- 1EPIGET LAB, Department of Clinical Sciences and Community Health, Università degli Studi di Milano, Milan, Italy
- 2Epidemiology Unit, Department of Preventive Medicine, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan, Italy
Malignant pleural mesothelioma (MPM) is a rare and aggressive cancer, which originates from the mesothelial cells of the pleura and is associated with asbestos exposure. In light of its aggressive nature, late diagnosis and dismal prognosis, there is an urgent need for identification of biomarkers in easily accessible samples (such as blood) for early diagnosis of MPM. In the last 10 years, epigenetic markers, such as DNA methylation and microRNAs (miRNAs), have gained popularity as possible early diagnostic and prognostic biomarkers in cancer research. The aim of this review is to provide a critical analysis of the current evidences on circulating epigenetic biomarkers for MPM and on their translational potential to the clinical practice for early diagnosis and for prognosis.
Introduction
Malignant Mesothelioma is a rare cancer originating from the mesothelial cells of pleura (i.e., Malignant Pleural Mesothelioma, MPM; 80–90%), peritoneum (10–15%), and pericardium (<5%). It is characterized by a long latency period (≥30–60 years) and non-specific symptoms, and thus often implicate late diagnosis and poor survival [1].
MPM presents with heterogeneous histological features, and can be classified in three main histological subtypes, depending on cellular morphology and biological markers: epithelioid mesothelioma represents the most common form (50–70% of cases) and shows polygonal, oval, or cuboidal cells similar to carcinomas; fibrous or sarcomatoid type (10–20%) is characterized by spindle cell morphology similar to those of sarcomas; the mixed or biphasic subtype (30%) presents both the epithelioid and sarcomatoid components, in different proportions [2]. High levels of cytokeratin 5 are expressed in most mesotheliomas regardless of subtype; epithelioid mesothelioma expresses high levels of calretinin, while sarcomatoid mesothelioma does not [3]. If compared to sarcomatoid mesothelioma, the epithelioid subtype is less aggressive, highly sensitive, and more responsive to chemotherapy, thus resulting in a longer survival [4, 5].
Although MPM is considered a rare malignancy (prevalence 1–9/100,000), about 40,000 deaths have been estimated to occur each year globally for asbestos-related exposures [6, 7]. Asbestos refers to a group of naturally occurring mineral silicate fibers classified into two main families, the serpentines and the amphiboles [1]. The serpentines consist of chrysotile with characteristic short and curly fibers, also called “white asbestos,” and account for 95% of asbestos in commercial use. The amphiboles, with straight and longer fibers, include crocidolite or “blue asbestos,” amosite, tremolite, actinolite, and anthophyllite [1, 8]. The World Health Organization estimates that 125 million people annually around the world are exposed to asbestos. The International Agency for Research on Cancer confirmed that all fibrous forms of asbestos are carcinogenic to humans, causing mesothelioma, cancer of the lung, larynx, and ovary. Evidences are limited in humans for pharynx, stomach, and colorectal cancers [9].
Exposure to asbestos other than in the occupational setting affects also the families of asbestos-workers, people living close to places where asbestos is mined or processed, and the populations exposed to the fragmentation of asbestos artefacts [10–13].
The observation that mesothelioma develops in a minority of asbestos-exposed individuals [14] suggests a sort of hyper-susceptibility to asbestos carcinogenic potential, probably due to the combination of environmental exposures and genetic susceptibility [15]. Various studies have shown an association between MPM and the oncogenic simian virus 40 [SV40; [16, 17]], suggesting a transforming synergic action of asbestos and SV40 [18, 19] however the evidence supporting this association is still controversial [20, 21]. Exposure to ionizing radiations seems to play a strong role in the development of mesothelioma [22, 23]. Other environmental exposures reported as risk factors for MPM are erionite in Turkey and fluoro-edenite in Italy [24]. On the other hand, a recent breakthrough in the study of mesothelioma susceptibility has emerged. Indeed, germline mutations in different genes mainly involved in DNA damage repair confer moderate-to-high genetic risk of MPM development [25]. The BAP1-tumor predisposition syndrome is the most studied genetic condition associated with MPM development and is caused by mutations in the BRCA1-associated protein 1 (BAP1) gene [14, 26–32]. This new evidence should be taken into account together with environmental/occupational exposures, as it may lead to a different risk stratification.
Although the etiology of MPM is well consolidated, patients are usually diagnosed in an advanced phase, when radical surgery cannot be performed, and the current available chemotherapy is not effective, thus leading to poor survival [6, 33].
Currently, diagnosis is invasive and relies mainly on morphological analysis, with malignant growths characterized by deep stromal invasion with dense cells and complex growth patterns [2]. Although immunohistochemical markers have improved in recent years, the lack of biomarkers able to discriminate between the different subtypes continues to hamper the diagnosis of MPM [34]. Recently, high-throughput analyses have uncovered key genomic and epigenomic alterations driving MPM [35, 36].
In this context, epigenetic markers such as DNA methylation and microRNAs (miRNAs) are emerging as promising biomarkers for several cancer types, including MPM. While genetic biomarkers may differ from case to case in most cancer patients (i.e., each patient may carry a different mutation within the same gene), different subjects show variable levels of epigenetic biomarkers in a specific target depending on health/disease status [37, 38].
All the above supports the need for identification of appropriate biomarkers that can be easily evaluated in accessible tissues, in order to detect the disease at earlier stages and improve prognosis. High-throughput molecular profiling of tumors, blood, and pleural fluids is currently shedding light on novel candidate biomarkers for early diagnosis, and revealing potential new therapeutic targets for improved treatment.
The purpose of this review is to provide a critical analysis of the available evidence on circulating epigenetic biomarkers for MPM and on their effective validity for early diagnosis or prognosis.
What Are Circulating Epigenetic Biomarkers?
The development of non-invasive methods to diagnose and monitor tumors is a major challenge in oncology. The analysis of liquid samples such as plasma, serum, urine, or cerebrospinal fluid (also known as liquid biopsy) is a suitable approach for the characterization of markers related to cancer progression, as these biological fluids are easy to collect [39–41].
An ideal biomarker for cancer detection should be easily and cheaply measurable and should allow to identify the disease at an early stage [42]. Circulating biomarkers can be released into the bloodstream by cells of different origins [e.g., Peripheral Blood Mononuclear Cells (PBMCs), and cancer cells] through various mechanisms, such as necrosis, apoptosis, or release of Extracellular vesicles (EVs) [43]. While circulating biomarkers derived from cancer cells could potentially allow the identification of a pathological condition with high specificity, those deriving from PBMCs may show higher sensitivity in pointing out the global response of the organism toward a detrimental condition.
Besides numerous protein markers largely investigated in MPM [mesothelin, osteopontin, fibulin, HMGB1 protein, etc.; [44–48]] also DNA, mRNA, and miRNA are released by cells and circulate in the blood of cancer patients. In this context, the unique properties of epigenetic biomarkers, expecially DNA methylation and miRNA expression, make them well suited as promising diagnostic and prognostic tools. DNA methylation is stable for a long time and miRNAs are particularly resistant to RNase-degradation. Moreover, aberrant peripheral epigenetic modifications have been frequently observed in early-stage cancer patients [38, 43, 49, 50]. Thus, circulating epigenetic biomarkers are promising and dynamic markers for early cancer detection, progression, and response to therapy.
DNA Methylation
DNA methylation involves the addition of a methyl group to the fifth carbon of the cytosine base, forming a 5-methyl-cytosine [51]. DNA methylation occurring in gene promoters can directly alter gene expression, by inhibiting the access to DNA of the transcriptional machinery function. This methylation is often referred to as “gene-specific methylation” [52]. On the other hand, methylation taking place in repetitive transposable elements (e.g., Alu, LINE-1, etc.) favors compaction of the chromatin structure, thus increasing DNA resistance to toxicants [53].
The patterns of DNA methylation are defined early during development, with two critical waves of methylation and demethylation occurring during embryogenesis [54]. Differentiated cells develop a stable and unique DNA methylation pattern that regulates tissue-specific gene transcription, and only cytosine followed by guanine can be efficiently methylated by DNA-methyltransferase [55]. Although DNA methylation is stable, it can be modified throughout life by several factors such as ageing, lifestyle, environmental exposures, and diseases. It thus represents an adaptive phenomenon linking environmental factors and the development of pathologic phenotypes such as cancers [56]. DNA methylation changes are considered to possibly play a role also in MPM development and progression, and have therefore been suggested as a potential tool for early diagnosis as well as for prognosis [57].
Several studies have been performed to evaluate alterations of DNA methylation in mesothelioma tumor samples [36, 50, 57], but very few have focused on alteration of DNA methylation in blood as circulating marker.
Fischer et al. [58] examined the methylation of nine gene specific promoters by two-stage methylation specific PCR in serum DNA of 43 patients with malignant mesothelioma (both pleural and peritoneal). Although the analysis of single gene methylation did not influence prognosis, the combined hypermethylation of the tumor-suppressors RARβ, DAPK, and RASSF1A was associated with shorter overall survival.
In the Arginine Deiminase and Mesothelioma (ADAM) study (a phase 2 randomized clinical trial), the role of “pegylated arginine deiminase” (ADI-PEG20), an arginine-lowering agent, was assessed in MPM patients. Indeed, arginine deprivation is synthetically lethal in cancers, such as mesothelioma, that are argininosuccinate synthetase 1 (ASS1)-negative. Among several molecular and biochemical parameters evaluated in plasma, promoter, and gene-body methylation of ASS1 was also assessed by methylation array. The authors reported that differential ASS1 gene-body methylation correlated with ASS1 protein levels, and longer arginine deprivation was associated with increased progression-free survival [59].
Guarrera et al. [60] evaluated DNA methylation levels in the whole blood as potential diagnostic markers for MPM. The study was conducted on asbestos exposed subjects, 163 MPM cases, and 137 non-MPM controls. Genome-wide methylation array allowed identifying differential methylation mainly in immune system–related genes. The top hypomethylated single-CpG island was detected in FOXK1 gene, an interactor of BAP1, which was found mutated in MPM tissue and constitutionally mutated in familial MPM, as described above. The analysis allowed to identify signatures of differential methylation in DNA from whole blood, between asbestos exposed MPM cases and controls.
The very few studies performed so far make it difficult to synthetically evaluate circulating DNA methylation as a potential biomarker for MPM. In addition, some biological factors must be taken into account. First, the amount of tumor-derived DNA fraction in blood is known to change according to the dimension and the state of the tumor [61]. For instance, the lung cancer-derived DNA fraction is very low in patients with relatively large tumors (e.g., 100 cm3) and it is generally undetectable in patients with smaller tumors, which is even more critical [62]. Second, DNA methylation in the cellular blood fraction might be only partially correlated to the methylation pattern of cancer cells, but rather represent a nonspecific marker related to the immune response toward the tumor. These factors greatly limit the use of circulating DNA methylation as diagnostic marker. Its possible role as prognostic marker needs to be further evaluated in larger populations.
miRNAs
miRNAs are short RNA molecules of about 22 nucleotides in length. miRNAs regulate gene expression at a post-transcriptional level, by silencing protein expression through cleavage and degradation of the mRNA transcript or by inhibiting translation [63]. A single miRNA is able to bind multiple mRNA targets (more than 100), in a sequence-specific manner, and a single mRNA target can thus be regulated by different miRNAs [64]. miRNAs play crucial roles in several physiological and pathological processes such as cell growth, differentiation, proliferation and metabolism, angiogenesis, stress response, tissue remodeling, disease, and malignancy development [65]. Unique miRNA expression profiles are associated with different cancer types [66–68], and it is noteworthy that about 50% of miRNA genes in the human genome are found in genomic regions associated with cancer susceptibility [69, 70]. miRNAs are involved also in cell-to-cell communications, as they can be released by active secretion in EVs, or more rarely by energy-free passive leakage of cellular miRNAs from disrupted cells [71].
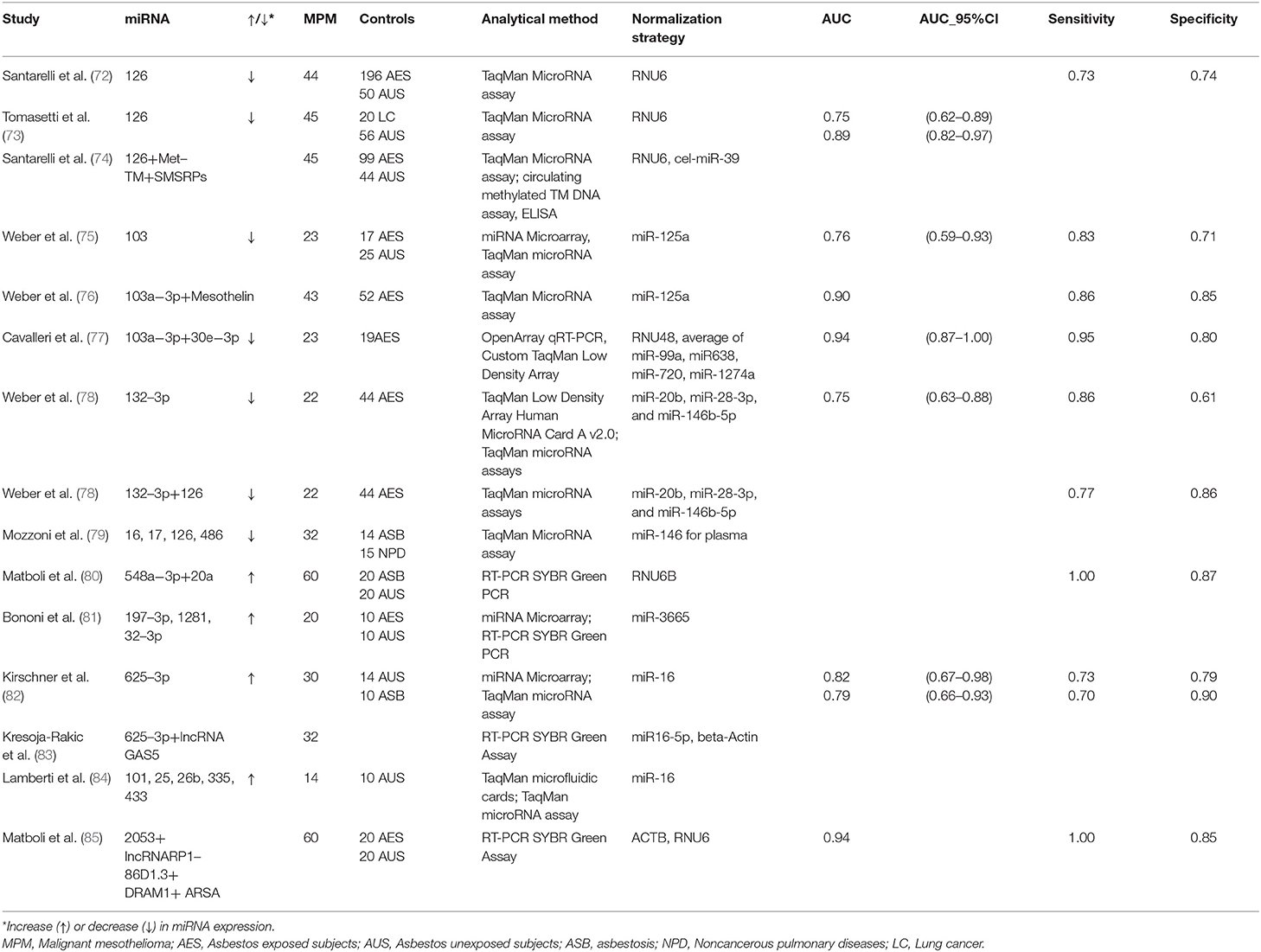
The possibility of considering circulating miRNAs as potential diagnostic and/or prognostic biomarkers for MPM has been explored for almost ten years. The main studies evaluating miRNAs are summarized in Table 1.
Santarelli et al. [72] showed a reduced miR-126 expression in serum samples of MPM patients in comparison with either asbestos exposed subjects or unexposed healthy controls. The discrimination power among groups was however moderate (Sensitivity = 60%, Specificity = 74%, and Sensitivity = 73%, Specificity = 74%, respectively). A subsequent study by Tomasetti et al. [73] confirmed miR-126 downregulation in serum from both 45 MPM and 20 Non-Small-Cell Lung Carcinoma (NSCLC) patients when compared to 56 healthy controls. Furthermore, low levels of miR-126 were strongly associated with worse prognosis in MPM patients.
miR-103 was reported to be significantly down-regulated in the blood cell fraction of 23 patients with MPM, compared to 17 subjects formerly exposed to asbestos, and 25 controls from the general population. The observed differential expression levels allowed discriminating between mesothelioma patients and asbestos-exposed controls with a sensitivity of 83% and a specificity of 71%. Sensitivity and specificity for discrimination between mesothelioma patients and healthy unexposed controls were 78 and 76%, respectively [75].
Our research group was the first to investigate miRNA expression in plasmatic EVs. EVs are membrane-surrounded structures released by all cell types under both physiological and pathological conditions. EVs facilitate intercellular communication processes, as they are able to transfer biologically active molecules such as DNA, RNA, miRNAs, proteins, and lipids [86, 87]. We investigated 23 MPM patients and 19 cancer-free subjects with past asbestos exposure, and found a two miRNA (miR-103a-3p and miR-30e-3p) signature able to discriminate the two groups with high sensitivity (95.5%) and specificity (80%) [77].
The expression of miR-625-3p was significantly higher in plasma/serum of 30 MPM patients and allowed to discriminate between cases and controls (the latter consisting of 14 healthy subjects and 10 subjects with asbestosis), in both the original and in an independent series of patients. miR-625-3p was also found upregulated in tumor samples from 18 MPM patients vs. nonmalignant pleura samples, suggesting a potential connection between circulating miRNAs and the tissue of origin [82].
Weber et al. [78] found reduced miR-132-3p expression levels in the plasma of 22 MPM patients compared to 44 asbestos-exposed controls, with a sensitivity of 86% and a specificity of 61%. In order to improve the marker performance, they also measured two miRNAs (miR-126 and miR625-3p) previously reported as possible biomarkers for MPM [72, 73]. Only miR-126 was significantly differentially expressed between the two groups. The combination of miR-132-3p and miR-126 improved the diagnostic performance.
Mozzoni et al. [79] quantified miRNA-16, miRNA-17, miRNA-126, and miRNA-486 in the plasma of 32 MPM patients, 14 subjects with asbestosis, and 15 subjects with other non-neoplastic pulmonary diseases. In addition, miRNA expression was evaluated in 24 formalin-fixed, paraffin-embedded tissues (FFPE) of the MPM subjects. All investigated miRNAs were downregulated in the plasma of subjects with MPM or asbestosis compared to the healthy subjects. Furthermore, the expression of miRNA-16 in both plasma and tissue was positively related with cumulative survival.
Serum miR-548a-3p and miR-20a levels were assessed in 60 newly diagnosed MPM patients, 20 asbestos exposed subjects without MPM and 20 healthy subjects. Significant overexpression of miR-548a-3p and miR-20a was observed in MPM compared with asbestos exposed and healthy control groups, and the combined serum miRNAs showed good sensitivity and specificity [80].
Bononi et al. [81] conducted a small study on 10 MPM patients, 10 past exposed workers, and 10 healthy controls. They found upregulated miR-197-3p, miR-1281, and miR-32-3p in the sera of patients with MPM compared to those of healthy controls, upregulated miR-197-3p and miR-32-3p in MPM compared to past-exposed workers, and upregulated miR-1281 in both MPM and exposed workers compared to healthy subjects.
Lamberti et al. [84] evaluated miRNA expression in the serum of 14 MPM patients, and 10 control subjects with non-neoplastic pleural effusions. Five miRNAs (miR-101, miR-25, miR-26b, miR-335, and miR-433) were upregulated, while two (miR-191, miR-223) were downregulated. miR-29a and miR-516 were expressed exclusively in MPM patients. Based on these findings, the authors proposed two miRNA signatures characterized by different combinations of down- and up-regulated miRNAs predicting histotype and survival, but the small sample size prevents firm conclusions.
Despite the intense research activity, the translation of these results from research to clinical practice is still problematic [88]. First of all, most studies were cross-sectional and involved late stage patients, leaving open the real role of miRNAs as early diagnostic markers. Moreover, other factors hamper the reproducibility of the results, such as the limited sample sizes and the different selection criteria of controls (unexposed healthy subjects, asbestos exposed subjects with benign diseases, other cancer patients, etc.). Further inconsistencies derive from the lack of standardization in laboratory methods, techniques (types of platform, standardization procedures, and validation) and the still poor knowledge of the factors that influence miRNAs (hemolysis, age, sex, BMI, etc.). In particular, the studies previously described took advantage of different types of analytical methods and platforms (Table 1). Moreover, each study adopted different normalization strategies, thus curbing the translational potential of the obtained results (Table 1). The majority of the cited studies used a single gene as normalizer. However, single reference miRNA is not sufficient to obtain reliable data, and the combination of several normalizers may be more appropriate [89]. For instance, some studies normalized data by using the small nucleolar RNA RNU6 [72, 73], generally expressed at low and variable levels in blood and known to be altered in chronic inflammation [89–91]. Two studies used miR-16 as normalizer [82, 84], although its expression is known to be dependent on the hemolysis of samples [82]. In order to overcome the biases due to normalization, various methods have been proposed. The so-called miRNA ratio approach is based on the ratio between up- and down-regulated miRNAs within the same sample [91]. Another normalization strategy named “Global Mean Normalization” uses the average expression level of all miRNAs detected in a sample as a normalization factor [92]. For a more detailed analysis of normalization strategies in circulating miRNAs, we refer to other reviews specifically focused on the topic [92, 93].
Very recently, Weber et al. [94] tried to overcome the limitation related to the cross-sectional design by examining blood samples collected before MPM diagnosis. They analyzed three circulating miRNAs (miR-132-3p, miR-126-3p, and miR-103a-3p), previously reported as differentially expressed among MPM patients and asbestos exposed subjects, using a nested case-control approach. Seventeen mesothelioma cases were identified in a German cohort of asbestos exposed workers [MoMAr cohort [95]] for which plasma samples were available before the date of diagnosis, with a median time between sample collection and diagnosis of about 9 months. Each case was matched with two cancer-free controls by age, gender, date of blood collection, and smoking status. None of the analyzed miRNAs was differentially expressed between the two groups, and no differences in miRNA expression was detected when cases were stratified by time between sample collection and date of diagnosis. Moreover, when using a high specificity cut-off (an important characteristic for early diagnostic markers to avoid false positive results), none of the cases was detected by the examined miRNAs.
These findings limit the use of miRNAs as early diagnostic markers and leave open the debate on miRNAs as suitable markers in clinical practice. Whether the detected miRNAs are specific of MPM or rather indicative of a disease status of the subjects remains an unsolved question. Just to make some examples, alteration of circulating miR-126 expression has been associated not only with MPM but also with other neoplastic conditions [e.g., NSCLC [96] or colorectal carcinoma [97]], and with non-neoplastic conditions such as diabetes [98]; in addition, altered expression of circulating miR-103a-3p was associated also with prostate cancer [99].
Moreover, most of the miRNAs altered in MPM were downregulated. The downregulation of circulating miRNAs can be strongly influenced by the tumor growth rather than representing an initial sign of the neoplastic lesion: this might limit the utility of miRNA-based liquid biopsy in detecting the presence of small cancer sites at early stages [100]. On the other hand, focusing on miRNAs which are unexpressed or low-abundant in normal conditions but become highly expressed in cancer cells could overcome this problem, allowing that even a small tumor could generate enough of a rare miRNA to be detected in blood [101].
Another puzzling issue is the concept of “circulating miRNA” itself, as different protocols of miRNA extraction result in a different selection of miRNA origins. Cell-free miRNAs circulating in the bloodstream have been found to be either associated with EVs or rather to exist in combination with a variety of proteins (e.g., lipoproteins, Ago2 protein, or other RNA binding proteins). While EV-associated miRNAs are in majority the result of an active process, in which they are sorted and packed with a functional role (either from cancer or immune cells), protein-bound miRNAs might also be the result of an uncontrolled cellular release (e.g., after cell necrosis) from any tissue within the body. This functional difference reflects the pattern of expression observed in the two components. The large majority of the studies available to date has used whole plasma or serum as a source of miRNAs: they therefore did not allow to specifically investigate cancer tissue miRNAs, but rather a generic response to tumor development.
Recently, it has been hypothesized that cancer-derived EVs may be selectively captured, in order to extract miRNA signatures representative of the cell of origin. This new perspective may be superior to whole plasma/serum analysis as it would dramatically increase the analytical specificity of the procedure and support its application as an early diagnostic tool.
Combination Of Circulating Molecular Biomarkers
Taking into account the limitations mentioned above, many authors [74, 83, 85, 94, 102] suggested to explore together markers of different origin to overcome the poor sensitivity and specificity of single markers. These studies combine proteins (e.g., mesothelin, fibulin, and osteopontin concentration) and different epigenetic biomarkers, including DNA methylation, expression of miRNA, and also expression of long-noncoding RNAs (lncRNAs), which are non-coding RNAs mostly involved in transcriptional regulation [103].
Santarelli et al. [74] evaluated two epigenetically regulated markers in MPM (miR-126 and methylated thrombomodulin promoter, and Met-TM) together with Soluble Mesothelin-Related Proteins (SMRPs) levels in blood serum of 45 MPM patients, 99 asbestos-exposed subjects, and 44 unexposed healthy controls. The combination of the three biomarkers only slightly improved the diagnostic performance in comparison to SMRPs alone.
Weber et al. [76] combined mesothelin and miR-103a-3p expression levels. Mesothelin concentration was evaluated in plasma, while miR-103a-3p in the cellular blood fraction. The study was conducted in 43 patients with MPM and 52 health individuals previously exposed to asbestos. The combination of mesothelin and miR-103a-3p showed a good sensitivity of 95% and a specificity of 81%.
Matboli et al. [85] measured a “MPM-specific RNA-based biomarker panel” in the serum of 60 MPM newly diagnosed patients, 20 healthy workers with past asbestos exposure, and 20 healthy subjects. The panel included the DNA damage regulated autophagy modulator 1 (DRAM1) and arylsulfatase A (ARSA), together with their epigenetic regulators: the microRNA (miR-2053) and the lncRNA RP1-86D1.3. MPM patients showed a higher expression of hsa-miRNA-2035 and lncRNA-RP1-86D1.3 and a lower expression of ARSA and DRAM1 (p < 0.001) compared to asbestos-exposed subjects and healthy controls. The diagnostic value of the combined expression analysis showed 100% sensitivity, 85% specificity, and 94% accuracy. Moreover, the authors reported that miR-2053 expression was an independent prognostic factor for progression-free survival.
Kresoja-Rakic et al. [83] evaluated the prognostic value of miR-625-3p together with the long-noncoding RNA GAS5 in the plasma of 36 MPM patients before and after platinum neo-adjuvant chemotherapy. The combination of increased expression of miR-625-3p and decreased expression of GAS5 was significantly associated with disease progression (p = 0.0393). Moreover, decreased levels of GAS5 were associated with shorter median overall and progression-free survival compared with that of patients with increased levels of GAS5 (p = 0.0308).
Conclusion
Due to the insidious onset of MPM characterized by non-specific symptoms, that often leads to a diagnosis in advanced stages and consequent poor prognosis, a great amount of studies has focused on the search for non-invasive biological indicators that might allow to shorten the diagnostic delay and be applied in high-risk subpopulations, such as those formerly exposed to asbestos.
The ideal marker (or a combination of several markers) should have some important features such as minimal invasiveness (i.e., it should be measurable in easily obtainable biological fluids such as blood), high specificity to avoid false positives in healthy subjects, sufficient sensitivity to identify subjects with MPM, and good ability to discriminate between MPM and other diseases. As a matter of fact, if used in the clinical setting for the purposes of diagnosis and differential diagnosis (i.e., not for screening), a good marker should have an excellent discriminating power between healthy and sick people and between different pathologies when applied in addition to imaging.
Notwithstanding the plethora of existing old and new biomarkers, none of the current available markers is sufficiently reliable to be used in the surveillance of subjects exposed to asbestos or in the early diagnosis of MPM. As underlined by the Helsinki 2014 Criteria, “At this point, no specific recommendations can be made regarding these biomarkers for screening or other purposes” [104]. The rarity of the pathology even in cohorts of subjects exposed to asbestos and the lack of clear evidence about effective treatments that lead to improvements in survival (reduction of mortality) in cases diagnosed earlier are other factors that limit the use of the markers described so far and make it problematic. Even their possible use as diagnostic markers that can guide in the selection of subjects needing further investigation to reach an early detection of the disease remains controversial.
All the issues discussed above highlight the need for validation studies on large populations, which focus on MPM in early stages, applying shared and standardized criteria to study design, sample collection and analysis, and laboratory methods. To this end, joint research programs are needed not only at national but also at transnational level, to integrate different (clinical, laboratory, epidemiological) competences and avoid an ineffective use of resources. Furthermore, taking into account the heterogeneity of malignant mesothelioma, particularly relevant could be the study of panels of markers of different biological significance, such as “mesomiRNAs” [88, 102], miRNAs so far associated with MPM, and other protein markers along with their evaluation in repeated measures that monitor symptomatic patients over time.
Author Contributions
LF prepared What Are Circulating Biomarkers? DNA Methylation, miRNAs, and Combination of Circulating Biomarkers paragraphs. MC prepared the Conclusion paragraph and supervised the final version of the manuscript. CM prepared the Introduction. AP supervised the preparation and revised the final version of the manuscript.
Funding
AP received support from Associazione Italiana per la Ricerca sul Cancro (AIRC) grant IG 2015 Id.17692.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Roe OD, Stella GM. Malignant pleural mesothelioma: history, controversy and future of a manmade epidemic. Eur Respir Rev. (2015) 24:115–31. doi: 10.1183/09059180.00007014
2. Husain AN, Colby TV, Ordonez NG, Allen TC, Attanoos RL, Beasley MB, et al. Guidelines for pathologic diagnosis of malignant mesothelioma 2017 update of the consensus statement from the International Mesothelioma Interest Group. Arch Pathol Lab Med. (2018) 142:89–108. doi: 10.5858/arpa.2017-0124-RA
3. Panou V, Vyberg M, Weinreich UM, Meristoudis C, Falkmer UG, Roe OD. The established and future biomarkers of malignant pleural mesothelioma. Cancer Treat Rev. (2015) 41:486–95. doi: 10.1016/j.ctrv.2015.05.001
4. Bille A, Krug LM, Woo KM, Rusch VW, Zauderer MG. Contemporary analysis of prognostic factors in patients with unresectable malignant pleural mesothelioma. J Thorac Oncol. (2016) 11:249–55. doi: 10.1016/j.jtho.2015.10.003
5. Montanaro F, Rosato R, Gangemi M, Roberti S, Ricceri F, Merler E, et al. Survival of pleural malignant mesothelioma in Italy: a population-based study. Int J Cancer. (2009) 124:201–7. doi: 10.1002/ijc.23874
6. Rossini M, Rizzo P, Bononi I, Clementz A, Ferrari R, Martini F, et al. New perspectives on diagnosis and therapy of malignant pleural mesothelioma. Front Oncol. (2018) 8:91. doi: 10.3389/fonc.2018.00091
7. Furuya S, Chimed-Ochir O, Takahashi K, David A, Takala J. Global asbestos disaster. Int J Environ Res Public Health. (2018) 15:E1000. doi: 10.3390/ijerph15051000
8. Pira E, Donato F, Maida L, Discalzi G. Exposure to asbestos: past, present and future. J Thorac Dis. (2018) 10(Suppl 2):S237–S45. doi: 10.21037/jtd.2017.10.126
9. Straif K, Benbrahim-Tallaa L, Baan R, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens–Part C: metals, arsenic, dusts, and fibres. Lancet Oncol. (2009) 10:453–4. doi: 10.1016/S1470-2045(09)70134-2
10. Bernstein DM, Rogers RA, Sepulveda R, Donaldson K, Schuler D, Gaering S, et al. The pathological response and fate in the lung and pleura of chrysotile in combination with fine particles compared to amosite asbestos following short-term inhalation exposure: interim results. Inhal Toxicol. (2010) 22:937–62. doi: 10.3109/08958378.2010.497818
11. Ferrante D, Bertolotti M, Todesco A, Mirabelli D, Terracini B, Magnani C. Cancer mortality and incidence of mesothelioma in a cohort of wives of asbestos workers in Casale Monferrato, Italy. Environ Health Perspect. (2007) 115:1401–5. doi: 10.1289/ehp.10195
12. Binazzi A, Marinaccio A, Corfiati M, Bruno C, Fazzo L, Pasetto R, et al. Mesothelioma incidence and asbestos exposure in Italian national priority contaminated sites. Scand J Work Environ Health. (2017) 43:550–9. doi: 10.5271/sjweh.3676
13. Marinaccio A, Binazzi A, Bonafede M, Corfiati M, Di Marzio D, Scarselli A, et al. Malignant mesothelioma due to non-occupational asbestos exposure from the Italian national surveillance system (ReNaM): epidemiology and public health issues. Occup Environ Med. (2015) 72:648–55. doi: 10.1136/oemed-2014-102297
14. Melaiu O, Gemignani F, Landi S. The genetic susceptibility in the development of malignant pleural mesothelioma. J Thorac Dis. (2018) 10(Suppl 2):S246–S52. doi: 10.21037/jtd.2017.10.41
15. Matullo G, Guarrera S, Betti M, Fiorito G, Ferrante D, Voglino F, et al. Genetic variants associated with increased risk of malignant pleural mesothelioma: a genome-wide association study. PLoS One. (2013) 8:e61253. doi: 10.1371/journal.pone.0061253
16. Rascoe PA, Jupiter D, Cao X, Littlejohn JE, Smythe WR. Molecular pathogenesis of malignant mesothelioma. Expert Rev Mol Med. (2012) 14:e12. doi: 10.1017/erm.2012.6
17. Qi F, Carbone M, Yang H, Gaudino G. Simian virus 40 transformation, malignant mesothelioma and brain tumors. Expert Rev Res Med. (2011) 5:683–97. doi: 10.1586/ers.11.51
18. Kroczynska B, Cutrone R, Bocchetta M, Yang H, Elmishad AG, Vacek P, et al. Crocidolite asbestos and SV40 are cocarcinogens in human mesothelial cells and in causing mesothelioma in hamsters. Proc Natl Acad Sci USA. (2006) 103:14128–33. doi: 10.1073/pnas.0604544103
19. Mazzoni E, Corallini A, Cristaudo A, Taronna A, Tassi G, Manfrini M, et al. High prevalence of serum antibodies reacting with simian virus 40 capsid protein mimotopes in patients affected by malignant pleural mesothelioma. Proc Natl Acad Sci USA. (2012) 109:18066–71. doi: 10.1073/pnas.1213238109
20. Lopez-Rios F, Illei PB, Rusch V, Ladanyi M. Evidence against a role for SV40 infection in human mesotheliomas and high risk of false-positive PCR results owing to presence of SV40 sequences in common laboratory plasmids. Lancet. (2004) 364:1157–66. doi: 10.1016/S0140-6736(04)17102-X
21. Shah KV. SV40 and human cancer: a review of recent data. Int J Cancer. (2007) 120:215–23. doi: 10.1002/ijc.22425
22. Carbone M, Yang H. Mesothelioma: recent highlights. Ann Transl Med. (2017) 5:238. doi: 10.21037/atm.2017.04.29
23. Goodman JE, Nascarella MA, Valberg PA. Ionizing radiation: a risk factor for mesothelioma. Cancer Causes Control. (2009) 20:1237–54. doi: 10.1007/s10552-009-9357-4
24. Attanoos RL, Churg A, Galateau-Salle F, Gibbs AR, Roggli VL. Malignant mesothelioma and its non-asbestos causes. Arch Pathol Lab Med. (2018) 142:753–60. doi: 10.5858/arpa.2017-0365-RA
25. Betti M, Aspesi A, Sculco M, Matullo G, Magnani C, Dianzani I. Genetic predisposition for malignant mesothelioma: a concise review. Mutat Res. (2019) 781:1–10. doi: 10.1016/j.mrrev.2019.03.001
26. Yoshikawa Y, Sato A, Tsujimura T, Emi M, Morinaga T, Fukuoka K, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. (2012) 103:868–74. doi: 10.1111/j.1349-7006.2012.02223.x
27. Neviere Z, Berthet P, Polycarpe F, Dubos-Arvis C, Do P, Gervais R. [Malignant mesothelioma and constitutional BAP1 gene mutations]. Rev Mal Respir. (2019) 36:241–8. doi: 10.1016/j.rmr.2017.11.014
28. Metintas S, Metintas M, Ucgun I, Oner U. Malignant mesothelioma due to environmental exposure to asbestos: follow-up of a Turkish cohort living in a rural area. Chest. (2002) 122:2224–9. doi: 10.1378/chest.122.6.2224
29. Ohar JA, Cheung M, Talarchek J, Howard SE, Howard TD, Hesdorffer M, et al. Germline BAP1 mutational landscape of asbestos-exposed malignant mesothelioma patients with family history of cancer. Cancer Res. (2016) 76:206–15. doi: 10.1158/0008-5472.CAN-15-0295
30. Betti M, Aspesi A, Biasi A, Casalone E, Ferrante D, Ogliara P, et al. CDKN2A and BAP1 germline mutations predispose to melanoma and mesothelioma. Cancer Lett. (2016) 378:120–30. doi: 10.1016/j.canlet.2016.05.011
31. Betti M, Aspesi A, Ferrante D, Sculco M, Righi L, Mirabelli D, et al. Sensitivity to asbestos is increased in patients with mesothelioma and pathogenic germline variants in BAP1 or other DNA repair genes. Genes, Chromosomes Cancer. (2018) 57:573–83. doi: 10.1002/gcc.22670
32. Emri SA. The Cappadocia mesothelioma epidemic: its influence in Turkey and abroad. Ann Transl Med. (2017) 5:239. doi: 10.21037/atm.2017.04.06
33. Nowak AK. Chemotherapy for malignant pleural mesothelioma: a review of current management and a look to the future. Ann Cardiothorac Surg. (2012) 1:508–15. doi: 10.3978/j.issn.2225-319X.2012.10.05
34. Galateau-Salle F, Churg A, Roggli V, Travis WD. The 2015 World Health Organization classification of tumors of the pleura: advances since the 2004 classification. J Thorac Oncol. (2016) 11:142–54. doi: 10.1016/j.jtho.2015.11.005
35. Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. (2016) 48:407–16. doi: 10.1038/ng.3520
36. Zhang X, Tang N, Rishi AK, Pass HI, Wali A. Methylation profile landscape in mesothelioma: possible implications in early detection, disease progression, and therapeutic options. Methods Mol Biol. (2015) 1238:235–47. doi: 10.1007/978-1-4939-1804-1_12
37. Gai W, Sun K. Epigenetic biomarkers in cell-free DNA and applications in liquid biopsy. Genes. (2019) 10. doi: 10.3390/genes10010032
38. Sage AP, Martinez VD, Minatel BC, Pewarchuk ME, Marshall EA, MacAulay GM, et al. Genomics and epigenetics of malignant mesothelioma. High Throughput. (2018) 7:20. doi: 10.3390/ht7030020
39. Rolfo C, Castiglia M, Hong D, Alessandro R, Mertens I, Baggerman G, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta. (2014) 1846:539–46. doi: 10.1016/j.bbcan.2014.10.001
40. Lin J, Ma L, Zhang D, Gao J, Jin Y, Han Z, et al. Tumour biomarkers-tracing the molecular function and clinical implication. Cell Proliferation. (2019) 52:e12589. doi: 10.1111/cpr.12589
41. Volckmar AL, Sultmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer. (2018) 57:123–39. doi: 10.1002/gcc.22517
42. Thomas ML, Marcato P. Epigenetic modifications as biomarkers of tumor development, therapy response, and recurrence across the cancer care continuum. Cancers. (2018) 10:E101. doi: 10.3390/cancers10040101
43. Tomasetti M, Amati M, Neuzil J, Santarelli L. Circulating epigenetic biomarkers in lung malignancies: from early diagnosis to therapy. Lung Cancer. (2017) 107:65–72. doi: 10.1016/j.lungcan.2016.05.023
44. Chen Z, Gaudino G, Pass HI, Carbone M, Yang H. Diagnostic and prognostic biomarkers for malignant mesothelioma: an update. Transl Lung Cancer Res. (2017) 6:259–69. doi: 10.21037/tlcr.2017.05.06
45. Bayram M, Dongel I, Akbas A, Benli I, Akkoyunlu ME, Bakan ND. Serum biomarkers in patients with mesothelioma and pleural plaques and healthy subjects exposed to naturally occurring asbestos. Lung. (2014) 192:197–203. doi: 10.1007/s00408-013-9526-9
46. Bonotti A, Foddis R, Landi S, Melaiu O, De Santi C, Giusti L, et al. A novel panel of serum biomarkers for MPM diagnosis. Dis Markers. (2017) 2017:3510984. doi: 10.1155/2017/3510984
47. Demir M, Kaya H, Taylan M, Ekinci A, Yilmaz S, Teke F, et al. Evaluation of new biomarkers in the prediction of malignant mesothelioma in subjects with environmental asbestos exposure. Lung. (2016) 194:409–17. doi: 10.1007/s00408-016-9868-1
48. Pei D, Li Y, Liu X, Yan S, Guo X, Xu X. Diagnostic and prognostic utilities of humoral fibulin-3 in malignant pleural mesothelioma: evidence from a meta-analysis. Oncotarget. (2017) 8:13030–8. doi: 10.18632/oncotarget.14712
49. Sekido Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis. (2013) 34:1413–9. doi: 10.1093/carcin/bgt166
50. McLoughlin KC, Kaufman AS, Schrump DS. Targeting the epigenome in malignant pleural mesothelioma. Transl Lung Cancer Res. (2017) 6:350–65. doi: 10.21037/tlcr.2017.06.06
51. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38:23–38. doi: 10.1038/npp.2012.112
52. Orphanides G, Reinberg D. A unified theory of gene expression. Cell. (2002) 108:439–51. doi: 10.1016/S0092-8674(02)00655-4
53. Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem. (2010) 52:145–67. doi: 10.1016/S0065-2423(10)52006-7
54. Ho SM, Cheong A, Adgent MA, Veevers J, Suen AA, Tam NNC, et al. Environmental factors, epigenetics, and developmental origin of reproductive disorders. Reprod Toxicol. (2017) 68:85–104. doi: 10.1016/j.reprotox.2016.07.011
55. Millar D, Holliday R, Grigg G. Five not four: history and significance of the fifth base. In: Beck S, Olek A, editors. The Epigenome, Molecular Hide and Seek. Weinheim: Wiley-VCH Verlag GmbH Co. KGaA (2003). p. 3–20.
56. Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. (2011) 3:267–77. doi: 10.2217/epi.11.22
57. Vandermeers F, Neelature Sriramareddy S, Costa C, Hubaux R, Cosse JP, Willems L. The role of epigenetics in malignant pleural mesothelioma. Lung Cancer. (2013) 81:311–8. doi: 10.1016/j.lungcan.2013.05.014
58. Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Kostrzewa M, et al. Promoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer. (2006) 54:109–16. doi: 10.1016/j.lungcan.2006.06.017
59. Szlosarek PW, Steele JP, Nolan L, Gilligan D, Taylor P, Spicer J, et al. Arginine deprivation with pegylated arginine deiminase in patients with argininosuccinate synthetase 1-deficient malignant pleural mesothelioma: a randomized clinical trial. JAMA Oncol. (2017) 3:58–66. doi: 10.1001/jamaoncol.2016.3049
60. Guarrera S, Viberti C, Cugliari G, Allione A, Casalone E, Betti M, et al. Peripheral blood DNA methylation as potential biomarker of malignant pleural mesothelioma in asbestos-exposed subjects. J Thorac Oncol. (2019) 14:527–39. doi: 10.1016/j.jtho.2018.10.163
61. Esposito A, Bardelli A, Criscitiello C, Colombo N, Gelao L, Fumagalli L, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev. (2014) 40:648–55. doi: 10.1016/j.ctrv.2013.10.003
62. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. (2017) 545:446–51. doi: 10.1038/nature22364
63. Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. (2016) 17:E1712. doi: 10.3390/ijms17101712
64. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
65. Filipow S, Laczmanski L. Blood circulating miRNAs as cancer biomarkers for diagnosis and surgical treatment response. Front Genet. (2019) 10:169. doi: 10.3389/fgene.2019.00169
66. Xue Z, Wen J, Chu X, Xue X. A microRNA gene signature for identification of lung cancer. Surg Oncol. (2014) 23:126–31. doi: 10.1016/j.suronc.2014.04.003
67. Wang F, Sun GP, Zou YF, Hao JQ, Zhong F, Ren WJ. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomarkers. (2012) 11:259–67. doi: 10.3233/CBM-2012-00284
68. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. (2005) 435:834–8. doi: 10.1038/nature03702
69. Rossi S, Sevignani C, Nnadi SC, Siracusa LD, Calin GA. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm. Genome. (2008) 19:526–40. doi: 10.1007/s00335-008-9119-8
70. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. (2004) 101:2999–3004. doi: 10.1073/pnas.0307323101
71. Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. (2013) 4:119. doi: 10.3389/fgene.2013.00119
72. Santarelli L, Strafella E, Staffolani S, Amati M, Emanuelli M, Sartini D, et al. Association of MiR-126 with soluble mesothelin-related peptides, a marker for malignant mesothelioma. PLoS One. (2011) 6:e18232. doi: 10.1371/journal.pone.0018232
73. Tomasetti M, Staffolani S, Nocchi L, Neuzil J, Strafella E, Manzella N, et al. Clinical significance of circulating miR-126 quantification in malignant mesothelioma patients. Clin Biochem. (2012) 45:575–81. doi: 10.1016/j.clinbiochem.2012.02.009
74. Santarelli L, Staffolani S, Strafella E, Nocchi L, Manzella N, Grossi P, et al. Combined circulating epigenetic markers to improve mesothelin performance in the diagnosis of malignant mesothelioma. Lung Cancer. (2015) 90:457–64. doi: 10.1016/j.lungcan.2015.09.021
75. Weber DG, Johnen G, Bryk O, Jockel KH, Bruning T. Identification of miRNA-103 in the cellular fraction of human peripheral blood as a potential biomarker for malignant mesothelioma–a pilot study. PLoS One. (2012) 7:e30221. doi: 10.1371/journal.pone.0030221
76. Weber DG, Casjens S, Johnen G, Bryk O, Raiko I, Pesch B, et al. Combination of MiR-103a-3p and mesothelin improves the biomarker performance of malignant mesothelioma diagnosis. PLoS One. (2014) 9:e114483. doi: 10.1371/journal.pone.0114483
77. Cavalleri T, Angelici L, Favero C, Dioni L, Mensi C, Bareggi C, et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS One. (2017) 12:e0176680. doi: 10.1371/journal.pone.0176680
78. Weber DG, Gawrych K, Casjens S, Brik A, Lehnert M, Taeger D, et al. Circulating miR-132-3p as a candidate diagnostic biomarker for malignant mesothelioma. Dis Markers. (2017) 2017:9280170. doi: 10.1155/2017/9280170
79. Mozzoni P, Ampollini L, Goldoni M, Alinovi R, Tiseo M, Gnetti L, et al. MicroRNA expression in malignant pleural mesothelioma and asbestosis: a pilot study. Dis Markers. (2017) 2017:9645940. doi: 10.1155/2017/9645940
80. Matboli M, Shafei AE, Azazy AE, Reda M, El-Khazragy N, Nagy AA, et al. Clinical evaluation of circulating miR-548a-3p and−20a expression in malignant pleural mesothelioma patients. Biomark. Med. (2018) 12:129–39. doi: 10.2217/bmm-2017-0224
81. Bononi I, Comar M, Puozzo A, Stendardo M, Boschetto P, Orecchia S, et al. Circulating microRNAs found dysregulated in ex-exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers. Oncotarget. (2016) 7:82700–11. doi: 10.18632/oncotarget.12408
82. Kirschner MB, Cheng YY, Badrian B, Kao SC, Creaney J, Edelman JJ, et al. Increased circulating miR-625-3p: a potential biomarker for patients with malignant pleural mesothelioma. J Thorac Oncol. (2012) 7:1184–91. doi: 10.1097/JTO.0b013e3182572e83
83. Kresoja-Rakic J, Szpechcinski A, Kirschner MB, Ronner M, Minatel B, Martinez VD, et al. miR-625-3p and lncRNA GAS5 in liquid biopsies for predicting the outcome of malignant pleural mesothelioma patients treated with neo-adjuvant chemotherapy and surgery. Noncoding RNA. (2019) 5:E41. doi: 10.3390/ncrna5020041
84. Lamberti M, Capasso R, Lombardi A, Di Domenico M, Fiorelli A, Feola A, et al. Two different serum MiRNA signatures correlate with the clinical outcome and histological subtype in pleural malignant mesothelioma patients. PLoS One. (2015) 10:e0135331. doi: 10.1371/journal.pone.0135331
85. Matboli M, Shafei AE, Ali MA, Gaber AI, Galal A, Tarek O, et al. Clinical significance of serum DRAM1 mRNA, ARSA mRNA, hsa-miR-2053 and lncRNA-RP1-86D1.3 axis expression in malignant pleural mesothelioma. J Cell Biochem. (2019) 120:3203–11. doi: 10.1002/jcb.27586
86. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. (2015) 4:27066.
87. Han L, Lam EW, Sun Y. Extracellular vesicles in the tumor microenvironment: old stories, but new tales. Mol Cancer. (2019) 18:59. doi: 10.1186/s12943-019-0980-8
88. Micolucci L, Rippo MR, Olivieri F, Procopio AD. Progress of research on microRNAs with diagnostic value in asbestos exposure: a call for method standardization. Bioscience Trends. (2017) 11:105–9. doi: 10.5582/bst.2016.01249
89. Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. (2015) 61:1333–42. doi: 10.1373/clinchem.2015.239459
90. Zhao H, Ma TF, Lin J, Liu LL, Sun WJ, Guo LX, et al. Identification of valid reference genes for mRNA and microRNA normalisation in prostate cancer cell lines. Sci Rep. (2018) 8:1949. doi: 10.1038/s41598-018-19458-z
91. Sharova E, Grassi A, Marcer A, Ruggero K, Pinto F, Bassi P, et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br J Cancer. (2016) 114:1362–6. doi: 10.1038/bjc.2016.151
92. Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J. (2015) 29:3853–62. doi: 10.1096/fj.15-271312
93. Faraldi M, Gomarasca M, Banfi G, Lombardi G. Free circulating miRNAs measurement in clinical settings: the still unsolved issue of the normalization. Adv Clin Chem. (2018) 87:113–39. doi: 10.1016/bs.acc.2018.07.003
94. Weber DG, Brik A, Casjens S, Burek K, Lehnert M, Pesch B, et al. Are circulating microRNAs suitable for the early detection of malignant mesothelioma? Results from a nested case-control study. BMC Res. Notes. (2019) 12:77. doi: 10.1186/s13104-019-4113-7
95. Johnen G, Burek K, Raiko I, Wichert K, Pesch B, Weber DG, et al. Prediagnostic detection of mesothelioma by circulating calretinin and mesothelin - a case-control comparison nested into a prospective cohort of asbestos-exposed workers. Sci Rep. (2018) 8:14321. doi: 10.1038/s41598-018-32315-3
96. Del Vescovo V, Grasso M, Barbareschi M, Denti MA. MicroRNAs as lung cancer biomarkers. World J Clin Oncol. (2014) 5:604–20. doi: 10.5306/wjco.v5.i4.604
97. Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1alpha-VEGF signaling pathway. Mol Cell Biochem. (2019) 454:177–89. doi: 10.1007/s11010-018-3462-1
98. Amr KS, Abdelmawgoud H, Ali ZY, Shehata S, Raslan HM. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br J Biomed Sci. (2018) 75:82–7. doi: 10.1080/09674845.2017.1402404
99. Mello-Grand M, Gregnanin I, Sacchetto L, Ostano P, Zitella A, Bottoni G, et al. Circulating microRNAs combined with PSA for accurate and non-invasive prostate cancer detection. Carcinogenesis. (2019) 40:246–53. doi: 10.1093/carcin/bgy167
100. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. (2015) 61:56–63. doi: 10.1373/clinchem.2014.221341
101. Leidner RS, Li L, Thompson CL. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. (2013) 8:e57841. doi: 10.1371/journal.pone.0057841
102. Micolucci L, Akhtar MM, Olivieri F, Rippo MR, Procopio AD. Diagnostic value of microRNAs in asbestos exposure and malignant mesothelioma: systematic review and qualitative meta-analysis. Oncotarget. (2016) 7:58606–37. doi: 10.18632/oncotarget.9686
103. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
Keywords: microRNA, DNA methylation, epigenetics, circulating biomarkers, mesothelioma
Citation: Ferrari L, Carugno M, Mensi C and Pesatori AC (2020) Circulating Epigenetic Biomarkers in Malignant Pleural Mesothelioma: State of the Art and critical Evaluation. Front. Oncol. 10:445. doi: 10.3389/fonc.2020.00445
Received: 01 October 2019; Accepted: 13 March 2020;
Published: 03 April 2020.
Edited by:
Yuen Yee Cheng, Asbestos Diseases Research Institute, AustraliaReviewed by:
Nobukazu Fujimoto, Okayama Rosai Hospital, JapanBen William Johnson, Asbestos Diseases Research Institute, Australia
Copyright © 2020 Ferrari, Carugno, Mensi and Pesatori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Cecilia Pesatori, YW5nZWxhLnBlc2F0b3JpJiN4MDAwNDA7dW5pbWkuaXQ=
 Luca Ferrari
Luca Ferrari Michele Carugno
Michele Carugno Carolina Mensi
Carolina Mensi Angela Cecilia Pesatori
Angela Cecilia Pesatori