- 1Department of Thoracic Oncology, National Cancer Center Hospital, Tokyo, Japan
- 2Department of Experimental Therapeutics, National Cancer Center Hospital, Tokyo, Japan
- 3Department of Respiratory Medicine, Mitsui Memorial Hospital, Tokyo, Japan
- 4Division of Translational Genomics, Exploratory Oncology Research and Clinical Trial Center, National Cancer Center, Tokyo, Japan
- 5Department of Laboratory Medicine, National Cancer Center Hospital, Tokyo, Japan
- 6Department of Pediatric Oncology, National Cancer Center Hospital, Tokyo, Japan
- 7Department of Hematology, National Cancer Center Hospital, Tokyo, Japan
- 8Division of Genome Biology, National Cancer Center Research Institute, Tokyo, Japan
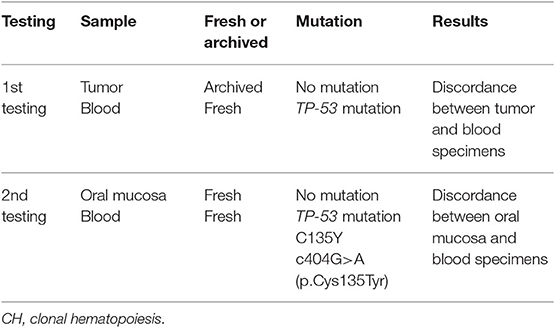
Reliable and accurate next generation sequencing (NGS) technologies are important in precision medicine. Analysis using currently available NGS genomic tests is conducted on cancer-derived DNA collected from tumor tissue, blood, or both. Clonal hematopoiesis (CH) produces a detectable somatic clonal mutation that is commonly associated with clonal expansion of hematopoietic cells with age and genomic analysis of blood samples can be used to detect CH. A 74-year-old Korean male had lung adenocarcinoma with a metastasis to the left scapula. He underwent palliative radiotherapy to the left scapula and received multi-line chemotherapies. After disease progression, he underwent re-biopsy of the metastatic tumor tissue from lung cancer and concomitant blood sampling. NGS genomic testing revealed no significant genomic mutation in the tumor tissue DNA but showed the TP53 mutation C135Y in peripheral blood DNA. To investigate the discordance between the genotyping results in tumor tissue and blood, we tested for the TP53 mutation using a target sequencing test in blood and normal oral mucosa. The TP53 mutation C135Y was only detected in the blood sample, confirming the presence of TP53-mutated CH. We should be aware of different characteristics in NGS genomic testing including sample type such as tumor, blood, or paired specimens. Performing genomic testing on paired tumor and blood samples is effective for discriminating mutations derived from CH from germline mutations and somatic mutations in tumor cells.
Background
Precision medicine is an emerging approach for providing patient-specific treatment based on genomic data using next generation sequencing (NGS). Reliable and accurate NGS technologies are essential in precision medicine (1). Several commercial genomic tests are available (Table 1). Each of these tests has different characteristics including the sequencing system; the number of tested genes; sample type such as tumor, blood, or paired specimens; and the curation and annotation system.
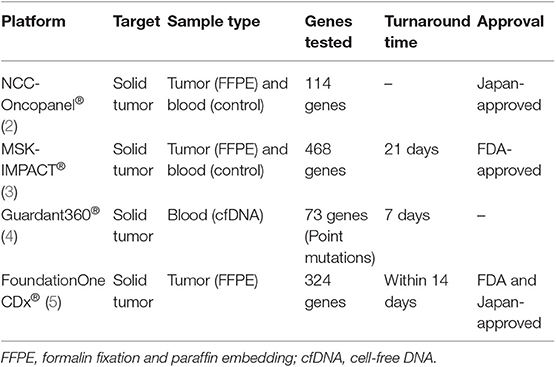
Table 1. Comparison of the genomic tests NCC-Oncopanel®, MSK-IMPACT®, Guardant360®, and FoundationOne CDx®.
Clonal hematopoiesis (CH) produces a detectable somatic clonal mutation that is commonly associated with clonal expansion of hematopoietic cells with age (6). As the use of NGS analysis has increased, studies have reported some discordances between genotyping results from tumor tissue and plasma specimens, which may be caused by tumor heterogeneity, variable shedding of tumor DNA into the plasma, and/or CH (7, 8). Here, we report a case of TP53-mutated CH in a patient with lung adenocarcinoma.
Case Presentation
A 74-year-old Korean man was referred to us for lung adenocarcinoma with left scapula metastasis. He had a normal complete blood count and no family history of malignant disease. Following palliative radiotherapy to the scapula, he had been treated with multi-line chemotherapies. However, his scapula metastasis had progressed. He provided written informed consent to undergo genomic testing. Undecalcified tumor tissue of the scapula metastasis from lung adenocarcinoma and a blood sample were submitted for NGS genomic testing using NCC-Oncopanel test, which was developed at the National Cancer Center in Japan. Testing of the blood sample detected a TP53 mutation (C135Y c404G>A [p.Cys135Tyr], allele frequency 29.8%), but there was no matching mutation in the tumor tissue (Figure 1). The criteria for myelodysplastic syndromes (MDS), Waldenström macroglobulinemia, IgM monoclonal gammopathy of undetermined significance (IgM-MGUS), and Li-Fraumeni syndrome were not met. We suspected CH as the cause of the discordance between findings in tumor tissue and blood. We therefore verified the presence of the TP53 mutation with a TP53 target sequencing test using the sequencing by synthesis method (Falco Holdings Co., Ltd.) in blood and normal oral mucosa. The TP53 mutation C135Y was only detected in the blood (Table 2).
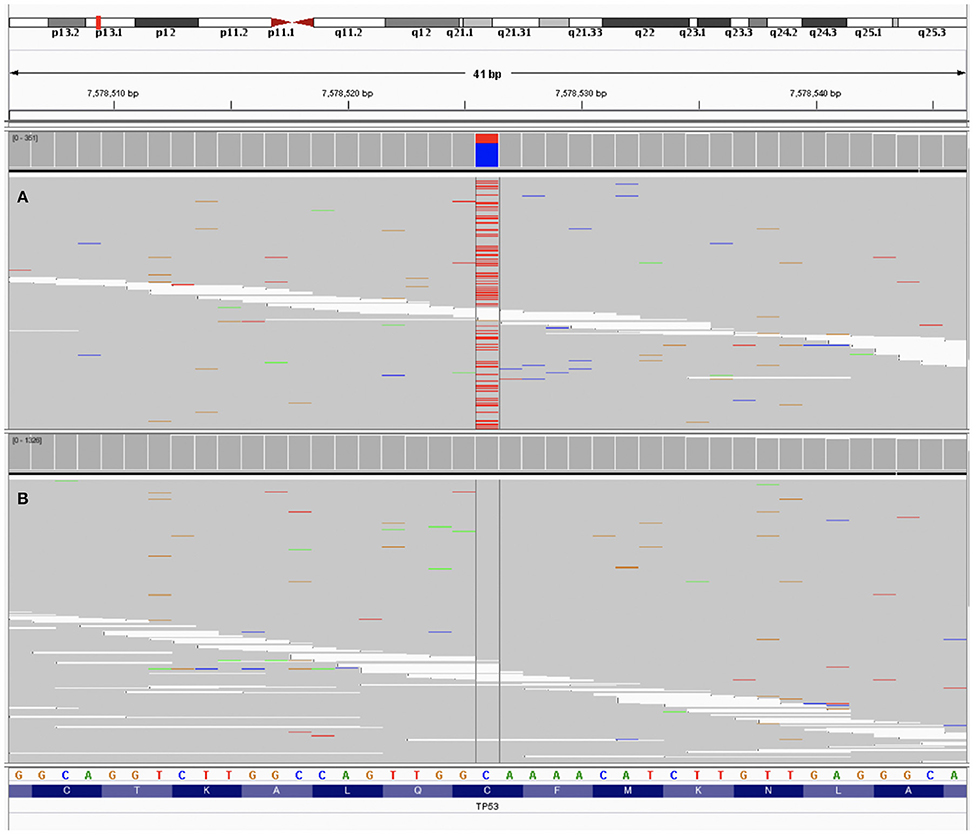
Figure 1. The data of patient's sample using next generation sequencing visualized by Integrative Genome Viewer (9). (A) Testing of the peripheral blood sample detected a TP53 mutation (C135Y c404G>A [p.Cys135Tyr], allele frequency 29.8%). (B) Testing of the tumor tissue showed no TP53 mutation.
Discussion and Conclusions
CH is an aging-related phenomenon in which hematopoietic stem cells or other early blood cell progenitors contribute to the formation of a genetically distinct subpopulation of blood cells (6). CH is considered to be associated with prior exposure to chemotherapy or irradiation in cancer patients as well as increased age (10). Plasma cell-free DNA genotyping detected that some mutations such as JAK2, TP53, and KRAS can be derived from CH and not from cancer (7).
The cause of the genotyping discordance between tumor tissue and blood is important for determining the treatment strategy. Although the TP53 mutation is not targetable, germline TP53 mutations result in a hereditary condition known as Li-Fraumeni syndrome and the KRAS mutation is a predictive marker of response to cetuximab therapy in colorectal cancer (11, 12). CH detected from cell-free DNA can cause misdiagnosis of the mutation origin and complicate the treatment strategy (7, 8). Pairing of plasma and tumor genomic tests is important, although paired sample testing is expensive.
We report a case of TP53-mutated CH in a lung adenocarcinoma patient in which target sequencing was used to confirm mutations derived from CH.
Ethics Statement
The studies involving human participants were reviewed and approved by Genomic profiling study (The TOP-GEAR study) was registered at UMIN Clinical Trials Registry (UMIN 000011141) and approved by Institutional Review Board at National Cancer Center Hospital on June 27th, 2013. The patients/participants provided their written informed consent to participate in this study.
Consent for Publication
This patient provided written informed consent to participate in TOP-GEAR study. The authors have obtained consent from the patient for publication of this case report.
Author Contributions
MI and YF drafted the manuscript. YF contributed to the management of the clinical case. MI, YF, TKub, HM, TKum, TS, KS, NY, and TKo reviewed the manuscript and participated in clinical data interpretation. All authors read and approved the final manuscript.
Funding
This study was supported by the Japan Agency for Medical Research and Development (AMED) (17lk1403003h0001 and 18lk1403003h0002) and National Cancer Center Research and Development Fund (30-A-6, and NCC Biobank).
Conflict of Interest
YF reports speaker's bureau from Sysmex. NY is a recipient of a research grant from Japan Agency for Medical Research and Development (AMED, 17lk1403003h0001, and 18lk1403003h0002). KS and TKo are recipients of a collaborative research grant from the Sysmex Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank all the medical staff who were involved in the care of the patient and the staff of NCC-Oncopanel. We thank Libby Cone, MD, MA, from DMC Corp. (www.dmed.co.jp) for editing drafts of this manuscript.
References
1. Kuderer NM, Burton KA, Blau S, Rose AL, Parker S, Lyman GH, et al. Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol. (2017) 3:996–8. doi: 10.1001/jamaoncol.2016.4983
2. Sunami K, Ichikawa H, Kubo T, Kato M, Fujiwara Y, Shimomura A, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: a hospital-based study. Cancer Sci. (2019) 110:1480–90. doi: 10.1111/cas.13969
3. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. (2017) 23:703–13. doi: 10.1038/nm.4333
4. Guardant Health. Guardant 360. (2019). Available online at: http://www.guardant360.com (accessed October 5, 2019).
5. Foundation Medicine. FoundationOne CDx. (2019). Available online at: https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx (accessed October 5, 2019).
6. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. (2014) 371:2477–87. doi: 10.1056/NEJMoa1409405
7. Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. (2018) 24:4437–43. doi: 10.1158/1078-0432.CCR-18-0143
8. Coombs CC, Gillis NK, Tan X, Berg JS, Ball M, Balasis ME, et al. Identification of clonal hematopoiesis mutations in solid tumor patients undergoing unpaired next-generation sequencing assays. Clin Cancer Res. (2018) 24:5918–24. doi: 10.1158/1078-0432.CCR-18-1201
9. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. (2011) 29:24–6. doi: 10.1038/nbt.1754
10. Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. (2017) 21:374–82.e4. doi: 10.1016/j.stem.2017.07.010
11. McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MH, Eeles RA, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol. (2014) 11:260–71. doi: 10.1038/nrclinonc.2014.41
Keywords: cell-free DNA, clonal hematopoiesis, next generation sequencing, precision medicine, TP53 mutation
Citation: Ito M, Fujiwara Y, Kubo T, Matsushita H, Kumamoto T, Suzuki T, Sunami K, Yamamoto N and Kohno T (2020) Clonal Hematopoiesis From Next Generation Sequencing of Plasma From a Patient With Lung Adenocarcinoma: A Case Report. Front. Oncol. 10:113. doi: 10.3389/fonc.2020.00113
Received: 12 November 2019; Accepted: 21 January 2020;
Published: 13 February 2020.
Edited by:
Joshua Michael Bauml, University of Pennsylvania, United StatesReviewed by:
Conor Steuer, Emory University, United StatesElena Levantini, Harvard Medical School, United States
Copyright © 2020 Ito, Fujiwara, Kubo, Matsushita, Kumamoto, Suzuki, Sunami, Yamamoto and Kohno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yutaka Fujiwara, ZnVqaXdhcmEteXV0YWthQG1pdHN1aWhvc3Aub3IuanA=
 Munehiro Ito
Munehiro Ito Yutaka Fujiwara
Yutaka Fujiwara Takashi Kubo
Takashi Kubo Hiromichi Matsushita5
Hiromichi Matsushita5 Noboru Yamamoto
Noboru Yamamoto Takashi Kohno
Takashi Kohno