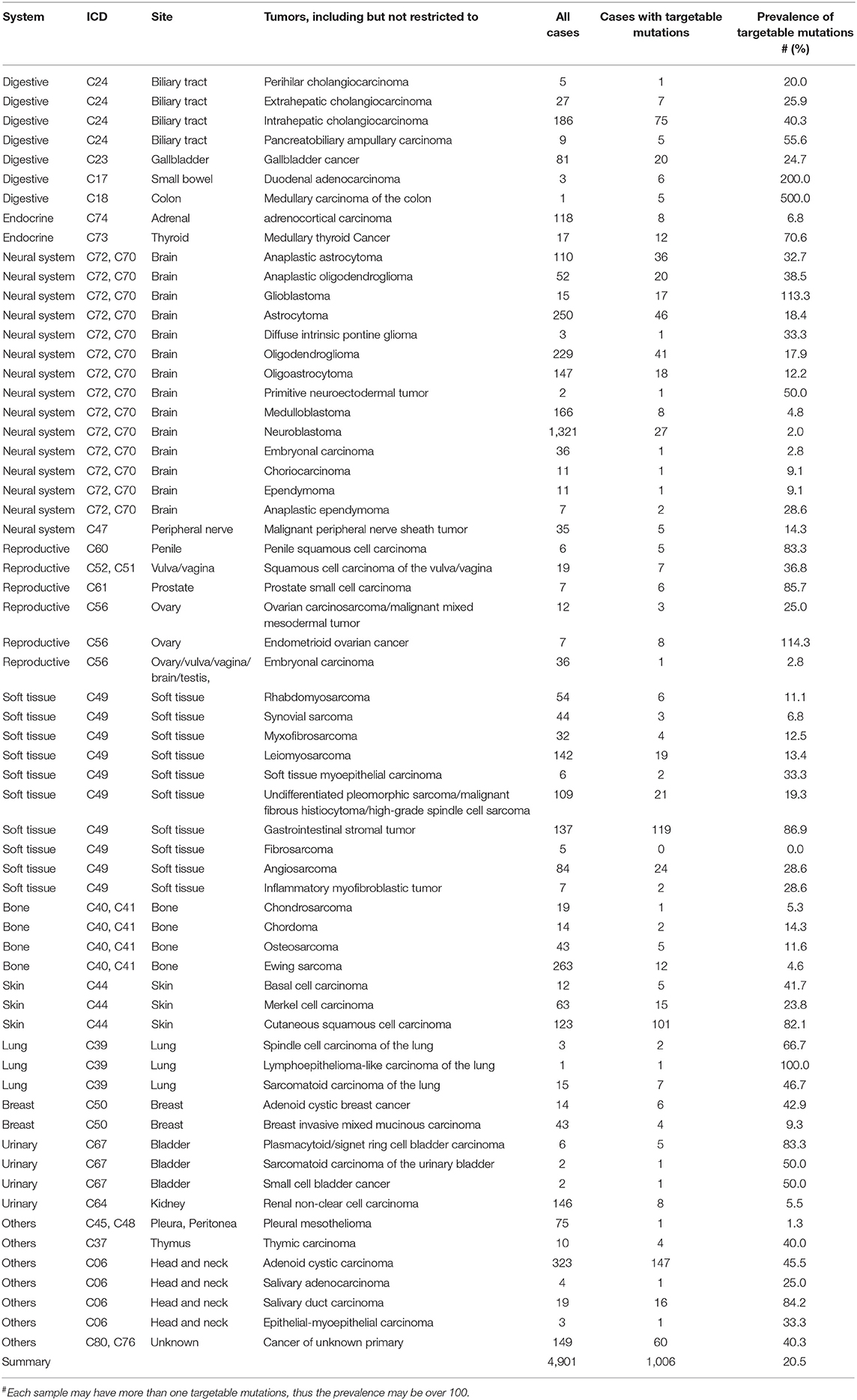
Within these 1,312 samples, a total of 7,998 alterations were identified in 712 genes (5,924 base substitutions, 1,206 gene amplifications or deletions, 840 short indels, and 28 gene rearrangements) for a mean of 4 alterations per tumor (Supplementary Table 6). Total 701 targetable mutations were identified in the 1,312 samples, with EGFR, KIT, CDKN2A, PIK3CA, PTEN, NF1, ERBB2, BRAF, BRCA2, and FGFR1/2/3 as the top 10 genes with targetable mutations identified in 266, 149, 119, 119, 112, 75, 66, 33, 33, 27, and 27 samples, respectively. Of the 1312 patients, 478 patients had at least 1 targetable mutation (Table 5 and Supplementary Table 7).
Consistencies and Discrepancies Between the Two Cohorts of Rare Tumors
Between the cBioPortal cohort and our independent cohort, there were 41 overlapping subtypes (41/63, cBioPortal; 41/67, our cohort) and 22 (cBioPortal) or 25 (our cohort) subtypes unique to each cohort (Table 6, Supplementary Figure 1).
We first compared the overall prevalence of TGAs in these two cohorts. The prevalence of targetable mutations was significantly higher in our cohort compared with the data from cBioPortal (53.4 vs. 20.4%, p < 0.001) (Table 6). Specifically, mutations or amplifications of BRAF, BRCA2, CDKN2A, EGFR, ERBB2, KIT, MET, NF1, ROS1 were 3 or more times more frequent in our cohort than in the cBioPortal cohort. Alterations of BRCA1, NTRK fusion were slightly more common in the cBioPortal cohort. When restricting analysis to the 41 overlapping subtypes, the difference of targetable mutations was still significant (54.1 vs. 26.1%, p < 0.001). We further focused on 4 rare tumors (gallbladder cancer, astrocytoma, gastrointestinal stromal tumor, and cancer of unknown primary) with more than 30 cases in both cohorts. We found the overall incidence rate of targetable mutations was higher in our cohort (Supplementary Table 8). For gallbladder cancer, ERBB2 and BRCA2 mutations were significantly more frequent in our cohort, while ATM mutation was enriched in the cBioPortal cohort (Figure 1A) (23). For astrocytoma, BRAF, ATM, CDKN2A, and EGFR mutations/amplifications were highly enriched in our cohort (Figure 1B). For gastrointestinal stromal tumor, the prevalence of the KIT mutation was similar between the two groups, but our cohort had a significantly higher prevalence of CDKN2A and NF1 (Figure 1C). For cancer of unknown primary, EGFR mutation and ALK fusion were highly enriched in our cohort, which indicate that those tumors might originate from lung (Figure 1D).
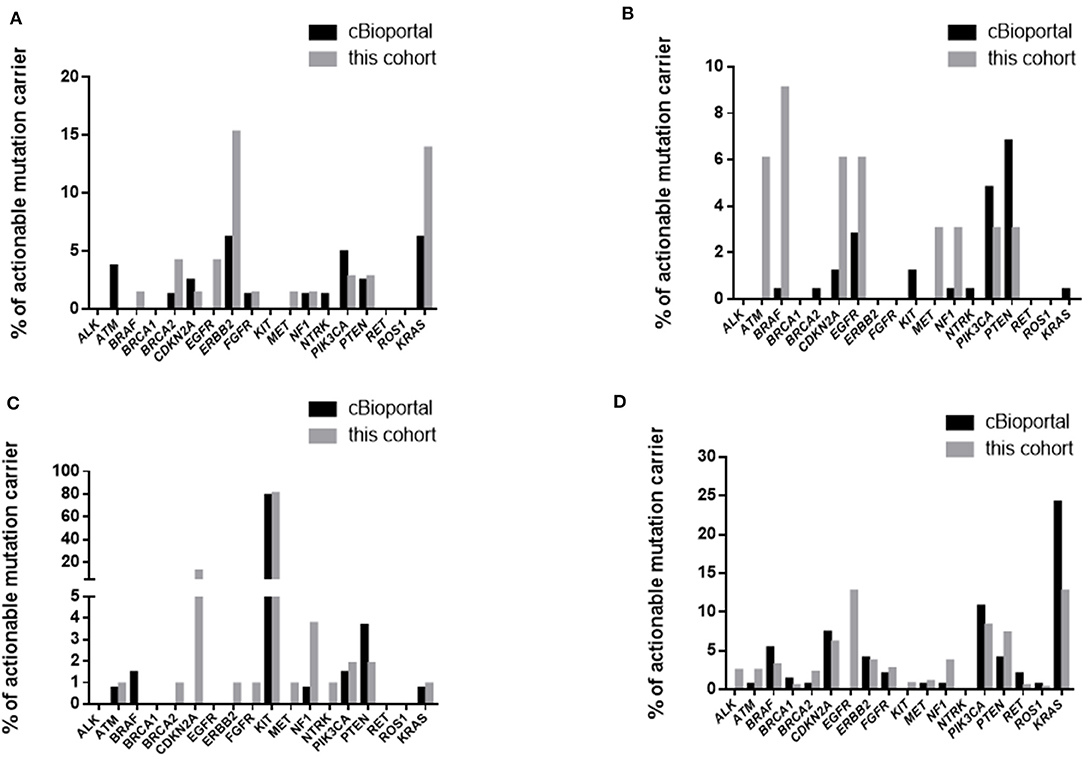
Figure 1. Comparison of targetable mutations in gallbladder cancer (A), astrocytoma (B), gastrointestinal stromal tumor (C), and cancer of unknown primary (D).
Discussion
This study focused on rare tumors in China and proposed a novel definition of rare tumors customized for China by jointly considering frequency and clinical characteristics to addresses the disparate requirements of clinical decision-making, clinical research, drug development, and health care services. Applying this new definition, a comprehensive list of rare tumors was explored for genetic biomarkers of response to targeted therapy both in the worldwide cBioPortal database and a mainland China-specific patient cohort mainly to explore potential novel treatment indications for those rare tumors in China. Results show that targetable gene alterations are frequently present in rare tumors, and that these mutations are enriched in Chinese population as compared to the general global population.
Most importantly, a definition of rare tumors in China was proposed for the first time based on the epidemiology data and availability of standard treatment in China. An incidence of ≤2.5/100,000 per year as a cut off value for rare tumor in China is novel and it is rigorous compared with those of the USA and Europe which is 15/100,000 and 6/100,000 respectively. The disparity should be mainly attributed to the facts that China has a larger population base, and a different epidemiological distribution for most types of tumors compared to western countries. We believe any threshold for rarity is artificial and should be considered as just indicative. We should always be aware that an incidence threshold rate as a line for rareness should be used with flexibility. The most important purpose of proposing the definition is to increase the attention from clinical practitioners and government personnel of China, as well as drug investigators all over the world, to promote the development of novel drugs and strategies for those rare tumors without consensus and guidelines for effective treatment in China, and finally to improve the outcome of rare tumor patients.
After applying our rare tumor criteria to patient data, we discovered the overall prevalence of TGAs in Chinese rare tumor patients' cohort was much higher than that of the cBioPortal cohort. We restricted our analysis of TGAs to genes having Level 1-4 evidence of being a cancer gene according to the OncoKB knowledge database. Using this framework, we identified mutations of ALK, ATM, BRAF, BRCA1, BRCA2, CDKN2A, EGFR, ERBB2, FGFR1,2,3, KIT, MET, NF1, NTRK1,2,3, PIK3CA, PTEN, RET, and ROS1 within our cohort. The cumulative prevalence of TGAs was significantly higher in Chinese cohort (53.43%) compared with general population worldwide (26.1%). This indicates that there might be higher possibilities those patients could benefit from targeted therapies. The underlying causes for the disparities in mutation prevalence were complicated as the two cohorts had significantly different compositions of tumor subtypes, as well as different numbers of patients in each subtype. The overall difference between the two cohorts was still significant (p < 0.001) if we only studied the shared 41 subtypes of rare tumor. This phenomenon is in agreement with the data showing that EGFR mutation rate in Asian NSCLC patients is higher than that of Caucasian patients. Our findings indicate that the classification of “rare tumor” is heterogeneous by ethnicity.
We also found that most common TGAs in both cohorts are actionable with available drugs. The top 5 targetable mutations found in Chinese patients cohort were EGFR, KIT, CDKN2A, PIK3CA, and PTEN; and in the cBioPortal cohort were PIK3CA, PTEN, KIT, CDKN2A, and ATM. Regarding the 4 shared targetable mutations, there is at least one targeted drug for each mutation (imatinib for KIT, palbociclib for CDKN2A, temsirolimus and everolimus for PIK3CA and PTEN) currently available in China (Table 6). This suggests that we have available effective treatment options for some rare tumor patients.
Finally, our data indicate that samples for genetic profiling of rare tumor are still inadequate. There are only 10.5% (4901/46566) tumor samples from rare tumors in cBioPortal database. Moreover, 52 out of 141 (36.9%) subtypes of rare tumors did not have genetic data available in cBioPortal or in our cohort (Supplementary Table 9). For most subtypes with data, the median number of samples was 19 in cBioPortal and 5 in our cohort. Considering the high prevalence of TGAs in the rare tumor population and the largely unmet medical needs of those patients, more attention and efforts should be applied in this field in the near future.
Conclusions
We defined rare tumor in China as ICD-specified tumors with incidence ≤2.5/100,000 per year in China, and subtypes of non-rare ICD-specified tumors with incidence ≤2.5/100,000 per year in China, and cancers of unknown primary. Genomic profiling of rare tumors matching this definition from cBioPortal and a Chinese cohort drawn from the Geneplus database demonstrated a substantial prevalence of targetable genomic alterations in these tumors, which was even higher in Chinese rare tumor patient population than in the general population. All of the above facilitates future drug investigations and treatment improvement for rare tumors.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
This study was approved by the ethics committees of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (NCC2019C-222). All patients signed written informed consent for further scientific analysis of genetic data.
Author Contributions
NL and XY conceived the study. SW and RC processed data, performed data analysis. YT, YY, YF, HH, DW, HF, YB, CS, AY, QF, and DG. contributed to data collection, generation of tumor list and scientific insights. SW and RC wrote the manuscript. SW, NL, and XY revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the patients for providing the valuable genetic data for scientific analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00536/full#supplementary-material
Abbreviations
NGS, Next generation sequencing; TGAs, targetable genomic alterations; cBioPortal, CBio Cancer Genomics Portal; ESMO, European Society for Medical Oncology; NCI, National Cancer Institute; FDA, Food and Drug Administration; ICD, International Classification of Diseases; CNKI, China National Knowledge Infrastructure; CUP, cancers of unknown primary; MSK, Memorial Sloan Kettering Cancer Center; SNV, Single nucleotide variants; InDels, insertions and deletions; CSF, cerebrospinal fluid.
References
1. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. (2004) 350:2129–39. doi: 10.1056/NEJMoa040938
2. Katzel JA, Fanucchi MP, Li Z. Recent advances of novel targeted therapy in non-small cell lung cancer. J Hematol Oncol. (2009) 2:2. doi: 10.1186/1756-8722-2-2
3. Muss HB. Targeted therapy for metastatic breast cancer. N Engl J Med. (2006) 355:2783–5. doi: 10.1056/NEJMe068260
4. Incorvati JA, Shah S, Mu Y, Lu J. Targeted therapy for HER2 positive breast cancer. J Hematol Oncol. (2013) 6:38. doi: 10.1186/1756-8722-6-38
5. Smalley KS, Nathanson KL, Flaherty KT. Genetic subgrouping of melanoma reveals new opportunities for targeted therapy. Cancer Res. (2009) 69:3241–4. doi: 10.1158/0008-5472.CAN-08-4305
6. Chang DZ, Kumar V, Ma Y, Li K, Kopetz S. Individualized therapies in colorectal cancer: KRAS as a marker for response to EGFR-targeted therapy. J Hematol Oncol. (2009) 2:18. doi: 10.1186/1756-8722-2-18
7. Siravegna G, Sartore-Bianchi A, Nagy RJ, Raghav K, Odegaard JI, Lanman RB, et al. Plasma HER2 (ERBB2) copy number predicts response to HER2-targeted therapy in metastatic colorectal cancer. Clin Cancer Res. (2019) 25:3046–53. doi: 10.1158/1078-0432.CCR-18-3389
8. Hsueh CT, Liu D, Wang H. Novel biomarkers for diagnosis, prognosis, targeted therapy and clinical trials. Biomark Res. (2013) 1:1. doi: 10.1186/2050-7771-1-1
9. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. (2018) 378:731–9. doi: 10.1056/NEJMoa1714448
10. Gaddipati H, Liu K, Pariser A, Pazdur R. Rare cancer trial design: lessons from FDA approvals. Clin Cancer Res. (2012) 18:5172–8. doi: 10.1158/1078-0432.CCR-12-1135
11. Munoz J, Kurzrock R. Targeted therapy in rare cancers–adopting the orphans. Nat Rev Clin Oncol. (2012) 9:631–42. doi: 10.1038/nrclinonc.2012.160
12. Greenlee RT, Goodman MT, Lynch CF, Platz CE, Havener LA, Howe HL. The occurrence of rare cancers in U.S. adults, 1995-2004. Public Health Rep. (2010) 125:28–43. doi: 10.1177/003335491012500106
13. DeSantis CE, Kramer JL, Jemal A. The burden of rare cancers in the United States. CA Cancer J Clin. (2017) 67:261–72. doi: 10.3322/caac.21400
14. Gatta G, van der Zwan JM, Casali PG, Siesling S, Dei Tos AP, Kunkler I, et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur J Cancer. (2011) 47:2493–511. doi: 10.1016/j.ejca.2011.08.008
15. Chi Le PL, Truong C, Le H. Incidence estimates and trends of nonmelanoma skin cancer in the United States, 1999-2016. Pharmacoepidemiol Drug Saf. (2019) 28:78. doi: 10.1001/archdermatol.2010.19
16. Chen Z, Zhang C, Pan Y, Xu R, Xu C, Chen Z, et al. T cell receptor β-chain repertoire analysis reveals intratumour heterogeneity of tumour-infiltrating lymphocytes in oesophageal squamous cell carcinoma. J Pathol. (2016) 239:450–8. doi: 10.1002/path.4742
17. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. (2017) 2017:1–16. doi: 10.1200/PO.17.00011
18. Luo J, Gu D, Lu H, Liu S, Kong J. Coexistence of a Novel PRKCB-ALK, EML4-ALK double-fusion in a lung adenocarcinoma patient and response to crizotinib. J Thorac Oncol. (2019) 14:e266–e268. doi: 10.1016/j.jtho.2019.07.021
19. Zhao J, Lin G, Zhuo M, Fan Z, Miao L, Chen L, et al. Next-generation sequencing based mutation profiling reveals heterogeneity of clinical response and resistance to osimertinib. Lung Cancer. (2019) 141:114–18. doi: 10.1016/j.lungcan.2019.10.021
20. Sun S, Liu Y, Eisfeld AK, Zhen F, Jin S, Gao W, et al. Identification of germline mismatch repair gene mutations in lung cancer patients with paired tumor-normal next generation sequencing: a retrospective Study. Front Oncol. (2019) 9:550. doi: 10.3389/fonc.2019.00550
21. Zhuo M, Guan Y, Yang X, Hong L, Wang Y, Li Z, et al. The prognostic and therapeutic role of genomic subtyping by sequencing tumor or cell-free DNA in pulmonary large-cell neuroendocrine carcinoma. Clin Cancer Res. (2019) 26:892–901. doi: 10.1158/1078-0432.CCR-19-0556
22. Yang X, Chu Y, Zhang R, Han Y, Zhang L, Fu Y, et al. Technical validation of a next-generation sequencing assay for detecting clinically relevant levels of breast cancer-related single-nucleotide variants and copy number variants using simulated cell-free DNA. J Mol Diagn. (2017) 19:525–36. doi: 10.1016/j.jmoldx.2017.04.007
Keywords: rare tumors, genomic profile, targetable genomic alterations, actionable mutation, NGS, China
Citation: Wang S, Chen R, Tang Y, Yu Y, Fang Y, Huang H, Wu D, Fang H, Bai Y, Sun C, Yu A, Fan Q, Gu D, Yi X and Li N (2020) Comprehensive Genomic Profiling of Rare Tumors: Routes to Targeted Therapies. Front. Oncol. 10:536. doi: 10.3389/fonc.2020.00536
Received: 31 December 2019; Accepted: 25 March 2020;
Published: 21 April 2020.
Edited by:
Lin Zhang, University of Pittsburgh, United StatesReviewed by:
Shi-Yong Sun, Emory University, United StatesDaniele Vergara, University of Salento, Italy
Copyright © 2020 Wang, Chen, Tang, Yu, Fang, Huang, Wu, Fang, Bai, Sun, Yu, Fan, Gu, Yi and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Yi, eWl4QGdlbmVwbHVzLm9yZy5jbg==; Ning Li, bGluaW5nQGNpY2Ftcy5hYy5jbg==
†These authors have contributed equally to this work
 Shuhang Wang
Shuhang Wang Rongrong Chen
Rongrong Chen Yu Tang
Yu Tang Yue Yu
Yue Yu Yuan Fang
Yuan Fang Huiyao Huang
Huiyao Huang Dawei Wu
Dawei Wu Hong Fang
Hong Fang Ying Bai
Ying Bai Chao Sun
Chao Sun Anqi Yu
Anqi Yu Qi Fan
Qi Fan Dejian Gu
Dejian Gu Xin Yi
Xin Yi Ning Li
Ning Li