- Department of Orthopedics, Huaihe Hospital, The First Affiliated Hospital of Henan University, Kaifeng, China
The epithelial-mesenchymal transition (EMT) is a vital step in osteosarcoma (OS) progression toward metastasis, but the specific molecular events governing this process are incompletely characterized, with miRNAs having increasingly been found to regulate the EMT. In this study, We assessed levels of miR-22 and its target, Twist1, via real-time PCR (qRT-PCR). We further used functional proliferation assays, measures of cell morphology, and western blotting to assess the functional relevance of miR-22 in OS and confirmed Twist1 as a miR-22 target via luciferase reporter assay. We observed a significant decrease in miR-22 levels in OS tumor samples relative to normal tissue, with such downregulating being significantly associated with tumor histological grade. When overexpressed, miR-22 impaired OS cell proliferation and EMT progression. We found Twist1 to be a direct miR-22 target, with levels of miR-22 and Twist1 mRNA being inversely correlated in patient samples. When overexpressed, miR-22 suppressed Twist1 translation and thereby attenuated the EMT in OS cells. These results clearly demonstrate that miR-22 can regulate the EMT in OS cells via targeting Twist1, thus highlighting a potentially novel pathway that can be therapeutically targeted in order to treat OS.
Introduction
Osteosarcoma (OS) is a highly prevalent form of bone cancer that most frequently impacts adolescents and younger children (1), in whom it is the second most prominent cancer-associated cause of death (2). Recent advances in surgical and chemo/radiotherapy have improved OS patient 5-year survival to 70% (3). The remaining 30% of patients, however, often exhibit high rates of local tumor recurrence, early development of tumor metastases, and limited tumor chemosensitivity, ultimately contributing to the poor outcomes in these individuals (4). There is therefore a clear need to explore the molecular mechanisms governing OS so as to highlight potential novel biomarkers of tumor development or progression, and to thereby develop better means of treating this deadly disease.
MicroRNAs (miRNAs) are small RNA molecules which lack coding potential, yet which are able to readily mediate the post-translational regulation of target mRNA expression via suppressing translation or inducing degradation (5). These miRNAs are capable of regulating a broad array of cellular processes, such as differentiation, apoptotic death, or proliferation with specific miRNAs serving to suppress or promote tumor function in a context-dependent manner (6–8). Epithelial-to-mesenchymal transition (EMT) is an essential process tightly associated with the tumor metastatic progression in a range of tumor types (9). This EMT process is associated with a reduction in the epithelial-like features of cancer cells and the acquisition of mesenchymal-like features that are necessary to mediate effective invasion and migration. ZEB1, Snail, and Twist1 are three key transcription factors known to regulate the EMT, with all three acting to repress the transcription of E-cadherin. Certain miRNAs have been shown to be capable of regulating the EMT, with miR-200 having been shown to be capable of repressing drug resistance and an epithelial phenotype in tumor cells (10, 11). Such evidence indicates that specific miRNAs are capable of modulating the EMT. Whether such miRNAs are able to modulate the EMT in the context of OS, however, remains incompletely understood, and as such further research into the role of miRNAs in this setting is vital as it has the potential to highlight novel therapeutic interventions for OS.
Several tumor types have been found to exhibit miR-22 dysregulation. For example, Zhang et al. determined that in breast cancer, miR-22 is able to target Sirt1 and thereby suppress oncongenesis and enhance cancer cell radiosensitivity (12). Xin et al. found miR-22 to target ATP citrate lyase and to thereby inhibit the growth and metastasis of OS, prostate cancer, lung cancer, and cervical cancer cells (13). The role of miR-22 in the progression of OS and in the EMT process, however, has yet to be assessed.
In the present study we explored the mechanistic role of miR-22 in OS. We determined that OS patient tissues exhibited significantly lower miR-22 levels than did normal paired tissue, and that this expression was negatively correlated with tumor stage. We further found that when overexpressed, miR-22 disrupted OS cell proliferation and the EMT via targeting the EMT-associated transcription factor Twist1. These results offer new insight into the molecular mechanisms whereby OS develops, and suggest that targeting this miR-22/Twist1 pathway may have therapeutic utility.
Materials and Methods
Human Samples
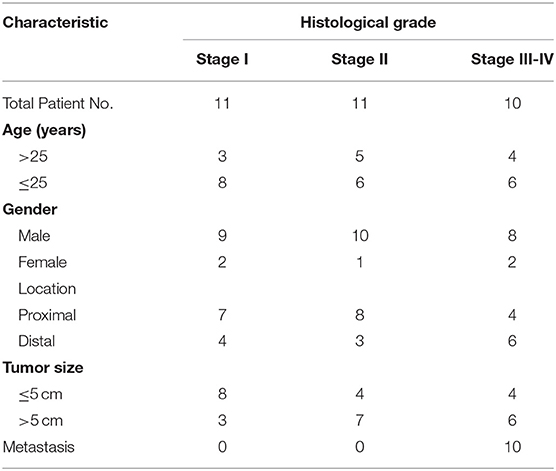
Pairs of human OS tissue samples and matched normal tissue controls were obtained from the Department of Orthopedics, Huaihe Hospital, the First Affiliated Hospital of Henan University. All samples were collected surgically, and were snap frozen with liquid nitrogen prior to analysis. We classified the samples according to Enneking system which is the common staging systems for malignant bone tumors (Table 1). The Institutional Review Board and Ethics Committee of Henan University approved this study, with patients having given written consent to participate.
Cell Culture and Transduction
The HOS and MG63 OS cell lines from ATCC were grown in RPMI-1640, whereas HEK-293T cells were grown in DMEM containing 10% FBS and penicillin/streptomycin at 37°C in a 5% CO2 incubator.
A lentiviral packaging kit (Open Biosystems, AL, USA) was used to produce lentiviruses bearing either miR-22 or negative control (NC) based on provided directions. HEK-293T cells were used for packaging. Supernatants were collected 48 h after co-transfection of these cells, and lentiviral supernatants were combined for 24 h with OS cell lines together with polybrene (2.5 μg/ml). Following transduction, stably transduced cells were selected using puromycin (1.5 μg/ml).
qRT-PCR
Trizol (Invitrogen, CA, USA) was used to isolate sample RNA according to provided direction. The stem-loop-specific primer approach was used for assessing miR-22 expression, with U6 used for normalization. Twist1 expression was assessed via using the RT Reagent Kit (Vazyme, China) to reverse transcribe total RNA, and GAPDH was used for normalizing gene expression. SYBR Green Master Mix (Vazyme, China) was used for all qRT-PCR reactions, with relative quantification (2−ΔΔCt) used for all fold change calculations.
Western Blotting
RIPA buffer containing protease inhibitors was employed to lyse cells, with supernatant protein levels being assessed via BCA assay (Beyotime, Jiangsu, China). For tissue samples, tissue was ground in liquid nitrogen before RIPA lysis, followed by processing as above. Protein samples were then separated via SDS-PAGE, transferred to a PVDF membrane (Roche, Switzerland), and subjected to Western blotting based on provided directions. The ECL Detection System (Thermo Scientific, IL, USA) was used for signal detection. Antibodies against E-cadherin and Twist1 were from Cell Signaling Technology (MA, USA). Anti-GAPDH was from Bioworld Technology (GA, USA), and anti-Vimentin was from Abcam (Cambridge, UK).
Luciferase Reporter Assay
Cells were plated in triplicate wells of 24-well plates for 24 h, after which the appropriate plasmids were transfected into the cells along with the pRL-TK Renilla plasmid using Lipofectamine 2,000 (Life Technologies). Following a 24 h incubation, the Dual-Luciferase Reporter Assay (Promega) was conducted based on provided directions.
Statistics
GraphPad Prism 5 was used for all analyses, with experiments independently performed three times. T-tests were used to compare all results, with P < 0.05 as the significance threshold. The Pearson's rank test was used to assess the relationship between miR-22 and Twist1 in human OS tissue samples.
Results
OS Tumors Exhibit Reduced miR-22 Expression Correlated With More Advanced Disease
We first assessed miR-22 expression in 32 paired human OS and normal tissue control samples via stem-loop qRT-PCR. We found that OS tissues exhibited a marked reduction in miR-22 expression relative to adjacent normal control samples (Figure 1A). We further found that there was a negative correlation between miR-22 expression and tumor histological grade (Figure 1B). This suggests that lower expression of miR-22 corresponds to a more advanced stage of OS.
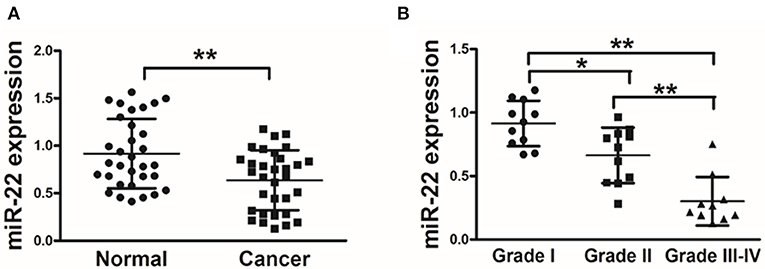
Figure 1. OS patient samples exhibit reduced miR-22 expression associated with more advanced disease. (A) qRT-PCR was used to assess miR-22 expression relative to U6 (for normalization) in 60 OS tissue pairs. (B) Relative miR-22 expression as a function of cancer stage. Data are means±SD of 3 replicates. *P < 0.05; **P < 0.01.
miR-22 Suppresses the Proliferation and EMT of OS Cells
We next assessed the effects of miR-22 on OS cell proliferation and metastasis via generating human OS cell lines (HOS and MG63) stably expressing miR-22 or negative control (Figure 2A). We found that miR-22 overexpression significantly reduced cell proliferation relative to NC controls not due to the effect on apoptosis (Figures 2B,C).
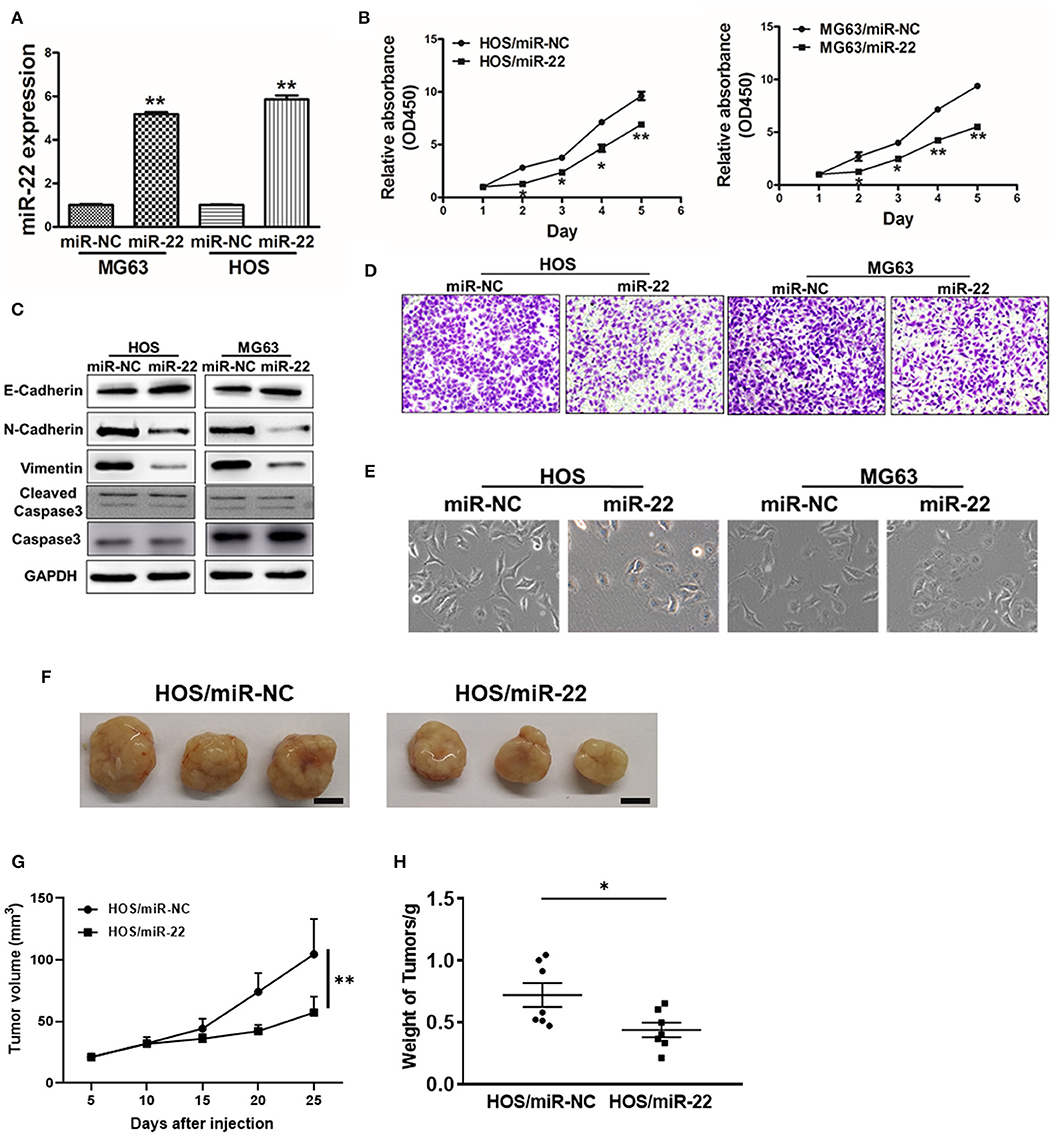
Figure 2. miR-22 suppresses proliferation and EMT in OS cells. (A) MG63 and HOS OS cell lines stably expressing miR-22 were assessed via qRT-PCR to confirm miR-22 expression. (B) A CCK8 assay was used to assess the proliferation of the indicated OS cells. (C) Western blotting was used to assess E-cadherin, N-cadherin, Vimentin, Caspase 3 and Cleaved caspase 3 levels in these cells. (D) Chambers of transwells covered with Matrigel were used for Invasion assays. (E) MG63 and HOS cells were assessed via phase contrast microscopy, with those overexpressing miR-22 exhibiting a shift from a spindle-like to a round/cobblestone morphology. (F–H) Female BALB/c nude mice were subcutaneously injected with 106 HOS cells harboring miR-NC or miR-22 overexpression. Tumor volume and weight were monitored over time as indicated, and the tumor was excised and weighed after 25 days. Bar = 10 mm. Data are means±SD of 3 replicates. *P < 0.05; **P < 0.01.
We further observed significant morphological changes in MG63 and HOS cells overexpressing miR-22, with a shift from a spindle-shaped morphology to a rounder/cobblestone appearance (Figure 2E). We then measured the EMT markers vimentin, N-cadherin and E-cadherin via western blotting, revealing them to be significantly decreased and increased, respectively, in OS cells overexpressing miR-22. Meanwhile, the invasion ability of OS cells expressing miR-22 is weaker to the control cells (Figure 2D). We also performed the in vivo assays, the results showed that miR-22 will indeed reduce cell proliferation abilities (Figures 2F,G). These results therefore suggested that miR-22 is capable of suppressing the proliferation and EMT of OS cells.
miR-22 Targets Twist1
To further explore the mechanisms whereby miR-22 regulates OS cell activity, we utilized the Targetscan tool to identify possible miR-22 target genes. One such predicted target was Twist1 (Figure 3A), which is a key transcription factor associated with the EMT and with metastasis. To confirm the ability of miR-22 to target Twist1, we generated luciferase reporter plasmids containing a WT or mutated version of the Twist1 3'-UTR containing the putative miR-22 binding site. We found that miR-22 was able to significantly reduce the luciferase activity of the WT but not mutated plasmid construct, consistent with its binding to the target region (Figure 3B). Western blotting further revealed that overexpressing miR-22 decreased Twist1 protein levels, but not snail levels which has been reported the miR-22-target in other cancer cell lines (Figure 3C). Together these findings show that miR-22 can specifically interact with the 3'-UTR of Twist1 to regulate its expression in OS cells.
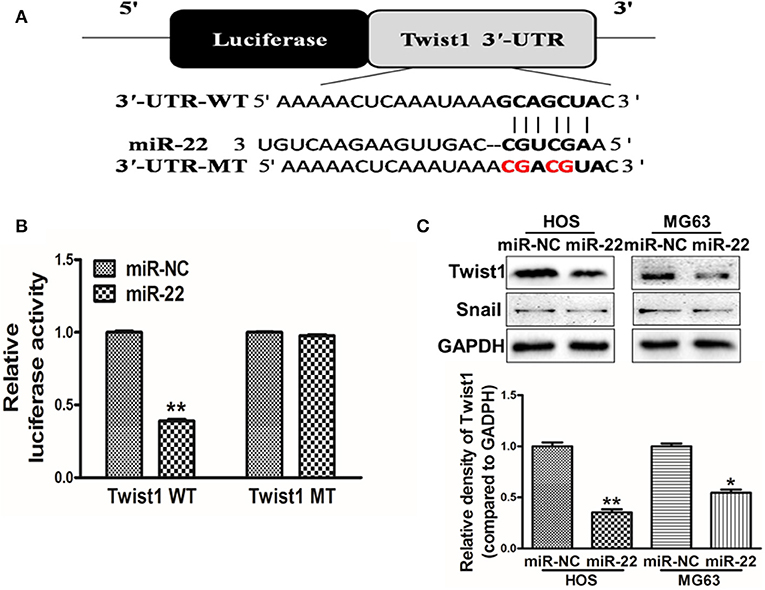
Figure 3. Twist1 is a miR-22 target. (A) The putative site of miR-22 binding within the Twist1 3'-UTR is show, with WT and mutant (MT; in red) constructs for these 3'-UTR regions being generated and cloned into a pMIR luciferase reporter vector. (B) The pMIR-Twist1-WT and pMIR-Twist1-MT vectors were transfected into HOS cells along with miR-22 or control, and after 48 h luciferase activity was assessed. (C) Twist1 protein levels in MG63/HOS cells were assessed via Western blotting. Data are means±SD of 3 replicates. *P < 0.05; **P < 0.01.
Twist1 and miR-22 Are Negatively Correlated in Human OS
In order to establish the clinical relevance of these results, we next explored Twist1 expression in human OS tissue samples, revealing a significant increase in Twist1 expression in tumor tissues relative to paired normal control tissues (Figure 4A). A Pearson's correlation analysis between Twist1 and miR-22 levels in OS samples further revealed that there was a strong negative correlation between the expression of these two factors (Pearson's correlation r = −0.7646) (Figure 4B).
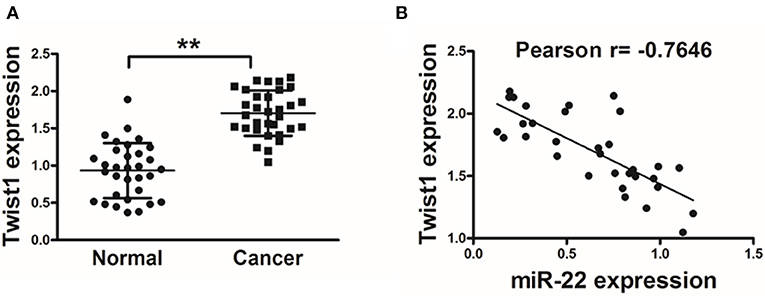
Figure 4. Twist1 expression negatively correlated with that of miR-22 in human OS. (A) qRT-PCR was used to assess Twist1 expression in human OS samples, with GAPDH used for normalization. (B) The association between Twist1 and miR-22 expression in OS samples was assessed via Pearson′s correlation analysis. Data are means±SD of 3 replicates. **P < 0.01.
miR-22 Regulates OS Cell Proliferation and EMT via Targeting Twist1
We finally sought to explore whether there was a direct functional link between miR-22 and Twist1 in the context of the regulation of OS cell proliferation and EMT. To test this, we co-transfected HOS cells using both miR-22 or NC along with a pCMV-Twist1 construct for 48 h. Overexpression of Twist1 lacking of the 3'-UTR miR-22 binding site was able to partially reverse the effects of miR-22 overexpression, with increased Twist1 expression (Figure 5A), enhanced proliferation (Figure 5B), increased invasion abilities (Figure 5D) and a restoration of normal cellular morphology (Figure 5C) and N-cadherin, vimentin protein levels upon Twist1 overexpression (Figure 5E). These results thus indicate that miR-22 is capable of targeting Twist1 in order to regulate the proliferation and EMT of OS cells.
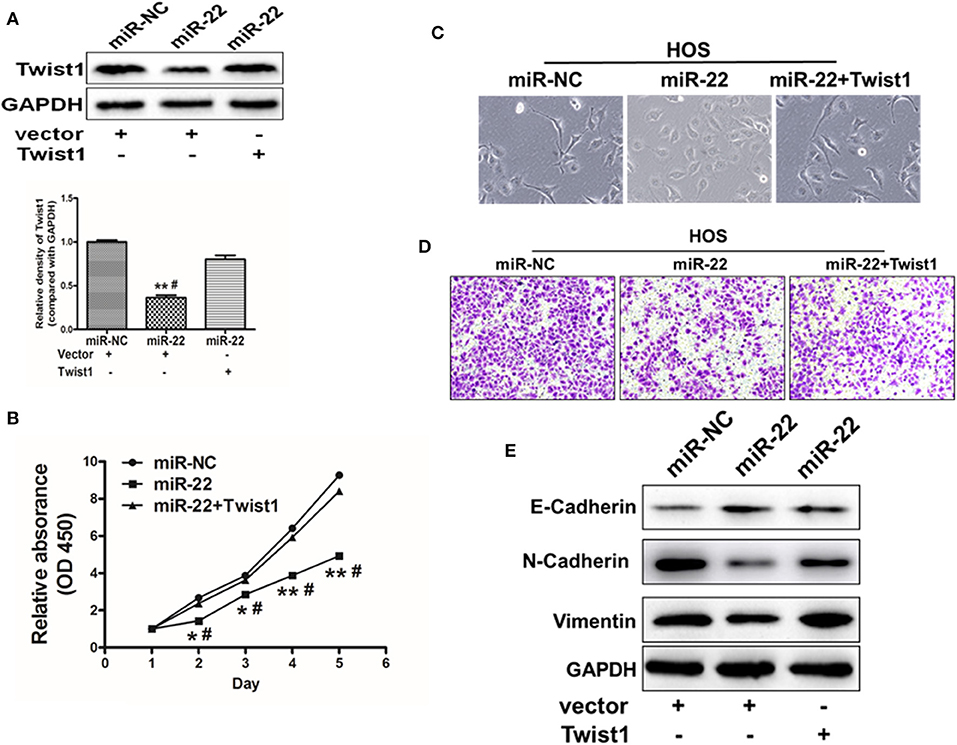
Figure 5. miR-22 targets Twist1 to regulate the EMT in OS cells. (A) Twist1 overexpression in HOS cells was partially able to reverse the effects of overexpressing miR-22. (B) A CCK8 assay was used to assess OS cell proliferation, revealing that Twist1 overexpression in HOS cells partially overcame the inhibition of proliferation induced upon miR-22 overexpression. (C) Images of changes in cell morphology in cells miR-22 mimics or control with our without Twist1 overexpression. (D) Chambers of transwell covered by Matrigel were used for invasion assays. (E) Western blotting was used to measure E-cadherin N-cadherin and Vimentin expression in HOS cells. Data are means ± SD of 3 replicates. *P < 0.05; **P < 0.01 vs. miR-NC control. #P < 0.05 vs. miR-22 + Twist.
Discussion
For past several decades, the survival of osteosarcoma patients has been mostly improved. Despite evidence of genomic instability and a high frequency of chromothripsis and kataegis, osteosarcomas carry few recurrent therapeutic gene targets. Meanwhile, the developments of targeted agents have also been generally disappointing (14). Thus, the underlying mechanism remain largely unknow. Several cancers have been found to exhibit miRNA dysregulation, and such altered miRNA activity has in turn been found to play a functional role in tumor progression, with particular miRNAs promoting or suppressing cancer progression. In the present report we found that human OS patient samples exhibited significantly reduced miR-22 expression relative to normal tissue controls, with decreased expression corresponding to increasing tumor histological grade. We further found that overexpressing miR-22 led to a significant repression of the EMT in OS cells, with Twist1 being a direct miR-22 target in these cells, thereby modulating EMT progression. This is the first study we are aware of demonstrating the ability of miR-22 to regulate Twist1 in OS, although further study of this signaling pathway is needed to validate these results.
The EMT is a key process in driving cancer metastasis (15), with several transcription factors including Twist1, Snail, and ZEB1 cooperating to control this complex process (15). The EMT is a common event that is observed in a wide range of cancer types undergoing metastasis (16). Epithelial-to-Mesenchymal Transition (EMT) is a dynamic, invertible and relative process. Many studies have indicated that, in tumor cells, “Epithelial, E” and “Mesenchymal, M” are not at the completely different poles. Oppositely, in most case, tumor cells can reside in an intermediate state termed “metastable” phenotype between the epithelial and mesenchymal stages enabling them to undergo EMT- or MET related processes under specific conditions, which likely contributes to their aggressive clinical behavior. Although osteosarcoma cells arise from cells that descend from the mesenchyme. The osteosarcoma cells still maintain partial epithelial characters, like some epithelial markers, which are necessary to mediate cohesiveness during migration, and allows resistance to the mechanical stress that cells experience during migration (17, 18). Although it remains largely unknown that how osteosarcoma cells with a metastable phenotype can shift EMT- and MET-related processes, several studies have shown first clues. For instance, in osteosarcoma cells, the EMT-TFs, such as Twist, Snails, and Zebs are over-activated (19) and the genes such as TIM3 (20), ST6GAL1 (21), TRIM66 (22), UHRF1 (23), and CYR61 (24) contribute to the positive regulation of EMT-TFs. Moreover, miRNAs are implicated in the suppression and induction of EMT-related processes in osteosarcoma (25, 26). For instance, miR-370 and miR-132 suppress EMT-related processes inhibiting tumor growth and metastasis via targeting fork head box protein M1 (FOXM1) and SRY-box 4 (SOX4), respectively (27, 28). In the present report we observed reduced Vimentin expression and increased E-cadherin expression in cells overexpressing miR-22, with this overexpression corresponding to a significant reduction in OS cell EMT capacity. We further found that miR-22 directly targeted Twist1 to mediate this repression of the EMT, and this was further confirmed to be a clinically relevant result as Twist1 expression was negatively correlated with that of miR-22 in patient samples. We have thus shown that miR-22 expression is closely linked with the regulation of the EMT process in OS cells.
Twist1 is a helix-loop-helix transcription factor that plays essential roles in osteoblasts, myoblasts, and mesodermal cells, regulating their differentiation nad migration activity (29–32). Twist1 further promotes tumor cell proliferation, invasion, and resistance to cell death, in addition to enhancing EMT progression and angiogenic development (33–35). Twist1 upregulation is observed in a variety of cancer types, including OS as well as breast, bladder, gastric, and ovarian cancer (36–40).
Conclusion
In conclusion, our results reveal that miR-22 is downregulated in human osteosarcoma in a manner that correlates with enhanced tumor progression an metastasis. We found that miR-22 was able to directly suppress its target Twist1, thereby impairing the proliferation and EMT of OS cells. These insights highlight the potential of this miR-22/Twist1 signaling axis to be used either as a prognostic biomarker or therapeutic target in OS.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by The Institutional Review Board and Ethics Committee of Henan University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YL and SZ designed the study, analyzed data, and drafted the manuscript. XW, JW, and GX participated in the manuscript preparation and carried out the experiments in vitro and in vivo. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol. (2008) 134:281–97. doi: 10.1007/s00432-007-0330-x
2. Di Fiore R, Guercio A, Puleio R, Di Marco P, Drago-Ferrante R, D'Anneo A, et al. Modeling human osteosarcoma in mice through 3AB-OS cancer stem cell xenografts. J Cell Biochem. (2012) 113:3380–92. doi: 10.1002/jcb.24214
3. Zhu H, Tang J, Tang M, Cai H. Upregulation of SOX9 in osteosarcoma and its association with tumor progression and patients' prognosis. Diagn Pathol. (2013) 8:183. doi: 10.1186/1746-1596-8-183
4. Wan J, Zhang X, Liu T, Zhang X. Strategies and developments of immunotherapies in osteosarcoma. Oncol Lett. (2016) 11:511–20. doi: 10.3892/ol.2015.3962
5. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. (2006) 103:2257–61. doi: 10.1073/pnas.0510565103
6. Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online (2014) 16:11. doi: 10.1186/1480-9222-16-11
7. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. (2010) 9:775–89. doi: 10.1038/nrd3179
8. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. (2006) 94:776–80. doi: 10.1038/sj.bjc.6603023
9. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. (2002) 2:442–54. doi: 10.1038/nrc822
10. Barbáchano A, Fernández-Barral A, Pereira F, Segura MF, Ordóñez-Morán P, Carrillo-de Santa Pau E, et al. SPROUTY-2 represses the epithelial phenotype of colon carcinoma cells via upregulation of ZEB1 mediated by ETS1 and miR-200/miR-150. Oncogene. (2016) 35:2991–3003. doi: 10.1038/onc.2015.366
11. Nishijima N, Seike M, Soeno C, Chiba M, Miyanaga A, Noro R, et al. miR-200/ZEB axis regulates sensitivity to nintedanib in non-small cell lung cancer cells. Int J Oncol. (2016) 48:937–44. doi: 10.3892/ijo.2016.3331
12. Zhang X, Li Y, Wang D, Wei X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol Res. (2017) 50:27. doi: 10.1186/s40659-017-0133-8
13. Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. (2016) 7:44252–65. doi: 10.18632/oncotarget.10020
14. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. (2014) 14:722–35. doi: 10.1038/nrc3838
15. Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol Res Pract. (2015) 211:557–69. doi: 10.1016/j.prp.2015.05.010
16. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. (2009) 119:1429–37. doi: 10.1172/JCI36183
17. Choi PW, Yang J, Ng SK, Feltmate C, Muto MG, Hasselblatt K, et al. Loss of E-cadherin disrupts ovarian epithelial inclusion cyst formation and collective cell movement in ovarian cancer cells. Oncotarget. (2016) 7:4110–21.
18. Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. (2014) 157:1146–59. doi: 10.1016/j.cell.2014.03.045
19. Yang G, Yuan J, Li K. EMT transcription factors: implication in osteosarcoma. Med Oncol. (2013) 30:697. doi: 10.1007/s12032-013-0697-2
20. Feng ZM, Guo SM. Tim-3 facilitates osteosarcoma proliferation and metastasis through the NF-κB pathway and epithelial-mesenchymal transition. Genet Mol Res. (2016) 15. doi: 10.4238/gmr.15037844
21. Meng Q, Ren C, Wang L, Zhao Y, Wang S. Knockdown of ST6Gal-I inhibits the growth and invasion of osteosarcoma MG-63 cells. Biomed Pharmacother. (2015) 72:172–8. doi: 10.1016/j.biopha.2015.04.020
22. Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J, et al. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget. (2015) 6:23708–19. doi: 10.18632/oncotarget.4291
23. Liu W, Qiao RH, Wang DM, Huang XW, Li B, Wang D. UHRF1 promotes human osteosarcoma cell invasion by downregulating the expression of Ecadherin in an Rb1dependent manner. Mol Med Rep. (2016) 13:315–20. doi: 10.3892/mmr.2015.4515
24. Hou CH, Lin FL, Hou SM, Liu JF. Cyr61 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol Cancer. (2014) 13:236. doi: 10.1186/1476-4598-13-236
25. Chen J, Yan D, Wu W, Zhu J, Ye W, Shu Q. MicroRNA-130a promotes the metastasis and epithelial-mesenchymal transition of osteosarcoma by targeting PTEN. Oncol Rep. (2016) 35:3285–92. doi: 10.3892/or.2016.4719
26. Tian K, Di R, Wang L. MicroRNA-23a enhances migration and invasion through PTEN in osteosarcoma. Cancer Gene Ther. (2015) 22:351–9. doi: 10.1038/cgt.2015.27
27. Duan N, Hu X, Yang X, Cheng H, Zhang W. MicroRNA-370 directly targets FOXM1 to inhibit cell growth and metastasis in osteosarcoma cells. Int J Clin Exp Pathol. (2015) 8:10250–60.
28. Liu Y, Li Y, Liu J, Wu Y, Zhu Q. MicroRNA-132 inhibits cell growth and metastasis in osteosarcoma cell lines possibly by targeting Sox4. Int J Oncol. (2015) 47:1672–84. doi: 10.3892/ijo.2015.3147
29. Mammoto T, Jiang A, Jiang E, Mammoto A. Role of twist1 phosphorylation in angiogenesis and pulmonary fibrosis. Am J Respir Cell Mol Biol. (2016) 55:633–644. doi: 10.1165/rcmb.2016-0012OC
30. Oh BY, Kim SY, Lee YS, Hong HK, Kim TW, Kim SH, et al. Twist1-induced epithelial-mesenchymal transition according to microsatellite instability status in colon cancer cells. Oncotarget. (2016) 7:57066. doi: 10.18632/oncotarget.10974
31. Palumbo-Zerr K, Soare A, Zerr P, Liebl A, Mancuso R, Tomcik M, et al. Composition of TWIST1 dimers regulates fibroblast activation and tissue fibrosis. Ann Rheum Dis. (2017) 76:244–51. doi: 10.1136/annrheumdis-2015-208470
32. Sozen B, Pehlivanoglu S, Demir N. Differential expression pattern of Twist1 in mouse preimplantation embryos suggests its multiple roles during early development. J Assist Reprod Genet. (2016) 33:1533–40. doi: 10.1007/s10815-016-0794-1
33. Weyemi U, Redon CE, Sethi TK, Burrell AS, Jailwala P, Kasoji M, et al. Twist1 and slug mediate H2AX-regulated epithelial-mesenchymal transition in breast cells. Cell Cycle. (2016) 15:2398–404. doi: 10.1080/15384101.2016.1198864
34. Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, et al. Twist1 promotes breast cancer invasion and metastasis by silencing foxa1 expression. Oncogene. (2017) 36:1157–66. doi: 10.1038/onc.2016.286
35. Yuan W, Li T, Mo X, Wang X, Liu B, Wang W, et al. Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget. (2016) 7:29507–19. doi: 10.18632/oncotarget.8789
36. Okamura H, Yoshida K, Haneji T. Negative regulation of TIMP1 is mediated by transcription factor TWIST1. Int J oncology. (2009) 35:181–6. doi: 10.3892/ijo_00000327
37. Roberts CM, Tran MA, Pitruzzello MC, Wen W, Loeza J, Dellinger TH, et al. TWIST1 drives cisplatin resistance and cell survival in an ovarian cancer model, via upregulation of GAS6, L1CAM, and Akt signalling. Sci Rep. (2016) 6:37652. doi: 10.1038/srep37652
38. Tseng WC, Chuang CW, Yang MH, Pan CC, Tarng DC. Kruppel-like factor 4 is a novel prognostic predictor for urothelial carcinoma of bladder and it regulates TWIST1-mediated epithelial-mesenchymal transition. Urol Oncol. (2016) 34:485.e15–24. doi: 10.1016/j.urolonc.2016.07.002
39. Wang L, Zhou R, Zhao Y, Dong S, Zhang J, Luo Y, et al. MACC-1 promotes endothelium-dependent angiogenesis in gastric cancer by activating TWIST1/VEGF-A signal pathway. PLoS ONE. (2016) 11:e0157137. doi: 10.1371/journal.pone.0157137
Keywords: miR-22, EMT, metastasis, twist1, osteosarcoma
Citation: Zhu S, Wang X, Wang J, Xi G and Liu Y (2020) Downregulation of miR-22 Contributes to Epithelial-Mesenchymal Transition in Osteosarcoma by Targeting Twist1. Front. Oncol. 10:406. doi: 10.3389/fonc.2020.00406
Received: 13 October 2019; Accepted: 09 March 2020;
Published: 24 April 2020.
Edited by:
Tao Liu, University of New South Wales, AustraliaReviewed by:
Hua Zhang, Guangdong Medical University, ChinaLuigi Ippolito, University of Florence, Italy
Copyright © 2020 Zhu, Wang, Wang, Xi and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Liu, hhyyly163@163.com
 Shu-tao Zhu
Shu-tao Zhu Yang Liu
Yang Liu