
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 31 January 2020
Sec. Cancer Molecular Targets and Therapeutics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00048
This article is part of the Research Topic The Role of ncRNAs in Solid Tumors Prognosis: From Laboratory To Clinical Utility View all 49 articles
Tumor immunosuppression may assist the immune escape of cancer cells, which promotes tumor metastasis and resistance to chemo-radiotherapy. The therapeutic strategies against tumor immunosuppression mainly focus on blocking immune checkpoint receptors, enhancing T-cell recognition and neutralizing inhibitory molecules. Although immunotherapies based on these strategies have improved the clinical outcomes, immunological nonresponse and resistance are two barriers to tumor eradication. Therefore, there is an urgent need to identify new biomarkers for patient selection and therapeutic targets for the development of combination regimen with immunotherapy. Recent studies have reported that non-protein-coding modulators exhibit important functions in post-transcriptional gene regulation, which subsequently modulates multiple pathophysiological processes, including neoplastic transformation. Differentiated from microRNAs, long non-coding RNAs (lncRNAs) are reported to be involved in various processes of the immune response in the tumor microenvironment (TME) to promote tumor immunosuppression. Currently, studies on tumor immunity regulated by lncRNAs are mainly confined to certain types of cancer cells or stromal cells. Additionally, the majority of studies are focused on the events involved in T cells and myeloid-derived suppressor cells (MDSCs). Although the reported studies have indicated the significance of lncRNAs in immunotherapy, the lack of comprehensive studies prevents us from exploring useful lncRNAs. In the current review, we have summarized the roles of lncRNAs in tumor immune response, and highlighted major lncRNAs as potential biomarkers or therapeutic targets for clinical application of immunotherapy.
The host immune surveillance evasion by the tumor cells enables them to proliferate, which results in tumorigenesis. The effector T lymphocytes and natural killer (NK) cells have limited efficacy in the tumor microenvironment (TME), due to the gene and/or protein alterations in the tumors and immune cells that are necessary for tumor cell recognition and killing. Additionally, the accumulation of immunosuppressive cells and/or cytokines may suppress the functions of effector immune cells. The TME, which exhibits immunosuppressive characteristics, may affect the outcomes of immunotherapy. Several studies indicated that long non-coding RNAs (lncRNAs) can affect the differentiation and functions of immune cells as well as the progression of tumors. This review summarizes the potential predictive value of lncRNAs for immunotherapy response.
Anticancer immunotherapies, especially immune checkpoint blockade (ICB) and adoptive cellular transfer, are associated with higher efficacy and tolerance than conventional therapies like targeted therapy (1–4). Particularly, the gene signatures indicative of a T cell-inflamed TME in solid tumors are positively correlated with clinical response to anti-programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) therapy (5). Several studies have proposed further stratification of TME to enhance the efficacy of ICBs. Prior to therapy initiation, TME can be classified into four subtypes according to the tumor mutational burden (TMB) and gene signatures for T cell-inflamed TME (5) or can be categorized into three basic immune profiles based on the localization of tumor-infiltrating lymphocytes (TILs) and the expression of PD-L1 in tumor biopsies (6, 7). Although the two stratifications have several differences, they emphasize the immune-inflamed phenotype of the tumors. The inflamed tumor profile is characterized by the infiltration of immune effector cells [e.g., CD8+ T cells and type 1 T helper (TH1) cells] and proinflammatory cytokines [e.g., interleukin (IL)-12, IL-1β, and type I and type II interferons (IFNs)] in the tumor parenchyma (5, 6). The inflamed tumor samples may be positive for PD-L1 staining. Furthermore, the TILs and immunosuppressive immune cell populations infiltrate inflamed tumors with low TMB. The primary immunosuppressive populations are regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs). The inflammatory TME may favor tumor growth and metastasis. The inflamed tumor profile, irrespective of the TMB status, indicates that the pre-existing antitumor immune response is markedly suppressed.
The PD-L1/PD-1 blockade deregulates the inhibitory signals transduced into the T cells and subsequently enhances the T cell-mediated cytotoxicity against tumor cells (8). The clinical response to PD-L1/PD-1 blockade ranges from 12 to 52% in the inflamed tumors, such as melanoma, bladder cancer, and mismatch repair-deficient colorectal cancer (9). Some individuals exhibiting high lymphocyte infiltrations and high PD-L1 expression were refractory to anti-PD-L1/PD-1 therapy (5, 10). This indicated that tumor-shrinkage occurs due to a combination of factors and that the immune cell infiltration and PD-L1 expression are necessary but insufficient for tumor response. Hence, several studies are ongoing to identify the biomarkers that can predict the response to PD-1 blockade (pembrolizumab) and to select patients most likely to benefit from immunotherapy. In addition to T cell-inflamed gene expression profile (GEP) and PD-L1 status, TMB and high microsatellite instability (MSI-H) may also act as predictive biomarkers for response to anti-PD-1 therapy (11). Previous studies have reported that TMB and T cell-inflamed GEP can independently predict the response to pembrolizumab. Furthermore, the prediction efficiency of TMB and T cell-inflamed GEP combination was stronger than that of TMB or T cell-inflamed GEP (11). Although TMB and inflammatory biomarkers, including T cell-inflamed GEP and PD-L1 status, have high predictive value for response to pembrolizumab, other hidden biomarkers need to be explored. The mechanisms underlying immunosuppression in inflamed tumors with low TMB are weakly associated with the PD-L1/PD-1 pathway and strongly associated with immunosuppressive immune cell populations. Strategies to relieve immunosuppression mediated by MDSCs, TAMs, and Tregs that activate the effector immune cells may play a major role in tumor regression in these patients. Thus, biomarkers and therapeutic targets focusing on immunosuppressive cells may enhance antitumor immune responses in such tumor types.
Both tumor-intrinsic and tumor-extrinsic mechanisms are employed by the tumors to evade immune surveillance. The generation of tumor antigens and the expression of major histocompatibility complex (MHC) class I molecules can be decreased to inactivate the T lymphocytes (12). Upon stimulation with IFN-γ, PD-L1 expressed on the tumor cells interacts with PD-1 expressed on the CD8+ T cells, which causes T cell exhaustion (13). In addition to the IFN-γ signaling pathway, the loss of sensitivity to tumor necrosis factor (TNF) has been demonstrated to promote tumor immune escape through the upregulation of anti-apoptotic proteins, such as BCL-2. Moreover, the tumor cells can also influence the functions of immune cells via the secretion of immunosuppressive mediators and exosomes that mediate cell-cell communication (13).
During tumor initiation and progression, the TME affects the tissue-resident and blood-derived cells and thus promotes the development of tumors. The following two types of myeloid cells are highly susceptible to environmental signals: dendritic cells (DCs) (antigen presentation) and macrophages (antigen degradation). These cells are influenced by cancer cells through tumor-derived soluble factors, such as IL-6, vascular endothelial growth factor (VEGF), macrophage colony-stimulating factor (M-CSF) (14). Additionally, the production and secretion of IL-1β, granulocyte/macrophage CSF (GM-CSF), prostaglandin E2 (PGE2), and VEGF by tumors promote the expansion of myeloid progenitor cells and immature myeloid cells. This is followed by the accumulation of these cells, which are termed as MDSCs (15). The accumulated DCs, macrophages, and MDSCs facilitate tumor growth by suppressing the proliferation of CD8+ T lymphocytes. In the lymphoid compartment, the key effector cells involved in balancing the tumor immunity are CD8+ T cells, NK cells, B cells, and CD4+ T cells. The CD8+ T cells are the predominant anticancer effector cells that give rise to cytotoxic T lymphocytes (CTLs) and kill the tumor cells which present a specific peptide-MHC complex (16). However, the CD8+ T cells recruited into the tumor beds also encounter numerous barriers, including the recognition of checkpoints, such as PD-L1/PD-1, CD28/CTLA-4, and immunoglobulin-like transcript receptors (17). Inhibitory molecules, such as T-cell immunoglobulin and mucin-domain containing-3 (TIM-3), lymphocyte activation gene (LAG-3), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) on the Tregs are critical to the suppressive function in antitumor immune response (18). TH1-polarized CD4+ T cells cooperate with the cytotoxic CD8+ T cells to promote the macrophage cytotoxic activities and enhance the antigen presentation by antigen-presenting cells (APCs). Contrastingly, TH2-polarized CD4+ T cells and other TH2 response-initiating cells, such as TH2-polarized monocytes and macrophages, and regulatory B cells (Bregs), are crucial pro-tumorigenic components that promote tumor cell survival and proliferation (19, 20). In addition to these cellular components mentioned above, the non-cellular components such as IL-6, IL-10, indoleamine 2,3-deoxygenase (IDO), and TGF-β are also indispensable for the regulation of intratumoral immunosuppression (20, 21).
LncRNAs, which are the most frequently expressed non-protein-coding transcripts, are localized in the cell nucleus, cytoplasm and exosomes (22) where they interact with various molecules such as DNA, RNA, and proteins. Some lncRNAs that are packaged into the exosomes can function as messengers for signal transduction between the cells (23). Several studies have reported that lncRNAs are involved in pathophysiological processes through the epigenetic, transcriptional, and post-transcriptional regulation of gene expression (24). Additionally, lncRNAs are reported to affect the differentiation and development of myeloid cells and the expression of inflammatory genes in immune cells (22, 25). Furthermore, lncRNAs are also involved in the regulatory circuit of the immune cells. In specific types of immune cells, the expression of lncRNAs is induced by intracellular signaling pathways (such as NF-κB) upon Toll-like receptor (TLR) activation or is suppressed upon cytokine receptor activation, which subsequently regulates the immune responses (25). Previous studies have suggested that some lncRNAs may promote tumor progression through the dysregulation of tumor proliferation, apoptosis (e.g., MA-LINC1, HOTAIR), metastasis (e.g., MALAT1), and angiogenesis (e.g., MIAT, MEG3). LncRNAs target the immune checkpoints and cytokines and promote the formation of the immunosuppressive microenvironment, which contributes to tumor progression and drug resistance. Specifically, lncRNAs are emerging new therapeutic targets and prognostic biomarkers for tissue- and clinical stage-specific cancers (26, 27).
To elucidate the detailed mechanisms underlying the interactions between immune system and tumor cells, researchers are gradually unveiling the roles of lncRNAs in immunosuppressive TME. Computational approaches such as LnCAR (28) and DriverLncNet (29) are developed to explore the functions of lncRNAs in tumor progression based on the causal relations from gene perturbation experiments. In a pan-cancer analysis, the lncRNAs identified by these tools were significantly correlated with the dysregulation of signatures associated with immune responses, including the activation and inhibition of T cells (28, 29). Thus, the strong correlations between lncRNAs and the immune response in cancers warrants further exploration of potential roles of lncRNAs as novel clinical predictors for the efficacy of checkpoint blockades.
Non-coding RNAs are RNA molecules that do not code for protein. LncRNAs are defined as non-coding RNAs with at least 200 nucleotides, a length cutoff that distinguishes lncRNAs from small regulatory RNAs, such as piRNA (Piwi-interacting RNAs) and other microRNAs (miRNAs). Genome-wide studies (tilling microarray, RNA sequencing, and chromatin marks) have reveled numerous non-coding transcribed bases in mammals (30). According to the current GENCODE Release (version 31) (https://www.gencodegenes.org/), about 30% of the known genes in human genome are transcribed as lncRNAs. The majority of lncRNAs with low conservation level were considered as transcriptional noises, which are by-products during the transcription and splicing of protein-coding genes. A study based on chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) revealed several functional lncRNAs (31). The chromatin associated-lncRNAs are usually transcribed by RNA polymerase II and are processed like messenger RNA with additional 5′-capping and 3′-polyadenylated tail (31). The high evolutionary conservation among these lncRNAs indicated that they exhibit biological functions in many pathophysiological activities, such as X-chromosome inactivation (XIST) (32) and imprinting (H19) (33). LncRNAs can be categorized into several types based on the distance between neighboring annotated genes. For example, intronic lncRNAs are transcribed from an intron in the genome, long intergenic lncRNAs (lincRNAs) are transcribed from DNA sequence located between protein-coding genes, and antisense lncRNAs are transcribed from the complementary DNA strand of protein-coding genes (30).
The diverse functions of lncRNAs are related to their subcellular localization and targets. In the cytosol, lncRNAs function predominately through RNA-RNA and RNA-protein interactions (Figure 1A). Some lncRNAs act as competing endogenous lncRNAs (ceRNAs) to sponge miRNAs, which results in miRNA dysfunction and subsequently affecting mRNA translation. Recently, lncRNA SNHG1 was reported to directly interact with miR-448 in the regulatory T lymphocytes, which could negatively regulate the expression of IDO (34). In addition to post-transcriptional regulation, several lncRNAs bind to the signaling molecules. For example, Lnc-BM (a lncRNA related to breast cancer brain metastasis), binds to JAK2 and modulates its kinase activity through the Lnc-BM/JAK2/STAT3/ICAM1 pathway (35). Similarly, long intergenic non-coding RNA for kinase activation (LINK-A), interacts with PtdIns(3,4,5) P3 and affects the activation of the AKT pathway in the breast cancer cells (26). LncRNAs play a critical role in the epigenetic and transcriptional regulation of gene expression by interacting with the chromatin (complexes that contain proteins and DNA molecules) within the nucleus. For example, XIST lncRNA acts in cis to silence the gene expression in the X-chromosome of a female by recruiting chromatin modifiers to the adjacent sites (36). Additionally, the DNA-binding domain and specific secondary structures of lncRNAs enable the interaction with one or more proteins and guide them to the specific DNA sites, where they function as decoys, guides and scaffolds to regulate gene expression at the transcriptional level. Examples of the functional modules of lncRNAs in the nucleus are illustrated in Figure 1B.
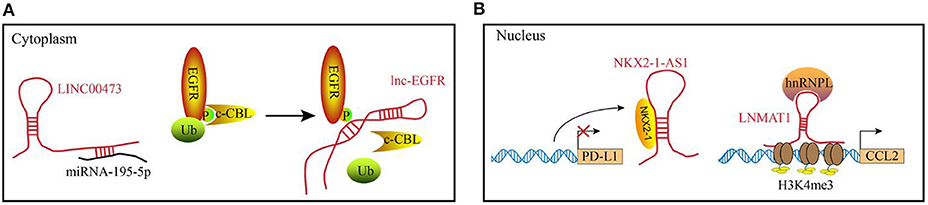
Figure 1. Examples of the functional mechanisms for long non-coding (lncRNAs). LncRNAs function through interacting with diverse molecules in (A) the cytoplasm or (B) cell nucleus. (A) In the cytoplasm, lncRNAs can interact with RNAs or proteins. For example, LINC00473 sponges miRNA-195-5p to reduce its expression level. lnc-EGFR binds to the phosphorylated EGFR to block the ubiquitination, which maintains the activation of EGFR pathway. (B) In the nucleus, lncRNA can act as decoys (e.g., NKX2-1-AS1) or guides (e.g., LNMAT1) to regulate the gene expression at transcriptional levels.
In the TME, lncRNAs are directly or indirectly involved in the regulation of tumor immunosuppression through multiple mechanisms (Figure 2). Some immunosuppressive cells are generated from their corresponding effector cells through dynamic expression of protein markers in the cell membrane as evidenced by the polarization of macrophages, exhaustion of CD8+ T cells, differentiation of helper T cells, and immunosuppressive function of MDSCs. This process can be regulated by endogenous lncRNAs, which regulate gene expression in response to specific stimuli. Moreover, lncRNAs in the tumor cells may facilitate signaling transduction to mediate the degradation of peptide-loading complex (PLC) components, and to sponge miRNAs to upregulate the expression of immune checkpoints, which contribute to decreased immunosurveillance. Furthermore, the lncRNAs encapsulated in the exosomes act as mediators for cell-cell communication in the tumor environment. Exosomes are small membrane-bound vesicles that deliver a specific cargo of proteins and nucleic acids from the parent cells to the recipient cells. Several studies indicate that exosome-transmitted lncRNAs secreted from tumor cells can promote the immunosuppressive function of stromal cells, such as cancer associated fibroblasts and macrophages (37–39). Additionally, the phenomena that exosomes being delivered from immune cells to tumor cells or other kinds of immune cells are observed. On the one hand, TAMs strengthen the aerobic glycolysis and apoptotic resistance of breast cancer cells through the transmission of extracellular vesicles (EVs) with myeloid-specific lncRNA (40). Meanwhile, the exhausted CD8+ T cells, which express TIM-3 and PD-1 during the antitumor immune responses, exhibit reduced proliferation and impaired anticancer activities. The regular CD8+ T cells uptake exosomes containing lncRNAs secreted by the exhausted CD8+ T cells, which leads to CD8+ T cell dysfunction (41). Furthermore, some lncRNAs expressed in the tumor cells are affected by immune cells. Tumors are highly organized tissues with numerous reciprocal interactions among distinct cell populations and between cells and soluble molecules. Cytokines, such as CCL5 and IL-8 can be secreted by macrophages and tumor-associated DCs, which interact with the receptors expressed in the tumor cells. The cytokine-mediated interaction enhances cancer progression through lncRNA-dependent pathways, such as the MALAT1/Snail pathway in colon cancer (42).
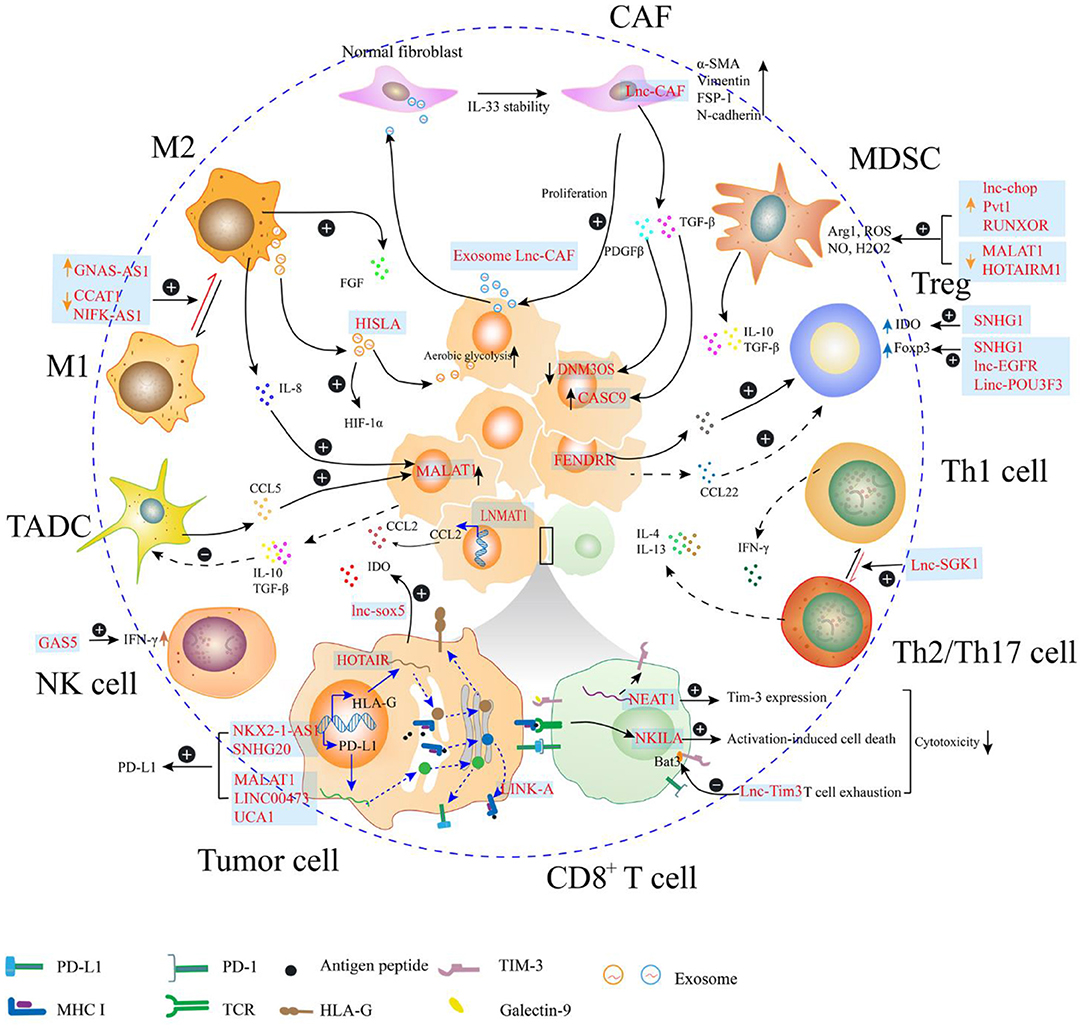
Figure 2. Long non-coding RNAs (lncRNAs) regulate the immunosuppression in the tumor microenvironment (TME). In the TME, lncRNAs regulate the expression of molecules (e.g., PD-L1, MHC I, and HLA-G) on the surface of the tumor cells, which may attenuate the function of effector T cell. Additionally, the cytotoxicity of T cell can be directly regulated by lncRNAs within T cell, through mediating activation-induced cell death or enhancing T cell exhaustion. LncRNAs can also participate in the phenotype transition of cells, such as helper T cell, fibroblast, and macrophage, which can contribute to the formation of immunosuppressive TME. In the myeloid-derived suppressor cell (MDSC), lncRNAs enhance the production of immunosuppressive molecules, such as Arg1 and reactive oxygen species (ROS). Th1, type 1 helper T cell; Th2, type 2 helper T cell; Th17, T helper cell17; Treg, regulatory T cell; MDSC, myeloid-derived suppressor cells; CAF, cancer-related fibroblast; M1, M1 macrophage; M2, M2 macrophage; TADC, tumor-associated dendritic cell; NK cell, natural killer cell.
In the following sections, we will discuss the mechanisms underlying lncRNA-mediated regulation of tumor immune responses. Additionally, we clarify the potential of lncRNAs as biomarkers for patient selection and the possibility to manipulate the expression of lncRNAs for clinical therapeutic applications.
LncRNAs are reported not only to mediate crucial signal transductions in cancer signaling pathways but also to affect the tumor immunity to promote tumor evasion from immunosurveillance. Denaro et al. have reviewed that immune cells, such as T cells, B cells, dendritic cells, macrophages, and myeloid cells, regulate cancer immunity through lncRNAs-related pathways. Some lncRNAs have been highlighted as theraputic targets and diagnostic markers in cancers (43). Thus, it is valuable to consider that lncRNAs participating in tumor immunosuppression have the potential for clinical applications. To systematically summarize the research results of lncRNAs regulating tumor immunosuppression, we retrieved literatures in the Pubmed database using the following combination of the search terms: “lncRNA or long noncoding RNA or long non-coding RNA,” “immune suppression or immunosuppressive,” and “tumor microenvironment.” Furthermore, studies on the pivotal immune checkpoints [PD-L1, TIM-3, and human leukocyte antigen (HLA)-G] and immunosuppressive cells (Tregs, MDSCs, and TAMs) that are reported to be regulated by lncRNAs in TME were also included. The studies were excluded based on the following criteria: (a) studies that have reported lncRNAs to regulate the function of immune cells but have not elucidated the roles of lncRNAs in cancers; (b) studies that have reported the role of lncRNAs in tumor metastasis or progression but have not directly demonstrated the interactions between lncRNAs and immune system; (c) studies that have reported the functions of lncRNAs in hematological tumor. The literature analysis indicated that the mechanisms underlying lncRNA-mediated regulation of tumor immunosuppression can be classified according to the type of cells expressing lncRNAs. In the tumor cells, some oncogenic lncRNAs regulate the immunogenicity of tumors by upregulating the expression of immune checkpoints (e.g., PD-L1 and IDO) and HLA-G, or directly by downregulating the generation of tumor antigens. Furthermore, lncRNAs within tumor cells may accumulate in the stromal cells that are recruited into the TME and secrete the suppressive molecules that affect the tumor cell-mediated immunosuppression. The reported studies mainly focused on immunosuppressive and immunoregulatory cells, such as M2 macrophages, MDSCs, and Tregs, which are associated with immunosuppressive TME. LncRNAs that regulate tumor immune escape and the corresponding target genes are described in detail below (Table 1).
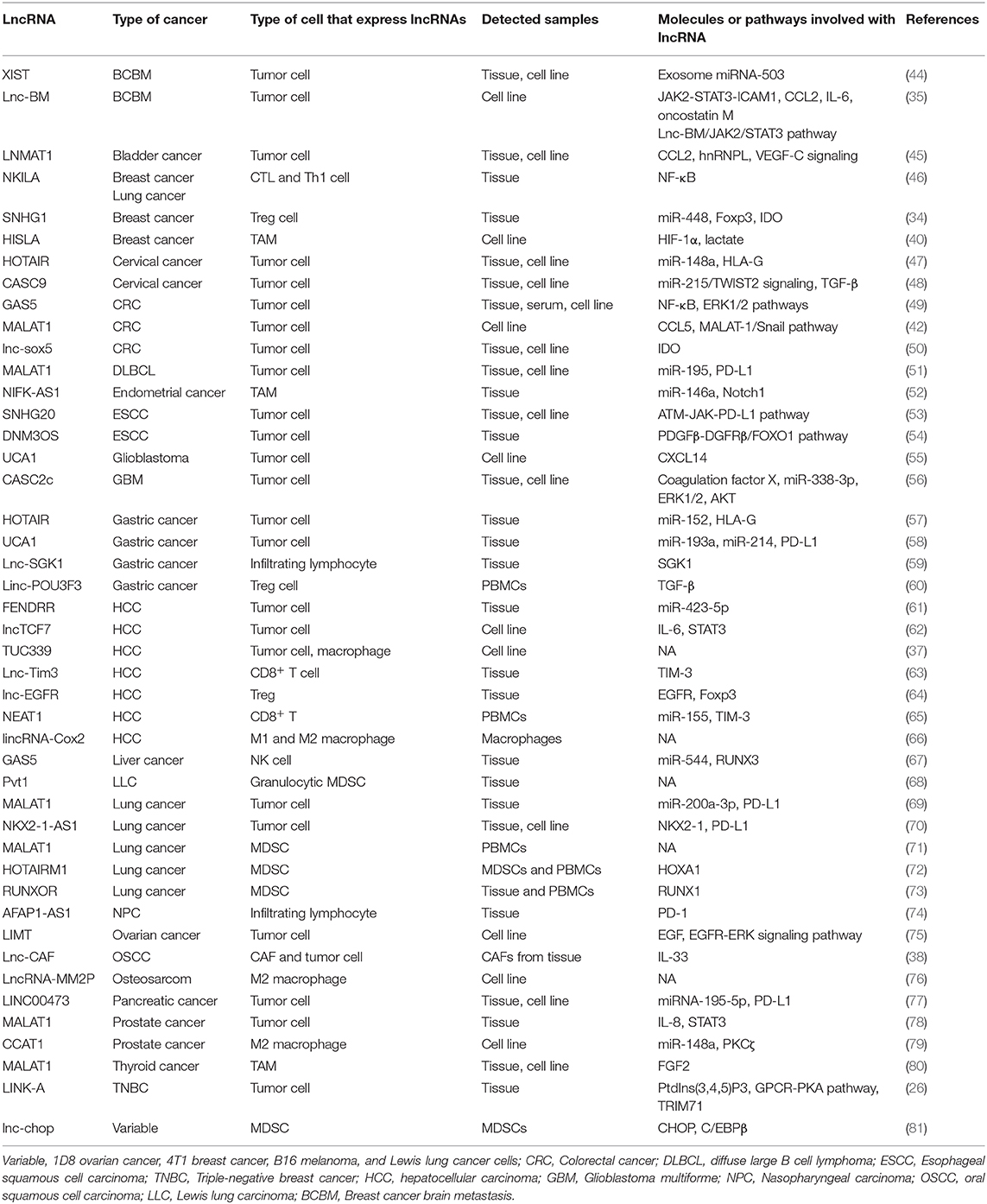
Table 1. Long non-coding RNAs (lncRNAs) and their respective molecules or pathways involved in the tumor microenvironment (TME) immunosuppression.
Mutation-derived tumor antigens are expressed on the membrane of cancer cells in the form of peptide-MHC class I complexes, which are recognized by cytotoxic CD8+ T lymphocytes that kill the tumor cells. During the antigen presentation of peptide-MHC complexes, the peptides are degraded from the cytosolic proteins by proteasomes. The degraded peptides are then transported into the lumen of the endoplasmic reticulum, where the antigen peptides bind to the MHC class I molecules and are transported to the cell surface (82). The dysregulated antigen presentation machinery in the tumor may facilitate cancer cells to escape the immunosurveillance. The components of PLC play an important role in the transportation and presentation of peptide-MHC complexes. The antigenicity of cancer cells may be reduced through the degradation of PLC components, which are modified by ubiquitin. The expression of LINK-A, a tissue-specific lncRNA, in the triple-negative breast cancer (TNBC) tissues was reported to be higher than that in the non-TNBC tissues. Additionally, LINK-A can predict poor prognosis in patients with breast cancers (83, 84). Hu et al. demonstrated that the expression of LINK-A was negatively associated with the abundance of APCs and CD8+ T cells in the basal-like breast cancers, which indicated a correlation between LINK-A and immunosuppression (26). In the transgenic MMTV-Tg (LINK-A) mouse model, LINK-A functions as an oncogenic lncRNA and initiate metastatic mammary gland tumors, which phenotypically resembled the human TNBC. Moreover, LINK-A could enhance the polyubiquitination-mediated degradation of the PLC components and tumor suppressors (Rb and p53) through the inhibitory GPCRs/PKA pathway (26). Treatment with the locked nucleic acids (LNAs) of LINK-A or GPCR antagonists in vivo increased the stability of MHC class I complexes and PLC components. Importantly, treatment with LNA did not affect the distribution of immune cells, such as CD8+ T cells, macrophages, and MDSCs in the regular mammary glands.
A recent study that tumor cells may upregulate non-classical HLA molecules, such as HLA-G, which can be modulated by cytokines like IL-10 and IFN-γ to evade immunosurveillance. HLA-G binds to the inhibitory receptors expressed on different immune cells, which results in the suppressive immune responses, such as the inhibition of cytotoxicity of CD8+ T cells and NK cells (85). Recent studies have reported that HOTAIR, a ceRNA, may modulate the expression of HLA-G by competitively binding to miR-152 (57) or miR-148a (47) in cancer cells. HOTAIR is overexpressed in different types of human malignancies and is involved in cancer progression and metastasis. In patients with cervical cancer, HOTAIR upregulation was correlated with more advanced clinical characteristics and shorter overall survival.
In the T cells, the reduction of tryptophan by indoleamine 2,3-dioxygenase 1 (IDO1) can activate the stress-response kinase GCN2, which inhibits T cell proliferation and induces the differentiation of naïve CD4+ T cells into Tregs. Therefore, IDO1 expression in tumors may contribute to immune evasion. Wu et al. reported that lnc-sox5 was upregulated during the tumorigenesis of colorectal cancer (CRC). Additionally, the absence of lnc-sox5 did not affect the growth of tumor cells in immunodeficient mice, but significantly suppressed tumorigenesis in immunocompetent mice (50). Flow cytometry analysis suggested that the knock down of lnc-sox5 promoted the infiltration and the cytotoxicity of CD3+CD8+ CTLs in tumors in immunocompetent mice. Furthermore, the frequency of Tregs was markedly suppressed. The expression of IDO1 is significantly reduced in Caco-2 cells and MC-38 cells upon lnc-sox5 knockdown. Therefore, lnc-sox5 may serve as a modulator of IDO1 in tumor cells and can be a potential therapeutic target for cancers.
PD-L1 expressed on the tumor cells interacts with PD-1 receptor expressed on the activated T cells, which transduce inhibitory signals for T cell proliferation and cytokine production. LncRNAs are reported to mediate the expression of PD-L1 on tumor cells through various mechanisms. LncRNAs can indirectly upregulate PD-L1 expression by sponging miRNAs. For example, lncRNA UCA1 repressed the expression of miR-193a, miR-26a/b, and miR-214 in gastric cancer through direct interactions and improved the expression of PD-L1 (58). Other studies also reported that lncRNA LINC00473 sponged miR-195-5p to enhance the expression of PD-L1 in prostate cancer (77), while lncRNA MALAT1 regulated tumor migration and immune evasion by modulating the miR-195/PD-L1 axis in diffuse large B-cell lymphoma (51) and the miR-200a-3p/PD-L1 axis in lung cancer (69), respectively. Soluble factors secreted by the immune cells also affect the expression of MALAT1. Kan et al. reported that CCL5 derived from tumor-associated DCs was associated with the up-regulation of MALAT1, which subsequently increased the expression of Snail to promote tumor progression (42). A recent study also reported that IL-8 secreted from M2 macrophages sufficiently promoted the expression level of MALAT1 by activating the STAT3 signaling pathway (78). These studies suggest that MALAT1 serves as a key regulator during tumor progression, especially during tumor immune evasion. LncRNAs can also regulate PD-L1 expression by interacting with proteins (53, 70). NKX2-1-AS1 is an antisense lncRNA that partially overlaps the NKX2-1/TTF1 gene. In lung adenocarcinomas, NKX2-AS1 and NKX2-1 were highly expressed, but NKX2-AS1 did not regulate the expression of NKX2-1 or nearby genes. NKX2-1-AS1 negatively regulated the transcriptional activity of PD-L1 by interfering with the binding of NKX2-1 protein to the promoter of PD-L1 by potentially functioning as a decoy molecule (70).
LncRNAs expressed in tumor cells may affect not only the tumor cells but also tumor-directed immune responses. For example, the tumor-suppressive growth arrest-specific transcript 5 (GAS5) lncRNA was reported to be associated with the expression of VEGF-A and IL-10 in the tumor cells (49). VEGF-A is a well-known proangiogenic molecule produced by the tumor cells. Additionally, VEGF-A plays a key role in the induction of immunosuppressive microenvironment by enhancing the expression of inhibitory checkpoint molecules in the CD8+ T cells, inhibiting the maturation of DCs, and promoting the differentiation of Tregs (86–88). IL-10 acts as an immunosuppressive cytokine to inhibit the maturation of DCs and the antigen cross-presentation to T cells (89). The tumor tissues exhibit lower GAS5 lncRNA expression than the corresponding normal tissue (90, 91). The knockdown of GAS5 in the CRC cells improved the expression of VEGF-A and IL-10 via the nuclear factor-κB (NF-κB) and Erk1/2 pathways, respectively (49). In the colitis-associated cancer mouse model, GAS5 was markedly down-regulated and was negatively associated with the expression of VEGF-A and IL-10 (49). Thus, VEGF-A and IL-10 cytokines suppressed by GAS5 may serve as targets for lncRNA-based therapeutic regimens against CRC.
Several studies suggest that tumor cell-derived lncRNAs may also be involved in the secretion of factors such as CCL2, coagulation factor X (FX), and exosomal miRNAs to promote tumor metastasis by influencing macrophage recruitment and polarization. Chen et al. reported that lymph node metastasis associated transcript 1 (LNMAT1), a novel lncRNA, was significantly upregulated in bladder cancers with lymph node metastasis. Moreover, enhanced LNMAT1 expression was significantly associated with more advanced clinicopathological characteristics, which indicated a poor survival for patients with bladder cancer (45). LNMAT1 is predominantly localized in the nucleus and recruits hnRNPL to the CCL2 promoter, which results in H3K4 tri-methylation and transcriptional activation (45). Subsequently, CCL2 activated by LNMAT1 recruits macrophages into the tumor mass and promotes the lymphatic metastasis via VEGF-C excretion (45). Lnc-BM, a metastasis-related lncRNA, may promote cancer progression in patients with breast cancer brain metastases (BCBMs) by promoting communication between macrophages and breast cancer cells in the brain TME (35). Tumor cell-derived Lnc-BM facilitated STAT3-dependent expression of CCL2 and ICAM1, which mediated macrophage recruitment and vascular co-option in the brain, respectively. The recruited macrophages secreted IL-6 and oncostatin M, which activated the Lnc-BM/JAK2/STAT3 pathway in breast cancer cells (35). Zhang et al. reported that lncRNA CASC2c interacts with miR-338-3p reciprocally to repress its expression, which increases the expression and secretion of FX. FX is involved in the recruitment and M2-polarization of macrophages in glioblastoma (56). LncRNA X-inactive-specific transcript (XIST) functions as a tumor suppressor in brain-metastatic breast cancer (44). XIST was significantly downregulated in brain-metastatic tumors of patients with breast cancer. The knockout of XIST in mammary glands of mice stimulated the growth of the primary tumor and brain metastases. Loss of XIST also enhanced the secretion of exosomal miRNA-503, which triggered the M2-polarization of microglia, and upregulated the immunosuppressive cytokines in microglia and subsequently suppressed T-cell proliferation (44). Tumor-derived exosomal lncRNAs have been indicated as signaling mediators that orchestrate the communications between tumor cells and macrophages in TME. Li et al. demonstrated that hepatocellular carcinoma (HCC) cell-derived exosomes contained overexpressed TUC339 lncRNA, which may be taken up by the non-polarized THP-1 macrophages (37). Additionally, in vitro experiments suggested that TUC339 is majorly involved in macrophage polarization (37).
In addition to macrophages, the functions of Tregs and fibroblasts can also be regulated by lncRNAs from tumor cells in TME. The tumor suppressor lncRNA FOXF1 Adjacent Non-Coding Developmental Regulatory RNA (FENDRR) is reported to be a favorable diagnostic biomarker for HCC (92). Microarray analysis suggested that the expression of FENDRR lncRNA in HCC samples was higher than that in the normal samples. Recent studies also suggested that FENDRR lncRNA suppressed the immune escape of HCC cells (61). The HCC cells transfected with siRNA of FENDRR lncRNA exhibited increased expression of TGF-β, IL-10, and VEGF. Furthermore, FENDRR lncRNA could competitively bind to miR-423-5p to upregulate the expression of growth arrest and DNA-damage-inducible beta (GADD45B). Previous studies have indicated that the loss of GADD45B could increase the number of Tregs (93), which may explain the lncRNA FENDRR-mediated inhibition of immune escape that was medicated by Tregs in the HCC cells (61). Ding et al. discovered a tumor cell-derived exosomal lncRNA, namely Lnc-CAF, which was markedly upregulated in the stromal fibroblasts. This novel stromal lncRNA reprogramed normal fibroblasts (NFs) to cancer-related fibroblasts (CAFs) through the Lnc-CAF/IL-33 pathway and promoted the progression of oral squamous cell carcinoma (OSCC) (38). In OSCC patients, the overexpression of Lnc-CAF and IL-33 was positively associated with higher TNM stages at diagnosis, which indicated worse outcomes.
In TME, the expression of lncRNAs in tumor cells could be modulated by some soluble immunosuppressive cytokines, which promoted tumor immune escape. This regulatory mode suggests a crosslink between stromal cells and cancer cells. In the cervical cancer cells, TGF-β secreted by CAFs can increase the expression of cancer susceptibility candidate 9 (CASC9) lncRNA, which promoted the migration of tumor cells by sponging miR-215 to up-regulate TWIST2 in vitro and in vivo (48). Furthermore, bioinformatics analysis has predicted the presence of complementary sequences between miR-215 and CASC9. Another study reported that the human esophageal cancer cells and tumor tissues exhibited higher DNM3OS lncRNA than normal samples. The expression of DNM3OS lncRNA can be promoted by CAF-derived PDGF-β in tumor cells through the PDGFβ/PDGFRβ/FOXO1 signaling pathway (54). In glioblastoma, CXCL14 in glioblastoma-associated stromal cells could induce glycolysis and invasion of glioma cells by regulating the UCA1/miR-182/PFKFB2 axis (55). M2-like TAMs may secret EGF to regulate the growth and migration of ovarian cancer cells and the metastasis of ovarian cancer. EGF secreted by M2-like TAMs may inhibit the expression of metastasis-inhibiting LIMT (lncRNA inhibiting metastasis) by activating the EGFR-ERK pathway to stimulate tumor progression in ovarian cancers (75). The proinflammatory cytokine IL-6 in TME may contribute to the development of HCC. IL-6 can activate STAT3, a transcription activator that binds to the promoter regions of lncTCF7, to induce the expression of lncTCF7 in a time- and dose-dependent manner. Additionally, STAT3 knockdown and inhibition of STAT3 activation decreased the expression of lncTCF7 (62).
Recent studies have reported that, lncRNAs are crucial regulators for the development and functions of several immune cell lineages, which have been elaborately reviewed elsewhere (25, 94). Furthermore, immune cells within the tumor mass usually undergo epigenetic changes to enhance the survival of tumor cells. LncRNAs derived from immune cells in TME are reported to be involved in the expression of membrane molecules and the secretion of cytokines, which are reviewed in detail below.
Recent studies have indicated that lncRNAs may influence the function of tumor-infiltrating T cells. Huang et al. reported that NF-κB-interacting lncRNA (NKILA) sensitized the T cells to activation-induced cell death (AICD), which was exploited by cancer cells to escape immunological destruction (46). Antitumor CTLs and TH1 cells were more sensitive to AICD than Tregs and TH2 cells in the breast and lung cancer microenvironments. Antigen-stimulated T cells increased the acetylation of histones at the NKILA promoter region, and subsequently enhanced STAT1-mediated transcription of NKILA (46).
In nasopharyngeal carcinoma (NPC), genome expression profiling data of tumor samples suggested that the expression of AFAP1-AS1 lncRNA was significantly associated with that of PD-1. The immunohistochemical analysis revealed that PD-1 and AFAP1-AS1 were co-expressed in the TILs of the NPC tissues. Patients with NPC exhibited poor prognosis when the tumor was positive for both AFAP1-AS1 and PD-1 (74). TIM-3 expressed on the CD8+ T lymphocytes is reported to be correlated with the phenotype of immune exhaustion. Lnc-Tim3 lncRNA specifically blocked the interaction between TIM-3 and Bat3 by binding to the intracellular domain of TIM-3, which suppressed the downstream signaling of the Lck/NFAT1/AP-1 pathway (63). In vitro experiments indicated that Lnc-Tim3 inhibited the production of IFN-γ and IL-2 in CD8+ T lymphocytes, and enhanced the expression of anti-apoptotic genes, such as Bcl-2 and MDM2. Overexpression of Lnc-Tim3 in the Jurkat T cells upregulated the exhaustion-associated markers, such as PRDM1, LAG-3, and PBX3 (63). Thus, Lnc-Tim3 could promote an exhausted-like phenotype in the CD8+ T lymphocytes. Yan et al. demonstrated that nuclear-enriched autosomal transcript 1 (NEAT1) lncRNA affected the expression of TIM-3 by modulating the expression of miR-155 (65). NEAT1 lncRNA was highly expressed in the peripheral blood mononuclear cells (PBMCs) and tumor tissues of patients with HCC. Downregulation of NEAT1 lncRNA inhibited the apoptosis of CD8+ T cells and enhanced their cytotoxic activity against tumor cells in vitro. In the HCC mouse models, the tumor growth was suppressed upon injection with NEAT1-silenced CD8+ T cells (65).
The differentiation and distribution of Tregs among PBMCs and tumor tissues of patients may be influenced by lncRNAs. Linc-POU3F3 expressed in Tregs could promote the distribution of Tregs among PBMCs by activating the TGF-β signaling pathway in patients with gastric cancer. In vitro co-culture experiments demonstrated that overexpression of linc-POU3F3 in Tregs promoted the proliferation of cancer cells (60). Jiang et al. demonstrated that lnc-EGFR expressed in the Tregs could affect the expression of Foxp3 in HCC cells and maintain the activation of EGFR by preventing the interaction with c-CBL and phosphorylated EGFR, which resulted in the reduction of EGFR ubiquitination (64). Similarly, SNHG1 lncRNA mediates tumor immune escape by affecting the differentiation of Tregs. Additionally, SNHG1 lncRNA regulated the expression of IDO by directly inhibiting the expression of miR-448. SNHG1 silencing in breast tumor cells inhibited tumor growth and downregulated the expression of SNHG1, IL-10, IDO, and Foxp3 (34). The high expression of Lnc-SGK1 lncRNA was correlated with preferable clinical characteristics in patients with gastric cancer having Helicobacter pylori (H. pylori) infection and high-salt diet. In vitro experiments indicated that H. pylori infection and high-salt diet could enhance the expression of Lnc-SGK1 by activating the SGK1/JunB signaling pathway, which induces the differentiation of helper T cells toward TH2 and TH17cells (59).
Macrophages exhibit a high plasticity, which endows them with diverse functions in response to microenvironment signals (95, 96). The classically activated macrophages (M1) are often generated upon stimulation with pathogen-associated molecular patterns (PAMPs), while the alternatively activated macrophages (M2) are generated upon stimulation with IL-4, IL-13, or IL-10 (97). Oppositely polarized macrophages differ in the expression of receptors, secretion of cytokines and chemokines, and effector functions (97). The M1 macrophages release pro-inflammatory factors, such as IL-6 and TNF-α and contribute to anti-tumoral immune responses (95). The M2 macrophages exhibit pro-tumoral functions by upregulating the expression of IL-10 and immune checkpoint molecules (89). Various strategies have been developed to target the macrophages in tumor mass. For example, therapies that reprogram M2 macrophages to adopt antitumor M1 phenotypes or enhance antigen-presenting capacity are demonstrated to enhance survival in preclinical models (98–100). Recently, several studies have focused on the regulation of macrophage polarization by lncRNAs. In non-small cell lung cancer (NSCLC), GNAS-AS1 lncRNA is highly expressed in the TAMs, tumor cell lines and clinical tumor samples. GNAS-AS1 promoted M2-polarization of macrophages and the progression of NSCLC cells by directly inhibiting miR-4319, which was downregulated by targeting the N-terminal EF-hand calcium binding protein 3 (NECAB3). The GNAS-AS1/miR-4319/NECAB3 pathway promoted tumor progression of NSCLC by shifting the polarization of macrophages (101). The expression of GNAS-AS1 lncRNA was negatively correlated to the overall survival of patients with NSCLC. Similarly, NIFK-AS1 lncRNA served as a ceRNA of miR-146a to modulate M2-polarization of macrophages in endometrial cancer (EC) (52). Additionally, NIFK-AS1 was downregulated in the TAMs. In vitro experiments have suggested that NIFK-AS1 overexpression suppresses the M2-polarization induced by IL-4, which further suppresses the proliferation, migration, and invasion of EC cells (52).
LincRNA-Cox2 is a long intergenic ncRNA (lincRNA) located downstream of the mouse Cox2 gene, which mediates the activation and repression of distinct classes of immune genes (102). LincRNA-Cox2 is reported to influence the functions of specific myeloid cell lineages such as macrophages via the TLR-dependent NF-κB pathway (25). Furthermore, Ye et al. reported that M1 and M2 macrophages exhibited differential expression of lincRNA-Cox2 in vitro (66). The expression of lincRNA-Cox2 in the M1 macrophages was higher than that in the non-polarized macrophages and M2 macrophages. The lncRNA silencing experiments revealed that the macrophages have a tendency toward an immunosuppressive phenotype, including the overexpression of IL-10 and arginase-1 (ARG-1), and the enhancement of pro-tumoral abilities. Thus, lincRNA-Cox2 may act as a suppressor of M2-polarization of macrophages to inhibit immune evasion and growth of tumor cells (66). Contrastingly, lncRNA-MM2P was reported to be involved in M2-polarization and M2 macrophage-mediated angiogenesis, which may have a potential role in macrophage-mediated tumorigenesis (76). Other lncRNAs, such as MIR155HG and CCAT1 have also been recognized as regulators of macrophage polarization (79, 103).
MDSCs are a population of heterogeneous cells comprising myeloid progenitors as well as immature mononuclear and polymorphonuclear cells (104). The chronic inflammation within the TME induced the accumulation of MDSCs, which exhibited immunosuppressive functions via the expression of ARG-1, IL-10, TGF-β, inducible NOS (iNOS), and COX2 (104–106) that promotes tumor growth.
Recent studies have suggested that lncRNAs play important roles in the immunosuppressive functions of MDSCs. The C/EBPβ and C/EBP homologous protein (CHOP) transcription factors are immunosuppressive regulators of MDSCs (107). Gao et al. identified a novel lncRNA, termed lnc-chop in MDSCs that interacts with CHOP and the C/EBPβ isoform, liver-enriched inhibitory protein. This interaction facilitates the activation of C/EBPβ and promotes the expression of immunosuppressive factors, such as NADPH oxidase 2, NO synthase 2, ARG-1, and cyclooxygenase-2 (107). Similarly, RUNXOR lncRNA was highly expressed in MDSCs isolated from tumor tissues of lung cancer. RUNXOR knockdown decreases the expression of immune suppressive molecules, such as arginase-1 in MDSCs (73). Moreover, hypoxia-induced HIF-1α could upregulate the expression of Pvt1 lncRNA in granulocytic MDSCs. Pvt1 lncRNA knockdown markedly impaires the immunosuppressive functions of G-MDSCs in vitro and in vivo (68). These findings indicate tumor-promoting roles of lncRNAs via regulation of the immunosuppressive function of MDSCs in TME (81).
Additionally, lncRNAs in MDSCs may also act as tumor suppressors. MALAT1 LncRNA is reported to play critical roles in tumorigenesis, angiogenesis, and metastasis. Recently, Zhou et al. reported that the expression of MALAT1 is downregulated in the PBMCs from patients with lung cancer, which was negatively correlated with the percentage of circulating MDSCs (71). In vitro experiments indicated that MALAT1 knockdown markedly increased the percentage of MDSCs (71). HOXA transcript antisense RNA myeloid-specific 1 (HOTAIRM1) lncRNA is reported to modulate the differentiation of myeloid cells by targeting HOXA1 (108). The expression of HOTAIRM1 in the PBMCs of patients with lung cancer was lower than that in the PBMCs of healthy controls. Additionally, the expression of HOTAIRM1 and its target gene HOXA1 were negatively associated with the ratio of MDSCs and ARG-1 levels in PBMCs of patients with lung cancer. Furthermore, HOTAIRM1 overexpression could decrease the immunosuppressive functions of MDSCs and suppress the induction of MDSCs from PBMCs of healthy control (72). Therefore, HOTAIRM1/HOXA1 might be a potential therapeutic target for lung cancer (72).
Although several studies have demonstrated the role of lncRNAs in the regulation of tumor progression and immune response, there are limited preclinical and clinical application of lncRNAs in cancer immunotherapy. We searched the clinicaltrials.gov website and only found one ongoing observational study on HOTAIR lncRNA as a biomarker for thyroid cancer. This study investigated the ability of HOTAIR in differentiating benign thyroid nodules from malignant nodules (NCT03469544). As reviewed above, HOTAIR upregulates the expression of HLA-G in tumor tissues (47, 57), which indicated the potential role of HOTAIR as a biomarker of tumor immune escape. Other lncRNAs that regulate the pivotal molecules and pathways that are targeted in clinical therapies, such as tumor vaccine, T cell-based therapy, and ICB, can also potentially predict response to tumor immunotherapy.
The efficiency of ICB therapy is essentially dependent on T cell-recognized neoantigens displayed by MHCs on tumor cells. The lack of tumor neoantigen recognition renders tumors insensitive to PD-L1/PD-1 pathway blockade therapy. Thus, neoantigen-based cancer vaccines, including neoantigen vaccine, peptide vaccine, DNA vaccine, RNA vaccine, and DC vaccine, are developed to overcome these limitations in cancer immunotherapy. Some of these vaccines have been investigated in clinical trials (109). In patients with PD-L1+ TNBC tissues, the overall response rate to ICB therapy ranges from 10 to 18.5% (110). In an ongoing Phase Ib clinical trial, HLA-A2+ Metastatic TNBC is treated using a combination of PVX-410 cancer vaccine and pembrolizumab (NCT03362060). Furthermore, lncRNAs involved in the regulation of antigen presentation may potentially serve as therapeutic targets in tumor vaccines. In the mouse models of breast cancer, LINK-A expression was induced in mammary gland tumors (26). The inhibition of LINK-A expression suppressed the tumor progression. Furthermore, the combinatorial therapy of LINK-A LNAs and ICB synergistically suppressed tumor growth and markedly prolonged the survival of the tumor-bearing mice (26). Therapeutic strategies that restore the tumor antigen presentation pathway may improve the sensitivity to ICB therapy in TNBCs that lack tumor antigenicity. Therefore, LINK-A has great potential as a valid biomarker in predicting the outcomes of patients with TNBC who receive ICB therapy. Additionally, LINK-A can be a potential therapeutic target that sensitizes breast tumors to immune checkpoint inhibitors.
Adoptive T cell therapies, especially chimeric antigen receptor (CAR) T cell therapies, have yielded remarkable clinical efficacies in the treatment of hematological malignancies (111–114). The U.S. Food and Drug Administration (FDA) has approved two CAR T cell products, Yescarta and Kymriah, for treating hematological malignancies (115). However, the efficacy of CAR T cell therapy for solid malignancies has been limited (116, 117). This may be partly due to the lack of CAR T cell trafficking to the tumor site, insufficient activation, and short-term survival of the transferred T cells, and/or the enhanced immunosuppressive TME in tumor mass (115, 117). Various innovative strategies have been explored to overcome these limitations. Here, we highlight the enhancement of the survival of transferred T cells. A recent review has summarized the strategies to improve T cell resistance to apoptosis, such as the use of co-stimulatory domains in CAR genes, the inhibition of specific death receptor pathways, and the insertion of genes for the anti-apoptotic molecules (BCL-XL or BCL-2) (117). For the epigenetic regulation of T cell apoptosis, lncRNAs may act as an adjuvant target of CAR T cell therapy. NKILA is reported to preferentially sensitize antitumor T cells to cell death upon activation by tumor antigens (46). In the mouse model established by Huang et al. CD8+ T cells transduced with NKILA shRNA were transferred into immunocompromised mice with established human breast cancer xenografts, which efficiently suppressed the tumor growth. The NF-κB activity, CTL cytotoxicity, and expression of anti-apoptotic genes in the experimental group were enhanced when compared to those in the tumors of the control group (46). NKILA silencing in the transferred TILs and CAR T cells may overcome tumor immune evasion by inhibiting their AICDs, and improve the efficacy of adoptive T cell therapy for cancers.
Recently, the post-transcriptional modulation of gene regulation by lncRNAs was reported to be a key factor in various pathophysiological processes. Analyzing the function and differentiation of immune cells provide us an insight into the functions of lncRNAs in the immunosuppressive TME. In this review, the role of lncRNAs in tumor immune escape or immune surveillance is illustrated. The functions of lncRNAs within tumor cells or immune cells and in mediating the cell-cell communications were discussed. The pivotal roles of lncRNAs in the regulation of immunosuppressive TME implied that lncRNAs can be employed as biomarkers for cancer diagnosis and prognostic evaluation. Additionally, lncRNAs are potential therapeutic targets for cancer.
The expression of some lncRNAs is tissue- or stage-specific. Thus, lncRNA expressions in a certain cancer type could not be validated in other types of cancers. Recently, some studies have reviewed the literatures related to lncRNAs that are associated with various immune cells and stromal cells in adaptive/innate immune system or in TME (22, 23, 25, 43). It is worth noting that most of them have great potential for cancer research in the respect of tumor immunotherapy. The literatures included in this review must be carefully examined. Except for some studies with elaborate design and solid evidence, most conclusions in the literature need to be considered prudently. However, these studies provide us a basis for exploring new biomarkers and therapeutic targets for the clinical application of immunotherapy.
Additionally, further studies are required to explore this novel but far-ranging field. New computational approaches must to be developed to explore the functions of lncRNAs. Moreover, lncRNA-specific animal models should also be established to enable a better understanding of the roles of lncRNAs within TME for clinical application.
YL, JYa, XM, and HS conceptualized this review. YL wrote and edited the manuscript. XL, CY, and JH reviewed the manuscript and provided feedback. YL and JYu created the figures. JYa, HS, and XM revised and edited the manuscript.
This work was supported by The National Key Research and Development Program of China [No. 2016YFC0906000 (2016YFC0906003)]; the funding from the National Natural Science Foundation of China (81773752, 11247018, 31870655, and 81902686); the joint International Cooperation grant from the Science and Technology department of Sichuan Province (2016HH0012); the Project of Chengdu Science and Technology bureau (2016-XT00-00023-GX); Key Program of the Science and Technology Bureau of Sichuan (Nos. 2017SZ00005 and 2018SZ0052); the Key Project of Sichuan Provincial Education Bureau (17ZA0194 and 17ZA0092); The Project of Sichuan Traditional Chinese Medicine Administration (2018KF006); China Postdoctoral Science Foundation Funded Project (2019M663511); The Projector of Sichuan Provincial Key Laboratory of nature product chemistry and small molecule catalysis (TRCWYXFZCH2016015 and TRCWYXFZCH2016014); The projector of Sichuan provincial Key Laboratory of Antibiotics Research and Re-evaluation (ARRLKF17-11). HS was supported by the grant from The Recruitment Program of Global Young Experts (known as the Thousand Young Talents Plan).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N Engl J Med. (2015)1627–39. doi: 10.1056/NEJMoa1507643
2. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. (2016) 375:1–12. doi: 10.1056/NEJMoa1602252
3. Mikkilineni L, Kochenderfer JN. Chimeric antigen receptor T-cell therapies for multiple myeloma. Blood. (2017) 130:2594–603. doi: 10.1182/blood-2017-06-793869
4. Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. (2012) 12:269–281. doi: 10.1038/nri3191
5. O'Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. (2018) 16:151–67. doi: 10.1038/s41571-018-0142-8
6. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. (2017) 541:321–30. doi: 10.1038/nature21349
7. Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. (2016) 22:1865–74. doi: 10.1158/1078-0432.CCR-15-1507
8. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. (2014) 515:568–71. doi: 10.1038/nature13954
9. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy : mechanisms, response biomarkers, and combinations. Sci Transl Med. (2016) 8:328rv4. doi: 10.1126/scitranslmed.aad7118
10. Pitt JM, Ve'Tizou M, Daille R, Roberti P, Yamazaki T, Pitt JM, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. (2016) 28:1255–69. doi: 10.1016/j.immuni.2016.06.001
11. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science. (2018) 362:eaar3593. doi: 10.1126/science.aar3593
12. Leone P, Shin E, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst. (2013) 105:1172–87. doi: 10.1093/jnci/djt184
13. Eichmüller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. (2017) 109:1–14. doi: 10.1093/jnci/djx034
14. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. (2012) 12:253–68. doi: 10.1038/nri3175
15. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. (2009) 9:162–74. doi: 10.1038/nri2506
16. Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. (2008) 14:623–8. doi: 10.1038/nm.f.1774
17. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. (2012) 12:252–64. doi: 10.1038/nrc3239
18. Toor SM, Sasidharan Nair V, Decock J, Elkord E. Immune checkpoints in the tumor microenvironment. Semin Cancer Biol. (2019) doi: 10.1016/j.semcancer.2019.06.021
19. Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. (2005) 7:211–7. doi: 10.1016/j.ccr.2005.02.013
20. Palucka AK, Coussens LM. The basis of oncoimmunology. Cell. (2016) 164:1233–47. doi: 10.1016/j.cell.2016.01.049
21. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. (2016) 27:1482–92. doi: 10.1093/annonc/mdw168
22. Atianand MK, Fitzgerald KA. Long non-coding rnas and control of gene expression in the immune system. Trends Mol Med. (2014) 20:623–31. doi: 10.1016/j.molmed.2014.09.002
23. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. (2018) 17:82. doi: 10.1186/s12943-018-0831-z
24. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
25. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. (2017) 35:177–98. doi: 10.1146/annurev-immunol-041015-055459
26. Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. (2019) 20:835–51. doi: 10.1038/s41590-019-0400-7
27. Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. (2013) 24:206–14. doi: 10.1016/j.devcel.2012.12.012
28. Xu J, Shi A, Long Z, Xu L, Liao G, Deng C, et al. Capturing functional long non-coding RNAs through integrating large-scale causal relations from gene perturbation experiments. EBioMedicine. (2018) 35:369–80. doi: 10.1016/j.ebiom.2018.08.050
29. Zhang Y, Liao G, Bai J, Zhang X, Xu L, Deng C, et al. Identifying cancer driver lncRNAs bridged by functional effectors through integrating multi-omics data in human cancers. Mol Ther. (2019) 17:362–73. doi: 10.1016/j.omtn.2019.05.030
30. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
31. Guttman M, Amit I, Garber M, French C, Lin MF, Huarte M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. (2009) 458:223–7. doi: 10.1038/nature07672
32. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. (1991) 349:38–44. doi: 10.1038/349038a0
33. Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. (1989) 10:28–36. doi: 10.1128/MCB.10.1.28
34. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. (2018) 118:24–30. doi: 10.1016/j.ijbiomac.2018.06.033
35. Wang S, Yang L, Lin C, Wang S, Liang K, Hu Q, et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. (2017) 127:4498–515. doi: 10.1172/JCI91553
36. Rocha ST, Heard E. Novel players in X inactivation : insights into Xist -mediated gene silencing and chromosome conformation. Nat Publ Gr. (2017) 24:197–204. doi: 10.1038/nsmb.3370
37. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. (2018) 19:1–19. doi: 10.3390/ijms19102958
38. Ding L, Ren J, Zhang D, Li Y, Huang X, Hu Q, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. (2018) 39:397–406. doi: 10.1093/carcin/bgy006
39. Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. (2016) 126:1216–23. doi: 10.1172/JCI81136
40. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. (2019) 21:498–510. doi: 10.1038/s41556-019-0299-0
41. Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8 + T cells impaired the anticancer function of normal CD8 + T cells. J Med Genet. (2019) 56:29–31. doi: 10.1136/jmedgenet-2018-105439
42. Kan J-Y, Wu D-C, Yu F-J, Wu C-Y, Ho Y-W, Chiu Y-J, et al. Chemokine (C-C Motif) ligand 5 cancer progression through dendritic cell-mediated colon is involved in tumor-associated non-coding RNA MALAT-1. J Cell Physiol. (2015) 230:1883–94. doi: 10.1002/jcp.24918
43. Denaro N, Merlano MC, Nigro C Lo. Long noncoding RNAs as regulators of cancer immunity. Mol Oncol. (2019) 13:61–73. doi: 10.1002/1878-0261.12413
44. Xing F, Liu Y, Wu S, Wu K, Sharma S, Mo Y, et al. Loss of XIST in breast cancer activates MSN-c-met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. (2018) 78:4316–30. doi: 10.1158/0008-5472.CAN-18-1102
45. Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. (2018) 9:3826. doi: 10.1038/s41467-018-06152-x
46. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. (2018) 19:1112–25. doi: 10.1038/s41590-018-0207-y
47. Sun J, Ji J, Huo G, Song Q, Zhang X. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int J Oncol. (2016)943–52. doi: 10.3892/ijo.2016.3589
48. Zhang J, Wang Q, Quan Z. Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-b expression. Biochem Biophys Res Commun. (2019) 515:644–50. doi: 10.1016/j.bbrc.2019.05.080
49. Li Y, Li Y, Huang S, He K, Zhao M, Lin H. Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget. (2017) 8:13690–702. doi: 10.18632/oncotarget.14625
50. Wu K, Zhao Z, Liu K, Zhang J, Li G. Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell Cycle. (2017) 16:1295–301. doi: 10.1080/15384101.2017.1317416
51. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. (2019) 231:116335. doi: 10.1016/j.lfs.2019.03.040
52. Zhou AY, Zhao W, Mao L. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int J Biochem Cell Biol. (2018) 104:25–33. doi: 10.1016/j.biocel.2018.08.017
53. Zhang C, Jiang F, Su C, Xie P, Xu L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem. (2019) 120:11642–50. doi: 10.1002/jcb.28444
54. Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X, et al. Cancer associated fibroblasts-promoted LncRNA DNM3OS confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clin Cancer Res. (2018) 25:1989–2000. doi: 10.1158/1078-0432.CCR-18-0773
55. He Z, You C, Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem Biophys Res Commun. (2018) 500:569–76. doi: 10.1016/j.bbrc.2018.04.091
56. Dominic A, Laurence J. Coagulation factor X regulated by CASC2c recruited macrophages and induced M2 polarization in glioblastoma multiforme. Front Immunol. (2018) 9:1557. doi: 10.3389/fimmu.2018.01557
57. Guan Z, Song B, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. (2015) 464:807–13. doi: 10.1016/j.bbrc.2015.07.040
58. Wang C, Zhu C, Xu J, Wang M, Zhao W, Liu Q, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. (2019) 18:115. doi: 10.1186/s12943-019-1032-0
59. Yao Y, Jiang Q, Jiang L, Wu J, Zhang Q. Lnc-SGK1 induced by Helicobacter pylori infection and high-salt diet promote Th2 and Th17 differentiation in human gastric cancer by SGK1/Jun B signaling. Oncotarget. (2016) 7:20549–60. doi: 10.18632/oncotarget.7823
60. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Am J Transl Res. (2015) 7:2262–9.
61. Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the regulatory T cells-mediated immune escape of hepatocellular carcinoma cells. Am Soc Gene Cell Ther. (2019) 17:516–29. doi: 10.1016/j.omtn.2019.05.027
62. Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W, et al. Long noncoding RNA lncTCF7, induced by IL-6 / STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res. (2015) 34:116. doi: 10.1186/s13046-015-0229-3
63. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. (2018) 9:478. doi: 10.1038/s41419-018-0528-7
64. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. (2017) 8:15129. doi: 10.1038/ncomms15129
65. Yan K, Fu Y, Zhu N, Wang Z, Hong J, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
66. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. (2017). 119:2951–963. doi: 10.1002/jcb.26509
67. Fang P, Xiang L, Chen W, Li S, Huang S, Li J, et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544 / RUNX3. Innate Immun. (2019) 25:99–109. doi: 10.1177/1753425919827632
68. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. (2019) 18:1–12. doi: 10.1186/s12943-019-0978-2
69. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol. (2019) 33:205873841985969. doi: 10.1177/2058738419859699
70. Kathuria H, Millien G, McNally L, Gower AC, Tagne J, Cao Y, et al. NKX2-1-AS1 negatively regulates CD274/PD-L1, cell-cell interaction genes, and limits human lung carcinoma cell migration. Sci Rep. (2018) 8:14418. doi: 10.1038/s41598-018-32793-5
71. Zhou Q, Tang X, Tian X, Tian J, Zhang Y, Ma J, et al. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J Cancer. (2018) 9:2436–42. doi: 10.7150/jca.24796
72. Tian X, Jie M, Ting W, Yue Z, Lingxiang M, Huaxi X, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1–HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol. (2018) 9:473. doi: 10.3389/fimmu.2018.00473
73. Tian X, Zhang Y, Mao L, Wang S, Ma J, Tian J, et al. Long non-coding RNA RUNXOR accelerates MDSC-mediated immunosuppression in lung cancer. BMC Cancer. (2018) 18:1–10. doi: 10.1186/s12885-018-4564-6
74. Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. (2017) 8:39001–11. doi: 10.18632/oncotarget.16545
75. Zeng X, Xie H, Yuan J, Jiang X, Yong J, Zeng D, et al. EGF promotes epithelial ovarian cancer metastasis via activating EGFR-ERK signaling and suppressing lncRNA LIMT expression. Cancer Biol Ther. (2019) 20:956–66. doi: 10.1080/15384047.2018.1564567
76. Cao J, Dong R, Jiang L, Gong Y, Yuan M, You J, et al. LncRNA-MM2P identified as a modulator of macrophage M2 polarization. Cancer Immunol Res. (2019) 7:292–305. doi: 10.1158/2326-6066.CIR-18-0145
77. Zhou W-Y, Zhang M-M, Liu C, Kang Y, Wang J-O, Yang X-H. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. (2019) 1:1–14. doi: 10.1002/jcp.28884
78. Zheng T, Ma G, Tang M, Li Z, Xu R. IL-8 secreted from M2 macrophages promoted prostate tumorigenesis via STAT3/MALAT1 pathway. Int J Mol Sci. (2019) 20:1–16. doi: 10.3390/ijms20010098
79. Liu J, Ding D, Jiang Z, Du T. Long non-coding RNA CCAT1/miR-148a/PKC ζ prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. (2019) 79:105–12. doi: 10.1002/pros.23716
80. Huang J, Song W, Lu B, Huang Y, Dong H, Zhu Z, et al. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem. (2017) 118:4821–30. doi: 10.1002/jcb.26153
81. Gao Y, Wang T, Li Y, Zhang Y, Yang R, Alerts E. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol. (2019) 200:2603–14. doi: 10.4049/jimmunol.1701721
82. Johnsen AK, Templeton DJ, Harding C V, Alerts E. Deficiency of transporter for antigen presentation (TAP) in tumor cells allows evasion of immune surveillance and increases tumorigenesis. J Immunol. (1999) 163:4224–31.
83. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L. The LINK-A lncRNA activates normoxic HIF1α signaling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. doi: 10.1038/ncb3295
84. Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, et al. The LINK-A lncRNA interacts with PtdIns (3,4,5) P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. (2017) 19:238–51. doi: 10.1038/ncb3473
85. Bukur J, Jasinski S, Seliger B. The role of classical and non-classical HLA class I antigens in human tumors. Semin Cancer Biol. (2012) 22:350–8. doi: 10.1016/j.semcancer.2012.03.003
86. Voron T, Colussi O, Marcheteau E, Pernot S, Nizard M, Pointet A, et al. VEGF-A modulates expression of inhibitory checkpoints on CD8 + T cells in tumors. J Exp Med. (2015) 212:139–48. doi: 10.1084/jem.20140559
87. Gabrilovich DI, Chen HL, Girgis KR, Cunningham HT, Meny GM, Nadaf S, et al. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat Med. (1996) 2:1096–103. doi: 10.1038/nm1096-1096
88. Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O, et al. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. (2013) 72:539–49. doi: 10.1158/0008-5472.CAN-12-2325
89. Ruffell B, Chang-strachan D, Chan V, Rosenbusch A, Ho CMT, Pryer N, et al. Macrophage IL-10 blocks CD8+T cell-dependentresponses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. (2014) 26:623–37. doi: 10.1016/j.ccell.2014.09.006
90. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. (2015) 51:E1–2. doi: 10.1002/mc.22120
91. Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. (2008) 28:195–208. doi: 10.1038/onc.2008.373
92. Mou Y, Wang D, Xing R, Nie H, Mou Y, Zhang Y. Identification of long noncoding RNAs biomarkers in patients with hepatitis B virus-associated hepatocellular carcinoma. Cancer Biomark. (2018) 23:95–106. doi: 10.3233/CBM-181424
93. Luo Y, Boyle DL, Hammaker D, Edgar M, Franzoso G, Firestein GS. Suppression of collagen-induced arthritis in growth arrest and DNA damage-inducible protein 45β-deficient mice. Arthritis Rheum. (2011) 63:2949–55. doi: 10.1002/art.30497
94. Carpenter S, Fitzgerald KA. Cytokines and long noncoding RNAs. Cold Spring Harb Perspect Biol. (2018) 10:1–20. doi: 10.1101/cshperspect.a028589
95. Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM) -targeted therapeutics. Adv Drug Deliv Rev. (2017) 114:206–21. doi: 10.1016/j.addr.2017.04.010
96. Murray PJ. Macrophage polarization. Annu Rev Physiol. (2017) 79:541–66. doi: 10.1146/annurev-physiol-022516-034339
97. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A, Mantovani A. Macrophage polarization : tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. (2002) 23:549–55. doi: 10.1016/S1471-4906(02)02302-5
98. Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. (2013) 19:1264–72. doi: 10.1038/nm.3337
99. Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, et al. “Re-educating” tumor-associated macrophages by targeting NF-κB. J Exp Med. (2008) 205:1261–8. doi: 10.1084/jem.20080108
100. Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1 / CSF1R blockade reprograms tumor-in fi ltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. (2014) 74:5057–69. doi: 10.1158/0008-5472.CAN-13-3723
101. Li Z, Feng C, Guo J, Hu X, Xie D. GNAS-AS1 / miR-4319 / NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics. (2019) 20:17–28. doi: 10.1007/s10142-019-00696-x
102. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediatesboth activation and repressionof immune response genes. Science. (2013) 341:789–92. doi: 10.1126/science.1240925
103. Li N. LncRNA MIR155HG regulates M1 / M2 macrophage polarization in chronic obstructive pulmonary disease. Biomed Pharmacother. (2019) 117:109015. doi: 10.1016/j.biopha.2019.109015
104. Marvel D, Gabrilovich DI, Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. (2015) 125:3356–64. doi: 10.1172/JCI80005
105. Bunt SK, Sinha P, Clements VK, Ostrand-rosenberg S, Bunt SK, Sinha P, et al. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. (2006) 176:284–90. doi: 10.4049/jimmunol.176.1.284
106. Bunt SK, Yang L, Sinha P, Clements VK, Leips J, Ostrand-rosenberg S. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. (2007) 67:10019–27. doi: 10.1158/0008-5472.CAN-07-2354
107. Thevenot PT, Sierra RA, Raber PL, Al-khami AA, Trillo-tinoco J, Zarreii P, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. (2014) 41:389–401. doi: 10.1016/j.immuni.2014.08.015
108. Corzo CA, Cotter MJ, Cheng P, Kusmartsev S, Sotomayor E, McCaffrey TV, et al. Mechanism regulating reactive oxygen species intumor-induced myeloid-derived suppressor cells. J Immunol. (2009) 182:5693–701. doi: 10.4049/jimmunol.0900092
109. Jiang T, Shi T, Zhang H, Hu J, Song Y, Wei J, et al. Tumor neoantigens : from basic research to clinical applications. J Hematol Oncol. (2019) 5:1–13. doi: 10.1186/s13045-019-0787-5
110. Jia AH, Truica CI, Wang B. Immunotherapy for triple-negative breast cancer: existing challenges and exciting prospects. Drug Resist Updat. (2017) 32:1–15. doi: 10.1016/j.drup.2017.07.002
111. Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. (2018) 378:449–59. doi: 10.1056/NEJMoa1709919
112. Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, et al. Chimeric antigen receptor–modified T cells for acute lymphoid leukemia. N Engl J Med. (2013) 368:1509–18. doi: 10.1056/NEJMoa1215134
113. Bagg A, June CH. Chimeric antigen receptor–modified T cells in chronic lymphoid leukemia. N Engl J Med. (2011) 365:725–33. doi: 10.1056/NEJMoa1103849
114. Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. (2014) 371:1507–17. doi: 10.1056/NEJMoa1407222
115. Mardiana S, Solomon BJ, Darcy PK, Beavis PA. Supercharging adoptive T cell therapy to overcome solid tumor – induced immunosuppression. Sci Transl Med. (2019) 11: eaaw2293. doi: 10.1126/scitranslmed.aaw2293
116. Ahmed N, Brawley VS, Hegde M, Robertson C, Ghazi A, Gerken C, et al. Human epidermal growth factor receptor 2 (HER2) –specific chimeric antigen receptor–modified T cells for the immunotherapy ofHER2-positive sarcoma. J Clin Oncol. (2015) 33:1688–96. doi: 10.1200/JCO.2014.58.0225
Keywords: immunosuppression, long non-coding RNA, tumor microenvironment, immune response, therapeutic target, biomarker
Citation: Luo Y, Yang J, Yu J, Liu X, Yu C, Hu J, Shi H and Ma X (2020) Long Non-coding RNAs: Emerging Roles in the Immunosuppressive Tumor Microenvironment. Front. Oncol. 10:48. doi: 10.3389/fonc.2020.00048
Received: 26 September 2019; Accepted: 13 January 2020;
Published: 31 January 2020.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Kevin Xueying Sun, Harbin Medical University, ChinaCopyright © 2020 Luo, Yang, Yu, Liu, Yu, Hu, Shi and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hubing Shi, c2hpaGJAc2N1LmVkdS5jbg==; Xuelei Ma, ZHJtYXh1ZWxlaUBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.